Significance
The energy-producing organelle mitochondrion contains its own compact genome, which is separate from the nuclear genome. In nearly all mammals, this mitochondrial genome is inherited exclusively from the mother, and transmission of paternal mitochondria or mitochondrial DNA (mtDNA) has not been convincingly demonstrated in humans. In this paper, we have uncovered multiple instances of biparental inheritance of mtDNA spanning three unrelated multiple generation families, a result confirmed by independent sequencing across multiple unrelated laboratories with different methodologies. Surprisingly, this pattern of inheritance appears to be determined in an autosomal dominantlike manner. This paper profoundly alters a widespread belief about mitochondrial inheritance and potentially opens a novel field in mitochondrial medicine.
Keywords: human genetics, mitochondria, biparental inheritance, paternal transmission, mtDNA
Abstract
Although there has been considerable debate about whether paternal mitochondrial DNA (mtDNA) transmission may coexist with maternal transmission of mtDNA, it is generally believed that mitochondria and mtDNA are exclusively maternally inherited in humans. Here, we identified three unrelated multigeneration families with a high level of mtDNA heteroplasmy (ranging from 24 to 76%) in a total of 17 individuals. Heteroplasmy of mtDNA was independently examined by high-depth whole mtDNA sequencing analysis in our research laboratory and in two Clinical Laboratory Improvement Amendments and College of American Pathologists-accredited laboratories using multiple approaches. A comprehensive exploration of mtDNA segregation in these families shows biparental mtDNA transmission with an autosomal dominantlike inheritance mode. Our results suggest that, although the central dogma of maternal inheritance of mtDNA remains valid, there are some exceptional cases where paternal mtDNA could be passed to the offspring. Elucidating the molecular mechanism for this unusual mode of inheritance will provide new insights into how mtDNA is passed on from parent to offspring and may even lead to the development of new avenues for the therapeutic treatment for pathogenic mtDNA transmission.
Mitochondria are energy-generating organelles that play a critical role in numerous cellular functions, including ATP production, cellular homeostasis, and apoptosis (1). Unlike nuclear DNA in which there are only two copies of each gene per cell, thousands of copies of mitochondrial DNA (mtDNA) are present in every nucleated cell. Typically, individuals harbor only one mtDNA genotype (that of the mother), and all mitochondrial genomes are approximately genetically identical (homoplasmy). In many mitochondrial diseases, however, wild-type and mutant maternal alleles coexist, and this is known as heteroplasmy. The extent of heteroplasmy may vary among tissues and contribute to mitochondrial disease severity. In this paper, we present work that shows that there are rare exceptions to a strict maternal inheritance pattern and that paternal contributions can be made to the mtDNA of the offspring.
In humans, since mitochondria (and thus mtDNA) are typically transmitted to subsequent generations exclusively through the maternal lineage (2), a clinically asymptomatic woman with low levels of a deleterious heteroplasmic mtDNA mutation may pass her disease-causing mutation to all of her offspring, resulting in mtDNA dysfunction and disease. The severity of clinical symptoms in affected children is often associated with the level of mtDNA heteroplasmy (i.e., the percentage of the deleterious mutation) (3). For example, the heteroplasmic mtDNA m.8993T > G mutation causes Leigh syndrome, a condition that is associated with regression in mental and motor skills, disability, and death due to seizures and respiratory failure (4, 5). When the mtDNA m.8993T > G mutation load is less than 30%, a child is expected to be asymptomatic. The probability of having severe symptoms is low until the mutant load reaches 60–70% for the m.8993T > G mutation (6).
Given their strict maternal inheritance, the options for treating pathogenic mtDNAs remain limited. Transmission of mtDNA mutations can potentially be avoided by using technologies, such as oocyte spindle transfer to reconstitute a carrier’s nucleus into the cytoplasm of enucleated donor oocytes that do not carry any mtDNA mutations. Once reconstituted, such oocytes could be in vitro fertilized and implanted using established in vitro fertilization procedures, resulting in a so-called “three-parent baby.” This process has already been successfully used to treat a m.8993T > G carrier with an extensive history of miscarriages and early death of offspring, resulting in the birth of a healthy child in early 2016 (7). However, most countries do not currently permit carrying embryos created through mitochondrial replacement therapy to term due to ethical controversies over mixing genetic material from three different individuals. In addition, the procedure is still very complicated and costly. In this paper, we provide evidence of a paternal contribution of mtDNA in humans and that it is consistent with control of the process by an autosomal dominant gene. These results open up a new frontier in the study of mtDNA genetics that may 1 d provide insights into alternative mechanisms for the treatment of inherited mitochondrial diseases.
Results
Whole mtDNA sequencing analysis of patients with suspected mitochondrial diseases has become a routine practice during the advancement of next-generation sequencing. However, the clinical laboratories that perform such analyses tend to report only previously reported pathogenic or likely pathogenic mutations. Unusual results are often ignored, especially when they do not involve likely pathogenic variants. This paper was initiated to examine just such an instance of an unusual result observed in a set of patients initially referred for clinical evaluation for mitochondrial disease.
The first proband identified in this paper (IV-2 from Family A) was a 4-y-old boy who was evaluated for fatigue, hypotonia, muscle pain, and ptosis at the MitoClinic at Cincinnati Children’s Hospital Medical Center (CCHMC). He was suspected to have a mitochondrial disorder. Other family members showed varying clinical symptoms but were not suspected to have mitochondrial disorders. His twin sister (IV-3) had speech delay but was otherwise healthy, and his grandfather (II-4) had suffered a heart attack but had no other conditions. His older sister (IV-1) was healthy. It should be noted that his mother (III-6) was diagnosed with neuropathy and leg pain that was suspected to be due to multiple sclerosis, but this too was not considered highly to be attributed to a mitochondrial disorder.
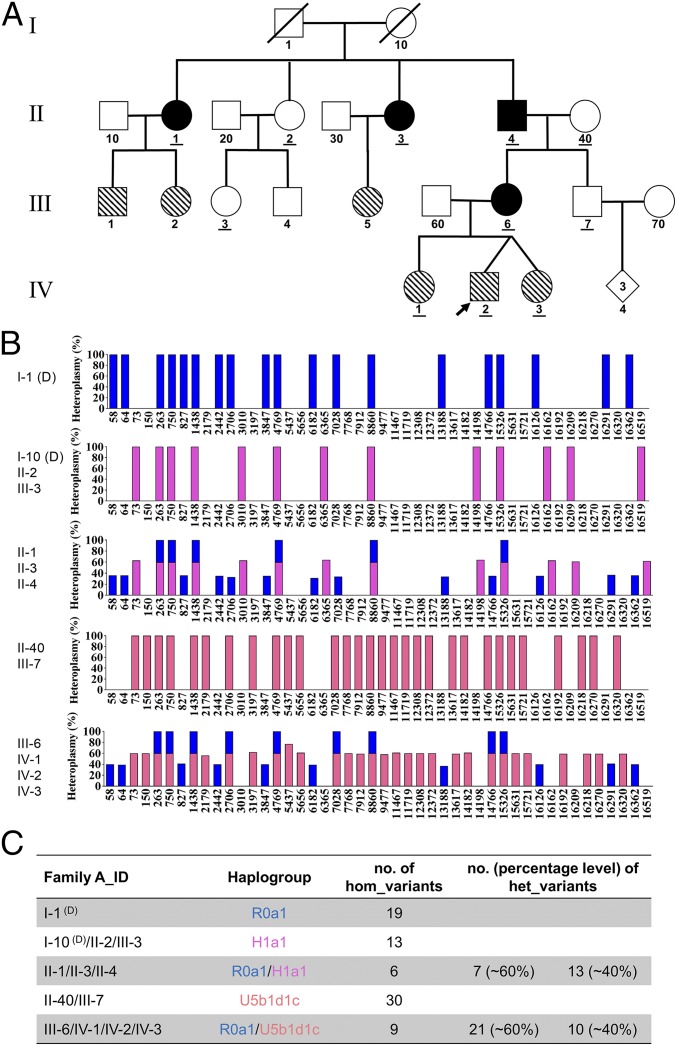
As the proband was suspected to be suffering from a mitochondrial disorder, whole mtDNA sequencing was performed and aligned to the human mitochondrial sequence reference (NC_012920.1). Although this analysis revealed no pathogenic or likely pathogenic mutations, nine homoplasmic variants (the orange-red plus blue bar in Fig. 1B, and IV-2 in Fig. 1C) and 31 heteroplasmic variants were identified. Among these 31 heteroplasmic variants, first blood sampling of the proband revealed an average heteroplasmy level of 29% for 10 variants (the blue bar in Fig. 1B) and reciprocally 71% for the other 21 variants (the orange-red bar in Fig. 1B and Dataset S1, IV-2, column d-E). Due to this abnormally high level of heteroplasmy, repeated sequencing and fresh blood sampling were performed to verify the authenticity of our findings and rule out the possibility of sample mix-up (Dataset S1, IV-2, column F-G and H-I). The repeat sequencing fully confirmed the initial results. IV-2’s two sisters were also tested and shared the same mtDNA heteroplasmy pattern (Dataset S1, IV-1 and Dataset S1, IV-3). Apparently, the proband and his sisters had inherited the mtDNA heteroplasmy pattern from their mother (Dataset S1, III-6) as they had the same heteroplasmy pattern with minor differences in heteroplasmy levels (40% vs. 60% in the mother, Fig. 1 B and C, III-6). A single row of histograms was used to represent all family members sharing the same mtDNA haplogroup (e.g., IV-1, IV-2, IV-3, and III-6) to avoid redundancy.
Fig. 1.
Biparental mtDNA inheritance pattern in Family A. (A) Pedigree of Family A. The black filled symbols indicate the four family members (II-1, II-3, II-4, and III-6) showing biparental mtDNA transmission, and the diagonal filled symbols indicate the six family members (III-1, III-2, III-5, IV-1, IV-2, and IV-3) carrying a high number and level of mtDNA heteroplasmy but with normal maternal transmission. The IDs of the 11 family members tested by whole mtDNA sequencing analysis have been underlined in the pedigree. (B) Schematic of the mtDNA genotypes defined by the homoplasmic and/or heteroplasmic variants aligned from the reference mitochondrial genome. Blue bars represent the genotype of paternally derived mtDNA, whereas purple-red and orange-red bars represent maternally derived mtDNA. Entries labeled (D) represent deduced mtDNA genotypes. (C) Summary of the haplogroup and mtDNA variant numbers in Family A.
To further explore the origin of this unique heteroplasmy pattern in III-6 of Family A, we examined the mtDNA in the grandmother (Dataset S1, II-40) and grandfather (Dataset S1, II-4) of the proband. Based on the position of their mtDNA variant sites, it was revealed that all 21 variants at around a 60% heteroplasmy level in III-6 were maternally inherited from II-40 (the orange-red bar, Fig. 1 B and C, II-40) with a haplogroup of U5b1d1c (SI Appendix, Fig. S1), whereas the other 10 variants at around the 40% heteroplasmy level were inherited from her father (the blue bar, Fig. 1 B and C, II-4). The nine homoplasmic variants should be inherited from both her parents. In other words, III-6’s heteroplasmy pattern could be well explained by a mixture of 30 variants (at the percentage of about 60%) maternally from II-40 and 19 variants (at the percentage of about 40%) paternally from II-4. By analyzing the segregation of mtDNA genotypes from II-4 and II-40 to III-6, we first demonstrated biparental mtDNA transmission in a human pedigree causing a high number and level of heteroplasmy in the offspring.
Testing in Additional Family Members.
Significantly, we demonstrated the same phenomenon within Family A by recruiting additional family members and examining their mtDNAs. For example, II-4 displayed a similar heteroplasmy pattern as in his daughter (III-6) with six homoplasmic variants and 20 heteroplasmic variants (Dataset S1, II-4). To infer II-4’s maternal mtDNA genotype (I-10), we further tested his sisters and niece (II-1, II-2, II-3, and III-3). Both II-1 (Dataset S1, II-1) and II-3 (Dataset S1, II-3) shared the same heteroplasmy pattern as II-4 but with minor differences in the heteroplasmy levels. However, III-3 (Dataset S1, III-3) only presented 13 homoplasmic variants that were passed maternally from II-2 (Dataset S1, II-2). It was thus deduced that I-10 would have the exact same mtDNA genotype as II-2 and III-3, based on maternal inheritance by default. Then, by matching these mtDNA variant sites, it was also discovered that all seven heteroplasmic variants at around the 60% heteroplasmy level in II-4 to be maternally inherited from I-10 (with a haplogroup of H1a1, SI Appendix, Fig. S2), whereas the other 13 variants at around the 40% heteroplasmy level came from his father (I-1 with a haplogroup of R0a1, SI Appendix, Fig. S3). The six homoplasmic variants should be inherited from both his parents. That is, II-40’s heteroplasmy pattern appeared to be caused by a mixture of 13 variants (at the percentage of about 60%) maternally from I-10 and 19 variants (at the percentage of about 40%) paternally from I-1. It is also oddly coincidental that these 19 variants are exactly the same variants passed from II-4 to his daughter (III-6), leading to a miscellaneous haplogroup of R0a1/H1a1 in II-4 and R0a1/U5b1d1c in III-6, respectively. Again, segregation of the mtDNA genotype demonstrated biparental mtDNA transmission with an autosomal dominant inheritance mode in this family. The alignment of a total of 46 differential mtDNA variants in this family can be seen in SI Appendix, Table S1, directly showing which ones are paternally or maternally inherited as well as their relative heteroplasmy levels.
Recruitment and Surveillance of Unrelated Families for Biparental Inheritance Patterns.
To confirm the findings in Family A, we recruited two additional families (Families B and C) whose probands initially presented with divergent pathologies that were suspected to have some level of mitochondrial involvement. The proband for Family B is a 35-y-old male with developmental delay, chronic fatigue, diabetes, congenital heart disease, supraventricular tachycardia (SVT), and status post ablation evaluated at MitoClinic at CCHMC. The proband for Family C, on the other hand, is a 46-y-old female who was diagnosed with Guillain-Barré syndrome at 6 y old and presented with prematurity, hyperextensibility, thin translucent skin, chronic fatigue, diffuse body pain, and possible periodic fever. This family was evaluated at the Mayo Clinic. Despite the differences in presentation between these probands, their families were recruited for this study because both individuals demonstrated a high number and level of mtDNA heteroplasmy when their mitochondrial genomes were sequenced (Figs. 2 and 3).
Fig. 2.
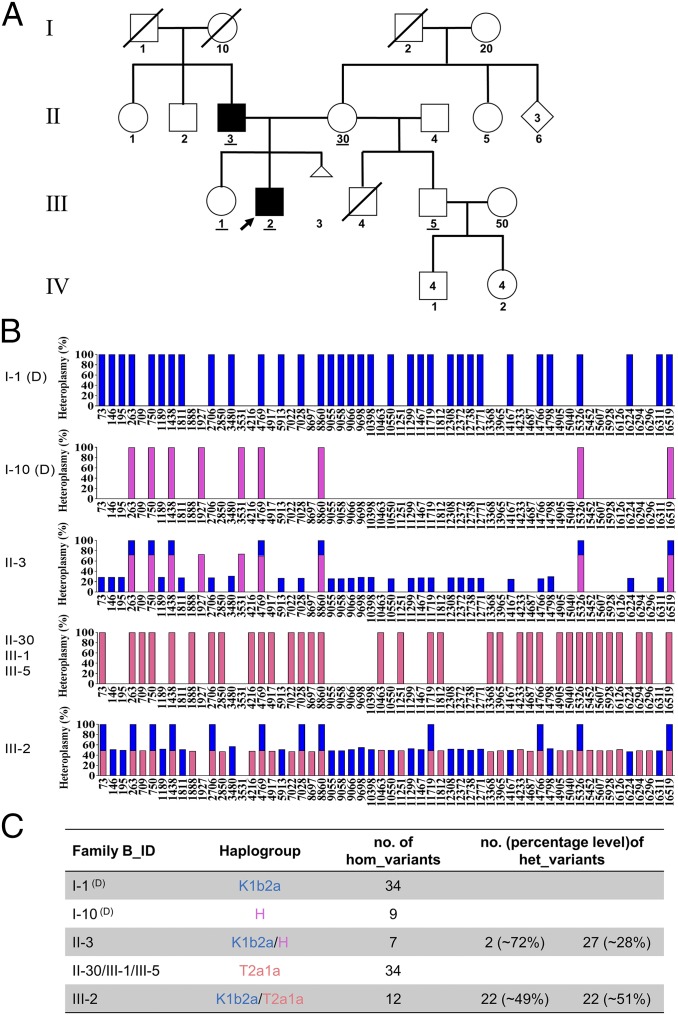
Biparental mtDNA inheritance pattern shown in Family B. (A) Pedigree of Family B. The black filled symbols indicate the two family members (II-3 and III-2) showing biparental mtDNA transmission. The IDs of five family members tested by whole mtDNA sequencing analysis have been underlined in the pedigree. (B) Schematic of the mtDNA genotype defined by the homoplasmic and/or heteroplasmic variants aligned from the reference mitochondrial genome. Blue bars represent the genotype of paternally derived mtDNA, whereas purple-red and orange-red bars represent maternally derived mtDNA. Entries labeled (D) represent deduced mtDNA genotypes. (C) Summary of the haplogroup and mtDNA variant numbers in Family B.
Fig. 3.
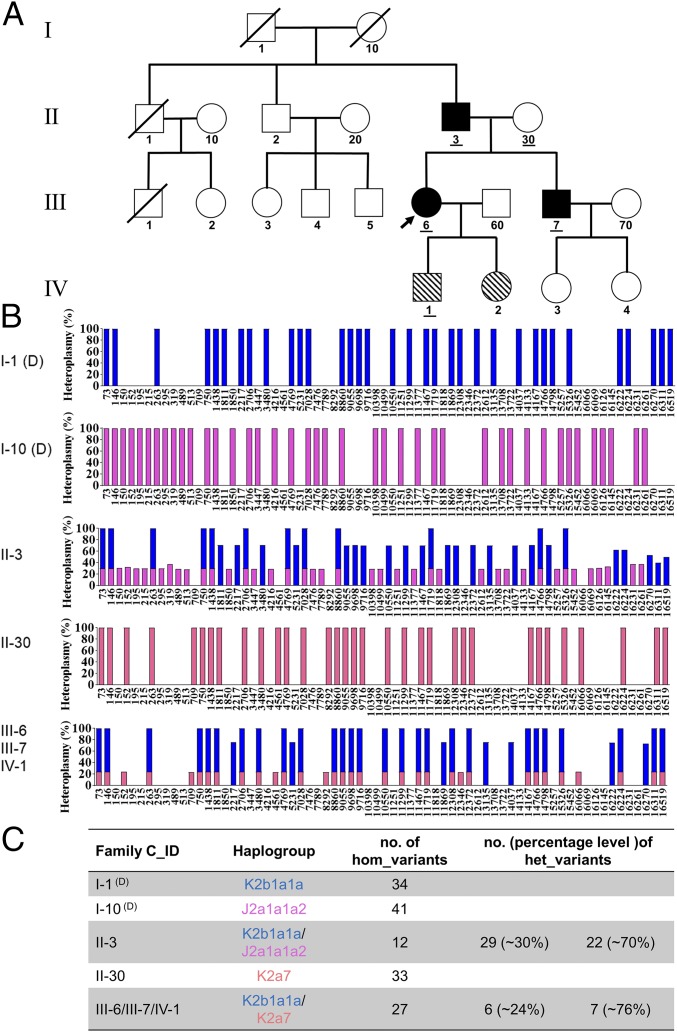
Biparental mtDNA inheritance pattern in Family C. (A) Pedigree of Family C. The black filled symbols indicate the three family members (II-3, III-6, and III-7) showing biparental mtDNA transmission, and the diagonal filled symbols indicate the two family members (IV-1 and IV-2) carrying a high number and level of mtDNA heteroplasmy but with normal maternal transmission. The IDs of the five family members tested by whole mtDNA sequencing analysis have been underlined in the pedigree. (B) Schematic of the mtDNA genotypes defined by the homoplasmic and/or heteroplasmic variants aligned from the reference mitochondrial genome. Blue bars represent the genotype of paternally derived mtDNA, whereas purple-red and orange-red bars represent maternally derived mtDNA. Entries labeled (D) represent deduced mtDNA genotypes. (C) Summary of the haplogroup and mtDNA variant numbers in Family C.
Compared with Family A, a strikingly similar mtDNA transmission pattern was demonstrated in Families B and C (see SI Appendix, Figs. S4–S6 and Table S2 and Dataset S2 for Family B and SI Appendix, Figs. S7–S9 and Table S3 and Dataset S3 for Family C). Taking Family B for illustration, II-3 having 29 heteroplasmic and seven homoplasmic variants (Dataset S2, II-3) should have inherited mtDNA from both his father (I-1, haplogroup of K1b2a, SI Appendix, Fig. S4) and his mother (I-10, haplogroup of H, SI Appendix, Fig. S5), who were supposed to possess 34 and nine homoplasmic variants, respectively (Fig. 2B). II-3 further transmitted his mtDNA that he inherited from I-1 to his son (III-2, Dataset S2, III-2), who also inherited all of his mother’s mtDNA (II-30, carrying 34 variants and a haplogroup of T2a1a) (Dataset S2, II-30 and SI Appendix, Fig. S6). However, III-2’s sister (III-1, Dataset S2, III-1) and half-brother (III-5, Dataset S2, III-5) only inherited the maternal mtDNA. Fresh blood sampling and repeated mtDNA sequencing in a second independent laboratory were also performed to rule out the possibility of sample mix-up for III-2 (Dataset S2, III-2, column F-G and H-I). Additionally, these samples were further verified using Pacific Bio single molecular sequencing (see Materials and Methods) and by restriction fragment length polymorphism (RFLP) analysis of Family A (SI Appendix, Fig. S10), and these results were fully consistent with the previous sequencing. Finally, it should be noted that our analysis failed to identify any pathogenic or likely pathogenic variants in either Family B or Family C, similar to what was observed in Family A.
Discussion
Our results clearly demonstrate biparental transmission of mtDNA in humans, counter to the central dogma of mitochondrial inheritance. This test has been confirmed in three separate lineages with multiple generations and by two other laboratories.
The Initial Discovery of Paternal mtDNA Transmission.
Strictly paternal mtDNA inheritance has been observed in algae and plants, and biparental mtDNA inheritance (via paternal leakage) is well documented in yeast (8). Paternal mtDNA leakage has also been observed occasionally in several species, such as Drosophila (9–12), mouse (13), and sheep (14). However, whether paternal mtDNA transmission exists in humans is controversial. Previous studies using human polyploid embryos generated by in vitro fertilization and intracytoplasmic sperm injection techniques have shown that paternal mtDNA can only be detected at the four-to-eight-cell stage by paternal allele-specific nested polymerase chain reaction (PCR) and restriction enzyme digestion analysis (15).
In 2002, Schwartz and Vissing published a report in NEJM describing a single male with mitochondrial myopathy and exercise intolerance who appeared to possess two different mtDNA haplotypes (16). One haplotype came from the mother, and the other came from the father (with the exception of a 2-bp deletion in the ND2 gene of the mtDNA, which was found only in the patient). This novel microdeletion arose sporadically on the paternal mtDNA background, although it cannot be ruled out that the father may also harbor this mutation at a low level in other tissues. Initially, direct sequencing of PCR-amplified mtDNA indicated that the patient’s muscle tissues were homoplasmic for the paternal mtDNA haplotype. However, by performing mtDNA analysis with solid-phase minisequencing and fragment analysis from different tissues in the index patient and his parents, the authors were able to demonstrate that the patient’s muscle tissues possessed a mtDNA sequence ∼90% identical to the paternal haplotype as well as 10% maternal (normal) mtDNA, suggesting biparental mtDNA inheritance. However, paternal mtDNA was present only in the patient’s skeletal muscle, whereas mtDNA from all other tissues studied in the patient showed no paternal mtDNA haplotype.
This single case report generated significant attention in the field as it was the first clue that biparental mtDNA transmission may be possible in humans. A separate group at Harvard Medical School was able to independently confirm the existence of the paternal haplotype in the patient from the NEJM paper, uncovering potential evidence of mtDNA recombination in the process (17). For over 16 y, however, no additional cases of biparental inheritance have been reported, despite efforts to turn up additional cases by several independent groups (18–20). With the advent of next-generation sequencing, this hypothesis was revisited. Ultradeep mtDNA resequencing revealed no evidence of paternal mtDNA haplotypes transmitted to offspring in humans (21). Furthermore, a separate review of the literature by Bandelt et al. (22) concluded that other instances of mixed haplogroups in 20 other publications were likely due to contamination or mislabeled samples, effectively ruling out both paternal mtDNA inheritance as well as the possibility of mtDNA recombination. Results, such as these, have led many in the field to conclude that the findings in the single case report may have been due to technical issues or sample mix-up and to move on to other topics of investigation.
A Resurgence of the Paternal Transmission Hypothesis.
The results presented in this paper make a robust case for paternal transmission of mtDNA. Here, we report biparental mtDNA inheritance (either directly or indirectly) in 17 members in three multigeneration families. Thirteen of these individuals were identified directly by sequencing of the mitochondrial genome, whereas four could be inferred based on preexisting maternal heteroplasmy caused by biparental inheritance in the previous generation.
Our developed methodology allowed the simultaneous: (i) characterization of complete nucleotide sequences of the mitochondrial genome, (ii) annotation of all heteroplasmic positions in mtDNA at very high sensitivity, and (iii) estimation of the mutant load at each position. In addition, this method does not require prior knowledge of the mtDNA sequence. The mtDNA heteroplasmy levels were readily calculated without complex calibration or computation. This method is sensitive for the detection and quantification of the heteroplasmy level with virtually no false positives according to our tests (23). These samples underwent whole mtDNA sequencing involving different batches, and each batch contains at least 12 samples in one pool to be loaded on the sequencer. None of the other samples in these batches have shown multiple heteroplasmy.
To further confirm these remarkable results and to exclude the possibility of sample mix-up and/or contamination, the whole mtDNA sequencing procedure was repeated independently in at least two different laboratories by different laboratory technicians with newly obtained blood samples. All results were reproducible, indicating no artifacts or contamination exist. More importantly, the multiple mtDNA variants that were paternally transmitted differ in both number and position among each of these three families as well as the related haplogroup (R0a1 in Family A, K1b2a in Family B, and K2b1a1a in Family C, respectively), providing two distinct forms of evidence supporting transmission of the paternal mtDNA.
Therefore, we have unequivocally demonstrated the existence of biparental mtDNA inheritance as evidenced by the high number and level of mtDNA heteroplasmy in these three unrelated multigeneration families. Most interestingly, the mixed haplogroups in these samples are very reminiscent of the mixed haplogroups found in the 20 studies that were dismissed by Bandelt et al. as due to contamination or sample mix-up. One is forced to wonder how many other instances of individuals with biparental mtDNA inheritance have been dismissed as technical errors, and whether Schwartz and Vissing’s original discovery has really been given the proper follow-up that it deserves. We suspect that these results will initiate a broader reassessment of the topic.
Possible Mechanisms of Biparental mtDNA Inheritance.
This unexpected paternal transmission of mtDNA raises several questions about how exactly paternal mtDNA can escape its normal fate of being eliminated from the embryo. Maternal transmission of mtDNA is the result of active elimination of paternal mitochondria, and the genes underlying this elimination process are intriguing candidates for the locus underlying the autosomal dominant inheritance pattern observed in our pedigrees.
Unfortunately, the molecular mechanisms underlying paternal mitochondrial elimination are only partially elucidated. In fact, it appears likely that a different combination of mechanisms operates depending on the species in question. In mice and C. elegans, the lysosomal pathway has been established to be very critical in this process (24). Autophagy is also thought to be critical in the elimination of paternal mitochondria in Caenorhabditis elegans (25) through the LC3-dependent autophagosome (26) and mitophagy (27). Recently, it has also been reported that mitochondrial endonuclease G relocates from the intermembrane space of paternal mitochondria to the matrix after fertilization where it proceeds to degrade or eliminate paternal mtDNA (28). It is not difficult to imagine how a defect in such an EndoG-like pathway in humans might produce a paternal contribution, such as the one observed here.
We propose that the paternal mtDNA transmission in these families should be accompanied by segregation of a mutation in one nuclear gene involved in paternal mitochondrial elimination and that there is a high probability that the gene in question operates through one of the pathways identified above. Taking Family A as an example, II-1, II-3, and II-4 should have inherited this mutated gene from I-1 thus causing a paternal mtDNA transmission, whereas II-2 inherited the normal allelic gene. II-4 further passed this mutation to III-6 but not III-7, causing a similar heteroplasmy pattern in III-6 but maternal transmission in III-7. IV-2 and his siblings (IV-1 and IV-3) inherited the exact heteroplasmic mtDNA pattern from III-6, displaying a normal maternal mtDNA transmission. This may give us a hint that, mechanistically speaking, the nuclear mutation will affect the paternal mitochondrial elimination from the paternal side.
The requirement that the nuclear allele be present on the paternal side may also suggest that the unidentified locus has some involvement in the control of mtDNA replication. The amount of paternal mitochondria passed on to the fertilized embryo is likely to be quite low relative to the maternal mitochondria already present in the oocyte (∼0.1% according to one estimate) (29). Even in the absence of active paternal mitochondrial elimination, the percentage of paternal mtDNA would remain undetectable without a mechanism that preferentially increases paternal mtDNA abundance relative to maternal mtDNA. This suggests a high likelihood that the gene in question also has some involvement in mtDNA replication or copy number control, particularly during the blastocyst stage when mtDNA replication resumes (30). A variety of nuclear-encoded factors mediate mtDNA replication, and overexpression of at least one of these proteins (the HMG box protein TFAM) has already been shown to increase the mtDNA copy number in vivo (31). There may even be a synergistic interaction between the particular mtDNA variants and the unidentified nuclear gene that only allows paternal transmission to occur when both elements are present. There is already precedence for mtDNA sequence variants—such as polymorphisms in the conserved sequence box II—leading to different replication rates between otherwise similar mtDNAs (32). Perhaps the unidentified nuclear gene exerts its influence on paternal mtDNA abundance by interacting with a similar variant or group of variants involved in mtDNA replication or copy number control. The key point that will need to be explained in any of these models is the means by which paternal mtDNA abundance is selectively increased while leaving the maternal mtDNA level untouched (or even decreased).
Summary.
The results presented in this paper provide clear and provocative evidence for the biparental transmission of mtDNA in three separate multigeneration families as confirmed by two independent laboratories. Clearly, these results will need to be brought in agreement with the fact that maternal inheritance remains absolutely dominant on an evolutionary timescale and that occasional paternal transmission events seem to have left no detectable mark on the human genetic record. Still, this remains an unprecedented opportunity in the field. Elucidation of the molecular mechanism by which this biparental transmission occurs will expand our fundamental understanding of the process of mitochondrial inheritance and may provide an alternative approach to minimize the consequences of the transmission of pathogenic maternal mtDNA in humans. Whatever the mechanism of this unusual phenomenon may be, it is clear that years of research will be required to fully understand and exploit the ramifications of this discovery.
Materials and Methods
Patients.
Three unrelated multigeneration families with high numbers and levels of mtDNA heteroplasmy were identified. Two families (Family A and Family B) were evaluated at the MitoClinic and referred to the Mitochondrial Diagnostic Laboratory at the Human Genetics Division of CCHMC, Family C was evaluated at the Mayo Clinic, and genetic testing was performed at the Diagnostic Laboratory at Baylor College of Medicine. To derive the origin of these unusual heteroplasmy events, written informed consent was obtained from all of the participating family members. This study was approved by the Institutional Review Board of CCHMC (approval Study ID: 2013–7868).
Family A: Proband (IV-2) was a 4-y-old boy who was evaluated for fatigue, hypotonia, muscle pain, and ptosis at the MitoClinic at CCHMC. He was suspected to have a mitochondrial disorder. His twin sister (IV-3) had speech delay but was otherwise healthy, and his older sister (IV-1) was healthy. His mother (III-6) was diagnosed with neuropathy and leg pain. She was suspected to have multiple sclerosis. His grandfather (II-4) had suffered a heart attack but had no other conditions.
Family B: Proband (III-2) is a 35-y-old male with developmental delay, chronic fatigue, diabetes, congenital heart disease, SVT, and status post ablation. He was suspected to have a mitochondrial disorder and was evaluated at MitoClinic at CCHMC. His sister (III-1) was diagnosed with cerebral palsy and was suspected to have mitochondrial disease.
Family C: Proband (III-6) is a 46-y-old female who was diagnosed with Guillain-Barré syndrome at 6 y old and presented with prematurity, hyperextensibility, thin translucent skin, chronic fatigue, diffuse body pain, and possible periodic fever. The family was evaluated at the Mayo Clinic.
Whole Mitochondrial Genome Sequencing Analysis.
The whole mitochondrial genome sequencing analysis for these three families was performed independently at two different CLIA-accredited laboratories, respectively, at CCHMC (Method 1) and Baylor College of Medicine (Method 2). Genomic DNA was isolated from the blood samples of the patients and their family members using the Gentra DNA extraction kit (Qiagen) according to the manufacturer’s instructions. Entire mtDNA was amplified by a single long range PCR as previously described (23, 33–35). One hundred nanograms of total genomic DNA isolated from blood were used as the template in a 50 μL PCR system.
For Method 1, the primers specifically recognize genuine mtDNA: F-2120 (GGACACTAGGAAAAAACCTTGTAGAGAGAG) and R-2119 (AAAGAGCTGTTCCTCTTTGGACTAACA). PCR amplifications were performed using TaKaRa LA Taq Hot Start polymerase (TaKaRa Biotechnology). PCR conditions were: 94 °C for 1 min; 98 °C for 10 s; and 68 °C for 16 min, 30 cycles; 72 °C for 10 min and held at 4 °C. The amplified mtDNA was used for library preparation by a Nextera XT DNA kit (Illumina). Sequencing was performed on the Illumina MiSeq platform (DNA Core Facility, CCHMC), and the data were analyzed using the NextGENe software. Briefly, sequence reads ranging from 100 to 200 bps were quality filtered and processed using NextGENe software and an algorithm similar to BLAT. The sequence error correction feature (condensation) was performed to reduce false positive variants and produce a sample consensus sequence and variant calls. Alignment without sequence condensation was used to calculate the percentage of the mitochondrial genome with depth of coverage of 1000×. Starting from quality FASTQ reads, the reads were quality filtered and were converted to the FASTA format. Filtered reads were then aligned to the human mitochondrial sequence reference NC_012920.1 followed by variant calling. Variant heteroplasmy was calculated by NextGENe software as follows: Base heteroplasmy (mutant allele frequency %) = mutant allele (forward + reverse)/total coverage of all alleles C, G, T, and A (forward + reverse) × 100. The clinical significance of the variants was analyzed with MitoMaster (https://www.mitomap.org/foswiki/bin/view/MITOMASTER/WebHome) (23, 33, 34).
For Method 2, the forward and reverse primers used for long range PCR were mt16426F (CCGCACAAGAGTGCTACTCTCCTC) and mt16425R (GATATTGATTTCACGGAGGATGGTG). PCR amplifications were performed using TaKaRa LA Taq Hot Start polymerase (TaKaRa Biotechnology). PCR conditions were: 95 °C for 2 min; 95 °C for 20 s; and 68 °C for 18 min, 30 cycles; 68 °C for 20 min and held at 4 °C (35). The amplified PCR products were fragmented to 200 bp, which were purified with AMPure XP beads. After end repair, 3′-adenylation, and Illumina InPE adapter ligation, DNA samples were enriched by PCR with Herculase II polymerase (Agilent Technologies). Twelve indexed DNA libraries were pooled together with equimolar ratios. Each pooled library was sequenced in a single lane of one flow cell on a HiSeq2000 with a 76 or 100 bp paired-end or single-end read chemistry. After demultiplexing with Illumina CASAVA software, the reads belonging to the same index were filtered to remove any reads with a median quality score below 25 by NextGENe software (SoftGenetics). Reads with one unknown assigned base call were also removed. The data were processed, and variant calls were made with two different heteroplasmy cutoff values, 20% and 1%. The aligned reads were examined with a NextGENe Viewer (35). To assure that every indexed sample was scored correctly during pool sequencing, an internal identity control system (InQC) was designed and incorporated into the analysis of each sample. Fourteen nuclear DNA regions containing polymorphic markers were amplified in a multiplex PCR, and the resulting DNA fragments were mixed with the long range-PCR–enriched mtDNA fragments from the same individual for indexing and massively parallel sequencing. In parallel, genotyping the same 14 nuclear markers of the same sample by either TaqMan assay or Sanger sequencing was performed independently. The genotyping result derived from the parallel testing was compared with those generated by massively parallel sequencing to verify sample identity. An unmatched result is always an indication of sample switching or cross-contamination during procedures, such as the library preparation process. Should these results not match, repeat analysis or another investigation is warranted. Also, to ensure the quantification of the heteroplasmy by massively parallel sequencing was reliable, a set of cloned synthetic 150-bp control DNAs was supplemented into each indexed sample as ExQCs.
To further verify the results of the previously described sequencing analysis, long range PCR products of the entire mtDNA were submitted for SMRT sequencing at the Institute of Genome Science, University of Maryland, which has the equipment and expertise needed for this experiment. Specifically, DNA libraries were constructed and prepared for sequencing using the SMRTbell Template Prep Kit 2.0 and the DNA/Polymerase Binding Kit 2.0 (Pacific Biosciences) according to the manufacturer’s instructions. Only 16.5 kb fragments were size selected with the BluePippin device (Sage Sciences) and were sequenced with the DNA Sequencing Kit 2.0 (Pacific Biosciences) using a 4 h movie collection. The data were analyzed using PacBio’s Long Amplicon Analysis module (Smrtanalysis 2.3.0) to find phased consensus sequences with the analysis only being run for subreads longer than 13 K bp. The resulting phased sequences were aligned to the mitochondrial reference genome, and variant calling was performed for each. The variants were then used to assign a genotype to each phased sequence by comparing them with variants found in the parental/ancestral mitochondrial genomes (36). Circular consensus sequence was also generated from the amplicon data and was used to calculate allele/haplogroup frequencies (37). The mitochondrial haplogroups were identified using MitoMaster as described previously.
For the RFLP analysis, PCR amplification was performed to amplify a 789 bp segment of mtDNA, including the hypervariable region HV2. PCR amplification was performed using TaKaRa LA Taq Hot Start polymerase (TaKaRa Biotechnology). PCR conditions were: 95 °C for 2 min; 95 °C for 20 s; and 68 °C for 1 min, 35 cycles; 68 °C for 10 min and held at 4 °C. The forward and reverse primers used for amplification were mt16426F (CCGCACAAGAGTGCTACTCTCCTC) and hmtR2 626 (TTTATGGGGTGATGTGAGCC). The resulting PCR products were then digested with the restriction endonucleases ApaLI and FokI (New England Biolabs) and were compared against the expected band sizes for each haplogroup.
Supplementary Material
Acknowledgments
We thank the patients and their families for participating in our study. We also thank the various DNA Cores where the next-generation sequencing was performed, Kejian Zhang, and Min-Xin Guan for their stimulating discussion. This work was supported, in part, by the Cincinnati Children’s Hospital Research Foundation and National Institute of Child Health and Human Development Grant 1R01HD092989-01A1 (to T.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810946115/-/DCSupplemental.
References
- 1.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace DC. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu Rev Biochem. 2007;76:781–821. doi: 10.1146/annurev.biochem.76.081205.150955. [DOI] [PubMed] [Google Scholar]
- 3.Freyer C, et al. Variation in germline mtDNA heteroplasmy is determined prenatally but modified during subsequent transmission. Nat Genet. 2012;44:1282–1285. doi: 10.1038/ng.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swalwell H, et al. Respiratory chain complex I deficiency caused by mitochondrial DNA mutations. Eur J Hum Genet. 2011;19:769–775. doi: 10.1038/ejhg.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nijtmans LG, Henderson NS, Attardi G, Holt IJ. Impaired ATP synthase assembly associated with a mutation in the human ATP synthase subunit 6 gene. J Biol Chem. 2001;276:6755–6762. doi: 10.1074/jbc.M008114200. [DOI] [PubMed] [Google Scholar]
- 6.White SL, et al. Genetic counseling and prenatal diagnosis for the mitochondrial DNA mutations at nucleotide 8993. Am J Hum Genet. 1999;65:474–482. doi: 10.1086/302488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, et al. Live birth derived from oocyte spindle transfer to prevent mitochondrial disease. Reprod Biomed Online. 2017;34:361–368. doi: 10.1016/j.rbmo.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Breton S, Stewart DT. Atypical mitochondrial inheritance patterns in eukaryotes. Genome. 2015;58:423–431. doi: 10.1139/gen-2015-0090. [DOI] [PubMed] [Google Scholar]
- 9.Kondo R, Matsuura ET, Chigusa SI. Further observation of paternal transmission of Drosophila mitochondrial DNA by PCR selective amplification method. Genet Res. 1992;59:81–84. doi: 10.1017/s0016672300030287. [DOI] [PubMed] [Google Scholar]
- 10.Sherengul W, Kondo R, Matsuura ET. Analysis of paternal transmission of mitochondrial DNA in Drosophila. Genes Genet Syst. 2006;81:399–404. doi: 10.1266/ggs.81.399. [DOI] [PubMed] [Google Scholar]
- 11.Nunes MD, Dolezal M, Schlötterer C. Extensive paternal mtDNA leakage in natural populations of Drosophila melanogaster. Mol Ecol. 2013;22:2106–2117. doi: 10.1111/mec.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dokianakis E, Ladoukakis ED. Different degree of paternal mtDNA leakage between male and female progeny in interspecific Drosophila crosses. Ecol Evol. 2014;4:2633–2641. doi: 10.1002/ece3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gyllensten U, Wharton D, Josefsson A, Wilson AC. Paternal inheritance of mitochondrial DNA in mice. Nature. 1991;352:255–257. doi: 10.1038/352255a0. [DOI] [PubMed] [Google Scholar]
- 14.Zhao X, et al. Further evidence for paternal inheritance of mitochondrial DNA in the sheep (Ovis aries) Heredity (Edinb) 2004;93:399–403. doi: 10.1038/sj.hdy.6800516. [DOI] [PubMed] [Google Scholar]
- 15.St John J, et al. Failure of elimination of paternal mitochondrial DNA in abnormal embryos. Lancet. 2000;355:200. doi: 10.1016/s0140-6736(99)03842-8. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz M, Vissing J. Paternal inheritance of mitochondrial DNA. N Engl J Med. 2002;347:576–580. doi: 10.1056/NEJMoa020350. [DOI] [PubMed] [Google Scholar]
- 17.Kraytsberg Y, et al. Recombination of human mitochondrial DNA. Science. 2004;304:981. doi: 10.1126/science.1096342. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz M, Vissing J. No evidence for paternal inheritance of mtDNA in patients with sporadic mtDNA mutations. J Neurol Sci. 2004;218:99–101. doi: 10.1016/j.jns.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Taylor RW, et al. Genotypes from patients indicate no paternal mitochondrial DNA contribution. Ann Neurol. 2003;54:521–524. doi: 10.1002/ana.10673. [DOI] [PubMed] [Google Scholar]
- 20.Filosto M, et al. Lack of paternal inheritance of muscle mitochondrial DNA in sporadic mitochondrial myopathies. Ann Neurol. 2003;54:524–526. doi: 10.1002/ana.10709. [DOI] [PubMed] [Google Scholar]
- 21.Pyle A, et al. Extreme-depth re-sequencing of mitochondrial DNA finds no evidence of paternal transmission in humans. PLoS Genet. 2015;11:e1005040. doi: 10.1371/journal.pgen.1005040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bandelt HJ, Kong QP, Parson W, Salas A. More evidence for non-maternal inheritance of mitochondrial DNA? J Med Genet. 2005;42:957–960. doi: 10.1136/jmg.2005.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang S, Huang T. Characterization of mitochondrial DNA heteroplasmy using a parallel sequencing system. Biotechniques. 2010;48:287–296. doi: 10.2144/000113389. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Q, Li H, Xue D. Elimination of paternal mitochondria through the lysosomal degradation pathway in C. elegans. Cell Res. 2011;21:1662–1669. doi: 10.1038/cr.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- 26.Djeddi A, et al. Sperm-inherited organelle clearance in C. elegans relies on LC3-dependent autophagosome targeting to the pericentrosomal area. Development. 2015;142:1705–1716. doi: 10.1242/dev.117879. [DOI] [PubMed] [Google Scholar]
- 27.Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Q, et al. Mitochondrial endonuclease G mediates breakdown of paternal mitochondria upon fertilization. Science. 2016;353:394–399. doi: 10.1126/science.aaf4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ladoukakis ED, Eyre-Walker A. Evolutionary genetics: Direct evidence of recombination in human mitochondrial DNA. Heredity (Edinb) 2004;93:321. doi: 10.1038/sj.hdy.6800572. [DOI] [PubMed] [Google Scholar]
- 30.Pikó L, Taylor KD. Amounts of mitochondrial DNA and abundance of some mitochondrial gene transcripts in early mouse embryos. Dev Biol. 1987;123:364–374. doi: 10.1016/0012-1606(87)90395-2. [DOI] [PubMed] [Google Scholar]
- 31.Ekstrand MI, et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 32.Kang E, et al. Mitochondrial replacement in human oocytes carrying pathogenic mitochondrial DNA mutations. Nature. 2016;540:270–275. doi: 10.1038/nature20592. [DOI] [PubMed] [Google Scholar]
- 33.Huang T. Next generation sequencing to characterize mitochondrial genomic DNA heteroplasmy. Curr Protoc Hum Genet. 2011;Chapter 19:Unit19.18. doi: 10.1002/0471142905.hg1908s71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma H, et al. Metabolic rescue in pluripotent cells from patients with mtDNA disease. Nature. 2015;524:234–238. doi: 10.1038/nature14546. [DOI] [PubMed] [Google Scholar]
- 35.Zhang W, Cui H, Wong LJ. Comprehensive one-step molecular analyses of mitochondrial genome by massively parallel sequencing. Clin Chem. 2012;58:1322–1331. doi: 10.1373/clinchem.2011.181438. [DOI] [PubMed] [Google Scholar]
- 36.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): Application and theory. BMC Bioinformatics. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.