Significance
Mutations in fused in sarcoma (FUS) contribute to a subset of familial amyotrophic lateral sclerosis (ALS). ALS-linked mutations cause a liquid–liquid phase separation of FUS protein, induce cytoplasmic inclusions in cells, and interfere with RNA metabolism pathways. Our proteomic analysis shows that proteins sequestered into inclusions are enriched in translation and RNA quality surveillance pathways. This study demonstrates that ALS mutations in FUS indeed suppress protein translation and affect the mRNA nonsense-mediated decay (NMD) pathway. NMD-promoting factors UPF1 and UPF3b increased, whereas the negative regulator UPF3a decreased in the cells of patients with ALS. The disruption in NMD regulation and suppression of protein biosynthesis likely contribute to neurodegeneration in ALS.
Keywords: amyotrophic lateral sclerosis, fused in sarcoma, protein translation, nonsense-mediated decay, RNA-protein granules
Abstract
Amyotrophic lateral sclerosis (ALS) is an incurable neurodegenerative disease characterized by preferential motor neuron death. Approximately 15% of ALS cases are familial, and mutations in the fused in sarcoma (FUS) gene contribute to a subset of familial ALS cases. FUS is a multifunctional protein participating in many RNA metabolism pathways. ALS-linked mutations cause a liquid–liquid phase separation of FUS protein in vitro, inducing the formation of cytoplasmic granules and inclusions. However, it remains elusive what other proteins are sequestered into the inclusions and how such a process leads to neuronal dysfunction and degeneration. In this study, we developed a protocol to isolate the dynamic mutant FUS-positive cytoplasmic granules. Proteomic identification of the protein composition and subsequent pathway analysis led us to hypothesize that mutant FUS can interfere with protein translation. We demonstrated that the ALS mutations in FUS indeed suppressed protein translation in N2a cells expressing mutant FUS and fibroblast cells derived from FUS ALS cases. In addition, the nonsense-mediated decay (NMD) pathway, which is closely related to protein translation, was altered by mutant FUS. Specifically, NMD-promoting factors UPF1 and UPF3b increased, whereas a negative NMD regulator, UPF3a, decreased, leading to the disruption of NMD autoregulation and the hyperactivation of NMD. Alterations in NMD factors and elevated activity were also observed in the fibroblast cells of FUS ALS cases. We conclude that mutant FUS suppresses protein biosynthesis and disrupts NMD regulation, both of which likely contribute to motor neuron death.
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease characterized by motor neuron death and progressive muscle wasting and paralysis. Approximately 15% of ALS cases are caused by inheritable genetic mutations, mostly in an autosomal dominant fashion. Mutations in the fused in sarcoma (FUS) gene were discovered to contribute to a subset of familial ALS (1, 2).
FUS is a DNA- and RNA-binding protein that is primarily localized to the nucleus, where it forms dynamic ribonucleoprotein granules. In contrast, ALS-related mutant FUS accumulates in the cytoplasm and forms stable ribonucleoprotein granules, which can lead to inclusion bodies and potentially contribute to neurotoxicity (3–5). FUS mutations have also been shown to impact many RNA metabolic processes, including transcription (6–8), splicing (9–11), mRNA transport (12), and stabilization (13), ultimately contributing to neuronal dysfunction. Recent studies demonstrated that mutations in FUS cause the liquid–liquid phase separation (LLPS) of FUS protein and the formation of self-assembled hydrogels (14) or liquid droplets in vitro (15, 16). It is noted that LLPS has also been reported for other RNA metabolic proteins involved in ALS, including TDP-43 (17), C9ORF72 dipeptide repeat (18), hnRNPA1 (17), and TIA1 (19). Thus, other cellular proteins are likely to be included in granules during LLPS in living cells, but the identities of such proteins remain to be determined.
This study started with testing the hypothesis that the identification of proteins associated with mutant FUS-dependent cytoplasmic granules is likely to provide critical insights into the toxic mechanism of mutant FUS. We developed a protocol to capture the dynamic mutant FUS-positive granules (3, 4) by membrane filtration and identified protein components by proteomic approaches. The bioinformatics analysis of proteins identified in wild-type (WT) and mutant FUS granules revealed multiple RNA metabolism pathways, among which protein translation and mRNA surveillance appeared to be novel.
We thus hypothesize that mutant FUS plays a role in protein translation. Two previous studies reported that ALS-linked FUS mutations were recruited to ribonucleoprotein granules; thus, FUS was speculated to be involved in protein translation (3, 20). However, protein translation was not measured in either study. Using three independent assays, we found that mutant FUS indeed impaired protein translation and that the cytoplasmic inclusions of mutant FUS were positive for stalled ribosomal complexes.
Mutations in FUS have been demonstrated to cause aberrant splicing (21); however, the molecular mechanism by which cells handle defective mRNA has not been explored in ALS. Nonsense-mediated decay (NMD) is a major mRNA surveillance system that is known to degrade defective mRNA and ∼3–20% of all mRNAs (22). NMD and protein translation are interrelated, as NMD utilizes the translocating ribosome as a proofreading mechanism for sensing defective mRNAs (23–25). We demonstrate that the phosphorylation level of a critical NMD regulator UPF1 (24), the NMD complex assembly, and the UPF1-mRNA binding all increased in the presence of mutant FUS, supporting that NMD is activated by mutant FUS. Additionally, two potent NMD-activating regulators [UPF1 and UPF3b (24, 26, 27)] were up-regulated, while the negative NMD regulator [UPF3a (28)] was down-regulated in skin fibroblast cells derived from a cohort of patients with ALS with FUS mutations compared with cells from control subjects, indicating disruption of the autoregulation of NMD. The hyperactivation of NMD was demonstrated using an NMD reporter assay in N2a cells and measuring endogenous mRNAs in the fibroblast cells of FUS ALS cases. Overall, the findings from this study thus provide an in-depth understanding of how RNA metabolism and protein translation are impacted by mutations in FUS and produce insights into the disease-causing mechanism of the mutant FUS subtype of ALS.
Results
Proteins Related to Translation and mRNA Surveillance Are Enriched in Mutant FUS Inclusions.
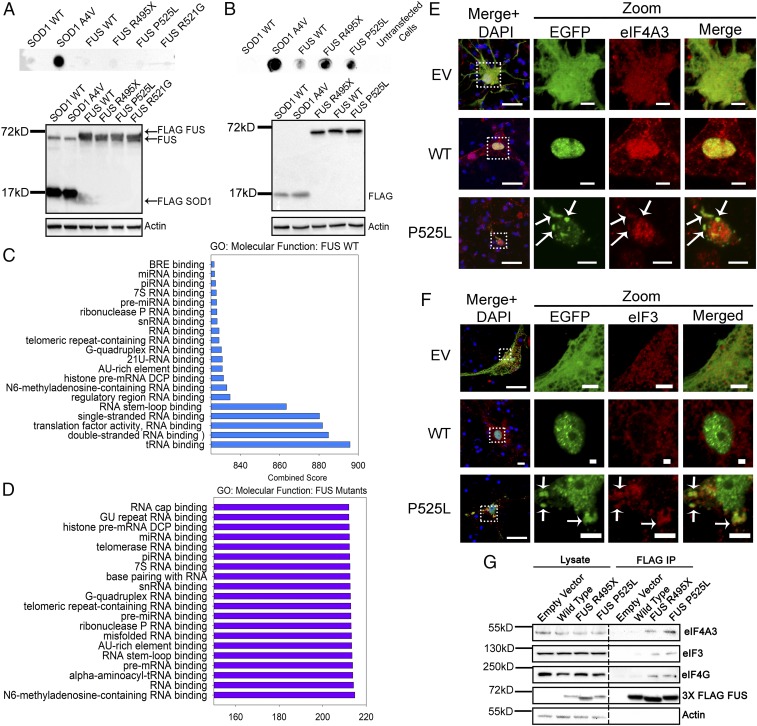
A more complete understanding of the protein composition that makes up the inclusions characteristic of mutant FUS-dependent ALS would provide a better understanding of the mechanism(s) driving the disease. To achieve this, we adapted a membrane filtration assay that was originally developed for detecting differentially soluble protein complexes (29). Utilizing previously described conditions in which radioimmunoprecipitation assay (RIPA) buffer was used to lyse cells (30), none of the WT, R495X, P525L, or R521G mutant FUS proteins were detected on the PVDF membrane (Fig. 1A). As a positive control and negative control, respectively, A4V mutant SOD1 was detected on the PVDF membrane filter, while WT SOD1 was not. The results suggest that, unlike A4V mutant SOD1-dependent cytoplasmic aggregates, mutant FUS cytoplasmic inclusions are dynamic and disassembled under the experimental conditions.
Fig. 1.
Proteomic identification, enrichment analysis, and validation of proteins in WT or mutant FUS inclusions isolated by membrane filtration. Membrane filtration, followed by dot blotting of granules isolated using the RIPA buffer (A) or the low-detergent hypotonic lysis buffer (B), was performed. The molecular function of proteins identified in WT (C) or mutant (D) FUS inclusions was analyzed using Enrichr software with the GO: Molecular Functions database. The top 20 most significant (P < 0.05) molecular functions are represented. Immunofluorescent staining of eIF4A3 (E) and eIF3 (F) in mouse primary cortical neurons was performed. Arrows indicate inclusions where proteins of interest are colocalized. (Scale bars: regular view, 20 μm; zoomed-in views, 5 μm.) (G) FUS IP, followed by Western blot for translation initiation factors (eIF4AIII, eIF3, and eIF4G) that were uniquely identified in mutant FUS inclusions.
A protocol using a hypotonic lysis buffer with a low detergent concentration was developed (SI Appendix, Fig. S1A). Using these conditions, P525L and R495X mutant FUS was detected on the membrane filter along with A4V mutant SOD1, whereas less WT FUS was detected and no WT SOD1 was detected (Fig. 1B). Native gel electrophoresis (4) also confirmed that FUS protein remained as an oligomeric species under these conditions, whereas FUS protein prepared in the RIPA buffer migrated faster (SI Appendix, Fig. S1B). We subjected the membrane “dots” to trypsin digestion followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using a published protocol (30). A total of 291, 268, and 278 proteins in FUS WT, R495X, and P525L, respectively, met the protein identification criteria described in SI Appendix and were considered significant identifications (Datasets S1–S3).
Proteins identified in the granules were subjected to functional enrichment analysis using the integrative tool, Enrichr (31, 32). The Gene Ontology (GO): Molecular Function database (33) and the DISEASES database (34) were utilized for analyzing protein functions and disease relevance, respectively. The top 20 most significant molecular function annotations aggregated from the GO: Molecular Function database by Enrichr revealed a variety of RNA-binding functions associated with WT (Fig. 1C) and mutant FUS (Fig. 1D). It is noted that several properties, including translation factor activity, tRNA binding, and RNA cap binding, are related to protein translation and mRNA surveillance mechanisms. The top 10 most significant results from the disease-gene association analysis for WT and mutant FUS (SI Appendix, Fig. S2) show a number of neurodegenerative diseases, including lateral sclerosis and Charcot–Marie–Tooth disease. Interestingly, the disease identified with a high significance was Diamond–Blackfan anemia, a severe ribosomopathy that results in defective protein synthesis (35). This association suggests that proteins identified in FUS granules are likely to be involved in protein translation. We therefore focused on testing the function of FUS in protein synthesis and related mRNA surveillance pathways.
We next examined the subcellular localization of critical proteins involved in protein translation (eIF3, eIF4A1, eIF4G, and rpS6) and mRNA surveillance (eIF4A3) that were identified in the MS results. In primary cortical neurons transfected with EGFP-tagged FUS, eIF4A3 (Fig. 1E), eIF3 (Fig. 1F), eIF4A1, eIF4G, and rpS6 (SI Appendix, Fig. S3 A–C, respectively) were all colocalized with cytoplasmic inclusions of mutant FUS. As a positive control, mutant FUS inclusions in primary neurons were positive for the stress granule marker G3BP1 (SI Appendix, Fig. S3D) as previously reported (4). The immunoprecipitation (IP) results also demonstrated that eIF3, eIF4G, and eIF4A3 interacted more with the mutant than WT FUS (Fig. 1G), validating the proteomic and colocalization results.
Protein Translation Is Impaired in the Presence of Mutant FUS.
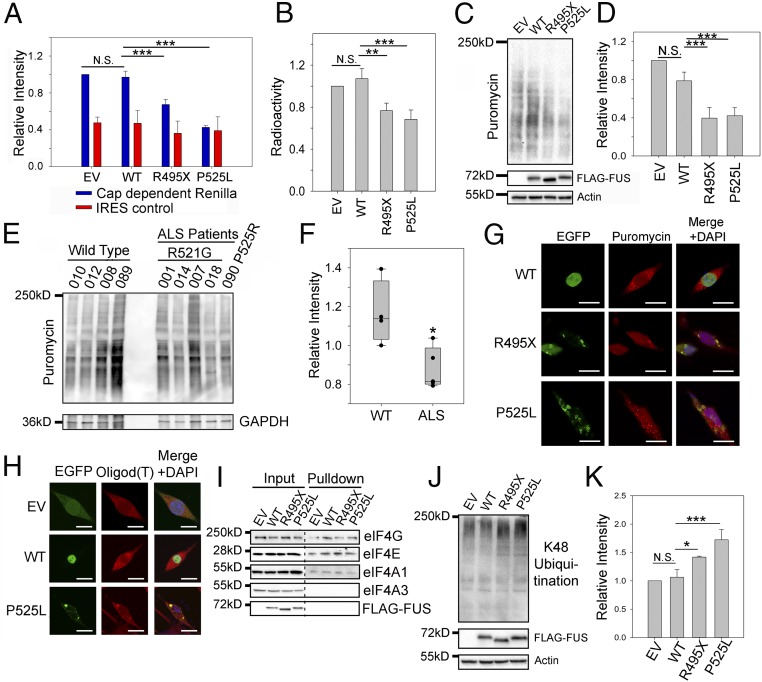
Based on the above proteomic identifications, GO enrichment analysis, and colocalization of translation machinery to the mutant FUS inclusions, we set out to test whether protein translation is impaired by ALS mutants of FUS using three independent assays. Utilizing a translation reporter assay (36), we examined how mutant FUS changed cap-dependent protein translation. Cotransfection of R495X or P525L mutant FUS with the reporter construct resulted in 50% and 70% reductions in the translation of the Renilla reporter gene, respectively (Fig. 2A). No detectable change was observed in cells transfected with empty vector (EV) or WT FUS. The control internal ribosomal entry site (IRES)-dependent translation of the luciferase gene was not changed (Fig. 2A). The second assay using an in vitro 35S-methionine (35S-Met) incorporation assay to measure translation of native mRNA showed that 35S-Met incorporation decreased by 25% and 35%, respectively, in the presence of mutant FUS compared with WT FUS or the EV control (Fig. 2B). Third, to examine endogenous protein translation, we used the surface sensing of translation (SUnSET) assay in which puromycin was used as a structural analog of aminoacyl tRNAs to prevent elongation after being incorporated into the nascent polypeptide chain (37). N2a cells were transfected with either EV, WT, or mutant FUS and treated with puromycin. Western blot analysis using an antipuromycin antibody showed reduced translation in cells expressing mutant FUS (Fig. 2 C and D). Finally, we carried out the SUnSET assay to examine protein translation in the skin fibroblast cells derived from patients with familial ALS who were carrying R521G or P525R (38) FUS mutations and from healthy controls with WT FUS (Fig. 2 E and F). Protein translation decreased by ∼30% in fibroblast cells from FUS ALS cases. The above results consistently support that mutant FUS represses protein translation.
Fig. 2.
Protein translation is impaired in the presence of mutant FUS. (A) Cap-dependent translation assay using the luciferase reporter in N2a cells expressing EV, WT, or mutant FUS. Blue and red bars represent the luminescence of the Renilla cap-dependent reporter and the Firefly luciferase transfection control, respectively. (B) In vitro 35S-Met incorporation assay using rabbit reticulocyte lysate mixed with N2a cell lysate containing EV, WT, or mutant FUS. After 1 h of incubation, proteins were precipitated and radioactivity was measured using a scintillation counter. Counts were normalized to the EV. (C and D) SUnSET assay measuring puromycin incorporated into proteins during translation in N2a cells expressing EV, WT, or mutant FUS. Western blots of puromycinylated proteins, FUS, and actin loading control are shown in C. Quantification in D was performed using the intensity of puromycinylated proteins in each lane to normalize against actin and EV. (E and F) SUnSET assay measuring puromycin incorporated into proteins during translation in fibroblast cells of patients with FUS ALS. Western blots of puromycinylated proteins and a GAPDH loading control are shown in E, and quantification results are shown in F. (G) Immunofluorescent staining of puromycinylated proteins in N2a cells expressing EV or EGFP-tagged WT or mutant FUS. Cells were incubated with puromycin for 30 min, fixed using paraformaldehyde (PFA), and stained with the antipuromycin antibody. (Scale bars: 20 μm.) (H) RNA FISH using a Cy3-tagged 21-mer oligo d(T) probe in N2a cells expressing WT or mutant FUS. (Scale bars: 20 μm.) (I) 7′MG pulldown to assess the cap binding and protein translation initiation in N2a cell lysate containing WT or mutant FUS. Various initiation complex members (eIF4E, eIF4G, and eIF4AI), FUS, and a negative control (eIF4AIII) were blotted. (J) K48 polyubiquitination in N2a cells expressing EV, WT, or mutant FUS with FUS expression and actin loading control. (K) Quantification of J using the K48 polyubiquitination intensity in each lane to normalize against actin and EV. Error bars in the figure represent SDs for three biological replicates. *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.001. N.S., not significant. ANOVA with a post hoc Tukey honest significant difference test was used to determine P values for multiple pairwise comparisons in A, B, D, and K. A Student’s t test was used to determine P values for simple pairwise comparison in F.
We next examined whether FUS inclusions contained puromycinlated proteins. Immunofluorescence using the same antipuromycin antibody showed that puromycinlated proteins were colocalized with mutant FUS inclusions (Fig. 2G), suggesting that translation complexes are localized in mutant FUS inclusions. Moreover, to test whether mRNA was also localized to the inclusion, we performed RNA fluorescence in situ hybridization (FISH) with two different RNA probes in N2a cells expressing WT or P525L mutant FUS. A generic Cy3 oligo d(T)21 probe was used to test mature polyadenylated mRNAs (Fig. 2H). Mature mRNA was distributed throughout the cytoplasm in cells with EV or WT FUS. However, a significant accumulation of mRNA in the mutant FUS inclusions was observed, which is consistent with previous observations (39). FUS has been reported to regulate the splicing of its own transcript (10); thus, we tested a probe specific to FUS transcript (exon 4) and found that FUS mRNA was also localized to the mutant FUS inclusions (SI Appendix, Fig. S3E). The results support that mutant FUS inclusions were colocalized with translation machinery and mature mRNAs.
We next examined whether translation initiation is impaired by mutant FUS. Binding of the initiation factor eIF4E to the 5′ cap of mRNA is the rate-limiting step in translation initiation where regulation often occurs (40). We performed a 7′-methylguanosine (7′MG) pulldown as an in vitro cap-binding assay (41) in the presence of WT or mutant FUS. After 7′MG pulldown, key members of the preinitiation complex were observed (Fig. 2I). Similar levels of eIF4G, eIF4E, and eIF4A1 were pulled down regardless of the presence of WT or mutant FUS. Neither WT nor mutant FUS bound to the 5′ cap. As a negative control, eIF4A3 also did not bind to the 5′ cap. The results suggest that mutant FUS does not interfere with the binding of the initiation complex to the 5′ cap structure in the translation initiation stage. Thus, it is likely that translation is disrupted by mutant FUS after the initiation step, resulting in premature termination.
We hypothesized that prematurely terminated polypeptides resulting from defective translation in the presence of mutant FUS will be polyubiquitinated and targeted for degradation. Therefore, we examined the level of K48-linked polyubiquitination in cells expressing WT or mutant FUS, since it is the major signal for targeting substrates for proteasomal degradation. Using an anti-K48 polyubiquitination tandem ubiquitin-binding entity (42), we found that the K48 polyubiquitination level increased ∼1.5- to 1.8-fold in cells expressing mutant FUS compared with WT FUS and the EV control (Fig. 2 J and K). In contrast, the level of K63-linked polyubiquitination, which is involved with other nonproteasome processes, did not change with either WT or mutant FUS (SI Appendix, Fig. S4). These results collectively support that ALS mutations in FUS cause defects in protein translation.
NMD Pathway Is Activated by Mutant FUS.
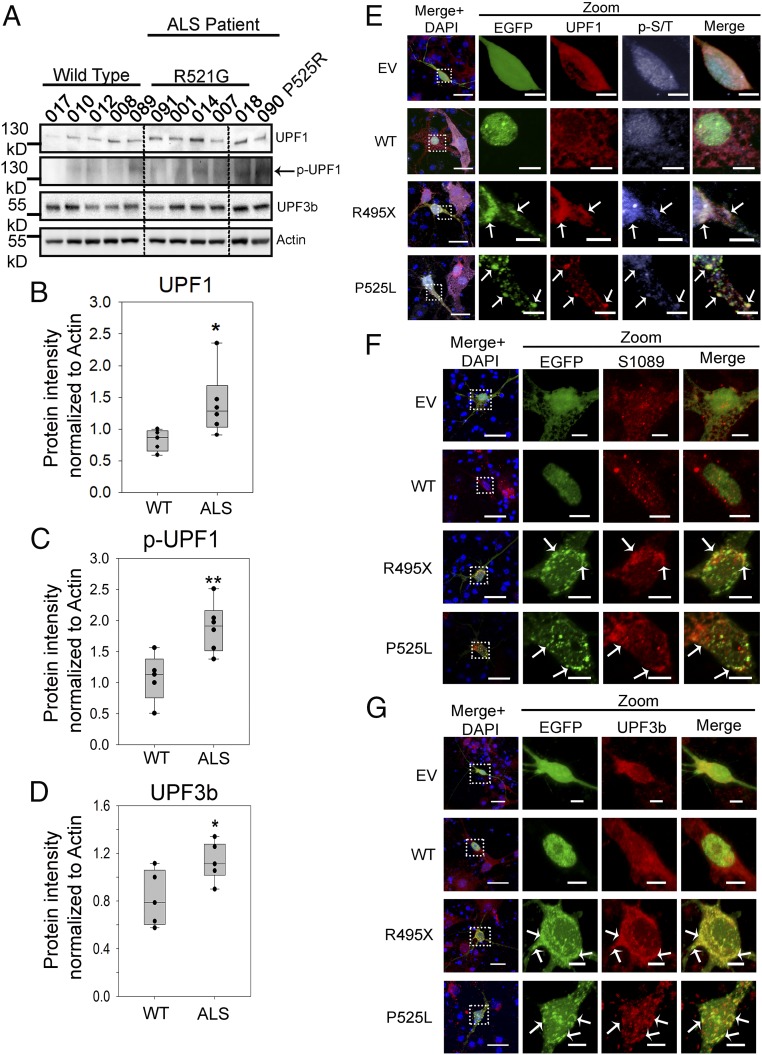
Protein translation and mRNA surveillance pathways are interrelated (23, 25). It has been reported that the inhibition of protein translation elongation by cycloheximide (43) can up-regulate NMD factors and activate NMD (44). Thus, we set to examine whether mutations in FUS can impact the mRNA degradative pathway NMD. UPF1 (24, 27) and UPF3b (28) are two critical positive regulators of NMD, and UPF1 phosphorylation (p-UPF1) is a critical step in NMD activation (24, 45). We evaluated the levels of UPF1, p-UPF1, and UPF3b in the skin fibroblast cells derived from six FUS ALS cases and five healthy controls (Fig. 3A). The antibody used for p-UPF1 was an anti–phospho-Ser/Thr ATM/AMR substrate (p-S/T) previously reported (46). Quantitative results show that the protein level of UPF1 (Fig. 3B), p-UPF1 (Fig. 3C), and UPF3b (Fig. 3D) increased in patients with ALS compared with healthy controls by 25%, 70%, and 35%, respectively. Similarly, N2a cells expressing P525L or R495X mutant FUS had increased levels of UPF1 and UPF3b (SI Appendix, Fig. S5) and p-UPF1 (SI Appendix, Fig. S6A). The results provide initial evidence that NMD activation is elevated in the presence of mutant FUS in the cells of patients with ALS with mutations in FUS.
Fig. 3.
Up-regulation of pro-NMD factors in cells of patients with familial ALS and in primary neurons expressing mutant FUS. (A–D) Levels of pro-NMD factors in patients with ALS carrying the R521G or P525R mutation and in control subjects with WT FUS. Western blots of UPF1, p-UPF1, UPF3b, and actin control were performed (A), and quantification of UPF1 (B), p-UPF1 (C), and UPF3b (D) was normalized against actin and obtained from three replicates. Error bars represent the SD between individuals. Quantifications were compared with healthy controls using a Student’s t test. *P ≤ 0.05; **P ≤ 0.005. (E–G) Immunofluorescent staining of UPF1, p-UPF1, and UPF3b in mouse primary neurons transfected with EV, EGFP-tagged WT, or mutant FUS at day 4 of in vitro culture. (E) Immunofluorescent staining of UPF1 and p-UPF1 using an anti–p-S/T ATM/AMR substrate antibody. (F) Immunofluorescent staining of p-UPF1 using an antibody against phosphor-S1089 in UPF1. (G) Immunofluorescent staining of UPF3b. Arrows indicate inclusions where proteins of interest are colocalized. (Scale bars: regular views, 20 μm; zoomed-in views, 5 μm.)
We next examined the subcellular localization of endogenous NMD factors in primary cortical neurons transfected with WT or mutant FUS. The colocalization of mutant FUS with UPF1 and p-UPF1 was demonstrated using two different p-UPF1 antibodies: the p-S/T antibody used in the Western blot analysis (Fig. 3E) and a different antibody against phosphor-S1089 of UPF1 (47) (Fig. 3F). Two additional NMD factors, UPF3b (Fig. 3G) and XRN1 (SI Appendix, Fig. S7A), were also localized in cytoplasmic inclusions of mutant FUS in primary neurons. SMG6 was not localized in mutant FUS inclusions serving as a control (SI Appendix, Fig. S7B). Similar results were obtained from N2a cells expressing WT or mutant FUS. Cytoplasmic inclusions of FUS were positive for UPF1 and p-UPF1, UPF3b, and XRN1 (SI Appendix, Fig. S6 A–C). SMG6 and eRF3b showed little localization to mutant FUS inclusions in N2a cells (SI Appendix, Fig. S6 D and E). The colocalization of NMD factors in mutant FUS inclusions further suggests that mutant FUS may impact the NMD pathway.
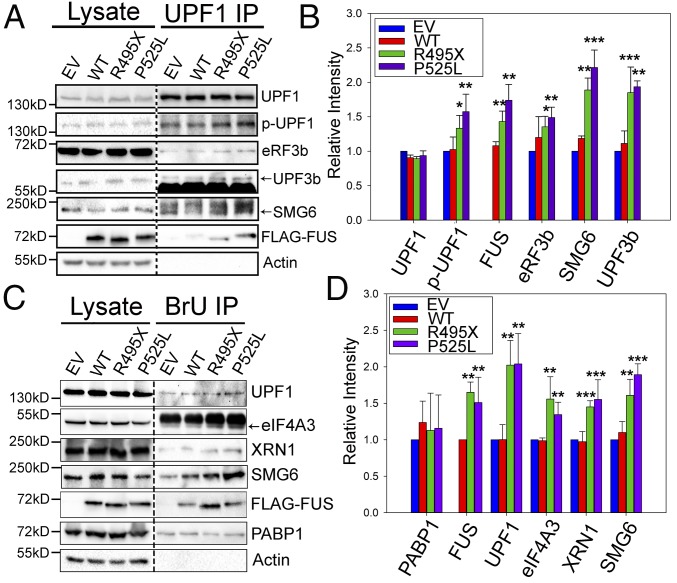
Since the assembly of key NMD factors with UPF1 is a critical aspect of pathway activation (48, 49), we next examined the assembly of NMD factors in the presence of WT or mutant FUS. Endogenous UPF1 IP followed by Western blots for various NMD factors is shown in Fig. 4A. Quantitative analysis showed that the UPF1 interaction with p-UPF1, UPF3b, the endonuclease SMG6, and FUS increased ∼1.5- to 2.0-fold, with statistical significance in the presence of mutant FUS (Fig. 4B). In addition, the translation termination factor eRF3b increased in the presence of mutant FUS (Fig. 4B). Enhanced assembly of NMD factors is additional evidence that NMD was activated by mutant FUS.
Fig. 4.
Interaction of NMD factors with UPF1 and RNAs increased in the presence of mutant FUS. (A and B) NMD factors coprecipitated with endogenous UPF1 from N2a cells expressing EV, WT, or mutant FUS. Immunoblots of UPF1, p-UPF1, eRF3b, UPF3b, SMG6, and 3× FLAG-FUS are shown in A, and quantitative results are shown in B. Protein intensities were normalized to corresponding UPF1 bands and compared with EV. (C and D) NMD factors coprecipitated with BrdU-containing RNAs. N2a cells expressing EV, WT, or mutant FUS were incubated with 1 μM BrdU, and RNAs were UV cross-linked to proteins. BrdU IP was performed using an anti-BrdU antibody, followed by Western blots for UPF1, eIF4A3, XRN1, SMG6, FUS, PABP1, and actin. BrU, bromouridine. Quantification of proteins in C is shown in D. Proteins were normalized to the loading control, PABP1, and compared with EV. The purple, green, blue, and red bars represent EV, WT FUS, R495X FUS, and P525L FUS, respectively. Error bars represent SDs for three biological replicates. *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.001. Quantifications were compared with EV using a Student’s t test.
We also tested the interaction of core NMD factors with mRNAs. We treated cells with bromouridine to label mRNAs, immunoprecipitated the labeled RNA with anti-BrdU antibody (50), and assessed NMD proteins by Western blot (Fig. 4C). Quantitative analysis showed that higher levels of NMD components responsible for triggering NMD (eIF4A3 and UPF1) and mRNA degradation (SMG6 and XRN1) were bound to RNAs in the presence of mutant FUS (Fig. 4D). An RNA-binding protein, PABP1, was used as a loading control and showed similar loading in all samples. The above results consistently support that the NMD pathway was activated by mutant FUS.
NMD Factors Are Dysregulated in Fibroblasts in FUS ALS Cases.
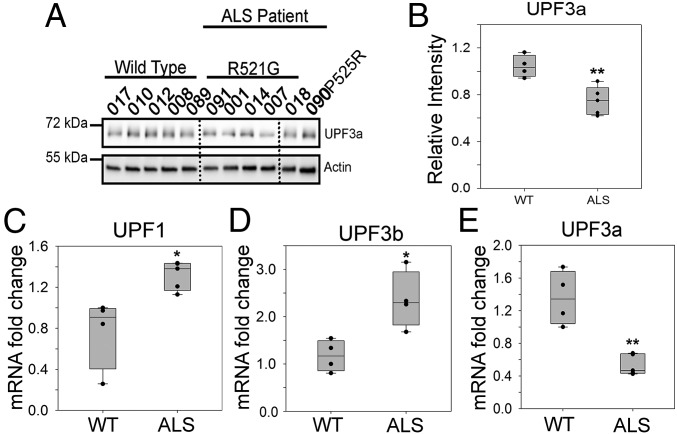
We demonstrated earlier that two positive regulators of NMD (UPF1 and UPF3b) and UPF1 phosphorylation increased in the skin fibroblast cells derived from six FUS ALS cases compared with five healthy controls (Fig. 3 A–D). The NMD pathway is tightly regulated by multiple mechanisms, including the molecular brake UPF3a (28). UPF3a competitively blocks the interaction of UPF3b with UPF2, thus delaying the activation of NMD. We examined protein levels of UPF3a in six FUS ALS cases (Fig. 5A) and found ∼30% lower levels of UPF3a protein in the fibroblast cells from these patients (Fig. 5B). Similarly, UPF3a protein levels decreased in N2a cells overexpressing the R495X and P525L mutants (SI Appendix, Fig. S8 A and B).
Fig. 5.
Down-regulation of the NMD negative regulator UPF3a in cells of patients with familial ALS. (A and B) Protein levels of the NMD negative regulator UPF3a in six patients with ALS and five control subjects, as shown in Fig. 3 A and B. Western blots of UPF3a and an actin control (A) and quantification of UPF3a normalized against actin (B) are shown. (C–E) Quantification of mRNA levels of dysregulated NMD factors. qPCR of UPF1 (C), UPF3b (D), and UPF3a (E) was performed using the cycle threshold method and is presented as the fold change in patients with ALS versus controls. Actin was used to normalize cycle threshold values. Error bars represent the SD between individuals. *P ≤ 0.05; **P ≤ 0.005. Quantifications were compared with healthy controls using a Student’s t test.
We further examined how the mRNA levels of UPF1, UPF3b, and UPF3a changed in the FUS ALS cases. The qPCR results show elevated levels of UPF1 (Fig. 5C) and UPF3b (Fig. 5D) and decreased levels of UPF3a (Fig. 5E) in the fibroblast cells of FUS ALS cases. Similar results were obtained for mRNA levels of these factors in N2a cells expressing WT or mutant FUS (SI Appendix, Figs. S5 C and D and S8C). Consistent changes in both mRNA and protein levels of the pro-NMD factors (UPF1 and UPF3b) and the negative regulator (UPF3a) illustrate a pattern of NMD dysregulation in the mutant FUS-linked familial patients with ALS, which will contribute to NMD hyperactivation.
UPF1-Mediated Autoregulation of NMD Is Impaired in FUS ALS Cases.
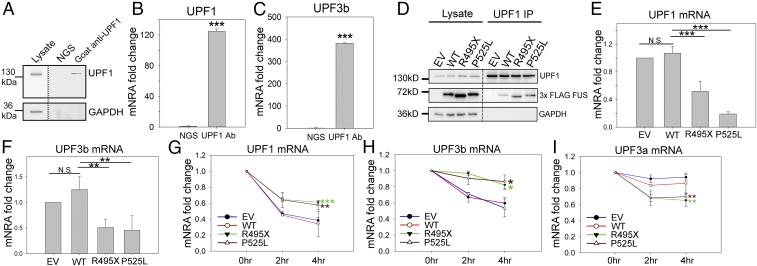
Core NMD factors, including UPF1 and UPF3b, are regulated through an intricate autoregulatory mechanism, by which their own mRNAs are targeted for NMD (44, 51). Given the dysregulation of NMD factors as shown above, we hypothesized that the ALS mutations in FUS disrupt the autoregulatory mechanism of NMD. To test this hypothesis, we first performed endogenous UPF1 IP followed by qPCR to examine whether the UPF1 protein binds its own mRNA and UPF3b mRNA. Using normal goat serum as a control, UPF1 protein was specifically pulled down by a UPF1 antibody (Fig. 6A). Along with the UPF1 protein, UPF1 mRNA (Fig. 6B) and UPF3b mRNA (Fig. 6C) were also pulled down. We then overexpressed WT, P525L, or R495X mutant FUS in N2a cells and performed a similar RNA IP experiment (Fig. 6 D–F). Quantitative analysis showed that, consistent with earlier results (Fig. 4A), higher levels of mutant FUS were pulled down with the UPF1 protein (Fig. 6D). More importantly, lower levels of UPF1 mRNA (Fig. 6E) and UPF3b mRNA (Fig. 6F) were pulled down along with the UPF1 protein in the presence of mutant FUS, suggesting that mutant FUS led to a lower turnover of UPF1 and UPF3b mRNA by NMD. The dampened autoregulatory mechanism through UPF1 binding supports the observation of increases in the mRNA and protein levels of UPF1 and UPF3b.
Fig. 6.
Disruption in the NMD autoregulation loop. Endogenous UPF1 IP from N2a cells was performed, followed by Western blot (A) and qPCR measurement of UPF1 (B) and UPF3b (C) mRNAs. N2a cell lysate was subjected to IP using normal goat serum (NGS) or goat anti-UPF1 antibody. The IP samples were aliquoted for Western blot (A) and qPCR quantification (B and C) comparing RNA coprecipitated with UPF1 protein versus NGS control. UPF1 IP from N2a cells expressing EV, WT, or mutant FUS was performed, followed by Western blot (D) and qPCR measurement of UPF1 (E) and UPF3b (F) mRNAs. UPF1, FUS, and GAPDH were assessed by Western blot, as shown in D. qPCR quantification was normalized to UPF1 protein precipitated and presented as fold change compared with EV. Turnover rates of UPF1 (G), UPF3b (H), and UPF3a (I) mRNAs in N2a cells expressing EV, WT, or mutant FUS are shown. Actinomycin D or DMSO control was added 2 or 4 h before harvesting for RNA isolation. Individual mRNAs of interest were quantified by qPCR, normalized against RPL13a, and presented as fold change versus DMSO treatment over time. Error bars represent the SD from three replicates. *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.001. N.S., not significant. ANOVA with a post hoc Tukey honest significant difference test was used in E–I, and a Student’s t test was used in B and C.
To further examine how decay is influenced by FUS mutations, we measured the UPF1, UPF3b, and UPF3a mRNA levels by qPCR after treating N2a cells expressing WT, P525L, or R495X mutant FUS with the transcriptional inhibitor actinomycin D. In cells expressing mutant FUS, the decay of UPF1 (Fig. 6G) and UPF3b (Fig. 6H) mRNA was significantly slower than in controls. In contrast, the mRNA decay of the NMD negative regulator UPF3a was significantly faster in cells expressing mutant FUS (Fig. 6I). The results collectively support that the stability of NMD factor mRNA was dysregulated by mutant FUS in a UPF1-dependent manner (i.e., the NMD autoregulatory circuit is impaired).
Enhanced Decay of NMD Substrates in the Presence of ALS Mutant FUS.
Based on the above findings on the dysregulation of NMD factors, we next tested whether the NMD activity is hyperactivated using four well-characterized NMD reporters [β-globin and GPX-1 with and without a premature stop codon (PTC)] (52), as well as a cohort of documented endogenous NMD substrates (53, 54). The levels of all four reporter transcripts (WT β-globin, WT GPX-1, PTC β-globin, and PTC GPX-1) were consistently lower in N2a cells expressing mutant FUS than in cells expressing WT FUS (SI Appendix, Fig. S9). It is noted that the transcript levels in WT FUS-expressing cells were unchanged compared with the EV control, with the exception of PTC GPX-1 (SI Appendix, Fig. S9D). The NMD reporter assays support higher NMD turnover of normal and PTC-containing mRNAs in the presence of mutant FUS.
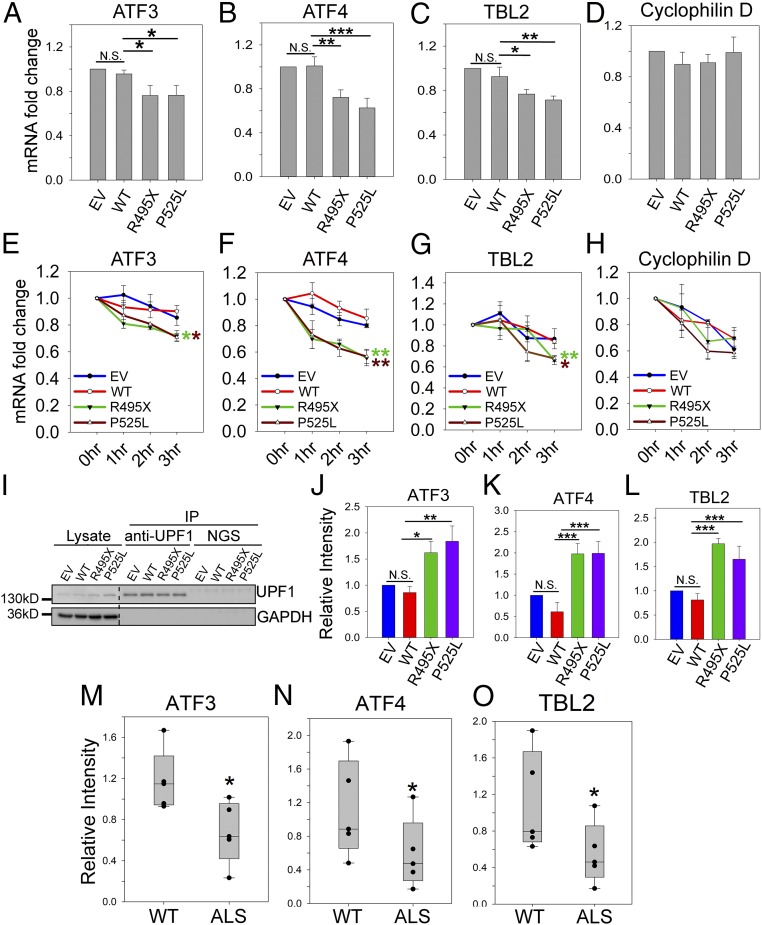
To better characterize NMD activity, we measured the mRNA levels of three NMD substrates: ATF3, ATF4, and TBL2 (53, 54). Total mRNA levels of ATF3, ATF4, and TBL2 (Fig. 7 A–C) decreased in N2a cells expressing R495X or P525L mutant FUS compared with cells expressing EV and WT FUS. As a control, mRNA levels of cyclophilin D did not change (Fig. 7D). We next measured the time course of mRNA levels after transcription inhibition by actinomycin D. The mRNA decay of ATF3, ATF4, and TBL2 (Fig. 7 E–G) was significantly faster in cells expressing R495X or P525L mutant FUS compared with cells expressing WT FUS. As a control, the decay of cyclophilin D did not differ between cells expressing mutant and WT FUS (Fig. 7H). Furthermore, we examined whether higher levels of these mRNAs were associated with UPF1. UPF1 protein IP was performed followed by qPCR to measure the amount of UPF1-bound mRNAs. While similar levels of UPF1 protein were immunoprecipitated (Fig. 7I), higher levels of ATF3, ATF4, and TBL2 mRNAs were bound to UPF1 in the presence of mutant FUS (Fig. 7 J–L). All three lines of evidence support the enhanced NMD decay of these endogenous substrates in cells expressing mutant FUS.
Fig. 7.
Enhanced NMD activity in the presence of mutant FUS. The mRNA levels of ATF3 (A), ATF4 (B), TBL2 (C), and cyclophilin D (D) in N2a cells transfected with EV, WT, or mutant FUS were determined. The levels of the indicated mRNA were quantified by qPCR using the cycle threshold (ΔΔCT) method, and the fold changes compared with WT are presented. Turnover rate of ATF3 (E), ATF4 (F), TBL2 (G), and cyclophilin D (H) mRNAs in N2a cells expressing EV, WT, or mutant FUS after treatment with actinomycin or DMSO control. Individual mRNAs of interest were quantified by qPCR, normalized against RPL13a, and presented as fold change versus DMSO treatment over time. Error bars represent the SD from three replicates. (I–L) Amount of ATF3, ATF4, and TBL2 mRNA bound to the UPF1 protein. N2a cells were cotransfected with EV, WT, or mutant FUS and an NMD reporter as indicated. After UPF IP, Western blot (I) demonstrates levels of UPF1 in lysate and IP samples. The levels of ATF3 (J), ATF4 (K), and TBL2 (L) mRNA in the UPF1 IP samples were quantified by qPCR using the ΔCT method. The fold changes normalized to WT are presented from three replicates. The mRNA levels of ATF3 (M), ATF4 (N), and TBL2 (O) in fibroblast cells derived from patients with familial ALS carrying FUS mutations and healthy WT controls are shown. *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.001. N.S., not significant. ANOVA with a post hoc Tukey honest significant difference test was used to determine P values in A–L. A Student’s t test was used to determine P values in M–O.
We next examined the mRNA levels of ATF3, ATF4, and TBL2 in fibroblast cells derived from patients with familial ALS. Indeed, the levels of all three mRNAs were lower in cells of patients with ALS than in healthy controls with WT FUS (Fig. 7 M–O). The results suggest that the NMD activity is induced in clinically relevant samples.
Discussion
FUS (14–16) and other proteins implicated in ALS (17–19, 55) have been reported to undergo LLPS and form liquid droplets, which facilitates the formation of membrane-less RNA-protein granules and inclusions (3–5). This study started with developing a method to isolate dynamic FUS-containing granules and identifying their protein compositions. Enrichment analysis of identified proteins implied that both WT and mutant FUS are involved in protein translation and mRNA surveillance (Fig. 1 C and D and SI Appendix, Fig. S2). Tight spatiotemporal regulation of protein synthesis in a motor neuron is critical for its function and survival (56), and reduced protein synthesis can be detrimental to normal neuronal function (57, 58). Moreover, mRNA surveillance is intimately integrated into protein translation (23). For instance, eIF4A3 is a core exon junction complex member that aids in initiating NMD (49). eIF3 is classically known as a critical initiation factor; however, it is also required for efficient translation termination in the event of NMD and promotes ribosomal recycling (59, 60). However, it is unknown how defects in mRNA surveillance are linked to suppression of protein synthesis by ALS mutation in FUS. The colocalization of eIF4A3 and eIF3 in mutant FUS inclusions (Fig. 1 E and F) led us to probe how protein translation and NMD are altered by mutant FUS and to discover the underlying mechanisms.
We used three independent assays to provide direct evidence that mutant FUS negatively impacted global protein production (Fig. 2 A–D). Furthermore, the SUnSET assay also detected significant reduction of protein translation in fibroblast cells derived from patients with familial ALS with two different FUS mutations (Fig. 2 E and F). In addition, mutant FUS inclusions were colocalized with mRNAs (Fig. 2H) and puromycinylated peptides (Fig. 2G), suggesting that such inclusions are sites of defective protein synthesis with stalled translation complexes. Mutant FUS inclusions have been reported as stress granule-like with stress granule markers such as G3BP1 and TIA1, but they display altered dynamics compared with heathy cells with endogenous WT FUS (61–64). We suggest that the impairment of protein translation as shown in this study is a functional consequence of the sequestration of the translation machinery in mutant FUS inclusions.
The above results raised the question how mRNAs resulting from impaired translation would be handled. It was reported that suppression of translation using cycloheximide up-regulated proteins involved in the NMD pathway, particularly UPF1 and UPF3b (44). Our proteomic analysis also suggested that proteins involved in the mRNA surveillance pathway were enriched in mutant FUS inclusions. We observed increased pro-NMD proteins UPF1 and UPF3b in fibroblast cells derived from a cohort of patients with familial ALS bearing two different FUS mutations, R521G and P525R (Fig. 3 A–D), as well as in N2a cells expressing mutant FUS (SI Appendix, Fig. S5). In addition, UPF1 phosphorylation (Fig. 3 E and F), NMD complex assembly (Fig. 4 A and B), and UPF1-mRNA binding (Fig. 4 C and D) all increased in the presence of mutant FUS, suggesting an elevated level of NMD activity as we demonstrated with an NMD reporter assay (SI Appendix, Fig. S9), in three endogenous NMD substrates in N2a cells expressing mutant FUS (Fig. 7 A–H), and in fibroblast cells from patients with FUS ALS (Fig. 7 M–O). We rationalized that higher levels of core NMD factors in mutant FUS inclusions would aid in the degradation of RNAs associated with prematurely terminated translation complexes, thus playing a protective role in FUS ALS. A yeast genetic screen identified that UPF1 rescued mutant FUS toxicity in Saccharomyces cerevisiae (65), and follow-up studies showed a similar protective effect of UPF1 overexpression in primary neurons (66) and TDP-43 rat models (67).
Different from UPF1 and UPF3b, UPF3a functions as a molecular brake by competing with UPF3b for interaction with UPF2 and delaying activation of the pathway (28). To our surprise, we found that both protein and mRNA levels of UPF3a decreased in the same cells of patients with familial ALS with mutations in FUS (Fig. 5). Loss of the down-regulatory mechanism could result in aberrant activation of NMD. Moreover, NMD is regulated by an intricate autoregulatory circuit to prevent overt activation of NMD. Specifically, the mRNA levels of NMD factors UPF1 and UPF3b are degraded through the NMD pathway itself (44, 51). Our results from mRNA decay experiments demonstrate the stabilization of the pro-NMD factors UPF1 and UPF3b and an increased degradation of the negative regulator UPF3a (Fig. 6 G–I), suggesting a disruption in the autoregulatory circuit. These results consistently support a model (Fig. 8) in which the NMD pathway is dysregulated and hyperactivated in the presence of mutant FUS. It was reported that UPF1 overexpression could increase the available pool of UPF1 to reactivate the autoregulatory feedback (51), thus enabling the degradation of UPF1 and UPF3b mRNAs and dampening the hyperactivation of NMD. This mechanism can provide an explanation of the reported protective effect of UPF1 overexpression in TDP-43 and FUS ALS models (65–67).
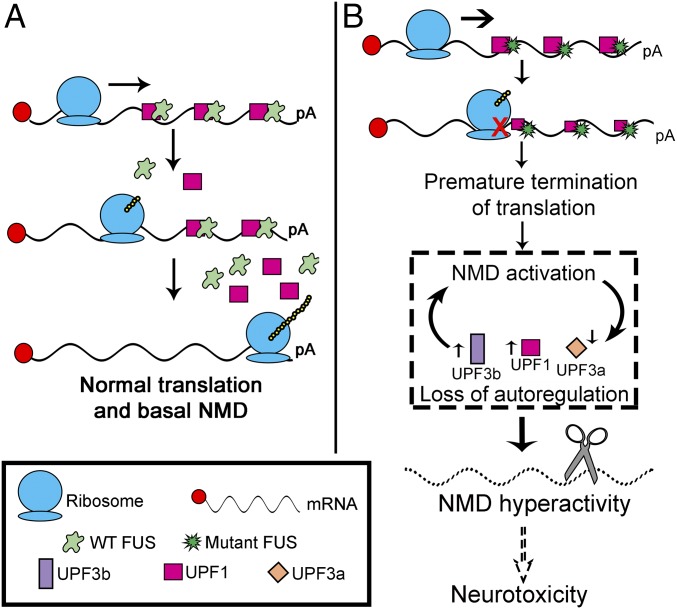
Fig. 8.
Model illustrating how mutant FUS impairs NMD regulation and suppresses protein translation. (A) Normal translation and basal NMD under physiological conditions with WT FUS. (B) Mutant FUS has greater binding ability to mRNAs and associated proteins, including UPF1, leading to ribosomal stalling, translation termination, and subsequent activation of NMD. Up-regulation of pro-NMD factors (UPF1 and UPF3b) and down-regulation of the molecular brake (UPF3a) cause the loss of autoregulation and hyperactivity of NMD. pA, poly(A) tail.
Dysregulation of NMD factors can, in turn, contribute to suppressing protein translation. For instance, besides its function of promoting NMD, UPF3b was reported to recruit termination factor eRFs to stalled ribosomes and to terminate protein translation (68). Interestingly, mutations in UPF3b can result in intellectual disabilities, autism spectrum disorder, and schizophrenia. These disorders are likely the consequence of defective NMD in dendrites and neurons, which results in deficient neuronal maturation and dendritic branching (69, 70).
In mutant FUS ALS, translation suppression and subsequent NMD activation appear to constitute a vicious cycle, as illustrated in Fig. 8. Increased translation termination events, potentially due to increased binding of mutant FUS to mRNAs, activate NMD at higher levels. Furthermore, the autoregulation of NMD is disrupted as the pro-NMD factors UPF1 and UPF3b increase and the molecular brake UPF3a decreases, contributing to the hyperactivation of NMD and increased degradation of natural NMD targets, such as ATF3, ATF4, and TBL2. This hyperactivity resulting from defects in translation termination may contribute to toxicity in motor neurons (Fig. 8). It is noted that critical steps in this model, including suppressed protein translation (Fig. 2 E and F); increased levels of UPF1, p-UPF1, and UPF3b protein (Fig. 3 A–D) and mRNA (Fig. 5 C and D); decreased levels of UPF3a protein (Fig. 5 A and B) and mRNA (Fig. 5E); and increased NMD degradation of ATF3, ATF4, and TBL2 mRNAs (Fig. 7 M–O), were consistently supported by results from fibroblast cells derived from patients with familial ALS carrying two different FUS mutations.
Although this study only demonstrated that mutant FUS suppressed global protein translation, it is conceivable that local translation in dendrites and axon terminals may also be impaired by mutant FUS. FUS has been demonstrated to be part of RNA transport granules and to be recruited to activated synapses (20, 71, 72). In cells bearing FUS mutations, however, there are defects in synaptic morphology and function (58, 71–75). Decreases in proteins required for synaptic maintenance and function may contribute to an ALS phenotype. Additionally, overactivation of NMD may produce deleterious effects in stress response pathways, including how cells respond to misfolded proteins, hypoxia, and DNA damage (22, 44). NMD also functions in fine-tuning the immune response by degrading mRNAs of proinflammatory factors (76). As neuroinflammation plays a role in ALS in a non–cell-autonomous fashion (77, 78), it is conceivable that dysregulation of NMD in astrocytes and microglia may also impact the immune response and contribute to the ALS phenotypes.
In summary, the mechanistic insights gained from this study begin to describe the role of FUS in protein translation and a critical mRNA quality control pathway, both of which are required for neuronal maintenance and function. Sequestration of UPF1 in mutant FUS inclusions, decrease in protein synthesis, NMD hyperactivation, or a combination of these events likely plays a role in neurodegeneration in ALS. It is noted that suppressed protein translation (Fig. 2 E and F), NMD activation (Fig. 3 A–D), disrupted NMD autoregulation (Fig. 5), and hyperactivity of NMD (Fig. 7 M–O) were consistently demonstrated in the fibroblast cells of patients with ALS with mutations in FUS. These mechanistic understandings support the notion that regulation of NMD and protein translation can serve as potential therapeutic targets for future development of new ALS treatment. The results also have a broader impact, since other RNA-binding proteins all undergo LLPS and form cytoplasmic granules, including TDP-43, C9ORF72 dipeptide repeat, hnRNPA1, and TIA1 (17–19, 55). Future studies will investigate whether these proteins, which are implicated in ALS, frontotemporal dementia, and related disorders, also influence the mRNA quality control pathway and impair protein translation.
Materials and Methods
Reagents, plasmids, oligonucleotide primers, and general methods for cell culture and transfection, primary neuron isolation and culture, skin fibroblast cell culture of patients with ALS, immunostaining, membrane filtration assay, LC-MS/MS, proteomics and protein functional enrichment analysis, IP, real-time RT-PCR, RNA FISH and confocal microscopy are described in SI Appendix, Materials and Methods. Critical protocols are briefly described below, and more details can be found in SI Appendix, Materials and Methods. Data are presented as means from three independent experiments. ANOVA with a post hoc Tukey honest significant difference test was used to determine P values for multiple pairwise comparisons.
Patient Skin Fibroblast Culture.
Human skin fibroblasts were prepared and maintained as previously described (38). Informed consent was obtained from all participants who donated a skin biopsy. Information on the 11 subjects (five patients with familial ALS with the R521G mutation, one patient with the P525R mutation, and five healthy controls with WT FUS) is shown in SI Appendix, Materials and Methods. The study was approved by the Institutional Review Board of the University of Kentucky. Details on the fibroblast cell culture are provided in SI Appendix, Materials and Methods.
Protein Translation Assays.
Protein translation efficiency was measured using three different assays: a cap-dependent translation reporter assay as previously described (36), an 35S-Met incorporation assay, and the SUnSET assay as previously described (37). The SUnSET assay uses puromycin as a structural analog of aminoacyl tRNAs to prevents elongation after being incorporated into the nascent polypeptide chain. Details on all three assays are provided in SI Appendix, Materials and Methods.
NMD Activity Assays.
NMD activity was assessed using an NMD reporter assay as previously described (52) or the qPCR of mRNA levels of endogenous NMD substrates (53, 54). Details are provided in SI Appendix, Materials and Methods.
Note Added in Proof.
During the production of this article, one group reported that FUS mutations suppress intraaxonal protein synthesis (79).
Supplementary Material
Acknowledgments
We thank Dr. Tianyan Gao, Dr. Jens Lykke-Andersen, and Dr. Jia Luo for the polio internal ribosomal entry site luciferase translation reporter plasmid, NMD reporters, and mouse breeding, respectively. We also thank Dr. Jozsef Gal for reading the manuscript. This study was supported, in part, by National Institutes of Neurological Disorder and Stroke Grant R01NS077284, Muscular Dystrophy Association Grant MDA352743, ALS Association Grant 6SE340, and Department of Veteran Affairs Merit Review Award I01 BX002149 (to H.Z.). Support from the Multidisciplinary Value Program initiative at the University of Kentucky College of Medicine is appreciated. M.K. is supported by National Institute of Environmental Health Sciences Training Grant T32ES007266 and the University of Kentucky College of Medicine Fellowship for Excellence in Graduate Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 12842.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810413115/-/DCSupplemental.
References
- 1.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 2.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murakami T, et al. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L, et al. Subcellular localization and RNAs determine FUS architecture in different cellular compartments. Hum Mol Genet. 2015;24:5174–5183. doi: 10.1093/hmg/ddv239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monahan Z, et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017;36:2951–2967. doi: 10.15252/embj.201696394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L, Gal J, Chen J, Zhu H. Self-assembled FUS binds active chromatin and regulates gene transcription. Proc Natl Acad Sci USA. 2014;111:17809–17814. doi: 10.1073/pnas.1414004111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo Y, et al. EWS and FUS bind a subset of transcribed genes encoding proteins enriched in RNA regulatory functions. BMC Genomics. 2015;16:929. doi: 10.1186/s12864-015-2125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz JC, et al. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 2012;26:2690–2695. doi: 10.1101/gad.204602.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogelj B, et al. Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci Rep. 2012;2:603. doi: 10.1038/srep00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y, Liu S, Liu G, Oztürk A, Hicks GG. ALS-associated FUS mutations result in compromised FUS alternative splicing and autoregulation. PLoS Genet. 2013;9:e1003895. doi: 10.1371/journal.pgen.1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishigaki S, et al. Position-dependent FUS-RNA interactions regulate alternative splicing events and transcriptions. Sci Rep. 2012;2:529. doi: 10.1038/srep00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 13.Udagawa T, et al. FUS regulates AMPA receptor function and FTLD/ALS-associated behaviour via GluA1 mRNA stabilization. Nat Commun. 2015;6:7098. doi: 10.1038/ncomms8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bao C, Lyu D, Huang S. Circular RNA expands its territory. Mol Cell Oncol. 2015;3:e1084443. doi: 10.1080/23723556.2015.1084443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel A, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 16.Murray DT, et al. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell. 2017;171:615–627.e16. doi: 10.1016/j.cell.2017.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molliex A, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boeynaems S, et al. Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell. 2017;65:1044–1055.e5. doi: 10.1016/j.molcel.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mackenzie IR, et al. TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron. 2017;95:808–816.e9. doi: 10.1016/j.neuron.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yasuda K, et al. The RNA-binding protein Fus directs translation of localized mRNAs in APC-RNP granules. J Cell Biol. 2013;203:737–746. doi: 10.1083/jcb.201306058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svetoni F, Frisone P, Paronetto MP. Role of FET proteins in neurodegenerative disorders. RNA Biol. 2016;13:1089–1102. doi: 10.1080/15476286.2016.1211225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karam R, Wengrod J, Gardner LB, Wilkinson MF. Regulation of nonsense-mediated mRNA decay: Implications for physiology and disease. Biochim Biophys Acta. 2013;1829:624–633. doi: 10.1016/j.bbagrm.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Celik A, He F, Jacobson A. NMD monitors translational fidelity 24/7. Curr Genet. 2017;63:1007–1010. doi: 10.1007/s00294-017-0709-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serdar LD, Whiteside DL, Baker KE. ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons. Nat Commun. 2016;7:14021. doi: 10.1038/ncomms14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karousis ED, Mühlemann O. Nonsense-mediated mRNA decay begins where translation ends. Cold Spring Harb Perspect Biol. June 11, 2018 doi: 10.1101/cshperspect.a032862. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan W-K, et al. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol. 2009;16:747–753. doi: 10.1038/nsmb.1612. [DOI] [PubMed] [Google Scholar]
- 27.Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell. 2010;143:938–950. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shum EY, et al. The antagonistic gene paralogs Upf3a and Upf3b govern nonsense-mediated RNA decay. Cell. 2016;165:382–395. doi: 10.1016/j.cell.2016.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gal J, et al. HDAC6 regulates mutant SOD1 aggregation through two SMIR motifs and tubulin acetylation. J Biol Chem. 2013;288:15035–15045. doi: 10.1074/jbc.M112.431957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamelgarn M, et al. Proteomic analysis of FUS interacting proteins provides insights into FUS function and its role in ALS. Biochim Biophys Acta. 2016;1862:2004–2014. doi: 10.1016/j.bbadis.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuleshov MV, et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen EY, et al. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pletscher-Frankild S, Pallejà A, Tsafou K, Binder JX, Jensen LJ. DISEASES: Text mining and data integration of disease-gene associations. Methods. 2015;74:83–89. doi: 10.1016/j.ymeth.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 35.Mills EW, Green R. Ribosomopathies: There’s strength in numbers. Science. 2017;358:eaan2755. doi: 10.1126/science.aan2755. [DOI] [PubMed] [Google Scholar]
- 36.Choo AY, Yoon S-O, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci USA. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 38.Kuang L, et al. Clinical and experimental studies of a novel P525R FUS mutation in amyotrophic lateral sclerosis. Neurol Genet. 2017;3:e172. doi: 10.1212/NXG.0000000000000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shelkovnikova TA, et al. Chronically stressed or stress-preconditioned neurons fail to maintain stress granule assembly. Cell Death Dis. 2017;8:e2788. doi: 10.1038/cddis.2017.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, et al. Snail determines the therapeutic response to mTOR kinase inhibitors by transcriptional repression of 4E-BP1. Nat Commun. 2017;8:2207. doi: 10.1038/s41467-017-02243-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hjerpe R, et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider-Poetsch T, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang L, et al. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell. 2011;43:950–961. doi: 10.1016/j.molcel.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamashita A. Role of SMG-1-mediated Upf1 phosphorylation in mammalian nonsense-mediated mRNA decay. Genes Cells. 2013;18:161–175. doi: 10.1111/gtc.12033. [DOI] [PubMed] [Google Scholar]
- 46.Durand S, Franks TM, Lykke-Andersen J. Hyperphosphorylation amplifies UPF1 activity to resolve stalls in nonsense-mediated mRNA decay. Nat Commun. 2016;7:12434. doi: 10.1038/ncomms12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Popp MW, Maquat LE. Attenuation of nonsense-mediated mRNA decay facilitates the response to chemotherapeutics. Nat Commun. 2015;6:6632. doi: 10.1038/ncomms7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okada-Katsuhata Y, et al. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012;40:1251–1266. doi: 10.1093/nar/gkr791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kashima I, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Imamachi N, et al. BRIC-seq: A genome-wide approach for determining RNA stability in mammalian cells. Methods. 2014;67:55–63. doi: 10.1016/j.ymeth.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 51.Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Mühlemann O. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA. 2011;17:2108–2118. doi: 10.1261/rna.030247.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee SR, Pratt GA, Martinez FJ, Yeo GW, Lykke-Andersen J. Target discrimination in nonsense-mediated mRNA decay requires Upf1 ATPase activity. Mol Cell. 2015;59:413–425. doi: 10.1016/j.molcel.2015.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 54.Viegas MH, Gehring NH, Breit S, Hentze MW, Kulozik AE. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the nonsense mediated decay pathway. Nucleic Acids Res. 2007;35:4542–4551. doi: 10.1093/nar/gkm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gopal PP, Nirschl JJ, Klinman E, Holzbaur EL. Amyotrophic lateral sclerosis-linked mutations increase the viscosity of liquid-like TDP-43 RNP granules in neurons. Proc Natl Acad Sci USA. 2017;114:E2466–E2475. doi: 10.1073/pnas.1614462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holt CE, Schuman EM. The central dogma decentralized: New perspectives on RNA function and local translation in neurons. Neuron. 2013;80:648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012;13:308–324. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu-Yesucevitz L, et al. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31:16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beznosková P, Wagner S, Jansen ME, von der Haar T, Valášek LS. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res. 2015;43:5099–5111. doi: 10.1093/nar/gkv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Protter DSW, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baron DM, et al. Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol Neurodegener. 2013;8:30. doi: 10.1186/1750-1326-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vance C, et al. ALS mutant FUS disrupts nuclear localization and sequesters wild-type FUS within cytoplasmic stress granules. Hum Mol Genet. 2013;22:2676–2688. doi: 10.1093/hmg/ddt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gal J, et al. Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol Aging. 2011;32:2323.e27–2323.e40. doi: 10.1016/j.neurobiolaging.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ju S, et al. A yeast model of FUS/TLS-dependent cytotoxicity. PLoS Biol. 2011;9:e1001052. doi: 10.1371/journal.pbio.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barmada SJ, et al. Amelioration of toxicity in neuronal models of amyotrophic lateral sclerosis by hUPF1. Proc Natl Acad Sci USA. 2015;112:7821–7826. doi: 10.1073/pnas.1509744112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jackson KL, et al. Preservation of forelimb function by UPF1 gene therapy in a rat model of TDP-43-induced motor paralysis. Gene Ther. 2015;22:20–28. doi: 10.1038/gt.2014.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neu-Yilik G, et al. Dual function of UPF3B in early and late translation termination. EMBO J. 2017;36:2968–2986. doi: 10.15252/embj.201797079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tarpey PS, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jolly LA, Homan CC, Jacob R, Barry S, Gecz J. The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum Mol Genet. 2013;22:4673–4687. doi: 10.1093/hmg/ddt315. [DOI] [PubMed] [Google Scholar]
- 71.Fujii R, et al. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr Biol. 2005;15:587–593. doi: 10.1016/j.cub.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 72.Belly A, Moreau-Gachelin F, Sadoul R, Goldberg Y. Delocalization of the multifunctional RNA splicing factor TLS/FUS in hippocampal neurones: Exclusion from the nucleus and accumulation in dendritic granules and spine heads. Neurosci Lett. 2005;379:152–157. doi: 10.1016/j.neulet.2004.12.071. [DOI] [PubMed] [Google Scholar]
- 73.Sephton CF, Yu G. The function of RNA-binding proteins at the synapse: Implications for neurodegeneration. Cell Mol Life Sci. 2015;72:3621–3635. doi: 10.1007/s00018-015-1943-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Swanger SA, Bassell GJ. Dendritic protein synthesis in the normal and diseased brain. Neuroscience. 2013;232:106–127. doi: 10.1016/j.neuroscience.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sephton CF, et al. Activity-dependent FUS dysregulation disrupts synaptic homeostasis. Proc Natl Acad Sci USA. 2014;111:E4769–E4778. doi: 10.1073/pnas.1406162111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hug N, Longman D, Cáceres JF. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016;44:1483–1495. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Geloso MC, et al. The dual role of microglia in ALS: Mechanisms and therapeutic approaches. Front Aging Neurosci. 2017;9:242. doi: 10.3389/fnagi.2017.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamanaka K, Komine O. The multi-dimensional roles of astrocytes in ALS. Neurosci Res. 2018;126:31–38. doi: 10.1016/j.neures.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 79.López-Erauskin JT, et al. ALS/FTD-linked mutation in FUS suppresses intra-axonal protein synthesis and drives disease without nuclear loss-of-function of FUS. Neuron. October 17, 2018 doi: 10.1016/j.neuron.2018.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.