Significance
Chromosomal double-strand breaks (DSBs) are cytotoxic forms of DNA damage that must be accurately repaired to maintain genome integrity. The conserved Mre11-Rad50-Xrs2NBS1 complex plays an important role in repair by functioning as a damage sensor and by regulating DNA end processing to ensure repair by the most appropriate mechanism. Yeast Sae2 is known to activate the Mre11 endonuclease to process DNA ends, and previous studies suggest an additional role to dampen checkpoint signaling. Here, we show Sae2 functions independently of the Mre11 nuclease to prevent Rad9 accumulation at DSBs. Excessive Rad9 binding inhibits DNA end processing by the Dna2-Sgs1 and Exo1 nucleases causing sensitivity of Sae2-deficient cells to DNA damaging agents.
Keywords: Sae2, Mre11, Rad9, DNA damage checkpoint, DNA repair
Abstract
The Mre11-Rad50-Xrs2NBS1 complex plays important roles in the DNA damage response by activating the Tel1ATM kinase and catalyzing 5′–3′ resection at DNA double-strand breaks (DSBs). To initiate resection, Mre11 endonuclease nicks the 5′ strands at DSB ends in a reaction stimulated by Sae2CtIP. Accordingly, Mre11-nuclease deficient (mre11-nd) and sae2Δ mutants are expected to exhibit similar phenotypes; however, we found several notable differences. First, sae2Δ cells exhibit greater sensitivity to genotoxins than mre11-nd cells. Second, sae2Δ is synthetic lethal with sgs1Δ, whereas the mre11-nd sgs1Δ mutant is viable. Third, Sae2 attenuates the Tel1-Rad53CHK2 checkpoint and antagonizes Rad953BP1 accumulation at DSBs independent of Mre11 nuclease. We show that Sae2 competes with other Tel1 substrates, thus reducing Rad9 binding to chromatin and to Rad53. We suggest that persistent Sae2 binding at DSBs in the mre11-nd mutant counteracts the inhibitory effects of Rad9 and Rad53 on Exo1 and Dna2-Sgs1–mediated resection, accounting for the different phenotypes conferred by mre11-nd and sae2Δ mutations. Collectively, these data show a resection initiation independent role for Sae2 at DSBs by modulating the DNA damage checkpoint.
Genomic integrity is constantly threatened by DNA damage that can result from exposure to exogenous sources, such as ionizing radiation, as well as from endogenous sources, including DNA replication errors and intermediates in excision repair or topoisomerase transactions. Cells respond to these insults by an elaborate network of surveillance mechanisms and DNA repair pathways, referred to as the DNA damage response (DDR) (1). DNA double-strand breaks (DSBs) are one of the most cytotoxic forms of DNA damage and can cause loss of genetic information, gross chromosome rearrangements, or even cell death in the absence of the appropriate response.
Typically, cells repair DSBs by nonhomologous end joining (NHEJ) or by homologous recombination (HR) (2). The Ku heterodimer (Ku70-Ku80), an essential NHEJ component, binds to DSB ends to protect them from degradation and recruits the DNA ligase IV complex to catalyze end ligation (3). HR employs extensive homology and templated DNA synthesis to restore the broken chromosome and is considered to be a mostly error-free mode of repair (4). HR initiates by nucleolytic degradation of DNA ends to generate long 3′ single-strand DNA (ssDNA) tails, a process termed end resection. RPA initially binds to the 3′ overhangs and is subsequently replaced by Rad51 to promote homologous pairing and strand invasion (5). Initiation of end resection is activated during S and G2 phases of the cell cycle when the sister chromatid is available as a repair template and is considered to be the main regulatory step in repair pathway choice (6–8).
In coordination with DNA repair mechanisms, cells respond to DSBs by a signaling cascade to halt cell cycle progression, induce transcription, and activate key repair proteins (1). Tel1ATM and Mec1ATR are the sentinel kinases that respond to DSBs in Saccharomyces cerevisiae (1, 9). Mre11-Rad50-Xrs2Nbs1 (MRXN) complex bound to ends recruits and activates Tel1ATM, whereas Mec1-Ddc2ATR-ATRIP binds to RPA-coated ssDNA generated by end resection. Yeast Tel1 and Mec1 redundantly phosphorylate multiple DNA repair proteins, as well as the downstream effector kinase, Rad53CHK2. Rad53 phosphorylation requires the Rad953BP1 adaptor protein, which is recruited to chromatin by methylated histone H3-K79, phosphorylated H2AH2AX (γH2A), and Dpb11TOPBP1 (1).
In addition to activating Tel1 kinase, MRX plays critical roles in tethering DNA ends and initiating end resection (4). The current model for end resection is for the Mre11 endonuclease to nick the 5′ strands internal to the DSB ends in a reaction stimulated by cyclin-dependent kinase (CDK)-phosphorylated Sae2CtIP (Fig. 1A) (10–13). Mre11 exonuclease then degrades in the 3′–5′ direction toward the break ends while more extensive processing of the 5′ strands is catalyzed by Exo1 or by Sgs1 helicase in concert with Dna2 endonuclease (14, 15). MRX also plays a noncatalytic role in end resection by recruiting Dna2 and Sgs1 to DSBs (16). In budding yeast, resection initiation by Mre11 nuclease and Sae2 is essential to remove covalently bound proteins, such as Spo11 from meiotic DSBs and hairpin-capped DNA ends, but is not essential for processing ends generated by endonucleases (15). In the absence of Mre11 nuclease (e.g., mre11-H125N mutant) or Sae2, resection of endonuclease-induced DSBs occurs primarily through the activity of Dna2-Sgs1. Thus, the mre11-H125N sgs1Δ double mutant exhibits greatly increased DNA damage sensitivity and delayed resection of an HO endonuclease-induced DSB relative to the single mutants, while the sae2Δ sgs1Δ double mutant is inviable (16–18). Exo1 can only efficiently promote resection at DNA ends when Ku is eliminated from cells. Consequently, deletion of YKU70 or YKU80 (encoding the Ku heterodimer) suppresses DNA damage sensitivity and end resection defects of mre11Δ, mre11-H125N, and sae2Δ/ctp1Δ mutants and also bypasses lethality of sae2Δ sgs1Δ cells, in an Exo1-dependent fashion (16, 17, 19–21).
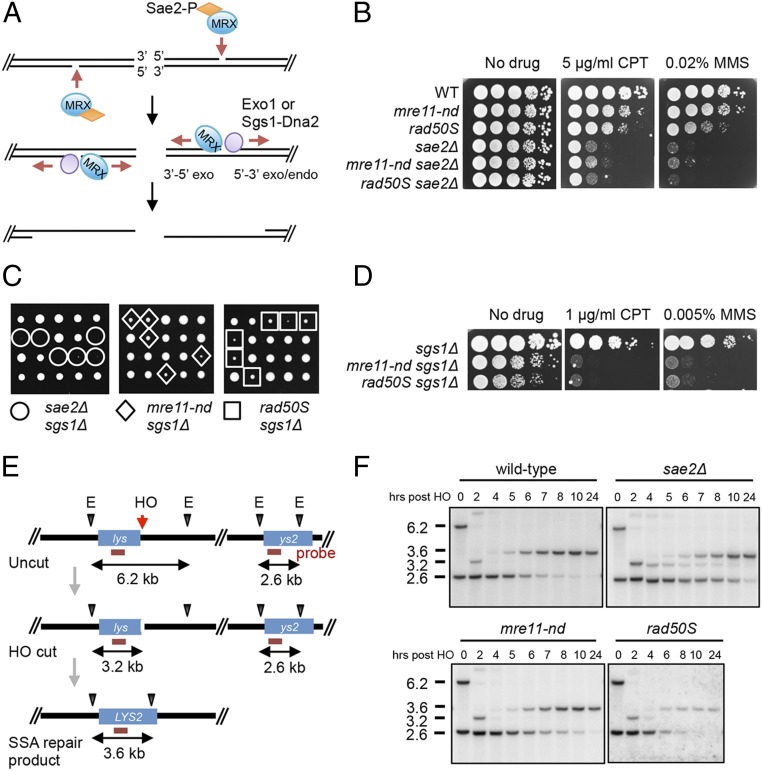
Fig. 1.
mre11-nd, sae2Δ, and rad50S mutants display different phenotypes. (A) Model for resection initiation at DSBs (see text for details). (B and D) Ten-fold serial dilutions of the indicated strains spotted on plates without drug, or plates containing CPT or MMS at the indicated concentrations. (C) SAE2/sae2Δ (or MRE11/mre11-nd or RAD50/rad50S) sgs1Δ/SGS1 heterozygous diploids were sporulated and tetrads were dissected on YPD plates. (E) Schematic of SSA assay showing location of the HO cut site and EcoRV (E) sites used to monitor DSB formation and deletion product. The horizontal red line indicated the sequences to which the probe hybridizes. (F) EcoRV-digested genomic DNA from the indicated strains, before and after HO induction was separated by agarose-gel electrophoresis, blotted, and hybridized with a fragment internal to the LYS2 gene.
In contrast to the mre11Δ mutant, which exhibits reduced Mec1 signaling in response to an unrepairable DSB due to resection defects, mre11-H125N and sae2Δ mutations confer normal Mec1 activation (22). Furthermore, phosphorylated Rad53 persists in sae2Δ cells, and overexpression (OE) of Sae2 diminishes Rad53 activation even though resection is unaffected, suggesting a role for Sae2 in attenuating DNA damage signaling (22). Mre11 persists at DSBs in sae2Δ cells due to a defect in end clipping, resulting in hyperactivation of the Tel1 checkpoint and suppression of mec1Δ sensitivity to genotoxic agents that cause replication-fork stalling (23–25). Consistent with findings in budding yeast, deletion of ctp1 suppresses fission yeast rad3Δ DNA damage sensitivity by hyperactivation of the Tel1-Chk1 checkpoint (26).
In efforts to further understand the role of Sae2 in the DNA damage response, several groups identified suppressor mutations that bypass sae2Δ camptothecin (CPT) sensitivity (27–30). One class of suppressors consists of point mutations within the N-terminal domain of Mre11 that broadly suppress sae2Δ sensitivity to a variety of DNA damaging agents (28–30). The mre11-H37Y and mre11-P110L mutations, which were characterized in detail, encode proteins with reduced DNA binding affinity and suppress Mre11 hyperaccumulation at DNA ends in the sae2Δ background, resulting in reduced checkpoint signaling. Another class of suppressors includes components of the DNA damage checkpoint. Rad9 accumulates close to DSBs in sae2Δ cells, and elimination of Rad9 restores Sgs1-Dna2–dependent end resection and DNA damage resistance to sae2Δ cells (31, 32). Additional sae2Δ suppressors include a gain-of-function SGS1 allele that overcomes the Rad9 inhibition to end resection, and tel1 and rad53 point mutations that reduce Rad9 binding, dampen checkpoint signaling and increase Dna2-Sgs1–dependent resection (27).
The goal of the current study was to investigate the differential sensitivity of sae2Δ and mre11-H125N mutants to DNA damaging agents. We provide evidence that Sae2 counteracts Rad9 binding to DSBs, independent of resection initiation, and the hyperaccumulation of Rad9 in the absence of Sae2 increases Rad53 activation and inhibits resection by Dna2-Sgs1 and Exo1.
Results
Differences Between Sae2 and Mre11 Nuclease-Deficient Cells.
By the current model for resection initiation (Fig. 1A), the mre11-H125N (hereafter referred to as mre11-nd) and sae2Δ mutants should exhibit similar DNA damage sensitivity. However, we find the sae2Δ mutant to be more sensitive to CPT and methyl methanesulfonate (MMS) than the mre11-nd mutant, and the double mutant exhibits the same sensitivity as the sae2Δ single mutant (Fig. 1B). As reported previously (17, 33), the mre11Δ mutant exhibits far greater sensitivity to CPT and MMS than mre11-nd or the sae2Δ mutant (SI Appendix, Fig. S1A). Furthermore, sae2Δ is lethal when combined with sgs1Δ, whereas the mre11-nd sgs1Δ double mutant is viable (Fig. 1C). The rad50-K81I (rad50S) mutant, which is also defective for Spo11 removal from meiotic DSBs and Mre11-catalyzed end clipping in vitro (11, 34), exhibits DNA damage sensitivity intermediate between mre11-nd and sae2Δ. The rad50S sgs1Δ double mutant is viable, but shows reduced proliferation and decreased DNA damage resistance relative to the single mutants, similar to mre11-nd sgs1Δ (Fig. 1 C and D).
We measured end resection in the mre11-nd, rad50S, and sae2Δ mutants by a single-strand annealing (SSA) assay. A strain was constructed with two fragments of the lys2 gene, which share 2.2-kb homology, separated by 20 kb on chromosome V (Fig. 1E). An HO endonuclease cut site was inserted at the junction of one lys2 repeat and the intervening sequence. Following DSB induction, the single-stranded regions of lys2 exposed by end resection anneal to restore LYS2, accompanied by deletion of the intervening sequence. Because there are no essential genes in the region deleted, the SSA product is viable. Additionally, the strains contain a galactose-inducible HO gene, and a rad51Δ mutation to prevent repair by break-induced replication. SSA was monitored by genomic blot hybridization. After DSB induction, the deletion product is detected by the appearance of a 3.6-kb fragment coinciding with disappearance of the 2.6-kb downstream lys2 fragment. Persistence of the HO cut fragment and delayed loss of the 2.6-kb fragment are indicative of reduced resection. Resection and deletion product formation are delayed in sae2Δ cells relative to wild type (WT) (Fig. 1F), as noted in previous studies (35, 36). By contrast, mre11-nd and rad50S cells show similar kinetics of repair to WT. Despite differences in timing of product formation, all of the strains exhibit similar survival on galactose-containing medium (SI Appendix, Fig. S1B). The requirement for Sae2 to complete repair by SSA is more prominent in the YMV80 strain background than we find for strains derived from W303 (35). The reason for this difference is currently unknown but could be due to relative usage of Exo1 and Dna2-Sgs1. We find SSA to be highly dependent on Exo1 in W303 (SI Appendix, Fig. S1C), whereas Dna2-Sgs1 is the primary mechanism for extensive resection in YMV80-derived strains (37). The delayed resection in the sae2Δ mutant correlates with the increased sensitivity to CPT and MMS and synthetic lethality with sgs1Δ, compared with mre11-nd and rad50S.
Accumulation of Rad9 at DSBs Is Suppressed by Sae2 and Is Independent of Mre11 Nuclease Activity.
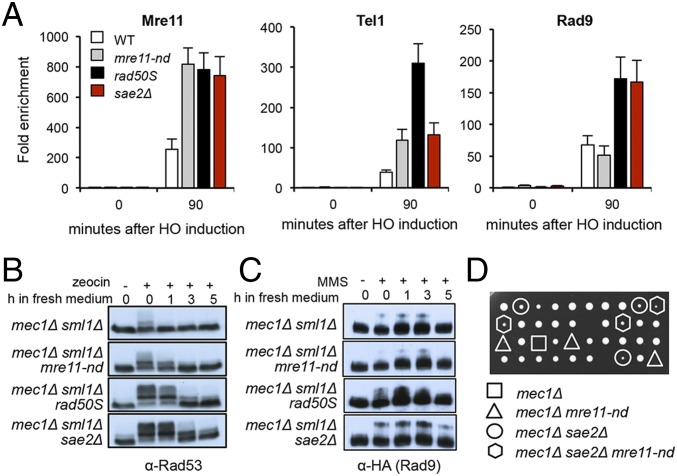
Previous studies have shown persistent Mre11 and Rad9 binding close to DSBs in the sae2Δ mutant leading to hyperactivation of Rad53 (22–24, 31, 32). Mre11 binding to DSBs is also increased in the absence of its nuclease activity (24). We compared Mre11, Tel1, and Rad9 binding in response to a single HO endonuclease-induced DSB by chromatin immunoprecipitation (ChIP) in mre11-nd, rad50S, and sae2Δ cells. While sae2Δ and rad50S cells show increased enrichment of Mre11, Tel1, and Rad9 close to the DSB, mre11-nd cells show Rad9 accumulation similar to WT cells, even though Mre11 and Tel1 levels are increased (Fig. 2A). Interestingly, Tel1 is increased to even higher levels in the rad50S mutant than observed in sae2Δ and mre11-nd mutants, and correlates with longer telomeres (SI Appendix, Fig. S2A)
Fig. 2.
Activation of the Tel1 checkpoint in sae2Δ and rad50S mutants. (A) The relative fold enrichment of Mre11, HA-Tel1, and Rad9-HA at 0.2 kb from the HO site was evaluated by qPCR after ChIP with anti-Mre11 and anti-HA antibodies. The error bars in all graphs indicate SD from three biological replicas. (B) Log phase cells (t = 0) from the indicated strains were treated with 30 μg/mL zeocin for 1 hour (h) and released into fresh YPD (t = 0–5). Protein samples from different time points before and after drug treatment were analyzed using anti-Rad53 antibodies. (C) Log phase cells (t = 0) from the indicated strains were treated with 0.015% MMS for 1 h and released into fresh YPD (t = 0–5), and protein samples from different time points before and after drug treatment were analyzed using anti-HA antibodies. (D) A SAE2/sae2Δ MRE11/mre11-nd mec1Δ/MEC1 sml1Δ/SML1 heterozygous diploid was sporulated and tetrads were dissected on YPD plates. Note that the highlighted segregants have the SML1 allele.
We anticipated that increased Tel1 and Rad9 binding to DSBs in rad50S and sae2Δ cells would result in enhanced activation of Rad53 phosphorylation relative to WT and mre11-nd cells. To specifically detect Tel1 activation, we monitored Rad53 phosphorylation in response to transient zeocin treatment of Mec1-deficient cells (all strains included sml1Δ to suppress lethality of mec1Δ). Rad53 activation and recovery are similar in mec1Δ and mec1Δ mre11-nd cells, whereas mec1Δ rad50S and mec1Δ sae2Δ cells show increased Rad53 phosphorylation and delayed recovery (Fig. 2B), in agreement with a previous study (23). Similar responses were observed following treatment of cells with MMS or CPT (SI Appendix, Fig. S2B). Consistent with Rad53 activation, Rad9 phosphorylation is increased in mec1Δ rad50S and mec1Δ sae2Δ mutants in response to MMS, compared with WT and mre11-nd cells (Fig. 2C). Surprisingly, while we were generating strains for these studies, we found that sae2Δ suppresses the lethality of mec1Δ SML1 cells, while mre11-nd and rad50S fail to suppress mec1Δ SML1 lethality (Fig. 2D and SI Appendix, Fig. S2C). sae2Δ suppression of mec1Δ lethality is Tel1 and Rad9 dependent (SI Appendix, Fig. S2D), indicating that it results from hyperactivation of the Tel1 checkpoint. Even though rad50S cells exhibit greater enrichment of Tel1 at DSBs than observed in sae2Δ cells, Rad53 activation is higher in the absence of Sae2, and this could account for suppression of mec1Δ lethality.
Rad9 Accumulation at DSBs and Rad53-Catalyzed Phosphorylation of Exo1 Contribute to sae2Δ DNA Damage Sensitivity.
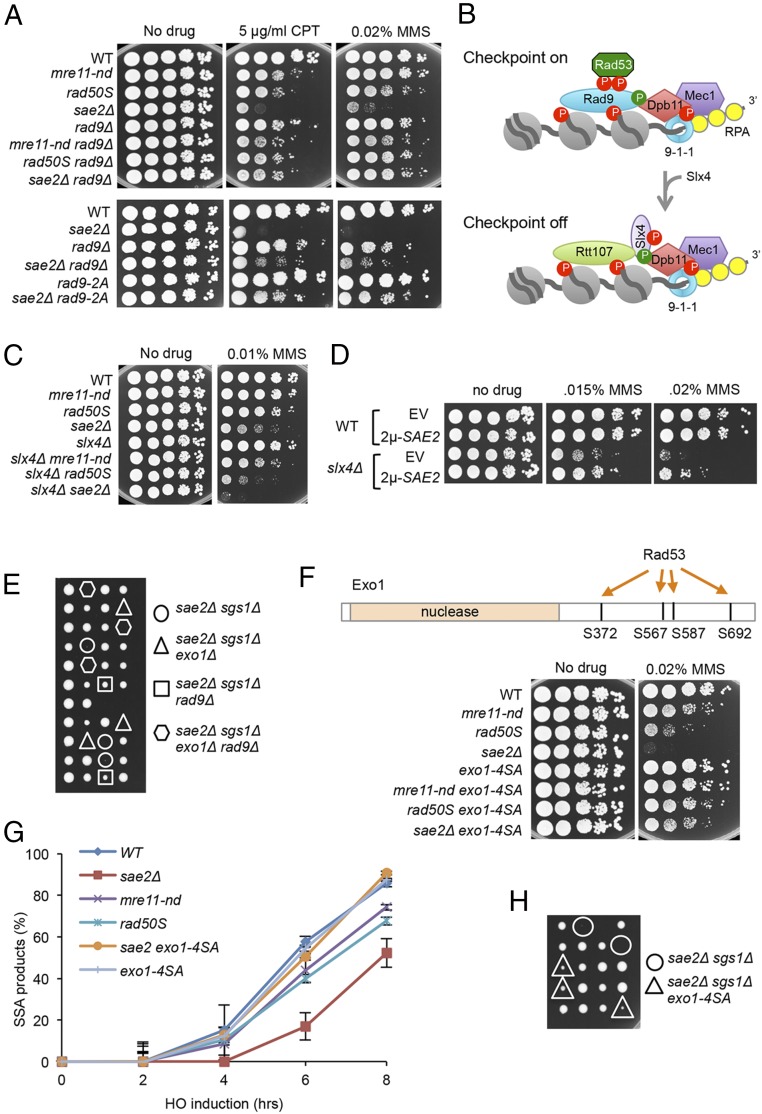
In agreement with published studies, we found that deletion of RAD9 suppresses the CPT and MMS sensitivity of the sae2Δ mutant (Fig. 3A) (27, 31). Surprisingly, rad9Δ had no suppressive effect in the mre11-nd background, and even resulted in higher sensitivity to CPT and MMS than the single mutants (Fig. 3A). Because the increased DNA damage sensitivity of sae2Δ cells, compared with mre11-nd, appears to result from Rad9 accumulation, we anticipated that by decreasing Rad9 binding, we should restore damage resistance to sae2Δ cells. Recruitment of Rad9 to chromatin requires Te11- and/or Mec1-phosphorylated H2A-S129 and Dot1-methylated H3-K79 (38–40). Consistently, we observe higher H2A phosphorylation (γH2A) close to the HO-induced DSB in sae2Δ cells compared with mre11-nd or WT (SI Appendix, Fig. S3A). We anticipated that dot1Δ and hta-S129A mutations would partially suppress sae2Δ DNA damage sensitivity because both have been shown to increase DNA end resection and the hta-S129A mutation was shown to partially suppress the end resection defect of sae2Δ cells (31, 32, 41–43). Surprisingly, the hta-S129A mutation fails to suppress sae2Δ DNA damage sensitivity (SI Appendix, Fig. S3B) (31, 32). The slight synergism between sae2Δ and hta-S129A mutations could be due to the role of γH2A in recruiting Rtt107, stabilizing replication forks and/or sister-chromatid recombination, coupled to the resection defect (44, 45). By contrast, dot1Δ partially suppresses sae2Δ DNA damage sensitivity. In addition to the nucleosome-dependent pathway for recruitment, Rad9 binds to Dpb11TopBP1, which is tethered to damage sites by the 9-1-1 clamp (Fig. 3B) (46). The Rad9 S462A and T474A mutations (hereafter referred to as rad9-2A) eliminate the interaction between Rad9 and Dpb11 (47). A previous study showed that the rad9-2A mutation suppresses the sae2Δ SSA defect (31); consistently, we find the rad9-2A mutation restores DNA damage resistance to sae2Δ cells (Fig. 3A).
Fig. 3.
Rad9 chromatin binding and Rad53 activation contribute to sae2Δ DNA damage sensitivity and sae2Δ sgs1Δ lethality. (A) Ten-fold serial dilutions of the indicated strains spotted on plates without drug, or plates containing indicated DNA damaging agents. (B) Schematic showing stabilization of Rad9 binding to chromatin by Dpb11 to activate the DNA damage checkpoint. CDK (green circles)- and Mec1 (red circles)-phosphorylated Slx4 competes with Rad9 for Dpb11 interaction, dampening the checkpoint. (C) Ten-fold serial dilutions of the indicated strains spotted on plates without drug, or plates containing indicated DNA damaging agents. (D) Ten-fold serial dilutions of the indicated strains spotted on plates without drug, or plates containing MMS. (E) Diploids heterozygous for the indicated mutations were sporulated and tetrads dissected on YPD plates. (F) Upper schematic showing Rad53-dependent phosphorylation sites in the C-terminal region of Exo1. Lower shows 10-fold serial dilutions of the indicated strains spotted on plates without drug, or plates containing indicated DNA damaging agents. (G) SSA kinetics were assessed by qPCR of genomic DNA from the indicated strains before and after HO induction. Error bars indicated SD from three independent trials. (H) Diploids heterozygous for the indicated mutations were sporulated and tetrads dissected on YPD plates.
Slx4 and its binding partner, Rtt107, compete with Rad9 for interaction with Dpb11 and γH2A, dampening DNA damage signaling (48). Since our studies indicate that Sae2 antagonizes Rad9 accumulation at DSBs, we tested genetic interaction between sae2Δ and slx4Δ. Consistent with a previous study slx4Δ synergizes with sae2Δ (49), and also with rad50S and mre11-nd (Fig. 3C). Moreover, expression of SAE2 from a high copy-number plasmid suppresses slx4Δ MMS sensitivity, suggesting that Sae2 can substitute for the checkpoint attenuation function of Slx4 (Fig. 3D).
Like rad9Δ, the tel1-kd mutation suppresses sae2Δ DNA damage sensitivity (32), but fails to suppress the CPT and MMS sensitivity of the mre11-nd mutant (SI Appendix, Fig. S3C). Elimination of Rad53 kinase activity, or Rad53 interaction with Rad9 (rad53-R605A), also results in suppression of sae2Δ CPT sensitivity (SI Appendix, Fig. S3D). Notably, elimination of Rad9, or Tel1, or Rad53 kinase activity, equalizes the DNA damage sensitivity of the mre11-nd, rad50S, and sae2Δ mutants, indicating that hyperactivation of the DNA damage checkpoint is responsible for the difference between sae2Δ and mre11-nd mutants.
Previous studies showed that rad9Δ suppression of the DNA damage sensitivity and resection defects of sae2Δ is by activation of Dna2-Sgs1, and not Exo1 (32). However, the finding that rad9Δ suppresses sae2Δ sgs1Δ lethality indicates that Exo1 must be activated. Indeed, we find that rad9Δ suppression of sae2Δ sgs1Δ lethality requires EXO1 (Fig. 3E). Exo1 has been identified as a substrate for the Rad53 kinase (50), and substitution of four serine residues in the C-terminal domain with alanine was shown to increase Exo1 activity at telomeres (51). To determine whether Rad53-catalyzed phosphorylation of Exo1 contributes to the down-regulation of resection observed in sae2Δ cells, we investigated genetic interaction between sae2Δ and exo1-4SA. The exo1-4SA allele suppresses sae2Δ MMS sensitivity, and sae2Δ sgs1Δ synthetic lethality (Fig. 3 F and H). The suppressive effect of exo1-4SA is also seen in mre11-nd and rad50S backgrounds. The exo1-4SA derivatives show different sensitivities to MMS, which we attribute to suppression of Dna2-Sgs1–catalyzed resection by Rad9, particularly in sae2Δ cells. We measured SSA by quantitative PCR (qPCR) and found the kinetics to be similar in WT and exo1-4SA cells (Fig. 3G). Notably, the sae2Δ exo1-4SA double mutant exhibits faster resection than sae2Δ. Thus, the DNA damage checkpoint inhibits resection in Sae2-deficient cells by Rad9 blockade of Dna2-Sgs1 and inhibitory phosphorylation of Exo1 by Rad53.
Overexpression of Sae2 Reduces Rad9 Binding at DSBs.
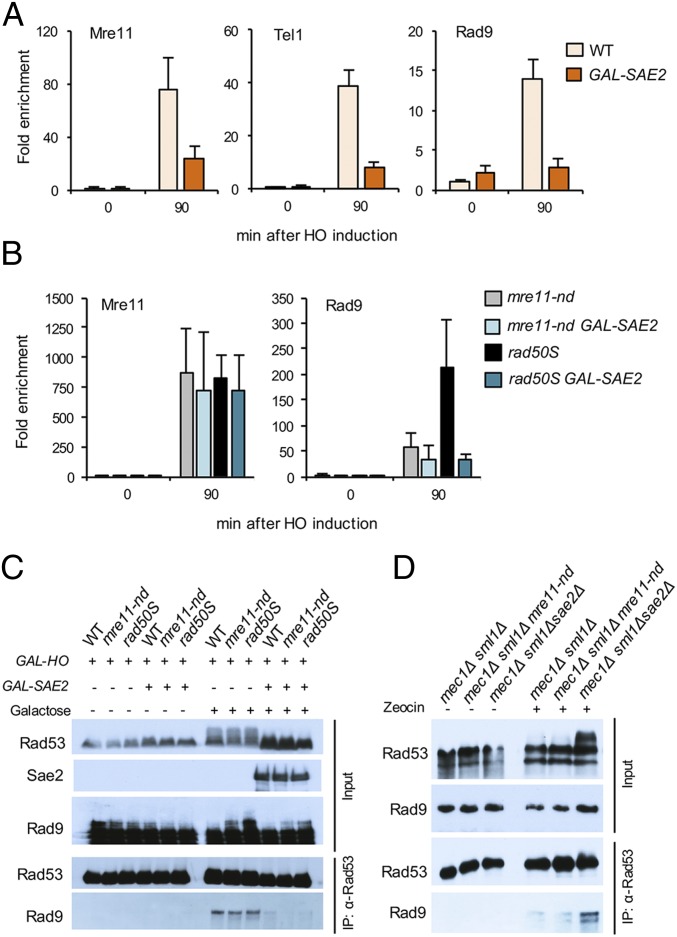
A previous study showed reduced Mre11 association with DSBs and attenuation of Rad53 activation when Sae2 is overexpressed (22). Sae2 OE does not diminish end resection, suggesting that the inhibitory effect of Sae2 on Rad53 activation is at a step subsequent to Mec1-Ddc2 recruitment to RPA-coated ssDNA (22). To determine whether Sae2 OE reduces Rad9 accumulation at DSBs we inserted the GAL promoter upstream of the SAE2 locus in a strain expressing a Sae2-Myc fusion. Following galactose induction to simultaneously induce HO cleavage and Sae2 expression, Mre11, Tel1, and Rad9 binding to sequences adjacent to the DSB were measured by ChIP. Consistent with the study by Clerici et al. (22), the amount of Mre11 bound to DSBs is decreased by ∼70% when Sae2 is OE (Fig. 4A); Tel1 and Rad9 binding are also significantly reduced. To determine whether reduced accumulation of Rad9 is a consequence of faster turnover of Mre11 at DSBs, we monitored Mre11 and Rad9 association with DSBs when Sae2 is OE in mre11-nd and rad50S backgrounds. Sae2 OE fails to remove Mre11 from ends when resection initiation is compromised; however, Rad9 levels are reduced (Fig. 4B). These data indicate separable functions of Sae2 in turnover of MRX at ends and inhibition of Rad9 binding to chromatin. The reduction in Rad9 binding when Sae2 is OE correlates with decreased Rad53 activation in response to the HO-induced DSB (Fig. 4C). We immunoprecipitated Rad53 from cells to determine whether the reduction in Rad9 binding caused by Sae2 OE prevents Rad53 interaction. As expected, Rad9 was recovered with Rad53 only after DSB induction. However, the Rad53-Rad9 interaction was greatly reduced in cells with Sae2 OE (Fig. 4C). Consistent with the negative effect of Sae2 on Rad53-Rad9 interaction, we find increased Rad53-Rad9 binding in mec1Δ sae2Δ cells, compared with mec1Δ or mec1Δ mre11-nd cells (Fig. 4D).
Fig. 4.
Sae2 OE reduces Rad9 binding at DSBs and to Rad53. (A) Relative fold enrichment of Mre11, Rad9-HA, and HA-Tel1 0.2 kb from the HO cleavage site was evaluated by qPCR after ChIP with anti-Mre11 or anti-HA antibodies. (B) Relative fold enrichment of Mre11 and Rad9 0.2 kb from the HO cut site was measured by qPCR in mre11-nd and rad50S cells with Sae2 OE. (C) Upper shows IP inputs and Lower shows Rad53 immunoprecipitates probed with α-Rad53, Myc (Sae2), or HA (Rad9) antibodies of the indicated strains, before or after galactose induction. (D) Upper shows IP inputs and Lower shows Rad53 immunoprecipitates probed with α-Rad53 or HA (Rad9) antibodies of the indicated strains, before or after zeocin treatment.
Sae2 Phosphorylation by Tel1 and/or Mec1 Dampens Checkpoint Signaling.
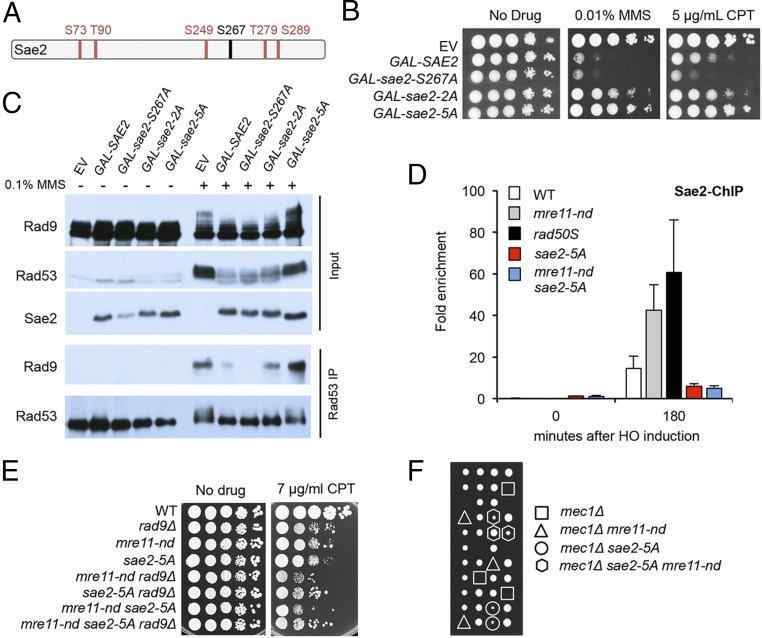
Mec1 and Tel1 phosphorylate Sae2 on multiple residues in response to DNA damage (Fig. 5A), but the physiological role of these modifications has not been firmly established (52, 53). Mutating five of the Mec1/Tel1 sites (S73, T90, S249, T279, and S289) to alanine, sae2-5A, prevents damage-induced phosphorylation and confers MMS sensitivity (52). A previous study showed that synthetic Sae2 peptides with phosphorylated T90 or T279 residues are able to interact with forkhead-associated domain 1 (FHA1) of Rad53 in vitro (54). Moreover, the sae2-T90A, T279A (sae2-2A) mutant is sensitive to MMS, and Rad53 remains hyperphosphorylated following MMS treatment (54).
Fig. 5.
Sae2 attenuates checkpoint signaling by competition for Tel1/Mec1 targets. (A) Schematic of Sae2 showing the main CDK (black) and Mec1/Tel1 (red) phosphorylation sites. (B) Ten-fold serial dilutions of WT cells with SAE2 or phosphorylation-defective sae2 alleles overexpressed from a GAL promoter were spotted on YPGal or YPGal with 0.01% MMS or 5 μg/mL CPT. (C) Western blot of inputs and Rad53 IP following OE of Sae2 or sae2 phosphorylation-site mutants. (D) Relative fold enrichment of Sae2-MYC 0.2 kb from the HO cleavage site was evaluated by qPCR after ChIP with anti-MYC antibodies. (E) Ten-fold serial dilutions of the indicated strains spotted on plates without drug, or plates containing indicated DNA damaging agents. (F) A SAE2/sae2-5A MRE11/mre11-nd mec1Δ/MEC1 sml1/SML1 heterozygous diploid was sporulated and tetrads were dissected on YPD plates. The highlighted segregants have the SML1 allele.
We expressed SAE2, sae2-5A, and sae2-2A alleles from the GAL promoter of a centromere-containing plasmid in WT cells. Sae2 OE results in sensitivity to CPT and MMS (Fig. 5B), whereas OE of the sae2-2A and sae2-5A variants does not sensitize cells to DNA damage. By contrast, OE of sae2-S267A, which prevents CDK-mediated phosphorylation of Sae2 (13), causes even greater CPT sensitivity than Sae2 OE. The effects of Sae2 OE are similar in WT and sae2Δ cells (SI Appendix, Fig. S4). Furthermore, OE of Sae2 or sae2-S267A reduces Rad53 and Rad9 phosphorylation in response to MMS, while sae2-5A OE does not (Fig. 5C). The reduction in Rad9 phosphorylation correlates with diminished interaction with Rad53, as measured by coimmunoprecipitation (Co-IP) (Fig. 5C). Thus, Mec1-Tel1 phosphorylation of Sae2 results in attenuation of Rad53 signaling, and this is independent of CDK-catalyzed phosphorylation and activation of Mre11 endonuclease.
Because the effects of Sae2 OE are consistent with sequestering Tel1 and Mec1 activity, we considered a model whereby a high local concentration of Sae2 when end clipping is abolished by the mre11-nd or rad50S mutation specifically affects Tel1 activity, dampening checkpoint activation. In agreement, we find greater enrichment of Sae2 at DSBs in mre11-nd and rad50S cells than observed for WT (Fig. 5D). The sae2-5A mutant protein shows decreased chromatin binding, consistent with the impaired checkpoint dampening function of sae2-5A. Furthermore, sae2-5A synergizes with mre11-nd for DNA damage sensitivity in a Rad9-dependent manner and suppresses mec1Δ lethality (Fig. 5 E and F). These data support the hypothesis that Tel1 and/or Mec1 phosphorylation of Sae2 is required for its role in checkpoint attenuation.
Discussion
Previous studies demonstrated increased DNA damage sensitivity and a greater delay in resection initiation caused by sae2Δ compared with mutations that inactivate the Mre11 nuclease activity (16, 17, 28, 33), indicating that Sae2 has a function in the DNA damage response that is independent of Mre11 endonuclease activation. We show here that the increased DNA damage sensitivity of sae2Δ cells is due to excessive Rad9 binding in the vicinity of DSBs and hyperactivation of the Rad53 checkpoint. While both sae2Δ and mre11-nd cells show elevated levels of Mre11 and Tel1 at DSBs as a consequence of delayed resection initiation, increased Rad9 binding is not seen in mre11-nd cells. We suggest that the delay in resection initiation caused by mre11-nd leads to a high local concentration of Sae2 in the vicinity of a DSB that competes with Rad9 for Tel1 activity, lowering the amount of Rad9 retained at DSBs and reducing Rad53 activation (Fig. 6). The increased accumulation of Rad9 at damaged sites in the sae2Δ mutant acts as a barrier to Dna2-Sgs1 resection, and hyperactivation of the Rad53 kinase results in inhibitory phosphorylation of Exo1. These two mechanisms contribute to the greater delay in end resection and higher DNA damage sensitivity of sae2Δ cells relative to mre11-nd cells.
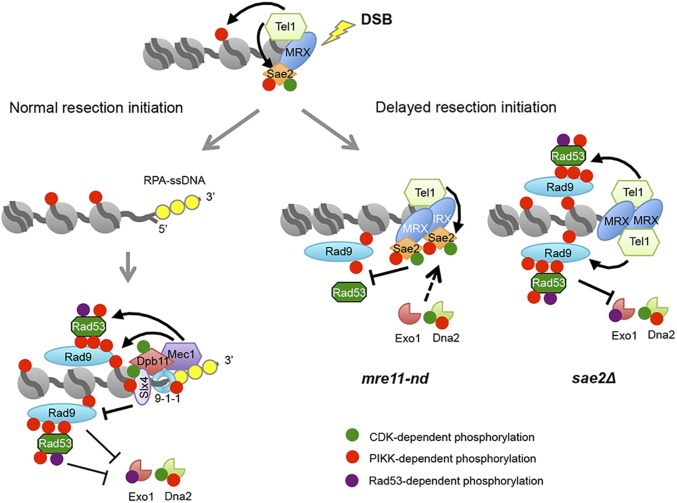
Fig. 6.
Sae2 controls short- and long-range resection pathways. Normally, resection initiation by Mre11 and Sae2 is fast resulting in low dwell time of Tel1 at DSBs, and consequently low activation of Tel1 kinase (for simplicity, only one side of a DSB is shown). After resection initiation, Mec1-Ddc2 is recruited to ssDNA overhangs, phosphorylating Rad9 and Rad53 to slow down resection by Dna2-Sgs1 and Exo1. Slx4-Rtt107 competes with Rad9 for Dpb11 binding to dampen the checkpoint and, consequently, increase extensive resection. When resection initiation is compromised by the mre11-nd mutation, MRX, Tel1, and Sae2 accumulate at DSBs and Sae2 competes with other Tel1 substrates for phosphorylation, reducing Rad9 binding, Rad53 activation, and allowing resection by Dna2-Sgs1 and Exo1. In the absence of Sae2, Tel1 is hyperactivated, causing increased Rad9 binding and Rad53 activation, thereby diminishing resection by Dna2-Sgs1 and Exo1.
The rad50S mutant behaves differently from mre11-nd and sae2Δ. While the DNA damage sensitivity and resection defects of rad50S cells are similar to mre11-nd, accumulation of Rad9 at DSBs resembles sae2Δ. If Rad9 accumulation is responsible for reduced resection in sae2Δ cells, then how do we explain the rad50S phenotype? Tel1 binding is much higher in rad50S than in sae2Δ cells and telomeres are longer (SI Appendix, Fig. S2A) (55), consistent with Tel1 hyperactivation; however, Rad53 activation (in the mec1Δ background) is slightly lower and rad50S fails to suppress mec1Δ lethality. We suggest that the high local concentration of Sae2 in rad50S cells attenuates Rad53 activation and this allows more efficient resection by Exo1. Indeed, the exo1-4SA mutation is more effective in suppressing rad50S DNA damage sensitivity than rad9Δ. We suggest that most of the checkpoint dampening role of Sae2 is through inhibition of Rad9 accumulation near DSBs, but cannot rule out additional roles via direct interaction with Rad53 and Dun1 (54).
Rad9 and its mammalian ortholog, 53BP1, have well-documented roles in preventing end resection (43, 56). In yeast, the Dna2-Sgs1 resection mechanism is the primary target of inhibition by Rad9 (57), particularly in sae2Δ cells (27, 31). In the absence of Sae2 and Mre11 endonuclease-catalyzed incision, there is no nick for Exo1 entry and Ku is highly effective in preventing Exo1 resection at ends; thus, resection in mre11-nd cells is mostly dependent on Dna2-Sgs1 (16–18). The increased Rad9 binding in sae2Δ compared with mre11-nd cells presumably creates a greater barrier for Dna2-Sgs1 accounting for the more severe phenotype of the sae2Δ mutant. By contrast to budding yeast, the increased resection observed in the absence of 53BP1 in mouse cells is largely dependent on CtIP (58). There are two possible explanations for this difference. First, CtIP might be required to recruit DNA2 to DSBs or function with DNA2-BLM/WRN in long-range resection; this idea is supported by studies showing epistasis between CtIP and DNA2 for resection defects and direct stimulation of DNA2-BLM activity by CtIP (59–62). Second, if 53BP1 recruitment to DSBs occurs before resection initiation it could potentially create a barrier to CtIP and MRN-catalyzed incision.
Resection initiation generates RPA-bound ssDNA, which is essential for Mec1-Ddc2 signaling, as well as creating a recessed 5′ end for loading the 9-1-1 damage clamp (1). Previous studies have shown that 9-1-1 has both positive and negative roles in regulating end resection (57), both of which are modulated by the multi-BRCT domain protein, Dpb11TOPBP1. Dpb11 interacts with the Ddc1 subunit of the 9-1-1 complex and Rad9 at damage sites to mediate Rad53 phosphorylation by Mec1. Thus, 9-1-1 negatively regulates resection via Rad9 and Rad53. Residual Rad9 binding to chromatin is observed in mutants deficient for 9-1-1, resulting in only partial derepression of resection at DSBs (57, 63). The positive function of 9-1-1 in resection is through recruitment of the Fun30SMARCAD1 chromatin remodeler (64), which counteracts the negative effect of Rad9 on resection (41, 42, 64, 65). In addition, Slx4-Rtt107 competes with Rad9 for binding to γH2A and to Dpb11. In the absence of Slx4-Rtt107, Rad9 recruitment to DSBs is increased and the Rad53 checkpoint is hyperactivated, resulting in reduced end resection (49, 66). When more Slx4 is bound to Dpb11 (rad9-2A mutant) the checkpoint is attenuated, resulting in suppression of sae2Δ DNA damage sensitivity. We suggest that Dna2-Sgs1 starts resection in mre11-nd and sae2Δ cells, generating a recessed 5′ end for 9-1-1 and Dpb11 loading; however, this initial resection is insufficient to displace MRX and Tel1, and when Sae2 is absent, Rad9 accumulates, slowing extensive resection by Dna2-Sgs1 and Exo1.
CDK and Mec1/Tel1-catalyzed phosphorylation events play critical roles in regulating resection nucleases. Sae2CtIP is licensed to activate Mre11 endonuclease when CDK activity is high in S- and G2-phase cells to ensure end resection occurs when a sister chromatid is available to template homology-dependent repair (11–13); similarly, Dna2 is positively regulated by CDK-catalyzed phosphorylation (67). Human EXO1 is also activated for end resection by CDK (68). The other positive action of CDK is by phosphorylation of Fun30 and Slx4, which is required for their interaction with Dpb11, and hence to 9-1-1 at the recessed 5′ end (48, 64, 69). While Mec1- and Tel1-mediated phosphorylation of Sae2 is important for resection (70), Mec1 and Tel1 act to repress extensive resection via recruitment of Rad9 and Rad53 to DSBs (71). As described above, Rad9 blocks extensive resection by Dna2-Sgs1, and Rad53 inhibits Exo1 activity. Here, we show that preventing CDK-catalyzed phosphorylation of Sae2 does not impact the checkpoint dampening function of Sae2, consistent with its role in activating Mre11 endonuclease. By contrast, Tel1- and/or Mec1-catalyzed phosphorylation of Sae2 in the vicinity of DSBs provide an additional way to regulate resection by attenuating Rad9 binding and Rad53 activation until resection initiates. This feedback control mechanism activates Dna2-Sgs1 and Exo1 if resection initiation is delayed.
In fission yeast, Mre11 nuclease and Ctp1Sae2 are more important for DNA damage resistance than observed in budding yeast (19, 72). As described above, resection in mre11-nd and sae2Δ mutants is highly dependent on Dna2-Sgs1. Langerak et al. (21) showed that Rqh1Sgs1 barely contributes to resection in fission yeast and instead Exo1 is largely responsible for long-range resection. The poor use of the Dna2-Rqh1 pathway at DSBs in fission yeast could be due to a stronger block by Crb2Rad9 (73). Although Mre11 nuclease and CtIP are both essential for proliferation of mammalian cells, Ctip−/− mouse embryos arrest at an earlier stage than Mre11H129N/H129N embryos, suggesting that CtIP, like Sae2 in budding yeast, has functions beyond stimulating Mre11 endonuclease (74, 75).
Materials and Methods
Media, Growth Conditions, and Yeast Strains.
Rich medium (1% yeast extract; 2% peptone; 2% dextrose) (YPD), synthetic complete (SC) medium and genetic methods were as described previously (76). CPT or MMS was added to SC or YPD medium, respectively, at the indicated concentrations. For survival assays, 10-fold serial dilutions of log-phase cultures were spotted on plates with no additive or the indicated amount of drug and incubated for 3 d at 30 °C. Diploids heterozygous for relevant mutations were sporulated and tetrads dissected to assess synthetic genetic interactions. Spores were manipulated on YPD plates and incubated for 3–4 d at 30 °C. The yeast strains used here, listed in SI Appendix, Table S1, are derived from W303 corrected for RAD5, and were constructed via crosses or by one-step gene replacement using PCR-derived DNA fragments. To generate the SSA assay system, we modified the BIR assay system described by Donnianni and Symington (77). A 5′-truncated lys2 fragment was inserted 20-kb telomere proximal to a 3′-truncated lys2 cassette and HO cut site in strain LSY2751 (77), such that the lys2 fragments have the same polarity on the left arm of Chr V. The SSA strains express GAL-HO, and RAD51 is deleted to prevent break-induced replication. Details of plasmid constructions are in SI Appendix, Supplementary Methods.
ChIP and Co-IP Assays.
Yeast cells were cultured in YP containing 2% lactate (YPL) or 2% raffinose (YPR) to log phase and arrested at G2/M phase by adding nocodazole (15 μg/mL) to the medium. For ChIP experiments, cells were collected 90 min or 180 min after adding galactose to 2% for HO induction. After formaldehyde cross-linking and chromatin isolation, Mre11, Rad9-HA, HA-Tel1, Sae2-MYC, or γH2A were immunoprecipitated using Mre11 polyclonal antibodies from rabbit serum, anti-HA antibodies (16B12, BioLegend), anti-MYC antibodies (9E10, Santa Cruz), or γH2A antibodies (ab15083, Abcam), respectively, and Protein A/G magnetic beads (Pierce) (33, 78). Protein A magnetic beads (Pierce) were used for the ChIP experiment shown in Fig. 4A and resulted in lower enrichments than observed with Protein A/G beads. Quantitative PCR was carried out using SYBR green real-time PCR mix (Biorad) and primers complementary to DNA sequences located 0.2 or 1 kb from the HO-cut site at the MAT locus. Reads were normalized to input signal and then normalized to the input/IP signal for DNA sequences located 66 kb from the HO-cutting site. For Co-IP experiments, cells were collected 1 h after galactose addition for Sae2-MYC induction for no DNA damage control and collected 3 h after MMS (0.1%) treatment or 2 h after zeocin treatment (30 μg/mL) with no nocodazole arrest step. Rad53 was immunoprecipitated from the cell extracts with anti-Rad53 antibodies (IL-7, gift from M. Foiani, IFOM, Milan), then immunoblotted with IL-7 and anti-HA antibodies to recognize Rad53 and Rad9-HA, respectively.
Western Blots.
Yeast cells were grown to 107 cells per milliliter in YPD, then CPT, MMS, or zeocin was added at the indicated final concentration for 60 min. Cells were released into fresh YPD medium and collected at the indicated time points for TCA precipitation. Cells were resuspended in 0.2 mL 20% TCA and then mechanically disrupted for 5 min using glass beads. Beads were washed twice with 0.2 mL 5% TCA each and pellets collected by centrifugation at 845 × g for 10 min. The pellet was resuspended in 0.15 mL SDS/PAGE sample buffer and proteins were separated by SDS/PAGE. Anti-Rad53 antibodies, anti-HA antibodies, and anti-MYC antibodies were used for immunoblots.
SSA Assay.
Cells containing the SSA reporter were grown for HO induction as described above. Aliquots of cells were removed before HO induction (0 h), and at 1- or 2-h intervals after addition of galactose to the media for isolation of genomic DNA. Genomic DNA was digested with EcoRV and the resulting blots were hybridized with a PCR fragment corresponding to LYS2 sequence by Southern blotting. SSA efficiency was also measured by qPCR. We designed primer pairs to amplify sequences 3 kb downstream of the HO cut site (HOcs) between two LYS2 homologies (3K_DS), and 3.2 kb upstream of the HOcs (3.2K_US). The Ct values for each primer pair were normalized to ADH1, and the SSA product was calculated by the ratio of 3K_DS/3.2K_US.
Supplementary Material
Acknowledgments
We thank M. P. Longhese, B. Pfander, and M. Smolka for yeast strains and plasmids, and M. Foiani for anti-Rad53 antibodies. This study was supported by National Institutes of Health Grants P01CA174653 and R35 GM126997.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816539115/-/DCSupplemental.
References
- 1.Finn K, Lowndes NF, Grenon M. Eukaryotic DNA damage checkpoint activation in response to double-strand breaks. Cell Mol Life Sci. 2012;69:1447–1473. doi: 10.1007/s00018-011-0875-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ceccaldi R, Rondinelli B, D’Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiruvella KK, Liang Z, Wilson TE. Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol. 2013;5:a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Symington LS, Rothstein R, Lisby M. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014;198:795–835. doi: 10.1534/genetics.114.166140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 6.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hustedt N, Durocher D. The control of DNA repair by the cell cycle. Nat Cell Biol. 2016;19:1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- 9.Gobbini E, Cesena D, Galbiati A, Lockhart A, Longhese MP. Interplays between ATM/Tel1 and ATR/Mec1 in sensing and signaling DNA double-strand breaks. DNA Repair (Amst) 2013;12:791–799. doi: 10.1016/j.dnarep.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Anand R, Ranjha L, Cannavo E, Cejka P. Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol Cell. 2016;64:940–950. doi: 10.1016/j.molcel.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 11.Cannavo E, Cejka P. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature. 2014;514:122–125. doi: 10.1038/nature13771. [DOI] [PubMed] [Google Scholar]
- 12.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huertas P, Cortés-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia V, Phelps SE, Gray S, Neale MJ. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011;479:241–244. doi: 10.1038/nature10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Symington LS. Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol. 2016;51:195–212. doi: 10.3109/10409238.2016.1172552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shim EY, et al. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 2010;29:3370–3380. doi: 10.1038/emboj.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29:3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Budd ME, Campbell JL. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS One. 2009;4:e4267. doi: 10.1371/journal.pone.0004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Limbo O, et al. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster SS, Balestrini A, Petrini JH. Functional interplay of the Mre11 nuclease and Ku in the response to replication-associated DNA damage. Mol Cell Biol. 2011;31:4379–4389. doi: 10.1128/MCB.05854-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langerak P, Mejia-Ramirez E, Limbo O, Russell P. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 2011;7:e1002271. doi: 10.1371/journal.pgen.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep. 2006;7:212–218. doi: 10.1038/sj.embor.7400593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Usui T, Ogawa H, Petrini JH. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol Cell. 2001;7:1255–1266. doi: 10.1016/s1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- 24.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 25.Kim HS, et al. Functional interactions between Sae2 and the Mre11 complex. Genetics. 2008;178:711–723. doi: 10.1534/genetics.107.081331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Limbo O, Porter-Goff ME, Rhind N, Russell P. Mre11 nuclease activity and Ctp1 regulate Chk1 activation by Rad3ATR and Tel1ATM checkpoint kinases at double-strand breaks. Mol Cell Biol. 2011;31:573–583. doi: 10.1128/MCB.00994-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonetti D, et al. Escape of Sgs1 from Rad9 inhibition reduces the requirement for Sae2 and functional MRX in DNA end resection. EMBO Rep. 2015;16:351–361. doi: 10.15252/embr.201439764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, et al. Sae2 promotes DNA damage resistance by removing the Mre11-Rad50-Xrs2 complex from DNA and attenuating Rad53 signaling. Proc Natl Acad Sci USA. 2015;112:E1880–E1887. doi: 10.1073/pnas.1503331112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puddu F, et al. Synthetic viability genomic screening defines Sae2 function in DNA repair. EMBO J. 2015;34:1509–1522. doi: 10.15252/embj.201590973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cassani C, et al. Structurally distinct Mre11 domains mediate MRX functions in resection, end-tethering and DNA damage resistance. Nucleic Acids Res. 2018;46:2990–3008. doi: 10.1093/nar/gky086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrari M, et al. Functional interplay between the 53BP1-ortholog Rad9 and the Mre11 complex regulates resection, end-tethering and repair of a double-strand break. PLoS Genet. 2015;11:e1004928. doi: 10.1371/journal.pgen.1004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gobbini E, et al. Sae2 function at DNA double-strand breaks is bypassed by dampening Tel1 or Rad53 activity. PLoS Genet. 2015;11:e1005685. doi: 10.1371/journal.pgen.1005685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krogh BO, Llorente B, Lam A, Symington LS. Mutations in Mre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex stability in addition to nuclease activity. Genetics. 2005;171:1561–1570. doi: 10.1534/genetics.105.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 35.Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem. 2005;280:38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- 36.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toh GW, et al. Histone H2A phosphorylation and H3 methylation are required for a novel Rad9 DSB repair function following checkpoint activation. DNA Repair (Amst) 2006;5:693–703. doi: 10.1016/j.dnarep.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 39.Wysocki R, et al. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura TM, Du LL, Redon C, Russell P. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol. 2004;24:6215–6230. doi: 10.1128/MCB.24.14.6215-6230.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X, et al. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature. 2012;489:576–580. doi: 10.1038/nature11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eapen VV, Sugawara N, Tsabar M, Wu WH, Haber JE. The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol Cell Biol. 2012;32:4727–4740. doi: 10.1128/MCB.00566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lazzaro F, et al. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J. 2008;27:1502–1512. doi: 10.1038/emboj.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mejia-Ramirez E, Limbo O, Langerak P, Russell P. Critical function of γH2A in S-phase. PLoS Genet. 2015;11:e1005517. doi: 10.1371/journal.pgen.1005517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie A, et al. Control of sister chromatid recombination by histone H2AX. Mol Cell. 2004;16:1017–1025. doi: 10.1016/j.molcel.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Granata M, et al. Dynamics of Rad9 chromatin binding and checkpoint function are mediated by its dimerization and are cell cycle-regulated by CDK1 activity. PLoS Genet. 2010;6:e1001047. doi: 10.1371/journal.pgen.1001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfander B, Diffley JF. Dpb11 coordinates Mec1 kinase activation with cell cycle-regulated Rad9 recruitment. EMBO J. 2011;30:4897–4907. doi: 10.1038/emboj.2011.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohouo PY, Bastos de Oliveira FM, Liu Y, Ma CJ, Smolka MB. DNA-repair scaffolds dampen checkpoint signalling by counteracting the adaptor Rad9. Nature. 2013;493:120–124. doi: 10.1038/nature11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dibitetto D, et al. Slx4 and Rtt107 control checkpoint signalling and DNA resection at double-strand breaks. Nucleic Acids Res. 2016;44:669–682. doi: 10.1093/nar/gkv1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci USA. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morin I, et al. Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J. 2008;27:2400–2410. doi: 10.1038/emboj.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baroni E, Viscardi V, Cartagena-Lirola H, Lucchini G, Longhese MP. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol Cell Biol. 2004;24:4151–4165. doi: 10.1128/MCB.24.10.4151-4165.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu Q, et al. Phosphorylation-regulated transitions in an oligomeric state control the activity of the Sae2 DNA repair enzyme. Mol Cell Biol. 2014;34:778–793. doi: 10.1128/MCB.00963-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang J, Suhandynata RT, Zhou H. Phosphorylation of Sae2 mediates forkhead-associated (FHA) domain-specific interaction and regulates its DNA repair function. J Biol Chem. 2015;290:10751–10763. doi: 10.1074/jbc.M114.625293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kironmai KM, Muniyappa K. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells. 1997;2:443–455. doi: 10.1046/j.1365-2443.1997.1330331.x. [DOI] [PubMed] [Google Scholar]
- 56.Zimmermann M, de Lange T. 53BP1: Pro choice in DNA repair. Trends Cell Biol. 2014;24:108–117. doi: 10.1016/j.tcb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ngo GH, Lydall D. The 9-1-1 checkpoint clamp coordinates resection at DNA double strand breaks. Nucleic Acids Res. 2015;43:5017–5032. doi: 10.1093/nar/gkv409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoa NN, et al. BRCA1 and CtIP are both required to recruit Dna2 at double-strand breaks in homologous recombination. PLoS One. 2015;10:e0124495. doi: 10.1371/journal.pone.0124495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peterson SE, et al. Activation of DSB processing requires phosphorylation of CtIP by ATR. Mol Cell. 2013;49:657–667. doi: 10.1016/j.molcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daley JM, et al. Enhancement of BLM-DNA2-mediated long-range DNA end resection by CtIP. Cell Rep. 2017;21:324–332. doi: 10.1016/j.celrep.2017.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paudyal SC, Li S, Yan H, Hunter T, You Z. Dna2 initiates resection at clean DNA double-strand breaks. Nucleic Acids Res. 2017;45:11766–11781. doi: 10.1093/nar/gkx830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Naiki T, Wakayama T, Nakada D, Matsumoto K, Sugimoto K. Association of Rad9 with double-strand breaks through a Mec1-dependent mechanism. Mol Cell Biol. 2004;24:3277–3285. doi: 10.1128/MCB.24.8.3277-3285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bantele SC, Ferreira P, Gritenaite D, Boos D, Pfander B. Targeting of the Fun30 nucleosome remodeller by the Dpb11 scaffold facilitates cell cycle-regulated DNA end resection. eLife. 2017;6:e21687. doi: 10.7554/eLife.21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Costelloe T, et al. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature. 2012;489:581–584. doi: 10.1038/nature11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y, et al. TOPBP1Dpb11 plays a conserved role in homologous recombination DNA repair through the coordinated recruitment of 53BP1Rad9. J Cell Biol. 2017;216:623–639. doi: 10.1083/jcb.201607031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen X, et al. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat Struct Mol Biol. 2011;18:1015–1019. doi: 10.1038/nsmb.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tomimatsu N, et al. Phosphorylation of EXO1 by CDKs 1 and 2 regulates DNA end resection and repair pathway choice. Nat Commun. 2014;5:3561. doi: 10.1038/ncomms4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen X, et al. Enrichment of Cdk1-cyclins at DNA double-strand breaks stimulates Fun30 phosphorylation and DNA end resection. Nucleic Acids Res. 2016;44:2742–2753. doi: 10.1093/nar/gkv1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cartagena-Lirola H, Guerini I, Viscardi V, Lucchini G, Longhese MP. Budding yeast Sae2 is an in vivo target of the Mec1 and Tel1 checkpoint kinases during meiosis. Cell Cycle. 2006;5:1549–1559. doi: 10.4161/cc.5.14.2916. [DOI] [PubMed] [Google Scholar]
- 71.Clerici M, Trovesi C, Galbiati A, Lucchini G, Longhese MP. Mec1/ATR regulates the generation of single-stranded DNA that attenuates Tel1/ATM signaling at DNA ends. EMBO J. 2014;33:198–216. doi: 10.1002/embj.201386041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Williams RS, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leland BA, Chen AC, Zhao AY, Wharton RC, King MC. Rev7 and 53BP1/Crb2 prevent RecQ helicase-dependent hyper-resection of DNA double-strand breaks. eLife. 2018;7:e33402. doi: 10.7554/eLife.33402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buis J, et al. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen PL, et al. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol Cell Biol. 2005;25:3535–3542. doi: 10.1128/MCB.25.9.3535-3542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 2005. [Google Scholar]
- 77.Donnianni RA, Symington LS. Break-induced replication occurs by conservative DNA synthesis. Proc Natl Acad Sci USA. 2013;110:13475–13480. doi: 10.1073/pnas.1309800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Donnianni RA, et al. Elevated levels of the polo kinase Cdc5 override the Mec1/ATR checkpoint in budding yeast by acting at different steps of the signaling pathway. PLoS Genet. 2010;6:e1000763. doi: 10.1371/journal.pgen.1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.