Significance
We describe a flavin-containing thioredoxin reductase (FFTR) from the anaerobic soil bacterium Clostridium pasteurianum. The enzyme differed from the well-known flavin thioredoxin reductase in its inability to use NADPH as an electron donor. Rather, FFTR used reduced ferredoxin as a reductant similar to photosynthetic ferredoxin−thioredoxin reductase, an iron−sulfur enzyme. As a founding member of a family of NADPH-independent flavin thioredoxin reductases, the enzyme opens a door to studies of redox metabolism in bacteria, including representatives of medical and biotechnological importance.
Keywords: flavoprotein, ferredoxin, protein conformational dynamics, redox-active disulfide, thioredoxin reductase
Abstract
Ferredoxin-dependent thioredoxin reductase was identified 35 y ago in the fermentative bacterium Clostridium pasteurianum [Hammel KE, Cornwell KL, Buchanan BB (1983) Proc Natl Acad Sci USA 80:3681–3685]. The enzyme, a flavoprotein, was strictly dependent on ferredoxin as reductant and was inactive with either NADPH or NADH. This early work has not been further pursued. We have recently reinvestigated the problem and confirmed that the enzyme, here designated ferredoxin-dependent flavin thioredoxin reductase (FFTR), is a flavoprotein. The enzyme differs from ferredoxin−thioredoxin reductase (FTR), which has a signature [4Fe−4S] cluster, but shows structural similarities to NADP-dependent thioredoxin reductase (NTR). Comparative amino acid sequence analysis showed that FFTR is present in a number of clostridial species, some of which lack both FTR and an archetypal NTR. We have isolated, crystallized, and determined the structural properties of FFTR from a member of this group, Clostridium acetobutylicum, both alone and in complex with Trx. The structures showed an elongated FFTR homodimer, each monomer comprising two Rossmann domains and a noncovalently bound FAD cofactor that exposes the isoalloxazine ring to the solvent. The FFTR structures revealed an alternative domain organization compared with NTR that enables the enzyme to accommodate Fdx rather than NADPH. The results suggest that FFTR exists in a range of conformations with varying degrees of domain separation in solution and that the stacking between the two redox-active groups for the transfer of reducing equivalents results in a profound structural reorganization. A mechanism in accord with the findings is proposed.
As major components of the redox system in most types of cells, thioredoxin (Trx) proteins are involved in the reduction of substrates such as ribonucleotides and in the reversible modification of the structural or functional features of multiple target proteins through dithiol−disulfide exchange reactions (1–4).
Two enzymes known to function in the reduction of Trx have been extensively studied: NADP−Trx reductase (NTR), a flavoprotein that uses NADPH as reductant (5, 6), and ferredoxin (Fdx)−Trx reductase (FTR), an iron−sulfur protein that uses photosynthetically reduced Fdx (7). The two proteins are not related in sequence or structure. Our knowledge of a third enzyme, a flavoprotein, that was identified 35 y ago and shown to function with Fdx in the reduction of Trx is limited (8).
To help fill this gap, we have isolated and characterized Fdx-dependent flavin Trx reductase (FFTR) from Clostridium acetobutylicum. The enzyme was confirmed to be a flavoprotein that is structurally similar to classical low-molecular-weight NTR (9), but differs in the absence of an NADPH binding site as well as in domain organization and protein conformational dynamics. The structures reveal details that define the substrate-binding properties of each of the enzymes. FFTR, NTR, and FTR have evolved sequences that enable them to interact in multiple Trx-dependent processes.
Results
FFTR Is Almost Exclusively Present in Members of the Genus Clostridium.
The availability of the Clostridium pasteurianum genome sequence led to the detection of two NTR homologs by amino acid sequence comparison (10). One of the proteins corresponded to the flavin enzyme reported by Hammel and coworkers (8, 10), FFTR. The second homolog was a protein called CP34, a dedicated peroxiredoxin reductase dependent on NADH (10) (SI Appendix, Fig. S1A). From the experimental characterization of the enzymes, it was concluded that C. pasteurianum lacks an authentic NTR. Similar conclusions are obtained when the genome of a closely related organism, C. acetobutylicum, was analyzed, except that this organism had two FFTR paralogues, CaFFTR1 (gene ID CA_C1548) and CaFFTR2 (gene ID CA_C3082) (SI Appendix, Fig. S1A).
FFTR is also present in Clostridia that produce some of the deadliest known neurotoxins, such as Clostridium difficile, Clostridium botulinum, Clostridium tetani, and Clostridium perfringens, and in several strains that naturally produce butanol (SI Appendix, Fig. S2 and Table S1). A complete genome sequence analysis showed that FFTR is widely distributed in the Firmicutes group of bacteria, with the exception of Bacillus species. Outside this group, FFTR was present but poorly represented in Spirochaetes (Treponema socranskii and Treponema brennaborense).
FFTR Adopts an Extended Conformation with the Flavin Cofactor Solvent-Exposed.
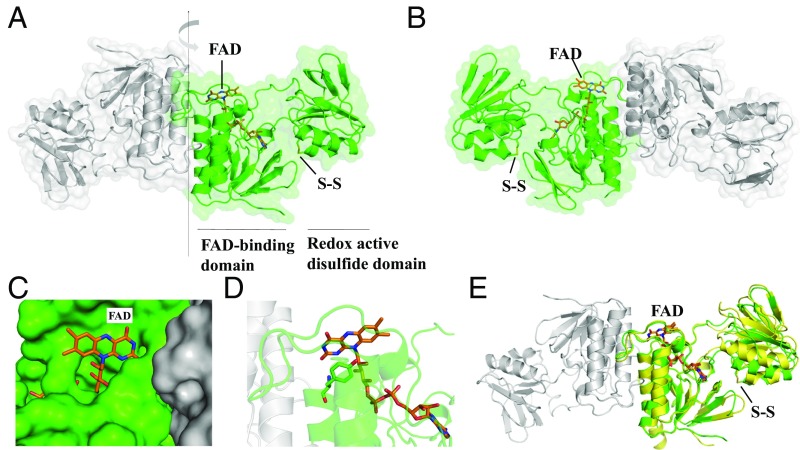
CaFFTR1, CaFFTR2, and CaTrx2 genes were cloned and heterologously expressed in Escherichia coli cells. The recombinant enzymes were shown to be active in Trx reduction (SI Appendix, Fig. S3). We solved the structures of the two C. acetobutylicum FFTR enzymes, CaFFTR1 and two CaFFTR2, at 1.6-, 1.9-, and 1.3-Å resolution, respectively (SI Appendix, Tables S1 and S2). Due to the high similarity of the CaFFTR1 and CaFFTR2 structures, we will henceforth refer to one of the CaFFTR2 structures unless otherwise noted. The results show that FFTR is a homodimer with a modular architecture (Fig. 1 A and B). Each monomer is composed of two Rossmann fold domains connected by two extended loops that are not in direct contact. The flavin adenine dinucleotide (FAD) cofactor is located at a distance of ∼30 Å from the disulfide redox active site (CxxC motif).
Fig. 1.
Crystal structure of C. acetobutylicum FFTR. (A) A ribbon drawing representation with semitransparent surface of the CaFFTR2 homodimer, with each monomer colored in green and gray. The two domains in one monomer are indicated, with the FAD cofactor and the disulfide shown as sticks. (B) A 180°-rotated view of A as indicated by the arrow in A, favoring the visualizing of the domain organization in FFTR. (C) FFTR is represented in surface, and FAD cofactor is shown in sticks (same view as in B). (D) Close-up view of the interaction between Tyr263 and the isoalloxazine ring of the flavin cofactor. (E) Superposition of FFTR1 (yellow) and FFTR2 (green). For the structural alignment, only the coordinates of the FAD-binding domain were matched. One monomer of FFTR1 is presented for simplification. The flavin and the redox-active Cys amino acids are depicted in stick representation.
One domain, the FAD-binding domain (residues 1 to 110 and 240 to 283) binds the FAD cofactor noncovalently and primarily participates in homodimer formation (Fig. 1A). The cofactor adopts a fully extended conformation and forms hydrophobic contacts, polar interactions, and hydrogen bonds with the polypeptide chain (SI Appendix, Fig. S4). The isoalloxazine ring is located on the surface of the FAD-binding domain, fully exposed to the solvent (Fig. 1 B and C). In the FFTR structures, a Tyr residue (Tyr266 in FFTR1 and Tyr263 in FFTR2) on the si face is engaged in a parallel π-stacking interaction with the pyrimidine moiety of the isoalloxazine. Its amide group establishes a hydrogen bond with the oxygen atom of the flavin ring (Fig. 1D). The Tyr forms part of the conserved amino acid motif “Pro260−Tyr−Gln−Tyr263” that accommodates the specificity-determining positions (SDPs) responsible for the unique functional attributes of the FFTR protein family (SI Appendix, Fig. S1A, SDP2).
The second domain (residues 120 to 217) contains the CxxC redox active motif here referred to as the redox-active disulfide domain. The cysteine residues of this motif are largely solvent-inaccessible and are oxidized in purified CaFFTR2 (Fig. 1B). By contrast, the electron density for these residues in CaFFTR1 is not clearly interpretable. Superimposing the CaFFTR1 structure onto CaFFTR2 reveals a slightly different orientation of the redox-active disulfide domain relative to the FAD-binding domain, suggesting inherent interdomain flexibility (Fig. 1E). We then used small-angle X-ray scattering (SAXS) to get information about the structure of FFTR in solution. The results showed good agreement between the scattering data in solution and the theoretical profiles estimated from our crystallographic structures (SI Appendix, Fig. S5 A and B). Therefore, it can be concluded that the conformations found in the crystals for CaFFTR1 and CaFFTR2 reliably represent those adopted in solution.
Trx Binding Alters FFTR Conformation.
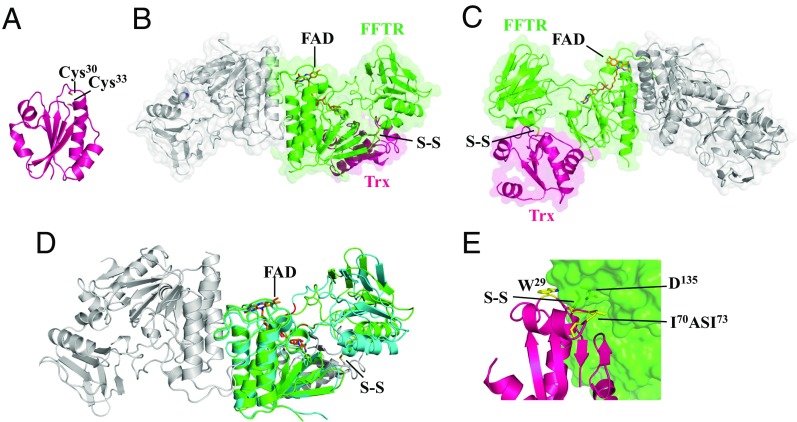
Biochemical experiments demonstrated that C. pasteurianum Trx (CpTrx) was functionally associated with CpFFTR (8). In C. acetobutylicum, a gene context analysis revealed that CaFFTR1 and CaFFTR2 are immediately adjacent to the Trx genes CaTrx1 (CA_C1547) and CaTrx2 (CA_C3083), respectively, suggesting that each form of FFTR is involved in the reduction of the Trx coded by its neighboring gene. In the experiments presently described, we used CaTrx2 because it contains the canonical WCGPC catalytic motif and shows the highest amino acid sequence identity to CpTrx (SI Appendix, Fig. S1B) (10). The crystal structure of CaTrx2 solved at 1.7-Å resolution shows a classical Trx fold (Fig. 2A) with a 0.93-Å root-mean-square deviation for the backbone atoms relative to E. coli Trx (EcTrx) (11). The two cysteines of the redox active motif (CxxC) are positioned at the N terminus of an α-helix and were found reduced in the CaTrx2 protein crystal.
Fig. 2.
Crystal structure of C. acetobutylicum Trx and FFTR−Trx complex. (A) Ribbon drawing representation of CaTrx2. (B) Ribbon drawing representations with semitransparent surface of the CaFFTR2−CaTrx2 complex, with one monomer of FFTR (green) bound to one Trx molecule (magenta) and the rest of the complex colored in gray. (C) A view rotated ∼180° about the vertical axis relative to B. Loops missing in the electron density map in Trx are indicated as dotted lines. (D) Comparison of FFTR free (blue) and in complex with Trx (FFTR in green and Trx in gray; the disulfide bond is indicated) for one monomer. Hinge region forming part of the two joining loops is shown in red. (E) Details of FFTR−Trx binding interface. Trx residues interacting with FFTR are shown as sticks and labeled. A hydrophobic loop [residues 70 to 73: Ile-Ala-Ser-Ile (IASI), in yellow] in CaTrx2 fits into a surface cleft of the redox-active disulfide domain of CaFFTR2. The FAD cofactor is represented in sticks, with carbons in orange.
We next prepared a covalent complex between cysteine-to-serine mutants of CaFFTR2 and CaTrx2 linked through an intermolecular disulfide bond. The mutation of the Cys was necessary for stabilization of the covalent complex (12). Crystals were obtained, and the structure of the complex was solved at 2.9-Å resolution. The results confirm the ratio of two Trxs per FFTR homodimer and the presence of a disulfide cross-link between CaFFTR2 and CaTrx2 (Fig. 2 B and C). The structural alignment of free CaFFTR2 and CaTrx2 onto the CaFFTR2−CaTrx2 complex indicates minimal conformational change in Trx but, by contrast, a structural domain rearrangement in FFTR (Fig. 2D). Binding of Trx is accompanied by a shift of the redox-active disulfide domain relative to the FAD domain that uncovers the CxxC motif in FFTR for interaction with Trx. The movement arises from a rotation of about 24° of the redox-active disulfide domain relative to the FAD-binding domain in the homodimer where the amino acid motifs Gly110−Met−Glu112 and Pro222−Pro223 act as the mechanical hinge elements (Fig. 2D and SI Appendix, Fig. S1A). Despite the different relative orientations of the domains in CaFFTR2−Trx2 and CaFFTR2, the isoalloxazine ring of FAD remains open to the solvent in both cases (Fig. 2C). We also performed SAXS experiments to further study the conformation of the complex in solution and found an excellent correlation between the theoretical profiles generated from the crystal structures and the experimental SAXS profiles (SI Appendix, Fig. S5). These data indicate that the structure of the CaFFTR2−CaTrx2 complex observed in the crystal reliably represents that in solution.
Recognition of Trx by FFTR2 is mainly due to two loops in Trx, one with the covalently bound Cys (Trp29−Pro32) and the other with a hydrophobic loop (Ile70−Pro74) that docks into a cleft of the redox-active disulfide domain formed by residues Lys122−Tyr139 (Fig. 2E). Specifically, Ile73 of Trx forms a hydrogen bond with Asp135 of FFTR2 that strongly stabilizes the interaction. Among the interacting amino acids, the Pro137−Leu−Tyr motif of FFTR2 constitutes SDPs in FFTR (SI Appendix, Fig. S1A, SDP1). Additional interactions seem to further stabilize the complex, namely, a hydrophobic contact between Trx (Phe91−Ser96) and the FAD-binding domain in FFTR (Asn83−Gly88) (SI Appendix, Fig. S6A).
FFTR and NTR Show Diverse Properties.
Several properties of the FFTR are similar to those previously reported for low-molecular-weight NTRs. Although the FAD domains involved in homodimerization and cofactor coordination are highly conserved, major differences were noted in the loops that connect the domains, as well as in the relative orientation of the domains. First, the redox-active disulfide domain of FFTR is a Rossmann fold, as in NTR, but has lost the GxGxxA consensus sequence and the characteristic “arginine motif” (HRRxxxR) important for NADPH binding (gray boxes in SI Appendix, Fig. S1). Second, two antiparallel beta-strands keep the two domains in NTR together, thereby tightly controlling the rotation of the domains that enables the NADPH or the CxxC redox motif to approach the isoalloxazine ring at its re face in the flavin-reducing (FR) or flavin-oxidizing (FO) conformations, respectively (12). By contrast, the two domains in FFTR are joined by two extended loops that pull them apart, resulting in exposure of the isoalloxazine ring of FAD to the solvent (SI Appendix, Fig. S7). Third, the relative orientation of the two domains is significantly different between FFTR and NTR. Considering that the CxxC redox motif should interact with FAD for disulfide reduction in the FO conformation but must be exposed to the solvent for Trx interaction in the FR conformation, the FFTR structures herein presented should represent the FR conformation.
The structures of the NTR−Trx complex from E. coli (12) and the FFTR−Trx complex from C. acetobutylicum (Fig. 2B) show comparable features involving the intermolecular disulfide bond at the interface (SI Appendix, Fig. S6B). However, previously reported experimental results showed that CpFFTR failed to reduce EcTrx (8). The structures reveal that specific contacts between Trx and FFTR are distinct at a second loop of interaction. In E. coli Trx, Arg73 has been found to be important for interacting with E. coli NTR (SI Appendix, Fig. S6B) (13). Structural alignment explains the marginal activity of FFTR with EcTrx: FFTR cannot properly accommodate the arginine side chain of EcTrx at the binding interface, indicating that the sequence of the second loop of Trx is a critical determinant for TR binding (SI Appendix, Fig. S6B).
A Putative Binding Pocket for Fdx in FFTR.
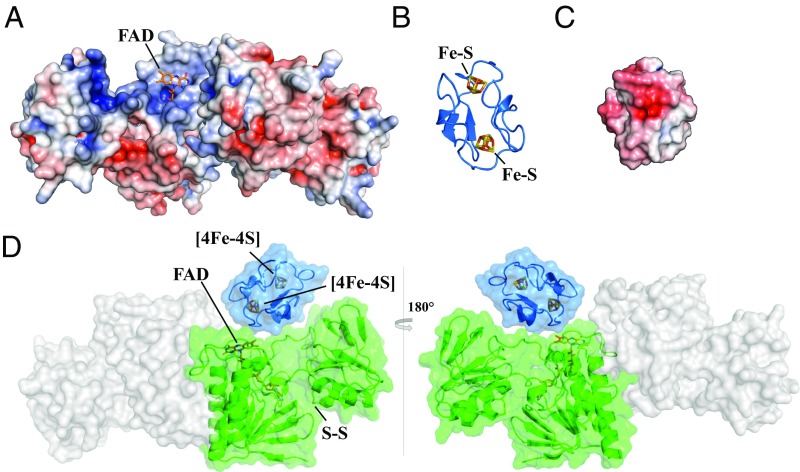
It is assumed that the structures represent the FR state, in which FFTR adopts a conformation suited for electron transfer from Fdx to FAD (SI Appendix, Fig. S7). Strikingly, the isoalloxazine ring of the flavin is solvent-exposed in the crevice formed by the two domains of the FFTR monomer that is of sufficient size and shape to accommodate Fdx. Indeed, electrostatic surface analysis of the structures revealed that the crevice presents regions of neutral and positive electrostatic potential (Fig. 3A)—a site expected for association with Fdx (14).
Fig. 3.
Model of Fdx interaction with FFTR. Electrostatic potential surfaces are colored in red for negative and blue for positive on a scale from −5 kT/e to +5 kT/e; uncharged residues are uncolored. (A) The electrostatic potential mapped onto the FAD solvent-accessible surface of CaFFTR2 in a view as in Fig. 1B. (B) A homology model of C. acetobutylicum Fdx using C. pasteurianum Fdx as template (Protein Data Bank ID code 1clf). Target and template proteins share 90% amino acid sequence identity. (C) The electrostatic potential map of CaFdx in a view as shown in B. (D) Fdx−FFTR complex model based on charge and shape complementary. One Fdx (blue ribbon diagram) is docked to a monomer of FFTR (green surface) at the interface of the two domains. The model is rotated 180° between Left and Right.
The functional Fdx of the CpFFTR system (8, 15) has been identified as a low-potential 2[4Fe−4S] cluster Fdx, and its 3D structure has been solved by NMR (16). We built a 3D model for the homologous Fdx from C. acetobutylicum (CaFdx; CA_C0303) (Fig. 3B) and visually examined a possible structure of the FFTR−Fdx complex. Here, we compared the binding pocket of positively charged residues of CaFFTR2 that could complement the surface size and regions of the negatively charged residues of CaFdx (Fig. 3C). Although the relative orientation of the Fdx protein in the complex could not be unambiguously determined, Fdx fits in a pocket of FFTR provided by amino acid residues of the two domains within a monomer (Fig. 3D). We also used a molecular docking tool, FRODOCK (17), to identify candidate binding sites for Fdx on the FFTR surface. The top 10 solutions for CaFFTR2 and CaFFTR2−Trx2 complex showed the most probable structure in which the Fe−S cluster of Fdx is positioned next to the FAD cofactor. Moreover, relative to the surface of the complete FFTR molecule, the region around FAD is recognized as a hot spot for the FFTR−Fdx interaction (SI Appendix, Fig. S8). One structural element that stands out is the loop region that is shorter in FFTR than in NTR (SI Appendix, Fig. S1A). This protein segment forms part of the cavity region within the redox-active disulfide domain and likely participates in substrate binding.
The Occurrence of Flavin TRs Is Diverse in Bacteria.
A structural homology search using the Dali server (18) identified the deeply rooted Trx reductase (DTR) protein (19) as the closest structural homolog to FFTR with an rmsd of 3.5 Å over 278 amino acids, followed by a recently described atypical TR from Desulfovibrio vulgaris named isolated Trx reductase (TRi) (rmsd of 4.2 Å over 275 amino acids) (20). Like FFTR, DTR and TRi are homodimers, with similar relative positions of the two domains and connecting loops but with variations in their local conformation (SI Appendix, Fig. S9). Notable differences include the presence of a C-terminal tail structural motif with a conserved aromatic residue that stacks over FAD at its re side in DTR, excluding the flavin from the solvent and likely playing a regulatory role (19). In FFTR, a tyrosine stacks over the FAD at its si face with an unknown function (Fig. 1D). According to amino acid sequence and 3D structure conservation, FFTR, DTR, Tri, and NTR belong to the same family of flavoenzymes and share a common evolutionary ancestor. The four groups have a similar two-domain architecture comprising a well-conserved flavin-binding domain together with a diverse substrate-binding domain (corresponding to the NADPH-binding domain in NTR) that contains the CxxC redox active motif.
When the occurrence and distribution of flavin TRs were determined, we found that NTR is widely distributed in bacteria although not confined to these organisms. On the contrary, the presence of FFTR, DTR, and TRi was restricted to specific groups. FFTR was identified mostly in Clostridium species. DTR was found in certain members of bacteria: Aquificace, Chloroflexi, Bacillus, Firmicutes, Chlorobi, Nitrospirae, and Cyanobacteria as well as in a few marine algae (19). TRi was detected in organisms of the order Desulfovibrionales (20), but its presence in Clostridium requires further investigation, as suggested by preliminary sequence analyses. Most organisms with FFTR, DTR, or TRi also contain NTR, but apparently FFTR, DTR, and TRi do not coexist (Table 1). Remarkably, in certain organisms, the Trx system seems to rely solely on FFTR or DTR. Examples include a large number of clostridial strains that naturally produce butanol (SI Appendix, Table S3); two pathogenic clostridia, C. tetani and C. perfringens (FFTR); a deeply branching thermophilic bacterium, Aquifex aeolicus (DTR); and an ancient cyanobacterium, Gloeobacter violaceus (DTR).
Table 1.
Gene content and distribution of NTR-related proteins in selected organisms
| Occurrence of Trx reductases | |||||
| FAD | [4Fe-4S] | ||||
| Bacterial species | NTR | FFTR | DTR | TRi | FTR |
| C. acetobutylicum | −(11) | +(11) | − | − | − |
| C. pasteurianum | −(11) | +(9) | − | − | − |
| C. botulinum Hall | + | + | − | − | − |
| C. tetani E88 | − | + | − | − | − |
| C. perfringens | − | + | − | − | − |
| C. difficile 630 | + | + | − | − | − |
| E. coli | +(47) | − | − | − | − |
| D. vulgaris | +(21) | − | − | +(21) | − |
| A. aeolicus | − | − | +(20) | − | − |
| G. violaceus | −(48) | − | +(20) | − | − |
| Synechocystis sp. | −(48) | − | − | − | +(49) |
Discussion
More than three decades ago, Hammel et al. (8) found that Trx reduction was driven by Fdx—and not NADPH—in C. pasteurianum (FFTR). The participating enzyme was a flavoprotein with properties that differed from the classical Fdx-dependent iron−sulfur TR enzyme (FTR) functional in oxygenic photosynthetic organisms. Conversely, FFTR showed structural similarities to NTR.
FFTR occurs exclusively in the Firmicute group, mostly in Clostridia. In most of these organisms, the cooccurrence of NTR and FFTR raises questions related to their functional redundancy and putative crosstalk between the relevant pathways. Surprisingly, there exist a number of organisms, including those with industrial and biomedical implications such as C. acetobutylicum and C. perfringens, in which the Trx system is composed exclusively of Fdx, FFTR, and Trx. Thus, in these cases Trx reduction depends exclusively on Fdx, rather than NADPH.
In the present work, we have investigated the molecular properties of FFTR and Trx of C. acetobutylicum using X-ray crystallography and SAXS, and obtained five high-resolution structures that provide insight into this mechanism of Fdx-dependent Trx reduction. The results show that FFTR and NTR share a similar domain architecture, but differ in the ways the domains interact to result in different functional properties. Nonetheless, we anticipate that the proteins share a common catalytic mechanism that involves the reduction of FAD with the subsequent transfer of reducing equivalents to the CxxC group that, in turn, reduces Trx by a dithiol–disulfide exchange reaction.
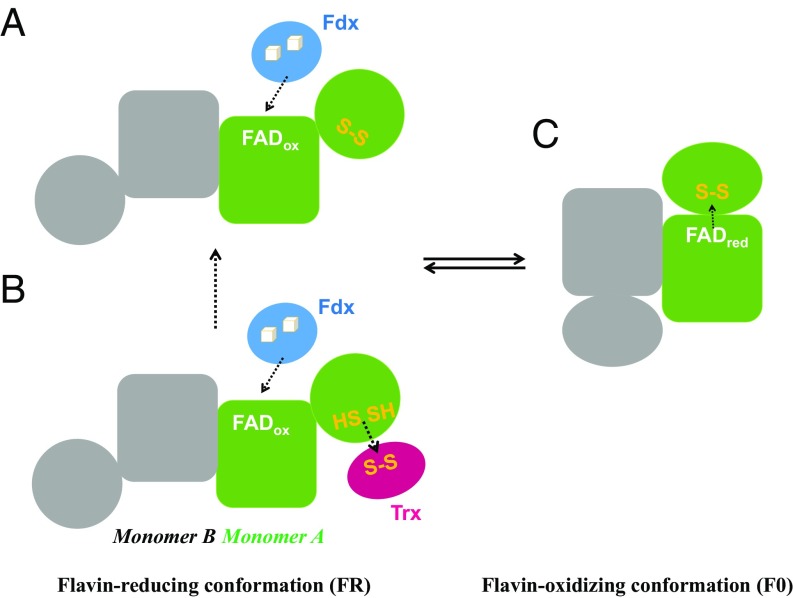
The structures of FFTR obtained in this work allowed us to envision the different steps of the catalytic cycle. FFTR is a homodimer composed of two Rossmann-fold domains linked by two extended loops. Interdomain flexibility seems to be essential in transducing the conformational changes induced by Fdx binding and Trx reduction, thus suggesting a pivotal role for the connecting region between the domains. A comparison of the three crystal structures (CaFFTR1, CaFFTR2, and CaFFTR2−CaTrx2 complex) reveals the modes of flexibility in FFTR associated with the connecting regions of the two domains that include both rotations and hinge-like opening and closing. Therefore, it is plausible to propose that FFTR adopts different conformations that modify the orientation of the disulfide redox-active domain in solution during the catalytic cycle (Fig. 4). The Clostridial FFTR-associated Fdx (21) contains two [4Fe-4S] clusters that supply two electrons to the enzyme, and it is expected to bind to the FR conformation of FFTR to access the oxidized isoalloxazine ring of the flavin (Fig. 4A). The current data do not allow us to draw a conclusion as to whether the binding of Trx would promote or modulate Fdx binding to the complex (Fig. 4B). Seemingly, the reduction of FAD and release of Fdx would trigger the switching from the FR to the FO conformation (Fig. 4C). Additional studies, such as a mutagenesis approach and high-resolution structures of the complex with Fdx and the structure of FFTR in the FO conformation, would ultimately confirm our proposed mechanism, and provide details about the sequence of events during the catalytic cycle.
Fig. 4.
FFTR working model. FFTR might explore different conformation states during its catalytic cycle. (A) In the FR state, the isoalloxazine group of the flavin cofactor is exposed for interaction with Fdx, and (B) the CxxC motif of FFTR can interact with oxidized Trx for reduction. (C) Reduction of the bound FAD cofactor would induce a conformational change that is propagated to the second domain in a redox-associated conformational change that results in the FO conformation. In this conformation, the disulfide of FFTR is reduced by FAD. Dashed arrows indicate hypothetical reducing equivalent flow for monomer A.
The structures presented here show that FFTR belongs to the flavin TR family that includes NTR, and the two NADPH-independent TRs recently described: DTR and the atypical TR in D. vulgaris named TRi (19, 20). Since DTR and TRi have not been experimentally characterized, a complete understanding of the functional diversity of the flavin TR family remains to be determined. Nevertheless, the catalytic properties of the enzymes are very different. From the sequence and 3D structure conservation, it can be inferred that FFTR, DTR, and TRi resulted from gene duplication events. It is possible that the protein structures have incorporated variations of the NTR fold and developed diverse functional properties during divergent evolution. These changes are valuable under certain conditions. Interestingly, the FAD-binding domains of the proteins remain essentially identical, but large variations are observed in the structure of the NADP-binding domain. A key structural motif in the evolution of the family is the interdomain connecting region that adopts either two beta-strands in NTRs that restrict the rotation of the two domains or extended linkers in FFTR, DTR, and TRi that are necessary for rotation and translation of the domains to accommodate substrate molecules such as Fdx in FFTR. Functional variation is thus achieved by structural adaptations that involve differences in domain interactions to accommodate the donor substrates. Apropos this point, a pyridine nucleotide-independent but F420-dependent TR has been described in archaea (22); the experimental 3D structure of the protein is awaited with interest.
Although the proteins are structurally different, FFTR functionally resembles FTR in catalyzing an Fdx-dependent reduction of Trx. The enzymes have, however, followed divergent evolutionary paths. Whereas FFTR uses flavin as cofactor, FTR contains a unique [4Fe-4S] cluster. Clostridia seem to have incorporated FFTR by gene duplication and molecular tinkering with an Fdx-dependent donor substrate, whereas the catalytic FTR subunit in Cyanobacteria evolved from deeply rooted Aquificae with later acquisition of the variable subunit (23). A question that arises is whether there are physiological or metabolic advantages for FTR vs. FFTR. The answer remains unknown, but it could be related to the fact that Fe−S clusters are very sensitive to oxygen, while flavins are versatile organic cofactors functional in different types of cells, including strict anaerobes. However, other reasons related to affinity, specificity, or cellular localization of the interacting partners should also be considered. From a functional point of view, the evolution of a Trx system dependent on Fdx in Clostridia is intriguing in that a parallelism between the two Fdx-linked mechanisms can be cited. In Cyanobacteria and chloroplasts, FTR transfers electrons to Trx from photosynthetically reduced Fdx, thus linking metabolism to light/dark cycles. In anaerobic bacteria such as C. pasteurianum and C. acetobutylicum, reduced Fdx is largely generated by the oxidation of pyruvate during fermentation. Thus, FFTR could divert a portion of the reducing equivalents formed to Trx under certain conditions. The identification of proteins linked to Trx in these organisms will help clarify the function of the system.
In sum, the present study (i) describes a previously uncharacterized family of NADPH-independent flavin Trx reductases that are widely distributed in anaerobes, including industrially and medically important species, and (ii) extends our knowledge on the diversity of TRs.
Materials and Methods
Genes encoding CaFFTR1, CaFFTR2, and CaTrx2 were amplified by PCR from C. acetobutylicum American Type Culture Collection 6013 genomic DNA and cloned into the pETEV15b vector (24). The resulting constructs yielded an N-terminal fusion of a histidine tag with a Tobacco Etch Virus (TEV) protease cleavage site under the control of the T7 promoter. The activity of the enzyme was measured using NADPH as electron donor, essentially as described in ref. 25. For FFTR−Trx complex formation, Cys29 in CaTrx2C32S was conjugated to 5-thio-2-nitrobenzoic acid (TNB) and reacted with Cys134 in CaFFTR2C131S to form an intermolecular disulfide according to refs. 12 and 26. Crystals of CaFFTR1, CaFFTR2, CaTrx2, and CaFFTR2−CaTrx2 complex were grown at room temperature by the vapor diffusion method from drops consisting of the solutions indicated in SI Appendix, Table S1. Diffraction data were collected using synchrotron radiation at the i24 (Diamond Light Source) and XALOC (27) (ALBA Synchrotron) beamlines. SAXS experiments were performed at beamline B21 of the Diamond Light Source. The Swiss-Model server (28) and the Frodock program were used for structural homology modeling and protein docking (17). Additional information on materials and methods can be found in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We thank Elena Andrés Galván and M. Gloria González Holgado for excellent technical assistance, and the beamline staff at the Diamond Light Source and ALBA Synchrotron for assistance in data collection. This work was supported by Spanish Ministerio de Ciencia, Innovación y Universidades Grants BFU2016-80343-P and BIO2016-75634-P. The research leading to these results received funding from the European Community’s Seventh Framework Program FP7/2007–2013 under BioStruct-X Grant Agreement 7687.
Footnotes
Conflict of interest statement: B.B.B. and I.A.B. are coauthors of a 2017 retrospective article.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6GN9, 6GNA, 6GNB, 6GNC, and 6GND).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812781115/-/DCSupplemental.
References
- 1.Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 2.Buchanan BB, Balmer Y. Redox regulation: A broadening horizon. Annu Rev Plant Biol. 2005;56:187–220. doi: 10.1146/annurev.arplant.56.032604.144246. [DOI] [PubMed] [Google Scholar]
- 3.Montrichard F, et al. Thioredoxin targets in plants: The first 30 years. J Proteomics. 2009;72:452–474. doi: 10.1016/j.jprot.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Schürmann P, Jacquot JP. Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:371–400. doi: 10.1146/annurev.arplant.51.1.371. [DOI] [PubMed] [Google Scholar]
- 5.Moore EC, Reichard P, Thelander L. Enzymatic synthesis of deoxyribonucleotides: V. Purification and properties of thioredoxin reductase from Escherichia coli B. J Biol Chem. 1964;239:3445–3452. [PubMed] [Google Scholar]
- 6.Holmgren A. Bovine thioredoxin system. Purification of thioredoxin reductase from calf liver and thymus and studies of its function in disulfide reduction. J Biol Chem. 1977;252:4600–4606. [PubMed] [Google Scholar]
- 7.Droux M, et al. Ferredoxin-thioredoxin reductase, an iron-sulfur enzyme linking light to enzyme regulation in oxygenic photosynthesis: Purification and properties of the enzyme from C3, C4, and cyanobacterial species. Arch Biochem Biophys. 1987;252:426–439. doi: 10.1016/0003-9861(87)90049-x. [DOI] [PubMed] [Google Scholar]
- 8.Hammel KE, Cornwell KL, Buchanan BB. Ferredoxin/flavoprotein-linked pathway for the reduction of thioredoxin. Proc Natl Acad Sci USA. 1983;80:3681–3685. doi: 10.1073/pnas.80.12.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuriyan J, et al. Convergent evolution of similar function in two structurally divergent enzymes. Nature. 1991;352:172–174. doi: 10.1038/352172a0. [DOI] [PubMed] [Google Scholar]
- 10.Reynolds CM, Meyer J, Poole LB. An NADH-dependent bacterial thioredoxin reductase-like protein in conjunction with a glutaredoxin homologue form a unique peroxiredoxin (AhpC) reducing system in Clostridium pasteurianum. Biochemistry. 2002;41:1990–2001. doi: 10.1021/bi011802p. [DOI] [PubMed] [Google Scholar]
- 11.Katti SK, LeMaster DM, Eklund H. Crystal structure of thioredoxin from Escherichia coli at 1.68 Å resolution. J Mol Biol. 1990;212:167–184. doi: 10.1016/0022-2836(90)90313-B. [DOI] [PubMed] [Google Scholar]
- 12.Lennon BW, Williams CH, Jr, Ludwig ML. Twists in catalysis: Alternating conformations of Escherichia coli thioredoxin reductase. Science. 2000;289:1190–1194. doi: 10.1126/science.289.5482.1190. [DOI] [PubMed] [Google Scholar]
- 13.Negri A, et al. Protein-protein interactions at an enzyme-substrate interface: Characterization of transient reaction intermediates throughout a full catalytic cycle of Escherichia coli thioredoxin reductase. Proteins. 2010;78:36–51. doi: 10.1002/prot.22490. [DOI] [PubMed] [Google Scholar]
- 14.Hase T, Schürmann P, Knaff DB. The interaction of ferredoxin with ferredoxin-dependent enzymes. In: Golbeck JH, editor. Photosystem I. Advances in Photosynthesis and Respiration. Vol 24. Springer; Dordrecht, The Netherlands: 2006. pp. 477–498. [Google Scholar]
- 15.Tanaka M, Nakashima T, Benson A, Mower H, Tasunobu KT. The amino acid sequence of Clostridium pasteurianum ferredoxin. Biochemistry. 1966;5:1666–1681. doi: 10.1021/bi00869a032. [DOI] [PubMed] [Google Scholar]
- 16.Kümmerle R, Kyritsis P, Gaillard J, Moulis J-M. Electron transfer properties of iron-sulfur proteins. J Inorg Biochem. 2000;79:83–91. doi: 10.1016/s0162-0134(99)00160-9. [DOI] [PubMed] [Google Scholar]
- 17.Ramírez-Aportela E, López-Blanco JR, Chacón P. FRODOCK 2.0: Fast protein-protein docking server. Bioinformatics. 2016;32:2386–2388. doi: 10.1093/bioinformatics/btw141. [DOI] [PubMed] [Google Scholar]
- 18.Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buey RM, et al. A new member of the thioredoxin reductase family from early oxygenic photosynthetic organisms. Mol Plant. 2017;10:212–215. doi: 10.1016/j.molp.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 20.Valette O, et al. Biochemical function, molecular structure and evolution of an atypical thioredoxin reductase from Desulfovibrio vulgaris. Front Microbiol. 2017;8:1855. doi: 10.3389/fmicb.2017.01855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisenstein KK, Wang JH. Conversion of light to chemical free energy. I. Porphyrin-sensitized photoreduction of ferredoxin by glutathione. J Biol Chem. 1969;244:1720–1728. [PubMed] [Google Scholar]
- 22.Susanti D, Loganathan U, Mukhopadhyay B. A novel F420-dependent thioredoxin reductase gated by low potential FAD: A tool for redox regulation in an anaerobe. J Biol Chem. 2016;291:23084–23100. doi: 10.1074/jbc.M116.750208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balsera M, et al. Ferredoxin:thioredoxin reductase (FTR) links the regulation of oxygenic photosynthesis to deeply rooted bacteria. Planta. 2013;237:619–635. doi: 10.1007/s00425-012-1803-y. [DOI] [PubMed] [Google Scholar]
- 24.de Pereda JM, Lillo MP, Sonnenberg A. Structural basis of the interaction between integrin α6β4 and plectin at the hemidesmosomes. EMBO J. 2009;28:1180–1190. doi: 10.1038/emboj.2009.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yonekura-Sakakibara K, et al. Analysis of reductant supply systems for ferredoxin-dependent sulfite reductase in photosynthetic and nonphotosynthetic organs of maize. Plant Physiol. 2000;122:887–894. doi: 10.1104/pp.122.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang PF, Veine DM, Ahn SH, Williams CH., Jr A stable mixed disulfide between thioredoxin reductase and its substrate, thioredoxin: Preparation and characterization. Biochemistry. 1996;35:4812–4819. doi: 10.1021/bi9526793. [DOI] [PubMed] [Google Scholar]
- 27.Juanhuix J, et al. Developments in optics and performance at BL13-XALOC, the macromolecular crystallography beamline at the ALBA synchrotron. J Synchrotron Radiat. 2014;21:679–689. doi: 10.1107/S160057751400825X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.