Significance
Isoprene is a climate-active gas, produced in huge amounts by trees, yet we know little about its biogeochemical cycle. Bacteria able to grow on isoprene have been isolated from soils and sediments, but the phyllosphere, the principal isoprene source, has remained unexplored. Using targeted cultivation-independent techniques, we show that the phyllosphere of an isoprene-emitting tree contains a diverse and active isoprene-degrading population. We reconstruct the genome of an isoprene-degrading Variovorax strain and show that it contains a functional isoprene monooxygenase. This detailed study targets isoprene degraders from the phyllosphere, applies metaomics to isoprene degradation, and isolates and sequences an isoprene-degrading member of the Proteobacteria.
Keywords: isoprene, microbiology, plant–microbe interactions, DNA-SIP, phyllosphere
Abstract
The climate-active gas isoprene (2-methyl-1,3-butadiene) is released to the atmosphere in huge quantities, almost equaling that of methane, yet we know little about the biological cycling of isoprene in the environment. Although bacteria capable of growth on isoprene as the sole source of carbon and energy have previously been isolated from soils and sediments, no microbiological studies have targeted the major source of isoprene and examined the phyllosphere of isoprene-emitting trees for the presence of degraders of this abundant carbon source. Here, we identified isoprene-degrading bacteria in poplar tree-derived microcosms by DNA stable isotope probing. The genomes of isoprene-degrading taxa were reconstructed, putative isoprene metabolic genes were identified, and isoprene-related gene transcription was analyzed by shotgun metagenomics and metatranscriptomics. Gram-positive bacteria of the genus Rhodococcus proved to be the dominant isoprene degraders, as previously found in soil. However, a wider diversity of isoprene utilizers was also revealed, notably Variovorax, a genus not previously associated with this trait. This finding was confirmed by expression of the isoprene monooxygenase from Variovorax in a heterologous host. A Variovorax strain that could grow on isoprene as the sole carbon and energy source was isolated. Analysis of its genome confirmed that it contained isoprene metabolic genes with an identical layout and high similarity to those identified by DNA-stable isotope probing and metagenomics. This study provides evidence of a wide diversity of isoprene-degrading bacteria in the isoprene-emitting tree phyllosphere and greatly enhances our understanding of the biodegradation of this important metabolite and climate-active gas.
Isoprene (2-methyl-1,3-butadiene) is emitted to the atmosphere at a rate of ∼500 Tg⋅y−1, on a par with methane and approximately one-third of the total volatile organic compound (VOC) emissions (1, 2). The vast majority originates from terrestrial plants (400–600 Tg⋅y−1), with a small but uncertain flux from marine algae (0.1–12 Tg⋅y−1) (2–4), bacteria, fungi, and animals (5, 6). A reactive diene, isoprene is rapidly photochemically oxidized (1), with a significant and complex effect on global climate (7). Hydroxyl (OH) and nitrate (NO3) radicals and ozone (O3) in the atmosphere react with isoprene depending on prevailing conditions (1). In pristine environments, isoprene reacts directly with ozone and hydroxyl radicals, resulting in ozone depletion. However, the high NOx levels typical of urban environments result in the formation of tropospheric ozone, with important negative effects on human health and on yields of ozone-sensitive crops (8). Globally, these reactions result in a net radiative forcing of 0.09 W⋅m−2, with an additional indirect effect since depletion of OH radicals increases the atmospheric lifetime of methane (9). The isoprene oxidation products form secondary organic aerosols and cloud condensation nuclei with implications for planetary albedo, air quality, and climate (10, 11).
Plants produce isoprene in the chloroplast and release it to the atmosphere from the abaxial surface of leaves via the stomata (12). Although isoprene production is not a universal trait among plants, it protects against heat and oxidative stress (13, 14) and has roles in plant/insect signaling and plant energy dynamics (5, 15). High-isoprene–emitting trees worldwide include oil palm (Elaeis guineensis), eucalyptus (Eucalyptus spp.) and, in temperate regions, English oak (Quercus robur), poplar (Populus spp.), and willow (Salix spp.), with reported emissions of 77 (English oak) and 175 (oil palm) µg⋅g−1 (dry leaves) h−1 (16). Due to its short atmospheric lifetime, isoprene concentrations vary from a few parts per billion by volume (ppbv) (17), to tens of ppbv in high-isoprene–emitting forests (18). Soils acted as a biological sink, both in closed chambers (19, 20) and in continuous systems using isoprene concentrations of 2–200 ppbv (21), suggesting that soil microbes act as a significant sink for isoprene, consuming up to 4% of global emissions.
Several isoprene-consuming bacterial strains have been isolated from soils and marine sediments (6, 22–25), and recently several strains have been characterized biochemically and genetically (26–30). Before this work, all strains characterized by sequence data were members of the Actinobacteria, and all terrestrial examples were rhodococci. They all contain six genes (isoABCDEF) encoding a multicomponent isoprene monooxygenase (IsoMO) (Fig. 1) with four additional genes, isoGHIJ, just upstream. van Hylckama Vlieg et al. (26, 27) showed that isoI encodes a glutathione S-transferase that conjugates glutathione with the epoxide product of IsoMO, and that isoH encodes a dehydrogenase that acts on the product of IsoI. IsoG and IsoJ are involved in the subsequent metabolic pathway but have yet to be characterized. In Rhodococcus AD45, the entire cluster isoGHIJABCDEF is cotranscribed as an operon (28). Significantly, all previously characterized isoprene gene clusters include the glutathione biosynthesis genes gshAB, suggesting a specific role for glutathione in isoprene metabolism. This small thiol is not usually found in Gram-positive bacteria (31), although a role in styrene metabolism has also been suggested recently (32).
Fig. 1.
The isoprene gene cluster in Rhodococcus sp. AD45. Genes present in all previously characterized isoprene degraders are shown in solid colors. In many isolates, aldh1 is located between isoJ and isoA. The horizontal line shows genes cotranscribed in Rhodococcus AD45 (28).
The plant microbiome plays an essential role in plant health and development (33), and the phyllosphere, although an unstable environment, constitutes an extensive habitat for microorganisms, mainly bacteria, the diverse community of which typically comprises 106–107 cells per cm2 (34). In the intercellular spaces of leaves, near the point of emission from stomata, isoprene concentrations reach 30 ppmv, approximately three orders of magnitude higher than atmospheric concentrations (12, 35–37). Apart from isoprene, plants produce a range of other VOCs. Although leaf-dwelling methylotrophs can reduce plant methanol emissions to the atmosphere (38), the extent to which the plant microbiome moderates release of other VOCs is not known (39, 40). Human intervention is likely to alter global emissions of isoprene, partly because emissions increase with temperature but are inversely related to atmospheric carbon dioxide concentrations, but also due to the development of high-isoprene–emitting agroforestry (41, 42). Therefore, our aim was to better understand the diversity of microbial taxa involved in isoprene consumption and their mechanisms of action. Specifically, this study addresses the following questions: Does the microbiome of a common isoprene-emitting tree harbor isoprene degraders able to take advantage of this abundant carbon source? And are novel genes expressed in response to isoprene?
Results and Discussion
Isoprene Degraders Isolated from Leaves and Soil.
Leaf washings from high-isoprene—emitting tree species (oak, poplar, willow) consumed isoprene in microcosms. After 15 d, 65% of the microcosms showed >30% depletion of added isoprene (SI Appendix, Table S1), confirming the presence of isoprene-degrading microbes. We obtained three Rhodococcus isolates that grew on isoprene as the sole source of carbon and energy—two strains (ACPA1 and ACPA4) from poplar leaves and one (ACS1) from soil beneath trees. All contained isoprene metabolic genes similar to those identified previously (28, 43).
Stable Isotope Probing Identifies Active Degraders.
To extend the revealed diversity beyond those amenable to cultivation, we used DNA-stable isotope probing (DNA-SIP) in two independent experiments. Since sun-exposed leaves may produce more isoprene than shaded leaves (44), leaves of white poplar (Populus alba) taken from locations on the tree exposed to full sun were used. Initially, microbial cells were dislodged from the leaves and incubated in minimal medium with 13C-labeled or unlabeled isoprene (headspace concentration ∼500 ppmv). Added isoprene was consumed rapidly, and cells were harvested after 7 or 8 d (SI Appendix, Fig. S1). Extraction, fractionation (based on incorporation of 13C label), and quantification of DNA showed that labeled DNA from the 13C-isoprene incubations migrated to “heavy” and “light” regions of the ultracentrifuge tubes depending on incorporation of label, whereas DNA from 12C-isoprene incubations was restricted (>99%) to light fractions (SI Appendix, Fig. S2A). Analysis of 16S rRNA gene amplicons showed that the unenriched bacterial community, highly consistent across six replicates, was composed mainly of Proteobacteria [Sphingomonas (mean relative abundance (RA) 36%), Pseudomonas (17%), members of the Oxalobacteraceae (8%), Methylobacterium (2%) and members of the Comamonadaceae (2%)], Bacteroidetes [Hymenobacter (17%) and Sphingobacteriaceae (1%)], and Actinobacteria (4%) (Fig. 2 and SI Appendix, Fig. S3), in agreement with previous studies of plant-associated bacteria (45, 46). Following enrichment, the active isoprene-degrading community was dominated by Rhodococcus (78 ± 8% mean RA ± SD), although several proteobacterial taxa were also labeled, principally members of the Xanthomonadaceae (8 ± 7%) and Comamonadaceae (3 ± 1.6%) (Fig. 2 and SI Appendix, Fig. S3). So far, all well-described terrestrial isoprene degraders in cultivation are members of the genus Rhodococcus, but we have previously noted the presence of sequences related to Proteobacteria in DNA-SIP incubations with labeled isoprene (29), despite the fact that no publicly available proteobacterial genomes contain recognizable isoprene-degradation–related gene sequences. We investigated these nonactinobacterial isoprene degraders in more detail, again using DNA-SIP.
Fig. 2.
Community profile of the unenriched (time-point zero), and unlabeled (light) and labeled (heavy) fractions of 13C-isoprene incubations from two DNA-SIP experiments, analyzed by 16S rRNA gene amplicon (16S) or shotgun metagenomic (MG) sequencing. For the first experiment (left-most four bars), the labeled and unlabeled bacterial communities were characterized by amplicon sequencing, using DNA extracted from unenriched time-point zero cells at the start of the experiment (U-E), DNA from light and heavy fractions of incubations with 13C-isoprene, and also by shotgun sequencing of the pooled DNA from heavy fractions of 13C-isoprene incubations. The microbial communities of the second experiment (right-most four bars) were analyzed by shotgun sequencing of the unenriched time-point zero DNA, pooled DNA from samples incubated without substrate (N-S), pooled DNA from the light fractions of 13C-isoprene incubations, and DNA from the heavy fractions of 13C-isoprene incubations. Taxa present at >10% are shown in boldface type. Taxa identified at higher levels comprise the sum of those not identified more specifically. The 16S and enriched samples show the mean of triplicates. For complete data, including individual replicates and 12C-isoprene controls, see SI Appendix, Fig. S3.
The second DNA-SIP experiment, with isoprene at 150 ppmv, included incubations without isoprene (no substrate) as an additional control. RNA was also extracted from the incubations, although not subjected to ultracentrifugation or separation into labeled and unlabeled fractions. Analysis showed that, as before, labeled and unlabeled DNA were efficiently separated (SI Appendix, Fig. S2B). In contrast to the first experiment, nucleic acids were analyzed using a shotgun approach. Community composition was assessed by clade-specific marker genes using MetaPhlAn2 (47) (Fig. 2 and SI Appendix, Fig. S3). Analysis of the same DNA, from the heavy fraction of the 13C-isoprene incubations of the first experiment, by both 16S rRNA amplicon and metagenomic sequencing, enabled direct comparison of the microbial community revealed by these two methods. Both confirmed the dominance of Rhodococcus sequences (78 and 94% RA by amplicon and shotgun sequencing, respectively), together with a contribution from Comamonadaceae (3 and 1.5%, respectively). However, the Xanthomonadaceae, identified as 8% among 16S rRNA amplicons, were not identified in significant numbers by the shotgun approach (<0.1% RA), possibly indicative of bias in the PCR-based method.
In the second SIP experiment, the time-point-zero community was dominated by the Bacteroidetes Hymenobacter and Pedobacter (50% RA). Sphingomonas, the most abundant genus of the first experiment, was a minor component (3%) in the second. Following isoprene enrichment, the labeled community was again dominated by Actinobacteria (mainly Rhodococcus, average 74% RA) together with, as before, Proteobacteria, notably Comamonadaceae, principally Variovorax, which averaged 16% of the labeled community, albeit with considerable intersample variability (SI Appendix, Fig. S3). In contrast, these taxa composed a small fraction (0.5–1%) of the DNA from the pooled light fractions. The community profile of control incubations without added substrate was similar to that of the light fractions, further confirming that the heavy fractions contained the labeled DNA from isoprene consumers (Fig. 2).
The transcriptionally active taxa were characterized by profiling the mRNA transcriptome reads using MetaPhlAn2 (SI Appendix, Fig. S4). This approach, albeit based on an extremely restricted subset of taxonomically informative marker genes, identified the Bacteroidetes genus Pedobacter and the Gammaproteobacterium Pseudomonas as transcriptionally active, not only in isoprene enrichments, but also in unenriched time-point-zero samples and in incubations without added substrate, suggesting that these taxa were able to scavenge nutrients from endogenous organic matter and/or dead microbial cells. Transcripts of Rhodococcus and Variovorax, as well as the Bacteroidetes Chryseobacterium and Riemerella were specifically enriched in isoprene incubations, indicating that these taxa were directly or indirectly stimulated by isoprene (SI Appendix, Fig. S4).
Assembly Identifies isoA Sequences.
To identify isoprene-degrading sequences, reads from pooled unenriched time-point-zero samples from each 13C-isoprene–enriched heavy fraction, from pooled light-fraction DNA, and from no-substrate incubations were coassembled, resulting in a total of 1.84 Gbp of sequence (SI Appendix, Table S3). Using, as query, the amino acid sequence of isoA (encoding the alpha subunit of IsoMO), we identified 11 sequences with 40–100% inferred amino acid identity to IsoA from Rhodococcus AD45 (Fig. 3). These scaffolds were examined in detail, and five contained sufficient sequence data to also identify one or more of the genes, specific for isoprene metabolism in Rhodococcus AD45 (28), immediately flanking the monooxygenase (Fig. 4) (36–100% amino acid identity with the Rhodococcus AD45 gene products), indicating a likely role in isoprene degradation. Similarly, reads originating from rRNA-depleted RNA were assembled, resulting in 220,637–454,247 transcripts containing 83.0–223.3 Mbp of sequence for each sample (SI Appendix, Table S4). Eighteen transcripts containing distinct isoA-related sequences were identified (Fig. 3), 8 of which contained other isoprene-related genes in addition to the IsoMO genes isoABCDEF (SI Appendix, Table S5). Many of these genes and transcripts were closely related to those of the Rhodococcus isolates obtained in this study, as well as to other strains identified previously (28, 29) (Figs. 3 and 4). Within-sample quantification of global transcript abundance showed that these Rhodococcus-like isoprene gene transcripts were among the most highly expressed, many among the top 0.2 or 1% of community-wide transcripts (Fig. 3 and SI Appendix, Table S5). In addition to sequences grouping with those of characterized isolates, three highly expressed transcripts, centered around metagenome scaffold MG_3829, formed a distinct cluster (97% identity to isoA from Rhodococcus opacus PD630), suggesting that there is yet more diversity to discover among these isoprene-degrading Actinobacteria. Apart from these Rhodococcus-like isoA genes, more divergent sequences were identified, as described below. Significantly, low-level transcription of isoA was also observed at time-point zero (unenriched) and in incubations without isoprene (no substrate), suggesting that there is a degree of constitutive expression of these isoprene-degradation genes.
Fig. 3.
The relationship between the isoA genes of known isoprene degraders (in boldface type), metagenome scaffold sequences (prefixed MG), and metatranscriptome sequences, together with other representative sequences from the databases. Transcripts are prefixed by “MT” followed by sample identification (Unenriched, time-point zero; No-subs, incubations without isoprene; 12C-1–12C-3, incubations with unlabeled isoprene; 13C-1–13C-3, incubations with labeled isoprene). Scaffolds or transcripts containing isoprene-related genes in addition to isoABCDEF are indicated with a double asterisk (**). For each sample, transcripts were ranked by normalized transcript abundance, and highly expressed transcripts are marked with four, three, two, or one red circle, indicating that the isoA-containing transcript was among the most abundant 0.2, 1, 10, or 50%, respectively, of all transcripts from that sample (SI Appendix, Table S4). Where identical isoA sequences were present on different transcripts from the same sample, only the most highly expressed is shown. Partial isoA sequences are indicated with the length in parentheses. The taxonomy of genome bins is shown after the scaffold identification. NA, not assigned; NB, not binned. Bootstrap values over 50% (1,000 replications) are shown as solid circles at the nodes. The scale bar indicates nucleotide substitutions per site.
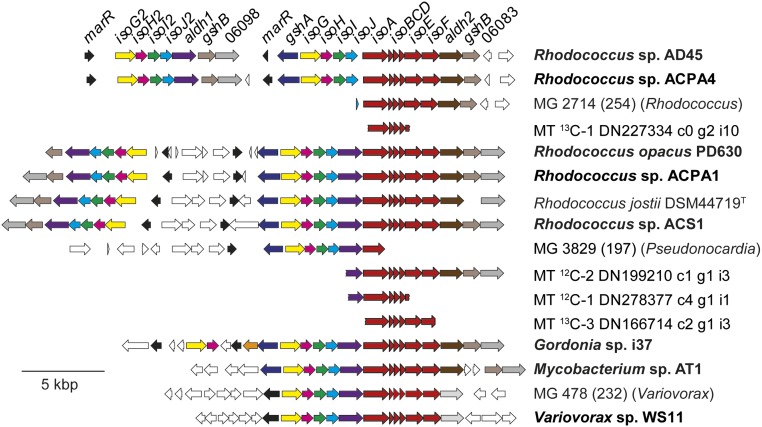
Fig. 4.
The isoprene metabolic gene clusters from known isoprene-degrading isolates (in boldface type) together with representative sequences from the assembled metagenome (prefix MG_ and including bin identification and predicted taxonomy) and metatranscriptomes (prefix MT_).
Genome Reconstruction Through Binning.
To reconstruct individual genomes from the metagenome, scaffolds were assigned to 266 bins based on abundance and nucleotide composition. Bin quality was assessed and refined, and taxonomy was assigned, resulting in 27 genome bins with predicted completeness >90% and contamination <5%. From the 266 bins, 18 were identified as of interest based on the presence of an isoA sequence or predicted RA of >2% in the labeled community, and these were examined in greater detail (SI Appendix, Tables S6 and S7). In total, nine isoA-containing scaffolds were assigned to genome bins, allowing taxonomy to be inferred for most of these sequences (Fig. 3). Several sequences, similar to those of characterized isolates, were assigned to actinobacterial bins (Fig. 3). The most abundant (average 31% RA across three 13C-isoprene enrichments), predicted to be 99% complete and with 1.1% contamination (SI Appendix, Table S6), had 98.7% average nucleotide identity to isolate Rhodococcus sp. ACPA4. Several other (less complete) Rhodococcus bins were also identified. However, since DNA-SIP revealed a considerable diversity of labeled organisms, we also looked at more dissimilar sequences. Genome bin 197, assigned to Pseudonocardia, contained two isoA homologs. The first of these, on scaffold MG_3829, contained a typical isoprene metabolic gene cluster with 75–93% amino acid identity to six isoprene metabolic gene products, including IsoA, from Rhodococcus AD45 (Fig. 4). The same genome also contained scaffold MG_720, with a less similar IsoA homolog (48% amino acid identity with IsoA from Rhodococcus AD45). This region of scaffold MG_720 contained genes with high sequence identity and an identical layout to Rhodococcus jostii DSM44719 and Pseudonocardia dioxanivorans CB1190 (48) annotated as toluene-4 monooxygenases (SI Appendix, Fig. S5). These monooxygenase genes are of low similarity and arranged in a different order to isoABCDEF from isoprene degraders and are not flanked by any other recognizable isoprene-related genes, suggesting that this region was not responsible for isoprene metabolism by known or predicted pathways. Interestingly, although they could not be identified as directly involved in isoprene degradation, these divergent genes on scaffold MG_720 of genome bin 197 were transcribed at moderately high levels (Fig. 3 and SI Appendix, Table S5). Genome bin 095 contained a sequence with homology to a putative toluene monooxygenase from Myxococcales bacterium 68–20 (OJY25058.1), with a similar gene layout. Again, although this genome contained no identifiable isoprene-related genes apart from the monooxygenase, the isoA homolog was represented in the transcriptome (Fig. 3 and SI Appendix, Table S5). These two examples suggest that isoprene is capable of inducing genes not central to its metabolism, that these monooxygenase genes are constitutively expressed at considerable levels, or that they form part of a novel and so-far-undescribed isoprene metabolic pathway.
Genome bin 232, assigned to the betaproteobacterial genus Variovorax, contained an isoA homolog (scaffold MG_478), which was represented by a moderately expressed transcript and aligned most closely with xamoA of the propylene-degrader Xanthobacter autotrophicus Py2 (Fig. 3 and SI Appendix, Table S5). In contrast to characterized isoprene degraders, however, X. autotrophicus Py2 does not use glutathione in its alkene metabolic pathway (49), does not contain any homologs of the isoprene metabolic genes surrounding isoA-F at the xamoA locus (50), and does not grow on isoprene (30). Scaffold MG_478 comprised 76,719 bp of contiguous DNA, allowing examination of the genomic context of isoA (Fig. 4). In addition to the six genes (isoABCDEF) encoding IsoMO, isoGHIJ and aldh1 were present in an identical layout to those of many isoprene-degrading isolates (Fig. 4), although sequence identity of the gene products with those of Rhodococcus AD45 ranged from 42–71%, much lower than those of characterized isolates. Glutathione biosynthesis genes gshA and gshB were not present in this isoprene cluster but were found on scaffold MG_3916 of this genome bin (85 and 76% amino acid identity to ACS17089.1 and ACS17095.1 from Variovorax paradoxus S110, respectively), perhaps reflecting the general use of glutathione in Gram-negative bacteria, as opposed to its often isoprene-specific use in Gram-positive strains.
The co-occurrence of both isoABCDEF and isoGHIJ has previously been successful in identifying bone fide isoprene degraders, but to verify that this sequence contained the genetic potential for isoprene oxidation, we cloned the putative IsoMO genes into a plasmid vector and induced expression in Rhodococcus AD45-ID, a strain of R. AD45 lacking the megaplasmid containing the iso genes, which is incapable of isoprene oxidation. When expressed, the IsoMO from bin 232 indeed oxidized isoprene, in contrast to controls (SI Appendix, Fig. S6).
Targeted Isolation of a Variovorax Strain.
By screening the 16S rRNA gene sequences of numerous isolates from isoprene enrichments, we obtained Variovorax sp. WS11, isolated from a soil enrichment, which grew on isoprene as the sole source of carbon and energy (SI Appendix, Fig. S7). The Variovorax sp. WS11 genome contains an isoprene gene cluster with identical layout to the metagenome-derived sequence of bin 232 (Fig. 4). Overall average amino acid identity (51) was compared between these genomes and 52 Variovorax genome assemblies available in GenBank. The metagenome-derived genome was most similar to Variovorax sp. CF079 (GCA_900101545.1) (82.3%) whereas strain WS11 was most similar to Variovorax sp. B2 (GCA_002891695.1) (88%), and similarity between bin 232 and V. sp WS11 was 79.5%, suggesting that both are novel species. All Variovorax genomes in GenBank were searched for isoprene-related gene sequences like those present in the Variovorax strains described here, but with negative results, suggesting that much diversity still exists outside the reference sequences.
Conclusions.
Although isoprene degraders are present in all isoprene-exposed environments tested (6), no studies have identified isoprene degraders residing on the leaves of isoprene-emitting trees at the source of emission. Here, the isoprene degraders retrieved from the poplar phyllosphere were dominated by Rhodococcus, but included other Actinobacteria (Pseudonocardia) and Proteobacteria (Variovorax). Taxonomy alone is insufficient to identify isoprene-degrading bacteria, with extremely closely related strains differing in terms of isoprene-degrading ability, and phylogeny based on 16S rRNA genes is not congruent with isoA-based analyses (30). These data suggest lateral transfer of the isoprene metabolic genes and imply that surveys that rely on 16S rRNA gene analysis are not able to identify isoprene degraders.
We used metagenomics to reconstruct the genome of a Variovorax strain and conventional methods to obtain related isolate Variovorax WS11. Both genomes contained the entire isoprene metabolic gene cluster. Despite being more similar in sequence to alkene monooxygenase from Xanthobacter autotrophicus Py2 than IsoMO from known isoprene degraders (Fig. 3), the presence of other genes unique to isoprene metabolism and the expression of the monooxygenase in a heterologous host proved that these are genuine isoprene metabolic gene clusters. Variovorax are metabolically versatile bacteria capable of degradation of natural products and xenobiotics, frequently plant-associated and with plant-growth–promoting effects (52, 53), and have been identified as part of the core bacterial microbiome of both Arabidopsis and poplar (54, 55).
Interestingly, we also detected a significant level of monooxygenase transcripts, similar or identical to 13C-labeled DNA scaffolds, which have not, so far, been implicated in isoprene degradation and are possibly indicative of novel isoprene metabolic pathways. This study shows that the leaves of an isoprene-emitting tree provide a habitat for taxonomically disparate isoprene degraders and forms a basis for continued development of molecular tools to detect isoprene degraders. This is a prerequisite for quantification of isoprene-related genes and transcripts and comparison of the activity of microbes associated with isoprene-emitting and nonemitting environments (including comparisons between high- and low-isoprene–emitting tree species), and hence to establishing the extent to which the tree microbiome is able to take advantage of, and mitigate the release of, this abundant carbon source and climate-active gas.
Materials and Methods
For full details, see SI Appendix.
Enrichment, Isolation, and Stable Isotope Probing.
Isoprene degraders were enriched from soil or from cells dislodged from leaves by ultrasound, and purified by standard methods. For SIP enrichments, cells were washed from 5 g leaves, resuspended in minimal medium, and supplied with 13C-labeled or unlabeled isoprene. Isoprene consumption was followed by gas chromatography (GC), and cells were harvested when they had consumed ∼11 or 6 µmol⋅ml−1 (isoprene C), for the first or second experiment, respectively. Each treatment was carried out in triplicate.
Nucleic Acid Extraction and Purification.
DNA and RNA were extracted using standard methods. Total RNA was depleted of rRNA using Ribo-Zero (Illumina). Labeled and unlabeled DNA were separated by density gradient ultracentrifugation and fractionation as described previously (29). Fractions containing labeled (“heavy”) and unlabeled (“light”) DNA were identified based on the data presented in SI Appendix, Fig. S2.
Sequencing of Nucleic Acids.
Communities profiled by 16S rRNA gene amplicons were sequenced using Illumina MiSeq and analyzed using Qiime (56). Metaomic libraries were sequenced using Illumina HiSeq (SI Appendix, Tables S3 and S4). Quality-filtered reads were taxonomically profiled using Metaphlan2 v2.5.0 (47), coassembled using IDBA-UD v1.1.1 (57) (SI Appendix, Table S3), binned using MaxBin v2.2 (58), and quality-checked and refined using CheckM v1.0.5 (59) and RefineM v0.0.23 (60). Filtered transcript reads were de novo assembled using Trinity v2.3.2 (61) (SI Appendix, Table S4). Local Blast databases were searched using tblastn v2.2.28 (62). Reads were mapped to assembled transcripts and quantified using kallisto v0.43.1 (63). Normalized expression levels, as transcripts per million (TPM), of each transcript were ranked for each sample as a percentile.
Expression of IsoMO.
The IsoMO genes isoABCDEF of metagenome bin 232 were PCR-amplified from the pooled heavy fractions of DNA-SIP enrichments and expressed from a vector in a strain of Rhodococcus (R. AD45-ID) cured of the megaplasmid that contains the isoprene metabolic genes. Isoprene uptake of IsoMO-expressing cell suspensions was quantified by GC.
Accession Numbers.
The genomes of Variovorax sp. WS11 and Rhodococcus AD45-ID have been deposited at DNA Database of Japan/European Nucleotide Archive/GenBank under accessions PXZZ00000000 and PYHL00000000. Versions described here are PXZZ01000000 and PYHL0100000, respectively. Sequence reads have been deposited at GenBank Sequence Read Archive (accession no. SRP101805) (SI Appendix, Tables S2–S4).
Supplementary Material
Acknowledgments
Plasmid pTipQC1 was a gift from Tomohiro Tamura. This work was funded by the Earth and Life Systems Alliance at the University of East Anglia; Natural Environment Research Council (NERC) Grants NE/J009725/1 (to J.C.M.) and NE/J009555/1 (to T.J.M.); European Research Council Advanced Grant 694578—IsoMet (to J.C.M.); and NERC Fellowship NE/L010771/1 (to J.P.). We acknowledge receipt of a Colciencias Colombian Government Scholarship (to N.L.L-M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genome sequences of Variovorax sp. WS11 and megaplasmid-cured Rhodococcus AD45-ID have been deposited at DNA Data Bank of Japan/European Nucleotide Archive/GenBank under accession nos. PXZZ00000000 and PYHL00000000. Versions described here are PXZZ01000000 and PYHL0100000, respectively. Sequence reads have been deposited at GenBank Sequence Read Archive (accession no. SRP101805).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812668115/-/DCSupplemental.
References
- 1.Atkinson R, Arey J. Gas-phase tropospheric chemistry of biogenic volatile organic compounds: A review. Atmos Environ. 2003;37(Suppl 2):197–219. [Google Scholar]
- 2.Guenther AB, et al. The model of emissions of gases and aerosols from nature version 2.1 (MEGAN2.1): An extended and updated framework for modeling biogenic emissions. Geosci Model Dev. 2012;5:1471–1492. [Google Scholar]
- 3.Shaw SL, Gantt B, Meskhidze N. Production and emissions of marine isoprene and monoterpenes: A review. Adv Meteorol. 2010;2010:1–24. [Google Scholar]
- 4.Dani SKG, et al. Relationship between isoprene emission and photosynthesis in diatoms, and its implications for global marine isoprene estimates. Mar Chem. 2017;189:17–24. [Google Scholar]
- 5.Sanadze GA. Biogenic isoprene emission as expression of dissipativity, a fundamental cell property. Russ J Plant Physiol. 2017;64:133–140. [Google Scholar]
- 6.McGenity TJ, Crombie AT, Murrell JC. Microbial cycling of isoprene, the most abundantly produced biological volatile organic compound on Earth. ISME J. 2018;12:931–941. doi: 10.1038/s41396-018-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pacifico F, Harrison SP, Jones CD, Sitch S. Isoprene emissions and climate. Atmos Environ. 2009;43:6121–6135. [Google Scholar]
- 8.Ashworth K, Wild O, Hewitt CN. Impacts of biofuel cultivation on mortality and crop yields. Nat Clim Chang. 2013;3:492–496. [Google Scholar]
- 9.Folberth GA, Hauglustaine DA, Lathière J, Brocheton F. Interactive chemistry in the Laboratoire de Météorologie Dynamique general circulation model: Model description and impact analysis of biogenic hydrocarbons on tropospheric chemistry. Atmos Chem Phys. 2006;6:2273–2319. [Google Scholar]
- 10.Fiore AM, et al. Global air quality and climate. Chem Soc Rev. 2012;41:6663–6683. doi: 10.1039/c2cs35095e. [DOI] [PubMed] [Google Scholar]
- 11.Carlton AG, Wiedinmyer C, Kroll JH. A review of Secondary Organic Aerosol (SOA) formation from isoprene. Atmos Chem Phys. 2009;9:4987–5005. [Google Scholar]
- 12.Fall R, Monson RK. Isoprene emission rate and intercellular isoprene concentration as influenced by stomatal distribution and conductance. Plant Physiol. 1992;100:987–992. doi: 10.1104/pp.100.2.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharkey TD, Wiberley AE, Donohue AR. Isoprene emission from plants: Why and how. Ann Bot. 2008;101:5–18. doi: 10.1093/aob/mcm240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeinali N, Altarawneh M, Li D, Al-Nu’airat J, Dlugogorski BZ. New mechanistic insights: Why do plants produce isoprene? ACS Omega. 2016;1:220–225. doi: 10.1021/acsomega.6b00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loivamäki M, Mumm R, Dicke M, Schnitzler J-P. Isoprene interferes with the attraction of bodyguards by herbaceous plants. Proc Natl Acad Sci USA. 2008;105:17430–17435. doi: 10.1073/pnas.0804488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kesselmeier J, Staudt M. Biogenic volatile organic compounds (VOC): An overview on emission, physiology and ecology. J Atmos Chem. 1999;33:23–88. [Google Scholar]
- 17.Greenberg JP, et al. Tethered balloon measurements of biogenic VOCs in the atmospheric boundary layer. Atmos Environ. 1999;33:855–867. [Google Scholar]
- 18.Wiedinmyer C, et al. Ozarks isoprene experiment (OZIE): Measurements and modeling of the “isoprene volcano.”. J Geophys Res Atmos. 2005;110:D18307. [Google Scholar]
- 19.Cleveland CC, Yavitt JB. Consumption of atmospheric isoprene in soil. Geophys Res Lett. 1997;24:2379–2382. [Google Scholar]
- 20.Pegoraro E, et al. The effect of elevated atmospheric CO2 and drought on sources and sinks of isoprene in a temperate and tropical rainforest mesocosm. Glob Change Biol. 2005;11:1234–1246. [Google Scholar]
- 21.Gray CM, Helmig D, Fierer N. Bacteria and fungi associated with isoprene consumption in soil. Elem Sci Anth. 2015;3:000053. [Google Scholar]
- 22.Srivastva N, Singh A, Bhardwaj Y, Dubey SK. Biotechnological potential for degradation of isoprene: A review. Crit Rev Biotechnol. 2018;38:587–599. doi: 10.1080/07388551.2017.1379467. [DOI] [PubMed] [Google Scholar]
- 23.Cleveland CC, Yavitt JB. Microbial consumption of atmospheric isoprene in a temperate forest soil. Appl Environ Microbiol. 1998;64:172–177. doi: 10.1128/aem.64.1.172-177.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Ginkel CG, de Jong E, Tilanus JWR, de Bont JAM. Microbial oxidation of isoprene, a biogenic foliage volatile and of 1,3-butadiene, an anthropogenic gas. FEMS Microbiol Lett. 1987;45:275–279. [Google Scholar]
- 25.Ewers J, Freier-Schroder D, Knackmuss HJ. Selection of trichloroethene (TCE) degrading bacteria that resist inactivation by TCE. Arch Microbiol. 1990;154:410–413. doi: 10.1007/BF00276540. [DOI] [PubMed] [Google Scholar]
- 26.van Hylckama Vlieg JET, Kingma J, Kruizinga W, Janssen DB. Purification of a glutathione S-transferase and a glutathione conjugate-specific dehydrogenase involved in isoprene metabolism in Rhodococcus sp. strain AD45. J Bacteriol. 1999;181:2094–2101. doi: 10.1128/jb.181.7.2094-2101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Hylckama Vlieg JET, Kingma J, van den Wijngaard AJ, Janssen DB. A glutathione S-transferase with activity towards cis-1,2-dichloroepoxyethane is involved in isoprene utilization by Rhodococcus sp. strain AD45. Appl Environ Microbiol. 1998;64:2800–2805. doi: 10.1128/aem.64.8.2800-2805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crombie AT, et al. Regulation of plasmid-encoded isoprene metabolism in Rhodococcus, a representative of an important link in the global isoprene cycle. Environ Microbiol. 2015;17:3314–3329. doi: 10.1111/1462-2920.12793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El Khawand M, et al. Isolation of isoprene degrading bacteria from soils, development of isoA gene probes and identification of the active isoprene-degrading soil community using DNA-stable isotope probing. Environ Microbiol. 2016;18:2743–2753. doi: 10.1111/1462-2920.13345. [DOI] [PubMed] [Google Scholar]
- 30.Johnston A, et al. Identification and characterisation of isoprene-degrading bacteria in an estuarine environment. Environ Microbiol. 2017;19:3526–3537. doi: 10.1111/1462-2920.13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson T, Newton G, Fahey R, Rawat M. Unusual production of glutathione in Actinobacteria. Arch Microbiol. 2009;191:89–93. doi: 10.1007/s00203-008-0423-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heine T, et al. On the enigma of glutathione dependent styrene degradation in Gordonia rubripertincta CWB2. Appl Environ Microbiol. 2018;84:e00154-18. doi: 10.1128/AEM.00154-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 34.Vorholt JA. Microbial life in the phyllosphere. Nat Rev Microbiol. 2012;10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 35.Brüggemann N, Schnitzler JP. Comparison of isoprene emission, intercellular isoprene concentration and photosynthetic performance in water-limited Oak (Quercus pubescens Willd. and Quercus robur L.) saplings. Plant Biol. 2002;4:456–463. [Google Scholar]
- 36.Sun Z, Hüve K, Vislap V, Niinemets Ü. Elevated [CO2] magnifies isoprene emissions under heat and improves thermal resistance in hybrid aspen. J Exp Bot. 2013;64:5509–5523. doi: 10.1093/jxb/ert318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singsaas EL, Lerdau M, Winter K, Sharkey TD. Isoprene increases thermotolerance of isoprene-emitting species. Plant Physiol. 1997;115:1413–1420. doi: 10.1104/pp.115.4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abanda-Nkpwatt D, Musch M, Tschiersch J, Boettner M, Schwab W. Molecular interaction between Methylobacterium extorquens and seedlings: Growth promotion, methanol consumption, and localization of the methanol emission site. J Exp Bot. 2006;57:4025–4032. doi: 10.1093/jxb/erl173. [DOI] [PubMed] [Google Scholar]
- 39.Junker RR, Tholl D. Volatile organic compound mediated interactions at the plant-microbe interface. J Chem Ecol. 2013;39:810–825. doi: 10.1007/s10886-013-0325-9. [DOI] [PubMed] [Google Scholar]
- 40.Bringel F, Couée I. Pivotal roles of phyllosphere microorganisms at the interface between plant functioning and atmospheric trace gas dynamics. Front Microbiol. 2015;6:486. doi: 10.3389/fmicb.2015.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenstiel TN, Potosnak MJ, Griffin KL, Fall R, Monson RK. Increased CO2 uncouples growth from isoprene emission in an agriforest ecosystem. Nature. 2003;421:256–259. doi: 10.1038/nature01312. [DOI] [PubMed] [Google Scholar]
- 42.Rosenkranz M, Pugh TAM, Schnitzler J-P, Arneth A. Effect of land-use change and management on biogenic volatile organic compound emissions: Selecting climate-smart cultivars. Plant Cell Environ. 2015;38:1896–1912. doi: 10.1111/pce.12453. [DOI] [PubMed] [Google Scholar]
- 43.Crombie AT, Emery H, McGenity TJ, Murrell JC. Draft genome sequences of three terrestrial isoprene-degrading Rhodococcus strains. Genome Announc. 2017;5:e01256-17. doi: 10.1128/genomeA.01256-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharkey TD, Loreto F, Delwiche CF. High carbon dioxide and sun/shade effects on isoprene emission from oak and aspen tree leaves. Plant Cell Environ. 1991;14:333–338. [Google Scholar]
- 45.Redford AJ, Bowers RM, Knight R, Linhart Y, Fierer N. The ecology of the phyllosphere: Geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ Microbiol. 2010;12:2885–2893. doi: 10.1111/j.1462-2920.2010.02258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laforest-Lapointe I, Messier C, Kembel SW. Tree phyllosphere bacterial communities: Exploring the magnitude of intra- and inter-individual variation among host species. PeerJ. 2016;4:e2367. doi: 10.7717/peerj.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Truong DT, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 48.Sales CM, et al. Genome sequence of the 1,4-dioxane-degrading Pseudonocardia dioxanivorans strain CB1190. J Bacteriol. 2011;193:4549–4550. doi: 10.1128/JB.00415-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen JR, Clark DD, Krum JG, Ensign SA. A role for coenzyme M (2-mercaptoethanesulfonic acid) in a bacterial pathway of aliphatic epoxide carboxylation. Proc Natl Acad Sci USA. 1999;96:8432–8437. doi: 10.1073/pnas.96.15.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krum JG, Ensign SA. Evidence that a linear megaplasmid encodes enzymes of aliphatic alkene and epoxide metabolism and coenzyme M (2-mercaptoethanesulfonate) biosynthesis in Xanthobacter strain Py2. J Bacteriol. 2001;183:2172–2177. doi: 10.1128/JB.183.7.2172-2177.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 2005;187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belimov AA, et al. Rhizosphere bacteria containing 1‐aminocyclopropane‐1‐carboxylate deaminase increase yield of plants grown in drying soil via both local and systemic hormone signalling. New Phytol. 2009;181:413–423. doi: 10.1111/j.1469-8137.2008.02657.x. [DOI] [PubMed] [Google Scholar]
- 53.Satola B, Wübbeler JH, Steinbüchel A. Metabolic characteristics of the species Variovorax paradoxus. Appl Microbiol Biotechnol. 2013;97:541–560. doi: 10.1007/s00253-012-4585-z. [DOI] [PubMed] [Google Scholar]
- 54.Bodenhausen N, Horton MW, Bergelson J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS One. 2013;8:e56329. doi: 10.1371/journal.pone.0056329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beckers B, Op De Beeck M, Weyens N, Boerjan W, Vangronsveld J. Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome. 2017;5:25. doi: 10.1186/s40168-017-0241-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuczynski J, et al. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Microbiol. 2011;27:1E.5.1–1E.5.20. doi: 10.1002/9780471729259.mc01e05s27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng Y, Leung HC, Yiu SM, Chin FY. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 58.Wu Y-W, Tang Y-H, Tringe SG, Simmons BA, Singer SW. MaxBin: An automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome. 2014;2:1–18. doi: 10.1186/2049-2618-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parks DH, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 61.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 63.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.