Significance
Xylan, the major hemicellulosic component of lignocellulose and the second most abundant polysaccharide after cellulose, contributes to the structural stability of wood and its recalcitrance to enzymatic digestion. The present study identifies Spirochaetes as primary agents of xylan degradation in the hindgut of wood-feeding higher termites, in contrast to the bovine rumen or the human colon, where Bacteroidetes are responsible for hydrolysis of xylan in grass or cereals. The presence of distinctive xylanases in Spirochaetes was so far undocumented to our knowledge. Their phylogenetic origin among gut bacteria of other phyla identifies horizontal gene transfer among the intestinal microbiota as an important driver in the evolutionary adaptation of higher termites to different lignocellulosic diets.
Keywords: metatranscriptome, xylanase, spirochetes, fiber-associated community, termite hindgut
Abstract
Symbiotic digestion of lignocellulose in wood-feeding higher termites (family Termitidae) is a two-step process that involves endogenous host cellulases secreted in the midgut and a dense bacterial community in the hindgut compartment. The genomes of the bacterial gut microbiota encode diverse cellulolytic and hemicellulolytic enzymes, but the contributions of host and bacterial symbionts to lignocellulose degradation remain ambiguous. Our previous studies of Nasutitermes spp. documented that the wood fibers in the hindgut paunch are consistently colonized not only by uncultured members of Fibrobacteres, which have been implicated in cellulose degradation, but also by unique lineages of Spirochaetes. Here, we demonstrate that the degradation of xylan, the major component of hemicellulose, is restricted to the hindgut compartment, where it is preferentially hydrolyzed over cellulose. Metatranscriptomic analysis documented that the majority of glycoside hydrolase (GH) transcripts expressed by the fiber-associated bacterial community belong to family GH11, which consists exclusively of xylanases. The substrate specificity was further confirmed by heterologous expression of the gene encoding the predominant homolog. Although the most abundant transcripts of GH11 in Nasutitermes takasagoensis were phylogenetically placed among their homologs of Firmicutes, immunofluorescence microscopy, compositional binning of metagenomics contigs, and the genomic context of the homologs indicated that they are encoded by Spirochaetes and were most likely obtained by horizontal gene transfer among the intestinal microbiota. The major role of spirochetes in xylan degradation is unprecedented and assigns the fiber-associated Treponema clades in the hindgut of wood-feeding higher termites a prominent part in the breakdown of hemicelluloses.
Lignocellulose is the most abundant biopolymer in terrestrial environments (1). It consists mainly of cellulose, hemicellulose, and lignin, and is remarkably recalcitrant to microbial degradation. The ability of termites to efficiently digest lignocellulose has a large impact on the global ecosystem (1). The elucidation of the underlying mechanisms has presented a formidable challenge to research for almost a century (2). Termites of phylogenetically basal lineages (“lower termites”) harbor symbiotic protists in their enlarged hindgut compartment that phagocytize wood particles and play a crucial role in cellulose and hemicellulose degradation (3). By contrast, termites of the phylogenetically most apical lineage (“higher termites”), which account for approximately two thirds of all termite species, are devoid of such eukaryotic symbionts (4). As a consequence, fiber digestion in the hindgut of higher termites must be attributed to their entirely prokaryotic microbial community (5).
The gut microbiota of higher termites comprises more than 1,000 bacterial phylotypes, which are organized into distinctive communities colonizing the microhabitats provided by the compartmentalized intestine, including the highly differentiated hindgut (6, 7). Of particular interest are the bacteria associated with wood particles in the dilated hindgut paunch of wood-feeding Nasutitermes species; these bacteria represent less than 30% of the total microbial population but contribute more than half of the cellulolytic activity in this compartment (8). Core members of the fiber-associated community are several so-far uncultured lineages of Fibrobacteres and the closely related candidate phylum TG3 (now classified as Fibrobacteria and Chitinivibrionia; ref. 9) and two lineages of uncultured Spirochaetes (Treponema Ic and If), which represent a separate line of descent that has been found exclusively in higher termites (8).
Previous metagenomic analyses (i.e., the comprehensive sequencing of the genetic material of an entire microbial community) had already reported the presence of diverse genes encoding putative cellulases and hemicellulases in the prokaryotic hindgut microbiota of higher termites (10–15). In the case of the wood-feeding Nasutitermes corniger, many of these genes were tentatively assigned to members of the Fibrobacteres and Spirochaetes (10). Metagenomic binning revealed that the genomes of Fibrobacteres encode an abundance of cellulase genes and more hemicellulase genes than the average number in other cellulolytic bacteria. However, these genomes lack the genes required to metabolize xylose (9), a major component of xylan and other hemicelluloses of wood (16). Moreover, none of the Fibrobacteres from termite guts have been cultured, and their hemicellulolytic ability remains to be elucidated.
Spirochaetes likely play a crucial role in reductive acetogenesis, the production of acetate from H2 and CO2 by anaerobic bacteria, which is an important reaction in the hindgut of lower and higher termites (17–19). Although metagenomes of hindgut contents of Nasutitermes spp. provided evidence that spirochetes carry genes encoding glycoside hydrolases (GHs) (10), a direct involvement of fiber-associated spirochetes in the degradation of cellulose or hemicelluloses has not been demonstrated. Thus, the major degraders of hemicellulose in the hindgut of higher termites remain unidentified.
The frequency of genes in the guts of the N. corniger and Amitermes wheeleri elucidated by metagenomic analyses does not necessarily correlate with their expression levels (12). For that reason, the role of bacteria in symbiotic digestion of lignocellulose inferred by metagenomics of the entire hindgut community has to be considered with caution. Metatranscriptomic analyses (i.e., the comprehensive profiling of the transcripts of a microbial community) of the bacteria that interact directly with wood fibers in the hindgut would help to elucidate the actual role of the bacterial gut microbiota in wood-feeding higher termites.
The situation is further obscured by an apparent division of the roles in lignocellulose degradation between higher termite hosts and their intestinal bacteria. Higher termites produce endogenous cellulases that are secreted in the midgut (20), and these activities are considerably higher than those of bacterial origin in the hindgut (21). The cellulase activities in the hindgut of higher termites are also surpassed by those in the hindgut of lower termites (22, 23); lower termite hindguts contain the bulk of the cellulolytic activities and most of the xylanolytic activities, but these are attributed to their flagellate protists and not to bacterial symbionts (22–25). In the flagellate-free gut of higher termites, the amount and location of hemicellulolytic activities, including xylanases, have not been studied.
To clarify the role of bacterial symbionts in lignocellulose degradation in higher termites, we investigated the distribution of xylanase activity among the different gut compartments of the wood-feeding higher termite Nasutitermes takasagoensis. By using a previously developed method to isolate wood fibers with their attached bacterial microbiota from the total hindgut contents (8), we conducted a meta-analysis of the transcripts of carbohydrate-active enzymes (CAZys) expressed by the fiber-associated bacterial community. We then used a combination of metagenomics, phylogenetic analyses, and indirect immunofluorescence microscopy to explore the origin and function of genes encoding the major hemicellulolytic enzymes in the hindgut. Our results shed light on the role in wood decomposition of the fiber-associated microbial community in the hindgut of higher termites and elucidate the unexpected major participants in hemicellulose degradation.
Results
Localization of Xylanase Activity in the Gut of N. takasagoensis.
To identify the site of xylan digestion in the gut of N. takasagoensis, we measured hydrolytic activity against beechwood xylan in soluble (supernatant) and particulate (pellet) fractions of homogenates of salivary glands and individual gut sections by using a previously described protocol (21, 25). Xylanase activity was essentially confined to the hindgut proper, and the activity in the particulate fraction (1.42 ± 0.19 U/g termite) exceeded that in the soluble fraction (0.36 ± 0.32 U/g termite) fourfold (Fig. 1), which suggests that most of the activity is associated with wood particles, bacterial cells, or both. Xylanase activities in salivary glands, foregut, and mixed segment were below the detection limit (0.02 U/g termite), whereas midgut sections occasionally showed trace activities in some preparations (0.03 ± 0.06 U/g termite; n = 5). When we compared xylanase activities with cellulolytic activities in the hindgut, we found that xylanase activities were in the same range as cellulolytic activities against carboxymethylcellulose (CMC) and more than an order of magnitude higher than activities against microcrystalline cellulose (Fig. 1). Moreover, activities against CMC and microcrystalline cellulose were more evenly distributed between the soluble and particulate fractions, which is in agreement with previous reports (8, 21). These results suggest that hindgut bacteria play an important role in degrading the xylan backbone of hemicellulose.
Fig. 1.
Enzyme activities against CMC, cellulose, and xylan in soluble and particulate fractions of hindgut homogenates of N. takasagoensis. The homogenates were separated into soluble and particulate fractions by centrifugation, and xylanase activities were determined in both fractions after detergent extraction. Values are means of replicate samples from five colonies; error bars denote SDs. One unit of enzyme activity is defined as the amount of enzyme that produces 1 μmol of reducing sugar (glucose equivalent for cellulose and xylose equivalent for xylan) per minute. “Cellulose” indicates microcrystalline cellulose (Sigmacell Type 20); “xylan” indicates beechwood xylan.
Metatranscriptomic Profiles of Genes Involved in Lignocellulose Degradation.
To identify genes involved in hemicellulose degradation activity detected in the hindgut compartment, we carried out a metatranscriptomic analysis. The three mRNA libraries prepared from replicate fractions of fiber-associated bacteria in the hindgut of N. takasagoensis each yielded approximately 4.5 Gbp of clean sequence reads (Table 1). De novo assembly of the reads resulted in 133,206 nonredundant gene clusters (here referred to as transcripts), from which 122,671 putative mRNA sequences were identified. The remaining transcripts are predicted as noncoding RNAs.
Table 1.
Properties of the metatranscriptomic libraries of the fiber-associated community in the hindgut of N. takasagoensis
| Sample | Raw data, Mbp | Clean data, Mbp | No. of transcripts | Total length, nt | Mean length, nt | N50 value | Fraction of mapped reads, % | No. of distinct mRNAs | Transcripts encoding CAZys* | Genes encoding CAZys* |
| Library 1 | 4,770 | 4,560 | 94,273 | 51,566,241 | 547 | 731 | 70.8 | — | — | — |
| Library 2 | 4,770 | 4,618 | 97,454 | 52,419,831 | 538 | 714 | 70.7 | — | — | — |
| Library 3 | 4,770 | 4,619 | 97,624 | 52,458,326 | 537 | 708 | 70.5 | — | — | — |
| Merged | — | — | 133,206 | 83,659,303 | 628 | 1,024 | — | 122,671 | 2,766 | 2,807 |
Reads were generated with Illumina HiSeq 2000.
Glycosyltransferases were excluded.
Database searches for CAZys on dbCAN with amino acid sequences (26) and dbCAN2 with nucleotide sequences (27) retrieved 2,766 transcripts with a total of 2,807 genes (Table 1). They encoded representatives of 62 GH families (1,553 genes), 12 carbohydrate esterase families (173 genes), 7 polysaccharide lyase families (39 genes), 3 families with auxiliary activities (21 genes), 52 families of carbohydrate-binding modules (CBMs; 1,111 genes), and 3 docking modules (92 genes; Dataset S1). Transcripts related to lignin degradation and oxidoreductive cellulases (e.g., auxiliary activity AA10 members; ref. 28) were absent in the dataset. The remaining genes were assigned to another 27 glycosyltransferase families that represent enzymes primarily involved in the biosynthesis of polysaccharides rather than in their degradation and were excluded from the present analysis.
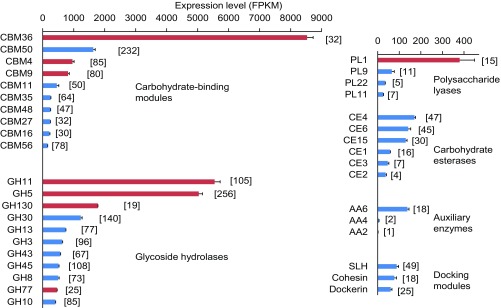
The short reads from each library were mapped onto the aforementioned identified transcripts, and the expression level of each transcript was determined as the number of fragments per kilobase of transcript per million mapped reads (FPKM), which calculates the abundance of paired-end reads and normalizes them to the length of the respective transcript (Fig. 2 and Dataset S1). In all replicates, the most highly expressed CAZy family genes were those of CBM36 (8,531 ± 195 FPKM), which consists primarily of xylan-binding domains; GH11 (5,548 ± 179 FPKM), which represents putative endo-β-1,4-xylanases (hereafter referred to as xylanases); and GH5 (5,037 ± 117 FPKM), which includes diverse cellulases and hemicellulases. Most transcripts assigned to GH5 were affiliated with subfamily 2 (74%) and subfamily 4 (14%), which consist predominantly of endo-β-1,4-glucanases or xyloglucanases and lichenases, respectively (29) (SI Appendix, Fig. S1). High expression levels were also observed for genes encoding GH130 (1,779 ± 4 FPKM) and GH30 (1,228 ± 43 FPKM), which represent hemicellulases (hydrolases and phosphorylases of mannosyl saccharides in GH130 and mostly β-xylosidases in GH30). The other abundant transcripts encode CBM50 proteins (1,632 ± 62 FPKM), which bind to bacterial peptidoglycan and chitin. The expression level of other CAZy family genes were typically far below 1,000 FPKM, and considering that they were represented by a large number of transcripts, most of them did not appear very prominent. These results indicate that the community of fiber-associated bacteria preferentially expresses genes involved in hydrolysis of hemicelluloses, particularly xylan, and cellulose.
Fig. 2.
Expression levels of CAZy family transcripts in the metatranscriptomic libraries of fiber-associated bacteria of N. takasagoensis grouped by the most relevant functions. Only the most highly expressed families in each group are shown (a full list is provided in Dataset S1). Values are means of replicate samples from three colonies; error bars represent SDs. Numbers in parentheses denotes the total numbers of distinct genes assigned to each CAZy family. Red bars indicate families with high expression levels per gene (Dataset S1).
Structure and Heterologous Expression of Putative GH11 Xylanases.
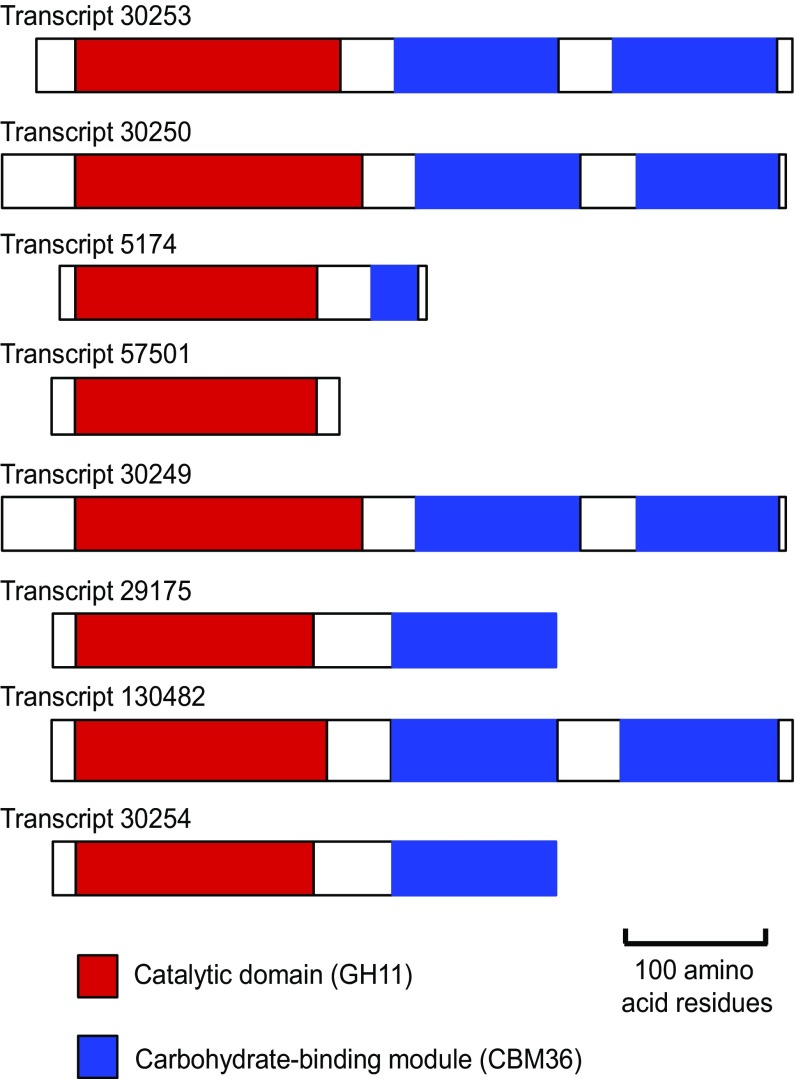
The 8 most abundant transcripts among the 99 transcripts assigned to family GH11 made up 88.9% of the metatranscriptomic reads assigned to this family (Dataset S2). With the exception of Transcript 57501, which encoded only a single catalytic domain, each transcript had a single gene that encoded a GH11 catalytic domain and one or two CBM36 domains (Fig. 3). The deduced amino acid sequences of the catalytic domains showed 95–100% identity to each other and 60–70% identity to those of bona fide GH11 xylanases from Firmicutes (as detailed later). An exception was Transcript 130482, whose catalytic domain showed only 81–82% identity to those of the other transcripts and 70–80% identity to those of bona fide GH11 xylanases from Firmicutes.
Fig. 3.
Domain structure of the GH11 xylanases predominantly expressed by the fiber-associated bacterial community in the hindgut of N. takasagoensis based on the deduced amino acid sequences of the eight most abundant transcripts.
To confirm that the predominantly expressed members of family GH11 encode functional xylanases, we amplified an entire GH11 gene from hindgut DNA of N. takasagoensis by using PCR primers designed to match the flanking regions of Transcript 30253, the homolog with the highest expression level (30.0% of the reads assigned to family GH11). The deduced amino acid sequence encoded by xylanase clone NtSymX11, determined by Sanger sequencing, showed the same constitution (a single catalytic domain and two CBM36 modules) and an amino acid identity of 96% in the catalytic domain to the product of Transcript 30253; the overall amino acid identity, including the CBM modules and their internal sequences, was approximately 87%. Phylogenetic analysis revealed that clone NtSymX11 falls into the monophyletic group formed by seven of the eight most abundant transcripts (as detailed later). After removal of a putative leader sequence [inferred by using the SignalP 4.1 server (30)], clone NtSymX11 was heterologously expressed in Escherichia coli. The partially purified polyhistidine-tagged (His-tag) recombinant enzyme (SI Appendix, Fig. S2A) showed high xylanase activity (592 U/mg protein) against beechwood xylan (SI Appendix, Fig. S2C).
Organismal Origin of the GH11 Xylanases.
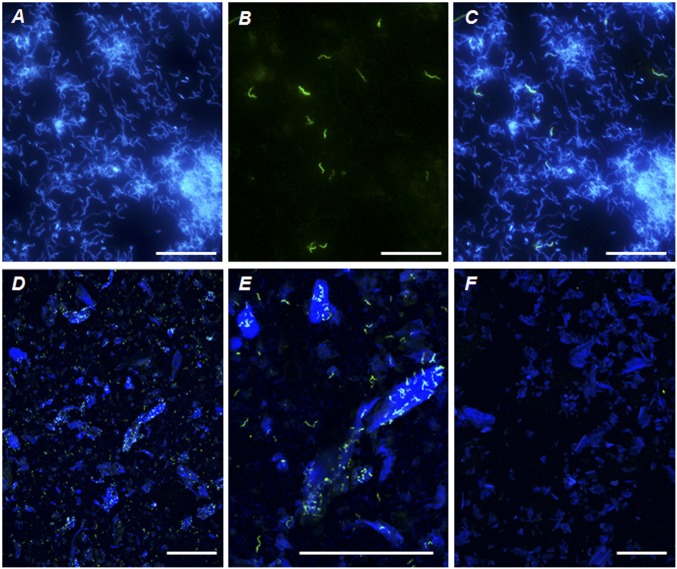
We raised monoclonal antibodies against components of the fiber-associated bacterial fraction. In Western blots, only 1 of 15 antibodies cross-reacted with recombinant NtSymX11 xylanase (SI Appendix, Fig. S2B). Indirect immunofluorescence microscopy of the P3 fluid of N. takasagoensis revealed that the antibody bound specifically to bacterial cells with a helical morphology, which occurred in an unattached state or in association with wood fibers (Fig. 4). As an undulate or helical morphology is encountered in the members of Fibrobacteres and Spirochaetes that colonize the hindgut of Nasutitermes spp. (31, 32), and attempts to colocalize the immunofluorescence signal with fluorescent rRNA-target oligonucleotide probes gave inconsistent results (SI Appendix, Supplementary Results), we investigated the organismal origin of the predominant xylanases by using a metagenomic approach.
Fig. 4.
Indirect immunofluorescence microscopy of the N. takasagoensis P3 content stained with a monoclonal antibody that cross-reacts with recombinant NtSymX11 xylanase (green) and counterstained with DAPI (blue). (A–C) The same field of view inspected with an epifluorescence microscope: (A) DAPI channel, (B) Alexa Fluor 488 channel (antibody), and (C) merged image. Merged images obtained with a confocal laser microscope at lower (D) and higher (E) magnification show the association of antibody-stained cells with autofluorescent wood particles. (F) Negative control without primary antibody treatment. (Scale bars: A–C, 20 µm; D–F, 100 μm.)
Metagenomic sequencing of the bacterial community associated with the fiber fraction yielded 126,962 contigs from more than 40 million reads (SI Appendix, Table S1). A local nucleotide BLAST search against this dataset identified 17 contigs with a significant similarity to Transcript 30253 (e-value < 10−10). Two of the contigs contained a complete GH11 gene (83% and 88% nucleotide sequence identity to Transcript 30253), in each case surrounded by genes with highest sequence similarity to those in Spirochaetes (SI Appendix, Fig. S3). Most of them were from Treponema primitia and Treponema azotonutricium, which are the only members of the Treponema I clade that have sequenced genomes and inhabit the hindgut of lower termites. The remaining contigs contained partial sequences of GH11 and CBM36, and one of the former was flanked as well by a gene with highest sequence similarity to a homolog in T. primitia (SI Appendix, Fig. S3).
Compositional binning of all metagenomic contigs with VizBin yielded three well-defined clusters (i.e., bins; SI Appendix, Fig. S4). BLAST analysis of 31 single-copy marker genes (33) indicated that bin 1 is composed almost exclusively of members of Fibrobacteria (phylum Fibrobacteres), whereas bin 2 is composed almost exclusively of Spirochaetes (Dataset S3). In bin 3, the top BLAST hit of each single-copy marker gene was almost exclusively represented by Chitinivibrio alkaliphilus, the first and only isolate of the Chitinivibrionia (formerly candidate phylum TG3; ref. 34) represented in public databases at the time of analysis. Hidden Markov model searches for the presence of pfam00457, the catalytic domain of GH11, identified a total of 30 CDSs in the three bins (2 in bin 1, 26 in bin 2, and 2 in bin 3); these CDSs encoded putative xylanases of family GH11. CBMs of CBM36, which were identified by using dbCAN because of the lack of pfam domains, were detected only in bin 2 (Dataset S4). These results indicate that the majority of genes involved in xylan hydrolysis originated from Spirochaetes.
Phylogenetic Analysis of GH11 Xylanases.
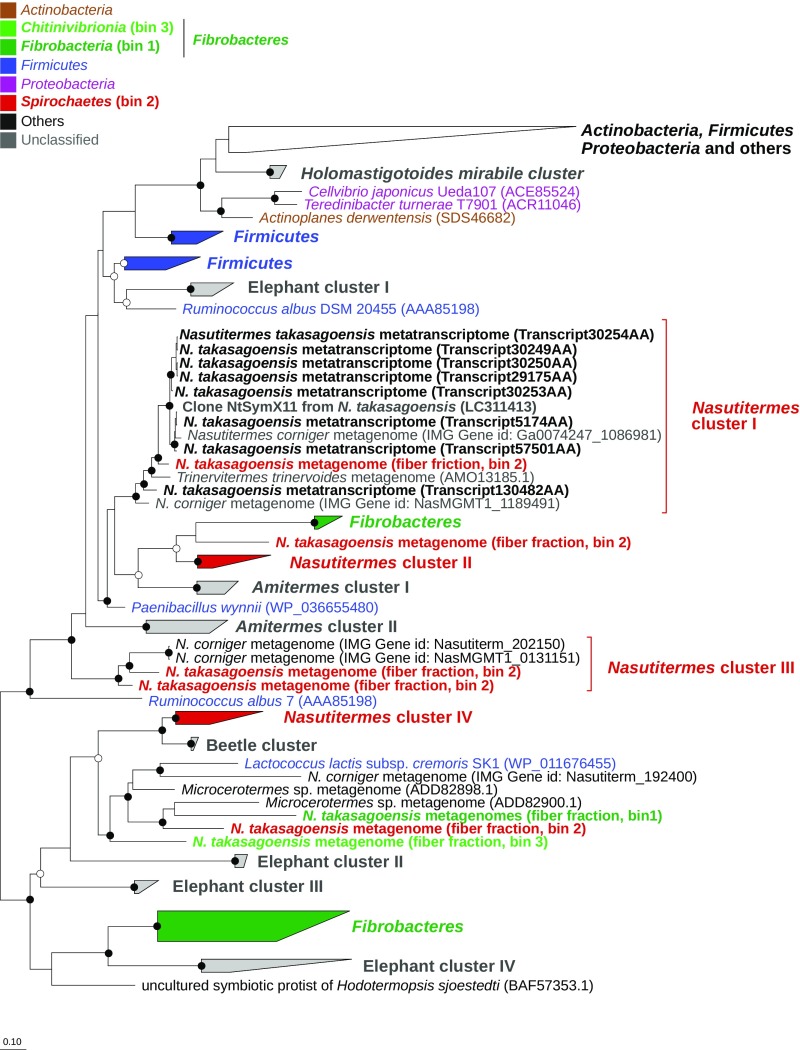
Phylogenetic analysis revealed that the GH11 homologs identified in the fiber-associated community of N. takasagoensis are very closely related to those of metagenomes of other Nasutitermes species. Together, they form four well-supported clusters (Nasutitermes clusters I–IV; Fig. 5). Nasutitermes cluster I comprises the eight predominantly expressed GH11 transcripts in the fiber fraction of N. takasagoensis and also several metagenomic sequences from the fiber fraction that were taxonomically assigned to Spirochaetes (bin 2) based on binning analysis. Nasutitermes cluster I also included sequences from previously published metagenomic analyses of other higher termite species (N. corniger and Trinervitermes trinervoides). It formed a larger monophyletic clade with another cluster of GH11 homologs from the fiber fraction of N. takasagoensis (Nasutitermes cluster II) and previously published sequences from metagenomes of A. wheeleri (12) and metagenome-assembled genomes of Fibrobacteres [from feces of herbivorous mammals (35)], with a GH11 homolog of a cultivated firmicute, Paenibacillus wynnii, in the most basal position. These sequences comprised a sister group with closely related GH11 homologs of other Firmicutes.
Fig. 5.
Phylogenetic relationship among bacterial GH11 members and phylum-level classification of homologs from fiber-associated bacteria in the hindgut of N. takasagoensis. The unrooted tree is based on 164 alignment positions and represents a consensus phylogeny obtained by using maximum-likelihood (ML) and Bayesian (BA) inference and the WAG model of protein evolution (60). Sequences obtained in this study are shown in bold, and text color indicates the taxonomic affiliation of homologs obtained from bacterial genomes and compositional bins. Node support was determined with the χ2 approximate likelihood ratio test (i.e., ML) and posterior probability (i.e., BA); confidence values are indicated by circles (open, ≥90% support in at least one method; filled, ≥90% support in both methods). The detailed tree, including all accession numbers and confidence values, is provided in SI Appendix, Fig. S5.
The remaining metagenomic sequences of spirochetal origin (bin 2) from N. takasagoensis fell into Nasutitermes clusters III and IV. Nasutitermes cluster III also included metagenomic sequences from N. corniger (12), again with a homolog of a cultivated member of the Firmicutes, Ruminococcus albus, in an ancestral position, which suggests multiple cross-phylum transfers of a GH11 xylanase gene from a firmicute to a spirochete. Nasutitermes cluster IV included previously published sequences from other Nasutitermitinae and from a moth [which had been obtained in the same study (36)]. They were most closely related to GH11 homologs from the gut of a beetle larva and loosely embedded in a larger clade of homologs from the gut metagenomes of various higher termites, which included sequences from the Fibrobacteres and Chitinivibrionia bins (bins 1 and 3) of the fiber fraction of N. takasagoensis. The only GH11 homolog in Nasutitermes cluster IV derived from a cultivated representative is again from a firmicute, Lactococcus lactis cremoris, but the ancestral positions were held by GH11 xylanases from the genome of Fibrobacter succinogenes and from metagenome-assembled genomes of various uncultivated members of Fibrobacteres in the gut of mammals (35).
Discussion
Xylans and other hemicelluloses are a major dietary component of the food of herbivorous animals. Most animals do not produce endogenous enzymes for the depolymerization of hemicelluloses but digest them with the help of their gut microbiota. In the bovine rumen and in the human colon, members of the phylum Bacteroidetes play a crucial role in this process (37), mainly through their GH10 endoxylanases (38). By contrast, the results of the present study show that, in the guts of higher termites, members of the phylum Spirochaetes are the main players in this process. This is remarkable for several reasons: (i) a function of Spirochaetes in xylan degradation is so far undocumented to our knowledge, (ii) the lineages responsible for the activity belong to a monophyletic clade that is specific for termites and that have coevolved with their termite hosts, and (iii) the GH11 xylanase genes of termite gut spirochetes were most likely acquired by horizontal gene transfer from other gut bacteria.
Although spirochetes are best known as human pathogens (39), they are also important pectin degraders in ruminants (40, 41) and play a key role in the production of acetate from H2 and CO2 in termite guts (17–19). An involvement of spirochetes in hemicellulose degradation has not been reported to our knowledge, and the genomes of termite gut treponemes isolated from lower termites (Treponema Ia) do not encode any xylanases of the GH11 family. Thus, these findings significantly advance our understanding of lignocellulose digestion in higher termites and shed light on the evolutionary history of termite gut spirochetes.
In lower termites, xylanase activity is localized predominantly in the hindgut (42), where it is most likely associated with symbiotic protists (25). The present study demonstrates that xylanase activity is also restricted almost exclusively to the hindgut in wood-feeding higher termites. Because higher termites lack such protists, the role of xylan (and cellulose) degradation has shifted to the symbiotic bacteria colonizing this gut compartment. Xylanase activity in the hindgut exceeds cellulase activity against CMC and microcrystalline cellulose, which is in agreement with the predominance of putative xylanases among the GH genes expressed by the fiber-associated community and the greater solubility of this substrate.
Previous metagenomic analyses identified 45–53 families of GH genes in the bacterial community in the luminal fluid of the hindgut paunch (P3 compartment) of Nasutitermes spp. (10, 12). Our transcriptomic analysis of N. takasagoensis detected transcripts of 62 GH families alone in the fiber fraction, which indicates that the GH repertoire of the fiber-associated bacterial community was comprehensively sampled. In all metatranscriptomic libraries, this enormous diversity was dominated primarily by members of family GH11, followed by members of family GH5. The GH11 family consists exclusively of xylanases that preferentially hydrolyze xylanosic bonds of heteroxylans, such as glucuronoxylans and arabinoxylans (43). The substrate specificities in the GH5 family are more diverse, but the majority of the transcripts were affiliated with GH5 subfamily 2, which consists predominantly of endo-β-1,4-glucanases (29). By contrast, other highly expressed GHs belonged to families GH130 and GH30 and GH5 subfamily 4, which again comprise various hemicellulases (44, 45). This result underscores that the primary role of the fiber-associated bacterial community is in the breakdown of hemicelluloses, especially the xylan backbone of wood, which link the cellulose fibrils to the lignin fraction and thus impede the access of endoglucanases and other cellulolytic enzymes to their substrate.
Although a function of spirochetes in xylan degradation has never been documented before to our awareness, earlier studies provided hints that xylanase genes found in the gut of higher termites are produced by spirochetes. An endoxylanase gene of family GH11 had been detected in bacterial DNA from a Nasutitermes sp. almost 15 y ago (35), but its origin remained obscure. Later, 4 of 14 GH11 xylanase genes recovered from the luminal content of the P3 compartment of a different Nasutitermes species had been assigned to termite gut treponemes based on phylogenetic binning (10), but neither their expression levels nor xylanase activities in the hindgut compartment were investigated. Our metatranscriptomic and enzymatic results indisputably corroborate the spirochetal origin of GH11 xylanases in N. takasagoensis. Although phylogenetic analysis underscores the close relationship of the GH11 xylanases to homologs in Firmicutes, the reference-independent assignment of the respective contigs, based on their genomic signatures, and the genomic neighborhood of the most highly expressed genes indicate that they are encoded by spirochetes. A misassembly of DNA fragments originating from Firmicutes with spirochete DNA in this and probably also previous studies is further excluded by our localization of the recombinant xylanase on the surface of fiber-associated helical cells by immunofluorescence microscopy.
The evolutionary origin of GH11 xylanases in termite gut spirochetes remains obscure. The genomes of Treponema spp. isolated from lower termites, which belong to the basal Treponema Ia subclade (46), do not encode homologs of this enzyme family. The GH11 xylanases in Nasutitermes cluster I, which encode the most highly expressed transcripts, and their homologs in the closely related Nasutitermes clusters II and III, are phylogenetically situated among xylanase gene sequences of Firmicutes. They represent separate lines of descent, with homologs from P. wynnii and R. albus as closest, albeit distant, relatives. By contrast, the GH11 xylanases in Nasutitermes cluster IV are embedded in a clade that is dominated by homologs of Fibrobacteres, including sequences from the corresponding metagenomic bins (bins 1 and 3) of N. takasagoensis and from the intestinal tracts of mammals (35, 47). In both cases, the most likely explanation for this scenario would be a transfer of the genes encoding GH11 xylanases from Firmicutes or Fibrobacteres to termite gut treponemes. In the radiation of Nasutitermes clusters I and II, the conspicuous presence of GH11 homologs of Fibrobacteres from mammalian guts [recovered from metagenome-assembled genomes (35)] suggests that such horizontal transfers of xylanase genes has also occurred in the opposite direction.
A horizontal transfer of the xylanase genes from Firmicutes to Spirochaetes (Nasutitermes cluster I) is supported by the results of metagenomic binning, which assigned the xylan-binding CBM36 modules associated with the corresponding GH11 xylanase genes or transcripts exclusively to Spirochaetes (bin 2). Most of the bacterial CBM36 modules in the CAZy database (79 of 89) originate from the genomes of Firmicutes. The remaining ones are from Bacteroidetes, Proteobacteria, Dictyoglomi, or environmental samples, but are not encountered among the genomes of Fibrobacteres or the corresponding metagenomic bins (1, 3). Together with the absence of GH11 and CBM36 homologs from all spirochetal genomes sequenced to date, including the Treponema I isolates in lower termites, these results suggest that the fiber-associated lineages in the gut of higher termites acquired the capacity for the hydrolysis of xylan by horizontal gene transfer.
It has been proposed that the ancestral loss of flagellates in higher termites was connected with a dietary shift from sound wood to fungus-infested or humified wood and plant litter, and that the return to a wood-feeding lifestyle occurred independently in several subfamilies (48). In the case of Nasutitermitinae, this change in lifestyle might be associated with the acquisition of GH11 xylanases by an ancestor of the Treponema Ic and If subclades. This would explain the presence of a GH11 homolog of Nasutitermes cluster I also in the hindgut of the grass-feeding T. trinervoides (14), which represents a sister group of wood-feeding Nasutitermes spp. and harbors members of Treponema Ic (46). The organismal origin of the GH11 homologs from the gut microbiota of Termitinae is more speculative. In the case of the dung-feeding A. wheeleri (12), they may derive from the clostridial lineages that are abundant in the gut of this termite, but, at least in the case of the Amitermes cluster I (a sister group of Nasutitermes cluster II), an ancestral transfer to termite gut treponemes also cannot be excluded. In the case of the wood-feeding Microcerotermes spp. (11, 35), whose gut microbiota encodes GH11 xylanases that are phylogenetically placed among homologs recovered from the metagenomic bins of fiber-associated Spirochaetes and Fibrobacteres from N. takasagoensis, the organismal origin remains to be determined.
Although the expression level of GH11 xylanases in the fiber fraction of the P3 compartment of N. takasagoensis was slightly higher than that of GHs of GH5 (as shown in the present study), a previous meta-analysis of the entire luminal contents of the hindgut paunch in N. corniger demonstrated that transcripts of family GH5 were considerably more abundant than those of family GH11 (12). It is not clear to which extent this reflects the interspecific differences between the microbiota of N. takasagoensis and N. corniger (12) or the composition of the total and fiber-associated bacterial community in the P3 compartment, which have been documented for both species (8). Moreover, it must be taken into account that the proportion of xylanase and cellulase activities in P3 compartment and fiber fraction will depend on the relative abundance of endoglucanases and hemicellulases in the transcripts of GH5. Also, the absolute activities of cellulases (determined with microcrystalline cellulose) and xylanases (determined with much more soluble xylan) have to be taken with caution, as indicated by the considerably higher cellulase activities obtained with soluble CMC (23).
In view of the large proportion of putative cellulases (e.g., GH5_2, GH8, GH45) in the metatranscriptomic libraries (SI Appendix, Fig. S1 and Dataset S1), the low cellulase activity obtained with the particulate fraction might be explained by a less efficient recovery of the cellulases of Fibrobacteres and other fiber-associated microbiota, which are most probably localized in the glycocalyx (49) or outer membrane vesicles (50). Therefore, the astonishingly high cellulase activities in the soluble fraction of the hindgut paunch may be artificial (Fig. 1).
Finally, it is important to consider that the enzyme activities in particulate and soluble fractions represent the GHs that are bound to microbial cells or released into the luminal fluid, whereas the metagenomic and metatranscriptomic analyses used only the DNA or RNA of cells obtained by the density-dependent enrichment of wood fibers. The fiber-associated Spirochaetes in the P3 compartment of Nasutitermes spp. are—in contrast to the firmly attached Fibrobacteres—also abundantly present in the fiber-free fraction (8). Considering that the most highly expressed GH11 genes from spirochetes encode CBMs but no protein structures known to anchor GHs to the cell surface, it is quite likely that at least some of xylanase activity in the soluble fraction derives from fiber-associated spirochetes (Fig. 1).
As in the rumen, where efficient degradation of lignocellulose involves colonization of dietary fibers by bacteria that hydrolyze structural polysaccharides to form soluble cellodextrins that are subsequently also utilized by the unattached bacterial populations (49, 51), the soluble sugars released by the hydrolytic activity of the fiber-associated community should serve at least in part also as carbon and energy sources of other, unattached populations. However, the average size of the wood particles in the hindgut of higher termites is considerably smaller (25 μm) than that of the forage in the rumen (200 µm) (52), and thereby creates a much larger surface area for bacterial colonization.
The dietary diversification of higher termites is considered a key element of their ecological and evolutionary success. In wood-feeding lineages, this involved the reversal from a detritivorous to a xylophagous lifestyle, although the trigger of this evolutionary transition of feeding habits remains elusive. Based on the correlation between diet and bacterial community structure in the hindguts of higher termites (53), the present study strongly suggests that the horizontal acquisition of novel digestive capacities by resident gut bacteria provided the termite host with the flexibility to exploit additional feeding substrates and eventually resulted in compositional changes in the entire hindgut microbiota.
Experimental Procedures
Colonies of N. takasagoensis were collected on Iriomote Island in Okinawa prefecture, Japan, and maintained as previously described (54, 55). Worker-caste termites were used for all experiments. Endo-β-1,4-xylanase (EC 3.2.1.8), endo-β-1,4-glucanase (EC 3.2.1.4), and hydrolytic activities acting on microcrystalline cellulose in salivary glands and gut sections (pooled preparations from 10 individuals each) were measured as previously described (21, 22, 25).
For metatranscriptomic analysis, fiber-associated bacteria were prepared by density-gradient centrifugation as previously described (8) and transferred to RNAprotect bacteria reagent (Qiagen) to stabilize RNA. Total RNA was extracted with the RNeasy Mini Kit (Qiagen) and shipped to BGI for library preparations and sequencing (Illumina HiSeq 2000). We annotated amino acid sequences provided by BGI by using dbCAN (26) to detect CAZys and relevant modules. Furthermore, we annotated nucleotide sequences of all contigs by using dbCAN2 (27) to find transcripts and genes that were missing in the dataset provided by BGI. Expression profiles of CAZy were analyzed by using a dataset for all expressed genes provided by BGI. For enzyme profiles, an amplified fragment of GH11 xylanase gene (NtSymX11) without a putative leader sequence was ligated with a pQE-1 expression vector and introduced into E. coli strain JM109. The resulting His-tagged enzyme was partially purified with a Ni-NTA spin column and examined by SDS/PAGE.
Monoclonal antibodies were raised against the bacteria in the fiber fraction following the method previously described (56). Antibodies that cross-reacted with the recombinant xylanase NtSymX11 were selected by Western blotting. For indirect immunofluorescence microscopy, the entire P3 fluid was mixed with the selected monoclonal antibody, followed by incubation with Alexa Fluor 488-labeled anti-mouse IgG, and then with DAPI to visualize DNA. Specimens were observed under a fluorescence microscope (BX41; Olympus) or a confocal laser microscope (C2; Nikon).
For metagenomic analysis, DNA was extracted from fiber-associated bacteria and purified by using the ISOPLANTII purification kit (Nippon Gene). Again, library preparation, sequencing, and assembly were done by BGI. We analyzed the contigs/scaffolds provided by BGI by using local BLAST search and Genetyx software (Genetyx) and compositional binning with Vizbin (57). Phylogenetic trees included an alignment of all publicly accessible GH11 sequences using maximum likelihood (phyml version 3.0.1; ref. 58) and Bayesian analysis (MrBayes version 3.2.1; ref. 59). Full details of the experimental procedures are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Karen A. Brune for linguistic improvements in the manuscript and Aki Kinjo for technical assistance. This study was supported by a research grant from the Institute for Fermentation, Osaka, Japan; by Grants-in-Aid for Scientific Research KAKENHI 26292177, 15K14900, and 17H01510 from the Japan Society for the Promotion of Science (JSPS); by University of the Ryukyus Research Incentive Grant 18SP03104; by FY2014 Bilateral Joint Research Program A/14/01075 between Germany (Deutscher Akademischer Austauschdienst) and Japan (JSPS); and by Deutsche Forschungsgemeinschaft in Collaborative Research Center SFB 987 (Microbial Diversity in Environmental Signal Response).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the sequence read archive of the DNA Data Bank Japan (DDBJ) under accession nos. DRA005983 (metatranscriptomes) and DRA005967 (metagenome). The complete sequence of the cloned GH11 gene (NtSymX11) was deposited in GenBank/European Nucleotide Archive/DDBJ database under accession no. LC311413.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810550115/-/DCSupplemental.
References
- 1.Cornwell WK, et al. Plant traits and wood fates across the globe: Rotted, burned, or consumed? Glob Change Biol. 2009;15:2431–2449. [Google Scholar]
- 2.Watanabe H, Tokuda G. Animal cellulases. Cell Mol Life Sci. 2001;58:1167–1178. doi: 10.1007/PL00000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ni J, Tokuda G. Lignocellulose-degrading enzymes from termites and their symbiotic microbiota. Biotechnol Adv. 2013;31:838–850. doi: 10.1016/j.biotechadv.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Lo N, Eggleton P. Termite phylogenetics and co-cladogenesis with symbionts. In: Bignell DE, Roisin Y, Lo N, editors. Biology of Termites: A Modern Synthesis. Springer; Dordrecht, The Netherlands: 2011. pp. 27–50. [Google Scholar]
- 5.Brune A. Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol. 2014;12:168–180. doi: 10.1038/nrmicro3182. [DOI] [PubMed] [Google Scholar]
- 6.Köhler T, Dietrich C, Scheffrahn RH, Brune A. High-resolution analysis of gut environment and bacterial microbiota reveals functional compartmentation of the gut in wood-feeding higher termites (Nasutitermes spp.) Appl Environ Microbiol. 2012;78:4691–4701. doi: 10.1128/AEM.00683-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikaelyan A, Meuser K, Brune A. Microenvironmental heterogeneity of gut compartments drives bacterial community structure in wood- and humus-feeding higher termites. FEMS Microbiol Ecol. 2017;93:fiw210. doi: 10.1093/femsec/fiw210. [DOI] [PubMed] [Google Scholar]
- 8.Mikaelyan A, Strassert JFH, Tokuda G, Brune A. The fiber-associated cellulolytic bacterial community in the hindgut of wood-feeding higher termites (Nasutitermes spp.) Environ Microbiol. 2014;16:2711–2722. [Google Scholar]
- 9.Abdul Rahman N, et al. A phylogenomic analysis of the bacterial phylum Fibrobacteres. Front Microbiol. 2016;6:1469. doi: 10.3389/fmicb.2015.01469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warnecke F, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 11.Nimchua T, Thongaram T, Uengwetwanit T, Pongpattanakitshote S, Eurwilaichitr L. Metagenomic analysis of novel lignocellulose-degrading enzymes from higher termite guts inhabiting microbes. J Microbiol Biotechnol. 2012;22:462–469. doi: 10.4014/jmb.1108.08037. [DOI] [PubMed] [Google Scholar]
- 12.He S, et al. Comparative metagenomic and metatranscriptomic analysis of hindgut paunch microbiota in wood- and dung-feeding higher termites. PLoS One. 2013;8:e61126. doi: 10.1371/journal.pone.0061126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poulsen M, et al. Complementary symbiont contributions to plant decomposition in a fungus-farming termite. Proc Natl Acad Sci USA. 2014;111:14500–14505. doi: 10.1073/pnas.1319718111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rashamuse K, et al. Metagenomic mining of glycoside hydrolases from the hindgut bacterial symbionts of a termite (Trinervitermes trinervoides) and the characterization of a multimodular β-1,4-xylanase (GH11) Biotechnol Appl Biochem. 2017;64:174–186. doi: 10.1002/bab.1480. [DOI] [PubMed] [Google Scholar]
- 15.Liu N, et al. Functional metagenomics reveals abundant polysaccharide-degrading gene clusters and cellobiose utilization pathways within gut microbiota of a wood-feeding higher termite. ISME J. August 16, 2018 doi: 10.1038/s41396-018-0255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albersheim P, Darvill A, Roberts K, Sederoff R, Staehelin A. Plant Cell Walls. Garland Science; New York: 2011. [Google Scholar]
- 17.Leadbetter JR, Schmidt TM, Graber JR, Breznak JA. Acetogenesis from H2 plus CO2 by spirochetes from termite guts. Science. 1999;283:686–689. doi: 10.1126/science.283.5402.686. [DOI] [PubMed] [Google Scholar]
- 18.Rosenthal AZ, Matson EG, Eldar A, Leadbetter JR. RNA-seq reveals cooperative metabolic interactions between two termite-gut spirochete species in co-culture. ISME J. 2011;5:1133–1142. doi: 10.1038/ismej.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohkuma M, et al. Acetogenesis from H2 plus CO2 and nitrogen fixation by an endosymbiotic spirochete of a termite-gut cellulolytic protist. Proc Natl Acad Sci USA. 2015;112:10224–10230. doi: 10.1073/pnas.1423979112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tokuda G, et al. Cellulolytic environment in the midgut of the wood-feeding higher termite Nasutitermes takasagoensis. J Insect Physiol. 2012;58:147–154. doi: 10.1016/j.jinsphys.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Tokuda G, Watanabe H. Hidden cellulases in termites: Revision of an old hypothesis. Biol Lett. 2007;3:336–339. doi: 10.1098/rsbl.2007.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tokuda G, et al. Major alteration of the expression site of endogenous cellulases in members of an apical termite lineage. Mol Ecol. 2004;13:3219–3228. doi: 10.1111/j.1365-294X.2004.02276.x. [DOI] [PubMed] [Google Scholar]
- 23.Tokuda G, Lo N, Watanabe H. Marked variations in patterns of cellulase activity against crystalline- vs. carboxymethyl-cellulose in the digestive systems of diverse, wood-feeding termites. Physiol Entomol. 2005;30:372–380. [Google Scholar]
- 24.Slaytor M, Sugimoto A, Azuma J, Murashima K, Inoue T. Cellulose and xylan utilisation in the lower termite Reticulitermes speratus. J Insect Physiol. 1997;43:235–242. doi: 10.1016/s0022-1910(96)00097-2. [DOI] [PubMed] [Google Scholar]
- 25.Arakawa G, Watanabe H, Yamasaki H, Maekawa H, Tokuda G. Purification and molecular cloning of xylanases from the wood-feeding termite, Coptotermes formosanus Shiraki. Biosci Biotechnol Biochem. 2009;73:710–718. doi: 10.1271/bbb.80788. [DOI] [PubMed] [Google Scholar]
- 26.Yin Y, et al. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, et al. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Book AJ, et al. Evolution of substrate specificity in bacterial AA10 lytic polysaccharide monooxygenases. Biotechnol Biofuels. 2014;7:109. doi: 10.1186/1754-6834-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aspeborg H, Coutinho PM, Wang Y, Brumer H, 3rd, Henrissat B. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5) BMC Evol Biol. 2012;12:186. doi: 10.1186/1471-2148-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen H. Predicting secretory proteins with SignalP. Methods Mol Biol. 2017;1611:59–73. doi: 10.1007/978-1-4939-7015-5_6. [DOI] [PubMed] [Google Scholar]
- 31.Hongoh Y, et al. Phylogenetic diversity, localization, and cell morphologies of members of the candidate phylum TG3 and a subphylum in the phylum Fibrobacteres, recently discovered bacterial groups dominant in termite guts. Appl Environ Microbiol. 2006;72:6780–6788. doi: 10.1128/AEM.00891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hongoh Y. Who digests the lignocellulose? Environ Microbiol. 2014;16:2644–2645. doi: 10.1111/1462-2920.12449. [DOI] [PubMed] [Google Scholar]
- 33.Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorokin DY, et al. Genome analysis of Chitinivibrio alkaliphilus gen. nov., sp. nov., a novel extremely haloalkaliphilic anaerobic chitinolytic bacterium from the candidate phylum Termite Group 3. Environ Microbiol. 2014;16:1549–1565. doi: 10.1111/1462-2920.12284. [DOI] [PubMed] [Google Scholar]
- 35.Parks DH, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 36.Brennan Y, et al. Unusual microbial xylanases from insect guts. Appl Environ Microbiol. 2004;70:3609–3617. doi: 10.1128/AEM.70.6.3609-3617.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dodd D, Mackie RI, Cann IKO. Xylan degradation, a metabolic property shared by rumen and human colonic Bacteroidetes. Mol Microbiol. 2011;79:292–304. doi: 10.1111/j.1365-2958.2010.07473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang M, et al. Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes. Proc Natl Acad Sci USA. 2014;111:E3708–E3717. doi: 10.1073/pnas.1406156111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta RS, Mahmood S, Adeolu M. A phylogenomic and molecular signature based approach for characterization of the phylum Spirochaetes and its major clades: Proposal for a taxonomic revision of the phylum. Front Microbiol. 2013;4:217. doi: 10.3389/fmicb.2013.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, et al. Monitoring the rumen pectinolytic bacteria Treponema saccharophilum using real-time PCR. FEMS Microbiol Ecol. 2014;87:576–585. doi: 10.1111/1574-6941.12246. [DOI] [PubMed] [Google Scholar]
- 41.Svartström O, et al. Ninety-nine de novo assembled genomes from the moose (Alces alces) rumen microbiome provide new insights into microbial plant biomass degradation. ISME J. 2017;11:2538–2551. doi: 10.1038/ismej.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slaytor M. Energy metabolism in the termite and its gut microbiota. In: Abe T, Bignell DE, Higashi M, editors. Termites: Evolution, Sociality, Symbioses, Ecology. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2000. pp. 307–332. [Google Scholar]
- 43.Paës G, Berrin JG, Beaugrand J. GH11 xylanases: Structure/function/properties relationships and applications. Biotechnol Adv. 2012;30:564–592. doi: 10.1016/j.biotechadv.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Cuskin F, et al. The GH130 family of mannoside phosphorylases contains glycoside hydrolases that target β-1,2-mannosidic linkages in Candida mannan. J Biol Chem. 2015;290:25023–25033. doi: 10.1074/jbc.M115.681460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valenzuela SV, Diaz P, Pastor FIJ. Modular glucuronoxylan-specific xylanase with a family CBM35 carbohydrate-binding module. Appl Environ Microbiol. 2012;78:3923–3931. doi: 10.1128/AEM.07932-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mikaelyan A, et al. Classifying the bacterial gut microbiota of termites and cockroaches: A curated phylogenetic reference database (DictDb) Syst Appl Microbiol. 2015;38:472–482. doi: 10.1016/j.syapm.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 47.Suen G, et al. The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist. PLoS One. 2011;6:e18814. doi: 10.1371/journal.pone.0018814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donovan SE, Eggleton P, Bignell DE. Gut content analysis and a new feeding group classification of termites. Ecol Entomol. 2001;26:356–366. [Google Scholar]
- 49.Weimer PJ. Why don’t ruminal bacteria digest cellulose faster? J Dairy Sci. 1996;79:1496–1502. doi: 10.3168/jds.S0022-0302(96)76509-8. [DOI] [PubMed] [Google Scholar]
- 50.Arntzen MØ, Várnai A, Mackie RI, Eijsink VGH, Pope PB. Outer membrane vesicles from Fibrobacter succinogenes S85 contain an array of carbohydrate-active enzymes with versatile polysaccharide-degrading capacity. Environ Microbiol. 2017;19:2701–2714. doi: 10.1111/1462-2920.13770. [DOI] [PubMed] [Google Scholar]
- 51.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martz FA, Belyea RL. Role of particle size and forage quality in digestion and passage by cattle and sheep. J Dairy Sci. 1986;69:1996–2008. doi: 10.3168/jds.S0022-0302(86)80626-9. [DOI] [PubMed] [Google Scholar]
- 53.Mikaelyan A, et al. Diet is the primary determinant of bacterial community structure in the guts of higher termites. Mol Ecol. 2015;24:5284–5295. doi: 10.1111/mec.13376. [DOI] [PubMed] [Google Scholar]
- 54.Tokuda G, Watanabe H, Matsumoto T, Noda H. Cellulose digestion in the wood-eating higher termite, Nasutitermes takasagoensis (Shiraki): Distribution of cellulases and properties of endo-β-1,4-glucanase. Zool Sci. 1997;14:83–93. doi: 10.2108/zsj.14.83. [DOI] [PubMed] [Google Scholar]
- 55.Tokuda G, Miyagi M, Makiya H, Watanabe H, Arakawa G. Digestive β-glucosidases from the wood-feeding higher termite, Nasutitermes takasagoensis: Intestinal distribution, molecular characterization, and alteration in sites of expression. Insect Biochem Mol Biol. 2009;39:931–937. doi: 10.1016/j.ibmb.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 56.Iwatani K, et al. Translocation of an 89-kDa periplasmic protein is associated with Holospora infection. Biochem Biophys Res Commun. 2005;337:1198–1205. doi: 10.1016/j.bbrc.2005.09.175. [DOI] [PubMed] [Google Scholar]
- 57.Laczny CC, et al. VizBin–An application for reference-independent visualization and human-augmented binning of metagenomic data. Microbiome. 2015;3:1. doi: 10.1186/s40168-014-0066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 59.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.