Significance
X-chromosome inactivation (XCI) is a female dosage compensation mechanism where one of the two X chromosomes is randomly silenced. However, some genes on the inactive X chromosome and outside the pseudoautosomal regions escape from XCI and are expressed from both alleles (escapees). By means of single-cell transcriptome we show that XCI is variable among individuals and from cells from the same individual. We discovered that XCI is indeed a cellular phenotype and that the expression of escapees is regulated by cell-cycle phases and by the expression of XIST. Additionally, in this context, we identified a list of undescribed escapee genes, and we validate many already suggested escapee genes with a direct measurement on single cells.
Keywords: single-cell transcriptomics, X inactivation, XIST, cell cycle, cell heterogeneity
Abstract
X-chromosome inactivation (XCI) provides a dosage compensation mechanism where, in each female cell, one of the two X chromosomes is randomly silenced. However, some genes on the inactive X chromosome and outside the pseudoautosomal regions escape from XCI and are expressed from both alleles (escapees). We investigated XCI at single-cell resolution combining deep single-cell RNA sequencing with whole-genome sequencing to examine allelic-specific expression in 935 primary fibroblast and 48 lymphoblastoid single cells from five female individuals. In this framework we integrated an original method to identify and exclude doublets of cells. In fibroblast cells, we have identified 55 genes as escapees including five undescribed escapee genes. Moreover, we observed that all genes exhibit a variable propensity to escape XCI in each cell and cell type and that each cell displays a distinct expression profile of the escapee genes. A metric, the Inactivation Score—defined as the mean of the allelic expression profiles of the escapees per cell—enables us to discover a heterogeneous and continuous degree of cellular XCI with extremes represented by “inactive” cells, i.e., cells exclusively expressing the escaping genes from the active X chromosome and “escaping” cells expressing the escapees from both alleles. We found that this effect is associated with cell-cycle phases and, independently, with the XIST expression level, which is higher in the quiescent phase (G0). Single-cell allele-specific expression is a powerful tool to identify novel escapees in different tissues and provide evidence of an unexpected cellular heterogeneity of XCI.
In eutherian mammals, X-chromosome inactivation (XCI) is a well-described mechanism of dosage compensation for the X chromosome in females (1–4). In female cells, only one X chromosome is transcribed [X-active (Xa)], whereas the second X chromosome is silenced [X-inactive (Xi)] (1). Marsupials have an imprinted pattern of XCI, and the paternal allele is predominantly inactive (5). In mice, an imprinted form of XCI occurs through early embryonic developmental stages (four- to eight-cell stage) (6–9), followed by inner-cell mass reactivation and random XCI in epiblast cells (10). In humans, the two X chromosomes are active during postzygotic stages, achieve gene dosage compensation by dampening their expression up to or even after late blastocyst formation, until one of the X chromosomes is randomly inactivated in each cell (11). In female somatic cells, random XCI is stable, resulting in a mosaicism for gene expression on the X chromosome, in which an average of 50% of cells express the active paternal X and 50% the active maternal X alleles.
Most of the genes on the Xi chromosome are transcriptionally repressed through multiple gene-silencing mechanisms that are still poorly understood (12). The process is initiated by the X-Inactivation Center (XIC) and spread along the Xi chromosome during early embryogenesis (1). The allele-specific expression of XIST, a long noncoding RNA (ncRNA) located in the XIC, is essential for mediating the establishment and maintenance of XCI in subsequent cycles of mitotic division by coating the Xi chromosome (13–16). The recruitment of various proteins [“Split-ends,” lamin B receptor, Wilms tumor 1-associated protein/Rbm15/Rbm15B, Polycomb repressive complex 1, and heterogeneous nuclear ribonucleoprotein U, SMCHD1 (17)] are important players in the initiation phase of XCI (12). Gene silencing is also mediated through Xi by nuclear and genomic reorganization with the occurrence of chromatin repressive modifications; all are important hallmarks of XCI propagation (18). Chromatin modifications include (i) depletion of RNA polymerase II, (ii) enrichment in histone repressive marks such as H3K9me and H4K20me3, recruitment of Polycomb remodeler with the marks, H3K27me3 and H2AK119ub1, macroH2A deposition, (iii) DNA hypermethylation at cpG islands on Xi, (iv) reshaping of the Xi with loss of topologically associating domains (TAD) structure, and (v) subnuclear spatial reorganization. Stable Xi exhibits features of constitutive heterochromatin, ensuring the persistance of the XIST-mediated transcriptional gene silencing. Mechanisms and protein factors implicated in this maintenance phase remain elusive.
However, not all X-linked genes are inactivated. In females, genes that escape from XCI (escapees) represent 15–25% of the X-linked genes, and a further 10% of escapees differ between individuals and cell types (19–22). Such genes have been associated with sex-specific traits and with clinical abnormalities in patients with X-chromosome aneuploidy, such as Turner and Klinefelter syndromes (23). Pathogenic variants in escapees also contribute to various disease phenotypes in women carriers, including Kabuki syndrome [KABUK1 (OMIM 147920) Online Mendelian Inheritance in Man] (24, 25), and to intellectual disabilities (26–31). Genes escaping XCI have been previously identified by whole-tissue studies using different approaches, such as X-linked gene expression comparisons between males and females (32), detecting allelic imbalance in clonal lymphoblast and fibroblast cell lines (19), identifying inactivated and active transcription start sites by methylation profiles (33), and among female individuals with X-chromosome aneuploidies (34). The ability to capture single cells and to study their allele-specific expression (ASE) (35) provides the opportunity to explore XCI patterns at the single-cell level and to identify escapee genes. Recent studies on mouse single cells have demonstrated the robust nature of this technology to monitor the dynamics of XCI through differentiation (36), mouse preimplantation female embryos (37) and in clonal somatic cells (38). Recently, Tukiainen et al. (39) performed an across-tissue study of X inactivation and partially validated their observation by performing shallow sequencing [1 million (Mio) reads per cell] on 940 single cells from lymphoblasts and dendritic cells. Here, using RNA-Seq at high sequencing depth (40 Mio reads per cell), we studied the X-linked ASE in 983 isolated, unsynchronized single fibroblast and lymphoblast cells and established the degree of XCI after the removal of potential confounding effects.
Results
Identification and Elimination of Confounding Doublets.
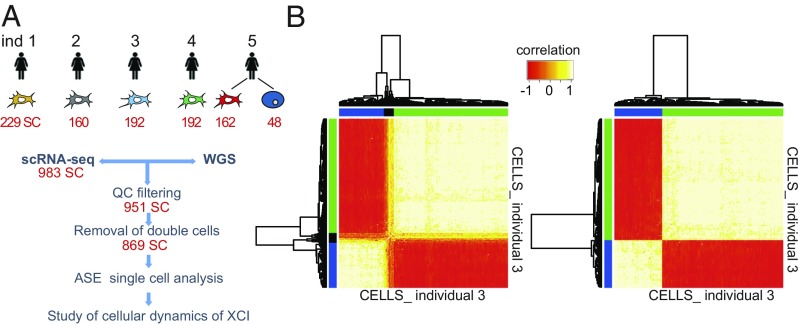
Genes located on the X chromosome of female cells express one allele from the randomly active chromosome, while escaping genes express both alleles (40) (SI Appendix, Fig. S1). Multiple cells (i.e., doublets) resulting from the simultaneous capture of two or more cells expressing discordant haplotypes obviously complicate the detection of escapee genes and potentially increase the number of false positives (41). After removing 32 cells because of low mapping quality (SI Appendix, Fig. S2), to eliminate all doublets with discordant haplotypes, we conducted a pairwise correlation analysis of X-linked ASE among all of the cells. We performed hierarchical clustering analysis and obtained three distinct clusters of cells (Fig. 1B and SI Appendix, Materials and Methods and Fig. S3). As expected, two clusters were populated by cells with one mutually inactivated X chromosome. The third cluster included cells with a biallelic expression pattern for all of the X chromosome genes, revealing the presence of doublets. Considering all five individuals (n = 983 cells), we identified a total of 82 doublets (∼8% of the total, consistent with Fluidigm manufacturer’s expectations), which were excluded from further analysis (Fig. 1B and SI Appendix, Fig. S3 and Table S1). Doublets expressing concordant haplotypes cannot be detected with this in silico approach; however, they do not inflate the number of false-positive escapees, and, consequently, they do not have an impact on escapee gene discovery and XCI analysis (see SI Appendix, Materials and Methods for details).
Fig. 1.
Identification and elimination of confounding doublets. (A) Flowchart of the study. Whole- genome sequencing for each individual identified the respective heterozygous sites. Single-cell RNA-seq provided RNA abundance for each single cell. (B) Heat map of unsupervised hierarchical clustering using cell–cell pairwise Pearson correlation coefficients of AR from single cells from individual 3 before (Left) and after (Right), excluding the doublets. The heat map separated the cells expressing one haplotype (blue and green cluster) from cells expressing two haplotypes (black cluster, doublets). Pearson correlations range from −1 (red) to 1 (white).
Identification of Escapee Genes.
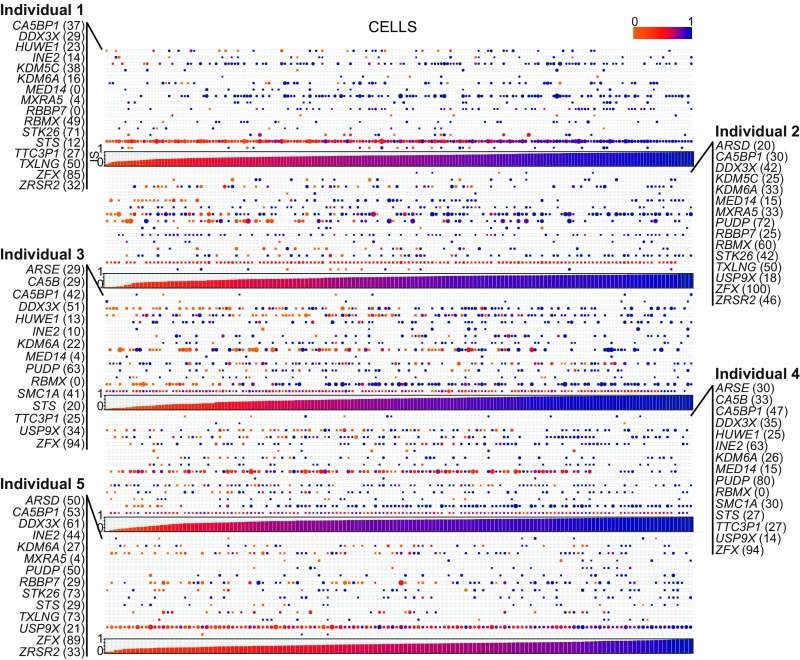
For each individual, we performed whole-genome sequencing to extract the informative heterozygous single-nucleotide variants (SNVs) and removed SNVs in repeated regions to avoid bias in ASE calculations (42). To estimate the allelic ratio (AR) for each gene in each cell, we calculated the ratio of the number of reads supporting the cell-specific expressed haplotypes over the total number of reads covering all SNVs of a gene (more details in SI Appendix, Materials and Methods). Fully inactivated genes displayed an AR equal to 1. In the relaxed discovery set of escapee genes, putative escapees were defined as having an AR ≤ 0.95 in at least one individual. The gene is considered as inactivated (i.e., exclusively expressed from the active chromosome) otherwise. As a proof of principle of the reliability of our approach, we first confirmed that XIST is expressed exclusively from the inactivated allele by analyzing its AR in all cells from individuals 3 and 4 (i.e., monozygotic twin samples), for which we were able to phase the haplotypes from parental genotyping (SI Appendix, Fig. S4). As an additional control, we examined the allele expression profile of genes in the PAR1 and PAR2 regions. As expected, all of these genes exhibited a balanced ASE across the two X chromosomes except VAMP7 in PAR2 where very few cells displayed an expression from the inactive chromosome (Fig. 2 and Dataset S1). The majority of chrX genes showed an inactivated status in all of the cells (SI Appendix, Fig. S5 and Dataset S1). From a total of 296 genes interrogated in at least one individual, we identified the relaxed set of 55 escapees (18.5%): 50 of them previously described to escape XCI in at least one study and five undescribed escapees [INE2 (antisense gene), STK26, UQCRBP1, LINC00630, and TTC3P1] (SI Appendix, Fig. S5). Of 203 genes with AR information from at least two individuals, we classified as robust escapees 22 genes (10.9%) that exhibit an escapee status in at least two individuals including three undescribed genes (INE2, STK26, and TTC3P1). As expected, the power to detect a gene escaping XCI is linearly related to the respective expression level (SI Appendix, Fig. S6). The number of overlapping escapees among all individuals is shown in a Venn diagram (SI Appendix, Fig. S7). Results from both relaxed and robust sets are consistent with the current understanding of XCI, in which it is predicted that 10–20% of X-chromosome genes escape XCI (20). In addition, we analyzed 48 single cells from a lymphoblastoid cell line derived from individual 5 to investigate the escapee concordance with the fibroblasts. After quality control and doublet removal, we were able to classify nine genes as escapee in lymphoblastoid cells with five of them being known escapees (DDX3X, KDM6A, MSL3, PUDP, ZFX), and four undescribed escapee genes (IDS, SLC9A7, STAG2, STK26). We observed that, although expressed and having an informative heterozygous site, the MSL3, IDS, SLC9A7, and STAG2 genes were not classified as escapees in fibroblasts. Conversely, several genes detected as escapees in fibroblasts were not escapees in lymphoblast cells. This could be partially ascribed to the previously observed tissue specificity of XCI (39, 43), to the recently discovered peculiar maintenance of XCI in lymphoblast cells (44), and to the relatively small number of lymphoblastoid single cells. Notably, the STK26 escapee status was clearly confirmed in both fibroblast and lymphoblastoid single cells.
Fig. 2.
Single-cell ASE profile of 22 robust escapee genes in human female fibroblasts. Composite figure of individual ARs per gene per cell (Top). AR profile of robust escapee genes (listed the rows) with a detectable expression in single cells (ordered along the columns) is shown. Every dot represents the AR of the respective gene in a cell. AR ranges from 0 (light orange) to 1 (dark blue). The size of the dot is proportional to the respective number of reads detected per cell. %, the percentage of cells where the respective gene is escaping XCI. (Bottom) Bar plot representing the IS (see text for details) per cell. IS ranges from 0 (light orange) to 1 (dark blue).
Heterogeneity of Escaping XCI.
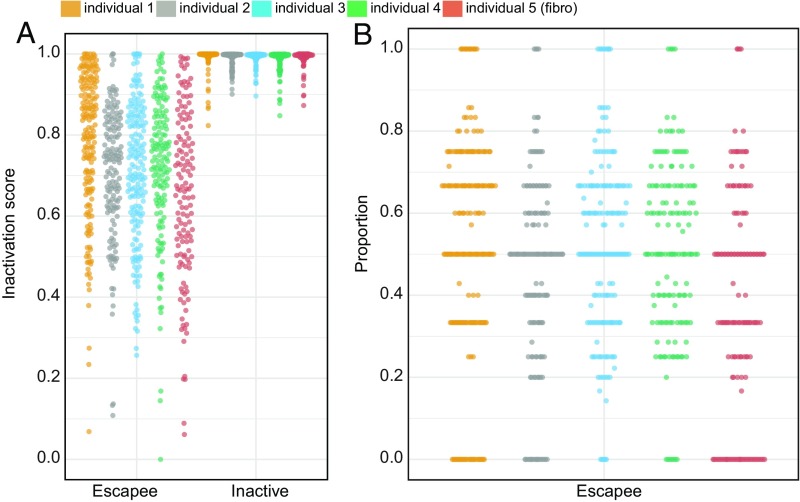
Among five individuals, the 22 robust escapee genes exhibited a heterogeneous AR profile, being inactivated in some cells and escaping XCI in others (Fig. 3). Specifically, we calculated a cellular escaping ratio per escapee gene as the proportion of cells escaping XCI with respect to the total number of cells expressing the gene. Some genes displayed a stable cellular escaping ratio among all of the individuals, while others were more variable. For example, CA5BP1 showed consistent cellular escaping ratios ranging from 30 to 53%; ZFX, a known constitutive escapee (45), presented with cellular escaping ratios ranging from 85 to 100%, while DDX3X had a broader range, from 29 to 61%. Overall, escapees had different cellular escaping ratios, thereby suggesting that each escapee gene is independently regulated. The observed cellular pattern of XCI of the escapees (Fig. 2) suggests a variable cellular ability of expressing genes from the inactivated allele. To investigate this hypothesis, we calculated the Inactivation Score (IS) for each cell defined as the mean AR of the escapee genes detected per cell (we considered only cells expressing at least two escapees; Xist was not included). First, we verified that neither AR nor IS is influenced by coverage depth and levels of gene expressions (SI Appendix, Fig. S8). For each individual, we ordered the cells according to the respective IS and discovered that the capacity to express from the inactive chromosome is a continuous variable (Figs. 2 and 3A). This suggested a cellular stratification that reflects the propensity of each cell to escape XCI, as confirmed by the proportion of escapee genes per cell (Fig. 3B). As expected, the IS is strongly negatively correlated with expression from the inactive X chromosome (SI Appendix, Fig. S9). Notably, in all five individuals, we observed two special groups of cells: one in which all of the detectable escapees behaved as inactivated and another in which all detectable escapees expressed both X alleles (Fig. 3B). These two extreme cell populations represent on average 15% of the total number of aggregate cells among the individuals [individual 1: 20%; individual 2: 14%; individual 3: 8%; individual 4: 7%; individual 5 (fibroblast): 26%]. As a control, we calculated the IS of the remaining inactivated genes per cell (all close to IS = 1, as expected. Fig. 3A, Inactivated). To validate this finding in an additional species, we analyzed data from 145 mouse single cells from a previous study (38). With the same procedure applied for human samples, but selecting as a robust set known mouse escapee genes (21, 46), we actually replicated the observations made in humans. We indeed observed the similar extent of cellular XCI heterogeneity and IS stratification (SI Appendix, Fig. S10). Overall, these results demonstrate that XCI is a complex intra- and interindividual heterogeneous process, and the ability to escape from X inactivation varies from gene to gene, from cell to cell, and also among individuals. The evidence of similar cell stratification in all individuals suggests the existence of a general regulatory mechanism that controls the propensity of a cell to express genes from the inactivated allele.
Fig. 3.
Cellular propensity to escape XCI. (A) Cells ranked by the IS calculated on all escapees in the robust set (Escapee) and on all inactivated genes (Inactive) per individual (color coded; see legend). Each dot represents a fibroblast cell. (B) Y axis: cells ordered according to the percentage of escapees displaying an inactive status (AR ≥ 0.95, with respect to all detectable escapees) per individual (x axis). Each dot represents a fibroblast cell.
Potential Drivers of Cellular XCI Heterogeneity.
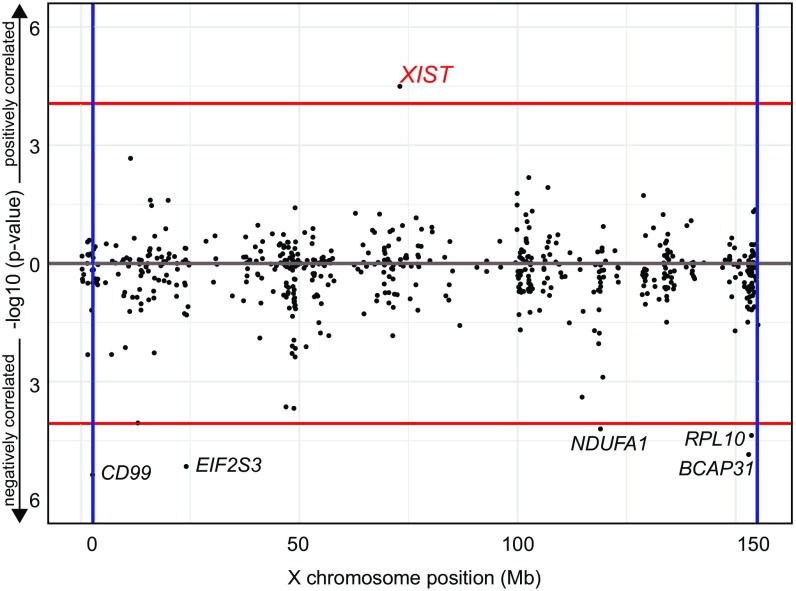
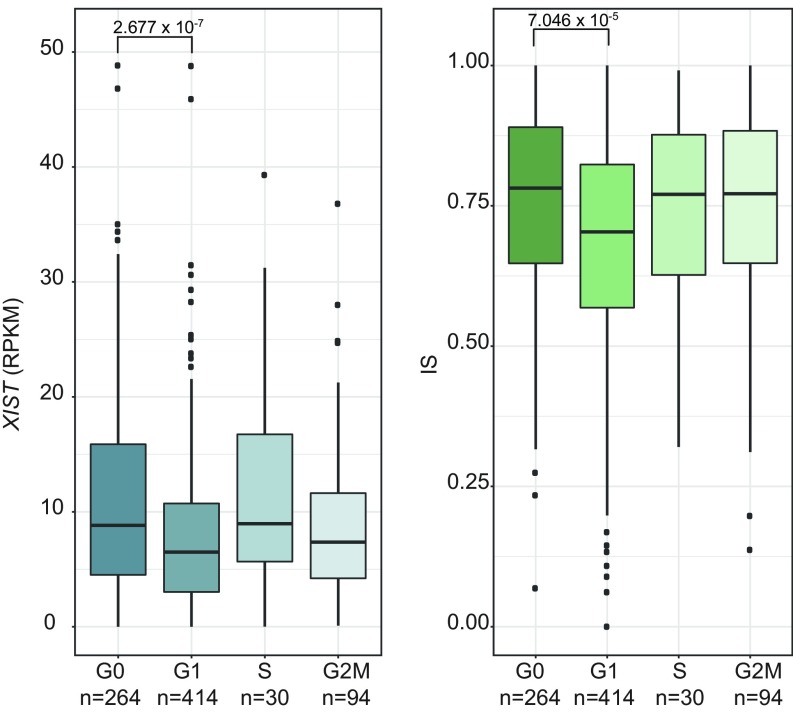
We hypothesized that the cellular XCI heterogeneity may be associated with the level of expression of genes on the X chromosome. To explore this hypothesis, we correlated the X-linked gene expression (RPKM > 1) with the IS. After FDR correction for multiple testing, genes were ranked based on the adjusted P value (Fig. 4). Notably, the only gene positively and significantly correlated with IS was XIST (nominal P value = 3.2 × 10−5; adj. P value = 3.0 × 10−3). XIST is a well-known ncRNA that regulates the establishment and the maintenance of XCI (14). Other significantly, albeit negatively, associated genes were EIF2S3, CD99, NDUFA1, RPL10, and BCAP31. To further investigate an eventual role for the cell cycle in XCI heterogeneity, we used the computational method Cyclone (47) to assign each cell to a specific cell-cycle phase according to the expression of the appropriate gene markers [from CycleBase (48)]. Cells not expressing MKI67 have been previously reported to be in G0 (49). A previous study indicated that XIST expression does not significantly change during cell-cycle phases (13). However, XIST expression in G0 was not evaluated in this study. Indeed, we observed a statistically significant difference between the populations of single cells in G0 and G1 regarding XIST expression (Fig. 5). A similar pattern, albeit not significant due to the small amount of cells, is detected in mouse data (SI Appendix, Fig. S11). Moreover, this finding is confirmed by a previous transcriptomics study in human cells arrested in G0/G1, S, and G2/M (50) (Gene Expression Omnibus accession no. GSE94479) where XIST expression in G0/G1 transition-arrested cells considerably decreases in the nonquiescent cell-cycle phase. Interestingly, in agreement with the hypothesis of a repressive XIST-dependent regulatory effect, the IS was also significantly higher in G0 than in G1 cells, on average (Fig. 5).
Fig. 4.
XIST expression association and correlation with IS and its repressive role for X-chromosome genes. Manhattan-like plot for Pearson correlation of X-linked gene expression with IS. The plot represents −log10 P value against the X-chromosome position. Vertical lines represent Pseudoautosomal Region (PAR) limits. Red horizontal lines represent Bonferroni significance thresholds for positively and negatively correlated genes, respectively.
Fig. 5.
XIST expression and IS dependency on the cell cycle. Distribution of XIST expression (Left) and of ISs per single cells (Right) according to cell-cycle phases (n = number of cells per stage). P values calculated with Mann–Whitney U test (see Materials and Methods for details).
Given these results, we explored the possibility that cell-cycle phasing influences the observed XCI heterogeneity in a way that is independent from XIST. The result of LASSO regression indicates that the cell cycle impacts IS (P = 6.33E-5) more significantly than XIST (P = 0.0018). To validate this observation, we inferred the directionality of the interactions IS–XIST IS–cell cycle and XIST–cell cycle with Bayesian networks (51). We found that the dominant model among all possible configurations is the one with the cell cycle independently influencing IS and XIST (SI Appendix, Fig. S12A). To further investigate the eventual genetic determinant of the cell-cycle effect on XCI, we extended the association analysis to all of the autosomal genes. No gene with positive correlation passed the significance threshold (Bonferroni correction for the number of genes: αTHR < 1.6E-6). (SI Appendix, Fig. S12B and Dataset S2). However, 37 negatively correlated genes passed the threshold (SI Appendix, Fig. S12C and Dataset S2). Gene ontology analysis with DAVID (52) revealed an enrichment for genes implicated in the mitochondrial process [adjusted P value (p.adj) < 6E10-7)], ribosome production (p.adj < 1.4E-5), rRNA processing (p.adj < 1.4E-4), and translational initiation (p.adj < 1.5E-5), all hallmarks of active (i.e., non-G0) cellular phases. These results together suggest that XCI heterogeneity is driven mainly by cell-cycle phases, in particular the transition between the quiescent and nonquiescent state. XIST ncRNA tends to be more expressed in the resting G0 phase than in G1 and seems to exert a repressive independent effect on the expression of the escapees from the inactive chromosome.
Discussion
Our study, using human single-cell RNA-Seq datasets, points to a pervasive heterogeneity in escaping XCI. We have shown that escapees have a different allelic expression profile in single fibroblasts from the same individual. More than 50% of the escapees had the tendency to be expressed mainly from Xa (Fig. 4), while ZFX and PUDP exhibited an overall biallelic expression in more than 70% of the cells. We also observed that some escapees exhibited a relatively stable cellular escaping ratio (proportion of cells in which the gene is escaping), i.e., ZFX and CA5BP1, whereas others, such as DDX3X, showed broader variability (Fig. 3). This finding explains, from a single-cell perspective, the previous observations of heterogeneous gene expression from Xi in cell lines derived from different individuals (53) and tissues (19, 46). A more recent study revealed the contribution of six escapee genes (TRX, CNKSR2, DDX3X, KDM5C, KDM6A, MAGEC3) to cancer sex bias (31). Here, we confirmed the escapee status for DDX3X, KDM5C, and KDM6A. We observed that the interindividual DDX3X inactivation profile is highly variable and thereby could potentially be associated with differences in cancer predisposition occurring among female individuals. Concerning the genes we detected as escapees in this study, mutations in HUWE1 have been found in individuals affected by Turner-type intellectual disability [MRX17 (MIM 300706)] (54, 55). Moreover, a recent study suggested the critical role of HUWE1 as a colonic tumor-suppressor gene (56). Pathogenic variants in RBMX have been associated with X-linked intellectual disability (57, 58). Expression of STK26 (alternatively named MST4) was recently shown to be significantly lower in patients affected by Graves’ disease, an autoimmune disorder characterized by abnormal thyroid function (59). It has also been shown that patients affected with Turner syndrome (XO) exhibit a high prevalence of hypothyroidism (60). We speculate that this phenotype may be related to the reduced expression of the single copy of STK26. Mutations and deletions of the escapee genes identified in the relaxed set, such as EDA, FHL1, IL1RAPL1, and FMR1, have been associated with different pathological conditions such as X-linked dominant tooth agenesis and X-linked ectodermal dysplasia (61–65), dominant reduced body myopathy (66) mental retardation (67, 68), and Fragile X syndrome (69), respectively. We hypothesize that the phenotypic variability of these diseases may be partially explained by the observed variable escapee status of their respective causative genes in the relevant tissues. It has been recently observed in a mouse study that some genes escape XCI in a fraction of cells only (38). We confirmed and extended this observation with the finding that XCI heterogeneity extends from genes to single cells in human and mouse cells. Inspired by a previous report describing the localization of XIST during the cell cycle (13), we explored the hypothesis that XIST expression and IS are related to the cell-cycle phases of individual cells. Our data indicate that the XIST gene is more expressed during the G0 than the G1 cell-cycle phase and that single cells in the G0 phase have an increased IS and therefore less propensity to express escapees from Xi. The transition from quiescent to nonquiescent cell-cycle phases seems to be the most prominent factor driving XCI heterogeneity whereas XIST seems to exert an independent repressive effect over the transcriptional activity of the inactive X chromosome. Overall, these results advance our understanding of X inactivation and suggest a potential influence of the heterogeneity of XCI in single cells on the phenotypic variability of X-linked single-gene disorders, whole X chromosome aneuploidies, and the observed female sex bias in cancer.
Materials and Methods
A complete description of methods, including statements of data availability and associated accession codes and references, is available in SI Appendix, Materials and Methods.
Ethical Statement.
The study was approved by the ethics committee of the University Hospitals of Geneva, and written informed consent was obtained from both parents of each individual before the study.
Samples.
We established six different cell lines from five female individuals: five primary fibroblast cell lines and one lymphoblastoid cell line (SI Appendix, Table S1). We captured 935 single-cell fibroblasts and 48 lymphoblastoid single cells. Lymphoblastoid cells were obtained from one of the five female individuals (Fig. 1A and SI Appendix, Table S1) (35, 70).
Cell Growth.
Cells were cultured in DMEM GlutaMAX (Life Technologies) supplemented with 10% FBS (Life Technologies) and 1% penicillin/streptomycin/fungizone mix (Amimed; BioConcept) at 37 °C in a 5% CO2 atmosphere.
Whole-Genome Sequencing.
Genomic DNA was extracted for all five individuals. Libraries were prepared with a TruSeq DNA kit (Illumina) and sequenced on an Illumina HiSEq. 2000 machine. For each individual, raw whole-genome DNA sequences were analyzed using an in-house pipeline previously described (35, 70). Briefly, we used the Burrows–Wheeler Aligner to align the sequencing reads (fastq) to the human reference genome (GRCh37/hg19). We used SAMtools to remove paired-end duplicates and pile up the remaining reads. BCFtools was used to call the SNVs and Annovar for the annotation. SNVs with a quality score <100 were excluded from the analysis. We used only uniquely mapped reads for SNV calling, and, in general, variants falling inside repeated regions such as segmental duplications or repeats (according to RepeatMasker) were filtered out (42).
Single-Cell Capture.
Single-cell capture was performed using the C1 single-cell auto prep system (Fluidigm) following the manufacturer’s procedure (35).
Single-Cell RNA-Seq.
The SMARTer Ultra Low RNA kit for Illumina sequencing (version 2; Clontech) was used for cell lysis and cDNA synthesis. Libraries were sequenced on an Illumina HiSeq2000 sequencer as 100-bp single-ended reads. RNA sequences were mapped with Genome Multitool (GEM) mapper (71). Uniquely mapping reads were extracted by filtering for mapping quality (MQ ≥ 150). Cells with less than 1 million uniquely mapped reads were excluded from further analysis (SI Appendix, Fig. S2).
Allele-Specific Expression and Classification of Escapee Genes.
For each gene on the X chromosome, the allelic ratio per cell was calculated based on the assumption that the active X allele is, on average, more transcribed than the inactive X (more details in SI Appendix, Methods S1). According to this metric, inactivated genes cluster around AR = 1 while known escapees appear as uniformly distributed between 0.5 ≤ AR ≤ 0.95 (SI Appendix, Figs. S13 and S14).
Haplotype and Multiple Cell (Doublet) Detection.
The estimated haplotype of each cell, calculated on the X chromosome, was compared with all others through pairwise correlation based on a hierarchical clustering procedure. Doublets simultaneously expressing both haplotypes (absolute correlation of 0.5) are identified and excluded from further analysis.
Annotation of the Escapee Genes.
We curated a list of 190 previously observed escapee genes in different cell types and tissues according to the literature (19, 20, 27, 32, 72–74) (Dataset S2) and have appended the results published in two studies (39, 75) in Dataset S3. Genes detected as escapees in our studies, in the absence of citation, have been labeled as undescribed escapee genes.
Cell-Cycle Phase Assignment.
G1, S, and G2M cell-cycle-stage–related gene markers were obtained from CycleBase (48) Cells not expressing MKI67 have been considered to be in G0 (76) The remaining cells were assigned to their respective cell-cycle phase according to the expression of CycleBase genes with Cyclone (47).
Accession Numbers.
Newly generated RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) data repository (accession no. GSE123028). Single-cell RNA sequences from limphoblasts and fibroblasts of individual 5 were obtained from European Genome-phenome Archive (accession no. EGAS00001001009).
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grant SNF-144082, European Research Council Grant ERC-249968, a ChildCare foundation grant (to S.E.A.), and Novartis Foundation Grant 18A052 (to F.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The newly generated RNA and DNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE123028). Single-cell RNA sequences from limphoblasts and fibroblasts of individual 5 were obtained from the European Genome-phenome Archive, https://www.ebi.ac.uk/ega/studies/EGAS00001001009 (accession no. EGAS00001001009).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806811115/-/DCSupplemental.
References
- 1.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 2.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 3.Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol. 2009;21:359–366. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 2011;3:a002592. doi: 10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharman GB. Late DNA replication in the paternally derived X chromosome of female kangaroos. Nature. 1971;230:231–232. doi: 10.1038/230231a0. [DOI] [PubMed] [Google Scholar]
- 6.Huynh KD, Lee JT. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature. 2003;426:857–862. doi: 10.1038/nature02222. [DOI] [PubMed] [Google Scholar]
- 7.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 8.Okamoto I, et al. Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature. 2005;438:369–373. doi: 10.1038/nature04155. [DOI] [PubMed] [Google Scholar]
- 9.Patrat C, et al. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc Natl Acad Sci USA. 2009;106:5198–5203. doi: 10.1073/pnas.0810683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mak W, et al. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- 11.Petropoulos S, et al. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell. 2016;167:285. doi: 10.1016/j.cell.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinheiro I, Heard E. X chromosome inactivation: New players in the initiation of gene silencing. F1000 Res. 2017;6:344. doi: 10.12688/f1000research.10707.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonkers I, et al. Xist RNA is confined to the nuclear territory of the silenced X chromosome throughout the cell cycle. Mol Cell Biol. 2008;28:5583–5594. doi: 10.1128/MCB.02269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown CJ, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 15.Brown CJ, et al. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 16.Ballabio A, Willard HF. Mammalian X-chromosome inactivation and the XIST gene. Curr Opin Genet Dev. 1992;2:439–447. doi: 10.1016/s0959-437x(05)80155-8. [DOI] [PubMed] [Google Scholar]
- 17.Wang CY, Jegu T, Chu HP, Oh HJ, Lee JT. SMCHD1 merges chromosome compartments and assists formation of super-structures on the inactive X. Cell. 2018;174:406–421.e25. doi: 10.1016/j.cell.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gendrel AV, Heard E. Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annu Rev Cell Dev Biol. 2014;30:561–580. doi: 10.1146/annurev-cellbio-101512-122415. [DOI] [PubMed] [Google Scholar]
- 19.Cotton AM, et al. Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome. Genome Biol. 2013;14:R122. doi: 10.1186/gb-2013-14-11-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 21.Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crowley JJ, et al. Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance. Nat Genet. 2015;47:353–360. doi: 10.1038/ng.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berletch JB, Yang F, Xu J, Carrel L, Disteche CM. Genes that escape from X inactivation. Hum Genet. 2011;130:237–245. doi: 10.1007/s00439-011-1011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lederer D, et al. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am J Hum Genet. 2012;90:119–124. doi: 10.1016/j.ajhg.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyake N, et al. KDM6A point mutations cause Kabuki syndrome. Hum Mutat. 2013;34:108–110. doi: 10.1002/humu.22229. [DOI] [PubMed] [Google Scholar]
- 26.Gropman A, Samango-Sprouse CA. Neurocognitive variance and neurological underpinnings of the X and Y chromosomal variations. Am J Med Genet C Semin Med Genet. 2013;163C:35–43. doi: 10.1002/ajmg.c.31352. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, et al. Genes that escape X-inactivation in humans have high intraspecific variability in expression, are associated with mental impairment but are not slow evolving. Mol Biol Evol. 2013;30:2588–2601. doi: 10.1093/molbev/mst148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones DT, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunford A, et al. Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat Genet. 2016;49:10–16. doi: 10.1038/ng.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yasukochi Y, et al. X chromosome-wide analyses of genomic DNA methylation states and gene expression in male and female neutrophils. Proc Natl Acad Sci USA. 2010;107:3704–3709. doi: 10.1073/pnas.0914812107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cotton AM, et al. Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum Mol Genet. 2015;24:1528–1539. doi: 10.1093/hmg/ddu564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sudbrak R, et al. X chromosome-specific cDNA arrays: Identification of genes that escape from X-inactivation and other applications. Hum Mol Genet. 2001;10:77–83. doi: 10.1093/hmg/10.1.77. [DOI] [PubMed] [Google Scholar]
- 35.Borel C, et al. Biased allelic expression in human primary fibroblast single cells. Am J Hum Genet. 2015;96:70–80. doi: 10.1016/j.ajhg.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen G, et al. Single-cell analyses of X chromosome inactivation dynamics and pluripotency during differentiation. Genome Res. 2016;26:1342–1354. doi: 10.1101/gr.201954.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borensztein M, et al. Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nat Struct Mol Biol. 2017;24:226–233. doi: 10.1038/nsmb.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reinius B, et al. Analysis of allelic expression patterns in clonal somatic cells by single-cell RNA-seq. Nat Genet. 2016;48:1430–1435. doi: 10.1038/ng.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tukiainen T, et al. GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fialkow PJ. X-chromosome inactivation and the Xg locus. Am J Hum Genet. 1970;22:460–463. [PMC free article] [PubMed] [Google Scholar]
- 41.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panousis NI, Gutierrez-Arcelus M, Dermitzakis ET, Lappalainen T. Allelic mapping bias in RNA-sequencing is not a major confounder in eQTL studies. Genome Biol. 2014;15:467. doi: 10.1186/s13059-014-0467-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng X, Berletch JB, Nguyen DK, Disteche CM. X chromosome regulation: Diverse patterns in development, tissues and disease. Nat Rev Genet. 2014;15:367–378. doi: 10.1038/nrg3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, et al. Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc Natl Acad Sci USA. 2016;113:E2029–E2038. doi: 10.1073/pnas.1520113113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider-Gädicke A, Beer-Romero P, Brown LG, Nussbaum R, Page DC. ZFX has a gene structure similar to ZFY, the putative human sex determinant, and escapes X inactivation. Cell. 1989;57:1247–1258. doi: 10.1016/0092-8674(89)90061-5. [DOI] [PubMed] [Google Scholar]
- 46.Berletch JB, et al. Escape from X inactivation varies in mouse tissues. PLoS Genet. 2015;11:e1005079. doi: 10.1371/journal.pgen.1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scialdone A, et al. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods. 2015;85:54–61. doi: 10.1016/j.ymeth.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 48.Santos A, Wernersson R, Jensen LJ. Cyclebase 3.0: A multi-organism database on cell-cycle regulation and phenotypes. Nucleic Acids Res. 2015;43:D1140–D1144. doi: 10.1093/nar/gku1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sobecki M, et al. Cell-cycle regulation accounts for variability in ki-67 expression levels. Cancer Res. 2017;77:2722–2734. doi: 10.1158/0008-5472.CAN-16-0707. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, et al. Transcriptional landscape of the human cell cycle. Proc Natl Acad Sci USA. 2017;114:3473–3478. doi: 10.1073/pnas.1617636114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scutari RNaM . Bayesian Networks in {R} with Applications in Systems Biology. Springer; Berlin: 2013. [Google Scholar]
- 52.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 53.Carrel L, Willard HF. Heterogeneous gene expression from the inactive X chromosome: An X-linked gene that escapes X inactivation in some human cell lines but is inactivated in others. Proc Natl Acad Sci USA. 1999;96:7364–7369. doi: 10.1073/pnas.96.13.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Froyen G, et al. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am J Hum Genet. 2008;82:432–443. doi: 10.1016/j.ajhg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Froyen G, et al. Copy-number gains of HUWE1 due to replication- and recombination-based rearrangements. Am J Hum Genet. 2012;91:252–264. doi: 10.1016/j.ajhg.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myant KB, et al. HUWE1 is a critical colonic tumour suppressor gene that prevents MYC signalling, DNA damage accumulation and tumour initiation. EMBO Mol Med. 2017;9:181–197. doi: 10.15252/emmm.201606684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shashi V, et al. A unique form of mental retardation with a distinctive phenotype maps to Xq26-q27. Am J Hum Genet. 2000;66:469–479. doi: 10.1086/302772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shashi V, et al. The RBMX gene as a candidate for the Shashi X-linked intellectual disability syndrome. Clin Genet. 2015;88:386–390. doi: 10.1111/cge.12511. [DOI] [PubMed] [Google Scholar]
- 59.Guo A, Tan Y, Liu C, Zheng X. MST-4 and TRAF-6 expression in the peripheral blood mononuclear cells of patients with Graves’ disease and its significance. BMC Endocr Disord. 2017;17:11. doi: 10.1186/s12902-017-0161-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.El-Mansoury M, et al. Hypothyroidism is common in turner syndrome: Results of a five-year follow-up. J Clin Endocrinol Metab. 2005;90:2131–2135. doi: 10.1210/jc.2004-1262. [DOI] [PubMed] [Google Scholar]
- 61.Kere J, et al. X-linked anhidrotic (hypohidrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nat Genet. 1996;13:409–416. doi: 10.1038/ng0895-409. [DOI] [PubMed] [Google Scholar]
- 62.Yotsumoto S, et al. A novel point mutation of the EDA gene in a Japanese family with anhidrotic ectodermal dysplasia. J Invest Dermatol. 1998;111:1246–1247. doi: 10.1046/j.1523-1747.1998.00443.x. [DOI] [PubMed] [Google Scholar]
- 63.Visinoni AF, de Souza RL, Freire-Maia N, Gollop TR, Chautard-Freire-Maia EA. X-linked hypohidrotic ectodermal dysplasia mutations in Brazilian families. Am J Med Genet A. 2003;122A:51–55. doi: 10.1002/ajmg.a.20276. [DOI] [PubMed] [Google Scholar]
- 64.Tao R, et al. A novel missense mutation of the EDA gene in a Mongolian family with congenital hypodontia. J Hum Genet. 2006;51:498–502. doi: 10.1007/s10038-006-0389-2. [DOI] [PubMed] [Google Scholar]
- 65.Tarpey P, et al. A novel Gln358Glu mutation in ectodysplasin A associated with X-linked dominant incisor hypodontia. Am J Med Genet A. 2007;143:390–394. doi: 10.1002/ajmg.a.31567. [DOI] [PubMed] [Google Scholar]
- 66.Liewluck T, et al. Unfolded protein response and aggresome formation in hereditary reducing-body myopathy. Muscle Nerve. 2007;35:322–326. doi: 10.1002/mus.20691. [DOI] [PubMed] [Google Scholar]
- 67.Tabolacci E, et al. A truncating mutation in the IL1RAPL1 gene is responsible for X-linked mental retardation in the MRX21 family. Am J Med Genet A. 2006;140:482–487. doi: 10.1002/ajmg.a.31107. [DOI] [PubMed] [Google Scholar]
- 68.Nawara M, et al. Novel mutation of IL1RAPL1 gene in a nonspecific X-linked mental retardation (MRX) family. Am J Med Genet A. 2008;146A:3167–3172. doi: 10.1002/ajmg.a.32613. [DOI] [PubMed] [Google Scholar]
- 69.Kremer EJ, et al. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991;252:1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- 70.Santoni FA, et al. Detection of imprinted genes by single-cell allele-specific gene expression. Am J Hum Genet. 2017;100:444–453. doi: 10.1016/j.ajhg.2017.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marco-Sola S, Sammeth M, Guigó R, Ribeca P. The GEM mapper: Fast, accurate and versatile alignment by filtration. Nat Methods. 2012;9:1185–1188. doi: 10.1038/nmeth.2221. [DOI] [PubMed] [Google Scholar]
- 72.Johnston CM, et al. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet. 2008;4:e9. doi: 10.1371/journal.pgen.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park C, Carrel L, Makova KD. Strong purifying selection at genes escaping X chromosome inactivation. Mol Biol Evol. 2010;27:2446–2450. doi: 10.1093/molbev/msq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sharp AJ, et al. DNA methylation profiles of human active and inactive X chromosomes. Genome Res. 2011;21:1592–1600. doi: 10.1101/gr.112680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Balaton BP, Cotton AM, Brown CJ. Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol Sex Differ. 2015;6:35. doi: 10.1186/s13293-015-0053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schonk DM, et al. Assignment of the gene(s) involved in the expression of the proliferation-related Ki-67 antigen to human chromosome 10. Hum Genet. 1989;83:297–299. doi: 10.1007/BF00285178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.