Abstract
Bacterial infections have been traditionally controlled by antibiotics and vaccines, and these approaches have greatly improved health and longevity. However, multiple stakeholders are declaring that the lack of new interventions is putting our ability to prevent and treat bacterial infections at risk. Vaccine and antibiotic approaches still have the potential to address this threat. Innovative vaccine technologies, such as reverse vaccinology, novel adjuvants, and rationally designed bacterial outer membrane vesicles, together with progress in polysaccharide conjugation and antigen design, have the potential to boost the development of vaccines targeting several classes of multidrug-resistant bacteria. Furthermore, new approaches to deliver small-molecule antibacterials into bacteria, such as hijacking active uptake pathways and potentiator approaches, along with a focus on alternative modalities, such as targeting host factors, blocking bacterial virulence factors, monoclonal antibodies, and microbiome interventions, all have potential. Both vaccines and antibacterial approaches are needed to tackle the global challenge of antimicrobial resistance (AMR), and both areas have the underpinning science to address this need. However, a concerted research agenda and rethinking of the value society puts on interventions that save lives, by preventing or treating life-threatening bacterial infections, are needed to bring these ideas to fruition.
Keywords: vaccines, AMR, antibiotics, bacterial infections
It has been well documented that Fleming predicted the potential for bacteria to develop resistance to penicillin soon after his discovery, and today we know that antimicrobial resistance (AMR) has developed to every approved antibiotic launched. Following the golden era of antibiotic development, when several different classes were discovered and optimization of known classes was rapid, the effective role of antibiotics addressing bacterial infection was taken for granted. Antibiotics reduce the global burden of bacterial infection. Multiple classes of antibiotics were developed and launched from the 1940s to 1980s, including the β-lactam antibiotics (including penicillin, cephalosporin, and carbapenem), aminoglycosides (including tobramycin), tetracyclines, macrolides (including erythromycin), glycopeptides (including vancomycin), polymyxins (including colistin), and fluoroquinolones (including ciprofloxacin). Since 1990, three novel-class antibiotics have been launched (pleuromutilins, lipoglycopeptides, and oxazolidinones), although many derivatives of older classes were also launched. From 1940 to 1990, resistance to new antibiotics took approximately two years to develop against the β-lactam classes and approximately nine to 16 years to develop against other classes. Since 1990, resistance developed against the oxazolidinone linezolid shortly after its launch (1).
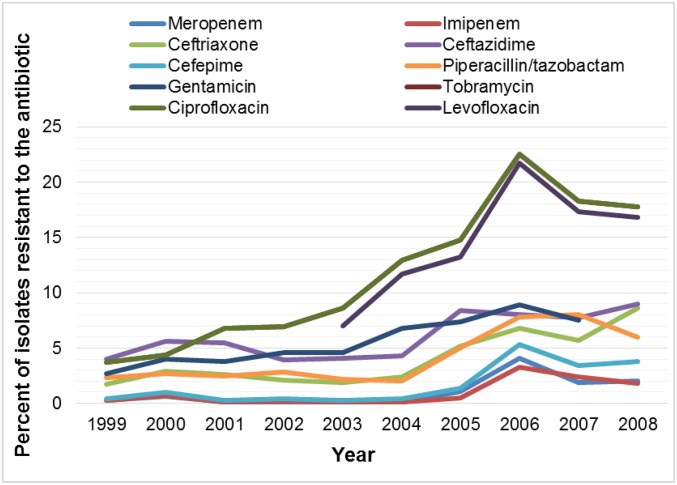
The use and overuse of antibiotics have created an environment where, to survive, bacteria must overcome our molecular weapons. Since the bulk of antibiotics have been used against gram-positive bacteria, it is logical that resistance to gram-positive agents were the first to rise to threat level, with methicillin-resistant Staphylococcus aureus (MRSA) perhaps the most well known. The spread of MRSA was recognized as a significant threat in the late 1990s to early 2000s, and drug developers spent significant effort on gram-positive discovery programs. This effort was mostly successful in providing health care professionals with additional options when faced with resistant gram-positive infections, but most of these new antibiotics were incremental improvements on old mechanisms. During this period, little attention was given to gram-negative resistance as there were several effective gram-negative antibiotics available. This changed dramatically in the early 2000s, when a surveillance study showed resistance to 10 antibiotics increased over just a few years. In 1999, resistance to each of the 10 antibiotics was less than 5% of the clinical isolates tested; however, by 2008, resistance to seven of those antibiotics had increased to between 6% and 18% of clinical isolates tested (Fig. 1) (2).
Fig. 1.
Percentage of Enterobacteriaceae strains from a US surveillance study that show increasing resistance to 10 antibiotics over a 10-year period. Data from Rhomberg and Jones (2).
Arguably, β-lactam antibiotics have been one of the more successful classes of antibacterial agents, but much of this can be attributed to research efforts to inhibit the main resistance mechanism, hydrolysis of the β-lactam ring by β-lactamases. As of 2016, the number of different β-lactamase alleles discovered exceeded 1,500 (ftp://ftp.ncbi.nlm.nih.gov/pathogen/betalactamases/Allele.tab; https://www.ncbi.nlm.nih.gov/pathogens/beta-lactamase-data-resources/), and they have been classified into four classes, A–D, based on their structural features. Classes A, C, and D are serine proteases, and class B is the metallo-β-lactamases. In the 1980s, development of β-lactamase inhibitors (BLIs) rescued β-lactam antibiotics. The concept was to coadminister a β-lactam antibiotic with a BLI and arrange the affinities such that the β-lactamase was inhibited before it could hydrolyze the β-lactam antibiotic, leaving the antibiotic free to kill the bacteria. The first BLI was clavulanic acid, which mainly inhibits class A β-lactamases, and other combinations have played a major role in our ability to treat resistant infections over the years. Currently, carbapenem resistance is the biggest threat to patients, which is mediated by class A (Klebsiella pneumoniae carbapenemase), class B (e.g., New Delhi metallo-β-lactamase), and class D (e.g., oxacillinase) β-lactamases. The approvals of avibactam + ceftazidime and vaborbactam + meropenem have provided new options for treating class A-mediated resistance, but neither provides broad coverage of the class B- and D-mediated carbapenem resistance. This approach has certainly provided a stopgap in our war against some multidrug-resistant (MDR) pathogenic bacteria, but new mechanism agents remain desperately needed as history has shown that the spread of plasmid-mediated β-lactamases can be formidable and further spread of metallo-β-lactamases remains a threat.
In the absence of new gram-negative antibiotics, physicians have been forced to return to the past. Colistin was first clinically available in the 1950s for gram-negative infections, but nephrotoxicity limited its utility, and it was quickly replaced when safer agents arrived. The lack of use may have contributed to the apparent lack of resistance. However, with limited options, colistin is often a last-line therapy. Unfortunately, it has widespread use in agriculture, and colistin resistance was recently identified (3).
The increased AMR and the challenge to discover new antibiotics force us to look for alternative ways to deal with bacterial infections. In this paper, we will review the state of the art of antibiotic discovery and the other technologies that may help in dealing with AMR. Particular attention will be paid to vaccines that, in addition to having a good track record in preventing some types of bacterial infections without generating AMR, face a “golden technological era” that may allow development of vaccines against a broader range of bacterial pathogens.
Challenges of Antibiotic Discovery
In the 1990s, the birth of bacterial genomics rapidly revealed a multitude of different antibacterial targets, expanding the approaches for antibacterials from the four to five mechanisms of established antibacterials to >100 different novel mechanisms. This knowledge led to massive industrial efforts in which single companies each ran high-throughput screens (HTSs) on 60–70 different approaches/targets. Pfizer, AstraZeneca, and GlaxoSmithKline ran a total of almost 200 HTSs; however, all three companies declared that none of these efforts resulted in a development candidate that was progressed (4, 5). Consequently, finding novel chemical starting points for antibacterials is hugely challenging, and this is illustrated by the fact that no gram-negative antibiotic with an entirely novel mechanism has been approved in more than 40 years. The failure to develop antibiotics with a novel mechanism was not considered to be due to the inability of research teams to create a potent inhibitor of a new essential bacterial target; indeed, finding inhibitors was most often very achievable. The failure was commonly found to be the challenge of developing these inhibitors into medicines with an appropriate balance of antibacterial activity, drug metabolism and pharmacokinetics properties, and safety. Since novel mechanism agents proved elusive, many companies that initially started looking for entirely novel classes of antibiotics switched their focus to making new versions of established classes, with proven drug-like properties, and although the resultant antibiotics address some aspect of unmet need, they all have limitations of the parent class.
So, what do we consider as the main scientific reason for this poor success? Regardless of whether it is developing a novel antibiotic or optimizing a known class, we think the fundamental challenge of antibiotic research and development is that antibiotics require higher exposures and, often, higher doses than those normally required for other medicines, which is the major cause of attrition in preclinical toxicity testing or in human trials. We consider the higher doses are attributable to three main reasons.
First, bacteria multiply and grow rapidly, so any new antibiotic must effectively penetrate and kill the bacteria quickly without generating resistance. Bacteria have established many defense systems to keep potentially toxic xenobiotics out or render them ineffective. These defenses include formidable and diverse membranes that require the antibiotics to have narrow and specific physicochemical properties to penetrate. Unfortunately, the properties required for effective penetration into the bacteria are not well known, although there are efforts to identify them (6). Many antibiotics bypass the membrane by traveling through porins, which are bacterial proteins that span the bacterial membranes and form aqueous channels to allow, for example, nutrients to penetrate into the cell. However, when exposed to an antibiotic, bacteria can down-regulate these porins, or mutate them to reduce their ability to transport the antibiotic into the bacteria. Once an antibiotic has successfully navigated into the bacteria, it will then face bacterial efflux pumps that eject these xenobiotics back outside the cell. Exposure of the bacteria to an antibiotic can also result in up-regulation of these efflux pumps to increase the rate of ejection. All of these mechanisms create a concentration gradient with much of the antibiotic on the outside of the cell and very little near its target. To be effective at the target, more antibiotic is needed on the outside, which dictates the need for a high dose to create sufficient exposure levels.
Second, lack of diagnostics means empirical therapy is needed to cover all likely causative pathogens of an infection. Our inability to rapidly and accurately diagnose the specific infecting pathogen at the point of care remains a challenge; current methods can take one to two days after the patient presents at the clinic. Until this changes, there is a need to develop broader spectrum antibiotics for empiric treatment so that they cover all of the key causative pathogens for a particular infection/indication being targeted; for example, for hospital-acquired pneumonia (HAP), the antibiotic ideally needs to cover MRSA, Pseudomonas aeruginosa, K. pneumoniae, and Acinetobacter. The antibiotic may have exquisite activity against one species (e.g., K. pneumoniae) but may be much less active against another (e.g., P. aeruginosa). Since the physician cannot rapidly pinpoint the infecting pathogen at the point of care, the physician often must assume the patient is infected by any of the bacteria common for that infection and may be the one that is least sensitive to the antibiotic [the one with the highest minimum inhibitory concentration (MIC)]. Classical pharmacokinetic (PK)/pharmacodynamic principles for selecting the dose of antibiotics are based on covering those high-MIC pathogens. The result of this is that it drives a higher dose to ensure coverage of the majority of pathogens/strains that can cause the infection being treated.
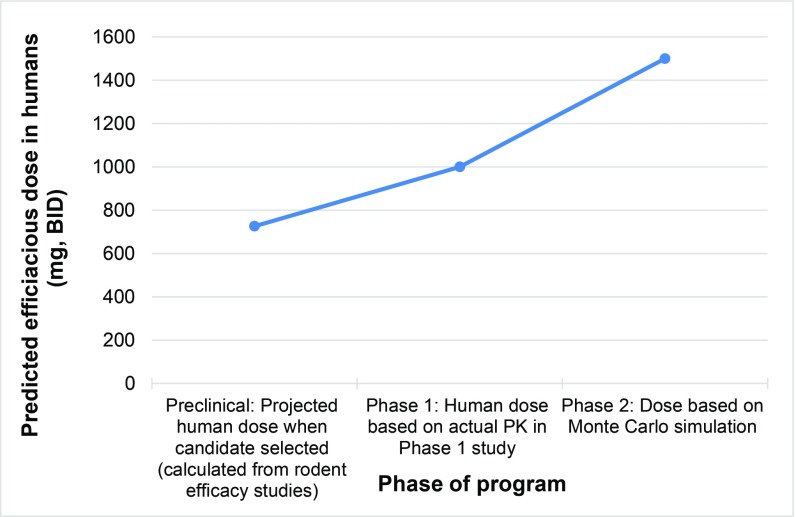
Third, a high dose is needed to cover the worst-case scenario. Development of the peptidyl deformylase inhibitor GSK1322322 illustrates how doses of an experimental antibiotic increase as data evolve and the need for an antibiotic to cover the worst-case scenario. In preclinical in vivo efficacy models of infections in rodents, it was determined that a dose of 75 mg/kg twice daily [bis in die (BID)] would be efficacious against the four common causative pathogens of community-acquired pneumonia. Lower doses would work against more sensitive pathogens, but it is important that the dose progressed was efficacious against the least sensitive strains. Using a study where a dose of 75 mg/kg of body weight was needed in the rat, allometric scaling [US Department of Health and Human Services Food and Drug Administration, Center for Drug Evaluation and Research Guidance for Industry, Estimating the Maximum Safe Starting Dose in Initial Clinical Trials for Therapeutics in Adult Healthy Volunteers, July 2005] gives a projected human dose for community-acquired pneumonia of 726 mg BID (Table 1). A phase 1 study was conducted, and based on actual exposures measured in humans, the target efficacious dose was predicted to be 1,000–1,500 mg BID. A Monte Carlo simulation (7–9) was employed to model a large patient population, taking into account PK variability across the population and the range of potential sensitivities of the infecting pathogen. This is conducted to determine the dose that will work in the worst-case scenario of a patient with the lowest PK exposure being infected with a strain that is least sensitive to the antibiotic. This led to a refined target dose of 1,500 mg BID (3,000 mg⋅d−1), which would ensure >90% of the population receive an exposure that would be effective at treating acute skin and skin structure infection. These are important considerations for determining the dose of antibiotics, but they drive up the dose given to every patient (Fig. 2). If the infecting pathogen could be rapidly and accurately diagnosed, the dose could be better tailored to the patient and the pathogen causing the infection.
Table 1.
In vivo models of infection demonstrating efficacious dose of GSK1322322 at time of candidate selection and the human equivalent dose
| Strain | MIC, μg/mL | Efficacious dose, mg/kg BID | Human equivalent dose (60 kg per person), mg BID |
| H. influenzae strain H128 | 1 | 75 (rat) | 726 |
| S. pneumoniae strain Ery-2 | 0.25 | 37.5 (rat) | 363 |
| S. aureus A-24 | 4 | 75 (mouse) | 366 |
| MRSA Panton–Valentine positive strain (PVL-2) | 4 | 37.5 (mouse) | 183 |
Fig. 2.
Increase in predicted efficacious dose of GSK1322322 from 726 mg BID (1.45 g⋅d−1) at candidate selection to 1,500 mg BID (3 g⋅d−1) by phase 2.
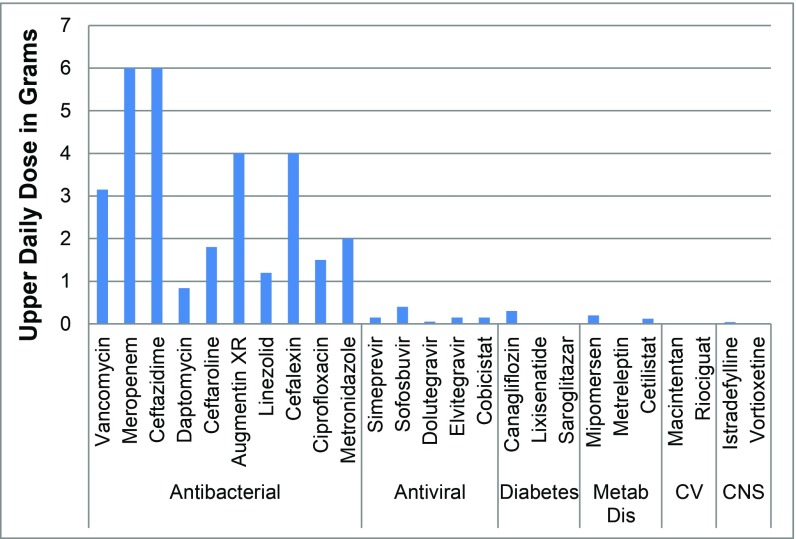
Therefore, these factors and the lack of rapid and accurate diagnostics to quickly identify the infecting pathogen result in all patients getting high doses to cover a multitude of eventualities; essentially, this is the opposite of personalized medicine approaches that are currently evolving in other therapy areas. The high dose of antibiotics is evident when comparing doses of antibiotics with doses of other recently approved agents from different therapeutic areas [antibacterial data from ref. 10, data for other therapy areas from Bronson et al. (11)] (Fig. 3). In addition, many patients are given combinations of antibiotics to cover a range of potential pathogens. For example, a patient with an intraabdominal infection with suspected MRSA could be given a combination of three drugs:
Piperacillin/tazobactam, 2.25 g every day, to cover gram-negative infections
Plus metronidazole, 500 mg three times daily, to cover anaerobes
Plus vancomycin, 1.4 g (monitoring required) BID, to cover MRSA and Enterococcus
Fig. 3.
Total upper daily dose of common antibiotics compared to drugs approved in 2013 from other therapy areas. Body weight of 70 kg was used for doses given as milligrams per kilogram of body weight (10, 11). CNS, central nervous system; CV, cardiovascular; Metab Dis, metabolic disease.
This adds up to 13.3 g of antibiotic per day, and considering this will be administered typically between five and 14 days, this totals up to 66–186 g of antibiotic that the patient will receive. In contrast, a yearly course of Crestor to reduce cholesterol totals 1.8–14.6 g (Crestor package insert), a yearly course of Zestril for heart disease totals 14.6 g (https://www.drugs.com/zestril.html), and a yearly course of Januvia for type 2 diabetes is 35.6 g (Januvia package insert).
Resistance, empirical therapy, and lack of new-class antibiotics have created quite a conundrum. To be successful, traditional antibiotics need to be given at high doses to cover PK variability and all of the causative pathogens; however, these high doses are given at or near their toxic threshold, and trying to balance these factors leads to high attrition during clinical development.
Strategies to Improve Antibiotics
Researchers have recognized and understood the above challenges and have moved toward identifying pioneering methods and disruptive technologies that have potential to transform the field. These include the following.
Potentiation Approaches.
To circumvent poor penetration due to the highly effective bacterial membranes and promiscuous pumps, researchers have investigated methods to increase the internal concentration of antibiotics through potentiation. Potentiation involves manipulating another typically nonessential part of the bacteria to make the organism more sensitive to the antibiotics. An example of potentiation includes efflux pump inhibitors. This has been a compelling approach as an effective efflux inhibitor has the potential to broaden the spectrum of a gram-positive agent to be effective against gram-negative infections or substantially increase the potency of a compound that already has some activity against gram-negative infections. However, although a lot of work has been conducted in this area across the industry, an efflux inhibitor has not yet progressed to phase 1. Our team worked with Mpex on that the area for three years; although effective inhibitors were identified in vitro, we were not able to overcome the in vivo toxicity. This remains a compelling approach and could also reduce the doses of new gram-negative antibiotics, and thereby overcome some of the “high-dose” challenges discussed previously. Broader research to enhance our understanding of how to effectively inhibit efflux pumps is needed, but at this time, we consider inhibition of efflux pumps to have a lower probability of success. Potentiator targets that are intracellular will suffer the same penetration challenges as traditional antibiotics; therefore, we recommend prioritizing potentiator mechanisms that affect targets on the outer surface of the bacteria, avoiding the need for the potentiator to penetrate into the periplasm or cytoplasm. A further challenge is the need for codevelopment of a potentiator with the antibiotic. This is quite achievable, as demonstrated by the BLI combinations; however, it does add complexity to the development.
Active Uptake Approaches.
In a different approach, researchers are investigating methods to increase influx by taking advantage of membrane transporters. This approach is validated by the approved antibiotic fosfomycin, which uses the bacterial sugar transporters to enter the cells. A popular transporter that has been a target for many years is iron uptake. Bacteria need iron to survive; however, free iron is extremely limited in biological systems, so bacteria have evolved methods to strip iron from host sources using siderophores. Bacteria release siderophores into their local environment, and once they have bound iron, the bacteria ingest the iron–siderophore complex through iron transporters. Since there are multiple siderophores and bacteria can also ingest siderophores from other bacterial species, the iron uptake transporters have a lower selectivity and high tolerance for larger structures. This allows researchers to add iron-binding siderophore mimetic groups to an antibiotic. A popular iron-binding group to use is a catechol, which has a reasonable affinity for iron and is featured in many natural siderophores. However, these can be oxidized through the metabolism, resulting in toxic byproducts, which has limited progress. The most advanced molecule utilizing this approach is a catechol-linked cephalosporin (cefiderocol, also known as S-649266) that is currently in clinical trials for gram-negative infections. To date, this approach has mostly been applied to known antibiotic classes, but applying this approach to novel antibacterial leads targeting novel targets could be an important strategy of overcoming the challenge that faces traditional lead optimization programs in designing molecules that optimally penetrate bacterial cells. Discovery work in this area has been limited and focused only on hijacking iron uptake, but there are numerous other transporters to evaluate. An analysis of the genomes of 11 different gram-positive agents showed that 13–18% of their genes encode transport proteins (12), and a concerted discovery strategy to exploit these systems with novel pharmacophores is needed. The European Innovative Medicines Initiative established a consortium called TRANSLOCATION (www.imi.europa.eu/projects-results/project-factsheets/translocation) that is researching passive and active penetration of small molecules into gram-negative bacteria to identify rules of penetration and increase our knowledge of transporters that may provide a source for future exploration (13). Active transport is likely to have a reasonable chance of success, especially given that fosfomycin and cefiderocol have somewhat validated this approach. The challenge will be to identify a transporter that can be exploited by a common method that is transferable to several antibiotics that have different mechanisms with minimal chemical optimization. This approach becomes less attractive if it requires significant customization and optimization to build the siderophore recognition features into the backbone pharmacophore of each antibiotic.
Vaccines and Their Role in AMR
There are several reasons to consider vaccines among the most promising preventative methods to address the challenges of AMR. First, vaccines can directly prevent infections caused by devastating AMR pathogens. In addition, they indirectly decrease the use of antibiotics by reducing the symptoms that usually trigger the use of antibiotics. Finally, the use of vaccines prevents the proliferation of bacteria, which do not reach the high numbers necessary for the establishment of resistant mutations (14).
Existing vaccines can already prevent infections caused by AMR pathogens, such as Streptococcus pneumoniae and Haemophilus influenzae. In addition, there are many vaccines in the clinical developmental phase that have the potential to prevent infections due to the major AMR bacterial pathogens, including Mycobacterium tuberculosis, Salmonella typhi, P. aeruginosa, S. aureus, pathogenic Escherichia coli, and Clostridium difficile. In the past decade, scientific advancement in the fields of immunology, genetics, structural biology, and microbiology has allowed the development of vaccine technologies with the potential to greatly improve the probability of success of novel vaccines to prevent infections caused by AMR pathogens (15).
History and Potential of Vaccine Development
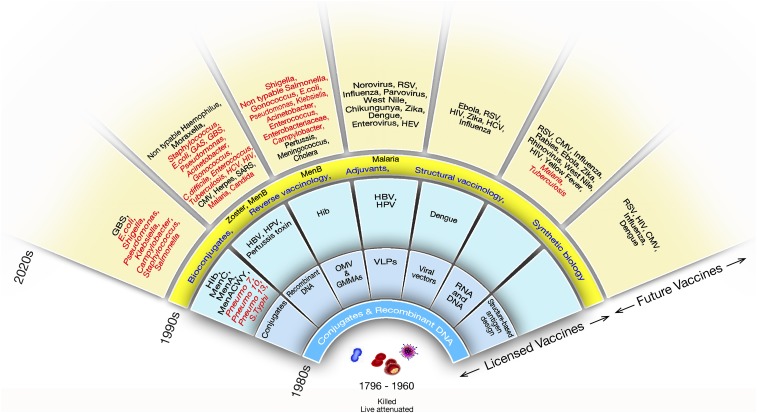
From Jenner in 1796 and Pasteur in the 1880s, to Maurice Hilleman in the 1960s, many life-saving vaccines were made using basic technologies that relied on growing bacteria, viruses, and parasites and on developing vaccines by killing them, attenuating them, or purifying some components from them. The vaccine field changed completely in the 1980s when recombinant DNA and glycoconjugation made it possible to develop vaccines that were impossible with previous technologies (Fig. 4).
Fig. 4.
Evolution of vaccine technologies and platforms. CMV, cytomegalovirus; GAS, group A Streptococcus; GBS, group B Streptococcus; HBV, hepatitis B virus; HCV, hepatitis C virus; HEV, hepatitis E virus; Hib, type B Haemophilus influenzae; HIV, human immunodeficiency virus; HPV, human papillomavirus; MenA, meningococcus A; MenACWY, meningococcus ACWY; MenB, meningococcus B; MenC, meningococcus C; Pneumo, pneumococcus; RSV, respiratory syncytial virus; SARS, severe acute respiratory syndrome; VLP, virus-like particles.
Recombinant DNA.
In the late 1970s, recombinant DNA allowed the production of hepatitis B vaccine in yeast, thus allowing the large-scale production of a vaccine that can eliminate the infection and the related liver cancer worldwide. In addition to the easy production of recombinant antigens, recombinant DNA allowed the manipulation of bacteria and viruses; during the next two decades, this enabled the development of novel platforms to develop and produce vaccines. These include the possibility to manipulate gram-negative bacteria to produce engineered outer membrane vesicles [OMVs; also named generalized modules for membrane antigens (GMMAs)], to make recombinant virus-like particles, and to engineer viral vectors.
Glycoconjugation.
A second technological revolution of the 1980s was the development of glycoconjugate vaccines composed of bacterial polysaccharides covalently linked to a protein carrier. This technology allowed the development of vaccines against H. influenzae type B; the development of 7-, 10-, and 13-valent vaccines against pneumococcus; and tetravalent vaccines against meningococcus. These vaccines alone have been able to prevent the deaths and hospitalization of millions of children during the past three decades. Today, the conjugation technology is looking at other targets, such as group B Streptococcus to prevent neonatal sepsis, E. coli to prevent urinary tract infections and sepsis, and Shigella to prevent bloody diarrhea and typhoid and paratyphoid fever.
Fig. 4 shows that new waves of technological innovation arrived in the 1990s. Some of the most important came from genomics, adjuvants, synthetic biology, and structural biology. Thanks to these technologies, 22 novel vaccines have been licensed since 1980s. Most of these vaccines represent a revolution in science and technology and had a huge impact on health globally. However, so far, we have just barely scratched the surface of the power of the new technologies. With a few exceptions, such as HIV, hepatitis C virus, tuberculosis, chronic infections, and perhaps universal influenza vaccines, all of the other vaccines shown in the outer circle of Fig. 4, in theory, can be developed using the technologies that are available today. Unfortunately, most of them will not be developed unless a sustainable market for them becomes available. The vaccines that can be technically developed include those against all major antimicrobial-resistant bacteria that are shown in red in Fig. 4. Below are some in-depth descriptions of the most promising technologies we have today.
Reverse vaccinology.
The ability to sequence the genome of pathogens allowed the setup of computer and experimental screening programs to discover genome-encoded antigens that would be very difficult or impossible to discover using conventional technologies (16). This technology was named reverse vaccinology because it was the first time in the two centuries of vaccine history that scientists discovered new vaccines starting from the information in a computer instead of starting from growing microorganisms. The development of the recently licensed four-component meningococcus B vaccine (4CMenB) has demonstrated that reverse vaccinology is an efficient way to select bacterial antigens that can provide broad protection against antigenically variable bacterial pathogens. In essence, the antigen selection process starts from the analysis of the genomic information on circulating strains. Thanks to the development of deep-sequencing technologies, this information is more complete than in the past, and it is now available for a large number of bacterial pathogens. Candidate antigens, selected based on sequence conservation and predicted surface exposure, are then expressed as recombinant proteins and tested in preclinical models for their ability to induce functional antibodies. Finally, more promising candidates are combined to achieve the best possible coverage based on molecular epidemiology data. In the case of 4CMenB, three recombinant antigens (fHbp, NadA, and NHBA) inducing serum bactericidal antibodies against strains expressing a conserved version of each respective protein have been combined with the OMV from New Zealand meningococcus B strain, inducing antibodies able to kill the strains expressing a conserved version of the outer membrane protein PorA. The 4CMenB has recently been introduced in the United Kingdom to all infants, demonstrating 82.9% effectiveness against all MenB strains (17). The success of 4CMenB suggests that a similar antigen selection approach can be applied to AMR bacterial pathogens, such as P. aeruginosa, S. aureus, E. coli, Acetinobacter, Shigella, Klebsiella, and gonococcus.
Vaccine adjuvants.
From 1924 to 1997, aluminum salts had been the only adjuvants licensed for human use. Improved knowledge of the molecular and cellular mechanisms of innate immunity has led to the discovery of new, more effective vaccine adjuvants with the ability to improve the speed, potency, and persistency of the immune response to vaccination. In 1997, an oil-in-water emulsion named MF59 was licensed to improve conventional influenza vaccines. Adjuvanted influenza vaccines showed 22% improved efficacy in the elderly and increased protection in children from 43 to 86% (18, 19). Since then, several novel adjuvants named AS01, AS03, and AS04 and synthetic oligonucleotides targeting Toll-Like Receptor 9 (TLR9) have been licensed as part of innovative vaccines (20). These include more powerful influenza vaccines, a vaccine against human papilloma virus, a vaccine against malaria, a vaccine against herpes zoster that has shown remarkable efficacy in elderly people, and an improved vaccine against hepatitis B. Probably, the most notable example is the adjuvant AS01, recently licensed for a vaccine against malaria and for a novel herpes zoster vaccine (21, 22). This adjuvant is composed of two immunostimulants, the saponin QS21 and monophosphoryl lipid A, targeting TLR4; both compounds are formulated in liposomes. While adjuvants so far have been licensed only for improved viral vaccines, such as influenza, and already provide an indirect contribution to AMR, preclinical data suggest that adjuvants will contribute directly to AMR in the near future by providing new vaccines against bacterial infections. For instance, AS01 has been used as a component of two experimental vaccines targeting bacterial pathogens in clinical testing: a vaccine to prevent reactivation of tuberculosis and a vaccine targeting nontypeable H. influenzae and Moraxella catharralis to prevent exacerbation in chronic obstructive pulmonary disease (COPD) (https://clinicaltrials.gov/ct2/show/NCT03281876?cond=COPD+vaccine&rank=6). Data generated in humans support the use of AS01 to improve responses to other vaccines based on bacterial recombinant proteins, particularly in the elderly. Additional adjuvants targeting the innate immune receptors TLR9 (DNA oligonucleotides) and TLR5 (Flagellin) have been tested in humans. A novel small-molecule adjuvant targeting TLR7 has been extensively tested in preclinical models with very promising results and is in the early clinical stage (23, 24). Other adjuvants targeting surface-exposed (TLR1/2), endosomal (TLR3), or cytoplasmic (Rig I, DNA sensors) innate immune receptors have been successfully tested in preclinical models and will be probably tested soon in humans. In the future, vaccine developers may have the opportunity to choose among different, clinically tested, adjuvant compounds tailored to the immune response that is needed to prevent the target infection.
Structural vaccinology.
The possibility to isolate protective human monoclonal antibodies (mAbs), combined with the ability to determine the structure of antigens and antigen–antibody complexes, opened a very promising new field in which the structural information can be used to engineer improved antigens (25). Although in its infancy, this technology has already delivered a revolutionary antigen for respiratory syncytial virus (RSV) by stabilizing the F protein of RSV in the prefusion conformation (26). This had been an impossible task using conventional technologies so far. Similar approaches are being used to improve the outlook for HIV and to develop universal vaccines against influenza. Human mAbs have been key to identify novel protective antigens, such as the CMV pentameric complex (27), and the prefusion conformation of the RSV F protein (26). Although most antigen design applications have been targeted to viral pathogens, the same technologies may also be applied to bacteria, allowing for identification of novel bacterial vaccine antigens with improved cross-protection among bacterial strains. Furthermore, advances in protein engineering now allow us to display recombinant antigens in multimeric, highly ordered structures using various protein scaffolds (28). Antigen display on nanoparticles can be rationally designed to optimize protein density and formation of the correct multimers in the correct conformation. The resulting protein nanoparticles are more immunogenic compared with soluble recombinant proteins and can be combined with novel adjuvants to obtain optimal antibody titers.
Bioconjugates.
Existing glycoconjugate vaccines require complex manufacturing based on the isolation of polysaccharides from the pathogens, followed by a nonselective chemical conjugation to a recombinant protein carrier. This makes the process expensive and unsuitable for very complex vaccines or for low-income countries. These limitations can be overcome by new bacterial metabolic engineering technologies that now allow the production of conjugate vaccines in one step by an engineered E. coli strain. The conjugation process occurs in the bacterial cell itself using the enzyme PglB, and it is site specific, preserving most of the antigenic properties of the carrier. In summary, through this new process called bioconjugation, a glycoconjugate vaccine can be produced in a single fermentation step and it is possible to use a protective antigen as a carrier without interfering with protective epitopes. Bioconjugates may play a fundamental role in the future to prevent infection caused by dangerous AMR pathogens expressing polysaccharide antigens, such as S. typhi, S. aureus, P. aeruginosa, K. pneumoniae, and Acinetobacter baumannii (29, 30).
Genetically modified OMV.
A vaccine based on a homologous bacterial OMV has been administered to effectively prevent meningococcus B infections during an outbreak in New Zealand in 2004/2008. The same New Zealand OMV is used as a component of a licensed 4CMenB vaccine. Recently, it has been shown that the immunization with New Zealand OMV resulted in a 31% reduction of gonococcal infections, one of the most prevalent and difficult to treat AMR bacteria (31). These data highlight the potential of OMV-based vaccines to prevent infection by AMR bacteria in the future. The New Zealand OMV is produced through detergent extraction from bacterial culture, is very immunogenic, and induces antibodies that kill bacteria through a complement-mediated mechanism. Interestingly, bacterial genetics have been successfully used to produce a new generation of improved bacterial OMVs. By metabolic engineering gram-negative bacteria, it is possible to induce the formation of naive OMVs without using detergents. The key to induce OMV release (overblebbing) is to target genes (e.g., tolR, tolB, nlpl) responsible for stabilizing the link between the bacterial outer membrane and peptidoglycan. Naive vesicles have the natural content of bacterial surface-exposed proteins in the correct conformation, and therefore have the potential to be more protective when used as vaccine components. In addition, genetic manipulation can be used to reduce LPS reactogenicity (by targeting genes responsible for LPS acetylation, such as lpxlM and lpxL1) and to overexpress protective antigens (32). Recently, this approach has been exploited to produce a novel vaccine based on Shigella OMV that induced high antibody titers against the LPS in preclinical models and clinical trials (33).
Nonconventional Technologies and Approaches
As in other therapy areas, consideration needs to be given to alternative approaches and modalities to address bacterial infections. Several promising approaches are highlighted below, and others are also under consideration (13).
Phage Therapy.
Phages are viruses that infect bacteria and are the most prominent life form on the planet. They were being used as antibiotics before penicillin was even discovered. There are many types of phage viruses, but the common approach for bacterial therapy involves lytic phages, which are phages that result in cell lysis, and thus death of the bacterial cell. Phages bind to bacteria, inject their DNA or RNA, and use the host enzymes and cofactors to replicate. Lytic phages contain a lysin enzyme that digests bacterial proteins, causing terminal damage. One advantage of using phages for treatment is that a very large number of phages can be administered in a very small dose and the bacteria causing the infection will replicate and produce more of the phages in situ. Also, phages are specific for bacteria, so there is no impact on mammalian cells. Unfortunately, their specificity is also their major limitation. One phage will typically target only a limited number of bacterial strains (e.g., a subset of strains of E. coli), which means multiple different phages are needed to treat one species. Given the lack of rapid diagnostics, dictating the need for empiric therapy, an impracticable number of different phages would currently be needed to cover all of the potential species of bacteria that could be involved in common infections like community-acquired bacterial pneumonia, urinary tract infections, and intraabdominal infections, for example. This limits phages to a later- or last-line therapy once the infecting organism is identified. Researchers are investigating the potential to broaden the specificity of phages (34), which could reduce the number of phages needed per species and transform this approach. The next major challenge is how to get the phages to the site of action since phages do not have favorable PK properties. To minimize these challenges, one group is developing a phage against P. aeruginosa to treat cystic fibrosis, where the phages are administered directly to the lungs by inhalation, and also a phage mixture to be taken orally for the treatment of C. difficile infection in the gastrointestinal tract (35, 36). These are compelling ideas, but efficacy in robust random controlled trials is needed to conclusively define the utility of the approach. Several studies to treat a variety of bacterial infections with phages delivered by oral, parenteral transdermal, topical, intranasal, and otic routes were recently reviewed (37). Another intriguing approach is to use phages as a delivery system, with one research group currently investigating the use of bacteriophage to deliver CRISPR CAS3 genes directly into bacteria as an alternative to antibiotics (www.locus-bio.com/). Phages have teased us with their antibiotic potential for decades, but no treatment has been approved, which can be interpreted as a signal for a low chance of success. However, we consider a breakthrough technology could quickly reverse this struggle and rapidly open this field for exploiting, although it is not clear what this technology will be and when it will be available.
Microbiome.
Over the past decade, it has become well accepted that the human gut microbiome has a major impact on the health and stability of the host. Broad-spectrum traditional antibiotics can eliminate large portions of the commensal bacteria, which can allow opportunistic pathogenic bacteria to establish in some circumstances. The most well-known example of this is infection by C. difficile (causing diarrhea, leading to dehydration and possibly death), which can follow broad-spectrum antibiotic therapy. C. difficile can be treated with antibiotics, such as metronidazole or vancomycin, but spores can remain in the gastrointestinal tract such that when antibiotic therapy stops, the spores can germinate and reinfect. Recurrent C. difficile is a challenging infection to treat, but there are approaches in development to restore the microbiome. Unpleasant, but successful, fecal matter transplants have been used to restore the microbiome of patients with C. difficile. The concept is to use a donor fecal sample (e.g., from a spouse) to colonize a patient’s GI flora such that the fecal bacteria recolonize the patient faster than the C. difficile spores can reestablish an infection. This procedure is quite successful and has encouraged research groups to identify better, more controlled methods to reestablish a patient’s microbiome. In one approach, a mixture of spores from several key bacteria isolated from healthy donor fecal samples is administered orally, and although initial efficacy was impressive, subsequent clinical trials did not confirm the initial result (38). In another approach, a single strain of a nontoxinogenic C. difficile (NTCD) is administered that can outcompete the infecting toxic C. difficile. Since the NTCD is not toxic to the host, the host can recover and the microbiome will eventually reestablish (39). In addition, groups are creating treatments that consist of a defined combination of bacterial strains from different species believed to be critical for a healthy microbiome that is resilient to reinfection or infection by C. difficile following treatment with broad-spectrum antibiotics (40). These approaches still require definitive phase 3 success, but progress is encouraging and this approach may also work to decolonize MDR pathogens from patients. The microbiome is likely to provide a rich source of therapies for a wide range of diseases. For AMR, we consider it highly likely that a microbiome treatment for C. difficile infection in the gastrointestinal tract will be available in the near term. The lung microbiome may also offer opportunity to prevent respiratory infections, such as HAP, ventilator-associated pneumonia, and Pseudomonas in cystic fibrosis.
Host Modulation.
Beyond agents that act on bacteria or bacterial products, research is building toward improving the host response to fight bacteria. As more and more is being discovered about the host immune and inflammation systems, opportunities arise for intervention to strengthen the host response to bacterial infection in situations of a weak immune response, or to dial-down the host response where it has overreacted (41). Computational methods are being used to research and filter through genome data to identify potential host targets that may impact bacterial infections, providing researchers with alternative targets to assess, as well as potential repurposing opportunities (42). Immunotherapeutics are thriving in other therapeutic areas but are relatively unexplored for bacterial infections; this has the potential to create a new generation of more effective and potentially resistance-proof antibacterial medicines, although there may be a need to use them in combination with traditional antibiotics. Host modulation offers great potential, but it will likely take a long time to realize its true opportunity since no targets are validated for bacterial infection. Breakthroughs may possibly come from fortuitous findings from host modulation agents under development for other indications that are found to strengthen the patient’s ability to resist infections compared with control patients. If a protective effect is recognized, a more robust approach will be required to demonstrate a conclusive connection between host modulation and treatment of bacterial infections so as to validate the target; dedicated trials will then be needed to repurpose such drugs, and new research programs must be initiated to identify more suitable compounds.
Monoclonal Antibodies.
Given that antibodies can target bacteria, it is not surprising that significant effort has been applied to develop antibody therapy as an alternative to antibiotics. Antibody therapy has been successfully applied to other therapeutic areas, such as inflammatory diseases, and broadly neutralizing monoclonal antibodies (mAbs) are recently having an impact on HIV research (43). One key challenge for antibodies to treat bacterial infections is that antibodies have a limited spectrum and target only a small number of strains of a specific species, whereas an antibody developed as an antibiotic must be capable of targeting a range of bacteria. New technology has allowed researchers to modify antibodies to increase the ability of a single antibody to bind two targets, a bispecific mAb, and to reduce the overall number of mAbs needed to treat an infection. This approach was demonstrated by Medimmune through the development of a bispecific mAb to target P. aeruginosa targets Psl and PcrV (44). Another approach is to target virulence factors (protein or toxin) that the bacteria produce such that they are no longer pathogenic to the host (45, 46). Significant research has led to a wealth of candidate antibodies for various bacterial infections, which have been reviewed (13, 47).
Some of the challenges that currently face this approach are the conduct of clinical trials, which mostly focus on treating a specific pathogen, and the cost of goods of mAbs. The use of mAbs to protect against many hospital-acquired infections, such as Acinetobacter, P. aeruginosa, or K. pneumoniae, in high-risk patients may be a more pragmatic approach than vaccination for these pathogens, where infection can be a relatively rare event in most people’s lives until they enter a higher risk environment, such as the intensive care unit (48). As antibody engineering and diagnostics technologies advance, coupled with manufacturing approaches that decrease the cost of goods, this could become an area of significant growth in the future. Of all of the approaches described, mAbs probably have the best opportunity to successfully treat AMR, although each mAb treatment will probably be limited to a specific species of bacteria. This will mean these treatments will be reserved for second- or third-line therapy once the infecting organism has been identified. The downside is each mAb will need to have its own clinical development path, which will greatly increase the cost of development. Technology or regulatory changes that reduce the cost or accelerate the development path will certainly catalyze the further development of mAbs.
Conclusions
AMR is eroding our ability to control infections with traditional antibiotics, and there are scientific challenges to develop new treatments at an equivalent rate. These challenges include the need to kill rapidly growing organisms that are adept at keeping out xenobiotics, lack of rapid diagnostics leading to empirical treatment of infections, and a need to administer high doses to cover worst-case scenarios. However, new innovations in vaccines and antibacterial approaches have potential to provide new tools to address this public health threat. Clearly, broader vaccination programs can play a bigger role in preventing bacterial infections and innovative platforms are available to create new vaccines against additional pathogens of concern. Furthermore, innovative approaches need to be explored for traditional small-molecule antibacterial discovery programs, and alternative approaches need to be robustly validated and progressed. Together, vaccines and antibiotics have played a key role in our ability to manage bacterial infections, which has enabled the advancement of medical science. However, this progress has been at risk for some time, despite the underpinning science and platforms that can address this global threat being available. Significant and coordinated investment is needed to broaden the application of innovative vaccine platforms to additional pathogens and to expand research around novel approaches that will improve the success of traditional and alternative antibacterial discovery. In conclusion, we believe that a coordinated effort in research and development of new antibiotics and vaccines that takes advantage of the opportunities provided by the new technologies, combined with appropriate policy measures, can greatly advance our ability to control AMR.
Acknowledgments
We thank Kelly Aubart, Nicole Scangarella-Oman, and Stephen Rittenhouse for their help in reviewing the manuscript and providing constructive comments, and Catherine Mallia for editorial assistance.
Footnotes
Conflict of interest statement: The authors are employees of GlaxoSmithKline.
This article is a PNAS Direct Submission.
References
- 1.Centers for Disease Control and Prevention 2013 Antibiotic resistance threats in the United States, 2013. Available at https://www.cdc.gov/drugresistance/threat-report-2013/index.html. Accessed August 1, 2018.
- 2.Rhomberg PR, Jones RN. Summary trends for the meropenem yearly susceptibility test information collection program: A 10-year experience in the United States (1999-2008) Diagn Microbiol Infect Dis. 2009;65:414–426. doi: 10.1016/j.diagmicrobio.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y-Y, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 4.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 5.Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov. 2015;14:529–542. doi: 10.1038/nrd4572. [DOI] [PubMed] [Google Scholar]
- 6.Richter MF, et al. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature. 2017;545:299–304. doi: 10.1038/nature22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong J, et al. Optimal dosing regimen of biapenem in Chinese patients with lower respiratory tract infections based on population pharmacokinetic/pharmacodynamic modelling and Monte Carlo simulation. Int J Antimicrob Agents. 2016;47:202–209. doi: 10.1016/j.ijantimicag.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 8.Muller AE, Schmitt-Hoffmann AH, Punt N, Mouton JW. Monte Carlo simulations based on phase 1 studies predict target attainment of ceftobiprole in nosocomial pneumonia patients: A validation study. Antimicrob Agents Chemother. 2013;57:2047–2053. doi: 10.1128/AAC.02292-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cojutti P, Pai MP, Pea F. Population pharmacokinetics and dosing considerations for the use of linezolid in overweight and obese adult patients. Clin Pharmacokinet. 2017;57:989–1000. doi: 10.1007/s40262-017-0606-5. [DOI] [PubMed] [Google Scholar]
- 10.Stanford Health Care 2017 Stanford Health Care antimicrobial dosing reference guide 2017. Available at https://www.scribd.com/document/368426639/Stanford-Health-Care-Antimicrobial-Dosing-Reference-Guide-2017. Accessed August 1, 2018.
- 11.Bronson J, Black A, Dhar M, Ellsworth B, Merritt JR. To market, to market-2013. Annu Rep Med Chem. 2014;49:437–508. [Google Scholar]
- 12.Lorca GL, et al. Transport capabilities of eleven Gram-positive bacteria: Comparative genomic analyses. Biochim Biophys Acta. 2007;1768:1342–1366. doi: 10.1016/j.bbamem.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czaplewski L, et al. Alternatives to antibiotics–A pipeline portfolio review. Lancet Infect Dis. 2016;16:239–251. doi: 10.1016/S1473-3099(15)00466-1. [DOI] [PubMed] [Google Scholar]
- 14.Rappuoli R, Bloom DE, Black S. Deploy vaccines to fight superbugs. Nature. 2017;552:165–167. doi: 10.1038/d41586-017-08323-0. [DOI] [PubMed] [Google Scholar]
- 15.Delany I, Rappuoli R, De Gregorio E. Vaccines for the 21st century. EMBO Mol Med. 2014;6:708–720. doi: 10.1002/emmm.201403876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delany I, Rappuoli R, Seib KL. Vaccines, reverse vaccinology, and bacterial pathogenesis. Cold Spring Harb Perspect Med. 2013;3:a012476. doi: 10.1101/cshperspect.a012476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parikh SR, et al. Effectiveness and impact of a reduced infant schedule of 4CMenB vaccine against group B meningococcal disease in England: A national observational cohort study. Lancet. 2016;388:2775–2782. doi: 10.1016/S0140-6736(16)31921-3. [DOI] [PubMed] [Google Scholar]
- 18.McElhaney JE, et al. Influence65 study group AS03-adjuvanted versus non-adjuvanted inactivated trivalent influenza vaccine against seasonal influenza in elderly people: A phase 3 randomised trial. Lancet Infect Dis. 2013;13:485–496. doi: 10.1016/S1473-3099(13)70046-X. [DOI] [PubMed] [Google Scholar]
- 19.Vesikari T, et al. Oil-in-water emulsion adjuvant with influenza vaccine in young children. N Engl J Med. 2011;365:1406–1416. doi: 10.1056/NEJMoa1010331. [DOI] [PubMed] [Google Scholar]
- 20.Reed SG, Orr MT, Fox CB. Key roles of adjuvants in modern vaccines. Nat Med. 2013;19:1597–1608. doi: 10.1038/nm.3409. [DOI] [PubMed] [Google Scholar]
- 21.Otieno L, et al. Safety and immunogenicity of RTS,S/AS01 malaria vaccine in infants and children with WHO stage 1 or 2 HIV disease: A randomised, double-blind, controlled trial. Lancet Infect Dis. 2016;16:1134–1144. doi: 10.1016/S1473-3099(16)30161-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunningham AL, et al. ZOE-70 Study Group Efficacy of the herpes zoster subunit vaccine in adults 70 years of age or older. N Engl J Med. 2016;375:1019–1032. doi: 10.1056/NEJMoa1603800. [DOI] [PubMed] [Google Scholar]
- 23.Wu TY, et al. Rational design of small molecules as vaccine adjuvants. Sci Transl Med. 2014;6:263ra160. doi: 10.1126/scitranslmed.3009980. [DOI] [PubMed] [Google Scholar]
- 24.Mancini F, et al. One dose of Staphylococcus aureus 4C-Staph vaccine formulated with a novel TLR7-dependent adjuvant rapidly protects mice through antibodies, effector CD4+ T cells, and IL-17A. PLoS One. 2016;11:e0147767. doi: 10.1371/journal.pone.0147767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rappuoli R, Bottomley MJ, D’Oro U, Finco O, De Gregorio E. Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J Exp Med. 2016;213:469–481. doi: 10.1084/jem.20151960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLellan JS, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macagno A, et al. Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128-131A complex. J Virol. 2010;84:1005–1013. doi: 10.1128/JVI.01809-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanekiyo M, et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature. 2013;499:102–106. doi: 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ihssen J, et al. Production of glycoprotein vaccines in Escherichia coli. Microb Cell Fact. 2010;9:61. doi: 10.1186/1475-2859-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riddle MS, et al. Safety and immunogenicity of a candidate bioconjugate vaccine against Shigella flexneri 2a administered to healthy adults: A single-blind, randomized phase I study. Clin Vaccine Immunol. 2016;23:908–917. doi: 10.1128/CVI.00224-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petousis-Harris H, et al. Effectiveness of a group B outer membrane vesicle meningococcal vaccine against gonorrhoea in New Zealand: A retrospective case-control study. Lancet. 2017;390:1603–1610. doi: 10.1016/S0140-6736(17)31449-6. [DOI] [PubMed] [Google Scholar]
- 32.Gnopo YMD, Watkins HC, Stevenson TC, DeLisa MP, Putnam D. Designer outer membrane vesicles as immunomodulatory systems–Reprogramming bacteria for vaccine delivery. Adv Drug Deliv Rev. 2017;114:132–142. doi: 10.1016/j.addr.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Launay O, et al. Safety profile and immunologic responses of a novel vaccine against Shigella sonnei administered intramuscularly, intradermally and intranasally: Results from two parallel randomized phase 1 clinical studies in healthy adult volunteers in Europe. EBioMedicine. 2017;22:164–172. doi: 10.1016/j.ebiom.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fairhead H, Wilkinson A, Severi E. 2017. US Patent WO 2017/174809, A1.
- 35.Sahota JS, et al. Bacteriophage delivery by nebulization and efficacy against phenotypically diverse Pseudomonas aeruginosa from cystic fibrosis patients. J Aerosol Med Pulm Drug Deliv. 2015;28:353–360. doi: 10.1089/jamp.2014.1172. [DOI] [PubMed] [Google Scholar]
- 36.Nale JY, et al. Bacteriophage combinations significantly reduce Clostridium difficile growth in vitro and proliferation in vivo. Antimicrob Agents Chemother. 2015;60:968–981. doi: 10.1128/AAC.01774-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rastogi V, et al. An overview on bacteriophages: A natural nanostructured antibacterial agent. Curr Drug Deliv. 2018;15:3–20. doi: 10.2174/1567201813666160406115744. [DOI] [PubMed] [Google Scholar]
- 38.Khanna S, et al. A novel microbiome therapeutic increases gut microbial diversity and prevents recurrent Clostridium difficile infection. J Infect Dis. 2016;214:173–181. doi: 10.1093/infdis/jiv766. [DOI] [PubMed] [Google Scholar]
- 39.Gerding DN, et al. Administration of spores of nontoxigenic Clostridium difficile strain M3 for prevention of recurrent C. difficile infection: A randomized clinical trial. JAMA. 2015;313:1719–1727. doi: 10.1001/jama.2015.3725. [DOI] [PubMed] [Google Scholar]
- 40.Vedanta Biosciences 2017 Vedanta Biosciences Announces Initiation of Phase 1a/1b Trial for New Drug Class of Rationally-Defined Bacterial Consortia Derived from the Human Microbiome. Available at https://www.vedantabio.com/news-media/press-releases/detail/2298/vedanta-biosciences-announces-initiation-of-phase-1a1b. Accessed August 1, 2018.
- 41.Pirofski LA, Casadevall A. Immunomodulators as an antimicrobial tool. Curr Opin Microbiol. 2006;9:489–495. doi: 10.1016/j.mib.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith SB, Magid-Slav M, Brown JR. Host response to respiratory bacterial pathogens as identified by integrated analysis of human gene expression data. PLoS One. 2013;8:e75607. doi: 10.1371/journal.pone.0075607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pegu A, Hessell AJ, Mascola JR, Haigwood NL. Use of broadly neutralizing antibodies for HIV-1 prevention. Immunol Rev. 2017;275:296–312. doi: 10.1111/imr.12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DiGiandomenico A, et al. A multifunctional bispecific antibody protects against Pseudomonas aeruginosa. Sci Transl Med. 2014;6:262ra155. doi: 10.1126/scitranslmed.3009655. [DOI] [PubMed] [Google Scholar]
- 45.Kummerfeldt CE. Raxibacumab: Potential role in the treatment of inhalational anthrax. Infect Drug Resist. 2014;7:101–109. doi: 10.2147/IDR.S47305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lowy I, et al. Treatment with monoclonal antibodies against Clostridium difficile toxins. N Engl J Med. 2010;362:197–205. doi: 10.1056/NEJMoa0907635. [DOI] [PubMed] [Google Scholar]
- 47.Hey A. History and practice: Antibodies in infectious diseases. Microbiol Spectr. 2015;3:AID-0026-2014. doi: 10.1128/microbiolspec.AID-0026-2014. [DOI] [PubMed] [Google Scholar]
- 48.Bagnoli F, Payne DJ. Reaction: Alternative modalities to address antibiotic-resistant pathogens. Chem. 2017;3:369–372. [Google Scholar]