Significance
To control adaptive responses to the ever-changing environment that plants are continuously exposed to, plant cells must integrate a multitude of information to make optimal decisions. Here, we reveal how plants can link information about the cellular energy status with the actual oxygen concentration of the cell to trigger a response reaction to low-oxygen stress. We reveal that oleoyl-CoA has a moonlighting function in an energy (ATP)-dependent signal transduction pathway in plants, and we provide a model that explains how diminishing oxygen availability can initiate adaptive responses when it coincides with a decreased energy status of the cell.
Keywords: low-oxygen stress, integrative signaling, acyl-CoA, ERFVII, ACBP
Abstract
Plant response to environmental stimuli involves integration of multiple signals. Upon low-oxygen stress, plants initiate a set of adaptive responses to circumvent an energy crisis. Here, we reveal how these stress responses are induced by combining (i) energy-dependent changes in the composition of the acyl-CoA pool and (ii) the cellular oxygen concentration. A hypoxia-induced decline of cellular ATP levels reduces LONG-CHAIN ACYL-COA SYNTHETASE activity, which leads to a shift in the composition of the acyl-CoA pool. Subsequently, we show that different acyl-CoAs induce unique molecular responses. Altogether, our data disclose a role for acyl-CoAs acting in a cellular signaling pathway in plants. Upon hypoxia, high oleoyl-CoA levels provide the initial trigger to release the transcription factor RAP2.12 from its interaction partner ACYL-COA BINDING PROTEIN at the plasma membrane. Subsequently, according to the N-end rule for proteasomal degradation, oxygen concentration-dependent stabilization of the subgroup VII ETHYLENE-RESPONSE FACTOR transcription factor RAP2.12 determines the level of hypoxia-specific gene expression. This research unveils a specific mechanism activating low-oxygen stress responses only when a decrease in the oxygen concentration coincides with a drop in energy.
Flooding contributes almost 60% to the worldwide cost and damage to crops provoked by natural disasters (1). Due to heavy precipitation and concomitant waterlogging or flooding events in large areas of the world, climate change will cause plants to be even more frequently exposed to oxygen-limiting conditions (hypoxia) in the near future (2).
In plants, subgroup VII ETHYLENE-RESPONSE FACTOR (ERFVII) transcription factors act as key regulators of hypoxic gene expression (3–6). During nonstress conditions, the ERFVII protein RELATED TO APETALA 2.12 (RAP2.12) is sequestered to the plasma membrane via direct interaction with ACYL-CoA BINDING PROTEIN (ACBP) (3, 7–9). Upon hypoxia, RAP2.12 is released from the plasma membrane and subsequently accumulates in the nucleus (3, 7, 9). Further, the stability of ERFVII proteins is tightly controlled in an oxygen-dependent manner employing the Cys branch of the N-end rule (3, 4). That is, ERFVII protein degradation is prevented under hypoxic conditions when N end rule-assisted degradation is impaired due to oxygen limitation (10). Although the homeostatic regulation of adaptive responses to low-oxygen stress in plants is well investigated (3, 4, 11), the identity of the initial trigger to release RAP2.12 from its membrane docking protein ACBP remains unknown and the existence of multiple signal queues that are integrated into low-oxygen specific responses is likely (12).
ACBPs represent an evolutionarily conserved protein family found in Escherichia coli, yeast, animals, and plants (13, 14) and participate in the regulation of unbound acyl-CoA levels by sequestration and transportation of acyl-CoAs (15, 16). The interaction between members of a protein family capable of reversibly binding acyl-CoAs with the ERFVII proteins RAP2.12 (3, 7) and RAP2.3 (8, 9) provided a first indication that acyl-CoAs can be involved in the release of ERFVII transcription factor protein during hypoxia. We elaborated this mode of action with experiments on RAP2.12 as a representative member of ERFVII transcription factors.
Acyl-CoAs are intermediates in both lipid catabolism and anabolism. In the catabolic pathway, fatty acids are activated in the cytosol by ACYL-CoA SYNTHETASES before their transport into mitochondria or peroxisomes where β-oxidation occurs. In plants, lipid anabolism occurs through two pathways: de novo fatty acid synthesis takes place in plastids and the generated fatty acids can be incorporated into complex lipids within the plastid by the so-called prokaryotic pathway. Alternatively, the fatty acid may be exported from the plastid to the cytosol to become substrate for the eukaryotic lipid biosynthesis pathway in the endoplasmic reticulum (ER). Transport of fatty acids from the plastid, through the cytosol into the ER is mainly mediated via palmitoyl-CoA (C16:0-CoA) and oleoyl-CoA (C18:1-CoA) that are produced from palmitic and oleic acid by the enzyme LONG-CHAIN ACYL-CoA SYNTHETASES (LACS) at the outer plastid membrane in root and shoot tissues (17–20). In addition to their involvement in lipid metabolism, acyl-CoAs are also known to modulate the activity of numerous enzymes, ion channels, and transcription factors in animals and microorganisms (15). Examples of acyl-CoAs directly affecting transcription factor activity by functioning as ligands have been reported for humans (HNF-4α) (21) and E. coli (FadR) (22). For plants, direct involvement of acyl-CoAs controlling transcription factor activity was not demonstrated yet, although an indirect regulatory role has been suggested (23).
Hypoxia has detrimental effects on the plant’s cellular homeostasis, in the first place, because oxidative phosphorylation in mitochondria is reduced, which ultimately leads to a decrease of the cellular energy charge. This results in ATP-consuming metabolic processes being attenuated, including fatty acid synthesis and processing (24). Consequently, the export of newly synthesized fatty acids from the plastid into the cytosol is affected, since the activation to acyl-CoAs is ATP dependent (18, 25). Therefore, an energy-related impact of hypoxia on acyl-CoA levels in the cytosol is expected and investigated here.
In this study, we reveal a combinatory signaling network by which the reduced energy level under low-oxygen stress is integrated into the ERFVII-dependent hypoxic signaling cascade. We show that dynamic responses of C18:1-CoA and C16:0-CoA levels to hypoxia constitute an early molecular trigger, leading to dissociation of the ACBP:RAP2.12 complex thereby activating the molecular low-oxygen response cascade. We describe an integrative signaling mechanism in which adaptive gene expression upon low-oxygen stress results from the specific combination of (i) low-energy triggered release of the transcription factor RAP2.12 from ACBP as mediated by an acyl-CoA signal; and (ii) low-oxygen dependent stabilization of the RAP2.12 protein according to the N-end rule of protein degradation.
Results
C18:1-CoA Promotes Dissociation of the ACBP:RAP2.12 Complex in Vitro and in Vivo.
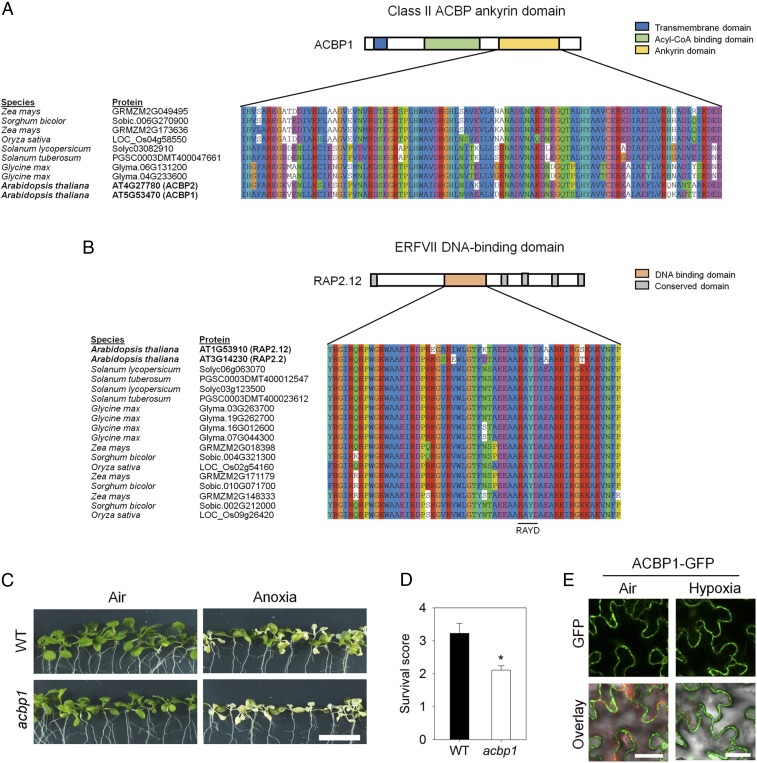
In Arabidopsis, the ERFVII transcription factor RAP2.12 is constitutively expressed but sequestered at the plasma membrane by binding to ACBP1 during normoxic conditions, while it accumulates in the nucleus under oxygen concentrations below 10% (vol/vol) (3, 7). The interaction domains of ACBP1 and RAP2.12 were previously identified in Arabidopsis (3, 26, 27) and appear to be well conserved among plant species (Fig. 1 A and B), indicating that complex formation of both proteins is a general feature in plants. Importantly, changing the expression of ACBP1 affects tolerance to low oxygen (28) (Fig. 1 C and D) similar to what was shown previously for RAP2.12 (3). During hypoxia GFP-tagged ACBP1 remains at the plasma membrane (Fig. 1E), while GFP-tagged RAP2.12 was shown to accumulate in the nucleus upon hypoxia (3, 7). This indicates that RAP2.12 dissociates from ACBP1 before its relocation to the nucleus. Consequently, the release of RAP2.12 from ACBP1 is considered as a trigger that activates adaptive gene expression in response to hypoxia.
Fig. 1.
ACBP is an important element of the low-oxygen stress response pathway in plants. (A) Multiple sequence alignment of class II ACBP ankyrin domain compared with ACBP1 and ACBP2 ankyrin domains in A. thaliana (27). (B) Multiple sequence alignment of ERFVII DNA-binding domain compared with RAP2.12 and RAP2.2 in A. thaliana (26). Sequences of homologous proteins were obtained from Phytozome v12.0 and aligned with Clustal X. (C) Eleven-day-old seedlings of wild type and acbp1 after 9 h of anoxia and 3-d recovery. (Scale bar: 1.0 cm.) (D) Survival scores for wild type and acbp1 after 9-h anoxia and 3-d recovery. Data are mean values ± SD; *P < 0.05, n = 4 (15 seedlings per replicate). (E) Effect of 2 h of hypoxia treatment (1% O2) on the localization of GFP-tagged ACBP1 in epidermal cells. Representative pictures are shown. (Scale bar: 10 µm.)
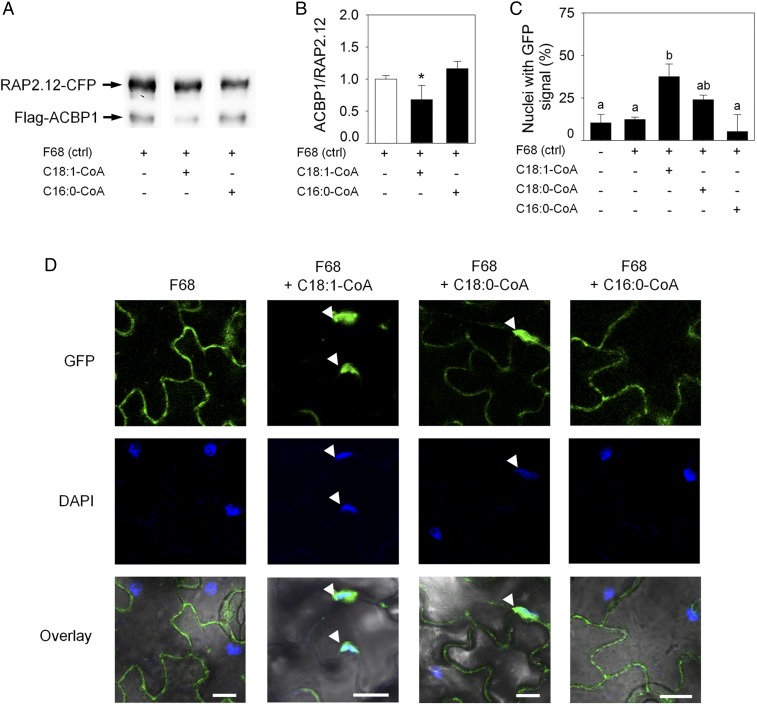
To investigate if acyl-CoAs interfere with the interaction between ACBP1 and RAP2.12, we performed an in vitro affinity assay (SI Appendix, Fig. S1) in the presence of either oleoyl-CoA (C18:1-CoA), which is a preferred substrate for ACBP1, or palmitoyl-CoA (C16:0-CoA) that is not a strongly interacting agent (29, 30). In vitro exposure of an ACBP1:RAP2.12 protein complex to C18:1-CoA, but not C16:0-CoA, significantly decreased the binding affinity between the two proteins as indicated by the reduced ratio of Flag-tagged ACBP1 and CFP-tagged RAP2.12 protein (Fig. 2 A and B). Apparently, interaction between ACBP1 and C18:1-CoA reduces the binding capacity of ACBP1 for the transcription factor RAP2.12. To confirm that C18:1-CoA–induced dissociation of RAP2.12 from ACBP1 also occurs in vivo, we exposed detached leaves of plants expressing 35S:RAP2.12-GFP to various acyl-CoAs under aerobic conditions. Application of C18:1-CoA, but not C18:0-CoA or C16:0-CoA, significantly induced nuclear accumulation of RAP2.12-GFP, indicating that RAP2.12 was released from ACBP1 in vivo after the application of C18:1-CoA, but not of C16:0-CoA (Fig. 2 C and D). Translocation of RAP2.12 to the nucleus was previously described to occur during hypoxic conditions (3, 7). Therefore, we tested if application of acyl-CoA to leaves might have induced hypoxia due to increased beta-oxidation or mitochondrial respiration. However, no increase of the oxygen consumption rate by leaf tissue after treatment with acyl-CoAs was observed, indicating that our experimental treatment did not affect the oxygen concentration of the tissue in this experiment (SI Appendix, Fig. S2). We concluded, remobilization of the transcription factor RAP2.12 from the plasma membrane into the nucleus can be triggered in vivo by increasing the level of C18:1-CoA.
Fig. 2.
Application of C18:1-CoA induces RAP2.12 relocalization into the nucleus. (A) Representative Western blot showing in vitro ACBP1:RAP2.12 complex stability after treatment with C18:1-CoA or C16:0-CoA. Pluronic F68 treatment served as control. (B) Quantification of ACBP1-to-RAP2.12-ratio as shown in A. Data are mean values ± SD *P < 0.05, n = 8. (C) Percentage of epidermal cells with nuclear localization of RAP2.12-GFP after treatment with different acyl-CoAs. Data are mean values ± SD; *P < 0.05. (D) Localization of RAP2.12-GFP in detached leaves incubated with 0.1% C18:1-CoA, C18:0-CoA or C16:0-CoA dissolved in 0.01% pluronic F68 under normoxic conditions for 3 h. Treatment with pluronic F68 only served as negative control. DAPI staining was used to identify nuclei. Arrows indicate nuclei with GFP signal. (Scale bar: 10 µm.)
Acyl-CoAs Provoke Distinct Transcript Responses.
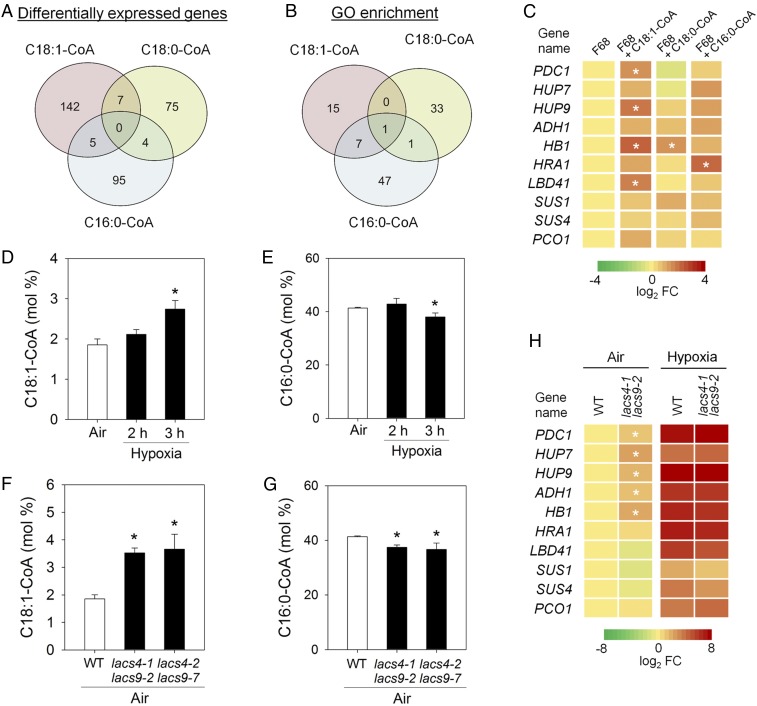
To determine if specific transcriptional responses are provoked by the application of different acyl-CoAs, RNA-Seq transcriptome analysis was performed on wild-type seedlings exposed to either 1 mM C18:1-CoA, C18:0-CoA, or C16:0-CoA. Uptake of these externally applied acyl-CoAs is mediated by ABCD transporters that first cleave the CoA group from the acyl chain, allowing the resulting fatty acid to cross lipid membranes. Once inside the cell, CoA is immediately reattached, which traps the acyl-CoA in the cellular compartment in which it has been imported (31). This analysis revealed that each of these acyl-CoAs modulates distinct sets of genes (Fig. 3A and Dataset S1). The high specificity of induced changes in gene expression underlines the eligibility of acyl-CoAs as signaling molecules in plants. To investigate the biological function of the differentially regulated genes, a Gene Ontology (GO) enrichment analysis (32) was carried out. While application of C18:0-CoA or C16:0-CoA mainly affected the expression of genes related to reproductive development and hormone signaling, C18:1-CoA mainly modulated the expression of genes associated with hypoxia and low-oxygen responses (Fig. 3B and Dataset S2). This result was confirmed by qPCR-assisted expression profiling executed on wild-type plants incubated with C18:1-CoA. Indeed, RAP2.12-regulated hypoxia-response genes were induced by C18:1-CoA treatment, while C18:0-CoA and C16:0-CoA had only minor effects on the expression of these genes (Fig. 3C and SI Appendix, Table S1). This observation is readily explained by our earlier observation that RAP2.12 relocalizes to the nucleus upon C18:1-CoA application (Fig. 2 C and D). Therefore, it is concluded that C18:1-CoA provides a specific cellular signal that is substantially involved in the control of gene expression by releasing RAP2.12 from ACBP1.
Fig. 3.
Changing oleoyl-CoA levels induces low-oxygen responsive gene expression. (A) Number of significantly differentially expressed genes in leaves after 1.5-h treatment with different acyl-CoAs as determined by RNA-Seq (FDR-adjusted P value <0.05). (B) Number of GO classes in which differentially expressed genes are significantly overrepresented under acyl-CoA treatments as shown in A. (C) qPCR analysis of differential expression of hypoxia-responsive genes after acyl-CoA treatment in air (reference: F68 only). Data are presented as mean values *P < 0.05, n = 5. (D) C18:1-CoA levels increase upon hypoxia in wild type. Data are mean values ± SD; *P < 0.05, n = 3. (E) C16:0-CoA levels decrease upon hypoxia in wild type. Data are mean values ± SD; *P < 0.05, n = 3. (F) C18:1-CoA levels are increased in lacs4lacs9 double mutants grown in air. Data are mean values ± SD; *P < 0.05, n = 3. (G) C16:0-CoA levels are lowered in lacs4lacs9 double mutants. Data are mean values ± SD; *P < 0.05, n = 3. (H) Expression data for hypoxia-responsive genes comparing wild type and lacs4-1 lacs9-2 in air or hypoxia (2 h 1% O2; mean values, *P < 0.05, n = 4).
Increase of C18:1-CoA to C16:0-CoA Ratio Induces Hypoxic Gene Expression in Vivo.
In a physiological context, C18:1-CoA–mediated dissociation of RAP2.12 from ACBP1 only makes sense when the endogenous acyl-CoA pool responds to hypoxia. Indeed, HPLC-assisted quantification of acyl-CoAs revealed a shift to elevated C18:1-CoA (Fig. 3D) and C20:0-CoA (SI Appendix, Fig. S3) levels with less C16:0-CoA (Fig. 3E) after 3 h of hypoxia, while no significant changes of the total pool of acyl-CoAs included in our analyses were observed (SI Appendix, Fig. S3). These dynamic responses of specific acyl-CoAs to changing environmental conditions as exemplified here for hypoxia are in line with the suggestion that acyl-CoAs in plants can play a role in stress signaling.
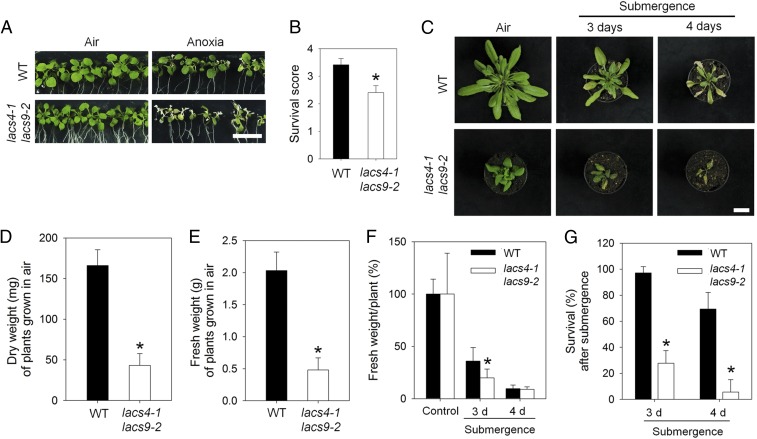
A similar shift of the acyl-CoA pool as observed during low-oxygen conditions was observed in transgenic plants in which two LACS genes, LACS4 and -9, were knocked out (33) (Fig. 3 F and G and SI Appendix, Fig. S4). Under aerobic conditions, lacs4lacs9 double knockout plants have elevated C18:1-CoA and reduced C16:0-CoA levels. To provide further proof that endogenous changes of the C18:1-CoA or C16:0-CoA level can have an effect on low-oxygen responses of plants, we tested the induction of hypoxic gene expression in lacs4lacs9 plants in air. Indeed, many RAP2.12-dependent hypoxia-response genes were activated under aerobic conditions in lacs4lacs9 (Fig. 3H and SI Appendix, Table S2). In addition, also tolerance to hypoxia was altered. Performance of lacs4lacs9 was impaired under both anoxia and submergence conditions compared with wild-type plants (Fig. 4). Similar observations were described for plants with constitutive activation of the hypoxia-stress response pathway (34), although it should be mentioned that the impact of constitutive activation of the ERFVII signaling pathway on plant performance under stress conditions also appears to depend on the experimental growth conditions and the recovery treatment (4, 35). Altogether, our data give rise to a model describing that the control of endogenous acyl-CoA levels through LACS activity is a prerequisite to properly induce hypoxia-tolerance responses in plants.
Fig. 4.
Decreased tolerance of lacs4 lacs9 knockout lines to anoxia and submergence. (A) Eleven-day-old seedlings of wild type and lacs4-1 lacs9-2 after 9 h of anoxia and 3-d recovery. (Scale bar: 1.0 cm.) (B) Survival scores for wild type and lacs4-1 lacs9-2 after 9-h anoxia and 3-d recovery. Data are mean values ± SD; *P < 0.05, n = 4 (15 seedlings per replicate). (C) Phenotype of wild type and lacs4 lacs9 mutant grown in air (control), or after 3- or 4-d submergence-induced hypoxic treatment. (Scale bar: 2 cm.) Photographs were taken 4 d after the submergence treatment. (D) Absolute dry weight of wild-type and lacs4-1 lacs9-2 plants grown in air. Data represent mean ± SD (three replicate experiments with every 12 plants per genotype). Asterisk indicates significant differences after one-way ANOVA (P < 0.05). (E) Absolute fresh weight of wild-type and lacs4-1 lacs9-2 plants grown in air. Data represent mean ± SD (three replicate experiments with every 12 plants per genotype). Asterisk indicates significant differences after one-way ANOVA (P < 0.05). (F) Relative fresh weight of wild-type and lacs4-1 lacs9-2 plants grown in air, or after 3 or 4 d of submergence followed by 4 d of recovery. Data represent mean ± SD (three replicate experiments with every 12 plants per genotype). Asterisk indicates significant differences after one-way ANOVA (P < 0.05). (G) Percentage of plants that survived the 3 or 4 d of flooding-induced hypoxia, respectively (mean values ± SD, three replicate experiments with every 12 plants per genotype). *P < 0.05 according to one-way ANOVA.
LACS Activity Is Reduced by Depletion of ATP.
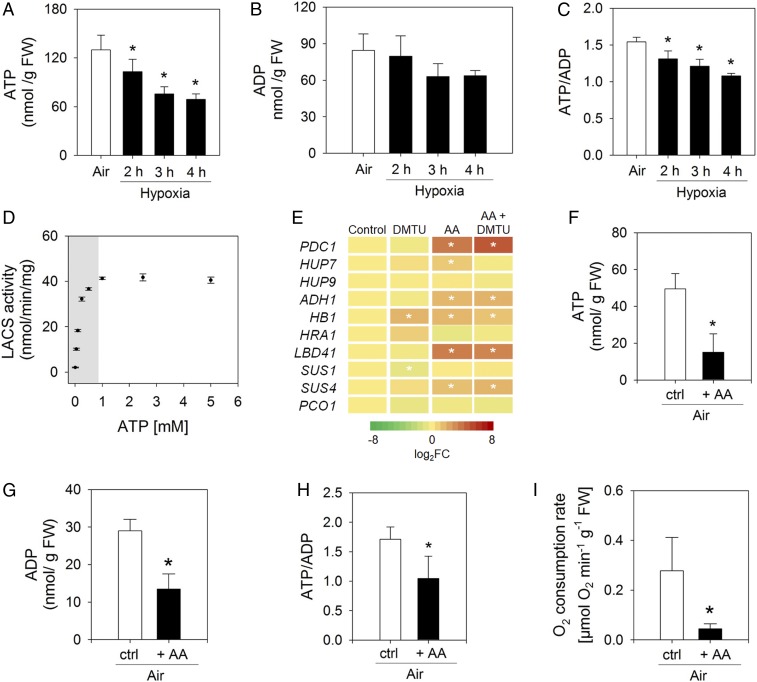
To explain the mechanism by which LACS activity in plants is affected by hypoxic conditions, we analyzed LACS activity in dependence of its substrate ATP. First, we monitored the cellular energy status in wild-type plants grown under hypoxia. Already after 2 h, the ATP level as well as the ATP-to-ADP ratio were significantly reduced compared with the air-treated control (Fig. 5 A–C). To examine at which ATP concentration LACS activity becomes substrate limited, an in vitro LACS activity assay was performed. This experiment showed that the activity of LACS4 was ATP dependent at ATP concentrations below 0.8 mM (Fig. 5D), which is close to the estimated cytosolic ATP concentration under aerobic conditions in planta (36–38). Since ATP concentrations in plant cells decrease during hypoxia (24), LACS activity is expected to diminish concomitantly. However, it should be mentioned that variations of the ATP concentration between tissues or even within a cell (between organelles) might lead to different local ATP-limiting conditions for LACS during hypoxic stress. Moreover, the hypoxia-induced low-energy status will also influence other processes that are not covered by the analyses that we describe here.
Fig. 5.
Decreasing the cellular ATP level constitutes limiting conditions for LACS activity and induces the expression of low-oxygen responsive genes. (A) ATP levels under hypoxia (mean ± SD, *P < 0.05, n = 5). (B) Concentration of ADP in wild-type seedlings grown under long-day conditions and exposed to hypoxia. Data shown are given in nanomoles per gram fresh weight and represent the mean ± SD of independent replicates (n = 5). (C) ATP-to-ADP-ratio under hypoxia (mean ± SD, *P < 0.05, n = 5). (D) In vitro LACS activity depends on ATP concentration (mean ± SD, *P < 0.05, n = 5). The gray area marks the ATP-concentration range usually determined in plant cells. (E) Differential expression of hypoxia-responsive genes after 3 h of 1 mM DMTU and/or 50 µM antimycin-A (AA) treatment under aerobic conditions (reference: mock-treated control). Data are presented as mean ± SD, *P < 0.05, n = 5. (F) ATP levels after 3 h of 50 µM AA treatment (mean ± SD, *P < 0.05, n = 5). (G) Concentration of ADP in wild-type seedlings exposed to 3 h of 50 µM AA treatment. Data represent mean ± SD (n = 5). Asterisk indicates significant differences after one-way ANOVA (P < 0.05). (H) ATP-to-ADP-ratio after 3 h of 50 µM AA treatment (mean ± SD, *P < 0.05, n = 5). (I) Oxygen consumption rate in wild-type leaves upon 3 h of 50 µM AA treatment. Data represent mean ± SD (n = 7). Asterisk indicates significant difference after Student’s t test (P < 0.05).
To verify that a drop in ATP can trigger hypoxia responses of plants in air, we chemically inhibited mitochondrial respiration using antimycin-A. Indeed, after 3 h of 50 µM antimycin-A treatment of rosettes in air, several hypoxia-responsive genes were significantly induced (Fig. 5E and SI Appendix, Table S3), while ATP levels and the ATP-to-ADP ratio were reduced (Fig. 5 F–H). Since application of antimycin-A strongly reduced the oxygen consumption rate of the tissue (Fig. 5I), it is unlikely that the induction of hypoxia-responsive genes is the consequence of low-oxygen concentrations here. Also an antimycin-A–induced ROS burst appeared unlikely to be responsible for the gene induction, since the induced genes responded similarly when in addition to antimycin-A also 1 mM of the hydrogen peroxide scavenger dimethylthiourea (DMTU) (39, 40) was supplied (Fig. 5E). Although it cannot be concluded that reduced ATP levels are exclusively responsible for triggering hypoxia responses in plants without performing dose–response analyses of individual and combined compounds, the data provide evidence that the cellular energy status is involved in the regulation of hypoxic gene expression.
Discussion
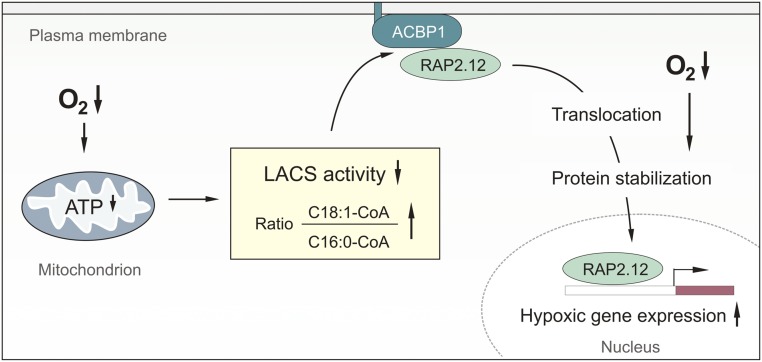
In this study, we demonstrate that acyl-CoAs provoke distinct transcriptional responses in plants, suggesting that they are involved in different signaling pathways. Specifically, binding of C18:1-CoA to ACBP triggers dissociation of the ACBP:RAP2.12 complex upon an hypoxia-induced energy crisis, resulting in mobilization of the transcription factor RAP2.12 into the nucleus. Consequently, RAP2.12-mediated gene expression is induced. Therewith we reveal the trigger of the ERFVII-mediated signaling cascade to activate cellular hypoxia-tolerance responses in plants (Fig. 6).
Fig. 6.
Triggering low-oxygen responses in plants integrates the cellular energy and oxygen status via modulation of oleoyl-CoA levels. Oxygen limitation reduces cellular ATP levels, which results in increased C18:1-CoA levels. Dissociation of ERFVII protein (as shown here for RAP2.12) bound to ACBP1 at the plasma membrane is promoted by C18:1-CoA. Free ERFVII protein is stable under low-oxygen conditions and relocalizes into the nucleus to activate hypoxic responses.
When the oxygen availability to cells diminishes, mitochondria produce less ATP due to a reduced activity of oxidative phosphorylation (41). Indeed, our plants showed a rapid decrease of their energy status upon hypoxia. Already within 2 h, the ATP-to-ADP ratio dropped significantly and remained decreasing throughout the rest of the experiment (Fig. 5C). Consequently, the activity of ATP-mediated reactions within the cell is expected to reduce too (24). Here, we show that ATP concentration-dependent LACS activity reaches its maximum at an ATP concentration of 1 mM (Fig. 5D) which resembles the concentration in a nonstressed plant cell (36–38). This means that any decrease of the cellular energy status is translated into a reduction of LACS activity. The enzyme LACS is among others located in the plastidial outer envelope where it is provided with C16:0 and C18:1 fatty acids from the stroma by thioesterases that are located in the inner envelope (42, 43). Using CoA and ATP as cosubstrates, LACS activates fatty acids and releases them as C16:0-CoA and C18:1-CoA into the cytosol (18). When the level of ATP drops and LACS activity decreases, the export rate of fatty acids will be reduced. As the elongation and rapid desaturation reactions in the plastidial stroma from C16:0 to C18:1 commence, the ratio C18:1 compared with C16:0 that is provided to LACS from the stroma is likely to increase through time under these conditions. As a consequence, the ratio of C18:1-CoA to C16:0-CoA that is released by LACS into the cytosol will increase and would readily explain why we observe an increased level of C18:1-CoA compared with C16:0-CoA in plants that were exposed to low oxygen (Fig. 3 D and E) as well as in lacs4lacs9 double knockout lines in air (Fig. 3 F and G).
Acyl-CoA fatty acid esters bind to ACBP proteins. Here, we show that specifically the interaction between C18:1-CoA with ACBP1 results in release of RAP2.12 from the ACBP1:RAP2.12 complex while C16:0-CoA does not affect the interaction between RAP2.12 and ACBP1 (Fig. 2). A shift of the ratio between C18:1-CoA and C16:0-CoA as described above, will therefore lead to the release of RAP2.12 from ACBP1. Indeed, application of C18:1-CoA, but not C16:0-CoA, increased the number of nuclei in which GFP-tagged RAP2.12 accumulated (Fig. 2 C and D). Consequently, also up-regulation of hypoxia-responsive marker genes was observed (Fig. 3 A–C).
Similar to this external application of acyl-CoAs, also an endogenous shift of the C18:1-CoA to C16:0-CoA ratio as observed in lacs4lacs9 mutant lines resulted in the up-regulation of hypoxia-responsive genes already during aerobic conditions (Fig. 3H). Altogether, these data describe how the ultimate trigger for release of RAP2.12 from ACBP1 is constituted by an energy crisis-provoked response of the acyl-CoA pool under hypoxia (Fig. 6).
Changes of the cellular energy status happen all of the time as most biotic and abiotic stress conditions affect energy metabolism in one way or another (44). It would be most detrimental for plant fitness, when each fluctuation of the ATP level immediately led to the activation of hypoxic gene expression (34). Therefore, the lifetime of ERFVII proteins depends on the actual cellular oxygen concentration via the Cys branch of the N-end rule for proteasomal protein degradation (3, 4, 11). Only when an energy crisis is provoked by low-oxygen conditions, the stabilization of RAP2.12 enables the protein to accumulate in the nucleus in a sufficient amount to activate hypoxic gene expression. However, when RAP2.12 is released from ACBP1 due to an energy deficit that is not related to low-oxygen stress, the protein will be rapidly degraded due to proteasomal activity. Therefore, we propose that the ACBP1:RAP2.12 complex forms the initial hub capable of integrating signal inputs related to the cellular energy charge with oxygen concentration-dependent determination of the lifetime of RAP2.12 protein (Fig. 6). Subsequently, RAP2.12 protein that is newly synthesized after the onset of hypoxia does still undergo N-end rule-assisted stabilization but may not be linked directly to the energy status of the cell.
Constitutive activation of the molecular stress response to low oxygen in the lacs4lacs9 mutant background led to reduced tolerance to low oxygen as well as to flooding stress (Fig. 4). This observation is congruent with earlier observations that constitutive activation of the hypoxia-stress response in plants via overexpression of a stable version of RAP2.12 protein reduced tolerance to hypoxia (34), although other studies indicate that the latter phenotype is likely conditional to growth conditions and recovery treatment too (4, 34). This underlines the importance of a timely control of stress responses that are optimally adjusted to the actual environmental conditions. The integration of (i) energy-dependent changes in C18:1-CoA levels as cellular trigger signal with (ii) the homeostatic control of the lifetime of RAP2.12 in an oxygen concentration-dependent manner provides a highly specific control mechanism to initiate hypoxic responses. The mechanism guarantees that a full low-oxygen response is activated only when hypoxia is detrimental for the plant’s energy status.
The ATP dependence of oleoyl-CoA synthesis by LACS in combination with the impact of oleoyl-CoA on the interaction between RAP2.12 and ACBP exposes mitochondrial activity as an early trigger for hypoxia signaling. Consistently, manipulating mitochondrial ATP synthesis using inhibitors of specific respiratory complexes like antimycin-A induced RAP2.12-controlled hypoxic gene expression even under aerobic conditions (45) (Fig. 5E). It is striking that the induction of hypoxic gene expression in air by acyl-CoAs (Fig. 3 C and H) or antimycin-A (Fig. 5E) was lower compared with the induction of these genes by hypoxic conditions (Fig. 3H). This observation stresses the impact of additional oxygen-dependent regulatory mechanisms, such as the N-end rule-mediated reduction of RAP2.12 lifetime in air. In this context, it is worth mentioning that the control of low-oxygen stress responses is not only linked to the oxygen and energy status of the cell, but it is also known to be influenced by other cellular factors such as nitric oxide (46, 47), hydrogen peroxide (48), calcium (49, 50), and potassium (51). In the near future, it will be intriguing to expand the mechanistic explanation of how the energy and oxygen status of a cell is integrated upon low-oxygen stress with these additional signaling components (12).
ACBPs are found in all kingdoms of life, while ERFVIIs are highly conserved among higher plants. Moreover, the interaction domains of both protein families are highly conserved in plants (Fig. 1). Therefore, the ACBP:ERFVII signaling hub as presented here may represent a universal mechanism in plants to initiate hypoxia-induced stress responses via the integration of multiple cellular signals. Moreover, the specific interaction of ACBPs with various acyl-CoAs on the one hand (Fig. 2) and the distinct cellular responses provoked by individual acyl-CoAs on the other hand (Fig. 3 A and B) suggest that many additional possibilities may exist of how acyl-CoAs can modulate cellular signaling pathways in plants.
Materials and Methods
Plant Materials.
Arabidopsis thaliana ecotype Col-0 was used as wild type for all analyses. The 35S:RAP2.12-GFP line and lacs4-1 lacs9-2 and lacs4-2 lacs9-7 double knockout lines were described previously (3, 33). The acbp1 knockout line (SAIL_683_C03) was obtained from the Nottingham Arabidopsis Stock Center (SI Appendix, Fig. S5).
Growth Conditions and Analysis of Oxygen Deprivation Response.
For testing anoxia tolerance of seedlings, seeds were sown on half-strength MS medium containing 0.5% (wt/vol) sucrose, stratified for 48 h at 4 °C, and germinated at 21 °C day/18 °C night with a photoperiod of 16 h light (150 μmol⋅m−2⋅s−1) and 8 h dark. At day 11, seedlings were exposed to full anoxia, by placing the culture plates in an environment containing 100% nitrogen, for 9 h in the dark to avoid photosynthetic oxygen production. After 3 d of recovery, the survival score was determined as previously described (4). For submergence assays, seeds were sown in moist soil, stratified at 4 °C in the dark for 48 h, and germinated at 21 °C day/18 °C night with a photoperiod of 16 h light and 8 h darkness. The 4-wk-old plants were submerged in water in 40-cm high plastic containers and kept in the dark to avoid photosynthetic oxygen production. Leaves stayed 10 cm under the water surface. After 3 or 4 d, water was removed from the boxes and plants were placed back under photoperiodic conditions (16 h/8 h, light/dark). Submergence tolerance was assayed after 4 d of recovery.
For acyl-CoA pool measurements, plants were grown on horizontal agar plates containing 0.8% agar in 1/2 MS medium (pH 5.7) with 15 mM sucrose for 2 wk under long-day conditions (16 h in 150 µmol photons⋅m−2⋅s−1 at 21 °C and 8 h in 0 µmol photons⋅m−2⋅s−1 at 19 °C). After 2.5 h in light, they were subjected to hypoxia in the dark by exposition to a stream of air containing 1% (vol/vol) oxygen, supplemented with nitrogen and 350 ppm carbon dioxide. Plants were harvested by freezing in liquid nitrogen after 2 h, 3 h, and 4 h of hypoxia. For normoxic control, untreated plants were harvested simultaneously with the start of hypoxic treatment.
For expression analysis after acyl-CoA treatment, wild-type seeds were sown in 24-well plates containing half-strength liquid MS medium, stratified for 48 h at 4 °C, and germinated at 21 °C day/18 °C night with a photoperiod of 16 h light (150 μmol⋅m−2⋅s−1) and 8 h dark. At day 14, seedlings were treated with different acyl-CoAs at a final concentration of 0.1% in 0.01% pluronic F68. Pluronic is a nonfatty acid-based detergent which means that the detergent properties of pluronic in solubilizing fatty acyls are not confounded by fatty acyls derived from the detergent itself (52).
Cloning of Constructs.
Coding sequences (CDSs) were amplified from a cDNA template using Phusion High Fidelity DNA polymerase (Thermo Fisher Scientific). The ACBP1 CDS was cloned into pENTR-D and recombined into pK7FWG2 (51) to fuse it with GFP. For in vitro expression, the CDS of ACBP1 was fused N terminally with a Flag tag by PCR and cloned into pF3A WG BYDV (Promega), while the CDS of RAP2.12 was fused C terminally to CFP by PCR and cloned into pF3A WG BYDV (Promega). A complete list of all primers used is provided in SI Appendix, Table S4.
Plant Transformation.
Transgenic ACBP1-GFP plants were generated by transforming wild-type plants with the vector pK7FWG2 (53) containing the ACBP1 CDS fused in frame to GFP at its C terminus. T0 seeds were screened for kanamycin resistance and the presence of GFP signals by confocal microscopy.
qRT-PCR.
RNA extraction, digestion of genomic DNA, cDNA synthesis, and qRT-PCR analysis were performed as described previously (54). For all experiments, four to five independent biological replicates were used, as indicated in the figure and table legends. For normalization, UBIQUITIN10 expression was used according to ref. 3. Primers for hypoxia core genes, LACS genes, and UBI10 are given in SI Appendix, Table S4.
RNA-Seq Analysis.
Illumina HiSeq sequencing was performed according to standardized protocols as described in detail in SI Appendix. Transcriptome analysis was performed by means of CLC Genomics Workbench v.6 using the A. thaliana reference sequence (Tair10). Expression values were normalized using quantile normalization and pairwise statistical analyses comparing the treatments performed using false discovery rate (FDR)-corrected P values based on Baggerly’s test (55).
Confocal Imaging.
For protein localization studies, GFP signals were imaged and analyzed with a Leica DM6000 TCS SP8 confocal microscope (Leica Microsystems). Nuclear staining was performed by using DAPI (molecular probes) according to the manufacturer’s instructions. For quantification of nuclear translocation of RAP2.12 after acyl-CoA treatment, 20 DAPI-stained nuclei per plant (five plants in total) were analyzed per treatment. Leaves of 5-wk-old soil-grown 35S:RAP2.12-GFP plants were incubated for 3 h with different acyl-CoAs at a final concentration of 0.1% in 0.01% pluronic F68 in a 24-well plate under continuous shaking in the light. The experiments were repeated three times.
In Vitro Binding Assay.
The method to determine whether acyl-CoAs affect the interaction between ACBP1 and RAP2.12 is explained in detail in SI Appendix, Supplementary Information Text and Fig. S1. In brief, both proteins were synthesized using wheat germ extract (54). The full CDS of RAP2.12 was fused C terminally with the CDS of CFP, and the CDS of ACBP1 was N-terminally fused with a FLAG tag. RAP2.12-CFP protein was bound to GFP–Trap-A beads (Chromotek) and incubated with ACBP1 protein overnight. Subsequently, beads were resuspended in wash buffer containing different acyl-CoAs at a final concentration of 0.1% in 0.01% pluronic F68, or as a mock control of only 0.01% F68. After one night of incubation, the buffer was replaced and the composition of the protein complex retained to the beads was analyzed on a Western blot.
Analysis of Acyl-CoA Esters.
Acyl-CoAs were extracted, derivatized, and analyzed using HPLC as described earlier (56). Detailed information about the method is provided in SI Appendix.
LACS4 in Vitro Enzyme Assay.
The in vitro LACS enzyme assay was carried out as described previously (17) using protein that was heterologously expressed in Escherichia coli (57). Details of the method are explained in SI Appendix.
ATP and ADP Quantification.
ATP and ADP were extracted with 16% trichloroacetic acid (33) and analyzed after derivatization by HPLC as described previously (58). Details of the procedure are described in SI Appendix.
Analysis of Oxygen Consumption Rates.
For the determination of oxygen consumption rates, 4-wk-old plants grown on soil under short-day conditions (8 h in 160 µmol photons m−2⋅s−1 at 20 °C and 16 h in 0 µmol photons m−2⋅s−1 at 16 °C) were used. Starting 3 h before measurement, the plants were sprayed every hour either with 50 mM Mes buffer (pH 6.5) containing 50 µM antimycin-A (59) or with 50 mM Mes buffer for control. This treatment was done in the last hour of dark phase and 2 h into the light phase, and then measurement of oxygen consumption rates at normoxic conditions was performed as described before (34) using the respective spraying solution. In a similar setup, the effect of different acyl-CoAs on the oxygen consumption rate was tested by incubating leaf disks in acyl-CoAs at a final concentration of 0.1% in 0.01% pluronic F68.
Statistical Analysis.
Statistical evaluation of significant variation between treatments or genotypes was done by performing Student’s t test or one-way ANOVA where appropriate.
Data Availability.
The RNA-Seq gene expression data are available in NCBI’s Gene Expression Omnibus (GEO) through GEO Series accession no. GSE97186.
Supplementary Material
Acknowledgments
We thank Sandro Parlanti and Frauke Augstein for valuable support. This work was supported by grants (to J.T.v.D.) (DO 1298/2-2) and (to P.G.) (GE 878/7-2) from the German Science Foundation (DFG). M.F. was supported by the DFG (Grant DFG FU 430/5-1).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE97186).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809429115/-/DCSupplemental.
References
- 1.Food and Agricultural Organization of the United Nations (FAO) 2015. The impact of disasters on agriculture and food security. (Food and Agricultural Organization of the United Nations, Rome) Report I5128E/1/11.15.
- 2.Hirabayashi Y, et al. Global flood risk under climate change. Nat Clim Chang. 2013;3:816–821. [Google Scholar]
- 3.Licausi F, et al. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature. 2011;479:419–422. doi: 10.1038/nature10536. [DOI] [PubMed] [Google Scholar]
- 4.Gibbs DJ, et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature. 2011;479:415–418. doi: 10.1038/nature10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bui LT, Giuntoli B, Kosmacz M, Parlanti S, Licausi F. Constitutively expressed ERF-VII transcription factors redundantly activate the core anaerobic response in Arabidopsis thaliana. Plant Sci. 2015;236:37–43. doi: 10.1016/j.plantsci.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Gasch P, et al. Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-Motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell. 2016;28:160–180. doi: 10.1105/tpc.15.00866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kosmacz M, et al. The stability and nuclear localization of the transcription factor RAP2.12 are dynamically regulated by oxygen concentration. Plant Cell Environ. 2015;38:1094–1103. doi: 10.1111/pce.12493. [DOI] [PubMed] [Google Scholar]
- 8.Li HY, Chye ML. Arabidopsis Acyl-CoA-binding protein ACBP2 interacts with an ethylene-responsive element-binding protein, AtEBP, via its ankyrin repeats. Plant Mol Biol. 2004;54:233–243. doi: 10.1023/B:PLAN.0000028790.75090.ab. [DOI] [PubMed] [Google Scholar]
- 9.Abbas M, et al. Oxygen sensing coordinates photomorphogenesis to facilitate seedling survival. Curr Biol. 2015;25:1483–1488. doi: 10.1016/j.cub.2015.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Dongen JT, Licausi F. Oxygen sensing and signaling. Annu Rev Plant Biol. 2015;66:345–367. doi: 10.1146/annurev-arplant-043014-114813. [DOI] [PubMed] [Google Scholar]
- 11.Weits DA, et al. Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat Commun. 2014;5:3425. doi: 10.1038/ncomms4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt RR, Weits DA, Feulner CFJ, van Dongen JT. Oxygen sensing and integrative stress signaling in plants. Plant Physiol. 2018;176:1131–1142. doi: 10.1104/pp.17.01394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grevengoed TJ, Klett EL, Coleman RA. Acyl-CoA metabolism and partitioning. Annu Rev Nutr. 2014;34:1–30. doi: 10.1146/annurev-nutr-071813-105541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng W, Su YC, Saunders RM, Chye ML. The rice acyl-CoA-binding protein gene family: Phylogeny, expression and functional analysis. New Phytol. 2011;189:1170–1184. doi: 10.1111/j.1469-8137.2010.03546.x. [DOI] [PubMed] [Google Scholar]
- 15.Neess D, Bek S, Engelsby H, Gallego SF, Færgeman NJ. Long-chain acyl-CoA esters in metabolism and signaling: Role of acyl-CoA binding proteins. Prog Lipid Res. 2015;59:1–25. doi: 10.1016/j.plipres.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Lung SC, Chye ML. Deciphering the roles of acyl-CoA-binding proteins in plant cells. Protoplasma. 2016;253:1177–1195. doi: 10.1007/s00709-015-0882-6. [DOI] [PubMed] [Google Scholar]
- 17.Shockey JM, Fulda MS, Browse JA. Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol. 2002;129:1710–1722. doi: 10.1104/pp.003269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koo AJ, Ohlrogge JB, Pollard M. On the export of fatty acids from the chloroplast. J Biol Chem. 2004;279:16101–16110. doi: 10.1074/jbc.M311305200. [DOI] [PubMed] [Google Scholar]
- 19.Li N, Xu C, Li-Beisson Y, Philippar K. Fatty acid and lipid transport in plant cells. Trends Plant Sci. 2016;21:145–158. doi: 10.1016/j.tplants.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 20.Troncoso-Ponce MA, Nikovics K, Marchive C, Lepiniec L, Baud S. New insights on the organization and regulation of the fatty acid biosynthetic network in the model higher plant Arabidopsis thaliana. Biochimie. 2016;120:3–8. doi: 10.1016/j.biochi.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 21.Hertz R, Magenheim J, Berman I, Bar-Tana J. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4alpha. Nature. 1998;392:512–516. doi: 10.1038/33185. [DOI] [PubMed] [Google Scholar]
- 22.DiRusso CC, Heimert TL, Metzger AK. Characterization of FadR, a global transcriptional regulator of fatty acid metabolism in Escherichia coli. Interaction with the fadB promoter is prevented by long chain fatty acyl coenzyme A. J Biol Chem. 1992;267:8685–8691. [PubMed] [Google Scholar]
- 23.Lung SC, et al. Arabidopsis ACYL-COA-BINDING PROTEIN1 interacts with STEROL C4-METHYL OXIDASE1-2 to modulate gene expression of homeodomain-leucine zipper IV transcription factors. New Phytol. 2018;218:183–200. doi: 10.1111/nph.14965. [DOI] [PubMed] [Google Scholar]
- 24.Geigenberger P. Response of plant metabolism to too little oxygen. Curr Opin Plant Biol. 2003;6:247–256. doi: 10.1016/s1369-5266(03)00038-4. [DOI] [PubMed] [Google Scholar]
- 25.Grattan RP, Roger SC. Acyl-CoA may be a neglected product in studies of fatty acid synthesis by isolated chloroplasts. FEBS Lett. 1981;135:182–186. [Google Scholar]
- 26.Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA. 1997;94:7076–7081. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du ZY, Chye ML. Interactions between Arabidopsis acyl-CoA-binding proteins and their protein partners. Planta. 2013;238:239–245. doi: 10.1007/s00425-013-1904-2. [DOI] [PubMed] [Google Scholar]
- 28.Xie LJ, et al. Arabidopsis acyl-CoA-binding protein ACBP3 participates in plant response to hypoxia by modulating very-long-chain fatty acid metabolism. Plant J. 2015;81:53–67. doi: 10.1111/tpj.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chye ML, Li HY, Yung MH. Single amino acid substitutions at the acyl-CoA-binding domain interrupt 14[C]palmitoyl-CoA binding of ACBP2, an Arabidopsis acyl-CoA-binding protein with ankyrin repeats. Plant Mol Biol. 2000;44:711–721. doi: 10.1023/a:1026524108095. [DOI] [PubMed] [Google Scholar]
- 30.Du ZY, Arias T, Meng W, Chye ML. Plant acyl-CoA-binding proteins: An emerging family involved in plant development and stress responses. Prog Lipid Res. 2016;63:165–181. doi: 10.1016/j.plipres.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Theodoulou FL, Carrier DJ, Schaedler TA, Baldwin SA, Baker A. How to move an amphipathic molecule across a lipid bilayer: Different mechanisms for different ABC transporters? Biochem Soc Trans. 2016;44:774–782. doi: 10.1042/BST20160040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Proost S, et al. PLAZA: A comparative genomics resource to study gene and genome evolution in plants. Plant Cell. 2009;21:3718–3731. doi: 10.1105/tpc.109.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jessen D, Roth C, Wiermer M, Fulda M. Two activities of long-chain acyl-coenzyme A synthetase are involved in lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis. Plant Physiol. 2015;167:351–366. doi: 10.1104/pp.114.250365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paul MV, et al. Oxygen sensing via the ethylene response transcription factor RAP2.12 affects plant metabolism and performance under both normoxia and hypoxia. Plant Physiol. 2016;172:141–153. doi: 10.1104/pp.16.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riber W, et al. The greening after extended darkness1 is an N-end rule pathway mutant with high tolerance to submergence and starvation. Plant Physiol. 2015;167:1616–1629. doi: 10.1104/pp.114.253088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gout E, Rébeillé F, Douce R, Bligny R. Interplay of Mg2+, ADP, and ATP in the cytosol and mitochondria: Unravelling the role of Mg2+ in cell respiration. Proc Natl Acad Sci USA. 2014;111:E4560–E4567. doi: 10.1073/pnas.1406251111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soccio M, Laus MN, Trono D, Pastore D. A new simple fluorimetric method to assay cytosolic ATP content: Application to durum wheat seedlings to assess modulation of mitochondrial potassium channel and uncoupling protein activity under hyperosmotic stress. Biologia. 2013;68:421–432. [Google Scholar]
- 38.van Dongen JT, Schurr U, Pfister M, Geigenberger P. Phloem metabolism and function have to cope with low internal oxygen. Plant Physiol. 2003;131:1529–1543. doi: 10.1104/pp.102.017202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dröse S, Brandt U. The mechanism of mitochondrial superoxide production by the cytochrome bc1 complex. J Biol Chem. 2008;283:21649–21654. doi: 10.1074/jbc.M803236200. [DOI] [PubMed] [Google Scholar]
- 40.Guo H, et al. Plastid-nucleus communication involves calcium-modulated MAPK signalling. Nat Commun. 2016;7:12173. doi: 10.1038/ncomms12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta KJ, Zabalza A, van Dongen JT. Regulation of respiration when the oxygen availability changes. Physiol Plant. 2009;137:383–391. doi: 10.1111/j.1399-3054.2009.01253.x. [DOI] [PubMed] [Google Scholar]
- 42.Dörmann P, Voelker TA, Ohlrogge JB. Cloning and expression in Escherichia coli of a novel thioesterase from Arabidopsis thaliana specific for long-chain acyl-acyl carrier proteins. Arch Biochem Biophys. 1995;316:612–618. doi: 10.1006/abbi.1995.1081. [DOI] [PubMed] [Google Scholar]
- 43.Jones A, Davies HM, Voelker TA. Palmitoyl-acyl carrier protein (ACP) thioesterase and the evolutionary origin of plant acyl-ACP thioesterases. Plant Cell. 1995;7:359–371. doi: 10.1105/tpc.7.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baena-González E, Sheen J. Convergent energy and stress signaling. Trends Plant Sci. 2008;13:474–482. doi: 10.1016/j.tplants.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner S, Van Aken O, Elsässer M, Schwarzländer M. Mitochondrial energy signaling and its role in the low-oxygen stress response of plants. Plant Physiol. 2018;176:1156–1170. doi: 10.1104/pp.17.01387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibbs DJ, et al. Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol Cell. 2014;53:369–379. doi: 10.1016/j.molcel.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vicente J, et al. The Cys-Arg/N-end rule pathway is a general sensor of abiotic stress in flowering plants. Curr Biol. 2017;27:3183–3190.e4. doi: 10.1016/j.cub.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pucciariello C, Parlanti S, Banti V, Novi G, Perata P. Reactive oxygen species-driven transcription in Arabidopsis under oxygen deprivation. Plant Physiol. 2012;159:184–196. doi: 10.1104/pp.111.191122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sedbrook JC, Kronebusch PJ, Borisy GG, Trewavas AJ, Masson PH. Transgenic AEQUORIN reveals organ-specific cytosolic Ca2+ responses to anoxia and Arabidopsis thaliana seedlings. Plant Physiol. 1996;111:243–257. doi: 10.1104/pp.111.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bose J, Pottosin II, Shabala SS, Palmgren MG, Shabala S. Calcium efflux systems in stress signaling and adaptation in plants. Front Plant Sci. 2011;2:85. doi: 10.3389/fpls.2011.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang F, Chen ZH, Shabala S. Hypoxia sensing in plants: On a quest for ion channels as putative oxygen sensors. Plant Cell Physiol. 2017;58:1126–1142. doi: 10.1093/pcp/pcx079. [DOI] [PubMed] [Google Scholar]
- 52.Poirier Y, Erard N, Petétot JM. Synthesis of polyhydroxyalkanoate in the peroxisome of Saccharomyces cerevisiae by using intermediates of fatty acid beta-oxidation. Appl Environ Microbiol. 2001;67:5254–5260. doi: 10.1128/AEM.67.11.5254-5260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karimi M, De Meyer B, Hilson P. Modular cloning in plant cells. Trends Plant Sci. 2005;10:103–105. doi: 10.1016/j.tplants.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 54.Schmidt R, et al. Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell. 2013;25:2115–2131. doi: 10.1105/tpc.113.113068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baggerly KA, Deng L, Morris JS, Aldaz CM. Differential expression in SAGE: Accounting for normal between-library variation. Bioinformatics. 2003;19:1477–1483. doi: 10.1093/bioinformatics/btg173. [DOI] [PubMed] [Google Scholar]
- 56.Larson TR, Graham IA. Technical advance: A novel technique for the sensitive quantification of acyl CoA esters from plant tissues. Plant J. 2001;25:115–125. doi: 10.1046/j.1365-313x.2001.00929.x. [DOI] [PubMed] [Google Scholar]
- 57.Overath P, Pauli G, Schairer HU. Fatty acid degradation in Escherichia coli. An inducible acyl-CoA synthetase, the mapping of old-mutations, and the isolation of regulatory mutants. Eur J Biochem. 1969;7:559–574. [PubMed] [Google Scholar]
- 58.Zhang L, et al. Overriding the co-limiting import of carbon and energy into tuber amyloplasts increases the starch content and yield of transgenic potato plants. Plant Biotechnol J. 2008;6:453–464. doi: 10.1111/j.1467-7652.2008.00332.x. [DOI] [PubMed] [Google Scholar]
- 59.De Clercq I, et al. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell. 2013;25:3472–3490. doi: 10.1105/tpc.113.117168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq gene expression data are available in NCBI’s Gene Expression Omnibus (GEO) through GEO Series accession no. GSE97186.