Abstract
Antibiotic resistance in bacteria has emerged as a global challenge over the past 90 years, compromising our ability to effectively treat infections. There has been a dramatic increase in antibiotic resistance-associated determinants in bacterial populations, driven by the mobility and infectious nature of such determinants. Bacterial genome flexibility and antibiotic-driven selection are at the root of the problem. Genome evolution and the emergence of highly successful multidrug-resistant clades in different pathogens have made this a global challenge. Here, we describe some of the factors driving the origin, evolution, and spread of the antibiotic resistance genotype.
Keywords: emergence, evolution, bacteria, multidrug resistance
The emergence of multidrug-resistant (MDR) bacteria is one of the most dramatic clinical and biological phenomena identified over the past century (1, 2). Since the discovery of antibiotics in the 1920s, each introduction of a new antibiotic class has been followed by the emergence of resistance, in some form or other, relatively quickly. Over the time period of the antibiotic era, the number of identified resistance mechanisms, and their abundance, has increased at an alarming rate. Molecules with the ability to kill or inhibit bacteria have likely been around since the first single-cell entities emerged, but the recent pace of evolution is still remarkable, indicating the key role for selective pressure in this process (3). There have been arguments about where this pressure is most intense—in the clinic, in agriculture, or in the environment—but it is likely that within each of these domains, pressure is being exerted that is driving resistance. Particular properties of the genes and mutations involved in resistance explain some aspects of their ability to evolve rapidly. Resistance determinants are largely components of the bacterial “accessory” genome where genetic flexibility is more readily tolerated and such determinants are often “mobile,” enhancing their ability to spread through infectious mechanisms (4). Hence, within species and between species evolution is exerting an influence.
A key feature of the antibiotic resistance era has been the recent emergence of highly successful clades or clones within bacterial species that have the ability to be resistant without any obvious impact on fitness. Indeed, there is evidence that such clades can be more aggressive in terms of their ability to transmit and cause clinical disease. Resistant clades have been identified in many of the common bacterial pathogens, such as Staphylococcus aureus and Salmonella enterica (5–7). Other clades have driven the emergence of newly recognized pathogens, including Clostridium difficile and Acinetobacter baumannii (8, 9). Some of these clades appear to have adapted to exploit modern human-created niches, such as the healthcare system or food chain. Some have the potential for global spread. Here, with a focus on the enteric bacteria, we will explore some of the factors driving the successful emergence of antibiotic resistance and the associated successful resistant clades.
A Brief Historical Perspective on MDR and Mobile Elements
Although the emergence of antibiotic resistance has attracted great attention in the past few years, it is not a new phenomenon and it is worth first considering some of the lessons from history. A comprehensive review by Watanabe (10) published in 1966 outlines the thoughts at that time (3). Although bacteria resistant to individual antibiotics were observed earlier, one of the first reports of MDR bacteria was of Shigella resistant to sulphonamides, streptomycin, chloramphenicol, and tetracycline in Hong Kong in 1955 (10). Escherichia coli with related resistance profiles were also isolated in the same setting and even within the same patient, suggesting the transferability of the MDR phenotype, which was subsequently proven experimentally. Even then, clinicians recognized that patients could be simultaneously colonized by both resistant and sensitive Shigella and that treatment could drive the consolidation of resistance. These early observations highlighted the fact that MDR itself was infectious and had the potential to move rapidly through bacterial species and even across species boundaries.
After the first wave signaling the emergence of MDR, investigators quickly identified a key role for plasmids (then named R Factors) as agents of the transferability of MDR and an era of plasmid biology began, where these extrachromosomal molecules were characterized both genetically and molecularly (11–13). Roles for conjugation, transformation, and transduction in the spread of MDR were demonstrated through laboratory studies, identifying the multiple potential mechanisms driving the spread of MDR. By the early 1970s molecular technologies improved to the point that plasmids and resistance determinants became tractable to detailed analysis. It was at this point that transposable genes were identified along with other forms of mobile genetic elements (14). The proof that “jumping genes” actually existed and that they encoded antibiotic resistance had enormous impact and the threat of antibiotic resistance was now being taken seriously. Stanley Falkow (15) produced a highly prophetic book entitled Infectious Multiple Drug Resistance that summarizes the state of the field at this time, and he strongly emphasized the potential dangers of neglecting the control of antibiotic usage (16). Soon after the description of transposons, other forms of mobile elements that contribute to the spread of MDR were also discovered, many of which were harbored within the chromosomes of MDR bacteria. These included integrons (17) and mobile elements that could drive or even parasitize the conjugative determinants present within bacteria (12). In addition to these plasmid-borne and transposable antibiotic-resistance genes, chromosomal mutations were identified that direct the expression of resistance mechanisms. Such mutations occur, for example, within genes encoding the targets of antibiotics, including gyrA and gyrB for fluoroquinolones and within genes that affect efflux pumps or porins that influence intracellular antibiotic levels (18). The accumulation of mutations in such core genes or similar regulatory elements thus significantly enhance resistance potential (3, 19).
Whole-Genome Sequences Provide the Genetic Blueprint of a Resistant Bacterium
These early phenotypic and molecular approaches allowed the description of individual or small clusters of resistance elements, but it was the whole-genome sequencing of bacterial isolates that started to provide a comprehensive blueprint against which to investigate how antibiotic resistance was evolving. The early generation of complete reference genomes for pathogens, such as Mycobacterium tuberculosis, Salmonella enterica serovar Typhi (S. Typhi), and Staphylococcus aureus, captured both the chromosomal and extrachromosomal components of the genome (20–22). Some of these early genome sequences were of MDR isolates, including the isolate CT18 of S. Typhi, facilitating the complete assembly of plasmids and their resistance elements. Even if the reference genome was antibiotic sensitive, comparative genomic analysis with resistant isolates facilitated the detailed mapping and cataloguing of resistance elements, including plasmids, transposons, insertions or deletions (indels), and single-nucleotide polymorphisms (SNPs). The invention of new sequencing technologies then facilitated the sequencing of whole bacterial populations, allowing detailed population structural analysis to be undertaken for the first time using thousands of genomes (23). Whole-genome–based phylogenetic and geophylogenetic analyses have transformed our ability to interrogate the evolution of resistance over time and space.
Over the past decade, through the whole-genome sequencing of hundreds of thousands of bacteria, we have a better understanding of the range of antibiotic-resistance genes (ARGs). ARGs can be divided into different classes based on the mechanism of resistance they encode, including antibiotic target replacement or protection, antibiotic inactivation, antibiotic efflux, or reduced permeability to antibiotics. Curated databases, such as the Comprehensive Antibiotic Resistance Database (CARD) (24) and ARG-Annotation (ARG-ANNOT) (25), are valuable resources that contain sequences of ARGS. At the time of writing, CARD contained 2,388 ARGs and ARG-ANNOT contained 1,808 entries. These databases can be used to quickly identify known resistance determinants in isolated bacteria using whole-genome sequencing and BLAST- or assembly-based bioinformatic tools, such as ARIBA (26) or SRST2 (27). Recent studies have indicated that these known ARGs are likely just the tip of the iceberg and there may be many more ARGs present in environmental and commensal bacteria. For example, using a new computational method, Berglund et al. (28) screened thousands of bacterial genomes and plasmids and over 5 terabases of metagenomic data obtained from the human microbiome and polluted environments, such as hospital effluent. The authors predicted 76 novel B1 metallo-β-lactamase genes, potentially more than doubling their number. Another study on a bacterium from an ancient, isolated cave ecosystem identified 5 novel—in addition to 13 known—ARGs. Further biochemical phenotyping revealed three mechanisms not previously shown to be involved in antibiotic resistance (29). As environmental sampling continues and computational methods improve, the number of known ARGs will undoubtedly grow further.
The Emergence of “Dominant” MDR Clades
An overriding feature of increased antibiotic resistance has been the emergence of MDR clades of different species that have spread extensively, often to different parts of the globe. These clades were initially identified using simple bacterial typing approaches, including phage typing, multilocus sequence typing (MLST), or subgenomic SNP typing, but were comprehensively defined using whole-genome sequence analysis. These clades are characterized by their remarkable genetic conservation, suggesting rapid recent population expansion. Often, hundreds of isolates covering large geographical regions harbor only a few hundred or even fewer SNPs that can be used to build phylogenetic history. Examples of such MDR clades include E. coli ST131 (30, 31), S. Typhi H58 (32), S. aureus ST239 (5), and C. difficile O27 (33, 34). Common features of dominant MDR clades are that they are genetically highly clonal with relatively limited core genome variation, have the ability to acquire and maintain antibiotic-resistance determinants, and demonstrate efficient transmission. Such clades counteract the dogma that harboring MDR can compromise fitness as they have significant virulence potential and possess the ability to rapidly spread globally.
E. coli ST131 began to emerge at the global level in the 2000s and was quickly recognized as a leading cause of E. coli extraintestinal infections in multiple settings across the world (35). E. coli ST131 are typically MDR, expressing extended spectrum β-lactamases (ESBLs), including bla CTX-M-15, conferring significant resistance to third-generation cephalosporins (e.g., ceftriaxone). ST131 E. coli also exhibit high levels of fluoroquinolone resistance. This clade appears to be clinically aggressive, causing a range of nosocomial infections, as well as being the leading cause of urinary tract infections (35). Phylogenetic analysis based on whole-genome sequencing data has been used to track the evolution of ST131 over the past decades and has identified several clades that mark this evolution. Founder clades A and B were precursors to a clade C type, which has been largely responsible for the global spread of ST131 (36–38). Throughout the recent evolution of ST131 there has been a series of gene acquisitions involving virulence-associated (e.g., fimbriae, iron acquisition, novel phages) and MDR-associated determinants (35), although none of these have been experimentally linked to the success of this clade. The C clade has evolved into several subclades that harbor MDR signatures. Of particular significance are fluoroquinolone-resistance mutations within gyrA/B and parC and an ESBL of type blaCTX-M-15 (39). Additionally, particular IncF-type plasmids have become “adapted” to ST131, and these exhibit evidence of microevolution within clades C1 and C2, frequently encoding blaCTX-M-15.
In common with E. coli ST131, fluoroquinolone-resistance mutations appear to have played a key role in influencing the emergence of some C. difficile clades (40). C. difficile has emerged over the past 40 years as a common cause of antibiotic-associated diarrhea, particularly in healthcare settings. Phylogenetic analysis has shown that multiple clades of C. difficile have emerged independently and spread rapidly, from different founder populations (40). C. difficile 027/NAP1/BI forms a particularly aggressive clade that quickly emerged in the past two decades, spreading out of North America into Europe and Asia. Two distinct lineages of C. difficile 027, known as FQR1 and FQR2, emerged in North America within a relatively short period after independently acquiring the same fluoroquinolone resistance mutation and a highly related conjugative transposon (40). Phylogenetic analysis of these MDR lineages has shown that as they have evolved they have spread geographically, with multiple transposons and ARGs being shuffled in and out of the clade. C. difficile 027 isolates have increased virulence in animal models compared with other lineages of C. difficile and, intriguingly, antibiotic treatment itself can enhance their transmissibility and may be both promoting spread and increasing the number of clinical cases (41).
MDR clades can dramatically influence the epidemiology of disease. The emergence of S. Typhi of haplotype H58 (also designated 4.3.1) over the past 30 years has arguably transformed the global epidemiology of typhoid. S. Typhi H58 was originally identified in a subgenomic SNP-based study on ∼100 S. Typhi isolates collected from around the world (6). This clade likely emerged in the Indian subcontinent in the 1980s (32). The clade has since spread into Southeast Asia and on at least two separate occasions into Africa, arriving in distinct waves into particular regions. Indeed, H58 can “replace” endemic S. Typhi isolates and can spread typhoid into areas where the disease was previously rare: for example, in Malawi (42). Many S. Typhi H58 harbor multiple ARGs located on a composite transposon present either on an IncHI1 plasmid or integrated into the chromosome (32). A recent large typhoid outbreak in Pakistan demonstrated a concerning progression when an MDR H58 isolate with a chromosomally integrated antibiotic-resistance cassette acquired a plasmid with additional ARGs, including an ESBL (43). MDR clades, such as H58, are thus poised to present even greater challenges to global health by acquiring additional AGRs, as the resulting isolates may be resistant to so many classes of antibiotics that they are difficult to treat. The H58 clade is remarkably conserved in terms of core genome content, but nevertheless may have acquired additional virulence traits, such as enhanced resistance to bile (44). It is not currently known what is driving the success of H58.
There is evidence that MDR may be helping drive emerging pathogens into new niches. S. enterica serovar Typhimurium (S. Typhimurium) is typically associated with gastroenteritis, with systemic disease being relatively rare. However, S. Typhimurium have been increasingly reported as a major cause of invasive (bacteraemia) disease in sub-Saharan Africa, with an estimated case fatality rate of ∼20% (45) and up to ∼50% in HIV+ individuals (46). Phylogenetic analysis of whole-genome sequences of S. Typhimurium isolates across the sub-Saharan region identified a dominant MLST clade named ST313. These S. Typhimurium ST313 are primarily localized in Africa, although distinct ST313 lineages have recently been identified in Europe (47). Within sub-Saharan Africa, invasive salmonellosis is typically associated with HIV-infected adults and children under 5 y of age, with malaria a common association (48, 49). These S. Typhimurium are highly antibiotic resistant, with the MDR determinants frequently located on a composite transposon harbored on the virulence-associated pSLT plasmid (7). Significantly, pSLT also harbors the invasion-associated spv locus, meaning that antibiotic exposure may also be influencing selection for this virulence factor. The ST313 epidemic in Africa involved an initial wave of so-called lineage I isolates followed by a second wave involving a distinct lineage II (50). Interestingly, although both lineages were MDR, the MDR cassette in lineage II isolates harbored an additional chloramphenicol-resistance gene not present in lineage I (7). The acquisition of chloramphenicol resistance by lineage II coincided with the widespread use of chloramphenicol in Africa, suggesting this was involved in shaping the second wave. More recently there have been sporadic cases in both Kenya and Malawi of S. Typhimurium ST313 harboring an additional antibiotic-resistance plasmid encoding an ESBL (51, 52).
It has been suggested that human-to-human rather than zoonotic transmission is driving the ST313-mediated epidemics in sub-Saharan Africa (53). Interestingly, the ST313 lineage exhibits several signatures that have been associated with host adaptation, including significant levels of genome degradation (gene inactivation) (7). ST313 isolates also exhibit increased virulence phenotypes compared with other S. Typhimurium, including increased serum resistance and altered animal pathogenicity. A recent study showed that a ST313 lineage II-associated SNP in the pgtE virulence gene led to increased expression of PgtE and contributed to this increased virulence phenotype, providing genetic evidence for adaptive SNPs (54).
Antibiotic-Associated Disease and Spread
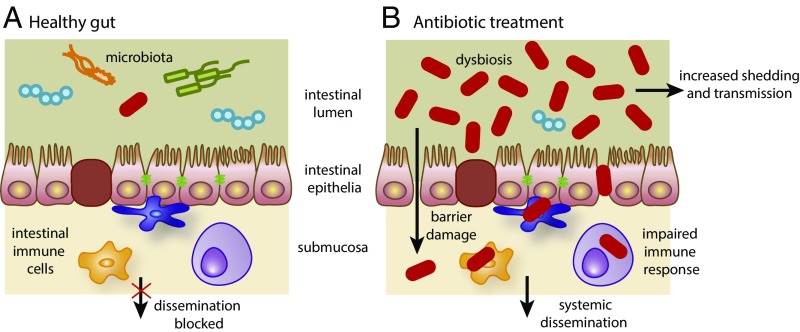
We now know that antibiotic treatment itself can cause clinical disease in certain situations. Individuals, particularly the elderly or immunocompromised, become susceptible to C. difficile diarrhea following treatment with antibiotics, such as ciprofloxacin or clindamycin. This is believed to be a consequence of the destruction of the beneficial microbiota in the intestine by the antibiotic (55). Individuals treated for C. difficile infection often relapse and restoration of a more diverse microbiota by fecal transplantation therapy can be more effective than antibiotic treatment, both in terms of recovery and prevention of relapse (56, 57). Indeed, individuals treated with antibiotics, but subclinically colonized with C. difficile, may become more infectious by secreting higher numbers of potentially infectious C. difficile spores (supershedders) (41). We do not know if infection or transmission by other MDR clades can be enhanced by antibiotic treatment. However, it may be that other clades are evolving to take advantage of the destruction of the beneficial microbiota during antibiotic treatment of humans. C. difficile exploits specialized toxins to break through the intestinal barrier during infection (58). However, this barrier is already compromised in many immunosuppressed individuals, in those undergoing bowel surgery, and in people with rare mutations that impact barrier function. Hence, such individuals may be more susceptible after antibiotic treatment to systemic infection when colonized by an aggressive MDR clade, such as E. coli ST131 (Fig. 1).
Fig. 1.
Disruption of the gut mucosa can lead to systemic infections by MDR bacteria following antibiotic treatment of immunocompromised patients. MDR bacteria are represented by red rod-shapes. (A) Healthy gut. The mucosa is intact, the microbiota is normal, and the growth of the MDR bacteria is restricted: they cannot spread systemically and are shed at very low levels. (B) With antibiotic treatment: the epithelial barrier is damaged, the microbiota has been disrupted or is dysbiotic and can no longer exert growth restriction on the MDR bacteria that consequently overgrow, spread systemically, and are shed at higher levels.
It is worth noting that most Vibrio cholerae bacteria, the cause of human cholera, isolated since 1980 are MDR. Because cholera is not usually treated with antibiotics it is not obvious why the MDR phenotype is so common in this pathogen. Most cases of cholera are caused by the pandemic clade known as El Tor, which has been spreading across the globe since the 1950s in multiple waves (59, 60). MDR V. cholerae El Tor is highly clonal, with the bacterial population frequently infected with STX integrative conjugative elements that harbor ARGs that have entered the clade on multiple occasions (60). It is intriguing to postulate that the MDR phenotype might be facilitating enhanced transmission from asymptomatic carriers, which is a testable hypothesis.
Origin and Transfer of Resistance Genes Between Environmental and Pathogenic Bacteria
Studies of bacteria from geologically isolated environments show that antibiotic resistance is ancient and ubiquitous in the environment (29, 61, 62). Environmental bacteria likely evolved antibiotic-resistance factors over eons as a defense against antimicrobials and chemicals present in the environment that are produced by other bacteria, fungi, or plants. Antibiotic resistance in bacterial pathogens was relatively rare until the recent wide-scale usage of antibiotics. While some surveys show distinct and separate clustering of ARGs derived from environmental and pathogenic sources, other studies show high levels of sequence homology (63–66) and indicate recent transfer of ARGs from environmental to pathogenic bacteria. A recent study even reported evidence supporting recent transfer between Gram-positive Actinobacteria and Gram-negative Proteobacteria (67). The authors proposed and experimentally tested a three-step mechanism for interphyla transfer that they call “carry-back” in which the Actinobacterial ARG is picked up by a Proteobacterial carrier sequence. This is significant because it suggests that genetic transfer can occur even between different phyla, allowing pathogens such as E. coli to access the large reservoir of ARGs in antibiotic-producing bacteria, such as Streptomyces that harbor numerous resistance genes.
The enormous reservoir of ARGs in environmental bacteria means that mechanisms of resistance may already exist even for antibiotics still in the developmental pipeline. Given the possibility of the mobilization of environmental ARGs, it is inevitable that resistance will continue to develop in bacterial pathogens in response to the introduction of new antibiotics. Clearly there is a need to develop new antimicrobial approaches, with the realization that they will not end the battle. To effectively fight infectious diseases in the long-term, we must use an integrative approach that includes preventive measures, such as vaccination and improved sanitation and hygiene strategies.
Acknowledgments
This work was supported by The Wellcome Trust and the Cambridge Biomedical Research Centre (National Institute for Medical Research, United Kingdom).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.The Review on Antimicrobial Resistance 2016 Tackling Drug-resistant Infections Globally: Final Report and Recommendations. Available at https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf. Accessed January 15, 2018.
- 2.World Health Organization . Antimicrobial Resistance: Global Report on Surveillance 2014. WHO; Geneva: 2014. [Google Scholar]
- 3.Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacLean RC, San Millan A. Microbial evolution: Towards resolving the plasmid paradox. Curr Biol. 2015;25:R764–R767. doi: 10.1016/j.cub.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roumagnac P, et al. Evolutionary history of Salmonella Typhi. Science. 2006;314:1301–1304. doi: 10.1126/science.1134933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kingsley RA, et al. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 2009;19:2279–2287. doi: 10.1101/gr.091017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott B, Androga GO, Knight DR, Riley TV. Clostridium difficile infection: Evolution, phylogeny and molecular epidemiology. Infect Genet Evol. 2017;49:1–11. doi: 10.1016/j.meegid.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 9.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: Emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe T. Infectious drug resistance in enteric bacteria. N Engl J Med. 1966;275:888–894. doi: 10.1056/NEJM196610202751607. [DOI] [PubMed] [Google Scholar]
- 11.Poulin-Laprade D, Carraro N, Burrus V. The extended regulatory networks of SXT/R391 integrative and conjugative elements and IncA/C conjugative plasmids. Front Microbiol. 2015;6:837. doi: 10.3389/fmicb.2015.00837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guédon G, Libante V, Coluzzi C, Payot S, Leblond-Bourget N. The obscure world of integrative and mobilizable elements, highly widespread elements that pirate bacterial conjugative systems. Genes (Basel) 2017;8:E337. doi: 10.3390/genes8110337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bañuelos-Vazquez LA, Torres Tejerizo G, Brom S. Regulation of conjugative transfer of plasmids and integrative conjugative elements. Plasmid. 2017;91:82–89. doi: 10.1016/j.plasmid.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Hedges RW, Jacob AE. Transposition of ampicillin resistance from RP4 to other replicons. Mol Gen Genet. 1974;132:31–40. doi: 10.1007/BF00268228. [DOI] [PubMed] [Google Scholar]
- 15.Falkow S. Infectious Multiple Drug Resistance. Pion Ltd; London: 1975. [Google Scholar]
- 16.Falkow S, Citarella RV, Wohlhieter JA. The molecular nature of R-factors. J Mol Biol. 1966;17:102–116. doi: 10.1016/s0022-2836(66)80097-9. [DOI] [PubMed] [Google Scholar]
- 17.Deng Y, et al. Resistance integrons: Class 1, 2 and 3 integrons. Ann Clin Microbiol Antimicrob. 2015;14:45. doi: 10.1186/s12941-015-0100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fàbrega A, Madurga S, Giralt E, Vila J. Mechanism of action of and resistance to quinolones. Microb Biotechnol. 2009;2:40–61. doi: 10.1111/j.1751-7915.2008.00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hülter N, et al. An evolutionary perspective on plasmid lifestyle modes. Curr Opin Microbiol. 2017;38:74–80. doi: 10.1016/j.mib.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Cole ST, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 21.Parkhill J, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 22.Kuroda M, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 23.Holt KE, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia B, et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta SK, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunt M, et al. ARIBA: Rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom. 2017;3:e000131. doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inouye M, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berglund F, et al. Identification of 76 novel B1 metallo-β-lactamases through large-scale screening of genomic and metagenomic data. Microbiome. 2017;5:134. doi: 10.1186/s40168-017-0353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pawlowski AC, et al. A diverse intrinsic antibiotic resistome from a cave bacterium. Nat Commun. 2016;7:13803. doi: 10.1038/ncomms13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolas-Chanoine MH, et al. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother. 2008;61:273–281. doi: 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 31.Coque TM, et al. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg Infect Dis. 2008;14:195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong VK, et al. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet. 2015;47:632–639. doi: 10.1038/ng.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuijper EJ, et al. Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Euro Surveill. 2008;13:18942. [PubMed] [Google Scholar]
- 34.Freeman J, et al. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev. 2010;23:529–549. doi: 10.1128/CMR.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pitout JD, DeVinney R. Escherichia coli ST131: A multidrug-resistant clone primed for global domination. F1000 Res. 2017;6:195. doi: 10.12688/f1000research.10609.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014;27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoesser N, et al. Modernizing Medical Microbiology Informatics Group (MMMIG) Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. MBio. 2016;7:e02162. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ben Zakour NL, et al. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. MBio. 2016;7:e00347–16. doi: 10.1128/mBio.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petty NK, et al. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci USA. 2014;111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He M, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawley TD, et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun. 2009;77:3661–3669. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feasey NA, et al. Rapid emergence of multidrug resistant, H58-lineage Salmonella Typhi in Blantyre, Malawi. PLoS Negl Trop Dis. 2015;9:e0003748. doi: 10.1371/journal.pntd.0003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klemm EJ, et al. Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. MBio. 2018;9:e00105–18. doi: 10.1128/mBio.00105-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson R, et al. Comparison of Salmonella enterica serovars Typhi and Typhimurium reveals typhoidal-specific responses to bile. Infect Immun. 2017;86:e00490-17. doi: 10.1128/IAI.00490-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uche IV, MacLennan CA, Saul A. A systematic review of the incidence, risk factors and case fatality rates of invasive nontyphoidal Salmonella (iNTS) disease in Africa (1966 to 2014) PLoS Negl Trop Dis. 2017;11:e0005118. doi: 10.1371/journal.pntd.0005118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gordon MA, et al. Non-typhoidal Salmonella bacteraemia among HIV-infected Malawian adults: High mortality and frequent recrudescence. AIDS. 2002;16:1633–1641. doi: 10.1097/00002030-200208160-00009. [DOI] [PubMed] [Google Scholar]
- 47.Ashton PM, et al. Public health surveillance in the UK revolutionises our understanding of the invasive Salmonella Typhimurium epidemic in Africa. Genome Med. 2017;9:92. doi: 10.1186/s13073-017-0480-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gordon MA, et al. Epidemics of invasive Salmonella enterica serovar enteritidis and S. enterica serovar Typhimurium infection associated with multidrug resistance among adults and children in Malawi. Clin Infect Dis. 2008;46:963–969. doi: 10.1086/529146. [DOI] [PubMed] [Google Scholar]
- 49.Brent AJ, et al. Salmonella bacteremia in Kenyan children. Pediatr Infect Dis J. 2006;25:230–236. doi: 10.1097/01.inf.0000202066.02212.ff. [DOI] [PubMed] [Google Scholar]
- 50.Okoro CK, et al. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet. 2012;44:1215–1221. doi: 10.1038/ng.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kariuki S, et al. Ceftriaxone-resistant Salmonella enterica serotype Typhimurium sequence type 313 from Kenyan patients is associated with the blaCTX-M-15 gene on a novel IncHI2 plasmid. Antimicrob Agents Chemother. 2015;59:3133–3139. doi: 10.1128/AAC.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feasey NA, et al. Drug resistance in Salmonella enterica ser. Typhimurium bloodstream infection, Malawi. Emerg Infect Dis. 2014;20:1957–1959. doi: 10.3201/eid2011.141175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kariuki S, et al. Lack of clonal relationship between non-typhi Salmonella strain types from humans and those isolated from animals living in close contact. FEMS Immunol Med Microbiol. 2002;33:165–171. doi: 10.1111/j.1574-695X.2002.tb00587.x. [DOI] [PubMed] [Google Scholar]
- 54.Hammarlöf DL, et al. Role of a single noncoding nucleotide in the evolution of an epidemic African clade of Salmonella. Proc Natl Acad Sci USA. 2018;115:E2614–E2623. doi: 10.1073/pnas.1714718115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelly CP, Pothoulakis C, LaMont JT. Clostridium difficile colitis. N Engl J Med. 1994;330:257–262. doi: 10.1056/NEJM199401273300406. [DOI] [PubMed] [Google Scholar]
- 56.Lawley TD, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leffler DA, Lamont JT. Clostridium difficile infection. N Engl J Med. 2015;373:287–288. doi: 10.1056/NEJMc1506004. [DOI] [PubMed] [Google Scholar]
- 58.Carter GP, et al. Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during Clostridium difficile infections. MBio. 2015;6:e00551. doi: 10.1128/mBio.00551-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weill FX, et al. Genomic history of the seventh pandemic of cholera in Africa. Science. 2017;358:785–789. doi: 10.1126/science.aad5901. [DOI] [PubMed] [Google Scholar]
- 60.Mutreja A, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477:462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perron GG, et al. Functional characterization of bacteria isolated from ancient arctic soil exposes diverse resistance mechanisms to modern antibiotics. PLoS One. 2015;10:e0069533. doi: 10.1371/journal.pone.0069533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.D’Costa VM, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 63.Forsberg KJ, et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Humeniuk C, et al. Beta-lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob Agents Chemother. 2002;46:3045–3049. doi: 10.1128/AAC.46.9.3045-3049.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poirel L, Rodriguez-Martinez JM, Mammeri H, Liard A, Nordmann P. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob Agents Chemother. 2005;49:3523–3525. doi: 10.1128/AAC.49.8.3523-3525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perry JA, Wright GD. The antibiotic resistance “mobilome”: Searching for the link between environment and clinic. Front Microbiol. 2013;4:138. doi: 10.3389/fmicb.2013.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang X, et al. Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat Commun. 2017;8:15784. doi: 10.1038/ncomms15784. [DOI] [PMC free article] [PubMed] [Google Scholar]