Significance
We are entering a new era in cell cartography, wherein proteins and multiprotein complexes will be mapped with nanoscale precision using advanced instrumentation. The central obstacle hindering progress in this area is the shortage of methods for labeling proteins for imaging by both fluorescence microscopy and electron microscopy. In this report, we describe a technology for tracking and mapping proteins by multiscale microscopy. To do this, we developed an innovative technology called VIPER. VIPER consists of a heterodimeric coiled-coil between a genetically encoded peptide tag (CoilE) and a reporter-conjugated peptide (CoilR). The key finding is that VIPER delivers a variety of chemical reporters, thus enabling effortless switching from fluorescence microscopy to high-resolution electron microscopy imaging without changing the genetically encoded tag.
Keywords: fluorescence microscopy, electron microscopy, coiled coil, biochemistry, chemical biology
Abstract
Many discoveries in cell biology rely on making specific proteins visible within their native cellular environment. There are various genetically encoded tags, such as fluorescent proteins, developed for fluorescence microscopy (FM). However, there are almost no genetically encoded tags that enable cellular proteins to be observed by both FM and electron microscopy (EM). Herein, we describe a technology for labeling proteins with diverse chemical reporters, including bright organic fluorophores for FM and electron-dense nanoparticles for EM. Our technology uses versatile interacting peptide (VIP) tags, a class of genetically encoded tag. We present VIPER, which consists of a coiled-coil heterodimer formed between the genetic tag, CoilE, and a probe-labeled peptide, CoilR. Using confocal FM, we demonstrate that VIPER can be used to highlight subcellular structures or to image receptor-mediated iron uptake. Additionally, we used VIPER to image the iron uptake machinery by correlative light and EM (CLEM). VIPER compared favorably with immunolabeling for imaging proteins by CLEM, and is an enabling technology for protein targets that cannot be immunolabeled. VIPER is a versatile peptide tag that can be used to label and track proteins with diverse chemical reporters observable by both FM and EM instrumentation.
Recent advances in imaging instrumentation and computational analysis have created an exciting opportunity for investigating the molecular basis of diseases with extraordinary detail. For example, in the area of fluorescence microscopy (FM), the development of superresolution microscopy (SRM) (1–3) has enabled new discoveries on the structure, organization, and dynamics of organelles (4–6). While SRM offers better resolution than conventional FM, it still falls short of obtaining the ultrastructural detail and cellular context afforded by electron microscopy (EM). EM is therefore more useful for imaging nanoscale subcellular features, including neuronal connections and components of the endocytic machinery. Correlative light and EM (CLEM) combines the best features of FM and EM (7, 8), but there are few methods for labeling and tracking cellular proteins across size scales and imaging platforms. New protein tags for multiscale microscopy need to be developed to fully exploit the potential of these technologies.
How can cellular proteins be labeled to take advantage of these new technologies? Immunolabeling is one of the only methods compatible with FM, EM, and CLEM. Antibodies can be conjugated to various chemical reporters. However, labeling proteins with antibodies has several drawbacks. The large size of antibodies reduces localization precision and labeling protocols can disrupt cellular ultrastructure (9). Scarce proteins and rare interactions can evade detection when immunolabeling is inefficient (9, 10). Many antibodies have poor specificity and cross-reactivity (11, 12), which can result in misleading observations. To summarize, issues with immunolabeling have led to widespread interest in having better genetically encoded tags for imaging cellular proteins.
Genetically encoded tags are widely available for FM, and a subset are compatible with SRM (1). However, most tags for FM are large (18–33 kDa), which can have negative consequences on protein folding, trafficking, and function (13, 14). Commonly used tags include fusions to fluorescent proteins, DNA alkyltransferases (15, 16), a dehalogenase (17), or dihydrofolate reductase (18). By comparison, there is a scarcity of genetically encoded tags for EM. There have been efforts to develop metal-chelating tags, but those tags have not been widely adopted due to multimerization, size, toxicity, and poor contrast (19–23). All other EM tags, including APEX and miniSOG (24–26), use the oxidation of diaminobenzidine (DAB) to form an insoluble polymer that is stained to generate contrast (27–29). DAB precipitation is difficult to control, which limits localization precision. A major shortcoming of the DAB-based tags is their reliance on the same chemical reaction to generate contrast.
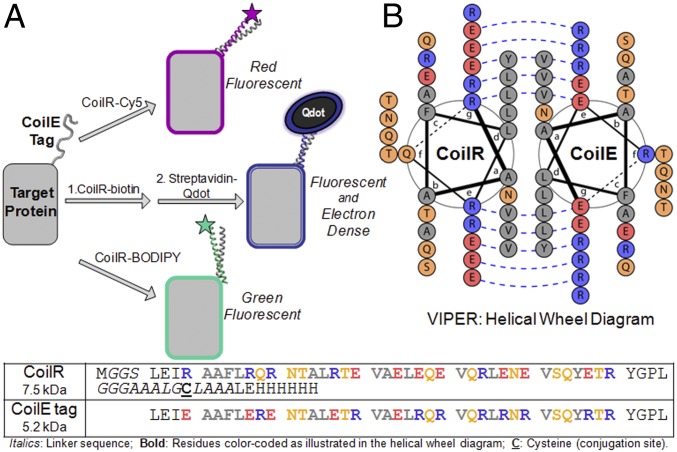
We report herein a technology that enables effortless switching from FM to high-resolution EM without changing the genetically encoded tag. In 2017, we published our first versatile interacting peptide (VIP) tag, named VIP Y/Z (30). Now we present VIPER, a distinct peptide tag that has high specificity in a miniaturized size. VIPER uses a heterodimeric coiled-coil between two peptides, a genetically encoded peptide tag (CoilE) and a reporter-conjugated peptide (CoilR), to label cellular proteins with several distinct chemical reporters (Fig. 1). The genetically encoded peptide, CoilE, is one of the smallest available peptide tags (5.2 kDa). We validated the specificity and versatility of VIPER by imaging CoilE-tagged proteins by both FM and EM.
Fig. 1.
VIPER is an enabling technology for multiscale microscopy. (A) A target protein is genetically tagged with the CoilE peptide. Then the tagged protein can be labeled by dimerization with a CoilR peptide covalently bound to various chemical reporters, including BODIPY, Sulfo-Cyanine5 (Cy5), or biotin for detection by a streptavidin-Qdot. (B) Helical wheel diagram of VIPER generated using DrawCoil 1.0. Sequences for the CoilE tag and the CoilR probe peptide are provided.
Results and Discussion
Design of VIPER, a Genetically Encoded Tag.
Most genetically encoded tags rely on large, complex protein structures to deliver contrast. Such tags are challenging and time-intensive to engineer. For example, it took 5 years to convert SNAP into CLIP (15) and 20 years to develop a satisfactory near-infrared fluorescent protein (31, 32). In contrast, VIP tags use an α-helical coiled-coil to label proteins. This is a simple structural motif amenable to design and optimization. Dimerization specificity and affinity are dictated by the peptide sequence (33–38). For VIP Y/Z (30), we adapted a heterodimeric coiled-coil reported by Keating and coworkers (35). That dimer had a reported dissociation constant (KD) of <15 × 10−9 M and a melting temperature (Tm) of 32 °C (35). VIP Y/Z precisely labeled protein targets in living cells with various chemical reporters, including fluorophores and quantum dots (Qdots) (30).
For the present work, we developed a distinct VIP tag with higher affinity. We selected a heterodimeric pair described by Vinson and coworkers (33): RR12EE345L and EE12RR345L. Dimerization between these two peptides is driven by a hydrophobic interface and optimized interstrand salt bridges, as shown in Fig. 1B. The result is a remarkably high-affinity dimer (KD 1.3 × 10−11 M; Tm 73 °C) (33). We used these peptides to create a CoilE tag and CoilR probe peptide, which dimerize to produce VIPER.
Homology-based gene assembly was used to introduce the CoilE tag into target proteins. CoilR probe peptides were generated by recombinant bacterial expression. The CoilR sequence included a hexahistidine tag for purification and a cysteine for site-specific labeling using thiol-maleimide chemistry. These features enabled us to rapidly generate a set of probe peptides: CoilR-biotin, CoilR-Cy5, and CoilR-BODIPY.
Localization of VIPER-Tagged Proteins to Distinct Subcellular Structures.
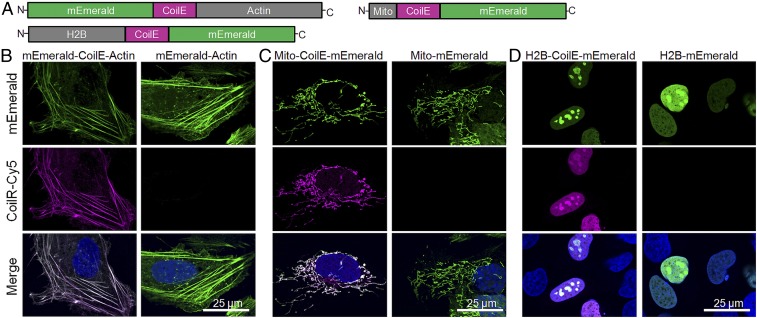
Our first priority was to establish that VIPER enabled selective labeling of cellular proteins. We selected three distinctive subcellular structures for labeling: the cytoskeleton (β-actin), nucleus (histone 2B; H2B), and the mitochondrial matrix (using a COX8 fragment encoding a localization sequence; “Mito”). We obtained mammalian expression vectors that encoded each target protein fused to a monomeric green fluorescent protein, mEmerald (39). We modified each vector to insert the CoilE sequence intragenically between the target protein and mEmerald (Fig. 2A). We transfected human osteosarcoma (U-2 OS) cells with vectors encoding tagged proteins, which we named mEmerald-CoilE-Actin, Mito-CoilE-mEmerald, and H2B-CoilE-mEmerald. We also transfected cells with proteins lacking the CoilE tag (mEmerald-Actin, Mito-mEmerald, H2B-mEmerald). Cells were fixed, permeabilized, and blocked before treatment with CoilR-Cy5.
Fig. 2.
Selective fluorescent labeling of cellular actin, mitochondria, and the nucleus using VIPER. (A) Representation of the CoilE-tagged proteins used to label the cytoskeleton (mEmerald-CoilE-Actin), mitochondria (Mito-CoilE-mEmerald), and the nucleus (H2B-CoilE-mEmerald). Transfected U-2 OS cells were labeled postfixation by treatment with CoilR-Cy5 (100 nM) and then imaged by confocal FM to observe the cytoskeleton (B), mitochondria (C), or the nucleus (D). CoilR-Cy5 labeling was specific for CoilE-tagged proteins, and the Cy5 (magenta) and mEmerald (green) signal colocalized. Green-magenta overlap appears white in the merged images and the nuclear stain (Hoechst 33342) is false-colored blue.
We used confocal FM to assess VIPER labeling and specificity in cells (Fig. 2). Transfected cells were identified using mEmerald fluorescence. We found that CoilR-Cy5 highlighted subcellular structures only in cells expressing CoilE-tagged proteins. For example, in cells expressing mEmerald-CoilE-Actin, CoilR-Cy5 fluorescence (magenta) colocalized with mEmerald fluorescence (green) (Fig. 2B). Similarly, cells expressing Mito-CoilE-mEmerald or H2B-CoilE-mEmerald had colocalized fluorescence in the mitochondria or nucleus, respectively (Fig. 2 C and D). CoilR-Cy5 signal in cells expressing the untagged mEmerald constructs was nearly undetectable. These results demonstrate that VIPER-labeling was selective and the CoilE tag did not change or disrupt the target protein’s localization. Our results showed that VIPER-labeling occurred with the CoilE tag inserted between two proteins, a useful feature for labeling proteins that do not tolerate tags at the N or C terminus.
We used a competition binding assay to assess VIPER labeling efficiency. Fixed cells were pretreated with increasing concentrations of unlabeled CoilR peptide (0, 100, 1000, 10,000, and 100,000 nM) to block subsequent Cy5 labeling of CoilE-tagged proteins. Then cells were treated with 100 nM CoilR-Cy5 to label the remaining unbound CoilE-tagged proteins. Pretreatment with 100 nM unlabeled CoilR peptide was sufficient to reduce the labeling by CoilR-Cy5 (SI Appendix, Fig. S1). Cy5 fluorescence became nearly undetectable after pretreatment with a 10-fold excess of unlabeled CoilR for cells expressing mEmerald-CoilE-Actin or Mito-CoilE-mEmerald. Cy5 signal localized to nucleoli was detected for cells pretreated with ≥1,000 nM CoilR, but the signal was reduced and became increasingly difficult to detect. H2B localized to a small, subnuclear volume, a feature that rendered CoilR-Cy5 locally concentrated and more detectable. Overall, our treatment conditions were sufficient to efficiently label most, but not all, of the CoilE-tagged targets in fixed cells.
Imaging Iron Uptake Using VIPER.
Next, we assessed VIPER by imaging two components of the iron uptake machinery: transferrin (Tf) and transferrin receptor 1 (TfR1). The TfR1 pathway is a well-described system for receptor-mediated endocytosis (40, 41). Briefly, iron-loaded Tf binds to TfR1 and the complex internalizes through clathrin-coated vesicles. These endosomes acidify, releasing iron from Tf. Reduced iron is transported into the cytosol, where it is used by iron-requiring proteins or stored. Then the apo–Tf/TfR1 complex recycles to the cell surface. Tf is released from TfR1, enabling the process to restart. Iron uptake is fast, with internalization of the Tf/TfR1 complex into early endosomes occurring within minutes of Tf binding and recycling of Tf-TfR1 back to the surface occurring in under 20 min (42).
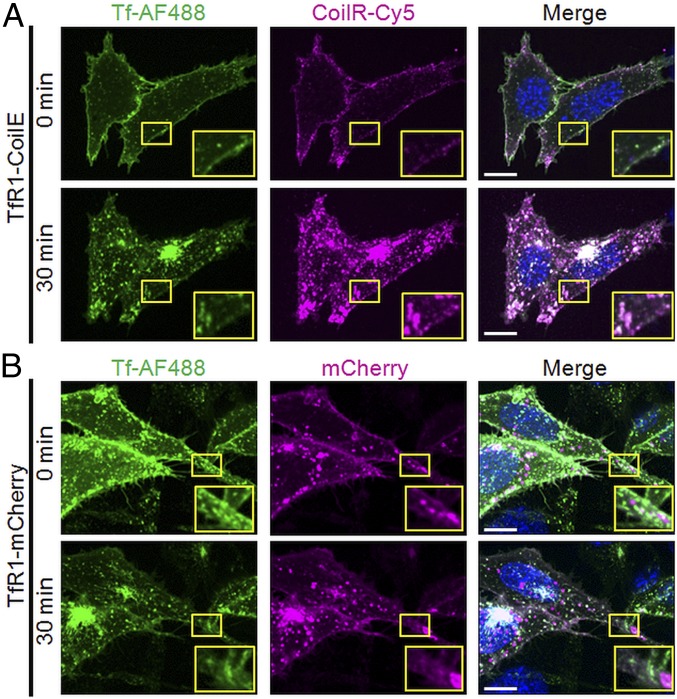
We used confocal FM to observe Tf and TfR1 localization and trafficking in living cells. We generated a vector with the CoilE tag at the extracellular, C-terminal domain of TfR1 (pcDNA3.1_TfR1-CoilE). For comparative analysis, we acquired a vector encoding TfR1 fused to the monomeric red fluorescent protein mCherry (pcDNA3_TfR1-mCherry; Addgene #55144). We used the Chinese hamster ovary (CHO) TRVb cell line for these studies, which does not express TfR1 or the closely related transferrin receptor 2 (TfR2) (43). We selected this cell line to ensure that all cellular TfR1 would be tagged by either CoilE or mCherry. Transfected cells were cooled to 4 °C to pause endocytosis and treated for 30 min with fluorescent ligand, Tf-AF488. Cells expressing TfR1-CoilE were simultaneously treated with CoilR-Cy5, while cells expressing TfR1-mCherry were not. Cells were washed, returned to 37 °C, and imaged immediately after labeling (0 min) and at 30 min.
At both time points, VIPER-tagged TfR1 colocalized with Tf-AF488 (Fig. 3), which provides strong evidence that tagged TfR1 retains its ligand-binding function. Most of the fluorescent signal from CoilR-Cy5 (receptor) and AF488 (ligand) was restricted to the cell surface at 0 min before appearing in bright, fluorescent endosomes at 30 min. At 30 min, some of the cell surface TfR1 no longer colocalized with Tf, consistent with recycling of the complex and release of Tf into the media. These results demonstrate that VIPER enables observation of receptor–ligand binding interactions, receptor endocytosis, and receptor recycling (indirectly). We found that the CoilR probe peptides were live-cell impermeant. As a result, CoilR-Cy5 labeling was restricted to the cell surface-localized TfR1-CoilE, which enabled us to follow the endocytosis of that pool of receptors.
Fig. 3.
VIPER-tagged transferrin receptor retains transferrin binding and endocytosis. (A) CHO TRVb cells expressing TfR1-CoilE were treated with CoilR-Cy5 and fluorescent ligand (Tf-AF488). In live cells, labeling by both Tf-AF488 and CoilR-Cy5 was localized to the cell surface at 0 min. After 30 min, AF488 and Cy5 signals from the Tf-TfR1 complex were observed together in endocytic vesicles. (B) Cells expressing TfR1-mCherry were treated with Tf-AF488. In A and B, yellow boxes delineate Insets, which provide a 2× magnified view. The merged images (Right column) include Tf-AF488 (green), nuclear stain (blue), and either mCherry (magenta) or CoilR-Cy5 (magenta). (Scale bars: 25 μm.)
Next, we imaged cells expressing TfR1-mCherry (Fig. 3B). Compared with VIPER, we observed less colocalization of green and red fluorescence at both time-points. We found that the Pearson’s correlation coefficient of Tf-AF488 with VIPER-labeled receptor (AF488 with Cy5) at both 0 min (81%) and 30 min (87%) was greater than that of Tf-AF488 with TfR1-mCherry, which was 65% at 0 min and 75% at 30 min (SI Appendix, Fig. S2). This study highlights a key feature of the VIPER tag: only VIPER enabled the unambiguous observation of cell-surface receptors being internalized following treatment with fluorescent ligand.
In a related experiment, we demonstrated that Tf and TfR1 internalization could be observed by time-lapse imaging. We acquired a 25-min time-course for CHO TRVb cells expressing either a tagged (TfR1-CoilE) or untagged (TfR1) receptor. VIPER labeling was highly specific, with CoilR-Cy5 signal only observed for cells expressing TfR1-CoilE and not for untagged TfR1. The complete time-course can be found in SI Appendix, Fig. S3.
Two-Color Pulse-Chase Labeling of TfR1.
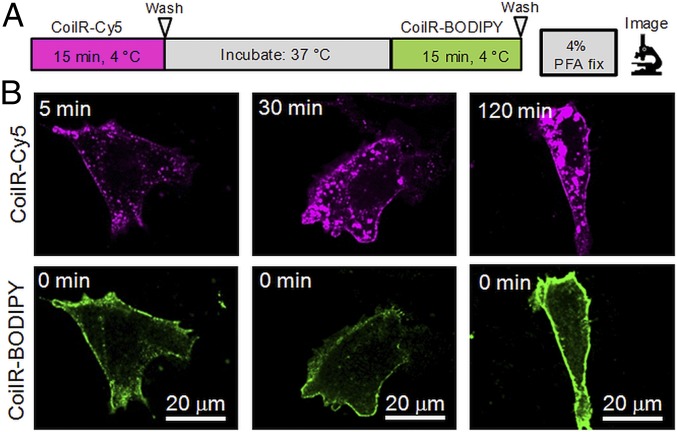
Pulse-chase labeling is an established method for sequentially labeling cells with distinguishable reporters. This method relies on fast labeling and live-cell compatibility to obtain two-color, time-resolved images of dynamic protein populations. We used VIPER to pulse-label a cell surface population of TfR1 with red-fluorescent Cy5 and then labeled a second population with green fluorescent BODIPY (Fig. 4). Briefly, CHO TRVb cells expressing TfR1-CoilE were cooled to 4 °C and then treated with CoilR-Cy5 to pulse-label receptors on the cell surface. We returned cells to 37 °C and allowed the Cy5-labeled receptors to distribute for 5, 30, or 120 min. Next, cells were treated with ice-cold CoilR-BODIPY to chase-label a second population of receptors. Cells were washed, fixed, and imaged. At each time-point, we observed the CoilR-Cy5–labeled TfR1 population (magenta) primarily within vesicles, consistent with rapid endocytosis of TfR1. In contrast, the CoilR-BODIPY–labeled TfR1 (green) was primarily localized to the cell surface. A small portion of BODIPY-labeled receptors appeared in fluorescent punctae, consistent with prior reports that some Tf-internalization occurs at 4 °C (40). This experiment was also performed with untagged TfR1, which verified that CoilR labeling was specific for TfR1-CoilE (SI Appendix, Fig. S4). This pulse-chase labeling experiment demonstrates that the VIPER technology can be used to track two distinct populations of receptors over time. Additionally, VIPER labeling was rapid, achieving sufficient labeling within 15 min of treatment.
Fig. 4.
Two-color pulse-chase labeling of TfR1. (A) Schematic of the pulse-chase labeling protocol. (B) Cells expressing TfR1-CoilE were pulse-labeled with CoilR-Cy5 (500 nM, 15 min), washed, and returned to 37 °C for 5, 30, or 120 min. TfR1-CoilE was then chase-labeled with CoilR-BODIPY (500 nM, 15 min), fixed, and imaged to detect both Cy5-labeled receptor (magenta) and BODIPY-labeled receptor (green).
VIPER Enables Protein Labeling for Multiscale Microscopy.
A new technology is required for labeling and observing proteins by both FM and EM. The instrumentation for multiscale microscopy is now available. We used a commercial CLEM instrument, the FEI CorrSight, which enables biological samples to be prescreened by FM before processing for EM (44). The FEI MAPS software facilitates the selection and tracking of cells across imaging platforms. Additionally, CLEM reporters are commercially available. Qdots are brightly fluorescent and electron dense, two essential features for multiscale microscopy (45).
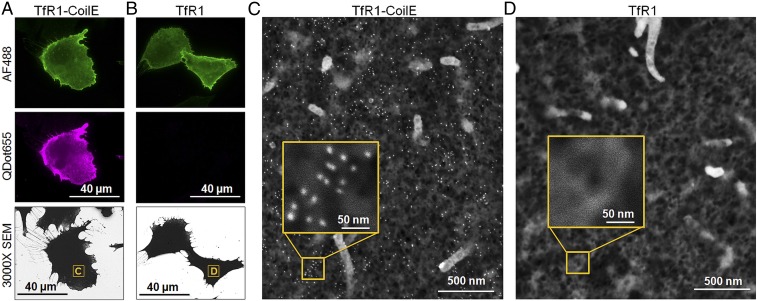
The VIPER tag can be used to label proteins with Qdots. We sought to determine if this unique feature could be used for multiscale microscopy of the iron uptake machinery. For these studies, live CHO TRVb cells were treated with both Tf-AF488 and biotinylated CoilR (CoilR-biotin), which we detected postfixation using streptavidin-Qdot655. We imaged cells first by FM and then by scanning EM (SEM) (Fig. 5). Fluorescence micrographs allowed us to identify transfected cells, which bound Tf-AF488. Additionally, we observed bright Qdot655 fluorescence associated with cells expressing TfR1-CoilE (Fig. 5A), but not for cells expressing untagged TfR1 (Fig. 5B). We used MAPS software to register the coordinates of fluorescent cells relative to the slide so that we could relocate the same cells for SEM imaging.
Fig. 5.
Imaging TfR1 by multiscale microscopy. Transfected CHO TRVb cells were treated with CoilR-biotin and Tf-AF488. After fixation, cells were treated with streptavidin-Qdot655 to detect biotinylated (VIPER-tagged) receptors. (A) Fluorescence micrographs of cells expressing TfR1-CoilE. Transfected cells were identified based on binding to Tf-AF488 (green) and labeling by Qdot655 (magenta). (B) Fluorescence micrographs of cells expressing untagged TfR1. We selected region C (in A) and region D (in B) for high-resolution SEM imaging. Samples were processed before imaging at 65,000× magnification. SEM micrographs of cells expressing TfR1-CoilE (C) showed selective Qdot labeling, while labeling was not observed on cells expressing untagged TfR1 (D). The Insets provide a magnified view of the boxed region.
Next, samples were dehydrated and carbon-coated for imaging by SEM. Micrographs were acquired on an FEI Helios Nanolab 660, which provided a topographical view of the cells preselected by FM. At 65,000× magnification, Qdots were observed as small, bright-white spheres on a dark gray background (see Fig. 5C, Inset for a magnified view of Qdots). Raised features on the cell surface, such as membrane protrusions, appear light gray or white. Micrographs revealed dense Qdot655 labeling for cells expressing TfR1-CoilE (Fig. 5C). The Qdot labeling enabled by VIPER appeared to be highly specific, with almost no nonspecific association of particles with TfR1 or cell surfaces (Fig. 5D).
Other EM tags use DAB precipitation to generate contrast (24–29), but the reaction product can be difficult to control. In contrast, Qdot-based target detection enables quantitative image analysis because labeling is stoichiometric. To demonstrate this feature, we algorithmically segmented and counted the number of Qdot655 particles per field-of-view in SEM micrographs. We captured 12 images (n = 6 cells) per condition. In VIPER-labeled cells, we identified single Qdots, dimers, and multimers, but found that most Qdots were distributed as monomers (SI Appendix, Table S1). SI Appendix, Figs. S5 and S6 provide representative micrographs with particle segmentation. We determined that there were 110 ± 34 Qdots/µm2 (mean ± SD) in cells expressing TfR1-CoilE; Qdot density ranged from 63 to 190 Qdots/µm2. We attribute this variation to receptor expression differences among transiently transfected cells. For cells expressing untagged TfR1, we observed an average of 0 Qdots/µm2. This CLEM study shows that VIPER is an effective EM tag that enables high-fidelity labeling of cell receptors with Qdots. We anticipate that the ability to identify a protein’s subcellular localization, clustering, and relative abundance will be useful for various applications in cell biology.
Comparison of VIPER with Immunolabeling.
Immunolabeling is widely used for labeling and imaging target proteins by FM, EM, and CLEM. For EM, proteins are typically treated with a primary antibody generated against the protein and a secondary antibody delivering an electron-dense reporter (e.g., colloidal gold or a Qdot). We selected three commercial antibodies against the extracellular domain of TfR1: 8D3 (46), Ab1086 (47), and Ab216665. Primary antibodies were detected by an antihost secondary antibody conjugated to Qdot655. We used Qdot655, instead of colloidal gold, to enable a direct comparison with our VIPER Qdot labeling. For these studies, we used TfR1-CoilE expressing CHO TRVb cells treated live with Tf-AF488. We selected cells to image based on Tf binding, attempting to match the Tf-AF488 green fluorescence intensity among samples. Fixed cells were then VIPER-labeled or immunolabeled.
For two of the antibodies, Ab1086 and Ab216665, we were unable to identify conditions for labeling the receptor, a common problem encountered by researchers using commercial antibodies. We saw no evidence of TfR1-CoilE labeling with Ab1086 and Ab216665 by FM or EM (SI Appendix, Fig. S7). However, we observed selective labeling of TfR1 by antibody 8D3, which was detected by a goat anti-rat IgG antibody conjugated to Qdot655 (Fig. 6). Qualitatively, immunolabeling with 8D3 (Fig. 6D) looked similar to VIPER labeling (Fig. 6C). However, quantitative analysis of six images per condition (n = 3 cells) indicated that 8D3 labeling (464 ± 97 Qdots/µm2) was more efficient than VIPER (270 ± 85 Qdots/µm2). However, it is also possible that indirect detection of the receptor resulted in multiple secondary antibodies bound to a single 8D3 primary.
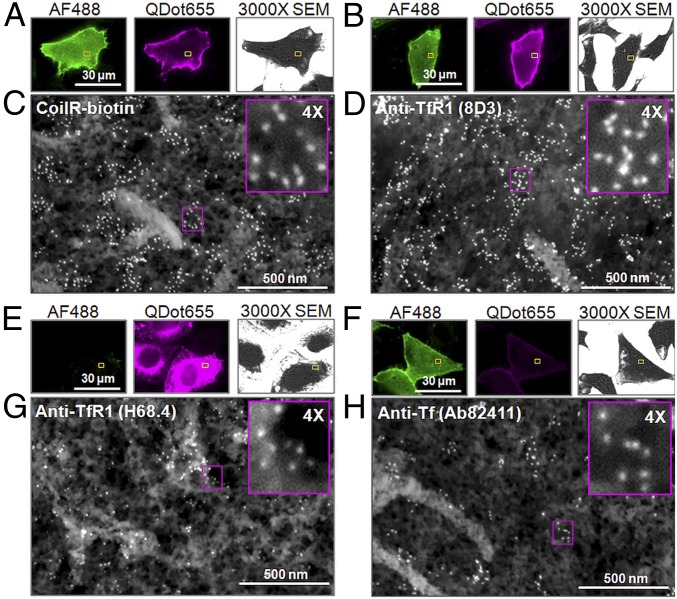
Fig. 6.
Target labeling and CLEM imaging by VIPER or immunolabeling. Cells expressing TfR1-CoilE were identified by binding to Tf-AF488. For VIPER labeling, fixed cells were treated with CoilR-biotin and streptavidin-Qdot655 (A). For immunolabeling, cells were treated with a primary antibody against Tf (Ab82411; F) or TfR1 [8D3 (B) or H68.4 (E)]. Primary antibodies were detected using secondary antibodies conjugated to Qdot655. Fluorescence micrographs of cells labeled with VIPER (A), 8D3 (B), H68.4 (E), and Ab82411 (F) were acquired and mapped for high-resolution SEM. After processing, we selected regions (yellow boxes) for SEM imaging at 100,000× magnification. The high-resolution view shows Qdot labeling of the cell surface (C, D, G, and H). Magenta Insets present a 4× magnification of the Qdots. For H68.4, detergent treatment caused membrane extraction, as observed by 3000× SEM, and the Tf-AF488 signal was reduced.
We next evaluated a widely used anti-TfR1 antibody: H68.4 (48). Cells had to be permeabilized postfixation to label the cytosolic domain of TfR1 with H68.4, which damaged the cell membrane (Fig. 6E). Moreover, loss of Tf-AF488 fluorescence occurred during permeabilization, presumably due to loss of membrane-associated Tf. With H68.4, we observed 182 ± 63 Qdots/µm2, substantially less than observed with 8D3 or VIPER. We also compared VIPER-labeling of TfR1 to immunolabeling of Tf (Fig. 6 F and H), which was inefficient (91 ± 19 Qdots/µm2) compared with VIPER, 8D3, or H68.4. Overall, VIPER surpassed immunolabeling for four of five antibodies evaluated.
Biotinylated Tf enabled us to do a final comparison with VIPER. Homodimeric TfR1 binds two iron-loaded Tf with low nanomolar affinity (49). Complete labeling of TfR1 with either CoilR-biotin or Tf-biotin would place two biotinylated ligands on each receptor complex. Therefore, streptavidin-Qdot655 detection of Tf-biotin should be comparable to detection of CoilR-labeled TfR1. We tested this hypothesis and observed selective Qdot655 labeling for both samples by FM and EM (SI Appendix, Fig. S8). We counted 210 ± 71 Qdots/µm2 for VIPER and 258 ± 65 Qdots/µm2 for Tf (SI Appendix, Table S2), a difference that is not statistically significant (P = 0.126). To summarize, our comparative analyses demonstrate that VIPER labeling is a selective and versatile alternative to commonly used methods for imaging proteins by EM or CLEM.
Conclusion
VIP tags are a new addition to the microscopy toolkit, joining immunolabeling, fluorescent proteins, and other genetically encoded tags. VIPER is a small tag, adding a peptide of less than 6 kDa to the target protein. We provide evidence that VIPER has high specificity for labeling various subcellular targets, including the cytoskeleton, mitochondria, and nucleus. Proteins can be labeled with various reporter chemistries either pre- or postfixation. We demonstrated the range of reporters by labeling TfR1-CoilE with CoilR-BODIPY, CoilR-Cy5, and CoilR-biotin. There are myriad reactive fluorophores and fluorescent sensors that could be site-specifically conjugated to CoilR, providing many other options for imaging applications.
VIPER is compatible with live-cell and dynamic imaging. We tagged TfR1 with CoilE to image iron uptake in cells. VIPER did not appear to affect protein localization, function (e.g., ligand binding), or trafficking. Importantly, VIPER enabled receptor populations to be dynamically observed and spatio-temporally resolved. Labeling was achieved within 15 min (Fig. 4). We did not evaluate shorter CoilR labeling times, but we believe that faster labeling is feasible.
We developed VIPER as a technology for multiscale microscopy. The CLEM studies presented here demonstrate that VIPER labeling offers a compelling alternative to immunolabeling. We evaluated four anti-TfR1 antibodies in direct comparison with VIPER. While both 8D3 and H68.4 selectively labeled TfR1, H68.4 labeling required processing that compromised the cell membrane. Half of the evaluated anti-TfR1 antibodies failed to label TfR1. Unlike VIPER, immunolabeling is reliant on antibodies with widely variable target specificity and affinity. For proteins that lack a specific antibody, VIPER creates an opportunity to observe those targets by multiscale microscopy.
We anticipate that the EM-compatibility of VIPER will be particularly useful for exploring the subcellular localization and assembly of cellular proteins. In future studies, VIPER-mediated biotinylation of receptors could be detected with other reporters, such as streptavidin-gold. Alternatively, CoilR could be direct-conjugated to gold for EM or bifunctionalized with a fluorophore plus gold for CLEM. These options should be explored with the goal of using multicolor imaging to study multiprotein assemblies with nanoscale precision.
VIPER’s compatibility with various chemical reporters creates a flexibility unmatched by the DAB-based tags. However, it is important to emphasize that VIPER does not replace or supersede all other genetically encoded tags. Rather VIPER augments other labeling methods, such as immunolabeling or fluorescent proteins, to enable researchers to tag and track multiple distinct targets at once. We anticipate that this enhanced microscopy toolkit will facilitate the generation of more detailed and informative maps of cellular proteins.
Materials and Methods
See SI Appendix, Materials and Methods for detailed VIPER-labeling protocols. The SI Appendix also provides a description of our quantitative image analysis. Supporting data are provided in SI Appendix, Figs. S1–S12 and Tables S1–S9.
Supplementary Material
Acknowledgments
We thank Dr. Michael Davidson (Florida State University) for depositing his fluorescent protein collection into Addgene; our colleagues at Oregon Health & Science University for their advice, particularly Drs. Joe Gray, Jim Korkola, Dan Zuckerman, Danielle Jorgens, Stefanie Kaech Petrie, and Crystal Chaw; and Dr. Timothy McGraw (Cornell University), who generously provided the Chinese hamster ovary TRVb cell line. Electron microscopy was performed at the Multiscale Microscopy Core. This research was supported by generous funding from Oregon Health & Science University and the National Institutes of Health (Grant R01 GM122854). J.K.D. was supported by the Achievement Rewards for College Scientists Foundation Oregon Chapter.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808626115/-/DCSupplemental.
References
- 1.Liu Z, Lavis LD, Betzig E. Imaging live-cell dynamics and structure at the single-molecule level. Mol Cell. 2015;58:644–659. doi: 10.1016/j.molcel.2015.02.033. [DOI] [PubMed] [Google Scholar]
- 2.Huang B, Bates M, Zhuang X. Super-resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Diezmann A, Shechtman Y, Moerner WE. Three-dimensional localization of single molecules for super-resolution imaging and single-particle tracking. Chem Rev. 2017;117:7244–7275. doi: 10.1021/acs.chemrev.6b00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson AD, Bewersdorf J, Toomre D, Schepartz A. HIDE probes: A new toolkit for visualizing organelle dynamics, longer and at super-resolution. Biochemistry. 2017;56:5194–5201. doi: 10.1021/acs.biochem.7b00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nixon-Abell J, et al. Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science. 2016;354:aaf3928. doi: 10.1126/science.aaf3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valm AM, et al. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature. 2017;546:162–167. doi: 10.1038/nature22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellisman MH, Deerinck TJ, Shu X, Sosinsky GE. Picking faces out of a crowd: Genetic labels for identification of proteins in correlated light and electron microscopy imaging. In: Müller-Reichert T, Verkade P, editors. Methods in Cell Biology. Vol 111. Academic; Waltham, MA: 2012. pp. 139–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Boer P, Hoogenboom JP, Giepmans BNG. Correlated light and electron microscopy: Ultrastructure lights up! Nat Meth. 2015;12:503–513. doi: 10.1038/nmeth.3400. [DOI] [PubMed] [Google Scholar]
- 9.Schnell U, Dijk F, Sjollema KA, Giepmans BNG. Immunolabeling artifacts and the need for live-cell imaging. Nat Methods. 2012;9:152–158. doi: 10.1038/nmeth.1855. [DOI] [PubMed] [Google Scholar]
- 10.Griffiths G, Lucocq JM. Antibodies for immunolabeling by light and electron microscopy: Not for the faint hearted. Histochem Cell Biol. 2014;142:347–360. doi: 10.1007/s00418-014-1263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradbury A, Plückthun A. Reproducibility: Standardize antibodies used in research. Nature. 2015;518:27–29. doi: 10.1038/518027a. [DOI] [PubMed] [Google Scholar]
- 12.Baker M. Reproducibility crisis: Blame it on the antibodies. Nature. 2015;521:274–276. doi: 10.1038/521274a. [DOI] [PubMed] [Google Scholar]
- 13.Huang L, Pike D, Sleat DE, Nanda V, Lobel P. Potential pitfalls and solutions for use of fluorescent fusion proteins to study the lysosome. PLoS One. 2014;9:e88893. doi: 10.1371/journal.pone.0088893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costantini LM, Snapp EL. Fluorescent proteins in cellular organelles: Serious pitfalls and some solutions. DNA Cell Biol. 2013;32:622–627. doi: 10.1089/dna.2013.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gautier A, et al. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Keppler A, et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 17.Los GV, et al. HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 18.Miller LW, Cai Y, Sheetz MP, Cornish VW. In vivo protein labeling with trimethoprim conjugates: A flexible chemical tag. Nat Methods. 2005;2:255–257. doi: 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Mercogliano CP, Löwe J. A ferritin-based label for cellular electron cryotomography. Structure. 2011;19:147–154. doi: 10.1016/j.str.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Morphew MK, et al. Metallothionein as a clonable tag for protein localization by electron microscopy of cells. J Microsc. 2015;260:20–29. doi: 10.1111/jmi.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diestra E, Fontana J, Guichard P, Marco S, Risco C. Visualization of proteins in intact cells with a clonable tag for electron microscopy. J Struct Biol. 2009;165:157–168. doi: 10.1016/j.jsb.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 22.Mercogliano CP, DeRosier DJ. Concatenated metallothionein as a clonable gold label for electron microscopy. J Struct Biol. 2007;160:70–82. doi: 10.1016/j.jsb.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clarke NI, Royle SJ. FerriTag is a new genetically-encoded inducible tag for correlative light-electron microscopy. Nat Commun. 2018;9:2604. doi: 10.1038/s41467-018-04993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam SS, et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods. 2015;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martell JD, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shu X, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaietta G, et al. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- 28.Kuipers J, et al. FLIPPER, a combinatorial probe for correlated live imaging and electron microscopy, allows identification and quantitative analysis of various cells and organelles. Cell Tissue Res. 2015;360:61–70. doi: 10.1007/s00441-015-2142-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liss V, Barlag B, Nietschke M, Hensel M. Self-labelling enzymes as universal tags for fluorescence microscopy, super-resolution microscopy and electron microscopy. Sci Rep. 2015;5:17740. doi: 10.1038/srep17740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zane HK, Doh JK, Enns CA, Beatty KE. Versatile interacting peptide (VIP) tags for labeling proteins with bright chemical reporters. Chembiochem. 2017;18:470–474. doi: 10.1002/cbic.201600627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez EA, et al. A far-red fluorescent protein evolved from a cyanobacterial phycobiliprotein. Nat Methods. 2016;13:763–769. doi: 10.1038/nmeth.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shcherbakova DM, Verkhusha VV. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat Methods. 2013;10:751–754. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moll JR, Ruvinov SB, Pastan I, Vinson C. Designed heterodimerizing leucine zippers with a ranger of pIs and stabilities up to 10(-15) M. Protein Sci. 2001;10:649–655. doi: 10.1110/ps.39401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woolfson DN. The design of coiled-coil structures and assemblies. In: David ADP, John MS, editors. Advances in Protein Chemistry. Vol 70. Academic; New York: 2005. pp. 79–112. [DOI] [PubMed] [Google Scholar]
- 35.Reinke AW, Grant RA, Keating AE. A synthetic coiled-coil interactome provides heterospecific modules for molecular engineering. J Am Chem Soc. 2010;132:6025–6031. doi: 10.1021/ja907617a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas F, Boyle AL, Burton AJ, Woolfson DN. A set of de novo designed parallel heterodimeric coiled coils with quantified dissociation constants in the micromolar to sub-nanomolar regime. J Am Chem Soc. 2013;135:5161–5166. doi: 10.1021/ja312310g. [DOI] [PubMed] [Google Scholar]
- 37.Yano Y, et al. Coiled-coil tag—Probe system for quick labeling of membrane receptors in living cell. ACS Chem Biol. 2008;3:341–345. doi: 10.1021/cb8000556. [DOI] [PubMed] [Google Scholar]
- 38.O’Shea EK, Lumb KJ, Kim PS. Peptide ‘Velcro’: Design of a heterodimeric coiled coil. Curr Biol. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 39.Day RN, Davidson MW. The fluorescent protein palette: Tools for cellular imaging. Chem Soc Rev. 2009;38:2887–2921. doi: 10.1039/b901966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayle KM, Le AM, Kamei DT. The intracellular trafficking pathway of transferrin. Biochim Biophys Acta. 2012;1820:264–281. doi: 10.1016/j.bbagen.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao N, Enns CA. Iron transport machinery of human cells: Players and their interactions. Curr Top Membr. 2012;69:67–93. doi: 10.1016/B978-0-12-394390-3.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciechanover A, Schwartz AL, Dautry-Varsat A, Lodish HF. Kinetics of internalization and recycling of transferrin and the transferrin receptor in a human hepatoma cell line. Effect of lysosomotropic agents. J Biol Chem. 1983;258:9681–9689. [PubMed] [Google Scholar]
- 43.McGraw TE, Greenfield L, Maxfield FR. Functional expression of the human transferrin receptor cDNA in Chinese hamster ovary cells deficient in endogenous transferrin receptor. J Cell Biol. 1987;105:207–214. doi: 10.1083/jcb.105.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.López CS, et al. A fully integrated, three-dimensional fluorescence to electron microscopy correlative workflow. In: Thomas M-R, Paul V, editors. Methods in Cell Biology. Vol 140. Academic; Cambridge, UK: 2017. pp. 149–164. [DOI] [PubMed] [Google Scholar]
- 45.Giepmans BNG, Deerinck TJ, Smarr BL, Jones YZ, Ellisman MH. Correlated light and electron microscopic imaging of multiple endogenous proteins using quantum dots. Nat Methods. 2005;2:743–749. doi: 10.1038/nmeth791. [DOI] [PubMed] [Google Scholar]
- 46.Kissel K, et al. Immunohistochemical localization of the murine transferrin receptor (TfR) on blood-tissue barriers using a novel anti-TfR monoclonal antibody. Histochem Cell Biol. 1998;110:63–72. doi: 10.1007/s004180050266. [DOI] [PubMed] [Google Scholar]
- 47.Ta H, et al. Mapping molecules in scanning far-field fluorescence nanoscopy. Nat Commun. 2015;6:7977. doi: 10.1038/ncomms8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thul PJ, et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 49.West AP, Jr, et al. Comparison of the interactions of transferrin receptor and transferrin receptor 2 with transferrin and the hereditary hemochromatosis protein HFE. J Biol Chem. 2000;275:38135–38138. doi: 10.1074/jbc.C000664200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.