Significance
First, our work provides critical biological interpretation of intermediate DNA methylation readouts at the nc886 differentially methylated region (DMR). nc886 was identified in multiple large-scale epigenome-wide association studies (EWAS) that did not recognize that this region acts as a contiguous DMR imposed by genomic imprinting, highlighting the need to reexamine several 450k data sets. Second, strict control of genomic imprinting was thought to be required for organismal viability. Reports of polymorphic imprinting are limited to specific tissue types such as placenta and brain. In blood and somatic tissues, we show nc886 imprinting is mosaic in the population and influenced by maternal environment.
Keywords: nc886, VTRNA2-1, polymorphic imprinting, epigenetic inheritance, DNA methylation
Abstract
Genomic imprinting mediated by DNA methylation restricts gene expression to a single allele determined by parental origin and is not generally considered to be under genetic or environmental influence. Here, we focused on a differentially methylated region (DMR) of approximately 1.9 kb that includes a 101-bp noncoding RNA gene (nc886/VTRNA2-1), which is maternally imprinted in ∼75% of humans. This is unlike other imprinted genes, which demonstrate monoallelic methylation in 100% of individuals. The DMR includes a CTCF binding site on the centromeric side defining the DMR boundary and is flanked by a CTCF binding site on the telomeric side. The centromeric CTCF binding site contains an A/C polymorphism (rs2346018); the C allele is associated with less imprinting. The frequency of imprinting of the nc886 DMR in infants was linked to at least two nongenetic factors, maternal age at delivery and season of conception. In a separate cohort, nc886 imprinting was associated with lower body mass index in children at 5 y of age. Thus, we propose that the imprinting status of the nc886 DMR is “tunable” in that it is associated with maternal haplotype and prenatal environment. This provides a potential mechanism for transmitting information, with phenotypic consequences, from mother to child.
Epigenome-wide association studies (EWASs) have used the Infinium Human Methylation 450 (HM450) array to identify CpG sites that are differentially methylated in the DNA of individuals with differing phenotypes or diseases. Whereas most studies are cross-sectional, some have sought to identify risk-associated epigenetic marks that hold predictive value (1, 2). It has been known for many years that genetic polymorphisms can alter the frequency of DNA methylation, that is, they can alter allele-specific DNA methylation (ASM) (3–5). DNA methylation is also associated with environmental perturbations (6–10): for example, maternal and paternal exposures such as nutrition and smoking are linked to the methylation states of CpG sites in children (11–15).
Genomic imprinting is a type of ASM that is usually associated with extended regions of parental-specific differentially methylated DNA [i.e., differentially methylated regions (DMRs)] and is established in the gametes. Most of the approximately 100 imprinted genes are monoallelically methylated at imprinting control regions, where the gene is expressed from the unmethylated allele in nearly all somatic tissues at a given developmental stage (16). The exceptions include a small subset of genes that are imprinted in a tissue- or isoform-specific manner, in brain and placenta, across most individuals (16, 17). Importantly, unlike nonimprinted ASMs, imprinting is not generally altered by genetic or nongenetic factors, but is instead fixed in the population (18). Tight control of nearly all imprinted genes across the human population is consistent with the known importance of imprinting in regulating embryonic development and neuronal function (19, 20). Disruptions in imprinting can lead to human disorders, including Beckwith–Wiedemann, Silver–Russel, Prader–Willi, and Angelman syndromes, all of which have severe phenotypes impacting human growth and development (21, 22).
Work performed in the P.A.J. and K.G. laboratories uncovered a unique pattern of DNA methylation at the promoter of a noncoding gene (nc886) whereby ∼75% of individuals exhibited ASM and 25% were completely unmethylated (23). Romanelli et al. (24) expanded on this finding to determine that this region of DNA was polymorphically imprinted in a small sample set. Before the conclusion that nc886 was polymorphically imprinted, interindividual variation of imprinting status had been reported only for placental-specific imprinted genes (25, 26).
Genomic imprinting is considered an all-or-none process, in which DMRs on the maternal or paternal autosomes cause monoallelic expression of the corresponding gene (27, 28). Imprinting is commonly, but not always, established by silencing of a single allele through DNA methylation (29). Maternally methylated imprinted genes are often associated with decreased size of the developing fetus (30), whereas paternally expressed imprinted genes are often associated with increased fetal weight (30). Importantly, maternal exposures during pregnancy shift the percentage of DNA methylation or level of expression at imprinted genes, primarily in the placenta (24, 31, 32). The best studied example of genetic and environmental impacts on imprinted genes is that of the H19/IGF2 locus, where quantitatively small but additive effects on levels of DNA methylation are seen (33). However, the possibility that a parental cue from the environment or parental genetics might shift the likelihood that genomic imprinting is established in a human child has yet to be investigated.
Here, we studied the frequency of imprinting of a RNA polymerase III-transcribed, noncoding RNA of 101 bp called nc886 (also referred to as VTRNA2-1), which can be silenced by DNA methylation (23, 34). The complete nc886 sequence is found in higher-order primates, although portions of it are present in other mammals (SI Appendix, Fig. S1), which may indicate its importance in primate evolution. ASM at the nc886 locus has previously been studied and described as a human “metastable epiallale,” which is an allele that shows epigenetic heterogeneity in a population and is sensitive to environmental conditions (6, 35). However, an appreciation of the imprinting biology of this locus has not been emphasized in prior studies. In this work, we show that the region surrounding nc886 acts as an imprinted DMR, as opposed to an intermediately methylated region. This locus represents an instance of nonplacental polymorphic imprinting in humans (24), and we show that variation of imprinting in the population is associated with prenatal environment.
nc886 Is Part of a 1.9-kb Polymorphically Imprinted Region
Genomic imprinting occurs as parental-specific DMRs that tightly regulate gene dosage (28). Nonimprinted ASMs, on the contrary, depend on genomic context rather than parental origin, and on local polymorphisms to affect CpG methylation states, which are generally confined to shorter regions of DNA (3, 18, 36). We and others have reported that nc886 expression is tightly regulated by DNA methylation and that the nc886 locus shows polymorphic imprinting in the human population when measured in peripheral blood cells: approximately 75% of individuals worldwide are monoallelically methylated and approximately 25% are biallelically unmethylated at nc886 (23, 24, 34, 37).
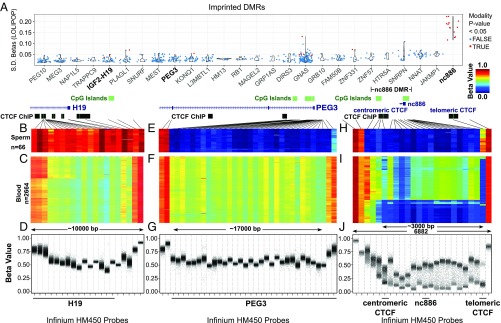
We first used HM450 data to examine DNA methylation at known imprinted DMRs (Fig. 1) (38). We calculated the variance of β-values in 26 imprinted DMRs that had a minimum of 5 HM450 probes by using data from peripheral blood of 2,664 European and Indian Asian subjects from the London Life Sciences Prospective Population (LOLIPOP) study (39). We confirm that all imprinted DMRs investigated, except for probes adjacent to nc886, have limited interindividual variance in this cohort. This variance is much higher for nc886 than for other included imprinted DMRs (Fig. 1A).
Fig. 1.
The nc886 DMR displays maternal polymorphic imprinting. (A) Populational variance for β-values in known imprinting control regions in peripheral blood. Each dot corresponds to one CpG interrogated by an HM450 probe. Populational SDs (y-axis) are plotted for CpGs located in known imprinting DMRs and nc886 (x-axis). Although all imprinting DMRs were examined, only imprinting control regions with more than five probes are included in the display and sorted by the mean SD in the population. Imprinting control regions were retrieved from an earlier study (38). Probes with multimodal distribution of β-values in the population are colored red. DNA methylation data were retrieved from the LOLIPOP study (39). (B, E, and H) DNA methylation data from the Infinium HM450 BeadChip platform in 66 sperm samples (GEO accession numbers GSE47627 and GSE64096) for a paternally methylated DMR for the H19 gene (B), a maternally methylated DMR for the PEG3 gene (E), and the nc886 DMR (H). Black lines point to the genomic location of each probe relative to the respective DMR. (C, F, and I) Heat map of HM450 β-values from peripheral blood for H19 (C), PEG3 (F), and nc886 (I); data obtained from Wahl et al. (39). (D, G, and J) β-Values of each sample are plotted for Infinium HM450 probes for H19 (D) PEG3 (G), and nc886 (J). HM450 probe IDs are provided in SI Appendix, Table S1. Only probes with sequence mapped optimally are included.
We chose to survey DNA methylation in more detail for known imprinted loci containing a maternally methylated DMR (PEG3), a paternally methylated DMR (H19), and, to expand our window of investigation of nc886, a polymorphically imprinted locus. We analyzed data from two independent studies of sperm DNA (GSE47627 and GSE64096) and also peripheral blood from the LOLIPOP study (39–41). The H19 DMR is fully methylated and the PEG3 DMR generally lacks DNA methylation in 66 sperm samples (Fig. 1 B and E, respectively). We found that the nc886 locus and its flanking regions were unmethylated in all sperm samples, similar to PEG3, a maternally methylated DMR (Fig. 1H, dark blue), with partial methylation at a CTCF site on the telomeric side. As expected of imprinted loci, we saw ∼50% methylation in all peripheral blood samples for H19 and PEG3 (Fig. 1 C and F, green). For nc886 in peripheral blood, the majority of samples showed approximately 50% methylation (green; Fig. 1I) and the remainder showed methylation of nearly 0% (blue; Fig. 1I). Based on these HM450 data, we defined the nc886 region as a maternally derived DMR in the human population with boundaries marked by two CTCF sites (spaced ∼3 kb apart; Fig. 1 H and I). Consistent with previous work from Monk and coworkers (24), whole-genome bisulfite sequencing data of various normal tissues from The Cancer Genome Atlas (TCGA) (42) and BLUEPRINT (43, 44) (SI Appendix, Fig. S2) better resolves the boundaries of the nc886 DMR, which is 1,979 bp in size and includes the nc886 locus. nc886 is overlapped by a CpG island, and the DMR is bounded on the centromeric side by repetitive sequences just outside a variably methylated CTCF site (Fig. 1 H and I) (24, 45). The nc886 DMR is flanked on the telomeric side by a CTCF site that is unmethylated in peripheral blood (Fig. 1I).
We noted that three type II probes (cg04515200, cg13581155, and cg11978884) within the nc886 DMR on the HM450 platform displayed a consistent bias toward hypomethylation relative to the rest of the probes in the DMR (Fig. 1H). We believe this is inherent to the Infinium probe design because we could still distinguish a separation in the methylation β-values for individuals who had monoallelic methylation at these probes, and, based on whole-genome bisulfite sequencing data, the three CpGs do not appear biased from an averaged methylation level of 0.5 (Fig. 1I and SI Appendix, Fig. S2). The high correlation of β-values among these three probes and probes across the nc886 DMR also support the idea that these three CpGs bear consistent methylation with the entire DMR and that this DMR is acting as a contiguous unit (SI Appendix, Fig. S3).
Imprinting is essentially dichotomous: DMRs are monoallelically methylated or not. As such, methylation data cannot be considered a continuous variable in which the methylation values of all of the assayed DNA molecules are averaged. We reexamined the LOLIPOP data set, taking this biology into account. For the H19 and PEG3 DMR, all data points were near a β-value of 0.5, as expected (Fig. 1 D and G). However, for nc886, the dichotomous nature of the data became clear, in that the nc886 DMR showed β-values primarily at 50% or near 0% with the exception of the three CpGs previously mentioned (Fig. 1J). Thus, by using the data from the large cohort (39), we have confirmed the previous observations that the nc886 DMR is polymorphically imprinted in humans (23, 24).
An A/C SNP Is Associated with Local DNA Methylation Density
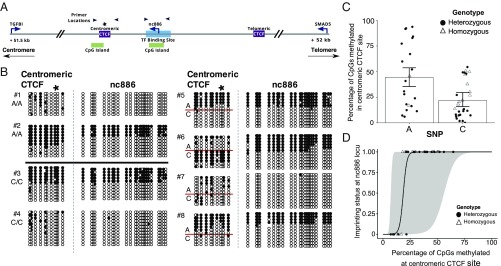
It has been unclear whether the polymorphic nature of imprinting of the nc886 DMR is governed by genetic or nongenetic factors, given the lack of evidence in the literature that genomic imprinting can be associated with the local haplotype (5). Similar to Van Baak et al. (35), we reanalyzed DNA methylation data from twins generated by Grundberg et al. (46). We confirmed their findings that monozygotic twins, but not dizygotic twins, are concordant for DNA methylation across the nc886 DMR (SI Appendix, Fig. S4), strongly suggesting that the nc886 DMR could be influenced by genetic factors. The nc886 locus is located within a haplotype block ranging in size from ∼5 kb to 35 kb, depending on the population (SI Appendix, Fig. S5). The 2 kb spanning the nc886 DMR has been examined for potentially causal SNPs without success or with the conclusion that SNPs did not impact DNA methylation (6, 23, 24, 37, 45). Given the central role of CTCF in imprinting, we focused on an A/C SNP (rs2346018; A allele frequency range, 31–45%; C allele frequency range, 54–68%; from the 1000 Genomes Project) located in the variably methylated centromeric CTCF binding site (36, 47, 48). By using the R package motifbreakR, we found that having a C SNP “breaks” the CTCF binding motif of the centromeric CTCF site and could therefore impact the ability of CTCF to bind its target (49). To determine if SNP status was associated with local DNA methylation density, we analyzed DNA methylation at the centromeric CTCF site and across the nc886 locus in peripheral blood DNA of 31 cancer-free individuals by using bisulfite conversion followed by clonal sequencing (primer locations shown in Fig. 2A). Eight examples are shown in Fig. 2B, with the data from the remaining 23 individuals shown in SI Appendix, Fig. S6. Clearly, the A and C alleles can be densely methylated in homozygotes and heterozygotes, but there was also sporadic methylation in predominantly methylated or unmethylated DNA strands (Fig. 2B).
Fig. 2.
An A polymorphism (rs2346018) is associated with higher local DNA methylation density at the centromeric CTCF site, which has a strong positive correlation to DNA methylation at nc886. (A) Diagram of the nc886 DMR. Black arrows indicate primer locations for bisulfite sequencing in Figs. 2 and 3 and SI Appendix, Fig. S6. (B) Bisulfite conversion and clonal sequencing of genomic DNA from white blood cells of eight individuals at the centromeric CTCF site and nc886. Asterisk indicates SNP location. (C) Percentage of CpGs methylated for each allele at the centromeric CTCF site in homozygotes and heterozygotes (n = 31; P < 1 × 10−9). (D) DNA methylation of the centromeric CTCF plotted against DNA methylation of nc886 as a discrete variable, with 0 being biallelically unmethylated and 1 being monoallelically methylated (n = 31; P = 4.7 × 10−7). Gray ribbon represents 95% CI.
We chose to test the hypothesis that genetic background could “tune” the likelihood that an individual displays imprinting at the nc886 DMR, thereby switching from imprinted to not imprinted. Examining data from heterozygous and homozygous individuals, we determined that the A allele has higher average methylation density than the C allele (Fig. 2C; P < 1 × 10−9). Furthermore, the odds ratio that a clone of the centromeric CTCF site is fully methylated in an individual with the A polymorphism (vs. C) was 2.86. These data demonstrate that DNA methylation of the centromeric CTCF site, 900 bp from nc886, correlates with a common SNP, with an A allele more likely to be methylated than a C allele. Given the role of CTCF in imprinting, the presence of the A allele might directly alter the likelihood of establishing DNA methylation at the nc886 DMR through reduced sequence binding affinity of CTCF or other DNA-binding proteins (e.g., DNMTs, ZFP57, Kaiso) (50, 51). Alternatively, the A allele might indirectly impact imprinting through increased DNA methylation density, and therefore binding affinity of CTCF (52) (see Fig. 5).
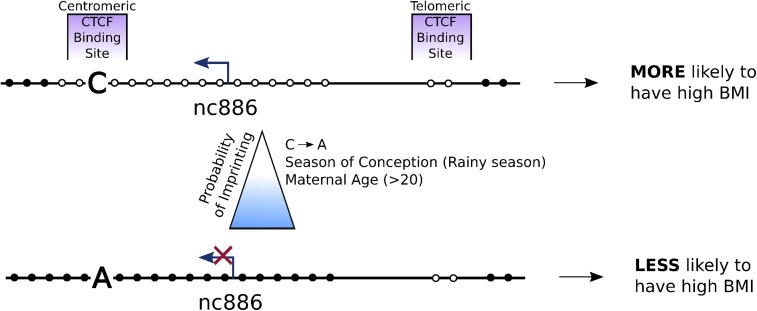
Fig. 5.
Working model for polymorphic imprinting of the nc886 locus. All individuals we examined have the single paternally derived unmethylated allele at the nc886 DMR. In 75% of the human population, the nc886 DMR is monoallelically methylated on the maternally derived allele (Bottom), with the remaining 25% having no methylation on either allele (Top). Here, we demonstrate that the presence of an A or C polymorphism in the centromeric CTCF binding site is associated with local DNA methylation density in the nc886 DMR. We hypothesize that the presence of the A allele or increased DNA methylation density reduces CTCF binding, allowing DNA methylation to spread into the DMR. We reanalyzed data from Silver et al. (6) and Markunas et al. (54) and found that maternal age and season of conception contribute to the likelihood that a child will have imprinting at the nc886 DMR. Therefore, we propose that a combination of maternal factors shifts the balance between imprinted and not-imprinted status in children, with downstream phenotypic consequences such as BMI.
Linked DNA Methylation of the Centromeric CTCF Binding Site and nc886
Previous work has shown that DNA methylation at the centromeric CTCF site serves as the boundary of ASM for the nc886 region (24, 45). Because the entire 1.9-kb region between the CTCF sites acts as a DMR (Fig. 1 and SI Appendix, Figs. S2 and S3), we hypothesized that DNA methylation of the centromeric CTCF site, or of any region within the DMR, might predict DNA methylation at nc886. Although other studies have suggested that the entire region can be imprinted (23, 32), they have not shown a lack of DNA methylation of this region in sperm, as we have (Fig. 1H), and it is technically challenging to show that DNA methylation is linked across a single DNA strand.
Fig. 2B shows clonal bisulfite sequencing analysis of genomic DNA from individuals who have monoallelic methylation or no methylation, as indicated by SNP status in the centromeric CTCF site. We classified 50% methylation, representative of monoallelic methylation at nc886, as being “1” and 0% methylation, or biallelic lack of methylation, as being “0,” and modeled the data against the percentage of DNA methylation at the centromeric CTCF site for all 31 individuals (Fig. 2D). Lack of DNA methylation at the centromeric CTCF site was associated with a very low probability of having DNA methylation at nc886. With increasing density of DNA methylation at the centromeric CTCF site, the probability of nc886 being classified as “imprinted” increased; thus, there was a strong positive correlation between DNA methylation at the centromeric CTCF site and at nc886 (P < 0.0001; R2 = 0.77; Fig. 2D). DNA methylation of the centromeric CTCF site therefore explains much of the variation in DNA methylation at nc886. Although we were not able to sequence individual DNA strands greater than 1 kb in length, these data and the high correlation between β-values across the nc886 DMR (SI Appendix, Fig. S3) suggest that DNA methylation of this region is indeed present on individual alleles (Fig. 2).
DNA Methylation of the Centromeric CTCF Binding Site Is Maternally Derived
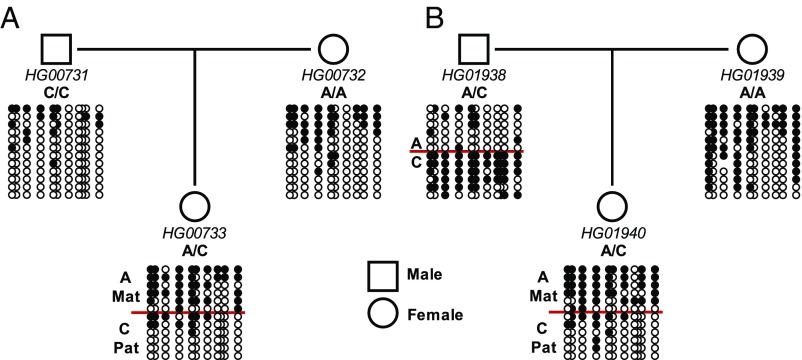
As summarized in SI Appendix, Table S2, previous studies analyzing SNPs have independently concluded that DNA methylation across the nc886 region is not dependent on genetic context but is maternally derived (23, 24, 37, 45). Additionally, when analyzing HM450 data from sperm, we find that the nc886 DMR lacks DNA methylation, indicating that methylation of this region is likely maternally derived (Fig. 1H). To confirm that DNA methylation of the centromeric CTCF site could be included in this region of maternally derived methylation, we used parent–offspring trios informative for the A/C SNP (rs2346018) and performed bisulfite conversion and clonal sequencing. As shown in Fig. 3, DNA methylation of the centromeric CTCF site was maternally derived. Thus, the existing literature and the lack of DNA methylation across the nc886 DMR in sperm (Fig. 1H), along with this result, supports that imprinting of the nc886 DMR is not paternally derived.
Fig. 3.
Maternal imprinting of the nc886 region extends to the centromeric CTCF site. Locus-specific bisulfite sequencing was performed on genomic DNA at the centromeric CTCF site. Genomic DNA was isolated from benign lymphoblastoid cell lines derived from parent–offspring trios (mother, father, child; A and B are representative of two different trios) of disease-free individuals, in which SNP status can be determined from the sequence. Two representative parent–offspring trios are shown. Clones are sorted based first on SNP status and then on DNA methylation.
Maternal Age and Nutrition Are Associated with the nc886 DMR
Recent EWAS studies have identified nc886 as a “metastable epiallele” that is altered in the population and is dependent on maternal nutritional status (6, 35, 53). However, these studies did not consider the data in terms of the percentage of each cohort demonstrating imprinting, which might account for the biology. Therefore, we reanalyzed the results from three independent studies in terms of percentage of each subpopulation that exhibits imprinting to determine if the likelihood that an individual is called “imprinted” shifts in each experimental group (SI Appendix, Fig. S7). To address global shifts in β-values introduced by different methods used to preprocess the HM450 data or by residual experimental batch effects, we applied unsupervised hierarchical clustering, using the DNA methylation β-values from the entire region to determine whether an individual had imprinting at the nc886 DMR and whether this call is conclusive.
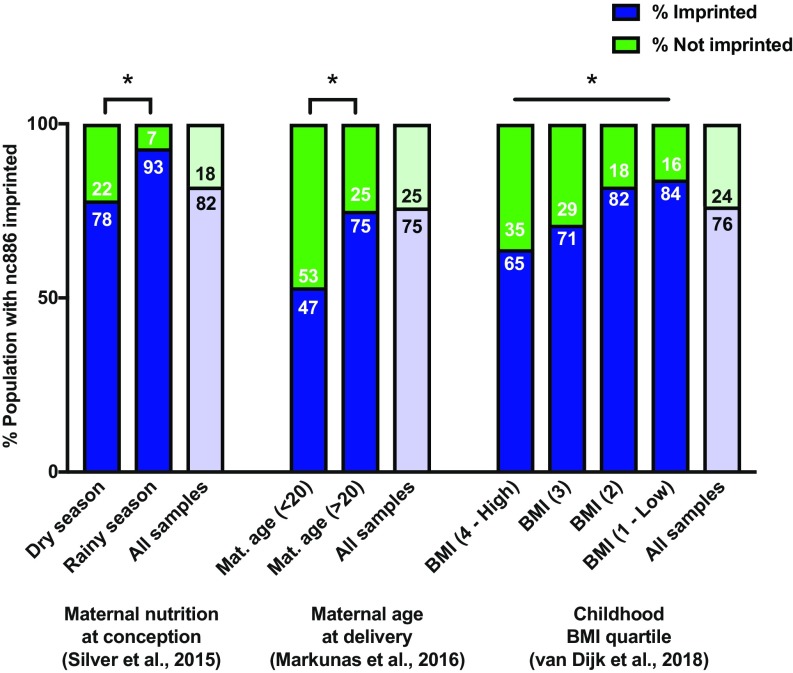
We used the valuable data from Silver et al. (6), who examined DNA methylation in 114 children in Gambia conceived in the rainy season (low-calorie, nutritionally rich) or dry season (high-calorie, nutritionally poor). Hierarchical clustering identified three groups of individuals, two of which showed the expected dichotomy of being imprinted or not imprinted. The third cluster, consisting of five children (4.4%) who had evidence of DNA methylation in only approximately half of the probes in the nc886 DMR, was classified as inconclusive (SI Appendix, Figs. S7A and S8B). When comparing imprinted individuals and nonimprinted individuals, we found that season of conception was significantly (P = 0.0417) associated with the frequency of imprinting of the nc886 DMR in infants (Fig. 4). In this data set, we performed likelihood ratio tests (LRTs) to adjust for available confounding factors, including exposure to aflatoxin (a common foodborne toxin in Africa) and sex. We found that neither variable was a significant predictor of imprinting at nc886, and, after adjusting for these factors, the significance of the season of conception slightly improves (P = 0.03902). Thus, season of conception and, by association, maternal nutrition, may contribute to “tuning” the likelihood that an individual has imprinting at the nc886 DMR. Furthermore, recent analysis of a Scottish birth cohort identified differential DNA methylation of nc886 in response to folate supplementation during pregnancy (53).
Fig. 4.
Maternal environment in utero impacts DNA methylation of nc886 in infants. HM450 data for the nc886 DMR was analyzed from three independent studies of peripheral blood of infants at birth. Each group is separated by the percentage of the population that is imprinted vs. not imprinted based on hierarchical clustering. Individuals with inconclusive evidence of imprinting have been removed from this analysis. Mat, maternal. *P < 0.05.
The same hierarchical clustering analysis was performed on data from Markunas et al. (54) for DNA methylation of the nc886 DMR in children born to 855 mothers of various ages (SI Appendix, Fig. S7 C and D). We found that children born to mothers younger than 20 y had significantly less imprinting than those born to older mothers (P = 0.0167; Fig. 4). After adjustment for sex, maternal age is still significantly associated with imprinting of the nc886 DMR (LRT P = 0.0281). Our data suggest that teenage pregnancy is associated with a decreased likelihood that a child will have imprinting at the nc886 DMR.
We replotted data from van Dijk et al. (55) in which HM450 analysis was performed on blood spots obtained from children at birth. This study identified lack of imprinting at nc886 measured at birth as being strongly associated with increased body mass index (BMI) in children assessed at 5 y of age. Hierarchical clustering of the nc886 DMR was used to define imprinted, not-imprinted, and inconclusive groups, and BMI was analyzed as a continuous variable (SI Appendix, Fig. S7 E and F). We confirmed that imprinting of the nc886 DMR was associated with lower BMI in children when analyzed as discrete quartiles (Fig. 4) and as a continuous variable (χ2 test P = 1.75 × 10−3 and logistic regression P = 1.94 × 10−3, respectively). We conclude that imprinting of the nc886 DMR is associated with a lower childhood BMI. Collectively, these three data sets demonstrate that maternal environment is linked to the establishment of imprinting across the nc886 DMR in children and that this epigenetic mark potentially impacts human phenotypes later in life.
Finally, we calculated the percentage of each population, across all samples, that exhibited imprinting (Fig. 4). When examining all samples from the BMI data set, we found that 76% of individuals were imprinted and 24% were not imprinted at the nc886 DMR, similar to the maternal age study and consistent with our previous findings in which 75% of individuals were imprinted and 25% were not (23). In the smallest data set, which examined maternal nutrition in Gambia, we found 82% of the population with imprinting. Whether this is a result of environmental differences or discrepancies in the genetic makeup of different populations needs to be investigated.
Discussion
We conclude that the nc886 DMR, when imprinted, is maternally methylated, given our evidence that this region behaves as a contiguous DMR that is not methylated in sperm (Fig. 1H). The fact that the entire 1.9-kb nc886 DMR is subject to tunable polymorphic imprinting has been largely overlooked, emphasizing the need to scrutinize data obtained from the HM450 platform for possible dichotomization. We find that some of the polymorphic nature of the imprinting can be explained by local genetic makeup, as measured by using an A/C SNP in the centromeric CTCF site. The mechanism by which this polymorphism alters the likelihood of imprinting is not completely clear but is possibly associated with chromatin conformation variation triggered by CTCF binding, given reports that CTCF binding sites are involved in genomic imprinting (56–58). This possibility is supported by the observation that mutations in CTCF binding sites in the XIST promoter alter CTCF binding efficiency and choice of X chromosome inactivation (51). Additionally, the presence of a genetic influence is supported by the fact that monozygotic twins are found to be more concordant in DNA methylation at the nc886 DMR than dizygotic twins (35). However, Van Baak et al. (35) concluded that DNA methylation at nc886 could not be explained by genetics alone and classified it as a region of “epigenetic supersimilarity.” Although our data provide high-resolution analysis of local DNA methylation in 31 individuals, a very recent paper by Zink et al. (37) found no genetic polymorphisms associated with imprinting of nc886 in an Icelandic population, suggesting a need for further genotyping and concurrent DNA methylation analysis in individuals from different populations and environmental contexts.
We find that the frequency of imprinting of the nc886 DMR in children is also associated with the mothers’ age and season of conception, indicating parental environment as potentially another mechanism for tuning the likelihood that the nc886 DMR will be imprinted (6, 54). Furthermore, the frequency of imprinting of the nc886 DMR at birth is maintained and is directly associated with the BMI of children at the age of 5 y (55). Collectively, these results suggest that genetic and environmental factors may affect the establishment of imprinting of a DMR, which is closely associated with human physiology (Fig. 5). Unfortunately, there are no known SNPs in nc886 that would allow us to examine tissues for allele-specific expression. However, we have shown that DNA methylation can silence transcription of nc886, making it very likely that differential methylation of this region is functionally important (23, 34).
A further understanding of causality from all of these observations will require an unraveling of potential biological roles of nc886 or the nc886 DMR. nc886 has been variously described as a regulator of the dsRNA-dependent protein kinase R (PKR) (23, 59–61) and Dicer (62). It was also suggested as a possible tumor suppressor (63) or oncogene (64). Alternatively, some other genetic or epigenetic factors within or around the 1.9-kb DMR sequence may be drivers of phenotypic effects associated with nc886. Thus far, EWAS studies have primarily focused on variations in DNA methylation levels without considering the essential role of imprinting in human development as we have done here.
Methods
DNA Methylation Analysis of Illumina HM450 Data.
Normalized β-values were downloaded from Gene Expression Omnibus (GEO), with accession numbers GSE47627 and GES64096 for sperm and GSE55763 for genomic DNA from peripheral blood of 2,664 individuals assayed on the Infinium HM450 platform (i.e., LOLIPOP data). Probes falling in the region chr5:135413937–135419936 (Genome Build GRCh37) were extracted, and their signals were visualized by using the R package SeSAMe (10). Genomic features were extracted from the University of California, Santa Cruz, Genome Browser. Probes annotated with potential cross-hybridization issues and SNP bias were excluded according to an earlier study (10, 65). This analysis was also performed on matched normal tissues from the TCGA database, as shown in SI Appendix, Fig. S2.
Locus-Specific Bisulfite Sequencing Analysis of the Centromeric CTCF Site and nc886.
Genomic DNA isolated from white blood cells of disease-free individuals was obtained from the Van Andel Research Institute Pathology and Biorepository Core. For trio analysis, the following DNA samples were obtained from the National Human Genome Research Institute Sample Repository for Human Genetic Research at the Coriell Institute for Medical Research: HG00731, HG00732, HG00733, HG01938, HG01939, and HG01940. Bisulfite conversion and clean-up of 2 μg of genomic DNA was performed by using the EZ DNA Methylation Kit (Zymo Research) according to the manufacturer’s protocol. Locus-specific PCR was performed by using primers specific for bisulfite-converted DNA and PCR products cloned by using the pGEM-T Easy vector and NEB 5-alpha–competent Escherichia coli (New England Biolabs). Colonies were screened for positive inserts by PCR and sequencing performed using the M13 promoter.
Statistical Analysis.
Determining whether SNP status is associated with DNA methylation in the centromeric CTCF region.
β-Binomial mixed-effects regression, weighted by the number of CpG sites in a region (10 for SNP A, 11 for C), was used to model data obtained from bisulfite sequencing and PCR via the R package (version 3.4.3; https://www.r-project.org/) glmmTMB (66). A random intercept for each individual was used to account for the paired heterozygous and homozygous measures; a second random intercept was included to account for DNA methylation of the nc886 region as determined by the presence of at least 10% of clones being completely methylated. β-Binomial was chosen because it is well-suited for modeling the binary methylated/unmethylated status of each CpG in the region, but the probability of a given site being methylated increases with each additional site methylated in the region. An LRT confirmed that weighted β-binomial was a better fit for these data than a weighted binomial, and Akaike information criterion and Bayesian information criterion showed this regression was better than negative binomial regression.
Determining the correlation between DNA methylation at the centromeric CTCF and nc886 regions.
The percentage of DNA methylation was calculated for each region analyzed by bisulfite conversion and sequencing, and the data were analyzed via logistic regression. Bisulfite sequencing was used to estimate DNA methylation in the centromeric CTCF and nc886 regions. Logistic regression was used to determine whether the probability of nc886 being methylated changed as the percentage of methylation of the CTCF region increased. The reported P value was calculated by using an LRT. The mean percent methylation of the centromeric CTCF site was estimated by using a mixed-effects negative binomial regression via the R package glmmTMB (66). Data were plotted as the mean change in probability per percent increase of methylation in the centromeric CTCF, with a 95% confidence band.
Population Analysis of DNA Methylation at nc886 from Published Datasets.
We downloaded from GEO Infinium HM450 data from genomic DNA of infants in studies of season of conception in Gambia (6) and maternal age at delivery (54) with accession numbers GSE59592 and GSE82273, respectively. For the childhood BMI study (55), the GEO Infinium HM450 data (accession no. GSE103657) was not optimal because of technical effects introduced by batch correction. Instead, the raw array data were renormalized, consistent with the methods of the published study, and BMI measurement data were accessed through the authors of the paper of van Dijk et al. (55).
Based on bisulfite sequencing analysis (SI Appendix, Fig. S6), which provides single-base resolution of individual DNA strands for DNA methylation, and previous studies, we concluded that there are only two possibilities for DNA methylation in this region: near 0% DNA methylation (biallelically unmethylated) or near 50% DNA methylation (monoallelically methylated) (6, 23, 24, 67). However, when data were plotted, it was clear that not all individuals exhibited this expected dichotomy of 50% methylated or not methylated (SI Appendix, Fig. S7). Upon further investigation, we found that individuals who did not fit this dichotomy comprised a third cluster based on hierarchical clustering via Manhattan distances. Therefore, we identified three clusters of individuals, which we called imprinted, not imprinted, or inconclusive based on partial methylation readouts throughout the region (SI Appendix, Fig. S7). To remain consistent with previously observed data, we restricted our analyses to focus on the patients not in this third, inconsistent cluster. Results without this restriction are reported in the SI Appendix.
Treating imprinting as the response variable, we applied logistic regression by using the glm function in R. The season of conception, maternal age (dichotomized by teenage pregnancy), and childhood BMI Z-scores were used as the regressors of the three data sets, respectively. P values for the coefficients were obtained by using a Wald test.
Supplementary Material
Acknowledgments
We thank the VARI Pathology and Biorepository Core for providing samples for bisulfite sequencing analysis, the VARI Bioinformatics and Biostatistics Core for providing statistical support, and Beverley Muhlhausler for curating and allowing access to BMI data from the DHA (docosahexaenoic acid) to Optimize Mother Infant Outcome (DOMInO) study. This study was supported by National Cancer Institute Grant R35CA209859 (to P.A.J.).
Footnotes
Conflict of interest statement: P.A.J. is a paid consultant for Zymo Research. In this study, Zymo Research reagents were used to analyze locus-specific DNA methylation patterns.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815005115/-/DCSupplemental.
References
- 1.Flanagan JM. Epigenome-wide association studies (EWAS): Past, present, and future. Methods Mol Biol. 2015;1238:51–63. doi: 10.1007/978-1-4939-1804-1_3. [DOI] [PubMed] [Google Scholar]
- 2.Lappalainen T, Greally JM. Associating cellular epigenetic models with human phenotypes. Nat Rev Genet. 2017;18:441–451. doi: 10.1038/nrg.2017.32. [DOI] [PubMed] [Google Scholar]
- 3.Chandler LA, Ghazi H, Jones PA, Boukamp P, Fusenig NE. Allele-specific methylation of the human c-Ha-ras-1 gene. Cell. 1987;50:711–717. doi: 10.1016/0092-8674(87)90329-1. [DOI] [PubMed] [Google Scholar]
- 4.Hitchins MP, et al. Dominantly inherited constitutional epigenetic silencing of MLH1 in a cancer-affected family is linked to a single nucleotide variant within the 5′UTR. Cancer Cell. 2011;20:200–213. doi: 10.1016/j.ccr.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Kerkel K, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 6.Silver MJ, et al. Independent genomewide screens identify the tumor suppressor VTRNA2-1 as a human epiallele responsive to periconceptional environment. Genome Biol. 2015;16:118. doi: 10.1186/s13059-015-0660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolinoy DC, Das R, Weidman JR, Jirtle RL. Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatr Res. 2007;61:30R–37R. doi: 10.1203/pdr.0b013e31804575f7. [DOI] [PubMed] [Google Scholar]
- 8.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Do C, et al. Mechanisms and disease associations of haplotype-dependent allele-specific DNA methylation. Am J Hum Genet. 2016;98:934–955. doi: 10.1016/j.ajhg.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou W, Triche TJ, Jr, Laird PW, Shen H. SeSAMe: Reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. July 31, 2018 doi: 10.1093/nar/gky691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soubry A, Hoyo C, Jirtle RL, Murphy SK. A paternal environmental legacy: Evidence for epigenetic inheritance through the male germ line. BioEssays. 2014;36:359–371. doi: 10.1002/bies.201300113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee HS. Impact of maternal diet on the epigenome during in utero life and the developmental programming of diseases in childhood and adulthood. Nutrients. 2015;7:9492–9507. doi: 10.3390/nu7115467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chango A, Pogribny IP. Considering maternal dietary modulators for epigenetic regulation and programming of the fetal epigenome. Nutrients. 2015;7:2748–2770. doi: 10.3390/nu7042748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joubert BR, et al. DNA methylation in newborns and maternal smoking in pregnancy: Genome-wide consortium meta-analysis. Am J Hum Genet. 2016;98:680–696. doi: 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joubert BR, et al. Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nat Commun. 2016;7:10577. doi: 10.1038/ncomms10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babak T, et al. Genetic conflict reflected in tissue-specific maps of genomic imprinting in human and mouse. Nat Genet. 2015;47:544–549. doi: 10.1038/ng.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prickett AR, Oakey RJ. A survey of tissue-specific genomic imprinting in mammals. Mol Genet Genomics. 2012;287:621–630. doi: 10.1007/s00438-012-0708-6. [DOI] [PubMed] [Google Scholar]
- 18.Tycko B. Allele-specific DNA methylation: Beyond imprinting. Hum Mol Genet. 2010;19:R210–R220. doi: 10.1093/hmg/ddq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartolomei MS. Genomic imprinting: Employing and avoiding epigenetic processes. Genes Dev. 2009;23:2124–2133. doi: 10.1101/gad.1841409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hitchins MP, Moore GE. Genomic imprinting in fetal growth and development. Expert Rev Mol Med. 2002;4:1–19. doi: 10.1017/S146239940200457X. [DOI] [PubMed] [Google Scholar]
- 21.Butler MG. Genomic imprinting disorders in humans: A mini-review. J Assist Reprod Genet. 2009;26:477–486. doi: 10.1007/s10815-009-9353-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eggermann T, et al. Imprinting disorders: A group of congenital disorders with overlapping patterns of molecular changes affecting imprinted loci. Clin Epigenetics. 2015;7:123. doi: 10.1186/s13148-015-0143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treppendahl MB, et al. Allelic methylation levels of the noncoding VTRNA2-1 located on chromosome 5q31.1 predict outcome in AML. Blood. 2012;119:206–216, and erratum (2013) 121:5104. doi: 10.1182/blood-2011-06-362541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romanelli V, et al. Variable maternal methylation overlapping the nc886/vtRNA2-1 locus is locked between hypermethylated repeats and is frequently altered in cancer. Epigenetics. 2014;9:783–790. doi: 10.4161/epi.28323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanna CW, et al. Pervasive polymorphic imprinted methylation in the human placenta. Genome Res. 2016;26:756–767. doi: 10.1101/gr.196139.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jinno Y, et al. Mosaic and polymorphic imprinting of the WT1 gene in humans. Nat Genet. 1994;6:305–309. doi: 10.1038/ng0394-305. [DOI] [PubMed] [Google Scholar]
- 27.Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 2011;3:a002592. doi: 10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reik W, Walter J. Genomic imprinting: Parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 29.Inoue A, Jiang L, Lu F, Suzuki T, Zhang Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature. 2017;547:419–424. doi: 10.1038/nature23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haig D. Parental antagonism, relatedness asymmetries, and genomic imprinting. Proc Biol Sci. 1997;264:1657–1662. doi: 10.1098/rspb.1997.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Susiarjo M, Sasson I, Mesaros C, Bartolomei MS. Bisphenol a exposure disrupts genomic imprinting in the mouse. PLoS Genet. 2013;9:e1003401. doi: 10.1371/journal.pgen.1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kappil M, Lambertini L, Chen J. Environmental influences on genomic imprinting. Curr Environ Health Rep. 2015;2:155–162. doi: 10.1007/s40572-015-0046-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tobi EW, et al. Prenatal famine and genetic variation are independently and additively associated with DNA methylation at regulatory loci within IGF2/H19. PLoS One. 2012;7:e37933. doi: 10.1371/journal.pone.0037933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helbo AS, Lay FD, Jones PA, Liang G, Grønbæk K. Nucleosome positioning and NDR structure at RNA polymerase III promoters. Sci Rep. 2017;7:41947. doi: 10.1038/srep41947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Baak TE, et al. Epigenetic supersimilarity of monozygotic twin pairs. Genome Biol. 2018;19:2. doi: 10.1186/s13059-017-1374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Do C, et al. Genetic-epigenetic interactions in cis: A major focus in the post-GWAS era. Genome Biol. 2017;18:120. doi: 10.1186/s13059-017-1250-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zink F, et al. Insights into imprinting from parent-of-origin phased methylomes and transcriptomes. Nat Genet. 2018;50:1542–1552. doi: 10.1038/s41588-018-0232-7. [DOI] [PubMed] [Google Scholar]
- 38.Court F, et al. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014;24:554–569. doi: 10.1101/gr.164913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wahl S, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541:81–86. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krausz C, et al. Novel insights into DNA methylation features in spermatozoa: Stability and peculiarities. PLoS One. 2012;7:e44479. doi: 10.1371/journal.pone.0044479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jenkins TG, et al. Intra-sample heterogeneity of sperm DNA methylation. Mol Hum Reprod. 2015;21:313–319. doi: 10.1093/molehr/gau115. [DOI] [PubMed] [Google Scholar]
- 42.Zhou W, et al. DNA methylation loss in late-replicating domains is linked to mitotic cell division. Nat Genet. 2018;50:591–602. doi: 10.1038/s41588-018-0073-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kulis M, et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47:746–756. doi: 10.1038/ng.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams D, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotechnol. 2012;30:224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- 45.Paliwal A, et al. Comparative anatomy of chromosomal domains with imprinted and non-imprinted allele-specific DNA methylation. PLoS Genet. 2013;9:e1003622. doi: 10.1371/journal.pgen.1003622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grundberg E, et al. Multiple Tissue Human Expression Resource Consortium Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet. 2013;93:876–890. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maurano MT, Wang H, Kutyavin T, Stamatoyannopoulos JA. Widespread site-dependent buffering of human regulatory polymorphism. PLoS Genet. 2012;8:e1002599. doi: 10.1371/journal.pgen.1002599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis A, Reik W. How imprinting centres work. Cytogenet Genome Res. 2006;113:81–89. doi: 10.1159/000090818. [DOI] [PubMed] [Google Scholar]
- 49.Coetzee SG, Coetzee GA, Hazelett DJ. motifbreakR: An R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics. 2015;31:3847–3849. doi: 10.1093/bioinformatics/btv470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prickett AR, et al. Genome-wide and parental allele-specific analysis of CTCF and cohesin DNA binding in mouse brain reveals a tissue-specific binding pattern and an association with imprinted differentially methylated regions. Genome Res. 2013;23:1624–1635. doi: 10.1101/gr.150136.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pugacheva EM, et al. Familial cases of point mutations in the XIST promoter reveal a correlation between CTCF binding and pre-emptive choices of X chromosome inactivation. Hum Mol Genet. 2005;14:953–965. doi: 10.1093/hmg/ddi089. [DOI] [PubMed] [Google Scholar]
- 52.Wang H, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richmond RC, et al. The long-term impact of folic acid in pregnancy on offspring DNA methylation: Follow-up of the Aberdeen Folic Acid Supplementation Trial (AFAST) Int J Epidemiol. March 12, 2018 doi: 10.1093/ije/dyy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Markunas CA, et al. Maternal age at delivery is associated with an epigenetic signature in both newborns and adults. PLoS One. 2016;11:e0156361. doi: 10.1371/journal.pone.0156361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Dijk SJ, et al. DNA methylation in blood from neonatal screening cards and the association with BMI and insulin sensitivity in early childhood. Int J Obes (Lond) 2017;42:28–35. doi: 10.1038/ijo.2017.228. [DOI] [PubMed] [Google Scholar]
- 56.Szabó PE, Tang SH, Silva FJ, Tsark WM, Mann JR. Role of CTCF binding sites in the Igf2/H19 imprinting control region. Mol Cell Biol. 2004;24:4791–4800. doi: 10.1128/MCB.24.11.4791-4800.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin S, Ferguson-Smith AC, Schultz RM, Bartolomei MS. Nonallelic transcriptional roles of CTCF and cohesins at imprinted loci. Mol Cell Biol. 2011;31:3094–3104. doi: 10.1128/MCB.01449-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 59.Lee K, et al. Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA. 2011;17:1076–1089. doi: 10.1261/rna.2701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jeon SH, et al. Characterization of the direct physical interaction of nc886, a cellular non-coding RNA, and PKR. FEBS Lett. 2012;586:3477–3484. doi: 10.1016/j.febslet.2012.07.076. [DOI] [PubMed] [Google Scholar]
- 61.Calderon BM, Conn GL. Human noncoding RNA 886 (nc886) adopts two structurally distinct conformers that are functionally opposing regulators of PKR. RNA. 2017;23:557–566. doi: 10.1261/rna.060269.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahn JH, et al. nc886 is induced by TGF-β and suppresses the microRNA pathway in ovarian cancer. Nat Commun. 2018;9:1166. doi: 10.1038/s41467-018-03556-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fort RS, et al. nc886 is epigenetically repressed in prostate cancer and acts as a tumor suppressor through the inhibition of cell growth. BMC Cancer. 2018;18:127. doi: 10.1186/s12885-018-4049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee EK, et al. nc886, a non-coding RNA and suppressor of PKR, exerts an oncogenic function in thyroid cancer. Oncotarget. 2016;7:75000–75012. doi: 10.18632/oncotarget.11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou W, Laird PW, Shen H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res. 2017;45:e22. doi: 10.1093/nar/gkw967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Magnusson A, et al. 2017 glmmTMB: Generalized Linear Mixed Models using Template Model Builder. R Package Version 0.1.3. Available at https://github.com/glmmTMB. Accessed May 16, 2018.
- 67.Waterland RA, Jirtle RL. Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.