Significance
Kinesins are microtubule-based molecular motors that can drive long-range intracellular transport. When not transporting cargoes, processive kinesin motors often adopt an autoinhibited state. The autoinhibited conformation of kinesin-3 is predominantly mediated by a coiled-coil segment that follows the neck and motor domains. In this study, we found that this coiled-coil segment associates with both the neck and motor domains and fastens them together. With the aid of the motor domain (or, namely, head), the autoinhibitory coiled-coil segment tightly locks down the entire neck domain and inhibits both the neck domain-mediated dimerization and the ADP release from the motor head. This “head-aided lockdown of neck” mechanism mediated by the internal coiled-coil segment may represent a paradigm for kinesin autoinhibition.
Keywords: intracellular transport, molecular motor, kinesin-3, autoinhibition, coiled-coil
Abstract
In kinesin-3, the coiled-coil 1 (CC1) can sequester the preceding neck coil (NC) for autoinhibition, but the underlying mechanism is poorly understood. Here, we determined the structures of the uninhibited motor domain (MD)-NC dimer and inhibited MD-NC-CC1 monomer of kinesin-3 KIF13B. In the MD-NC-CC1 monomer, CC1 is broken into two short helices that unexpectedly interact with both the NC and the MD. Compared with the MD-NC dimer, the CC1-mediated integration of NC and MD not only blocks the NC dimer formation, but also prevents the neck linker (NL) undocking and the ADP release from the MD. Mutations of the essential residues in the interdomain interaction interface in the MD-NC-CC1 monomer restored the MD activity. Thus, CC1 fastens the neck domain and MD and inhibits both NC and NL. This CC1-mediated lockdown of the entire neck domain may represent a paradigm for kinesin autoinhibition that could be applicable to other kinesin-3 motors.
Kinesins are a superfamily of microtubule (MT)-based molecular motors that can convert the chemical energy of ATP hydrolysis into the mechanical power to drive long-range intracellular transport or dictate MT dynamics (1–5). To transport cellular cargoes, in addition to the core motor domain (MD; referred to as the “head”) that can bind to MTs and undergoes nucleotide-dependent conformational changes, processive kinesin motors (kinesin-1 to -3) require a characteristic neck domain [containing a neck linker (NL) and a neck coil (NC)] and a globular tail domain (2, 6). The neck domain forms a dimeric “neck” for assembling a functional “two-headed” motor and controlling the directionality and processivity, while the tail domain is responsible for binding to cargoes or cargo adaptors (7–9). However, in the absence of cargoes, most of these processive kinesin motors exist in an autoinhibited state to avoid futile ATP hydrolysis and the potential congestion of MT tracks (10). In kinesin-1 (the conventional kinesin), the compact autoinhibited conformation is predominantly mediated by its tail domain that folds back to directly interact with the MD (11, 12). This “tail-to-head” intramolecular interaction interferes with either the MT-binding capacity or the ATPase activity of the MD (or both) and keeps the motor in a folded inactive state (13). Similar tail-to-head intramolecular interaction happens in kinesin-2 to maintain the motor in an autoinhibited conformation as well (14, 15). The tail domain-mediated autoinhibition of kinesin-1 and -2 would prevent their improper cargo loading and erroneous movement and thus ensure the precise spatial and temporal regulation of motor motility (10).
Kinesin-3 (including KIF1A/B/C, KIF13A/B, and KIF16A/B in mammals) is a unique family of kinesin motors for intracellular transport with superprocessivity (2, 3, 16). In comparison with other kinesins, kinesin-3 is characterized by a family-specific forkhead-associated (FHA) domain that is sandwiched between two coiled-coils [coiled-coil 1 (CC1) and CC2] and, together, follows the neck domain (Fig. 1A). When not transporting cargoes, most of kinesin-3 motors also adopt an autoinhibited conformation (10). The founding member KIF1A was found to exist as a globular compact monomer that is largely inactive (only with a capacity of slow diffusive motion) and needs the cargo-mediated dimerization for fast processive movement (17–19). The monomeric autoinhibited conformation of KIF1A is partly contributed by the intramolecular interaction between CC2 and the FHA domain that led to the decreased propensity of dimerization and the inhibition of MT binding (20). Moreover, in KIF13B and KIF16B, a central stalk region immediately following CC2 was found to directly interact with the MD (21, 22), and this “stalk-to-head” intramolecular interaction prevents the binding of MTs and keeps the motor in a folded inactive conformation. In addition to the inactivation of the MD, the neck domain of kinesin-3 is also inhibited by following CC1. The earlier studies of UNC-104 (KIF1A homolog in Caenorhabditis elegans) demonstrated that CC1 can fold back to associate with the NC to form a compact self-folded helix bundle that sequesters the NC and prevents its dimerization (23). The intramolecular interaction between the NC and CC1 was suggested to be able to control the monomer–dimer transition of KIF1A/UNC-104 by switching of the NC between the intramolecular monomer and intermolecular dimer (16, 24). More intriguingly, the CC1-mediated inhibition of the NC was also found in KIF13A, KIF13B, and KIF16B, and the cargo-mediated dimerization of these motors resulted in their superprocessive motions (16), supporting that the CC1-mediated autoinhibition is highly conserved in kinesin-3 motors and plays a prominent role in regulating the motor motility, although the mechanism of inhibition remains poorly understood.
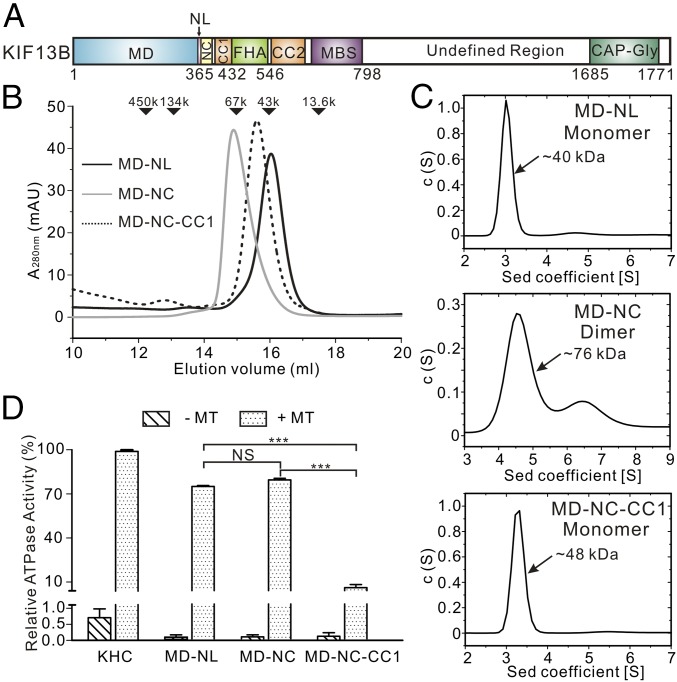
Fig. 1.
Biochemical characterization of the various MD-containing fragments of KIF13B. (A) Domain organization of kinesin-3 KIF13B. KIF13B contains an N-terminal MD, a neck domain (containing the NL and NC), a CC1-FHA-CC2 tandem followed by a MBS domain, and a C-terminal CAP-Gly domain. The residue numbers for each domain are indicated below the domain organization. (B) Analytical gel-filtration analysis of the various KIF13B fragments, including MD-NL (solid black lines), MD-NC (solid gray lines), and MD-ND-CC1 (dotted lines). The elution volumes of molecular mass markers are indicated at the top of the elution profile. mAU, milliabsorbance unit. (C) Analytical ultracentrifugation characterization of the various KIF13B fragments. The calculated molecular masses for MD-NL, MD-NC, and MD-NC-CC1 are indicated. (D) ATPase activities of the various KIF13B fragments in the presence (+) or the absence (−) of MTs. Data were normalized by the MT-stimulated ATPase activity of KHC as 100%. Each bar represents the mean value ± SD. NS, no significant differences. ***P < 0.001 (Student’s t test).
In this study, we determined the structures of the uninhibited MD-NC dimer and the inhibited MD-NC-CC1 monomer of the kinesin-3 motor KIF13B [that plays a prominent role in transporting neuronal cargoes and controlling endocytosis (25, 26)]. In the MD-NC dimer, the NC forms a coiled-coil dimer for assembling an uninhibited two-headed motor. In the MD-NC-CC1 monomer, CC1 is broken into two short helices (CC1a and CC1b) that unexpectedly interact with both the NC and the MD to form a compact self-folded monomer. CC1a folds back to pack with the NC in an antiparallel manner, and this NC-CC1a two-helix bundle further leans against the MD to integrate the three domains together and fix both the NC and NL, while CC1b does not contact with the NC, but anchors at a distinct site of the MD to stabilize the integrated monomer. The CC1-mediated integration of the NC and the MD not only inhibits the NC dimer formation but also prevents the NL undocking and the ADP release from the MD. Mutations of the essential residues in the interdomain interaction interface in the MD-NC-CC1 monomer restored the MD activity. Thus, CC1 fastens the neck domain and MD and sequesters both the NC and NL for kinesin-3 autoinhibition.
Results
Biochemical Characterization of the MD-Containing Fragments of KIF13B.
To investigate the mechanism underlying the CC1-mediated autoinhibition of kinesin-3, we initiated this work with biochemically characterizing the various fragments of KIF13B. We focused on the N-terminal MD-containing fragments with or without CC1 [i.e., MD-NL, MD-NC, and MD-NC-CC1 (Fig. 1A)] and characterized the oligomeric states of these fragments by using the analytical gel-filtration and ultracentrifugation analysis (Fig. 1 B and C). As expected, the MD (with the NL) predominantly exists in a monomeric state in solution (Fig. 1 B and C). Extension of the MD to the NC induced dimerization of the MD (Fig. 1 B and C). However, the NC-mediated dimerization was blocked by further inclusion of CC1, and the resulting MD-NC-CC1 tandem adopted a stable monomeric state in solution (Fig. 1 B and C). Thus, consistent with the previous studies of KIF13B (16), CC1 is an inhibitory segment capable of interfering with the NC-mediated dimerization.
In addition to the oligomeric states, we also performed the MT-simulated ATPase assay to characterize the ATPase activities of these KIF13B fragments. Without MTs, the basal ATPase activities of MD-NL, MD-NC, and MD-NC-CC1 are extremely low (Fig. 1D), consistent with the established critical role of MTs in the ATP hydrolysis cycle of kinesin motors (27). Upon adding MTs, both the MD (with the NL) and the MD-NC tandem shows the similar high ATPase activities (Fig. 1D), indicating that extension of the MD to the NC has little impact on its ATPase activity. In contrast, the MD-NC-CC1 tandem shows the significantly lower ATPase activity compared with the MD-NL and -NC tandems (Fig. 1D), suggesting that further inclusion of CC1 inhibits the MT-simulated ATPase activity of the MD. Taken together, all of the biochemical data demonstrated that CC1 prevents the NC dimer formation as well as the MT-simulated ATPase activity of the MD.
Structure of the Uninhibited MD-NC Dimer.
The NC can mediate dimerization of the MD, and the MD-NC dimer is in an uninhibited state (Fig. 1 B–D). To dissect the NC-mediated dimerization, we performed the crystal screening of the MD-NC tandem. After extensive trials, the diffraction-quality crystals were obtained. The structure of the MD-NC tandem was solved by the molecular-replacement method and refined to 2.6 Å (Fig. 2 and SI Appendix, Table S1). Although one molecule was found in the asymmetric unit of the crystal, there exists a twofold symmetry-related MD-NC dimer (Fig. 2 A and B). In the MD-NC dimer structure, the MD adopts a canonical kinesin-motor fold with eight β-strands (β1–β8) to form a central β-sheet that is surrounded by six α-helices (α1–α6), The NL adopts a linear conformation and docks onto the MD, and the NC forms a coiled-coil dimer that brings the two MDs together to assemble an uninhibited two-headed motor (Fig. 2 B and C). Although no extra nucleotides were added during crystallization, ADP and Mg2+ were still found to reside in the nucleotide-binding pocket of the MD based on their well-traced electron densities (Fig. 2 A and B).
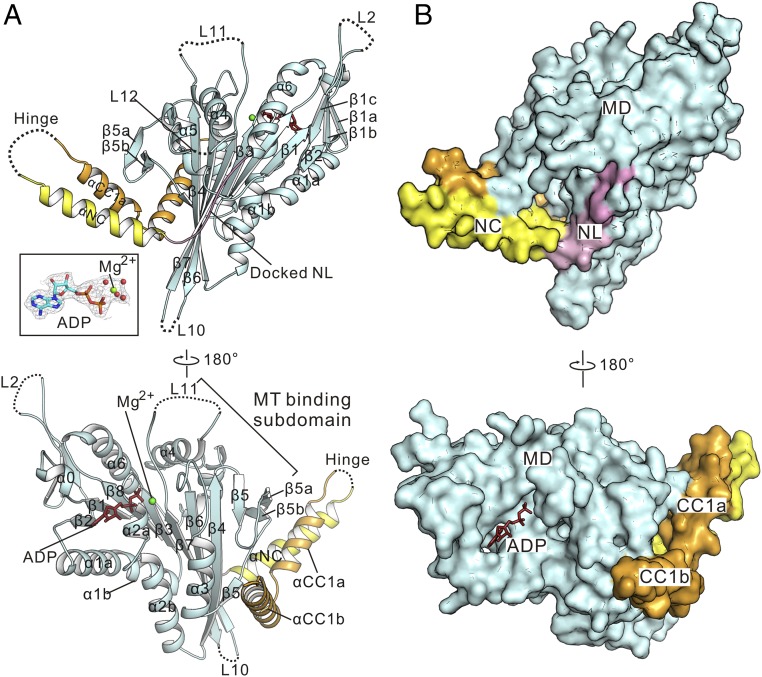
Fig. 2.
Structure of the MD-NC dimer of KIF13B. (A) A ribbon diagram of the structure of the MD-NC subunit. The MD, NL, and NC are colored in pale cyan, pink, and yellow, respectively. Missing loops in the structure are marked with dashed lines. A, Inset shows the omit electron-density maps of ADP and Mg2+ (contoured at 1.5σ level). (B and C) A ribbon diagram (B) and surface representation (C) of the structure of the MD-NC dimer. The MD-NC dimer is assembled by the NC coiled-coil dimer (highlighted by a dashed box in B). (D) A combined ribbon and stick diagram showing the interhelical packing between the NC coiled-coil dimer. The side chains of the interhelical packing residues are shown as sticks. (E) Structural comparison of the dimeric MD-NC structures of kinesin-1/-3. Here, kinesin-1 KIF5C (PDB ID code 3KIN) is colored in pink, and kinesin-3 KIF13B is colored in pale cyan. The distance between the nucleotide-binding pockets of the two MDs for both kinesin-1 and -3 is labeled.
In the MD-NC dimer structure, the NC initiates from P363 that produces a characteristic turn-like structure capping at the N terminus of the coiled-coil dimer (Fig. 2D). Consistent with the previous structural studies of the NC (28), the interhelical packing between the NC coiled-coil dimer (buried with a surface area of ∼1,390 Å2) is largely contributed by the hydrophobic residues from the a and d sites of the NC [based on the heptad repeat pattern (a–g) analysis]—that is, I367, I368, L371, V375, L378, and L382 form the hydrophobic packing core of the coiled-coil dimer (Fig. 2D). Moreover, the electrostatic interactions between E374 and R379 from the two NC helices further stabilize the coiled-coil formation (Fig. 2D).
We compared the structure of the uninhibited MD-NC dimer of kinesin-3 with that of kinesin-1 [Protein Data Bank (PDB) ID code 3KIN; in which the NL also docks onto the MD] (SI Appendix, Fig. S1). From the overall structure, the MD-NC dimer of kinesin-3 largely resembles that of kinesin-1 (SI Appendix, Fig. S1), indicating that the NC-mediated dimerization is a similar mechanism in processive kinesin motors. However, the distance between the two domains (both upon NL docking) is somewhat different between the two dimer structures—that is, ∼70 Å in kinesin-3 vs. ∼90 Å in kinesin-1 (based on the distance between the nucleotide-binding pockets of the two MDs) (Fig. 2E and SI Appendix, Fig. S1). In the MD-NC dimer of kinesin-3, the two MDs are related by a rotation of ∼180° rather than ∼120° observed in that of kinesin-1 (SI Appendix, Fig. S1). Upon superimposition of one subunit in the dimer, the other one of kinesin-3 undergoes a rotation of ∼60° and moves closer to the superimposed one (Fig. 2E). These structural differences between the two MD-NC dimers of kinesin-1/-3 are likely caused by the different lengths of NL that link the two MDs through the NC coiled-coil dimer. Taken together, the structure of the MD-NC dimer of KIF13B demonstrated the NC-mediated dimerization of the MD and revealed the potential active conformation of kinesin-3 motors.
Crystallization of the Inhibited MD-NC-CC1 Monomer.
In contrast to the uninhibited MD-NC dimer, the MD-NC-CC1 tandem forms an inhibited monomer in which both the NC-mediated dimerization and the MT-simulated ATPase activity of the MD are blocked (Fig. 1). To investigate the mechanism of inhibition, we performed the crystal screening of the MD-NC-CC1 tandem. However, all of the crystals of this fragment were diffracted poorly, despite extensive efforts. We then had to resort to the MD-NC-CC1 tandem with a Y73C mutation in the MD for crystallization. In our previous studies, the Y73C mutation was demonstrated to be capable of enhancing the crystal packing with little impact on the MD (29), suggesting that this point mutation would not interfere with the inhibited monomeric state of the MD-NC-CC1 tandem. Supporting this hypothesis, the MD-NC-CC1-(Y73C) mutant adopted a monomeric conformation in solution with the low MT-simulated ATPase activity similar to that of the MD-NC-CC1 tandem (SI Appendix, Fig. S2A). With the Y73C mutation in the MD-NC-CC1 tandem, we successfully obtained the diffraction-quality crystals that are suitable for structural determination.
Structure of the Compact MD-NC-CC1 Monomer.
The structure of the MD-NC-CC1 tandem with the Y73C mutation was solved by the molecular-replacement method and refined to 2.4 Å (Fig. 3 and SI Appendix, Table S1). Consistent with the monomeric state in solution (Fig. 1 and SI Appendix, Fig. S2A), only one molecule was found in the asymmetric unit of the crystal. In the structure of the MD-NC-CC1-(Y73C) mutant, the Y73C mutation is located in loop 3 of the MD, and this position is far away from the binding sites for contacting with NC and CC1 (SI Appendix, Fig. S2B), supporting the fact that the Y73C mutation has limited effect on the overall structure of the MD-NC-CC1 tandem and does not interfere with the interdomain interactions between the MD, the NC, and CC1. Based on the crystal-packing analysis and structural comparison, the Y73C mutation stabilizes the flexible loop 3 and extends the following helix α1a that happens to sit at the intermolecular packing interface of the crystal (SI Appendix, Fig. S2 C and D). The electrostatic interactions between the extended helix α1a and the neighboring molecule significantly enhance the crystal packing (SI Appendix, Fig. S2D). Thus, both the biochemical data and the structural analysis demonstrate that the Y73C mutation facilitates the crystal packing with minimal impact on the overall structure and motor activity of the MD-NC-CC1 tandem, and the structure of the MD-NC-CC1-(Y73C) mutant can be reasonably used to represent that of the wild-type inhibited MD-NC-CC1 monomer.
Fig. 3.
Structure of the MD-NC-CC1-Y73C monomer of KIF13B. (A) A ribbon diagram of the structure of the MD-NC-CC1-Y73C monomer. MD, NL, NC, and CC1 are colored in pale cyan, pink, yellow, and light orange, respectively. Missing loops in the structure are marked with dashed lines. The omit electron-density maps of ADP, Mg2+, and the coordinated water molecules are shown in A, Inset and contoured at the 1.5σ level. (B) A surface representation of the structure of the MD-NC-CC1-Y73C monomer. The color scheme follows that in A. This shows that the MD-NC-CC1 tandem forms a compact self-folded monomer.
In the structure of the MD-NC-CC1 monomer, the MD adopts a canonical kinesin-motor fold, the NL linearly docks onto the MD, and the NC forms a single α-helical structure rather than a coiled-coil dimer (Fig. 3A). Additionally, ADP and Mg2+ were also well traced in the nucleotide-binding pocket of the MD, even without extra nucleotides being added during crystallization (Fig. 3A). Instead of forming an extended α-helix, CC1 is broken into two short α-helices (CC1a and CC1b) around the middle that unexpectedly interact with both the NC and the MD to assemble a compact, self-folded monomer (Fig. 3). The N-terminal helix CC1a folds back to pack with the NC in an antiparallel manner, and together they form a two-helix bundle that directly sequesters the NC and prevents its coiled-coil dimer formation (Fig. 3A). Moreover, this two-helix bundle further leans against the MD and interacts with a short β-hairpin (formed by β5a and β5b) from the MT-binding subdomain to integrate the three domains together (Fig. 3). On the other hand, the C-terminal helix CC1b (perpendicular to CC1a) does not contact with the NC, but anchors onto the MD at a distinct site (Fig. 3A), which would further stabilize the self-folded conformation of the MD-NC-CC1 tandem.
To avoid the potential artifacts in the MD-NC-CC1 monomer caused by the crystal packing, we further performed the molecular dynamics simulations of this monomer structure in solution (SI Appendix, Fig. S3). The overall structure of the MD-NC-CC1 monomer underwent little conformational change during the simulations, supporting that the compact MD-NC-CC1 monomer is relatively stable (SI Appendix, Fig. S3). However, in comparison with the core structure of the MD, several loops (loops 2 and 10–12) of the MD and the hinge between the NC and CC1 exhibit more dynamic conformations (SI Appendix, Fig. S3), suggesting that these regions are intrinsically flexible. Consistent with this feature, in the MD-NC-CC1 monomer structure, loops 2 and 10–12 of the MD and the NC/CC1 hinge could not be clearly defined due to the poor electron-density maps (as indicated by the dashed lines in Fig. 3A).
Interdomain Interaction Interface in the MD-NC-CC1 Monomer.
In the self-folded MD-NC-CC1 monomer, the interdomain interaction interface (buried with a surface area of ∼2,327 Å2) between the three domains is highly conserved in KIF13B and can be further divided into two sites (sites I and II) (Fig. 4A and SI Appendix, Fig. S4). Site I is contributed by the MD, NC, and CC1a that form the primary tripartite interaction interface (Fig. 4A). In this tripartite interface, the interhelical packing between the NC and CC1a is largely mediated by the hydrophobic residues from the a and d sites [based on the heptad repeat pattern (a–g) analysis]—that is, I367, I368, L371, V375, L378, and L382 from NC and L393, L397, I404, and M407 from CC1a (Fig. 4B and SI Appendix, Fig. S5). In addition to the hydrophobic packing, the electrostatic and hydrogen-bonding interactions between NC and CC1a further stabilize the antiparallel two-helix bundle formation (Fig. 4B). The further interdomain packing between the NC-CC1a two-helix bundle and the MD is mediated by a combined network of the electrostatic, hydrogen-bonding, and hydrophobic interactions (Fig. 4C). Specifically, R174 from β5a and Y182 and D184 from β5b of the MD form the electrostatic and hydrogen-bonding interactions with E399 and K402 from CC1a; H176 from β5a and S177 from loop 8b of the MD form the specific hydrogen bonds with S400 and R396 from CC1a, respectively; and V178 and L179 from loop 8b and Y182 from β5b of the MD make the hydrophobic contacts with I367 and L371 from NC and L403 from CC1a (Fig. 4 C and E).
Fig. 4.
Interdomain interaction interface in the compact MD-NC-CC1 monomer. (A) A ribbon diagram of the structure of the compact MD-NC-CC1 monomer. The interdomain interaction interface between the MD, NC, and CC1 can be divided into two sites (sites I and II, highlighted by boxes). (B–D) A combined ribbon-and-stick model showing the interdomain interface in site I (B and C) and site II (D). Site I is contributed by the interhelical packing between the NC and CC1a (B) and the interaction interface between the NC-CC1a and the MD (C), and site II is contributed by the interaction interface between CC1b and the MD (D). The side chains of the key residues involved in the interdomain interface packing are shown as sticks. (E) Structure-based sequence alignment of the binding regions between the MD and NC-CC1 from different species of KIF13B. The identical and highly conserved residues are colored in red and green, respectively. (E, Upper) The secondary structures and residue numbers of KIF13B are marked at the top. (E, Lower) The hydrophobic and polar residues responsible for the interdomain interaction are marked with yellow dots and blue triangles, respectively.
On the other hand, site II is constructed by the MD and CC1b that form the additional bipartite interaction interface (Fig. 4A). CC1 is broken around V409 and T410 that forms a tight turn between the two perpendicular helices (CC1a and CC1b), and CC1b interacts with the MD at a distinct site formed by β4, β5, α3, and loop 8a (Fig. 4 A and E). In the N-terminal part of this bipartite interface, W411 from CC1b is buried and sandwiched between K145 from β4 of the MD and L415 from CC1b; K414 from CC1b forms the electrostatic interaction with E147 from β4 of the MD; and T418 from CC1b forms the specific hydrogen bond with the backbone of A190 from β5 of the MD (Fig. 4 D and E). In the C-terminal part of this bipartite interface, R425 from CC1b forms the hydrogen-bonding interaction with S199 from α3 of the MD, and I421 from CC1b makes the hydrophobic contacts with L189 from β5 and P164 from loop 8a of the MD (Fig. 4 D and E). Taken together, the extensive electrostatic, hydrogen-bonding, and hydrophobic interactions between the MD, NC, and CC1 in both sites I and II integrate the three domains together to form a compact monomer. Moreover, most of the essential residues involved in the interdomain interaction interface are also highly conserved in other kinesin-3 motors (SI Appendix, Fig. S5).
Both the NC and NL of the Neck Domain Are Sequestered in the MD-NC-CC1 Monomer.
With the structures of the uninhibited MD-NC dimer and inhibited MD-NC-CC1 monomer, we next dissected the molecular mechanism for CC1-mediated autoinhibition. The neck domain of kinesin motors (composed of the NC and NL) plays an obligatory role in controlling the motor activity—that is, the NC mediates dimerization of the MD, while the NL docks onto or undocks from the MD to mediate the chemomechanical coupling and determine the directionality and processivity (6, 7, 27). In the uninhibited MD-NC dimer, the NC forms a coiled-coil dimer for the MD dimerization, and the NL is free to dock onto or undock from the MD without restriction (Fig. 5A). In contrast, in the inhibited MD-NC-CC1 monomer, the NC is sequestered by CC1a to form an antiparallel two-helix bundle and is unable to form a coiled-coil dimer for dimerization of the MD (Fig. 5B). Thus, the NC-mediated dimerization is blocked due to the direct intramolecular interaction between the NC and CC1, consistent with the previous studies and biochemical characterization.
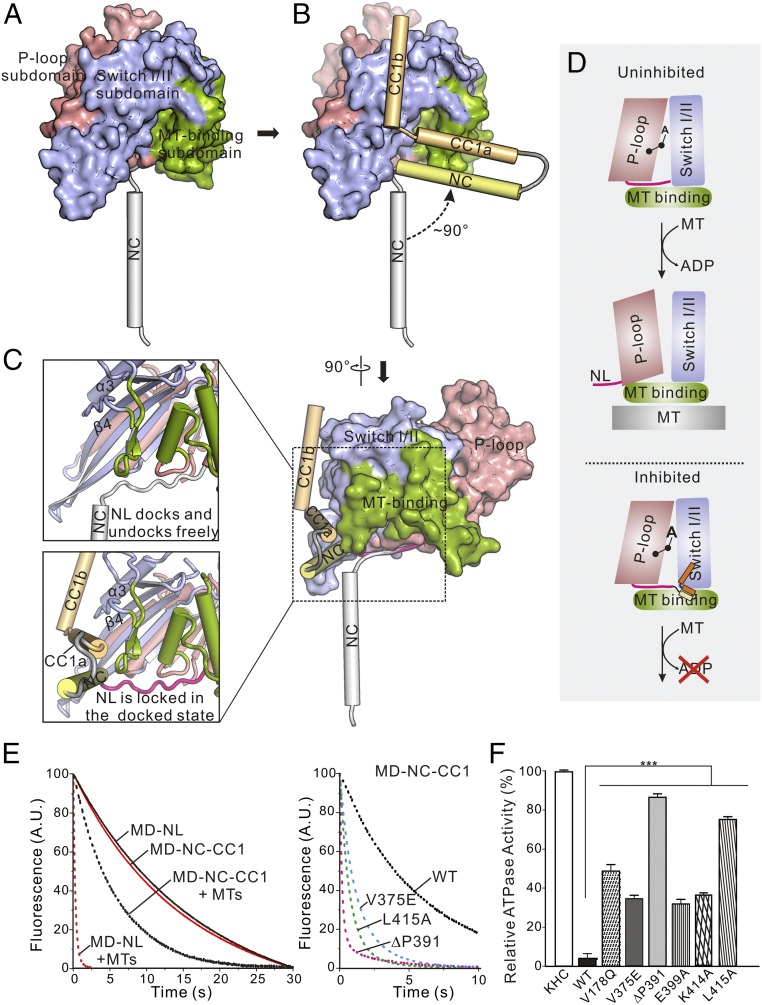
Fig. 5.
Both NC and NL of the neck domain are sequestered in the MD-NC-CC1 monomer. (A) A combined ribbon-and-surface representation of the MD-NC subunit from the dimer. The MD is shown in surface and can be divided into the P-loop, SwitchI/II, and MT-binding subdomains colored in salmon, slate, and green, respectively, and the NC is in a cylinder representation and colored in gray. (B) Structural comparison of the MD-NC and MD-NC-CC1 tandems. The MD and NC of the MD-NC tandem are colored as in A, and NC and CC1 of the MD-NC-CC1 tandem are colored in yellow and orange, respectively. After superimposition of the two structures, the NC undergoes dramatic conformational changes. (C) A close-up view of the NL regions of the MD-NC and MD-ND-CC1 structures. The NL can dock/undock freely in the MD-NC tandem, but is locked in the docked state in the MD-NC-CC1 tandem. (D) A schematic working model for explaining the conformational changes between the three subdomains coupled with the NL undocking and ADP release. The color scheme for the three subdomains follows that in A. The NL is shown as a pink line, and the NC and CC1 are colored in yellow and orange, respectively. (E) Kinetics of the mant-ADP release from the MD-NL (red), the MD-NC-CC1 (black), and the various MD-NC-CC1 mutants (V375E, blue; ΔP391, pink; and L415A, green). Normalized fluorescence variations on release of ADP from the different fragments in the absence of MTs (solid lines) or the presence of MTs (dashed lines) are shown. (F) MT-stimulated ATPase activity of the wild type and various mutants of the MD-NC-CC1 tandem of KIF13B. Each bar represents the mean value ± SD. ***P < 0.001 (Student’s t test).
More intriguingly, the NC-CC1a bundle and CC1b further interact with the MD, which would tether and fix the NC onto the MD firmly (Fig. 3). With superimposition of the MD, the binding of the NC-CC1a bundle and CC1b to the MD drives the NC to undergo a rotation of ∼90° toward the MD (Fig. 5B). Thus, in addition to preventing the NC coiled-coil dimer formation, CC1 also restrains its freedom of motion through binding to the MD. Given that the NC immediately follows the NL, restriction of the NC motion subsequently sequesters the NL and locks it in a docked conformation (Fig. 5C), suggesting that the NL would hardly undock from the MD in the inhibited MD-NC-CC1 monomer. Taken together, CC1 fastens the neck domain and MD by interacting with the two domains and sequesters both the NC and NL of the neck domain for autoinhibition.
Release of ADP from the MD Is Inhibited in the MD-NC-CC1 Monomer.
The initial biochemical characterization of the MD-NC-CC1 tandem demonstrated that its MT-stimulated ATPase activity is also inhibited (Fig. 1). The decreased activity may be caused by the direct interference of binding to MTs. However, in the MD-NC-CC1 monomer structure, the NC-CC1a bundle only leans against the periphery of the MT-binding subdomain of the MD and the major binding site of this subdomain is completely exposed (Fig. 3), indicating that the MD-NC-CC1 tandem is still capable of interacting with MTs. To test this hypothesis, we performed the MT-binding assay for the MD (with the NL) and the MD-NC-CC1 tandem. As expected, the MD-NC-CC1 tandem can bind to MTs similarly to the MD-NL tandem (SI Appendix, Fig. S6), demonstrating that the MT-binding capacity of the MD in the MD-NC-CC1 tandem is not severely impaired. Thus, the decreased MT-stimulated ATPase activity of the MD-NC-CC1 tandem is unlikely to be caused by interfering with the MT-binding property of the MD.
Based on previous studies, the MD can be divided into three subdomains—that is, the P-loop, SwitchI/II, and MT-binding subdomains (30) (Fig. 5A). The intramolecular structural changes between the three subdomains are intimately coupled with the ATP hydrolysis cycle of the MD and its ATPase activity (27, 30). Upon docking onto the MD, the NL anchors between the P-loop and MT-binding subdomains, and its undocking allows the intersubdomain rearrangement required for the MT-stimulated ADP release from the MD (Fig. 5D). However, in the inhibited MD-NC-CC1 monomer, the NL is sequestered in a docked state, which would block the movement between the P-loop and MT-binding subdomains and prevent the subsequent MT-stimulated ADP release (Fig. 5D). To test this hypothesis, we performed the MT-stimulated mant-ADP release assay for the MD (with the NL) and the MD-NC-CC1 tandem. Consistent with the essential role of MTs for the ADP release, the ADP release from the MD-NL tandem is slow without MTs but is significantly accelerated upon adding them (Fig. 5E and SI Appendix, Table S2). In the absence of MTs, the ADP release from the MD-NC-CC1 tandem is also slow and remains slow after adding MTs compared with the MD-NL tandem (Fig. 5E and SI Appendix, Table S2), indicating that the MT-stimulated ADP release is severely impaired in the MD-NC-CC1 monomer. Taken together, consistent with the structural analysis, the MT-stimulated ADP release from the MD in the MD-NC-CC1 tandem is inhibited by sequestering the NL, which would cause the decreased ATPase activity.
Disruptions of the MD-NC-CC1 Monomer Restore the MD Activity.
Based on the structural analysis of the inhibited MD-NC-CC1 monomer, we introduced a series of the point mutations in the MD, NC, and CC1 to disrupt the interdomain interaction interface—that is, V178Q in the MD, V375E in the NC, and E399A, K414A, and L415A in CC1—and checked the MT-stimulated ATPase activity of these mutants. We also removed a proline from the NC/CC1 hinge (ΔP391), which was previously demonstrated to disrupt the folded-back conformation of CC1 (16, 28). All of the mutations did not have major effects on the secondary structures of the MD-NC-CC1 tandem, as indicated by the similar profile in the CD spectra (SI Appendix, Fig. S7A). However, most of the mutations in the interdomain interaction interface destabilized the MD-NC-CC1 tandem in the limited proteolysis assay (SI Appendix, Fig. S7B), suggesting that the compact self-folded structure is disrupted by these mutations. As expected, in comparison with the wild-type MD-NC-CC1 tandem, the mutations in the interdomain interaction interface or in the NC/CC1 hinge all significantly enhanced the MT-simulated ATPase activity (Fig. 5F), indicating that disruptions of the contacts between the three domains or the folded-back conformation of CC1 restore the MD activity. Consistent with the increased ATPase activity, the MT-stimulated ADP release of the MD-NC-CC1 tandem was also markedly accelerated by the mutations (V375E, ΔP391, and L415A that keep the MD intact) (Fig. 5E and SI Appendix, Table S2), further implicating the loosening of the NL for the ADP release. On the other hand, disruptions of the interactions between the three domains or the folded-back conformation of CC1 would also release the NC for dimerization of the MD. Consistent with this hypothesis, the ΔP391-mutant adopts a dimeric conformation, and most of the other mutants (V178Q, E399A, K414A, and L415A with the intact NC) have a tendency to form a dimer and exist in a monomer/dimer equilibrium in solution (SI Appendix, Fig. S8). In contrast, the V375E mutant [with the mutation in the NC that disrupts the MD-NC-CC1 monomer as well as the NC coiled-coil dimer (Fig. 2D)] remains as a monomer (SI Appendix, Fig. S8), further supporting the essential role of the NC for dimerization of the MD.
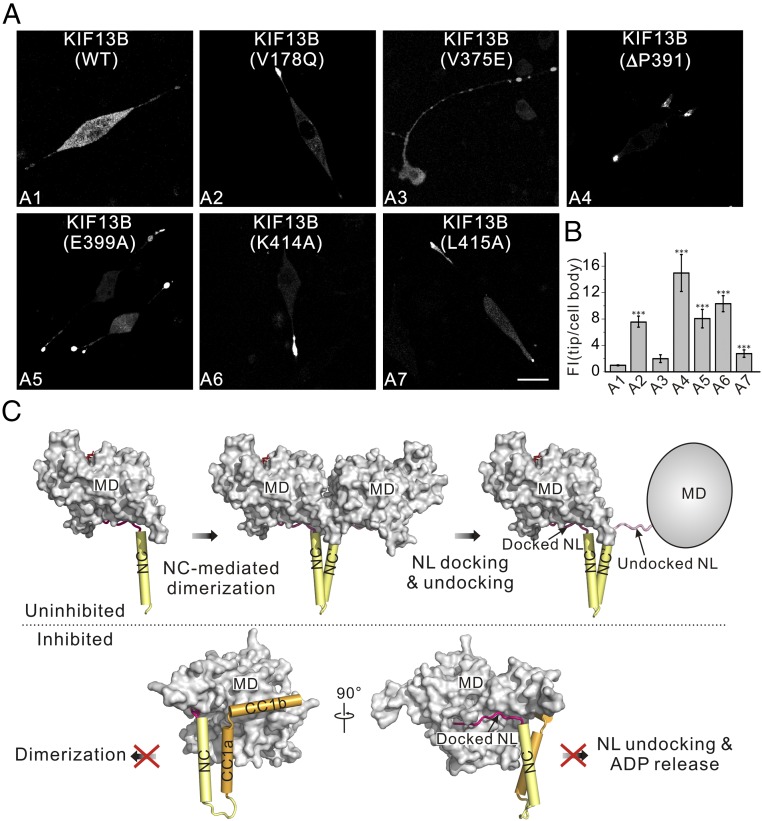
Finally, we assessed the effects of these mutations on the full-length KIF13B in vivo by using a cell-based assay (Fig. 6A). Consistent with previous results (16), the wild-type KIF13B adopts an inactive state and was largely localized in the cell body (Fig. 6 A and B). Similar to the wild-type KIF13B, the Y73C–KIF13B mutant was also largely localized in the cell body (SI Appendix, Fig. S2 E and F), supporting the minimal impact of this mutation on the full-length protein. As expected, most of the KIF13B mutants (V178Q, ΔP391, E399A, K414A, and L415A) showed the enhanced accumulations at the cell tips in comparison with the wild-type KIF13B (Fig. 6 A and B), indicating the restored motor motility by these mutations. In contrast, the V375E–KIF13B mutant did not show the significantly increased tip accumulations (Fig. 6 A and B), also consistent with the essential role of the NC-mediated dimerization for the motor activity. Taken together, all of the biochemical and cellular data demonstrated that disruptions of the MD-NC-CC1 monomer release the sequestered neck domain and restore the MD activity.
Fig. 6.
Disruptions of the MD-NC-CC1 monomer restore the motor activity of KIF13B. (A) Cellular localizations of the full-length KIF13B and its various mutants in the N2A cells. The wild-type KIF13B was largely localized in the cell body (A1). Most of the mutants showed the significantly increased accumulations at the cell tips, except for the V375E mutant (with the mutation in NC) (A2–A7). (Scale bar: 20 μm.) (B) Quantification of the cellular distribution data shown in A. The ratio of the tip to cell body average FI was quantified for each construct for >15 cells (n > 15). Each bar represents the mean value ± SEM. ***P < 0.001 (Student’s t test). (C) A working model for the CC1-mediated autoinhibition of kinesin-3. In the uninhibited state (C, Upper), the NC mediates dimerization of the MD to assemble a dimeric motor, and the NL freely docks onto or undocks from the MD for processive movement. The MD is in the surface representation (colored in gray), and the NL and NC are in the ribbon representation (colored in pink and yellow, respectively). The gray oval represents the MD with the NL undocking. In the inhibited state (C, Lower), CC1 (colored in orange) folds back to interact with both the NC and the MD, which not only inhibits the NC-mediated dimerization, but also prevents the NL undocking and the ADP release from the MD.
Discussion
Intrinsic Structural Versatility of CC1 for Autoinhibition.
In kinesin-3 motors, the coiled-coil domain CC1 is a well-known autoinhibitory segment that folds back to sequester the preceding NC for inhibiting the motor activity (2, 16, 23), but the underlying mechanism is not well understood. In this study, we determined the structure of the inhibited MD-NC-CC1 monomer of KIF13B in which CC1 is broken into two short α-helices (CC1a and CC1b) that interact with both the NC and the MD to sequester the entire neck domain (Figs. 3 and 4). Thus, instead of forming an extended coiled-coil, CC1 adopts a distinct two-helix conformation for autoinhibition. Consistent with this unexpected structural feature, our previous studies of the NC-CC1-FHA tandem of KIF13A demonstrated that CC1 is not a perfect coiled-coil for dimerization (28). Moreover, CC1 contains several intrinsic unusual features (i.e., the potential defects in the interhelical packing and a “stutter” break in the middle) that can distort the coiled-coil formation and break the extended helix (28) (SI Appendix, Fig. S5), supporting the propensity of CC1 to form a two-helix conformation in the autoinhibited state. Taken together, CC1 of kinesin-3 motors is not a canonical coiled-coil but possesses the intrinsic structural versatility to adopt a distinct conformation for autoinhibition.
In the MD-NC-CC1 monomer, CC1a folds back to pack with the NC in an antiparallel manner to form a two-helix bundle with a short hinge (Figs. 3 and 4), consistent with previous structural prediction (16). Removal of the signature proline (P391) from the short hinge disrupted the folded-back conformation of CC1 and promoted the motor dimerization (SI Appendix, Fig. S8), also consistent with the previous studies (16, 28). It has been proposed that, for different kinesin-3 motors, the length of the hinge between the NC and CC1 may determine their packing orientation (16)—i.e., for the long hinge, CC1 forms an extended helix to pack with the NC in a parallel manner based on the electron-microscopy studies of UNC-104 (23), while for the short hinge, CC1 interacts with the NC in an antiparallel manner, as demonstrated in this study (Fig. 3). However, given the high sequence similarity of CC1 among different kinesin-3 motors (SI Appendix, Fig. S5), CC1 in the long-hinged kinesin-3 motors may also be capable of adopting the two-helix conformation for sequestering the NC. Moreover, due to the short length of the NC, the half of CC1 (CC1a) is sufficient to cover the whole NC, as shown in the MD-NC-CC1 monomer structure (Fig. 4 and SI Appendix, Fig. S5). Thus, we may not exclude the possibility that the antiparallel packing between CC1a and the NC for autoinhibition could also exist in other long-hinged kinesin-3 motors.
The essential role of CC1 for regulating the NC-mediated dimerization was described in the work of Al-Bassam et al. (23). They worked on the different constructs of UNC-104 with MTs by the cryo-EM method at low resolution and found that, upon binding to MTs, AMPPNP (the nonhydrolyzable ATP analog) appears to trap the MD-NC-CC1 tandem in a monomeric state (23). In this work, the similar MD-NC-CC1 tandem of KIF13B adopts a monomeric conformation in solution and is also capable of binding to MTs (Fig. 1 and SI Appendix, Fig. S6), consistent with the work of Al-Bassam et al. work. However, the MD of the MD-NC-CC1 monomer of KIF13B was tapped in an ADP-bound state (since we did not add extra nucleotides during the protein purification and crystallization) (Fig. 3). The discrepancy of the MD-NC-CC1 monomer with different nucleotide-bound states between the two studies might be caused by different experimental conditions—i.e., in the study of Al-Bassam et al., the work was carried out with MTs, whereas in this study, the work was largely performed without MTs.
The MD Is Unexpectedly Involved in the CC1-Mediated Autoinhibition.
In addition to the broken CC1, the other unexpected structural feature in the MD-NC-CC1 monomer is that the MD is involved in the CC1-mediated autoinhibition (Fig. 3). The MD contains two distinct sites (in the β4-α3 region, referred to as the NC-CC1-binding region) to capture the NC-CC1a bundle and CC1b, respectively, to form a compact self-folded structure (Fig. 4). In comparison with the MD-NC dimer, the binding of the NC-CC1a bundle and CC1b induced some local conformational changes in the β5a/β5b region of the MD, while no significant structural changes were found in the Switch I and II elements (since the MDs from the two structures were both trapped in the similar ADP-bound state) (SI Appendix, Fig. S9). The two site-mediated interactions between the MD and NC-CC1a/CC1b not only tether the NC to the MD, but also sequester the NL, which ultimately leads to the complete lockdown of the entire neck domain from the NL to the NC (Fig. 5). Without binding to the MD, both the NC-CC1a bundle and CCb1 would be flexible and unstable in solution [as indicated in the limited proteolysis assay (SI Appendix, Fig. S7B)], and the neck domain would be only partially inhibited due to the fact that the NL could freely undock from the MD without restraints (Fig. 5). Thus, with two distinct binding sites, the MD tightly packs with NC-CC1a/CC1b and facilitates CC1 to inhibit the entire neck domain. Consistent with this hypothesis, mutations of the residues of the MD (that are in the interdomain interaction interface) disrupted the MD-NC-CC1 monomer and restored the MD activity (Figs. 5 and 6). Intriguingly, most of the essential residues in the NC-CC1-binding region of the MD are highly conserved in other kinesin-3 motors (SI Appendix, Fig. S5), suggesting that the MD in these motors may also participate in the CC1-mediated autoinhibition. Moreover, mutations of the MD of kinesin-3 KIF1A are coupled with a number of human neuronal disorders, and some of the disease-related mutations are located in the NC-CC1-binding region (31–33) (SI Appendix, Fig. S5), indicating that the identified NC-CC1-binding region of the MD of kinesin-3 motors may also be of functional importance.
CC1 Fastens the Neck Domain and MD for Kinesin-3 Autoinhibition.
Based on the structures of the uninhibited MD-NC dimer and inhibited MD-NC-CC1 monomer, the molecular mechanism for the CC1-mediated autoinhibition could be proposed (Fig. 6C). In the uninhibited state, the NC mediates dimerization of the MD to assemble a two-headed motor, and the NL freely docks onto or undocks from the MD without restraints (Fig. 6C). In the CC1-mediated autoinhibited state, CC1 folds back to interact with both the NC and the MD to fasten the neck domain and the MD, which not only inhibits the NC-mediated dimerization but also prevents the NL undocking and the ADP release from the MD (Fig. 6C). In contrast, the MT-binding capacity of the MD is not severely impaired by CC1 due to the exposure of the MT-binding subdomain (Fig. 3 and SI Appendix, Fig. S6). Thus, with the aid of the MD, CC1 sequesters both the NC and NL of the neck domain without interfering with the MD (Fig. 6C). This dual inhibition of the neck domain by CC1 would secure the inactive state of the motor without any futile ATP hydrolysis. The CC1-mediated lockdown of the entire neck domain may represent a paradigm for kinesin autoinhibition and could be applicable to other kinesin-3 motors.
The CC1-mediated prevention of the NL undocking and the ADP release found in kinesin-3 is also reminiscent of the mechanism of kinesin-1 autoinhibition. In kinesin-1, a short peptide from the tail domain was found to directly bind to the MD (11–13). The structure of the motor-tail complex of kinesin-1 demonstrated that a single tail peptide can cross-link the two MDs and, together with the NC dimer, prevents the movement of the two MDs that is required for the NL undocking and the ADP release (34, 35). In kinesin-3, the CC1-mediated fastening of the neck domain and MD locks down the entire neck domain, and the NL is prevented from undocking from the MD for the ADP release (Fig. 6C). Thus, different approaches used by kinesin-1 and -3 (tail peptide-mediated vs. CC1-mediated) come to the same result, and prevention of the NL undocking and the ADP release may be a general mechanism for kinesin autoinhibition.
In the full-length motor, in addition to the key autoinhibitory segment CC1, other regions have been shown to further facilitate the autoinhibited conformation of kinesin-3. In KIF1A, CC2 was shown to be able to fold back to interact with the FHA domain (20), and in KIF13B, the membrane-associated guanylate kinase homolog binding stalk (MBS) segment immediately following CC2 was found to interact with the MD (21). On the other hand, in our previous work, we found that the CC1-FHA tandem of kinesin-3 can form an extended dimer that would release the CC1-mediated inhibition and couple with the NC dimer to assemble a dimeric motor for processive movement (28, 36). Moreover, the activation and dimerization of kinesin-3 were demonstrated to be chiefly mediated by cargo binding (16, 19, 21). Based on all of the previous studies and this work, a working model for the cargo-mediated transition of the full-length KIF13B from an autoinhibited monomer to an uninhibited dimer could also be proposed (SI Appendix, Fig. S10). In the monomeric autoinhibited state, CC1 interacts with both NC and the MD, CC2 interacts with the FHA domain, and the MBS segment binds to the MD (SI Appendix, Fig. S10). Upon binding to cargoes or cargo adaptors likely through the MBS segment in KIF13B (21), the FHA domain could be somehow released to promote the formation of the CC1-FHA dimer that would further release the CC1-mediated inhibition of the neck domain (SI Appendix, Fig. S10). In the dimeric uninhibited state, the FHA domain, CC1, and the NC would work together to form a stable dimer that brings the two MDs together for walking along MTs (SI Appendix, Fig. S10).
Materials and Methods
Protein Expression and Purification.
DNA sequences encoding the rat and human KIF13B MD-NL (1–371), MD-NC (1–389), MD-NC-CC1 (1–432), and various mutants were each cloned into a modified pET32a vector with a C-terminal His6-tag. Point mutations were introduced by using the standard PCR-based mutagenesis method and confirmed by DNA sequencing. Recombinant proteins were expressed in Escherichia coli BL21 (DE3) host cells at 16 °C. The His6-tagged fusion proteins were purified by Ni2+-Sepharose 6 Fast Flow (GE Healthcare) affinity chromatography followed by size-exclusion chromatography (Superdex-200 26/60; GE Healthcare) in the buffer containing 50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 2 mM MgCl2, 1 mM EGTA, and 1 mM DTT. For the analytical gel-filtration analysis, protein samples were concentrated to ∼5.0 mg/mL and loaded onto the Superdex-200 10/300 GL column (GE Healthcare).
Analytical Ultracentrifugation.
Sedimentation velocity experiments were performed on a Beckman XL-I analytical ultracentrifuge equipped with a four-cell An-60 Ti rotor (Beckman Coulter) under 220,000 × g at 25 °C. The partial specific volume of protein samples and the buffer density were calculated by using the program SEDNTERP (bitcwiki.sr.unh.edu/index.php/Main_Page). The final sedimentation velocity data were analyzed and fitted to a continuous sedimentation coefficient distribution model by using the program SEDFIT.
Crystallization.
Crystals of the MD-NC tandem (∼25 mg/mL) were obtained in 1.4 M Na/KPO4 (pH 6.9). Crystals of the MD-NC-CC1-Y73C mutant (∼20 mg/mL) were obtained in 0.2 M proline, 0.1 M Hepes (pH 7.5), and 14% (wt/vol) PEG 3350. All of the crystals were obtained by using the sitting-drop vapor-diffusion method at 16 °C. Before being flash-frozen in liquid nitrogen, crystals were soaked in the mother liquor supplemented with 15% (vol/vol) ethylene glycol for cryoprotection.
Data Collection, Structural Determination, and Refinement.
All of the diffraction datasets were collected at the beamline BL17U1 and BL19U of the Shanghai Synchrotron Radiation Facility with a wavelength of 0.989 Å at 100 K (37). Datasets were integrated and scaled with HKL2000 (38). The structures of the MD-NC tandem and the MD-NC-CC1-Y73C mutant were determined by the molecular-replacement methods using the MD of KIF13B (PDB ID code 5ZBR) as the searching model with Phaser (39). Additional missing residues were manually modeled into the structures according to the 2Fo − Fc and Fo − Fc electron-density maps using COOT (40). The structures were further refined and validated with PHENIX (41). The statistics for the data collection and structural refinement are summarized in SI Appendix, Table S1. The protein structure figures were prepared by using the program PyMOL (https://pymol.org/2/).
MT-Stimulated ATPase Assay.
Measurements of the MT-stimulated ATPase activities of various KIF13B fragments were performed by using the HTS Kinesin ATPase Endpoint Assay Biochem Kit (Cytoskeleton, Inc.). Briefly, all of the measurements were based on the malachite green phosphate assay to probe inorganic phosphate generated during the reaction. A standard curve of phosphate was made to estimate the amount of phosphate generated. The basal ATPase activities (without MTs) of these KIF13B fragments were also measured by using the same method. Each protein sample had three replicates, and each measurement was repeated at least three times independently. The kinesin-1 heavy chain (KHC) supplied in the kit was used as the control. All of the data were analyzed by using the GraphPad Prism5 program.
Kinetics of MT-Stimulated Mant-ADP Dissociation.
The bound ADPs in the MD (with the NL) and the MD-NC-CC1 tandem of KIF13B were first exchanged for mant-ADPs by equilibration with a 10-fold excess of mant-ADP at 25 °C for 30 min. The excess mant-ADPs were removed by exchanging into the reaction buffer containing 80 mM Pipes (pH 6.9), 1 mM MgCl2, 1 mM EGTA, and 0.1% Tween 20 using a Sephadex G-25 column. Release of mant-ADP was started by mixing the mant-ADP–loaded protein with a 50-fold excess of unlabeled ATP in the same buffer. The reaction was monitored by using a SX20 Stopped-Flow spectrometer at 25 °C. Excitation was at 365 nm, and fluorescence was recorded by using a 400-nm long-pass filter. For the MT-stimulated mant-ADP dissociation experiments, additional preprepared 1 μM MTs and 100 μM taxol were supplemented in the buffer. All of the data were analyzed and fitted by using the GraphPad Prism5 program.
Cell Culture, Imaging, and Data Analysis.
The full-length human KIF13B and various mutants were each cloned into a pEGFP-N3 vector. N2A cells were cultured in DMEM containing 10% (vol/vol) FBS and were grown at 37 °C. The cells were transfected with the wild-type KIF13B and various mutants by Lipofectamine 3000 (Invitrogen), according to the manufacturer’s instructions. Fluorescence images were obtained by using Olympus FV1000 Laser Scanning Confocal Microscopy with a 60× (NA = 1.42) oil objective. Confocal settings used for image capture were held constant in comparison experiments. For the cellular distribution data analysis, the specific regions of the cell body (excluding the nucleus) and the tip of each cell were chosen, and the average fluorescence intensities (FIs) were calculated, respectively. All of the fluorescence images were processed and analyzed by ImageJ (NIH). The final quantification graphs were generated by the GraphPad Prism5 program.
Accession Numbers.
The atomic coordinates of the MD-NC dimer and the MD-NC-CC1-Y73C monomer have been deposited in the Protein Data Bank, with PDB ID codes 6A1Z and 6A20, respectively.
Supplementary Material
Acknowledgments
We thank Dr. Yong Zhang for the help of molecular dynamics simulations. We thank the beamline BL17U of the Shanghai Synchrotron Radiation Facility for the beam time. This work was supported by National Key R&D Program of China Grant 2017YFA0503501; National Major Basic Research Program of China Grant 2014CB910202; and National Natural Science Foundation of China Grants 31470746, 31770786, 31600608, and 31600622.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates of the MD-NC dimer and the MD-NC-CC1-Y73C monomer have been deposited in the Protein Data Bank [PDB ID codes 6A1Z (MD-NC dimer) and 6A20 (MD-NC-CC1-Y73C monomer)].
See Commentary on page 12845.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811209115/-/DCSupplemental.
References
- 1.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 2.Verhey KJ, Kaul N, Soppina V. Kinesin assembly and movement in cells. Annu Rev Biophys. 2011;40:267–288. doi: 10.1146/annurev-biophys-042910-155310. [DOI] [PubMed] [Google Scholar]
- 3.Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 4.Cross RA, McAinsh A. Prime movers: The mechanochemistry of mitotic kinesins. Nat Rev Mol Cell Biol. 2014;15:257–271. doi: 10.1038/nrm3768. [DOI] [PubMed] [Google Scholar]
- 5.Walczak CE, Gayek S, Ohi R. Microtubule-depolymerizing kinesins. Annu Rev Cell Dev Biol. 2013;29:417–441. doi: 10.1146/annurev-cellbio-101512-122345. [DOI] [PubMed] [Google Scholar]
- 6.Vale RD, Fletterick RJ. The design plan of kinesin motors. Annu Rev Cell Dev Biol. 1997;13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- 7.Woehlke G, Schliwa M. Walking on two heads: The many talents of kinesin. Nat Rev Mol Cell Biol. 2000;1:50–58. doi: 10.1038/35036069. [DOI] [PubMed] [Google Scholar]
- 8.Endow SA. Determinants of molecular motor directionality. Nat Cell Biol. 1999;1:E163–E167. doi: 10.1038/14113. [DOI] [PubMed] [Google Scholar]
- 9.Gennerich A, Vale RD. Walking the walk: How kinesin and dynein coordinate their steps. Curr Opin Cell Biol. 2009;21:59–67. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verhey KJ, Hammond JW. Traffic control: Regulation of kinesin motors. Nat Rev Mol Cell Biol. 2009;10:765–777. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- 11.Friedman DS, Vale RD. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat Cell Biol. 1999;1:293–297. doi: 10.1038/13008. [DOI] [PubMed] [Google Scholar]
- 12.Coy DL, Hancock WO, Wagenbach M, Howard J. Kinesin’s tail domain is an inhibitory regulator of the motor domain. Nat Cell Biol. 1999;1:288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- 13.Hackney DD, Stock MF. Kinesin’s IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat Cell Biol. 2000;2:257–260. doi: 10.1038/35010525. [DOI] [PubMed] [Google Scholar]
- 14.Imanishi M, Endres NF, Gennerich A, Vale RD. Autoinhibition regulates the motility of the C. elegans intraflagellar transport motor OSM-3. J Cell Biol. 2006;174:931–937. doi: 10.1083/jcb.200605179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammond JW, Blasius TL, Soppina V, Cai D, Verhey KJ. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J Cell Biol. 2010;189:1013–1025. doi: 10.1083/jcb.201001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soppina V, et al. Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc Natl Acad Sci USA. 2014;111:5562–5567. doi: 10.1073/pnas.1400759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81:769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- 18.Tomishige M, Klopfenstein DR, Vale RD. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 2002;297:2263–2267. doi: 10.1126/science.1073386. [DOI] [PubMed] [Google Scholar]
- 19.Klopfenstein DR, Tomishige M, Stuurman N, Vale RD. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 2002;109:347–358. doi: 10.1016/s0092-8674(02)00708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JR, et al. An intramolecular interaction between the FHA domain and a coiled coil negatively regulates the kinesin motor KIF1A. EMBO J. 2004;23:1506–1515. doi: 10.1038/sj.emboj.7600164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada KH, Hanada T, Chishti AH. The effector domain of human Dlg tumor suppressor acts as a switch that relieves autoinhibition of kinesin-3 motor GAKIN/KIF13B. Biochemistry. 2007;46:10039–10045. doi: 10.1021/bi701169w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farkhondeh A, Niwa S, Takei Y, Hirokawa N. Characterizing KIF16B in neurons reveals a novel intramolecular “stalk inhibition” mechanism that regulates its capacity to potentiate the selective somatodendritic localization of early endosomes. J Neurosci. 2015;35:5067–5086. doi: 10.1523/JNEUROSCI.4240-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Bassam J, et al. Distinct conformations of the kinesin Unc104 neck regulate a monomer to dimer motor transition. J Cell Biol. 2003;163:743–753. doi: 10.1083/jcb.200308020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammond JW, et al. Mammalian kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 2009;7:e72. doi: 10.1371/journal.pbio.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horiguchi K, Hanada T, Fukui Y, Chishti AH. Transport of PIP3 by GAKIN, a kinesin-3 family protein, regulates neuronal cell polarity. J Cell Biol. 2006;174:425–436. doi: 10.1083/jcb.200604031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanai Y, Wang D, Hirokawa N. KIF13B enhances the endocytosis of LRP1 by recruiting LRP1 to caveolae. J Cell Biol. 2014;204:395–408. doi: 10.1083/jcb.201309066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cross RA. Review: Mechanochemistry of the kinesin-1 ATPase. Biopolymers. 2016;105:476–482. doi: 10.1002/bip.22862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren J, et al. Structural correlation of the neck coil with the coiled-coil (CC1)-forkhead-associated (FHA) tandem for active kinesin-3 KIF13A. J Biol Chem. 2016;291:3581–3594. doi: 10.1074/jbc.M115.689091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren J, et al. Structural delineation of the neck linker of kinesin-3 for processive movement. J Mol Biol. 2018;430:2030–2041. doi: 10.1016/j.jmb.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Cao L, et al. The structure of apo-kinesin bound to tubulin links the nucleotide cycle to movement. Nat Commun. 2014;5:5364. doi: 10.1038/ncomms6364. [DOI] [PubMed] [Google Scholar]
- 31.Citterio A, et al. Variants in KIF1A gene in dominant and sporadic forms of hereditary spastic paraparesis. J Neurol. 2015;262:2684–2690. doi: 10.1007/s00415-015-7899-9. [DOI] [PubMed] [Google Scholar]
- 32.Hotchkiss L, et al. Novel de novo mutations in KIF1A as a cause of hereditary spastic paraplegia with progressive central nervous system involvement. J Child Neurol. 2016;31:1114–1119. doi: 10.1177/0883073816639718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JR, et al. De novo mutations in the motor domain of KIF1A cause cognitive impairment, spastic paraparesis, axonal neuropathy, and cerebellar atrophy. Hum Mutat. 2015;36:69–78. doi: 10.1002/humu.22709. [DOI] [PubMed] [Google Scholar]
- 34.Hackney DD, Baek N, Snyder AC. Half-site inhibition of dimeric kinesin head domains by monomeric tail domains. Biochemistry. 2009;48:3448–3456. doi: 10.1021/bi8022575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaan HY, Hackney DD, Kozielski F. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science. 2011;333:883–885. doi: 10.1126/science.1204824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huo L, et al. The CC1-FHA tandem as a central hub for controlling the dimerization and activation of kinesin-3 KIF1A. Structure. 2012;20:1550–1561. doi: 10.1016/j.str.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q-S, et al. Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nucl Sci Tech. 2018;29:68. [Google Scholar]
- 38.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.