Significance
An early event during cellular activation in many biologic systems is recruitment of the cytosolic enzyme phospholipase-Cγ1 to the plasma membrane where, on activation, it cleaves its lipid substrates into functional second messengers. T cell antigen receptor engagement during the immune response leads to rapid formation of a multiprotein complex that brings phospholipase-Cγ1 to the plasma membrane with three adapter molecules. We reconstituted this quaternary complex in vitro and used biophysical techniques to determine stoichiometry and measure the affinities of the interacting proteins and the energetics of formation. We observe cooperative formation of a circular loop of interactions associated with a significant entropic penalty facilitating reversibility. Such instability might be a target of regulation during T cell activation.
Keywords: phospholipase C, protein complex, signal transduction, T cell antigen receptor, tyrosine phosphorylation
Abstract
The T cell antigen receptor encounters foreign antigen during the immune response. Receptor engagement leads to activation of specific protein tyrosine kinases, which then phosphorylate multiple enzymes and adapter proteins. One such enzyme, phospholipase-Cγ1, is responsible for cleavage of a plasma membrane lipid substrate, a phosphoinositide, into two second messengers, diacylglycerol, which activates several enzymes including protein kinase C, and an inositol phosphate, which induces intracellular calcium elevation. In T cells, phospholipase-Cγ1 is recruited to the plasma membrane as part of a four-protein complex containing three adapter molecules. We have used recombinant proteins and synthetic phosphopeptides to reconstitute this quaternary complex in vitro. Extending biophysical tools to study concurrent interactions of the four protein components, we demonstrated the formation and determined the composition of the quaternary complex using multisignal analytical ultracentrifugation, and we characterized the thermodynamic driving forces of assembly by isothermal calorimetry. We demonstrate that the four proteins reversibly associate in a circular arrangement of binding interfaces, each protein interacting with two others. Three interactions are of high affinity, and the fourth is of low affinity, with the assembly of the quaternary complex exhibiting significant enthalpy–entropy compensation as in an entropic switch. Formation of this protein complex enables subsequent recruitment of additional molecules needed to activate phospholipase-Cγ1. Understanding the formation of this complex is fundamental to full characterization of a central pathway in T cell activation. Such knowledge is critical to developing ways in which this pathway can be selectively inhibited.
Three decades of investigation have resulted in a description of T cell antigen receptor (TCR) engagement and the biochemical events that follow. The binding of the TCR ligand, an antigenic peptide presented by a protein encoded by the major histocompatibility complex, leads to the activation of associated and recruited protein tyrosine kinases (PTKs) (1). Critical substrates of these PTKs include the adapter molecules, linker for activation of T cells (LAT), and Src homology 2 (SH2) domain-containing leukocyte protein of 76kDa (SLP-76) (2, 3). The phosphorylation of tyrosine residues on these adapters initiates binding of many other adapters and enzymes to generate molecular complexes of great heterogeneity (4). In this manner enzymes such as phospholipase C-γ1 (PLC-γ1) and son of sevenless homolog 1 (SOS1) are recruited to the plasma membrane where they encounter their substrates. The products that are a consequence of these enzyme activations—diacylglycerol, elevated intracellular calcium, and activated Ras—have complex intracellular effects during the process of T cell activation (5, 6).
Sites of protein–protein interactions between these various signaling molecules were initially identified by coimmunoprecipitation experiments using wild-type and mutant proteins expressed in various cells (7). Detailed binding studies using expressed domains or regions of the SLP-76 adapter molecule in vitro have been the subject of our research (8–10). The SLP-76 domain containing three phosphorylatable tyrosine residues was initially shown by others to be the site of interaction with the adapter Nck, the guanine nucleotide exchange factor Vav, and the PTK, IL-2–inducible T cell kinase (ITK) (11–13). In our studies the affinities of Vav and Nck interactions with SLP-76 were measured, the presence of Nck-Vav dimers colocalizing at SLP-76 was identified, and the relationship of these binding events to actin polymerization was demonstrated with fluorescence microscopy (9). In another study we showed that the proline-rich region of SLP-76 binds with high affinity to the C-SH3 domain of the adapter protein Gads and with low affinity to the SH3 domain of PLC-γ1 (8, 14, 15). The latter interaction is allosterically enhanced in a ternary complex. Further, the interactions and the affinities of Grb2, Gads, and PLC-γ1 SH2 domains with LAT were described (8). The detailed analysis of the LAT–Grb2–SOS1 interaction revealed that phosphorylated LAT could bind multiple Grb2 molecules, each capable of simultaneously engaging SOS1, which, in turn, could bind two Grb2 molecules (16). These binding events can lead to oligomerization of signaling complexes, which were shown by others to lead to phase transitions (17) and in our experiments were required for optimal signaling in cells (16). Inhibition of oligomerization by targeted mutation of SOS1 in mice leads to functional consequences in vivo (18). Finally, the C-terminal SH2 domain of SLP-76 was also shown to enable oligomerization of LAT-based signaling complexes by its binding to multiple sites on another adapter molecule, the adhesion and degranulation-promoting protein (ADAP) (10).
The formation of the multiprotein complex comprised of LAT, Gads, SLP-76, and PLC-γ1 is required for the recruitment of PLC-γ1 to the plasma membrane where it is subsequently activated to cleave the phosphoinositide phosphatidylinositol 4, 5-bisphosphate (PIP2) into two critical signaling molecules, diacylglycerol and inositol 1,4,5-trisphosphate (IP3) (15, 19). Despite the studies that mapped the mutual interactions of the four proteins that form this structure (15, 20), much remains to be learned about the heterogeneous protein–protein interactions formed within this quaternary complex. Previously, protein interactions were explored using expressed protein domains and synthetic phospho-peptides forming only binary or ternary assemblies (8). In the current study, highly purified full-length proteins or large polypeptides were generated in a detailed study of this important multiprotein complex.
We reconstituted the quaternary complex containing LAT, Gads, SLP-76, and PLC-γ1 in vitro and determined its stoichiometry by analytical ultracentrifugation. Calorimetric analysis allowed us to measure thermodynamic driving forces of the various interacting proteins in the assembly. The data confirm that each protein in the quaternary complex interacts with two different binding partners. These simultaneous binding events likely create spatial constraints that could contribute to the specificity of this multiprotein quaternary complex. In this model the multiple protein–protein interactions generate a circular arrangement of binding interfaces. The quaternary complex is associated with a significant entropic penalty, which facilitates the reversibility of the quaternary complex. We hypothesize that this potential instability of the complex can be the target of regulation and thus enable the dynamic control of signaling in a cellular setting.
Results
Assembly of a Four-Molecule Complex.
The choice of protein constructs is critical for studying the formation of a multiprotein complex and for comparison with previous results. In our original study of mutual interactions between components of the LAT, Gads, SLP-76, and PLC-γ1 complex, we used 18- to 20-a.a. LAT peptides with a single phosphorylated tyrosine as the central residue (8). The sequence of these phosphopeptides corresponded to each of four LAT interaction sites (Y132, Y171, Y191, and Y226). The Y132 site, when phosphorylated, binds the N-SH2 domain of PLC-γ1, while the other three sites bind the SH2 domains of either Gads or Grb2 (4). These short, singly phosphorylated peptides do not allow us to model multiple LAT-binding events. To confirm simultaneous binding of Gads and PLC-γ1, we generated a 54-a.a. polypeptide derived from the LAT sequence (LAT 125–178) with phosphotyrosine residues corresponding to LAT Y132 and/or Y171 (pLAT). For some studies this polypeptide was labeled at the N terminus with DyLight 488 after peptide synthesis (pLAT488). Schematics of the protein constructs and their domain organization and the interactions in the quaternary complex are shown in SI Appendix, Fig. S1 A–E. For our initial studies we used a previously reported full-length Gads construct (GadsFL) and a His-tagged fragment of PLC-γ1 (8). This fragment contains only the two adjacent SH2 domains and one SH3 domain of PLC-γ1 (PLC-γ1tr). Both constructs were expressed in bacteria, refolded, and affinity purified.
For our first experiments we prepared full-length SLP-76 (SLP-76FL) containing two affinity tags, an N-terminal His tag and a C-terminal Flag tag, using a bacterial expression system. The double-affinity purification scheme overcame protein degradation and improved purification of the full-length protein (SI Appendix, Fig. S2A). The CD spectrum was consistent with significant unstructured fractions reported previously (SI Appendix, Fig. S2B) (8). We tested SLP-76FL for its ability to bind purified GadsFL by isothermal titration microcalorimetry (ITC). The Kd for SLP-76FL/GadsFL binding was 9 nM (SI Appendix, Fig. S2C), which is slightly lower than, although within experimental uncertainty, and comparable to the Kd value of 19 nM measured previously between GadsFL and a polypeptide containing the proline-rich region of SLP-76 (residues 158–421) (8). Additionally, we tested the capacity of purified SLP-76FL to bind a tyrosine-phosphorylated peptide derived from the sequence of HPK, a binding partner of the SLP-76 C-terminal SH2 domain (SI Appendix, Fig. S2D). The measured Kd value was 11 nM, which corresponds closely to the value of 8 nM (range, 3–17 nM) obtained for the SLP-76 SH2 domain binding to the same HPK phosphopeptide (10).
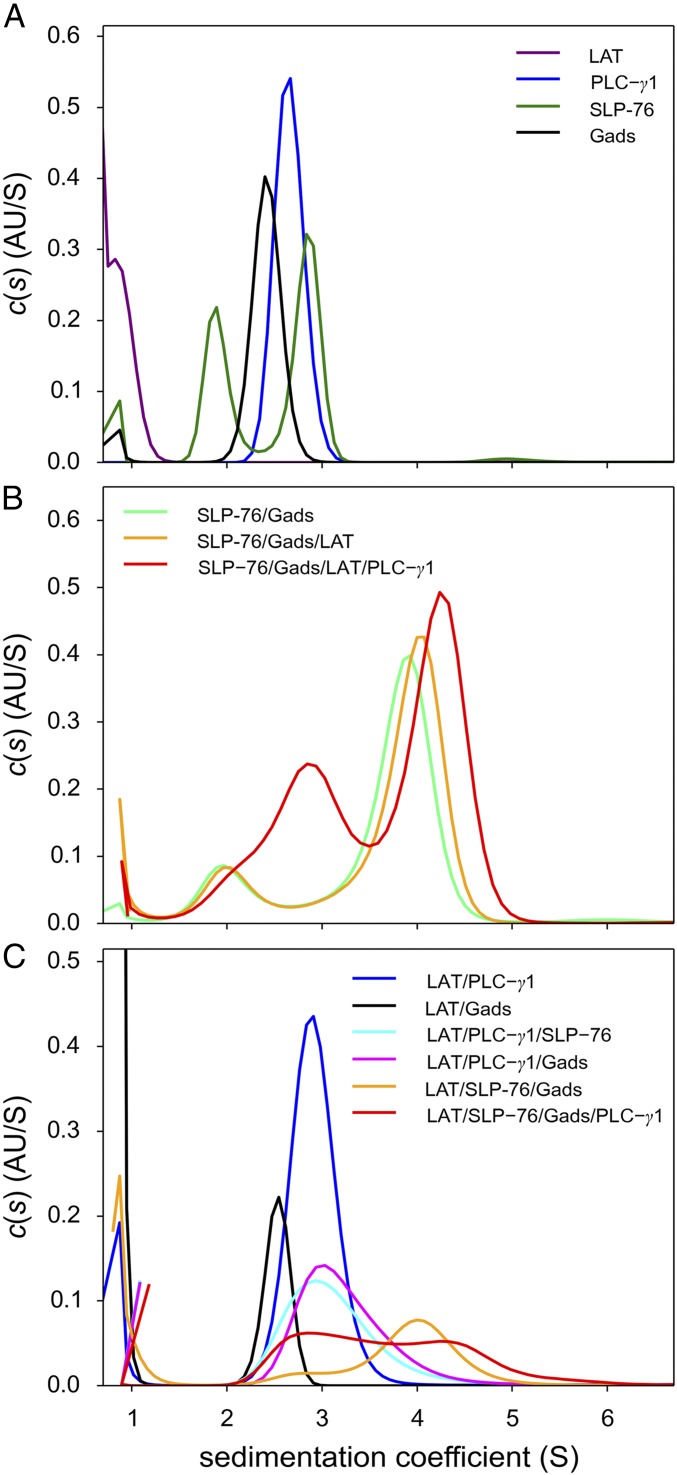
Sedimentation velocity analytical ultracentrifugation (SV-AUC) was used to study the reversible formation of complexes by SLP-76FL, GadsFL, PLC-γ1tr, and pLAT488 (Fig. 1). To enable greater yield and purity, we converted to preparations of SLP-76FL made using baculoviral expression in Sf9 cells. Sedimentation coefficient distributions [c(s)] of the individual proteins demonstrate the hydrodynamic properties and oligomeric states of the individual proteins. GadsFL (Fig. 1A, black line) and PLC-γ1tr (Fig. 1A, blue line) each sedimented in a single boundary at 2.5 S and 2.8 S, respectively, consistent with monomeric and moderately extended proteins with frictional ratios of ∼1.6. Small peaks observed at s-values <1 S are consistent with sedimentation of optically unmatched buffer components. Due to its small size and high friction, the pLAT488 peptide sedimented at ∼0.9 S (Fig. 1A, purple line), poorly resolved from smaller contaminating species likely representing free dye. SLP-76FL (Fig. 1A, dark-green line) exhibited two peaks with 2.9 S and 1.9 S, respectively. The faster-sedimenting species is consistent with the expected size of SLP-76FL and a best-fit frictional ratio of 1.8, whereas the slower species with an estimated mass of ∼30 kDa likely represents a degradation product.
Fig. 1.
Formation of the SLP-76FL/GADSFL/PLC-γ1tr pLAT488 complex by SV. The c(s) distributions were calculated from absorbance boundaries at 50,000 rpm, 20 °C, observed at 280 nm or 488 nm, respectively. All proteins were at 3.5 μM. (A) Distributions obtained for free protein and peptide components from absorbance data at 280 nm for SLP-76FL (dark-green line), GADSFL (black line), and PLC-γ1tr (blue line) and from absorbance at 488 nm for pLAT488 (purple line). (B) Distributions obtained from a binary mixture of SLP-76FL and GADSFL (light-green line), a ternary mixture of SLP-76FL, GADSFL, and pLAT488 (orange line), and a quaternary mixture of SLP-76FL, GADSFL, pLAT488, and PLC-γ1tr (red line), all observed at 280 nm. (C) From the same run, data acquired at 488 nm showing exclusively pLAT488–containing complexes in mixtures of pLAT488 with PLC-γ1tr (blue line), pLAT488 with GADSFL (black line), pLAT488 with PLC-γ1tr and SLP-76FL (cyan line), pLAT488 with PLC-γ1tr and GADSFL (magenta line), and pLAT488 with SLP-76FL and GADSFL (orange line) and in a quaternary mixture of pLAT488 with SLP-76FL, GADSFL, and PLC-γ1tr (red line).
We then used SV-AUC to study binary, ternary, and quaternary complex formation. The high affinity of the Gads–SLP-76 interaction has been noted above, and the ability to detect a complex containing the Gads with a proline-rich polypeptide derived from the SLP-76 sequence was previously reported (8, 14). Consistent with expectations, using full-length versions of both Gads and SLP-76 in equimolar mixtures at 3.5 μM (well above the Kd), we observed a faster sedimenting boundary at 3.9 S (Fig. 1B, light-green line). The estimated molecular weight and frictional ratio (1.8) are consistent with a 1:1 complex with an extended hydrodynamic shape. Similarly, SV-AUC of equimolar 3.5-μM mixtures of pLAT488 with GadsFL (Fig. 1C, black line) and of pLAT488 with PLC-γ1tr (Fig. 1C, blue line) demonstrated the formation of binary complexes between the LAT phosphopeptide and protein. In this case, due to the low molecular weight of the peptide, complex formation is revealed exclusively by detection of LAT and its complexes at 488 nm (Fig. 1C), exhibiting 2.5 S and 3 S species for pLAT488/GadsFL and pLAT488/PLC-γ1tr, respectively, both sedimenting significantly faster than free phospho-LAT.
Triple-protein complex formation is expected between pLAT, SLP-76, and Gads, as pLAT binds to the SH2 domain of Gads, and Gads in turn simultaneously binds to the proline-rich region of SLP-76. As shown in Fig. 1 B and C (orange lines), equimolar mixtures of pLAT488, GadsFL, and SLP-76FL at 3.5 μM exhibited only a slight increase in the s-value compared with the GadsFL/SLP-76FL complex (Fig. 1B, green line), again due to the relatively small molecular weight of pLAT. However, sedimentation data exclusively recording pLAT488-containing species (Fig. 1C, orange line) show pLAT488 cosedimenting in the 4 S boundary that can be identified as a SLP-76FL/GadsFL/pLAT488 ternary complex. (A small peak at ∼2.8 S reveals a binary pLAT488/GadsFL complex from a small stoichiometric excess of GadsFL.)
Finally, the addition of PLC-γ1tr resulted in a further increase of the s-value of the fastest peak to 4.2 S (red lines in Fig. 1 B and C). Considering that the PLC-γ1tr/pLAT488 complex sediments only at 3 S (Fig. 1C, blue line), this increased SV of the quaternary mixture compared with the ternary mixture lacking PLC-γ1tr demonstrates that PLC-γ1tr interacts with the SLP-76FL/GadsFL/pLAT488 ternary complex. Generally, in binary and ternary mixtures when high-affinity interactions lead to highly populated or very long-lived complexes, these will dominate the velocity of the fastest sedimentation boundary. By contrast, more weakly associating and transient complex formation produces sedimentation boundaries that reflect the coupled migration of dynamically exchanging free and bound species in the mixture, and thus the largest complex will not separate into a faster-sedimenting boundary (21). Rather, an increase in the population of complexes with concentration will lead only to a change in the observed time-average s-value of the interacting system. Accordingly, the small increase in the s-value in the quaternary mixture compared with the ternary SLP-76FL/GadsFL/pLAT488 mixture demonstrates the formation of a quadruple complex but does not reflect its s-value or size. Hydrodynamic characterization of the quaternary complex requires higher saturation.
To increase the extent of complex formation and to facilitate specific detection of its components, we introduced several changes. Attempts to add chromophoric labels to Gads, SLP-76, and PLC-γ1 by incubation with reactive dyes were unsatisfactory. The addition of dye to SLP-76 induced dimerization (SI Appendix, Fig. S3), and labeling of Gads and PLC-γ1 reduced their solubility to a degree that preparation of quaternary mixtures at sufficient concentration became impractical. To resolve these problems, we expressed these three proteins using baculoviral constructs and Sf9 insect cells. To improve the properties of the SLP-76 protein, we eliminated the N-terminal SAM domain, which contributes to dimerization (22), and removed the adjacent unstructured tyrosine-containing domain, which in vivo is required for binding of Nck, Vav, and Itk to produce SLP-76tr. We also converted S159 to cysteine to enable labeling with the maleimide conjugate of DyLight 405 and converted C524 to serine to eliminate unwanted labeling and to limit oxidation-induced dimerization (SLP-76tr-405). For Gads, we added the HaloTag to the N terminus (Halo-GadsFL), which increased solubility and allowed us to incorporate the dye JF646 for specific detection of Gads (Halo-GadsFL-646) (23, 24). The addition of the HaloTag also adds increased mass, thus improving resolution in SV-AUC experiments. Finally, we expressed nearly full-length PLC-γ1 protein (PLC-γ1FL), which lacked only the terminal 1220–1291 residues of this protein, eliminating aggregation problems, as previously described (25).
In SV-AUC, the individual proteins sedimented as single species (SI Appendix, Fig. S4A), with c(s) distributions from absorbance data at 488 nm, exhibiting a 0.9 S peak for free pLAT488; at 646 nm, exhibiting a 3.9 S peak for Halo-GadsFL-646; at 405 nm, exhibiting a 2.2 S peak for SLP-76tr-405; and a sharp peak at 6.7 S in the interference optics for PLC-γ1FL. Control experiments for the binary and ternary complexes show sedimentation patterns analogous to those discussed above, but at higher s-value as a consequence of the described changes in Gads and PLC-γ1 (SI Appendix, Fig. S4 B and C): the Halo-GadsFL-646/SLP-76tr-405 complex sediments at 4.7 S; the pLAT488/Halo-GadsFL-646 complex sediments at 4.0 S; the pLAT488/PLC-γ1FL complex sediments at 6.8 S; and for the ternary complexes we observe pLAT488/Halo-GadsFL-646/SLP-76tr-405 at ∼5.0 S and PLC-γ1FL/pLAT488/Halo-GadsFL-646 at ∼7 S. As above, in any of these LAT-containing complexes the presence of LAT was confirmed via pLAT488-specific 488-nm detection, and, similarly, the presence of Gads and SLP-76 was confirmed at their respective wavelengths. Interestingly, no binary or ternary complexes were observed that would suggest SLP-76tr-405 binding to PLC-γ1FL (SI Appendix, Fig. S4). The absence of such complexes is compatible with experiments that demonstrate that the interaction of the PLC-γ1 SH3 domain with proline residues in SLP-76 is of low affinity. This association has previously been demonstrated only at 4 °C using changes in CD and ITC and at higher protein concentrations in the presence of Gads by SV-AUC (8).
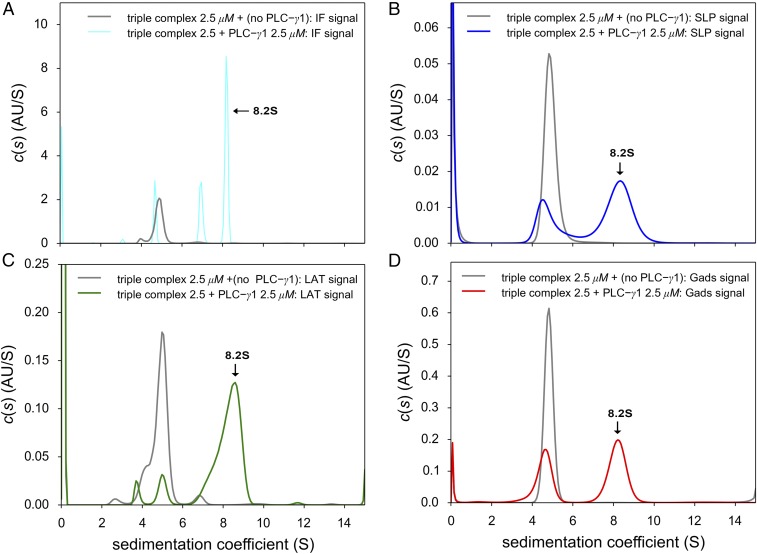
Finally, to detect a quaternary complex, pLAT488, Halo-GadsFL-646, and SLP-76tr-405 were mixed with and without PLC-γ1FL at equimolar concentrations (2.5 μM) and were subjected to SV-AUC with multisignal detection (Fig. 2). While the ternary mixture lacking PLC-γ1FL (Fig. 2, gray lines) exhibited a 5.0 S peak, corresponding to a high-affinity pLAT488/Halo-GadsFL-646/SLP-76tr-504 complex, the addition of PLC-γ1FL led to detection of a unique 8.2 S sedimentation boundary (raw data, fits, and statistical confidence bands are shown in SI Appendix, Figs. S5 and S6). Analysis of the 8.2 S peak in the c(s) distributions based on absorbance data recording the individual specific chromophoric tags of pLAT488 (Fig. 2C), Halo-GadsFL-646 (Fig. 2D), and SLP-76tr-405 (Fig. 2B), and accounting for their contributions to the interference signal (Fig. 2A) (which is dominated by PLC-γ1FL due to its greater mass), demonstrates the presence of each of the protein components in the fast peak. In addition, there was a residual 4.5–5.0 S peak detectable by absorption of each of the three dyes, indicating a coexisting SLP-76tr-405/Halo-GadsFL-646/pLAT488 complex, and interference optics additionally detected a 6.7 S peak consistent with monomeric PLC-γ1FL. This suggests that quadruple complex formation is not complete at 2.5 μM.
Fig. 2.
SV experiments for the detection of the quaternary complex pLAT488/Halo-GadsFL-646/SLP-76tr-405/PLC-γ1FL. The formation of a quaternary complex can be discerned from the appearance of a fast-sedimenting 8.2 S peak (arrows) in the c(s) distribution for the equimolar quadruple mixture compared with the high-affinity ternary SLP-76tr-405/Halo-GadsFL-646/pLAT488 complex at 5.0 S in the triple mixture not containing PLC-γ1FL (gray line in all panels). (A) Data recorded by interference optics (cyan line), which reports on all protein components, including free PLC-γ1FL at 6.8 S. (B) Absorbance data at 405 nm (blue line) tracking only SLP-76tr-405–containing species through its DyLight 405 label. (C) Absorbance data at 488 nm (green line) reporting exclusively on free pLAT488 and pLAT488-containing complexes through its DyLight 488 label. (D) Absorbance data at 646 nm (red line), reporting exclusively on Halo-GadsFL-646–containing species through its JF646 tag. All proteins were equimolar at 2.5 μM. Raw data and detailed statistical information of the c(s) fits can be found in SI Appendix, Figs. S5 and S6.
To further confirm that formation of the quadruple complex depends on cooperative binding via doubly phosphorylated LAT, we attempted complex formation with singly phosphorylated pLAT132488 or pLAT171488 peptides. PLC-γ1FL can successfully form binary complexes with pLAT132488 and pLAT488 but interacts poorly with pLAT171488 detected at ∼6.8 S (SI Appendix, Fig. S7A). Moreover, SLP-76tr-405/Halo-GadsFL-646 can successfully form ternary complexes with pLAT171488 and pLAT488 but does not form complexes well with pLAT132488 detected at ∼4.5–5 S (SI Appendix, Fig. S7B). Four-protein mixtures of 2.5 µM SLP-76tr-405/Halo-GadsFL-646/PLC-γ1FL with pLAT132488, pLAT171488, or doubly phosphorylated LAT (SI Appendix, Fig. S7C, purple, blue, and cyan lines, respectively) were subjected to SV-AUC to test for quadruple complex formation. An 8.2 S peak was again observed when doubly phosphorylated LAT was used (SI Appendix, Fig. S7C, cyan line), but no quadruple complexes were formed when either singly phosphorylated LAT peptide was included in the four-protein mixture, as no peak at ∼8.2 S was observed (SI Appendix, Fig. S7C, purple and blue lines). Thus, simultaneous binding of pY132 by PLC-γ1 and pY171 by Gads is required for quaternary complex formation.
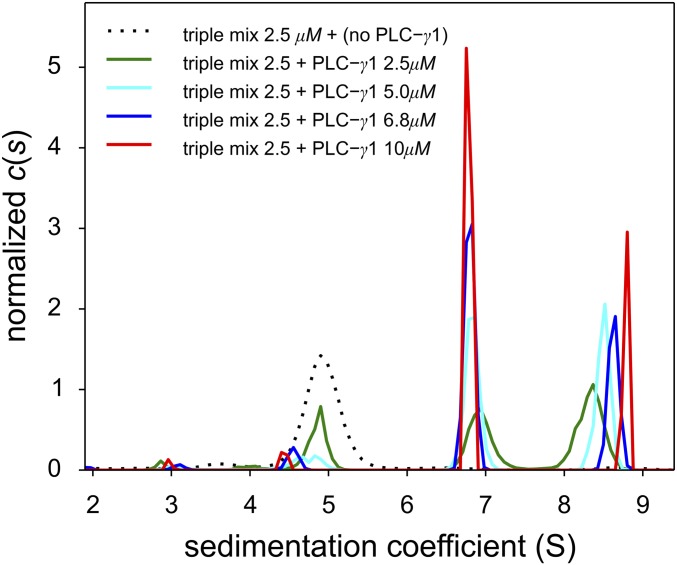
Our hypothesis is that the quaternary complex requires four protein–protein binding interactions to generate a cyclic or ring-like structure despite the weak interaction between SLP-76 and PLC-γ1. To overcome this low-affinity binding, we reasoned that increasing the relative amount of PLC-γ1FL would enhance formation of the quaternary complex. We carried out a titration series and increased PLC-γ1FL from 0 to 10 µM in the presence of 2.5 µM each of pLAT488, Halo-GadsFL-646, and SLP-76tr-405. Using interference optics, we observed a progressive increase in the s-value of the quaternary complex from 8.2 to 8.8 S (Fig. 3) accompanied by a decrease in the signal amplitude from the SLP-76tr-405/Halo-GadsFL-646/pLAT488 complex at ∼4.5–5 S and an increase in the signal of free PLC-γ1FL at ∼6.8 S. Corroborating our observations from the quaternary mixture of smaller constructs discussed above (Fig. 1), this concentration-dependent increase in the SV of the fast boundary with the increasing concentration of PLC-γ1 is consistent with a reaction boundary of a dynamic mixture rapidly interconverting on the time scale of sedimentation, which is known to be on the order of 1,000 s (21, 26, 27). From observing the dynamic exchange, we can deduce the lifetime of the quaternary complex to be shorter than the characteristic sedimentation time, i.e., less than 1,000 s. At the highest obtainable PLC-γ1FL concentration (10 μM), the integration of the c(s) peak obtained at different signals leads to a LAT:Gads:SLP-76:PLC-γ1 ratio of 1:0.92:1.11:1.05. This result is within the error of extinction coefficients and is consistent with a 1:1:1:1 complex. With regard to the shape of the complex, even though saturation was not yet achieved, an estimate of the hydrodynamic frictional ratio assuming an asymptotic s-value of 9 S, together with the molar mass of the complex would lead to a value of ∼1.6, which is similar to the average determined for the individual proteins (1.54).
Fig. 3.
Titration series of a ternary mixture of equimolar pLAT488, Halo-GadsFL-646, and SLP-76tr-405 with various concentrations of PLC-γ1FL reveals increasing saturation of the quaternary complex. Shown are the c(s) distributions from interference optical SV-AUC data with pLAT488, Halo-GadsFL-646, and SLP-76tr-405, each at 2.5 μM, without PLC-γ1FL (dotted black line), and with PLC-γ1FL at 2.5 μM (green line), 5 μM (cyan line), 6.8 μM (blue line), and 10 μM (red line).
Energetics and Cooperativity of the Four-Molecule Complex.
Having identified the oligomeric states of the four proteins and the stoichiometry of their complexes, it is possible to extract thermodynamic binding parameters from ITC experiments of binary, ternary, and quaternary mixtures and to perform an analysis of the coupled equilibria between all coexisting species in solution (SI Appendix, Supplementary Methods). The systematic characterization of the eight complexes formed, achieved by titrating either pLAT or SLP-76tr from the syringe into a cell containing single or multicomponent protein solutions, is shown in SI Appendix, Fig. S8. The enthalpy change (ΔH) and the change in entropy (ΔS) determined for binary interactions (Fig. 4 A–C and Table 1) were similar to those measured previously for truncated proteins (8), with the exception of PLC-γ1FL binding pLAT, for which we obtain a similar Kd but significantly less favorable binding enthalpy compared with the LATp171 peptide (84 nM vs. 62 nM and −5.4 kcal/mol vs. −15.7 kcal/mol, respectively) (10). In addition, in the previous work with LAT phospho-peptides, both pY132 and pY171 showed binding for the PLC-γ1 fragment, while only a 1:1 interaction was detected in the current study, highlighting the different binding modes for the full-length PLC-γ1 (8). Therefore, despite the challenges in generating functional large constructs, the full-length constructs can offer more insights into the physiological function of the molecule. Consistent with previous data (8) and with the SV-AUC results in the present work, we could not detect significant binding between SLP-76tr and PLC-γ1FL by ITC at 20 °C.
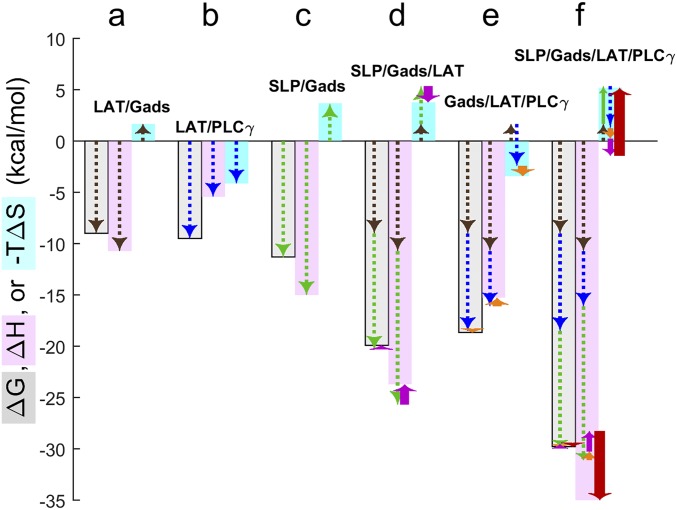
Fig. 4.
Thermodynamic driving forces of multiprotein complex formation and contributions of individual binding interfaces and cooperativity. Analysis of ITC experiments relies on a model of the equilibrium populations of free proteins and protein complexes and the associated heats produced by population changes during the titration. This analysis reveals the magnitude of favorable or unfavorable total free-energy change (ΔG, gray-shaded bars), enthalpy changes (ΔH, pink-shaded bars), and entropy changes (–TΔS, cyan-shaded bars) driving the formation of the different two-, three-, and four-molecule complexes in solution. (A–E) Specifically, results from binary titrations reveal the energetics of the pLAT/Halo-GadsFL–binding interface (A, brown dashed arrows), the pLAT/PLC-γ1FL interface (B, blue dashed arrows), and the SLP-76tr/Halo-GadsFL interface (C, green dashed arrows). These can be used to interpret the measured energetics of the three-protein complexes; observed deviations from the additivity of the constituent binary interfaces reveal small cooperativity contributions [ΔΔG, ΔΔH, and –TΔΔS] in the SLP-76tr/Halo-GadsFL/pLAT complex (D, magenta arrows) and the Halo-GadsFL/pLAT/PLC-γ1FL complex (E, orange arrows). (To illustrate the sum of contributions graphically, arrows are sequentially stacked with the end point of previous contribution forming the starting point of the next, offset sideways for clarity.) (F) Finally, these cooperativity contributions, in addition to the direct contributions all binding interfaces, can be incorporated in the dissection of the energetics of quaternary complex formation, revealing small additional ΔΔGquad and large opposing ΔΔHquad and –TΔΔSquad contributions to the formation of the SLP-76tr/Halo-GadsFL/pLAT/PLC-γ1FL complex (bold red arrows).
Table 1.
Thermodynamic binding parameters of binary interactions determined from ITC experiments at 20 °C, pH 7.4
| Peptide (syringe) | Protein (cell) | Kd, nM | ΔH, kcal/mol | −TΔS, kcal/mol | ΔG, kcal/mol |
| pLAT | Halo-GadsFL | 213 | −10.7 | 1.7 | −9.0 |
| pLAT | PLC-γ1FL | 84 | −5.4 | −4.1 | −9.5 |
| pLAT | SLP-76tr | ND | ND | ND | ND |
| SLP-76tr | Halo-GadsFL | 3.6 | −15.0 | 4.6 | −10.4 |
| SLP-76tr | PLC-γ1FL | ND | ND | ND | ND |
The binary titration of PLC-γ1FL/Halo-GadsFL was not performed due to the low concentration of protein stocks. ND, not detected.
For the higher-order complexes, the ITC experiments primarily report on the population and total ΔH of complexes, thereby revealing total ΔG, ΔH, and ΔS of assemblies as depicted in the bar graphs of Fig. 4. These, in turn, can be dissected into contributions from known constituent binary interfaces studied separately (LAT/Gads, brown arrows; LAT/PLC-γ1, blue arrows; and SLP-76/Gads, green arrows in Fig. 4 A–C) and cooperativity as recognized from the deviation from additivity of the constituent binding interfaces (purple, orange, and red arrows in Fig. 4). For example, the pLAT/Halo-GadsFL/SLP-76tr complex has a small detectable difference in total ΔH and ΔS compared with the sum of its two separate binding interfaces, resulting in low, unfavorable estimates of ΔΔH (2.0 kcal/mol) and favorable −TΔΔS estimates of −1.71 kcal/mol (purple arrows in Fig. 4D and Table 2). [This negative cooperativity eluded detection in previous work with truncated proteins (16).] Due to the high affinity of both interfaces, we can predict this complex will be highly populated in subsaturated quaternary mixtures. The Halo-GadsFL/pLAT/PLC-γ1FL ternary complex is only slightly less stable than a noncooperative complex with the same binary interfaces, with slightly unfavorable ΔΔH of 0.84 kcal/mol and slightly favorable –TΔΔS of −1.0 kcal/mol, at a total free-energy change (ΔG) of −18.6 kcal/mol (Fig. 4E). Due to the weak interaction between PLC-γ1FL and SLP-76tr, neither of the ternary complexes containing the two molecules could be studied (Table 2).
Table 2.
Thermodynamic cooperativity parameters in ternary and quaternary complexes
| Peptide (syringe) | Protein (cell) | ΔΔH, kcal/mol | –TΔΔS, kcal/mol | ΔΔG, kcal/mol |
| pLAT | Halo-GadsFL/PLC-γ1FL | 0.8 | −1.0 | −0.2 |
| pLAT | Halo-GadsFL/SLP-76tr | 2.0 | −1.7 | 0.3 |
| pLAT | Halo-GadsFL/PLC-γ1FL/SLP-76tr* | −6.8 | 6.6 | −0.1 |
Cooperativity values are assessed as deviations from additivity of constituent binding interfaces, as depicted in Fig. 4.
The ΔΔG, ΔΔH, and ΔΔS values are based on a measured total ΔGquad of −29.8 kcal/mol and ΔHquad of −35.0 kcal/mol, subtracting the ΔG or ΔH values, respectively, of the binary interfaces, and the independently measured cooperativity parameters ΔΔH and ΔΔG in the triple complexes lacking the PLC-γ1FL/SLP-76tr interface.
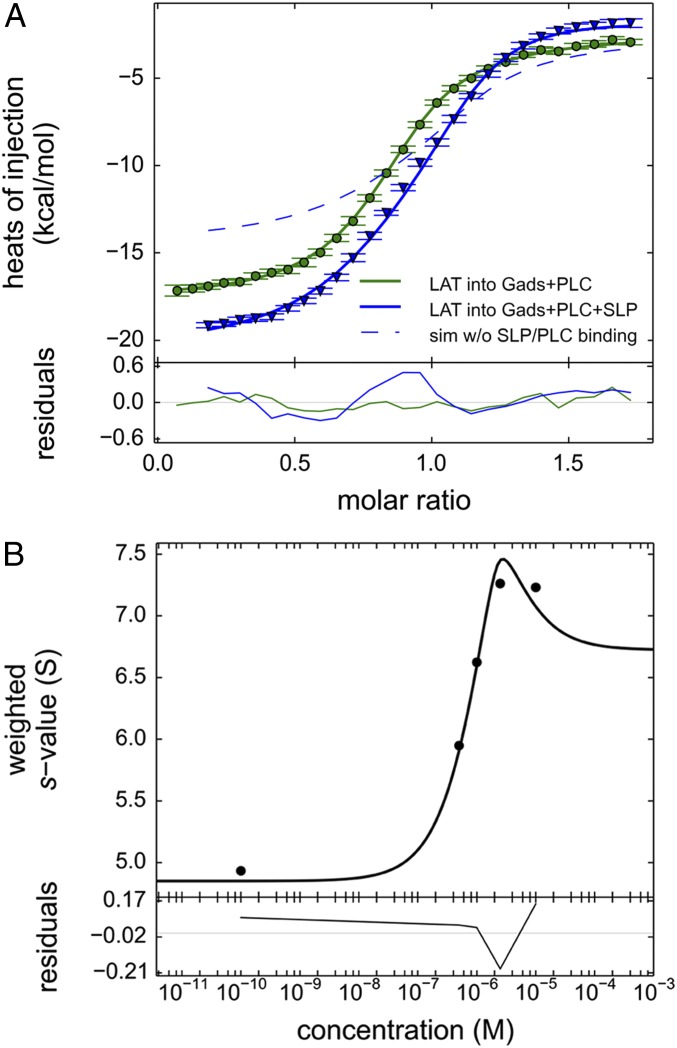
The energetics of the quaternary complex were determined from experiments titrating pLAT into a mixture of Halo-GadsFL, SLP-76tr, and PLC-γ1FL (Fig. 4F and blue triangles in Fig. 5A). For comparison, consider first a control experiment (green circles in Fig. 5A) in which pLAT was titrated into the Halo-GadsFL and PLC-γ1FL mixture at the same concentrations, monitoring all binding with the exception of SLP-76tr interactions. The raw data in this experiment record the heat from both high-affinity pLAT/Halo-GadsFL and pLAT/PLC-γ1FL binding. The data can be well modeled by including a small cooperativity between Halo-GadsFL and PLC-γ1FL on pLAT (green line in Fig. 5A). Using this as a reference point, we consider the addition of SLP-76tr to the cell before the pLAT titration to allow the quaternary complex formation. (As an experimental control, the final contents of the ITC cells after this titration were studied by SV-AUC, as shown in SI Appendix, Fig. S9, to verify the presence of the expected complexes.) In the presence of SLP-76tr, we expect the formation of high-affinity SLP-76tr/Halo-GadsFL complexes already in the cell, but this heat will not be detected since it dissipates during the equilibration of the calorimeter cell before the titration. Additionally, from ternary complex formation experiments with SLP-76tr/Halo-GadsFL/pLAT, we also know that SLP-76tr causes an unfavorable cooperative ΔH on the Halo-GadsFL/pLAT interaction with +2.0 kcal/mol (Fig. 4D and Table 2). Accounting for this negative cooperativity, and assuming there was no further interaction of SLP-76tr, we would expect to see an isotherm following the blue dashed line in Fig. 5A. However, the experimental data of the quaternary titration show significantly larger heat of binding (blue triangles in Fig. 5A), indicating more favorable binding enthalpy. Clearly, this gain in enthalpic driving force must originate from additional contacts of SLP-76tr in the quadruple complex. Quantitatively, a best-fit total ΔGquad of −29.8 kcal/mol (95% CI: −30.0 to −29.7 kcal/mol) and total ΔHquad of −35.0 kcal/mol (95% CI: −35.3 to −34.1 kcal/mol) was obtained for the quadruple complex in a model of linked equilibria (blue line in Fig. 5A), with contributions of the additional contacts of SLP-76tr amounting to a large enthalpy change, ΔΔHquad, of −6.8 kcal/mol but only little free-energy change, ΔΔGquad, of −0.1 kcal/mol (Table 2), thus revealing a large unfavorable −TΔΔSquad of +6.6 kcal/mol. These values are graphically illustrated in the bar graph of Fig. 4F, with the bold red arrows depicting the new energetic contributions arising in the four-molecule complex. This nonadditivity in the binding energetics of the quaternary complex reveals cooperativity, manifested predominantly through compensating entropic and enthalpic components.
Fig. 5.
Comparison of ITC binding isotherms titrating pLAT to form ternary and quaternary complexes. (A) pLAT at 50 μM in the syringe was injected into an equimolar 5-μM mixture of Halo-GadsFL and PLC-γ1FL (green circles), which allows the formation of a ternary complex, or into an equimolar 5-μM mixture of SLP-76tr, Halo-GadsFL, and PLC-γ1FL (blue triangles) allowing the formation of a quaternary complex. The data were fitted to a binding model with three or four coupled equilibria, respectively (solid lines), as described in detail in SI Appendix, Supplementary Methods and schematics in SI Appendix, Fig. S1. The model for the ternary mixture (green line) was constrained to the ΔH and ΔS values for the binary interfaces determined in separate experiments (Table 1), refining the estimates ΔΔH and ΔΔS for the enthalpic and entropic contributions from cooperativity in the Halo-GadsFL/pLAT/PLC-γ1FL ternary complex (Table 2). The cooperativity parameters of the ternary complex, in turn, were fixed in the model for the quaternary mixture (blue line), now additionally including equilibrium populations of the quaternary complex SLP-76tr/Halo-GadsFL/pLAT/PLC-γ1FL, from which best-fit parameters for ΔΔHquad and ΔΔSquad as well as the total heat change and entropy change of the quaternary complex ΔHquad and ΔSquad were determined. For comparison, the theoretically expected titration curve for ΔΔHquad = 0 and ΔΔSquad = 0, i.e., in the absence of any SLP-76tr interactions other than those observed in binary complexes with GADS and ternary complexes with GADS and pLAT, is shown as a dashed blue line. (B) Weighted-average c(s) of mixtures of pLAT488, Halo-GadsFL, and SLP-76tr, each at 2.5 μM, as a function of PLC-γ1FL (black circles) and theoretical model of coupled equilibria (black line) based on binding and cooperativity constants predetermined by ITC.
The binding affinities from ITC analysis should be consistent with the SV-AUC data, which offer orthogonal information of the complex formation by monitoring the size and shape change. For example, let us consider the titration series of PLC-γ1FL into equimolar mixtures of SLP-76tr-405, Halo-GadsFL-646, and pLAT488 (Fig. 3, green line). Hypothetically, if the SLP-76tr contacts causing ΔΔHquad were due to interfaces also contributing significantly to complex stability (absent of compensatory entropy changes), then at the micromolar concentrations used for the SV-AUC experiments the quaternary complex would have to form stoichiometrically. By contrast, the green line in Fig. 3 demonstrates the coexistence of free PLC-γ1FL (at 6.8 S), the high-affinity triple complex SLP-76tr-405/Halo-GadsFL-646/pLAT488 (4.5–5 S), and the quadruple complex (>8 S) at equimolar (2.5-μM) concentrations of all proteins. In fact, the changing population ratios observed in SV-AUC at low-micromolar concentrations suggests an “effective Kd”—in a simplifed picture of PLC-γ1FL binding stable SLP-76tr/Halo-GadsFL/pLAT complex—in the low- micromolar or high-nanomolar range, which would correspond to a contribution of approximately −8 to −9 kcal/mol to the total free energy of the quadruple complex. This is qualitatively consistent with the ΔGquad from ITC (recognized as the difference in the size of the gray bar in Fig. 4 D and F) and independently supports the very small ΔΔGquad. For a more quantitative analysis, the PLC-γ1FL concentration-dependence of the weighted-average s-value of quadruple mixtures is shown in Fig. 5B (circles). The resulting binding isotherm is fitted well with an interaction model of coupled equilibria between all free species and binary, ternary, and quaternary complexes using predetermined thermodynamic parameters from the ITC results (Fig. 5B, solid line). This shows the consistency between the two orthogonal methods for describing the interactions using the quaternary binding model, both leading to the same conclusions.
Neither ITC nor AUC provides microscopic insights to localize the origin of the observed ΔΔHquad in the four-molecule complex, although from the experimental design in Fig. 5 the simplest explanation, short of allostery across two proteins, would be a binding interface of SLP-76 with PLC-γ1, as has been detected previously between fragments of the two proteins. However, as mentioned above, the interaction between SLP-76tr and PLC-γ1FL could not be detected by either ITC or AUC within the practical range of protein concentrations. In ITC this could be due to low binding enthalpy or lack of free energy of binding, and no further conclusion can be drawn. However, in AUC, the lack of binding at low micromolar concentrations offers a lower limit for the ΔG of this interface of −4 kcal/mol [knowing that the Kd must be in the low micromolar range or higher, given the sensitivity of SV for interactions (21)]. If this interface is indeed contributing to ΔΔGquad in the four-molecule complex, it follows that additional unfavorable contributions must exist within this complex to explain the low measured ΔΔGquad value of −0.1 kcal/mol. In this regard, it is interesting to calculate the stability of a quadruple complex in the hypothetical scenario that the SLP-76tr/PLC-γ1FL interface contributes −4 kcal/mol to the overall free energy of binding but in the absence of compensating unfavorable interactions. The free energy of binding can be converted to the corresponding binding constants and used in the mass action law to predict species populations. For equilibrium equimolar mixtures of constituent proteins at 100 nM, these would be mostly (>80%) in the assembled quadruple complex, in contrast to <7% for the experimentally measured quadruple complex stability. Thus, in the context of the four-molecule complex, weak binding interfaces and their compensating unfavorable entropy can bring about dramatic changes in the population of assembled vs. disassembled states.
Discussion
Phospholipase-C activation drives a central signaling pathway in eukaryotic cells. Cleavage of phosphoinositides by this enzyme results in calcium elevation and diacylglycerol production, which control many intracellular processes (5, 6). The PLC-γ isoforms are activated by tyrosine kinases and contain a unique array of domains including a split PH domain, two SH2 domains, and an SH3 domain (28). These domains have regulatory functions and are central to mediating protein–protein interactions. In the case of growth factor receptor tyrosine kinases such as the EGF receptor, one SH2 domain of PLC-γ1 binds to the cytosolic tail of the EGFR after EGFR kinase activation (29). Activation of the EGFR-bound PLC-γ1 is thought to be mediated by the intrinsic EGFR kinase. Activation of nonreceptor tyrosine kinases following TCR engagement involves a different PLC-γ1 recruitment mechanism. As modeled in the current work, the transmembrane adapter molecule LAT becomes tyrosine phosphorylated on multiple sites, and both PLC-γ1 and a bound pair of cytosolic adapters, Gads and SLP-76, bind simultaneously.
The phosphorylation of LAT and these consequent binding events happen rapidly. Biochemical studies have demonstrated that these proteins are phosphorylated within 1 min of TCR engagement. Confocal microscopy using genetically tagged versions of these signaling molecules also shows the rapidity of their recruitment to observable microclusters (30). These microcluster structures are the site of tyrosine kinase activation and substrate phosphorylation (31, 32). Their formation follows TCR engagement within 1 min and is coincident with calcium elevation in the cell (31). Importantly, mutations in LAT and SLP-76 that affect protein–protein interactions impair signaling and microcluster formation (7, 33). These various biochemical and imaging studies relate directly to results in this work: The formation of the quaternary LAT/Gads/SLP-76/PLC-γ1 complex occurs rapidly and is regulated by precise protein–protein interactions.
From biophysical studies of binary, ternary, and quaternary mixtures of purified LAT, Gads, SLP-76, and PLC-γ1 in solution, we can deduce an assembly in which each protein component simultaneously binds two different neighbors in a closed loop of interactions. As previously hypothesized, this can increase the specificity of assembly through spatial constraints and thereby overcome the promiscuity of individual binding interfaces. In principle, such an architecture with each protein being held in place at two interfaces could pose a problem of excessive stability, considering that strong enhancement of binding is a hallmark of multivalent interactions. However, the detailed study of thermodynamic driving forces and cooperativity in the LAT/Gads/SLP-76/PLC-γ1 assembly reveals an entropic control mechanism whereby the significant favorable enthalpy of SLP-76 interactions within the four-molecule complex (in addition to those with its tight binding partner Gads) is nearly completely compensated by large unfavorable entropy changes. While we cannot isolate contributions from the SLP-76/PLC-γ1 interface, it will likely contribute modest additional favorable free energy, which must be compensated in the quaternary complex to explain the measured small ΔΔGquad.
The observed compensation of binding enthalpy with unfavorable entropy in the quaternary complex ties in with current ideas about the role of highly flexible and unstructured protein regions in cell biology and signaling. In the context of natively unstructured proteins, unfavorable binding entropy (resulting from binding-induced folding) was previously recognized as a mechanism to uncouple binding specificity from the free energy of binding, so as to provide reversibility to multiprotein complexes and signaling complexes (34, 35). Similarly, short complex lifetimes despite high specificity has been proposed as a general feature of intrinsically disordered signaling proteins that undergo entropically unfavorable transitions upon binding (35), which is consistent with the dynamic exchange observed in our SV experiments. Although we are presently unable to identify a particular interface or protein domain as the source of the entropic penalty, PLC-γ1 is known to exhibit conformational flexibility, as does the proline-rich region of SLP-76, and LAT is known to be unstructured (14, 36, 37). Either one or all of these polypeptide chains may be conformationally restricted in the multiprotein complex, forming an entropic spring. In this way, the quaternary architecture depicted in SI Appendix, Fig. S10 with two binary interfaces for each protein not only can enhance specificity but also can offer mechanisms for efficient and regulated assembly and disassembly. Likely, such a mechanism can be further modulated by events such as phosphorylation to cause long-range modulation of flexibility and accessibility (38) as well as allosteric interactions between binding sites, fine-tuning the energetics and oligomeric states of intermediates before full assembly. In fact, due to the membrane anchoring of LAT in vivo, it is conceivable that the interaction is modulated by PLC-γ1 simultaneously interacting with phospholipids of the cell membrane, by multivalent interactions of additional ligands, and through spatial partitioning within the signaling microclusters.
We have focused on just part of the process of PLC activation in the T cell, the recruitment of the cytosolic PLC to the plasma membrane by its binding to phosphorylated LAT and the Gads/SLP dimer. Presumably, formation of this complex is required to bring the enzyme to its substrate, PIP2, which resides in the membrane. However, we have not addressed the subsequent recruitment to SLP-76 of Nck, Vav, and Itk. In this system Nck and Vav are thought to have a scaffolding function, and Itk, a member of the Tec family of PTKs, is required to phosphorylate PLC-γ1 on tyrosine 783, an event needed for the activation of the enzyme (39, 40). In addition to the requirement for these binding events for PLC-γ1 activation, it is possible that these additional molecules might affect the interaction and dynamics of the four-molecule complex that is the subject of the current study.
Expanding the study to include additional molecules will be very challenging. In the present work we have extended the previously developed multisignal SV approach to four different protein components and extended the thermodynamic modeling in SEDPHAT to interactions of four proteins with 13 complexes of various composition. We took advantage of the fact that SV and ITC are highly synergistic: SV offers a high size resolution and can exploit spectral signatures of the molecules (21) to establish the binding model and thereby the molecular states, information that is required for interpreting the ITC titrations in terms of the binding energetics. Both can be conducted to corroborate the affinities and can be subjected to global multimethod analysis (41). We believe this combined approach will also be helpful for other dynamic multiprotein interactions, to overcome the difficulty of transient complex formation, structural flexibility, and structural polymorphism that can hamper traditional structural methods (42).
Materials and Methods
Protein and Peptide Purification.
Proteins were expressed in bacteria or insect cells, purified by Ni-affinity (on a 5-mL HisTrap HP column; GE Healthcare) and/or Flag-tag affinity chromatography, and further purified with size-exclusion chromatography (SEC) using a Superdex-75 or Superdex-200 10/300 GL column (GE Healthcare) with 1× PBS, 2.5 mM DTT, 1 mM EDTA, pH 7.4 elution buffer (supplemented with 50 mM arginine for GadsFL and PLC-γ1tr). Protein purity was assessed by SDS/PAGE with Coomassie blue, and concentration was determined spectrophotometrically.
SLP-76FL (His-SLP-76–10G-Flag) was purified either from bacteria or baculovirus expression systems. The coding sequence for full-length SLP-76 was amplified by PCR with primers containing DNA sequences for 10Gly-Flag, followed by a base frame shift to clone into the MCS-1 of the bacterial expression vector pETDuet-1 (Novagen). For bacterial expression, we used a His-SLP-76–10G-Flag/pETDuet-1 plasmid and Rosetta 2(DE3) pLys (Novagen) cells. The cells were cultured in LB medium to an OD600 of 0.7, and protein overexpression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 1.5 h at 37 °C. Harvested cells were immediately resuspended in PBS, pH 7.4, supplemented with 2.5 mM DTT, 5% glycerol, and 2× EDTA-free protease inhibitor mixture (buffer A), and were disrupted with a French press at 770 psi. Cell debris was removed by centrifugation at 30,597 × g for 30 min. The supernatant was loaded onto a 5-mL HisTrap HP column (GE Healthcare) using an ÄKTA FPLC system and was eluted in 1× PBS, 2.5 mM DTT, 5% glycerol, pH 7.5, with a 0–500 mM imidazole gradient. cOmplete EDTA-free protease inhibitor mixture (catalog no. 11873580001; Sigma) and EDTA were added to the eluted protein fractions to a final concentration of 1 mM. The protein was further purified by ANTI-FLAG M2 Affinity Agarose Gel (A2220-5ML; Sigma) according to the manufacturer’s protocol.
For expression of SLP-76FL in insect cells, the cDNA encoding SLP-76 was cloned into the pDest vector, expressed in Sf9 insect cells, and harvested (Leidos Biomedical Research, Inc.). Insect cells were disrupted in lysis buffer (20 mM Hepes, 300 mM NaCl, 2.5 mM DTT, 5% glycerol) containing cOmplete protease inhibitor (Sigma) and subjected twice to disruption in a French press at 770 psi. Cell debris was removed, and protein was purified by a His Trap HP column as above. Positive fractions were determined from the chromatogram at 280 nm and pooled. EDTA-free protease inhibitor mixture and EDTA were added to a final concentration of 1 mM.
The truncated version SLP-76tr (His-GST-TEV-SLP-76159-533AA-Gly10-Flag) was also cloned and expressed using the baculovirus expression system, as above (Leidos Biomedical Research, Inc.). Cells were disrupted as above, and the cell supernatant was loaded onto a Glutathione Sepharose-4B 10 mL column (catalog no. 17075601; GE Healthcare) preequilibrated in buffer B (20 mM Hepes, 300 mM NaCl, 2.5 mM DTT) with 1 mM EDTA added. The column was washed with five column-volumes of buffer B supplemented with EDTA-free protease inhibitor mixture, followed by four column-volumes of buffer B. The column-bound protein was cleaved on the column for 2 h at room temperature with His-tagged tobacco etch virus (TEV) protease in a final volume of 10 mL. The cleaved protein and the TEV protease eluted with the flowthrough after cleavage. The column was washed with 10 mL of buffer B and was mixed with the flowthrough collected in the previous step. To this protein solution, EDTA-free protease inhibitor mixture was added to a final 2× concentration. The cleaved protein fragment was purified by Ni-affinity chromatography using 20 mM Hepes, 300 mM NaCl, 1 mM TCEP, pH 7.3, with a 0–500 mM imidazole gradient, taking advantage of a low affinity toward the Ni column despite the lack of His tags. Immediately after elution of the protein, EDTA was added to a final concentration of 1 mM.
GadsFL was overexpressed in Rosetta 2(DE3) pLys cells transformed with GADS/pET28a plasmid, cultured in LB medium to an OD600 of 0.5, and induced by 1 mM IPTG for 3 h at 37 °C (8). The cells were then harvested by centrifugation at 3,795 × g for 15 min. The cells were suspended in 30 mL buffer A and disrupted by a French press at 770 psi. Then supernatant and pellet were separated by centrifugation at 26,892 × g for 45 min. The pellet was resuspended in PBS containing 2.5 mM DTT, 5% glycerol, 6 M Urea, pH 7.4. Ni-affinity chromatography followed using the same buffer with a 0–500 mM imidazole gradient. The purified denatured protein was refolded by dialysis in PBS, 2.5 mM DTT, and 1 mM EDTA supplemented with 50 mM arginine, pH 7.4.
Full-length Gads was amplified by PCR and cloned into the pHTN HaloTag CMV-neo vector (Promega). The cDNA for Halo-Gads was cloned into the pDest vector, expressed in Sf9 insect cells, and harvested (Leidos Biomedical Research, Inc.). Cells were lysed and disrupted with a French press, and cell debris was removed by centrifugation as above. Protein was purified by Ni-affinity chromatography using the protocol for SLP-76FL above.
PLC-γ1tr protein (amino acids 550–846) was also overexpressed and purified from Rosetta 2(DE3) pLys cells and was transformed with a PLC-γ1 (amino acids 550–846)/pET28a plasmid, following the protocol used for GadsFL (8).
PLC-γ1FL protein was expressed in insect cells as described above. Cells were lysed and disrupted in a French press, and cell debris was removed by centrifugation as above. The protein was purified with the HaloTag Protein Purification System (Promega).
The numbering of the human LAT protein corresponds to that of isoform b, National Center for Biotechnology Information, NP 001014989.2. The phospho-peptide corresponding to residues 125–178 of human LAT protein was synthesized with two phospho-tyrosines at positions 132 and 171 by solid-phase peptide synthesis using fluorenylmethyloxycarbonyl (Fmoc) chemistry. Similarly, two singly phosphorylated peptides were synthesized, one at phospho-tyrosine position 132 and the other at phospho-tyrosine position 171. All phospho-peptides were purified by a reverse-phase HPLC system using a C4 reverse-phase column (Vydac) and a water/acetonitrile gradient containing 0.1% TFA; all the purified peptides were analyzed and the sequence was confirmed by MALDI-TOF mass spectrometry.
Protein and Peptide Labeling.
Proteins were labeled after Ni-affinity chromatography before SEC. For SLP-76tr (amino acids 159–533) the N-terminal cysteine of SLP-76tr was labeled with Dylight-405 (no. 46600; Thermo Fisher Scientific) at pH 7.3 for 2 h at room temperature using maleimide chemistry keeping the protein/fluorophore molar ratio at 1:10. For Halo-Gads, Janelia Farms-646 Halo ligand fluorophore (24) was reacted at room temperature for 15 min on a rotating wheel. For both proteins, this was followed by SEC as described above, and the labeling ratio was calculated spectrophotometrically (DeNovix).
For pLAT, the N-terminal amine of HPLC-purified pLAT was labeled in the solution phase with DyLight-488 NHS ester, taking advantage of the lack of lysine residues in pLAT. After labeling, the ligated peptide pLAT488 was separated from excess fluorophore by SEC using a G25 column and was further purified by HPLC reverse-phase chromatography using a C4 reverse-phase column (Vydac) and a water/acetonitrile gradient containing 0.1% TFA solvent system. Singly phosphorylated peptides were labeled and further purified similarly. Purity and labeling efficiency were assessed by HPLC and spectrophotometry, and mass was confirmed by MALDI-TOF mass spectrometry.
ITC.
ITC measurements were performed in a VP-ITC calorimeter (Malvern). Experiments were carried out at 20 °C unless noted otherwise. The initial concentration of all proteins in the cell was kept at 5 µM, and the peptide in the syringe was kept at 50 µM, both in 1× PBS containing 2.5 mM DTT and 1 mM EDTA, and was degassed. After equilibration, thermograms were recorded for 28 8-μL injections at 180-s intervals, integrated using NITPIC, and modeled in SEDPHAT (43, 44). For the current work, SEDPHAT was extended to allow modeling of reactions with four different components (version 15.03, available for download at https://sedfitsedphat.nibib.nih.gov/software/default.aspx).
Modeling followed principles previously described (44). Briefly, the populations of all free and complex species of all components in chemical equilibrium were calculated based on mass action laws on mass conservation, by numerically solving the linked equilibria for each step in the titration. For experiments comprising all four protein components the linked equilibria involved four 1:1 complexes, four 1:1:1 complexes, and one 1:1:1:1 complex, as suggested independently by SV. The full set of coupled equilibria is described in the SI Appendix, Supplementary Methods. For experiments comprising only two or three protein components, the number of complexes modeled was reduced accordingly. Driven by changes in the solution composition in the cell over the course the titration, the changes in complex populations are associated with the release or uptake of heat, depending on the complex molar enthalpy changes (ΔHk), and the sum of these reaction heats from all complexes is fitted to the experimentally measured change in the total heat content of the solution. In this fit, molar enthalpy changes, ΔHk, along with association constants (Kk) of various complexes (k) are refined or constrained to predefined values determined previously in this study. For convenience, the possibility for subdivision of the cumulative association constants and complex enthalpy changes was embedded into the model (e.g., KABC = kAB × kBC × kcoopABC and ΔHABC = ΔHAB + ΔHBC + ΔΔHcoopABC) to allow fixing contributions of previously characterized interfaces within a complex and directly assessing cooperativity contributions (Fig. 4). Binding-incompetent fractions, likely arising from unavoidable concentration errors, were added to the model, with values of 0–10% LAT, 5–11% PLC-γ1FL, and 18–21% GadsFL. Statistical analysis of parameter CIs was based on F-statistics (45).
SV Analytical Ultracentrifugation.
SV experiments were performed in Optima XL-I/A analytical ultracentrifuges (Beckman Coulter) following standard protocols (21, 46, 47). Briefly, samples were loaded into cell assemblies with 12-mm Epon double-sector centerpieces and sapphire windows. As control, samples of individual proteins were included in all runs side-by-side with the protein mixtures (see Fig. 1A and SI Appendix, Fig. S4A for representative results). Placed in the rotor, the cell assemblies were subjected to 2–3 h of temperature equilibration at 20 °C at rest before acceleration to 50,000 rpm. Data acquisition used absorbance detection at 405 nm, 488 nm, and 646 nm and Rayleigh interferometry. The c(s) analysis was applied in SEDFIT (https://sedfitsedphat.nibib.nih.gov/default.aspx) to determine the diffusion-deconvoluted c(s) distribution of species adhering to the hydrodynamic scaling law of compact particles, using maximum entropy regularization (48). Raw scan data were fit with this model to within the noise of data acquisition (SI Appendix, Fig. S5), confirming that the c(s) analysis based on a single best-fit average frictional ratio exhausts the information content of the data, as expected due to the typically low information content of SV on polydispersity of diffusion (49). Irrespective of this approximation, this approach allows deconvolution of diffusion to arrive at high-resolution c(s) distributions (50). Statistical confidence limits for the signal weighted-average s-values of peaks of interest were calculated using Monte Carlo analysis of integrated c(s) peaks (48), which were on the order of 0.1–1%.
For quantitative analysis of binding isotherms, the c(s) distributions were integrated over the entire s range. As proved in detail elsewhere (48, 51) integrals over s × c(s) rigorously correspond to signal-weighted average sedimentation coefficients, sw, which were loaded into SEDPHAT (21) as a function of protein concentration. Similar to the modeling of ITC above, SEDPHAT was extended to allow analysis of sw isotherms of four-component interacting systems, using the same algorithm for numerical solutions to the simultaneous mass action laws and mass conservation in the four-component system, accounting for the free species, four binary 1:1 complexes, four ternary 1:1:1 complexes, and the 1:1:1:1 quadruple complex. For the analysis of the sw isotherm, the binding constants were fixed to the values derived from ITC analysis. Sedimentation coefficients of the free species and high-affinity complexes were taken from peak s-values of experimental data, while the s-value of the quaternary complex was refined within constraints from 9 S to 10 S. The s-values of complexes known to be poorly populated were fixed at hydrodynamic estimates based on the mean frictional ratio of the constituent species.
CD Spectroscopy.
The ellipticity spectrum of SLP-76 from 200–250 nm was measured at 25 °C with a J-810 CD spectrometer (JASCO Corp.) in PBS buffer and was corrected for buffer signals.
Supplementary Material
Acknowledgments
We thank Dr. Lisa Jenkins for assistance with MALDI-mass spectrometry, Drs. Nathan Coussens and Paul Randazzo for advice and discussions, and Drs. David Margulies and Sharlyn Mazur for critically reviewing the manuscript. This research was supported by the Intramural Research Programs of the NIH, the National Cancer Institute Center for Cancer Research, and the National Institute of Biomedical Imaging and Bioengineering.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.E.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817142115/-/DCSupplemental.
References
- 1.Chan AC, Desai DM, Weiss A. The role of protein tyrosine kinases and protein tyrosine phosphatases in T cell antigen receptor signal transduction. Annu Rev Immunol. 1994;12:555–592. doi: 10.1146/annurev.iy.12.040194.003011. [DOI] [PubMed] [Google Scholar]
- 2.Zhang W, Sloan-Lancaster J, Kitchen J, Trible RP, Samelson LE. LAT: The ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell. 1998;92:83–92. doi: 10.1016/s0092-8674(00)80901-0. [DOI] [PubMed] [Google Scholar]
- 3.Jordan MS, Koretzky GA. Coordination of receptor signaling in multiple hematopoietic cell lineages by the adaptor protein SLP-76. Cold Spring Harb Perspect Biol. 2010;2:a002501. doi: 10.1101/cshperspect.a002501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balagopalan L, Coussens NP, Sherman E, Samelson LE, Sommers CL. The LAT story: A tale of cooperativity, coordination, and choreography. Cold Spring Harb Perspect Biol. 2010;2:a005512. doi: 10.1101/cshperspect.a005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh-hora M. Calcium signaling in the development and function of T-lineage cells. Immunol Rev. 2009;231:210–224. doi: 10.1111/j.1600-065X.2009.00819.x. [DOI] [PubMed] [Google Scholar]
- 6.Kortum RL, Rouquette-Jazdanian AK, Samelson LE. Ras and extracellular signal-regulated kinase signaling in thymocytes and T cells. Trends Immunol. 2013;34:259–268. doi: 10.1016/j.it.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beach D, Gonen R, Bogin Y, Reischl IG, Yablonski D. Dual role of SLP-76 in mediating T cell receptor-induced activation of phospholipase C-gamma1. J Biol Chem. 2007;282:2937–2946. doi: 10.1074/jbc.M606697200. [DOI] [PubMed] [Google Scholar]
- 8.Houtman JC, et al. Binding specificity of multiprotein signaling complexes is determined by both cooperative interactions and affinity preferences. Biochemistry. 2004;43:4170–4178. doi: 10.1021/bi0357311. [DOI] [PubMed] [Google Scholar]
- 9.Barda-Saad M, et al. Cooperative interactions at the SLP-76 complex are critical for actin polymerization. EMBO J. 2010;29:2315–2328. doi: 10.1038/emboj.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coussens NP, et al. Multipoint binding of the SLP-76 SH2 domain to ADAP is critical for oligomerization of SLP-76 signaling complexes in stimulated T cells. Mol Cell Biol. 2013;33:4140–4151. doi: 10.1128/MCB.00410-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J, Motto DG, Koretzky GA, Weiss A. Vav and SLP-76 interact and functionally cooperate in IL-2 gene activation. Immunity. 1996;4:593–602. doi: 10.1016/s1074-7613(00)80485-9. [DOI] [PubMed] [Google Scholar]
- 12.Bunnell SC, et al. Biochemical interactions integrating Itk with the T cell receptor-initiated signaling cascade. J Biol Chem. 2000;275:2219–2230. doi: 10.1074/jbc.275.3.2219. [DOI] [PubMed] [Google Scholar]
- 13.Dombroski D, et al. Kinase-independent functions for Itk in TCR-induced regulation of Vav and the actin cytoskeleton. J Immunol. 2005;174:1385–1392. doi: 10.4049/jimmunol.174.3.1385. [DOI] [PubMed] [Google Scholar]
- 14.Liu Q, et al. Structural basis for specific binding of the Gads SH3 domain to an RxxK motif-containing SLP-76 peptide: A novel mode of peptide recognition. Mol Cell. 2003;11:471–481. doi: 10.1016/s1097-2765(03)00046-7. [DOI] [PubMed] [Google Scholar]
- 15.Yablonski D, Kadlecek T, Weiss A. Identification of a phospholipase C-gamma1 (PLC-gamma1) SH3 domain-binding site in SLP-76 required for T-cell receptor-mediated activation of PLC-gamma1 and NFAT. Mol Cell Biol. 2001;21:4208–4218. doi: 10.1128/MCB.21.13.4208-4218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houtman JC, et al. Oligomerization of signaling complexes by the multipoint binding of GRB2 to both LAT and SOS1. Nat Struct Mol Biol. 2006;13:798–805. doi: 10.1038/nsmb1133. [DOI] [PubMed] [Google Scholar]
- 17.Su X, et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016;352:595–599. doi: 10.1126/science.aad9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kortum RL, et al. The ability of Sos1 to oligomerize the adaptor protein LAT is separable from its guanine nucleotide exchange activity in vivo. Sci Signal. 2013;6:ra99. doi: 10.1126/scisignal.2004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, et al. Association of Grb2, Gads, and phospholipase C-gamma 1 with phosphorylated LAT tyrosine residues. Effect of LAT tyrosine mutations on T cell angigen receptor-mediated signaling. J Biol Chem. 2000;275:23355–23361. doi: 10.1074/jbc.M000404200. [DOI] [PubMed] [Google Scholar]
- 20.Berry DM, Nash P, Liu SK, Pawson T, McGlade CJ. A high-affinity Arg-X-X-Lys SH3 binding motif confers specificity for the interaction between Gads and SLP-76 in T cell signaling. Curr Biol. 2002;12:1336–1341. doi: 10.1016/s0960-9822(02)01038-2. [DOI] [PubMed] [Google Scholar]
- 21.Schuck P, Zhao H. Sedimentation Velocity Analytical Ultracentrifugation: Interacting Systems. CRC, Taylor & Francis; Boca Raton, FL: 2017. [Google Scholar]
- 22.Liu H, et al. SLP-76 sterile α motif (SAM) and individual H5 α helix mediate oligomer formation for microclusters and T-cell activation. J Biol Chem. 2013;288:29539–29549. doi: 10.1074/jbc.M112.424846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.N Peterson S, Kwon K. The HaloTag: Improving soluble expression and applications in protein functional analysis. Curr Chem Genomics. 2012;6:8–17. doi: 10.2174/1875397301206010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimm JB, Brown TA, English BP, Lionnet T, Lavis LD. Synthesis of Janelia fluor HaloTag and SNAP-tag ligands and their use in cellular imaging experiments. Methods Mol Biol. 2017;1663:179–188. doi: 10.1007/978-1-4939-7265-4_15. [DOI] [PubMed] [Google Scholar]
- 25.Gresset A, Hicks SN, Harden TK, Sondek J. Mechanism of phosphorylation-induced activation of phospholipase C-gamma isozymes. J Biol Chem. 2010;285:35836–35847. doi: 10.1074/jbc.M110.166512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert GA, Jenkins RC. Boundary problems in the sedimentation and electrophoresis of complex systems in rapid reversible equilibrium. Nature. 1956;177:853–854. doi: 10.1038/177853a0. [DOI] [PubMed] [Google Scholar]
- 27.Schuck P. Diffusion of the reaction boundary of rapidly interacting macromolecules in sedimentation velocity. Biophys J. 2010;98:2741–2751. doi: 10.1016/j.bpj.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hajicek N, Charpentier TH, Rush JR, Harden TK, Sondek J. Autoinhibition and phosphorylation-induced activation of phospholipase C-γ isozymes. Biochemistry. 2013;52:4810–4819. doi: 10.1021/bi400433b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 30.Balagopalan L, Kortum RL, Coussens NP, Barr VA, Samelson LE. The linker for activation of T cells (LAT) signaling hub: From signaling complexes to microclusters. J Biol Chem. 2015;290:26422–26429. doi: 10.1074/jbc.R115.665869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bunnell SC, et al. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J Cell Biol. 2002;158:1263–1275. doi: 10.1083/jcb.200203043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito T, Yokosuka T. Immunological synapse and microclusters: The site for recognition and activation of T cells. Curr Opin Immunol. 2006;18:305–313. doi: 10.1016/j.coi.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 33.Bunnell SC, et al. Persistence of cooperatively stabilized signaling clusters drives T-cell activation. Mol Cell Biol. 2006;26:7155–7166. doi: 10.1128/MCB.00507-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flock T, Weatheritt RJ, Latysheva NS, Babu MM. Controlling entropy to tune the functions of intrinsically disordered regions. Curr Opin Struct Biol. 2014;26:62–72. doi: 10.1016/j.sbi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 35.Zhou HX. Intrinsic disorder: Signaling via highly specific but short-lived association. Trends Biochem Sci. 2012;37:43–48. doi: 10.1016/j.tibs.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harkiolaki M, et al. Structural basis for SH3 domain-mediated high-affinity binding between Mona/Gads and SLP-76. EMBO J. 2003;22:2571–2582. doi: 10.1093/emboj/cdg258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nag A, Monine MI, Faeder JR, Goldstein B. Aggregation of membrane proteins by cytosolic cross-linkers: Theory and simulation of the LAT-Grb2-SOS1 system. Biophys J. 2009;96:2604–2623. doi: 10.1016/j.bpj.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang M, Yang T, Paretsky JD, Berry JF, Schomaker JM. Inverting steric effects: Using “attractive” noncovalent interactions to direct silver-catalyzed nitrene transfer. J Am Chem Soc. 2017;139:17376–17386. doi: 10.1021/jacs.7b07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bogin Y, Ainey C, Beach D, Yablonski D. SLP-76 mediates and maintains activation of the Tec family kinase ITK via the T cell antigen receptor-induced association between SLP-76 and ITK. Proc Natl Acad Sci USA. 2007;104:6638–6643. doi: 10.1073/pnas.0609771104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Devkota S, Joseph RE, Min L, Bruce Fulton D, Andreotti AH. Scaffold protein SLP-76 primes PLCγ1 for activation by ITK-mediated phosphorylation. J Mol Biol. 2015;427:2734–2747. doi: 10.1016/j.jmb.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao H, Schuck P. Global multi-method analysis of affinities and cooperativity in complex systems of macromolecular interactions. Anal Chem. 2012;84:9513–9519. doi: 10.1021/ac302357w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu H, Fuxreiter M. The structure and dynamics of higher-order assemblies: Amyloids, signalosomes, and granules. Cell. 2016;165:1055–1066. doi: 10.1016/j.cell.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brautigam CA, Zhao H, Vargas C, Keller S, Schuck P. Integration and global analysis of isothermal titration calorimetry data for studying macromolecular interactions. Nat Protoc. 2016;11:882–894. doi: 10.1038/nprot.2016.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houtman JC, et al. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: Application to adaptor protein complexes in cell signaling. Protein Sci. 2007;16:30–42. doi: 10.1110/ps.062558507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson ML. Why, when, and how biochemists should use least squares. Anal Biochem. 1992;206:215–225. doi: 10.1016/0003-2697(92)90356-c. [DOI] [PubMed] [Google Scholar]
- 46.Schuck P. Basic Principles of Analytical Ultracentrifugation. CRC, Taylor & Francis; Boca Raton, FL: 2015. [Google Scholar]
- 47.Chaturvedi SK, Ma J, Zhao H, Schuck P. Use of fluorescence-detected sedimentation velocity to study high-affinity protein interactions. Nat Protoc. 2017;12:1777–1791. doi: 10.1038/nprot.2017.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuck P. Sedimentation Velocity Analytical Ultracentrifugation: Discrete Species and Size-Distributions of Macromolecules and Particles. CRC, Taylor & Francis; Boca Raton, FL: 2016. [Google Scholar]
- 49.Schuck P. On computational approaches for size-and-shape distributions from sedimentation velocity analytical ultracentrifugation. Eur Biophys J. 2010;39:1261–1275. doi: 10.1007/s00249-009-0545-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.