Significance
About 38 million people are infected with human immunodeficiency virus (HIV) worldwide. While antiretroviral therapy suppresses HIV in patients, it does not eradicate the virus, and the regimen requires lifelong adherence. The main obstacle for a cure is the persistent reservoir of latent provirus in resting T cells. Different approaches, including viral reactivation and induced deep latency, depend on targeting proviral transcription. We determined the crystal structure of the best defined regulatory component of proviral transcription, the super elongation complex (SEC), in complex with viral Tat protein and an oligonucleotide model for the viral transactivating response region (TAR) RNA. This structure provides mechanistic insights into transcriptional regulation of the HIV provirus and detailed information on an important target for finding an HIV cure.
Keywords: HIV-1 TAR, Tat, super elongation complex, crystal structure, transcriptional regulation
Abstract
Promoter-proximal pausing by RNA polymerase II (Pol II) is a key regulatory step in human immunodeficiency virus-1 (HIV-1) transcription and thus in the reversal of HIV latency. By binding to the nascent transactivating response region (TAR) RNA, HIV-1 Tat recruits the human super elongation complex (SEC) to the promoter and releases paused Pol II. Structural studies of TAR interactions have been largely focused on interactions between the TAR bulge and the arginine-rich motif (ARM) of Tat. Here, the crystal structure of the TAR loop in complex with Tat and the SEC core was determined at a 3.5-Å resolution. The bound TAR loop is stabilized by cross-loop hydrogen bonds. It makes structure-specific contacts with the side chains of the Cyclin T1 Tat-TAR recognition motif (TRM) and the zinc-coordinating loop of Tat. The TAR loop phosphate backbone forms electrostatic and VDW interactions with positively charged side chains of the CycT1 TRM. Mutational analysis showed that these interactions contribute importantly to binding affinity. The Tat ARM was present in the crystallized construct; however, it was not visualized in the electron density, and the TAR bulge was not formed in the RNA construct used in crystallization. Binding assays showed that TAR bulge-Tat ARM interactions contribute less to TAR binding affinity than TAR loop interactions with the CycT1 TRM and Tat core. Thus, the TAR loop evolved to make high-affinity interactions with the TRM while Tat has three roles: scaffolding and stabilizing the TRM, making specific interactions through its zinc-coordinating loop, and making electrostatic interactions through its ARM.
Despite the remarkable progress in developing novel antiretroviral therapies (ARTs) against the human immunodeficiency virus-1 (HIV-1), HIV-1 remains an incurable infection due to the virus’s ability to persist in a latent state mainly in resting memory CD4+ T lymphocytes. Upon interruption of ART, the reactivated latent virus can reinitiate rounds of viral infection.
Different approaches to a cure for HIV-1 infection include elimination of all viral reservoirs following reactivation of the latent provirus (referred to as shock and kill), immune control without reservoir eradication, or a combination of both (1, 2). An alternative approach to a functional cure aims to lock out proviruses in a deep latency that prevents viral reactivation by inhibiting viral transcription (3–5), thereby suppressing the residual viremia arising from reactivation of latently infected cells. In this so-called lock-and-block approach, transcriptional inhibitor treatment combined with ART could potentially reduce the size of the viral reservoir by blocking ongoing viral replication, as well as reactivation events, that replenish the viral latent reservoir under current ART.
Understanding and manipulation of transcriptional regulation of the HIV-1 viral genome is central to both approaches. Biochemical and structural studies have in recent years revealed a clearer picture of many of the critical factors and interactions regulating Tat-dependent transcription of the HIV-1 genome (6, 7). HIV transcription is regulated at the levels of chromatin organization, transcription initiation, polymerase recruitment, and transcription elongation, with Tat playing a central role in the latter. During viral transcription, RNA polymerase II (Pol II) is recruited to the HIV promoter and initiates transcription, which stalls after a short 50–60-nucleotide transcript containing the Tar region has been formed. The HIV Tat protein bound to a host super elongation complex (SEC) binds to TAR and releases the paused polymerase (8, 9). The host SEC consists of positive elongation factor b (P-TEFb), composed of CDK9 and Cyclin T1 (CycT1), the transcriptional elongation factors ELL2 and ENL/AF9, and the ∼1,200-amino acid scaffold proteins AFF1 and/or AFF4. Tat recruits the SEC to Pol II by simultaneously binding to CycT1, AFF1/4, and the nascent TAR RNA (10–12). A recent 5.9-Å resolution model of TAR bound to Tat:AFF4:P-TEFb provided a first glimpse of the overall binding mode between TAR and the SEC (10) and showed that the TAR loop contacted the CycT1 Tat-TAR recognition motif (TRM) and the Zn2+ coordinating loop of Tat. The analysis of this structure was limited by the available resolution. Thus, the nature of interactions with individual nucleobases and amino acids was not defined and the origins of TAR recognition were unclear.
We have now determined the crystal structure of TAR with Tat:AFF4:P-TEFb to 3.5-Å resolution. At this resolution, the interaction surfaces between the TAR loop and Tat/CycT1 are delineated, with CycT1 contributing roughly two-thirds and Tat contributing one-third of the protein interaction surface. The CycT1 TRM is flexible in the absence of TAR (11), and its structure could not be defined at the resolution of the previous TAR complex. Here, we have been able to visualize the ordered TRM and find that it contributes the majority of direct interactions with TAR nucleobases. Based on this improved structure and the additional details it provides, it is now possible to account for recognition of the TAR loop by the TRM and the core Zn2+-coordinating loop of Tat.
Results
Crystal Engineering.
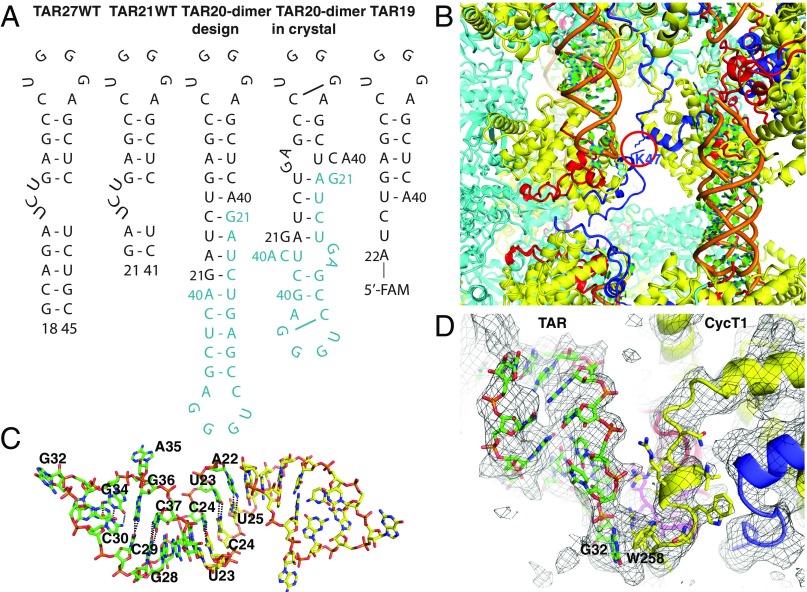
The major obstacle to understanding sequence-specific recognition of TAR by the Tat-SEC complex has been a long-standing lack of well-ordered, well-diffracting crystals. We carefully analyzed the packing of a previous crystal form that diffracted anisotropically to 5.9 Å in the best direction (10). This crystal form contained a 21-nucleotide RNA (TAR21 WT; Fig. 1A). This RNA oligonucleotide had been designed to have blunt ends (Fig. 1A); however, in the crystal a dimer was formed through base pairing between apparent overhanging ends. The RNA rearrangement also appeared to eliminate the TAR bulge, although it was not possible to be certain given the limited order in this region and the limited resolution. We sought to generate a more stable RNA dimer to promote more stable and ordered crystallization. This led to the TAR20 construct (Fig. 1A), which was designed to have a normal loop, no bulge, and seven interstrand Watson-Crick base pairs. We also noted potential charge repulsion between twofold symmetry-related AFF4 Lys47-residues at a lattice contact (Fig. 1B) and therefore engineered the mutation AFF4 K47Y.
Fig. 1.
Crystal optimization. (A) Synthetic TAR oligonucleotides used for cocrystallization with Tat:AFF4:P-TEFb, EMSAs, and FP experiments. Symmetry-related molecules for TAR20 crystallization design and experimental result are shown in blue. (B) Crystal packing of the initial low-resolution crystal of the TAR-SEC complex. TAR (orange and green) forms extensive contacts in the direction of the threefold symmetry axis (vertical direction). CDK9 (cyan), CycT1 (yellow), and Tat (red) provide additional contacts. AFF4 (blue) makes unfavorable contacts between symmetry-related K47, indicated by the red circle. (C) Symmetry-related TAR molecules form a dimer with a continuous helical structure in the crystal environment. (D) 2Fo-Fc density (1.1σ) of refined TAR20 complex with Tat:AFF4:P-TEFb shows clear density for the TAR phosphate backbone and many of the nucleobases, as well as for the protein model.
Cocrystallization with the optimized TAR20 and AFF432–67-Y47 yielded crystals that diffracted to 3.5-Å resolution with a much improved anisotropic ΔB = 21 Å2 compared with ΔB = 110 Å2 for the previous analysis. The protein-RNA complex refined to an R/Rfree 0.248/0.281 at 3.5-Å resolution with clearly resolved electron density for the TAR loop and CycT1 TRM region (Fig. 1D and SI Appendix, Table S1). As expected, two TAR20 molecules form a dimer in the crystal structure, although the details of the interactions are different from the initial design (Fig. 1A). TAR nucleotides A22U23C24U25 interact with A22U23C24U25 of the twofold symmetry-related molecule through edge-on hydrogen bonds in two canonical Watson-Crick (A-U) and two noncanonical (C-U) base pairs (Fig. 1 A and C) (13). Thus, five sets of base pairs were formed, instead of the expected seven, with G21, G26A27, and A40 bulged out. This crystallographic TAR dimerization mode results in a more compact RNA dimer (Fig. 1A), thus stabilizing crystal formation in the context of the SEC complex. We emphasize that structural conclusions can be drawn only with respect to the TAR loop from this structure, given the noncanonical nature of the RNA stem and the absence of the U23C24U25 bulge.
Structure of TAR20 and Its Interactions with the SEC.
The crystal structure of TAR20 in complex with Tat:AFF4:P-TEFb confirms the overall location and orientation of TAR relative to CycT and Tat (Figs. 1D and 2 A and B). However, at 3.5-Å resolution, this new structure reveals interactions between specific TAR loop nucleotides and CycT1 TRM and Tat amino acid residues (Fig. 2 C and D). Although true atomic details of these interactions are still missing, we can now infer which residues and bases are hydrogen bond donors and acceptors at reasonable distances.
Fig. 2.
HIV-1 TAR interactions with Tat:AFF4:P-TEFb. (A) Ribbon diagram showing an overview of the complex. (B) Surface representation of TAR (Left) and CycT1 (Right) with Coulombic surface coloring from −10 kcal/(mol) (red) to 10 kcal/(mol) (blue) using a dielectric constant of 4.0 in Chimera (47). (C and D) Cartoon and ball-and-stick diagram of the TAR contact site (orange and green) with Tat (red) and CycT1 (yellow). AFF4 is shown in dark blue.
The TAR loop conformation is stabilized by Watson-Crick hydrogen bonds between nucleobases C30 and G34, with the G33 base also pointing into the loop. The G33 N2 atom is in hydrogen-bonding distance to C30 O2 and N3 atoms (Figs. 1C and 2D). This base cluster appears to stabilize the loop structure, while nucleotides U31, G32, and A35 point away from the TAR loop center (Fig. 2C). Despite pointing outward, nucleobase G32 is well-defined in the electron density map due to a G32 base interaction with the CycT TRM residue W258 (Fig. 1D). In contrast, missing density for the A35 purine base and weak density for U31 are consistent with their orientation both outside the TAR loop and away from the CycT1 or Tat interaction surface. This TAR loop structure leads to an extended flat molecular surface of 355 Å2 (14), which faces a similarly flat molecular surface composed of CycT1 TRM (233 Å2) and the Tat cysteine-rich domain (121 Å2) (Fig. 2B). The buried surface area for the loop contacts (700 Å2) combined with the buried surface from the bulge contacts lies within the range of observed values for other RNA-protein complexes (15).
CycT1 TRM, the major component of the protein interaction surface, adopts a helical conformation for residues 252–256 surrounded by more extended peptide conformations for residues 250–251 and 257–261. The TRM is anchored by coordination between CycT1 C261 and a Tat Zn2+ ion, as also seen in the absence of TAR. The difference is that residues 251–260 are much less well-defined in the absence of TAR. This conformation of the TRM positions the Arg251, Arg254, and Arg259 side chains in the vicinity of TAR backbone phosphates, where they can form electrostatic interactions with the negatively charged phosphate backbone (Fig. 2 C and D). The orientation of CycT1 Arg251 could potentially also allow contacts with the TAR base G33. In addition to arginine electrostatic interactions, the CycT1 Trp258 and Arg259 side chains contribute multiple van der Waals contacts with the flipped-out G32 TAR base to strengthen the TAR complex.
The CycT1 TRM conformation is stabilized by its interactions with TAR and two subunits of the SEC complex, AFF4 and Tat. CycT1 residues Leu255 and Trp256 contact AFF4 Leu56 and Gly57, and the whole TRM folds on top of the Cys-rich domain of Tat. Thus, the Tat core contributes in two ways to TAR binding: indirectly, by providing a scaffold for CycT1 TRM folding, and directly, through hydrogen bonds between the Tat Zn2+-binding loop, Asn24 to Tyr26, and TAR G33 (Fig. 2 C and D). AFF4 itself has no direct contacts with TAR (10, 11).
Effect of CycT1 Mutations on TAR Binding Affinity.
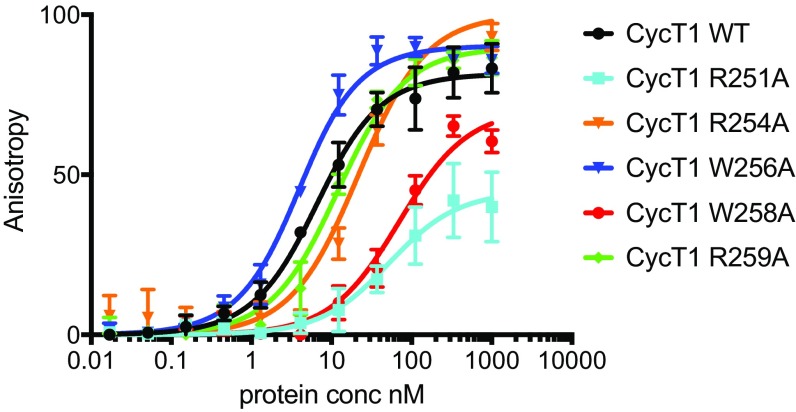
To measure the contribution of CycT1 TRM residues to TAR loop binding affinity and to validate the TAR complex structure, we determined dissociation constants (Kd) for Tat1–57:AFF42–73:P-TEFb binding to a synthetic fluorescein-labeled TAR loop oligo (TAR19; Fig. 1A) in fluorescence polarization (FP) experiments (Fig. 3 and SI Appendix, Figs. S1 and S2). We determined dissociation constants for the protein complex with WT CycT1 (Kd = 5.5 nM, SD = 0.66) and with five TRM mutants: CycT1 R251A (51 nM, SD = 13.35), R254A (20.7 nM, SD = 4.5), W256A (3 nM, SD = 0.31), W258A (69 nM, SD = 9.88), and R259A (11 nM, SD = 1.09). The results confirm TRM Arg251 and Trp258, with 10–12-fold increases in Kd when mutated, as key residues for TAR loop binding. This is consistent with the structural observation that their side chains interact with the phosphate backbone and the TAR loop base G32 (Fig. 2 C and D). In contrast, the CycT1 W256A mutant has a slightly higher affinity than the WT, and CycT1 R254A and R259A have a four- and twofold reduced affinity, respectively, compared with the WT. In the crystal structure, Trp256 points away from the CycT1-TAR interface and the Arg259 side chain, although in contact distance to TAR nucleotide G32, is not well-defined in the electron density. Cys261 in the CycT1 TRM region was previously shown to be essential for HIV-1 Tat binding to human CycT1 and hence indirectly essential for TAR binding (16). Collectively, these results amount to a highly consistent dataset accounting for the central role of the TRM in TAR loop recognition.
Fig. 3.
Effect of CycT1 TRM mutations on binding affinity for the HIV-1 TAR loop. Binding of Tat:AFF4:P-TEFb with WT CycT1 or with mutant CycT1 to 2 nM of fluorescently labeled TAR19 is monitored by the change in relative fluorescence anisotropy. Error bars reflect the SD from three experimental replicates. Control experiments are shown in SI Appendix, Fig. S1.
Effect of TAR Mutations on Tat:AFF4:P-TEFb Binding.
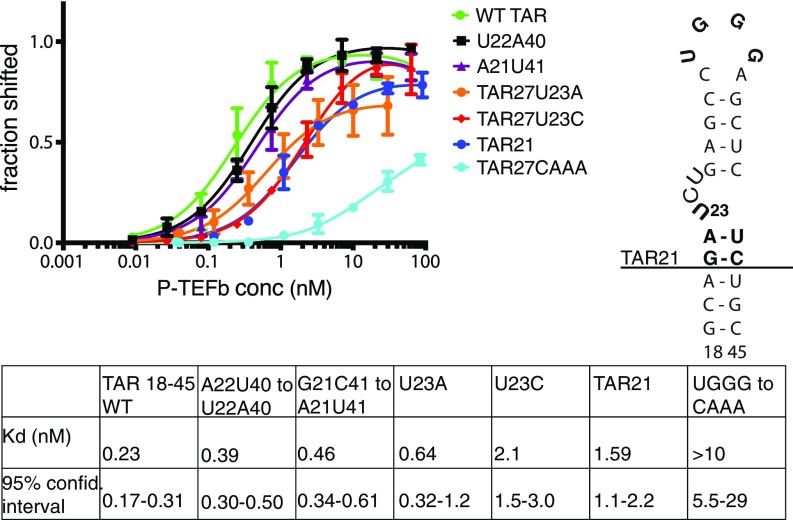
Early studies of TAR binding to isolated Tat peptides identified TAR bases important for Tat binding to the TAR bulge (17, 18). Later experiments focused on the study of TAR loop interactions in the ternary CycT1-Tat-TAR complex (19, 20) and suggested critical nucleotides for TAR loop stability and for interactions with CycT1 and Tat. However, productive transcription of the integrated HIV-1 genome requires recruitment of the complete Tat:AFF4:P-TEFb complex to TAR, and the presence of AFF1/4 has a profound effect on TRM folding and function (11, 21). We analyzed the binding affinity of this quaternary complex to WT TAR27 and to TAR mutants in the stem, bulge, and loop regions of TAR, using electrophoretic mobility shift assay (EMSA) (Fig. 4 and SI Appendix, Fig. S3). The TAR mutants were chosen with reference to earlier studies to quantify the relative contribution of the TAR stem, bulge, and loop to Tat:AFF4:P-TEFb affinity. The results from EMSAs show that mutations in the lower TAR stem region (U22A40, A21U41) adjacent to the bulge lead to just 1.7- and 2-fold reductions in Tat:AFF4:P-TEFb binding. A mutation of the TAR bulge nucleotide U23 to A or C results in threefold and ninefold reductions in binding affinity, and a destabilized bulge structure with WT sequence in TAR21 shows a sevenfold reduced affinity (Fig. 4). However, replacement of TAR loop nucleotides 31–34 UGGG with CAAA drastically reduces binding affinity to Tat:AFF4:P-TEFb, by more than 50-fold, indicating that TAR loop interactions are providing the majority of high-affinity interactions. TAR bulge interactions contribute to a substantial but lesser degree of affinity, and the contribution of stem nucleobases is negligible. These observations are completely consistent with the structural finding that interactions with the ordered core of the SEC occur primarily through the TAR loop.
Fig. 4.
HIV-1 TAR binding to Tat:AFF4:P-TEFb. Binding of WT TAR and TAR mutants to WT Tat:AFF4:P-TEFb was measured in EMSAs with 100 pM 32P-labeled TAR27. Error bars reflect the SD from three experimental replicates. TAR mutations are indicated in bold letters in the schematic depiction of TAR (right).
Discussion
HIV-1 Tat functions as key activator of viral transcription by recruiting the host cell SEC to the nascent TAR RNA (8, 9). Seminal studies showed that Tat binds through its transactivation domain to CycT1 (22–24) and to the AFF4 subunit of the SEC (11, 21), thereby inducing a conformational state that allows the CycT1 subunit of SEC to bind to the TAR loop (24). While both the TAR bulge and the TAR loop are essential for Tat-mediated transactivation (25–27), structural information on TAR loop interactions with Tat and the SEC has been lacking. Our recent integrative structure of the TAR loop in complex with Tat:SEC provided a first low-resolution outline of the contact regions (10). However, a detailed structural description of how the TAR loop is recognized has been elusive.
The crystal structure we present in this study describes nucleobase-level details of the bound TAR loop structure and its interactions with Tat:AFF4:P-TEFb. The bound TAR loop is stabilized by cross-loop hydrogen bonds between C30 and G34 (Figs. 1C and 2D) and additional contacts with G33, which also points toward the loop center. The remaining loop residues G32, A35, and U31 point outward from the loop. This stable loop structure places two nucleobases, G32 and G33, in a position to make contacts with CycT1 Trp258 and Tat Tyr26. The TAR loop conformation confirms indirect evidence for a cross-loop hydrogen bond from molecular dynamics (MD) studies and CycT1 binding studies (19, 20, 28), where mutations in C30 or G34 showed large reductions in CycT1 binding but could be rescued by also mutating the hydrogen-bonding nucleobase to restore hydrogen-bonding capabilities. The structure is also in agreement with combined NMR/MD studies of the free TAR stem-loop that showed high flexibility of the loop region with transient formation of a C30-G34 Watson-Crick base pair and increased stability for G33 (29). These observations support that the TAR loop conformation in this TAR20-SEC complex faithfully represents the functional conformation in HIV-1 transactivation.
In previous structures of P-TEFb, either the CycT1 TRM was poorly ordered, despite its tethering through Cys261 to the tricoordinate Tat-bound Zn2+ ion (11, 30), or it was stabilized by crystal contacts with symmetry-related Tat molecules (12). Even in our previous 5.9-Å structure of the Tat-SEC complex with TAR, the conformation could not be established with confidence and was modeled as an ensemble of multiple possible states (10). The present structure provides a striking clarification of this situation. CycT1 TRM adopts an alpha helical structure in the TAR complex, similar to the TRM conformation in the apo-SEC, stabilized by crystal contacts (12). The stable structure of the TRM in the TAR complex explains our previous observation that this was one of two regions of the protein complex with the largest decreases in hydrogen-deuterium exchange upon TAR binding (10).
Folded RNAs are typically read out at the structural level rather than through a series of individual nucleobase recognition sites (15, 31, 32). The TAR loop-TRM interaction fits this pattern. The TAR loop contacts a mostly flat, positively charged protein interaction surface composed of Tat and the CycT1 residues (Fig. 2B). Only two TAR bases, G32 and G33, contact the protein complex directly. However, extensive contacts between the TAR sugar phosphate backbone and residues in the CycT1 TRM, especially Arg251, Arg254, and Arg259 (Fig. 2 C and D), are observed. Thus, SEC recognition of TAR is predominantly based on readout of the structure as opposed to the sequence.
The TAR loop complex structure with Tat:AFF4:P-TEFb described in this study provides the missing part of TAR interactions with Tat-SEC, but is lacking information on Tat ARM interactions with TAR, which have been reported in previous NMR structures of a homologous TAR stem loop with linear Tat peptides (33) and cyclic peptide Tat mimetics (34, 35). These puzzle pieces can now be conceptually combined into one structure by superimposing the structures on the common TAR molecules (Fig. 5).
Fig. 5.
Combined structure of the TAR-peptide complex and the TAR loop-SEC complex. The TAR-peptide complex (PDB ID 2KX5; TAR: slate; peptide: magenta) and the TAR loop-SEC complex (coloring as in Fig. 2) are aligned on the common TAR component. Tat and CycT1 TRM contact the TAR loop. The TAR bulge is positioned in a pocket formed by CycT1 helices H1, H3, and H4 and Tat residue G48, adjacent to the missing Tat ARM, whose binding site is indicated by the Tat-peptide mimetic bound in the major groove of TAR. Views A and B are rotated 180°.
The combined structures clearly show that Tat ARM-peptide binding in the major groove of TAR and Tat/CycT1 binding to the TAR loop are compatible with each other. The last visible Tat residue in the crystal structure, G48, is positioned close to the TAR bulge and the major groove of the superimposed TAR-peptide complex so that the adjacent Tat residues in the ARM region can easily interact with nucleotides in the TAR bulge and the major groove, as seen in the NMR structures with cyclic Tat mimetics. Such a two-point binding mode increases binding specificity and affinity for TAR and has been observed in other RNA-binding proteins (32, 36) (Fig. 5). The two-point binding mode also explains the detrimental effect of amino acid insertions between the Tat transactivation domain and the ARM on transactivation (37), because insertions lead to misalignments between the TAR bulge and loop and the corresponding protein binding sites. Thus, the composite X-ray/NMR model of the TAR loop complex with Tat-SEC and the TAR complex with Tat-peptide provides a holistic model of the complete TAR interactions with the Tat-SEC complex. While details of side chain conformations and hydrogen bonding are still incomplete at the present resolution, these data have clarified how the TAR loop is recognized when the SEC is hijacked by Tat, substantially resolving a question of 20 years’ standing.
Materials and Methods
Protein Expression and Synthesis.
Human AFF42–73 was cloned into a modified pET28 plasmid (SI Appendix, Fig. S4). The recombinant protein includes an N-terminal TEV-protease-cleavable His-tag (21). An AFF4 peptide 32–67 with acetylated and amidated termini was synthesized at the University of Utah DNA/Peptide Facility.
P-TEFb and P-TEFb-Tat1–57 were expressed in High Five insect cells using recombinant baculovirus infections (SI Appendix, Fig. S4). We coexpressed human His-tagged CDK9 1–330 and human CycT1 1–264 with and without untagged codon-optimized HIV-1 Tat 1–57. Baculovirus generation and High Five cell infections have been described in detail (21). AFF4 fragments 2–73 and 32–67 with an N-terminal TEV-protease-cleavable His-tag and a His6-GST-tag, respectively, were expressed in E. coli.
TAR RNA.
Synthetic TAR fragments of WT and mutant TAR27, encompassing nucleotides 18–45, and TAR20 (nucleotides 21–40: rGrArUrCrUrGrArGrCrCrUrGrGrGrArGrCrUrCrA) were purchased from IDT. The RNA was annealed at 0.1 mg/mL in 20 mM Na-Hepes pH 7.3, 100 mM KCl, and 3 mM MgCl2. Best results were obtained by heating the RNA at 75 °C for 2 min followed by rapid cooling on ice. The purity of the RNA, analyzed by denaturing and native 10% polyacrylamide gel electrophoresis, was at least 95%.
Protein Purification.
Tat-P-TEFb and AFF42–73 were purified separately following recently described procedures (21). AFF432–67 was purified as GST-fusion protein over glutathione Sepharose (GSTrap FF; GE Healthcare) followed by TEV cleavage and a second purification step over a HisTrap column (GE Healthcare). The flow-through fractions of the HisTrap column containing cleaved AFF432–67 were concentrated and, in a final step, purified over a Superdex S200-size exclusion column. Tat-P-TEFb and AFF432–67 were combined at a 1:1.4 (mol/mol) ratio, concentrated to 0.6 mL, and injected onto an analytical Superdex S200-size exclusion column equilibrated with 25 mM Na-Hepes pH 7.4, 0.2 M NaCl, 0.05 M KCl, and 1 mM TCEP.
Crystallization and Structure Determination.
Purified Tat1–57:AFF432–67K47Y:P-TEFb was combined with an annealed TAR20 fragment, nucleotides 21–40, at a 1:1.3 (mol/mol) ratio and concentrated to 7 mg/mL in 25 mM Na-Hepes pH 7.3, 0.2 M NaCl, 0.05 M KCl, 0.1 M ammonium sulfate, 3 mM MgCl2, and 0.5 mM TCEP. Crystals were grown in sitting drops from 0.8 μL protein-TAR complex combined with 0.5 μL reservoir solution. The drops were equilibrated against 50 mM Tris 8.5, 0.2 M ammonium acetate, 6 mM MgCl2, and 8% PEG 4K at 18 °C. Single needle-shaped crystals grew to a size of about 0.05 × 0.05 × 0.25 mm.
Crystals were soaked in 0.1 M Na-Hepes pH 8.0, 50 mM NaCl, 100 mM ammonium acetate, 6 mM MgCl2, 15% PEG 4K, 30% glycerol, and 2 mM DTT for cryoprotection, and flash-frozen in liquid nitrogen. X-ray data were collected at beamline 8.3.1 at the Lawrence Berkeley National Laboratory’s (38) Advanced Light Source using a Pilatus 3 6M detector (Dectris AG). The reflections were processed using XDS (39), AIMLESS, and CTRUNCATE (40) (SI Appendix, Table S1). Reported anisotropic ΔB values were calculated using AIMLESS/CCP4 (40). The Rmerge for the whole dataset was relatively high due to the inclusion of very weak reflections. Based on their Pearson correlation coefficient between random half datasets (CC1/2) (41), these weak reflections contributed significant information and were included in structure refinement. The mean I/SD was greater than 2.0 at 3.74-Å resolution.
The structure was determined by molecular replacement with PHENIX (42) using the Tat:AFF4:P-TEFb complex (PDB ID 4OGR) as the search model, followed by refinement of the protein complex in PHENIX. Electron density maps showed some strong extra density close to CycT TRM and extending from there into the crystal solvent channel. The dimensions and strength of the electron density were consistent with the presence of TAR in this location. Since it is very difficult to build a de novo RNA structure into relatively low resolution maps, we superimposed the NMR structures of TAR, bound to a Tat peptide (pdb ID 1ARJ) (33), onto the extra electron density and chose the best fitting model #8 of the ensemble as a starting point for further complex refinement. The most critical parts of the structure, CycT1 TRM and TAR, went through multiple cycles of manual rebuilding into omit maps using COOT (43), followed by automatic refinement in PHENIX. RNA-specific tools for rebuilding and refinement, RCrane (44) and Erraser (45), were used to guide RNA modeling. The TAR complex structure was refined at 3.5-Å resolution to an Rfree value of 28.12% (SI Appendix, Table S1) with good geometry based on Molprobity scores. The atomic coordinates and structure factors (PDB ID 6CYT) are available at the Protein Data Bank (www.rcsb.org). Figures were prepared with PyMOL (46) and Chimera (47).
Electrophoretic Mobility Shift Assay.
Refolded synthetic TAR (nucleotides 18–44) was radioactively labeled with 32P-γ−ATP using T4-polynucleotide kinase. A 10-µl reaction was prepared with 200 nM TAR, 0.3 mCi 32P-γ−ATP (7,000 Ci/mmol; MP Biomedicals), and 10 units of T4-polynucleotide kinase (New England BioLabs) in 70 mM Tris/HCl pH 7.6, 10 mM MgCl2, and 2 mM DTT. After incubating at 37 °C for 1 h, 25 µl annealing buffer (20 mM Na-Hepes pH 7.3, 100 mM KCl, and 3 mM MgCl2) were added to the reaction. The mixture was purified twice over Illustra G25 spin columns (GE Healthcare) to remove free nucleotides. The purified labeled TAR was diluted to 10 nM (3,000–5,000 cpm/µl) with annealing buffer for storage and use in EMSAs.
Binding reactions (10 μL) were carried out in 20 mM Na-Hepes pH 7.3, 100 mM KCl, 3 mM MgCl2, 1 mM DTT, 4% glycerol with 12 units RNasin (Promega), 10 µg/mL BSA, and 5 µg/mL Poly(I:C) (Invivogen). Each reaction contained 100 pM labeled TAR RNA. Reactions were incubated at 20 °C for 30 min, and RNA-binding complexes were separated on a prerun 6% polyacrylamide gel in 0.5× TBE (100 V, 1 h at 4 °C). Gels were dried, exposed to storage phosphor screens, and measured on a Typhoon phosphorimager (GE Healthcare). Each EMSA was repeated two to three times and analyzed with GraphPad Prism Version 7.
Fluorescence Polarization Assay.
Tat-P-TEFb WT and Tat-P-TEFb with CycT1 single-site mutation R251A, R254A, W256A, W258, or R259A were expressed and purified as described above. AFF42–73 was expressed and purified as described previously (21). Protein stocks at 5–100 μM concentration were flash-frozen and stored at −80 °C. The sequence of 5′-6-Fam-TAR19 was chosen to validate the crystal structure, which included TAR20 without a bulge. The TAR19 molecule was expected to have a low nanomolar Kd suitable for fluorescence polarization experiments with a fluorescein-labeled ligand.
5′-6-FAM labeled ssRNA TAR19 (rArUrCrUrGrArGrCrCrUrGrGrGrArGrCrUrCrA) (Integrated DNA Technologies) (Fig. 1A) was dissolved in water to 100 μM. The TAR19 RNA was diluted to 15 μM in annealing buffer (20 mM Na-Hepes 7.5, 100 mM KCl, 3 mM MgCl2) and refolded by incubating the RNA at 75 °C for 2 min followed by a quick transfer to ice.
All further dilutions of protein and TAR19 were made in FP-buffer (25 mM Na-Hepes pH 7.3, 100 mM NaCl, 10% glycerol, 0.05% Nonidet P-40, 0.5 mM TCEP). WT and mutant Tat-P-TEFb were preincubated for 15 min with twofold molar excess AFF4 2–73 before making serial threefold dilutions in FP-buffer. Diluted protein was combined with 5′-FAM-TAR19 in a final assay volume of 90 μL and at a final RNA concentration of 2 nM. Three times 20 μL of each solution were transferred to a Greiner 384 flat-bottom, black small-volume plate.
Fluorescence anisotropy was measured at 30 °C with a Synergy Neo2 reader (Biotek) with an excitation wavelength of 485 nm and an emission wavelength of 528 nm. All measurements for one experiment were done in triplicate, and each experiment was repeated three times and analyzed with GraphPad Prism Version 7. Binding curves were fit with a single-site quadratic binding equation (48, 49):
where Bmax is the maximum specific binding, L is the concentration of nucleic acid, x is the concentration of Tat:AFF4:P-TEFb, and Kd, app is the apparent dissociation constant for Tat:AFF4:P-TEFb and nucleic acid. Error bars are representative of the SD from the mean of three experimental replicates.
Supplementary Material
Acknowledgments
We are grateful for thoughtful discussions of our results with Dr. Gabriele Varani and Dr. Matthew D. Shortridge. This work was supported by NIH Grant P50GM0882250 (to J.H.H.). Beamline 8.3.1 at the Advanced Light Source is operated by the University of California Office of the President, Multicampus Research Programs and Initiatives Grant MR-15-328599, National Institutes of Health Grants R01 GM124149 and P30 GM124169, Plexxikon Inc., and the Integrated Diffraction Analysis Technologies program of the US Department of Energy Office of Biological and Environmental Research. The Advanced Light Source (Berkeley, CA) is a national user facility operated by the Lawrence Berkeley National Laboratory on behalf of the US Department of Energy under Contract DE-AC02-05CH11231, Office of Basic Energy Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6CYT).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806438115/-/DCSupplemental.
References
- 1.Cillo AR, Mellors JW. Which therapeutic strategy will achieve a cure for HIV-1? Curr Opin Virol. 2016;18:14–19. doi: 10.1016/j.coviro.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Margolis DM, Garcia JV, Hazuda DJ, Haynes BF. Latency reversal and viral clearance to cure HIV-1. Science. 2016;353:aaf6517. doi: 10.1126/science.aaf6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mousseau G, et al. An analog of the natural steroidal alkaloid cortistatin A potently suppresses Tat-dependent HIV transcription. Cell Host Microbe. 2012;12:97–108. doi: 10.1016/j.chom.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mousseau G, Mediouni S, Valente ST. Targeting HIV transcription: The quest for a functional cure. Curr Top Microbiol Immunol. 2015;389:121–145. doi: 10.1007/82_2015_435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darcis G, Van Driessche B, Van Lint C. HIV latency: Should we shock or lock? Trends Immunol. 2017;38:217–228. doi: 10.1016/j.it.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Mousseau G, Valente ST. Role of host factors on the regulation of Tat-mediated HIV-1 transcription. Curr Pharm Des. 2017;23:4079–4090. doi: 10.2174/1381612823666170622104355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schiralli Lester GM, Henderson AJ. Mechanisms of HIV transcriptional regulation and their contribution to latency. Mol Biol Int. 2012;2012:614120. doi: 10.1155/2012/614120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He N, et al. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell. 2010;38:428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sobhian B, et al. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulze-Gahmen U, et al. Insights into HIV-1 proviral transcription from integrative structure and dynamics of the Tat:AFF4:P-TEFb:TAR complex. eLife. 2016;5:e15910. doi: 10.7554/eLife.15910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schulze-Gahmen U, Lu H, Zhou Q, Alber T. AFF4 binding to Tat-P-TEFb indirectly stimulates TAR recognition of super elongation complexes at the HIV promoter. eLife. 2014;3:e02375. doi: 10.7554/eLife.02375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu J, et al. Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4. Cell Cycle. 2014;13:1788–1797. doi: 10.4161/cc.28756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 15.Bahadur RP, Zacharias M, Janin J. Dissecting protein-RNA recognition sites. Nucleic Acids Res. 2008;36:2705–2716. doi: 10.1093/nar/gkn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garber ME, et al. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Churcher MJ, et al. High affinity binding of TAR RNA by the human immunodeficiency virus type-1 tat protein requires base-pairs in the RNA stem and amino acid residues flanking the basic region. J Mol Biol. 1993;230:90–110. doi: 10.1006/jmbi.1993.1128. [DOI] [PubMed] [Google Scholar]
- 18.Weeks KM, Crothers DM. RNA recognition by Tat-derived peptides: Interaction in the major groove? Cell. 1991;66:577–588. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- 19.Richter S, Cao H, Rana TM. Specific HIV-1 TAR RNA loop sequence and functional groups are required for human cyclin T1-Tat-TAR ternary complex formation. Biochemistry. 2002;41:6391–6397. doi: 10.1021/bi0159579. [DOI] [PubMed] [Google Scholar]
- 20.Richter S, Ping Y-H, Rana TM. TAR RNA loop: A scaffold for the assembly of a regulatory switch in HIV replication. Proc Natl Acad Sci USA. 2002;99:7928–7933. doi: 10.1073/pnas.122119999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulze-Gahmen U, et al. The AFF4 scaffold binds human P-TEFb adjacent to HIV Tat. eLife. 2013;2:e00327. doi: 10.7554/eLife.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 23.Garber ME, Wei P, Jones KA. HIV-1 Tat interacts with cyclin T1 to direct the P-TEFb CTD kinase complex to TAR RNA. Cold Spring Harb Symp Quant Biol. 1998;63:371–380. doi: 10.1101/sqb.1998.63.371. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, et al. HIV-1 TAR RNA enhances the interaction between Tat and cyclin T1. J Biol Chem. 2000;275:34314–34319. doi: 10.1074/jbc.M006804200. [DOI] [PubMed] [Google Scholar]
- 25.Calnan BJ, Biancalana S, Hudson D, Frankel AD. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 1991;5:201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- 26.Feng S, Holland EC. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988;334:165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- 27.Berkhout B, Jeang KT. Trans activation of human immunodeficiency virus type 1 is sequence specific for both the single-stranded bulge and loop of the trans-acting-responsive hairpin: A quantitative analysis. J Virol. 1989;63:5501–5504. doi: 10.1128/jvi.63.12.5501-5504.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulinski T, et al. The apical loop of the HIV-1 TAR RNA hairpin is stabilized by a cross-loop base pair. J Biol Chem. 2003;278:38892–38901. doi: 10.1074/jbc.M301939200. [DOI] [PubMed] [Google Scholar]
- 29.Dethoff EA, et al. Characterizing complex dynamics in the transactivation response element apical loop and motional correlations with the bulge by NMR, molecular dynamics, and mutagenesis. Biophys J. 2008;95:3906–3915. doi: 10.1529/biophysj.108.140285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tahirov TH, et al. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature. 2010;465:747–751. doi: 10.1038/nature09131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stefl R, Skrisovska L, Allain FH-T. RNA sequence- and shape-dependent recognition by proteins in the ribonucleoprotein particle. EMBO Rep. 2005;6:33–38. doi: 10.1038/sj.embor.7400325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serganov A, Patel DJ. Towards deciphering the principles underlying an mRNA recognition code. Curr Opin Struct Biol. 2008;18:120–129. doi: 10.1016/j.sbi.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aboul-ela F, Karn J, Varani G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J Mol Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 34.Davidson A, Patora-Komisarska K, Robinson JA, Varani G. Essential structural requirements for specific recognition of HIV TAR RNA by peptide mimetics of Tat protein. Nucleic Acids Res. 2011;39:248–256. doi: 10.1093/nar/gkq713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davidson A, et al. Simultaneous recognition of HIV-1 TAR RNA bulge and loop sequences by cyclic peptide mimics of Tat protein. Proc Natl Acad Sci USA. 2009;106:11931–11936. doi: 10.1073/pnas.0900629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walden WE, et al. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science. 2006;314:1903–1908. doi: 10.1126/science.1133116. [DOI] [PubMed] [Google Scholar]
- 37.Luo Y, Peterlin BM. Juxtaposition between activation and basic domains of human immunodeficiency virus type 1 Tat is required for optimal interactions between Tat and TAR. J Virol. 1993;67:3441–3445. doi: 10.1128/jvi.67.6.3441-3445.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacDowell AA, et al. Suite of three protein crystallography beamlines with single superconducting bend magnet as the source. J Synchrotron Radiat. 2004;11:447–455. doi: 10.1107/S0909049504024835. [DOI] [PubMed] [Google Scholar]
- 39.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Keating KS, Pyle AM. RCrane: Semi-automated RNA model building. Acta Crystallogr D Biol Crystallogr. 2012;68:985–995. doi: 10.1107/S0907444912018549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chou F-C, Echols N, Terwilliger TC, Das R. RNA structure refinement using the ERRASER-Phenix pipeline. Methods Mol Biol. 2016;1320:269–282. doi: 10.1007/978-1-4939-2763-0_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.The PyMOL Molecular Graphics System 2016. PyMol Version 1.8.2.1 (Schrödinger, LLC, New York)
- 47.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 48.Vos SM, et al. Architecture and RNA binding of the human negative elongation factor. eLife. 2016;5:e14981. doi: 10.7554/eLife.14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang X, Aulabaugh A. Application of fluorescence polarization in HTS assays. Methods Mol Biol. 2009;565:127–143. doi: 10.1007/978-1-60327-258-2_6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.