Significance
Mass spectrometry-based proteogenomics of patient-derived xenografts (PDXs) identified dihydropyrimidinase-like-3 (DPYSL3) as a multilevel (RNA/protein/phosphoprotein) expression outlier specific to a claudin-low (CLOW) PDX. DPYSL3 has established functions in neural cell migration and axon outgrowth but is understudied in breast cancer. Here, we demonstrate that loss of DPYSL3 promotes cell-cycle arrest, multinucleation, and collapse of the vimentin microfilament network associated with increased phospho-vimentin. DPYSL3 is also a negative regulator of p21-activated kinase (PAK) and suppresses epithelial-to-mesenchymal transition (EMT). In turn, EMT regulators induce DPYSL3, suggesting that DPYSL3 provides negative feedback on EMT. DPYSL3 therefore serves as a biomarker for CLOW tumors that exhibit PAK-dependent motility and EMT and is also susceptible to therapeutic approaches that promote vimentin phosphorylation during mitosis.
Keywords: DPYSL3, CRMP4, multinucleation, EMT, claudin-low breast cancer
Abstract
A Clinical Proteomic Tumor Analysis Consortium (CPTAC) proteogenomic analysis prioritized dihydropyrimidinase-like-3 (DPYSL3) as a multilevel (RNA/protein/phosphoprotein) expression outlier specific to the claudin-low (CLOW) subset of triple-negative breast cancers. A PubMed informatics tool indicated a paucity of data in the context of breast cancer, which further prioritized DPYSL3 for study. DPYSL3 knockdown in DPYSL3-positive () CLOW cell lines demonstrated reduced proliferation, yet enhanced motility and increased expression of epithelial-to-mesenchymal transition (EMT) markers, suggesting that DPYSL3 is a multifunctional signaling modulator. Slower proliferation in DPYSL3-negative () CLOW cells was associated with accumulation of multinucleated cells, indicating a mitotic defect that was associated with a collapse of the vimentin microfilament network and increased vimentin phosphorylation. DPYSL3 also suppressed the expression of EMT regulators SNAIL and TWIST and opposed p21 activated kinase 2 (PAK2)-dependent migration. However, these EMT regulators in turn induce DPYSL3 expression, suggesting that DPYSL3 participates in negative feedback on EMT. In conclusion, DPYSL3 expression identifies CLOW tumors that will be sensitive to approaches that promote vimentin phosphorylation during mitosis and inhibitors of PAK signaling during migration and EMT.
Breast cancer is classified into four common intrinsic subtypes based on gene-expression profiling (luminal A, luminal B, HER2-enriched, and basal-like), each of which are associated with different patterns of recurrence and therapeutic responses (1, 2). Further gene expression studies have identified additional but less-frequent breast cancer intrinsic subtypes. For example, Herschkowitz et al. reported the claudin-low (CLOW) subtype, through the combined analysis of mouse mammary tumor models and human breast tumors (3). CLOW tumors are characterized by low expression of claudin 3, 4, and 7 and E-cadherin; low expression of luminal markers; and high levels of mesenchymal markers (3). The incidence of CLOW tumors is reported to be between 7% and 14% of breast cancers, and there is an association with metaplastic features whereby breast cancers show squamous or mesenchymal differentiation patterns (4, 5). Sabatier et al. reported the 5-y disease-free survival (DFS) of CLOW subtype was lower to that observed in the luminal A subtype, similar to that observed in the luminal B subtype, and trended to be better than that observed in the two other poor-prognosis subtypes (basal-like and HER2-enriched) (6). The pathological complete response rate of CLOW tumors after neoadjuvant chemotherapy was closer to that of basal-like and HER2-enriched tumors and significantly higher than the rates observed for luminal A and B tumors (6). As far as we are aware, there are no targeted therapeutic agents specifically for CLOW tumors, and there is only a limited understanding of their unique biological features.
Patient-derived xenografts (PDXs) generated in immunosuppressed mice strains provide a useful setting to analyze the biological properties of the intrinsic subtypes of breast cancer because this approach effectively captures the biological diversity of the disease (7). The Clinical Proteomic Tumor Analysis Consortium (CPTAC) generated quantitative iTRAQ mass spectrometry-based proteomics and phosphoproteomics data from 21 breast cancer PDXs that was combined with RNA sequencing (RNA-seq) and DNA sequencing to provide integrated proteogenomic profiles (8). Here, we further explored these data to identify outliers in the proteogenomic data with the intent of discovering subtype specific therapeutic hypotheses. These analyses identified dihydropyrimidinase-like 3 (DPYSL3) as a highly phosphorylated protein selectively expressed in a CLOW PDX referred to as WHIM12.
DPYSL3 [also referred to as collapsing response mediator protein 4 (CRMP4)] is a member of the DPYSL gene family that shares 50–70% sequence homology (9–12). DPYSL3 is highly expressed in developing and adult nervous systems (12–14) and functions in a variety of cellular processes, including cell migration, differentiation, neurite extension, and axonal regeneration (15–18). DPYSL3 regulates the actin cytoskeleton in neuroblastoma cells to inhibit cell migration (19). DPYSL3 is also reported to play a role in cell migration and metastasis suppression in prostate cancer. However, in pancreatic cancer, DPYSL3 is positively associated with liver metastasis and poor outcome (20–24). A PubMed informatics tool indicated no functional studies of DPYSL3 in breast cancer reported to date and no known associations with the CLOW subset of the disease.
The objective of the study described herein is to characterize the functional consequences of the DPYSL3 expression by CLOW breast cancer and assess its potential significance regarding the therapeutic vulnerabilities of this breast cancer subtype.
Results
DPYSL3 Expression Is Enriched in CLOW Breast Cancer.
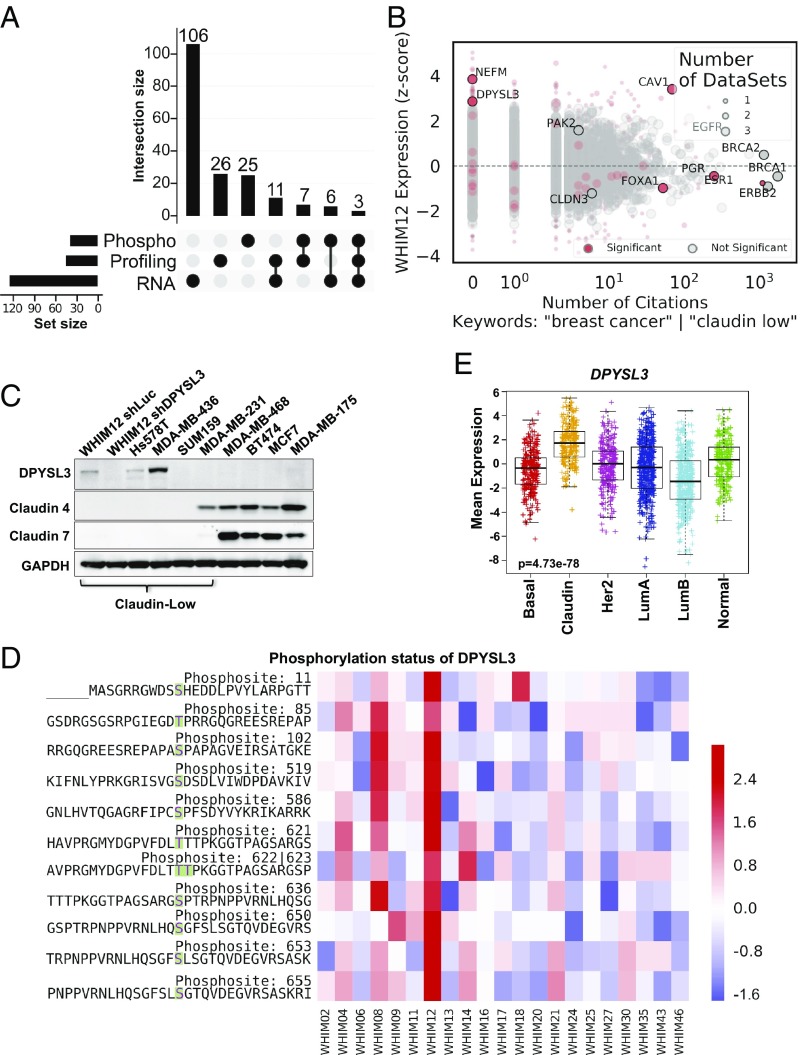
Integration of mass spectrometry-based proteomic profiling, phosphoprotein profiling, and RNA-seq across the WHIM series of PDX tumors identified differentially expressed genes in the WHIM12 breast cancer PDX, previously classified as a high-confidence CLOW tumor based on transcriptomic profiling (8). ANOVA was performed for each data source to identify differential genes across breast cancer intrinsic subtypes. After further filtering for genes whose expression levels were two standard deviations above or below the mean in WHIM12, only three genes (NEFM, DPYSL3, and CAV1) were highly and differentially expressed in all three datasets (Fig. 1A and Dataset S1). Next, a total 5,588 genes, of which 875 were significant on the ANOVA model, were aggregated from the three datasets and mapped to PubMed IDs (PMIDs) by using PubMed ELink, and each abstract was downloaded by using PubMed EFetch. After removing papers reporting high-dimensional data (>50 genes per PMID), abstracts were filtered by the presence of the keywords “breast cancer” or “claudin-low” to obtain the number of publications corresponding to each gene related to breast cancer. Despite the extremely high Z-score for CLOW across all three ‘omics datasets, an absence of citations regarding DPYSL3 and NEFM in breast cancer indicated the lack of research on these gene products (Fig. 1B and Dataset S2).
Fig. 1.
DPYSL3 is enriched in CLOW WHIM12 PDX tumors. (A) Upset plot showing the intersection of significantly differentially expressed genes (adjusted P < 0.05) in WHIM12 across three datasets (Phospho, Profiling, and RNA) representing outlier expression levels at least two standard deviations mean expression values. The set size shows the total number of genes which meet these criteria within each dataset, and the intersection size indicates the number of overlapping genes across datasets as indicated by the darkened circles. RNA-seq (RNA) shows the largest number of outlier genes at 126, followed by proteomic profiling (Profiling) with 47 outlier genes and phosphoproteomics (Phospho) with 41 outlier genes. A subset of these outlier genes (11 genes) are shared between the Profiling and RNA datasets, 7 outlier genes are shared between Profiling and Phospho datasets, 6 outlier genes are shared between Phospho and RNA datasets, and 3 outlier genes are shared across all three datasets. (B) Comparison between differentially (red circles) and nondiffrerentially (gray circles) expressed outlier genes in WHIM12 and number of citations related to breast cancer. The size of each dot corresponds to the number of datasets that gene is found at either a significant or not significant level. (C) Western blotting showing DPYSL3, Claudin 4, and Claudin 7 expression in CLOW and non-CLOW breast cancer cell lines of various intrinsic subtypes. GAPDH was used as a loading control. (D) Heatmap showing 11 highly phosphorylated residues (in bold) in DPYSL3 protein from CLOW WHIM12 PDX compared with other breast cancer PDXs of various subtypes from phosphoproteomic profiling (8). Accompanying information is presented in Dataset S2. (E) Box and whiskers plots with center line representing mean mRNA expression levels of DPYSL3 gene expression across intrinsic subtypes from breast samples in the METABRIC dataset (25, 26). Box extends to interquartile range (IQR), and whiskers extend to 1.5 × IQR of mean gene expression. basal-like (Basal), CLOW (Claudin), HER2-enriched (Her2), luminal A (LumA), luminal B (LumB) and normal-like (Normal). P value was determined by Kruskal–Wallis test. Accompanying information is presented in SI Appendix, Fig. S1 D and E.
To validate whether DPYSL3 or NEFM levels were specific to CLOW tumors, expression levels of these genes were analyzed across breast cancer cell lines from the Broad Institute Cancer Cell Line Encyclopedia (CCLE). Non-CLOW and CLOW cell lines expressed low levels of NEFM (SI Appendix, Fig. S1A). On the other hand, expression analysis demonstrated that 28.6% of CLOW cell lines highly expressed DPYSL3, compared with 14.3%, 10%, and 6.3% of basal-like, HER2-enriched, and luminal cell lines, respectively (SI Appendix, Fig. S1B). Among the CLOW cell lines, Hs578T and MDA-MB-436 abundantly expressed DPYSL3 mRNA, suggesting useful experimental model systems. Up-regulation of DPYSL3 protein in these cell lines, and in a cell line derived from the WHIM12 PDX, was confirmed via Western blotting (Fig. 1C). In the Western-blotting expression screen, DPYSL3 protein expression could only be detected when the relative mRNA expression level was greater than +2 on a log2-based scale in the CCLE data (SI Appendix, Fig. S1 A and B). DPYSL3 protein could not be detected in a number of cell lines with expression levels less than +2 (Fig. 1C). Additionally, Western blotting across WHIM PDX tumors confirmed WHIM12-specific DPYSL3 expression (SI Appendix, Fig. S1C). Phosphoproteomic mass-spectrometry-based analysis revealed 11 highly phosphorylated DPYSL3 phosphosites in WHIM12, more than any other PDX examined (8) (Fig. 1D). Breast tumor expression profiling confirmed higher DPYSL3 mRNA expression in CLOW tumors in comparison with other breast cancer subtypes (25, 26) (Fig. 1E) and two other clinical datasets were consistent in this regard (SI Appendix, Fig. S1 D and E).
These data suggested high levels of DPYSL3 transcript and protein expression, as well as hyperphosphorylation, were associated with CLOW breast cancer, and thus this protein may regulate some of the unique biological features of this subtype. Since DPYSL3 expression was only identified in the single example of a CLOW PDX amongst the 23 PDXs examined, CLOW cell lines were added to the experimental design to further characterize the functional properties of DPYSL3.
DPYSL3 Is Required for Proliferation in DPYSL3-Expressing CLOW Breast Cancer.
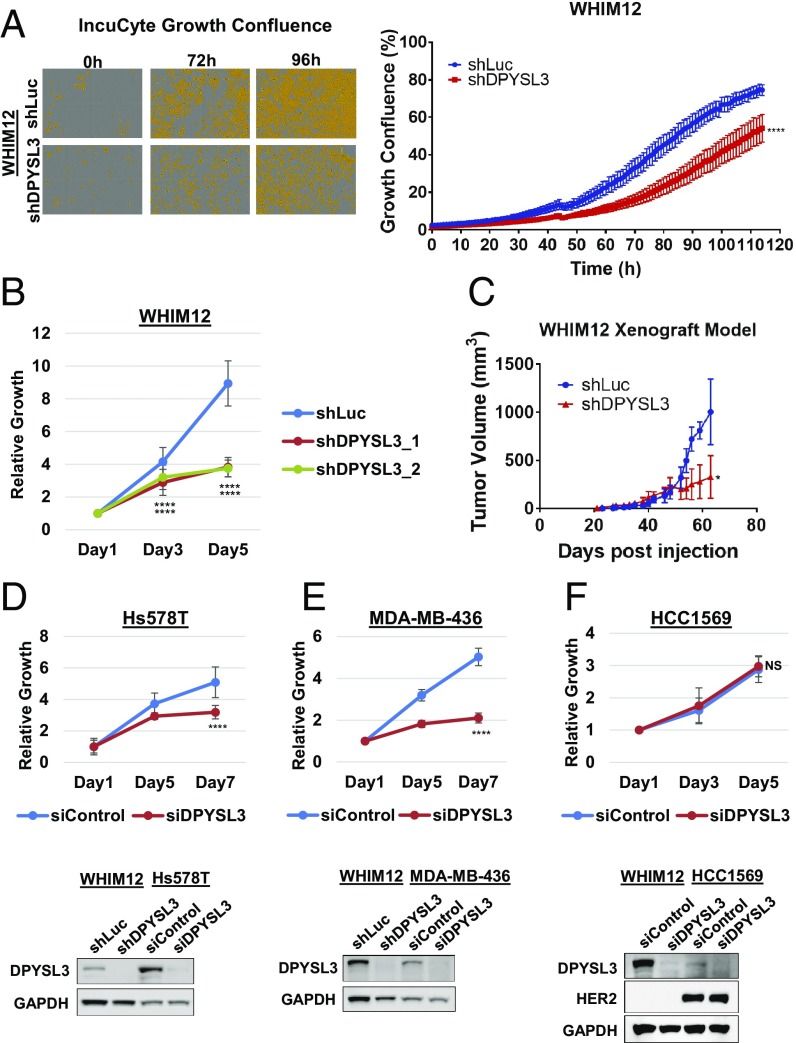
To investigate the role of DPYSL3 in CLOW breast cancer, expression was stably knocked down by using two different shRNA constructs in the WHIM12 cell line (SI Appendix, Fig. S2 A and B). Proliferation rates were determined by using an IncuCyte imaging system. DPYSL3 knockdown cells showed significantly lower proliferation and confluency levels compared with the shLuc control cells. The difference between confluency level in WHIM12 shLuc control and WHIM12 shDPYSL3 knockdown cells reached >20% after 72 h (P < 0.0001) (Fig. 2A). Alamar blue-based growth analysis of shLuc control and two different shRNAs of DPYSL3 (shDPYSL3_1 and shDPYSL3_2) in WHIM12 cells confirmed that both DPYSL3 knockdown constructs strongly decreased cellular proliferation (P < 0.0001) (Fig. 2B). WHIM12 shLuc and shDPYSL3 cells were injected into mice mammary fat pads, and the tumor volumes were measured over 2 mo. Tumors in both arms were detectable at 20 d after injection. shLuc control tumor volume increased rapidly after 50 d, while shDPYSL3 exhibited a much lower increase over the course of the experiment (P < 0.05) (Fig. 2C). Ki67 levels were examined by immunohistochemistry (IHC) in shLuc and shDPYSL3 xenograft tumor sections. Ki67 levels in shDPYSL3 xenograft tumors were significantly lower relative to shLuc tumors (P < 0.0001), suggesting that proliferating cells were predominant in shLuc tumors (SI Appendix, Fig. S2C).
Fig. 2.
DPYSL3 drives growth in CLOW breast cancer experimental models. (A, Left) Representative images of vessel wells acquired on IncuCyte of stable WHIM12 shLuc and shDPYSL3 cells at indicated time points. Cells are pseudocolored orange to aid visualization. (A, Right) Quantification of A, Left, depicting vessel well confluency over the course of the experiment. Error bars indicate SEM. ***P < 0.001 (Student t test comparing confluency at the last time point measured). (B) Growth of stable WHIM12 shLuc, shDPYSL3_1, and shDPYSL3_2 cell lines. Data are averages from three independent experiments ± SEM. ****P < 0.001 (ANOVA, Tukey’s multiple comparisons test). (C) Tumor growth of WHIM12 xenografts. Data are shown as averages of four mice per group ± SEM. *P < 0.05 (Student t test comparing tumor volume at the last time point measured). (D–F) Growth of indicated cell lines transfected with siControl or siDPYSL3. Data are averages from three independent experiments ±SEM (D–F, Upper). ****P < 0.001 (Student t test comparing last time point measured). D–F, Lower show Western blots to compare DPYSL3 expression levels. GAPDH was used as a loading control.
To extend these results, further cell lines were examined. First, two additional DPYSL3-expressing CLOW breast cancer cell lines Hs578T and MDA-MB-436 were transfected with DPYSL3 siRNA (Fig. 2 D and E). The CLOW cell line SUM159 and the cells MCF7 and ZR75.1 had undetectable levels of DPYSL3 protein and were examined as negative controls for the knockdown regents (SI Appendix, Fig. S2 F–H). The line HCC1569, which expresses modest levels of DPYSL3 (Fig. 2F), was examined to determine the potential role of DPYSL3 expression in a non-CLOW cell line. DPYSL3 siRNA had no effect on SUM159, MCF7, and ZR75.1, supporting the specificity of the reagents. In contrast, DPYSL3 siRNA inhibited growth in both Hs578T and MDA-MB-436, similar to the effects observed with the WHIM12 cell line (Fig. 2 D and E and SI Appendix, Fig. S2 E–H). In addition, overexpression of DPYSL3 in the SUM159 cell line promoted significantly greater proliferation (SI Appendix, Fig. S2D). The proliferation of HCC1569 line was unperturbed by siDPYSL3 knockdown despite DPYSL3 expression (Fig. 2F). These data indicate that the requirement for DPYSL3 for normal growth is restricted to DPYSL3 expressing CLOW cell lines within the confines of the cell line data examined.
Loss of DPYSL3 Induces Multinucleation and Polyploidy.
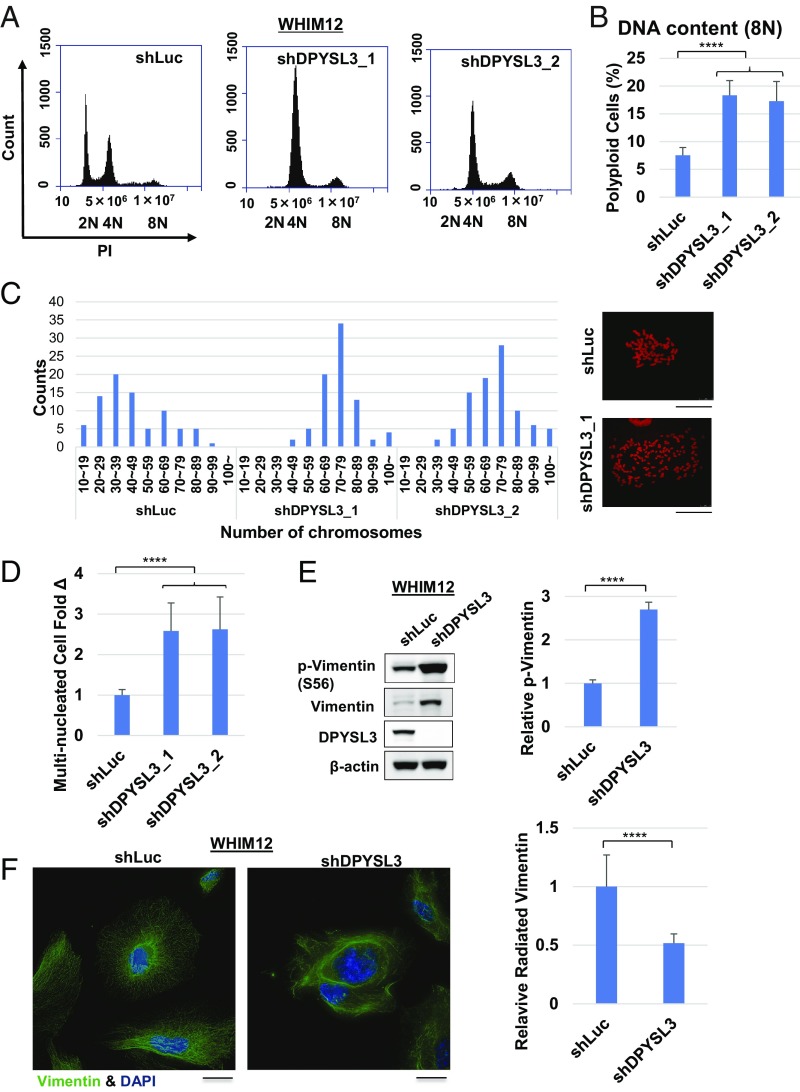
Flow-cytometric analysis demonstrated that WHIM12 shDPYSL3_1 cells had an abnormal cell-cycle distribution of cells with a proportion of cells in G1, S, and G2/M phase at 70.5%, 10.8%, and 14.5%, respectively, but also two peaks at 4N and 8N (Fig. 3A). In contrast, WHIM12 shLuc cells had proportions of cells in G1, S, and G2/M phase at 47.0%, 19.7%, and 29.0%, respectively, and two peaks at 2N and 4N (Fig. 3A). Another shRNA construct of DPYSL3, shDPYSL3_2, showed a similar distribution of cell cycle as shDPYSL3 (shDPYSL3_1) (Fig. 3A). WHIM12 shDPYSL3_1 and shDPYSL3_2 had a significant increase in the percentage of polyploid cells with 8N, compared with shLuc cells (P < 0.0001) (Fig. 3B). Additionally, cell-cycle analysis in Hs578T cells stably transfected with the same shLuc and shDPYSL3 constructs (Hs578T shLuc and Hs578T shDPYSL3, respectively) also indicated a shift in the cell-cycle profile in association with loss of DPYSL3 expression (SI Appendix, Fig. S3A). Mitotic spreads demonstrated that WHIM12 shDPYSL3 cells had approximately twice as many chromosomes as the WHIM12 shLuc control (Fig. 3C). Since these data suggested multiple nucleation and polyploidy in DPYSL3-depleted cells, the number of multiple nucleated cells in shLuc, shDPYSL3_1, and shDPYSL3_2 were quantified. The percentage of multiple nucleation of WHIM12 shDPYSL3_1 and shDPYSL3_2 cells was >2.5 times higher than that of shLuc (P < 0.0001) (Fig. 3D and SI Appendix, Fig. S3B).
Fig. 3.
Multiple nucleation and polyploidy are induced in the absence of DPYSL3 in CLOW breast cancer. (A) Representative histograms depicting cell cycle distribution of stable WHIM12 shLuc, shDPYSL3_1, and shDPYSL3_2 cell lines by flow cytometry. (B) Quantification of 8N polyploid cells from A. WHIM12 shDPYSL3_1 and shDPYSL3_2 cells have a significantly higher percentage of 8N for polyploid cells compared with WHIM12 shLuc cells. ****P < 0.0001 (Student t test). (C) Mitotic spread of WHIM12 shLuc, WHIM12 shDPYSL3_1, and shDPYSL3_2 cells. Cells were stained with propidium iodide (PI), and the number of chromosomes was counted by using a confocal microscope or an Eclipse Ti microscope. (D) Bar graphs depicting fold change of multinucleated cells in WHIM12 shDPYSL3_1 and shDPYSL3_2 cells compared with shLuc, quantified from immunofluorescence. Data are averages from three independent experiments ± SEM. ****P < 0.0001 (derived from Student t test). (E, Left) Western blotting was performed with the indicated antibodies in WHIM12 shLuc and shDPYSL3 cells. (E, Right) Bar graphs depicting average phosphorylated vimentin levels from three independent experiments ± SEM. ****P < 0.0001 (derived from Student t test). (F, Left) Representative confocal microscopy immunofluorescence images of stable WHIM12 shLuc and shDPYSL3 cells stained with vimentin (pseudocolored in green) and DAPI (pseudocolored in blue). (F, Right) Bar graphs depicting average number of cells with radiated vimentin structure ± SEM from three independent experiments. ****P < 0.0001 (derived from Student t test).
Next, DPYSL3 was transiently knocked down in WHIM12, with knockdown validation assessed by Western blotting (SI Appendix, Fig. S3C). The fold increase in multiple nucleation in the WHIM12 siDPYSL3 cells was 2.6 on day 4 that was sustained at 2.1 on day 8. However, multinucleated cells of the WHIM12 siControl cells had no increase by day 8 (SI Appendix, Fig. S3D). Hs578T and MDA-MB-436 transiently transfected with siControl and siDPYSL3 were used to extend these results. The percentage of multinucleated cells after 4 d upon transfection in Hs578T and MDA-MB-436 siDPYSL3 cells was twice as high as that of the control (SI Appendix, Fig. S3 E and F). As a negative control, there was no change in multiple nucleation between siControl vs. siDPYSL3 in SUM159, MCF7, and ZR75.1 (SI Appendix, Fig. S3 F and G). These results demonstrated that loss of DPYSL3 induces multinucleation, and this is consistent with previous reports that DPYSL3 can affect microtubule function during mitotic spindle formation (27).
A role for vimentin was also examined since DPYSL3 mRNA levels correlate positively with VIM levels in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) and The Cancer Genome Atlas datasets (SI Appendix, Fig. S4 A and B). Furthermore, a recent report demonstrated that increased vimentin phosphorylation causes multinucleation (28). We therefore investigated phosphorylation levels at serine 56 of vimentin in WHIM12 and Hs578T shLuc and shDPYSL3 cells. Vimentin was found to be up-regulated and highly phosphorylated when expression of DPYSL3 was suppressed (P < 0.0001), consistent with the multinucleation mechanism observed with a small molecule disruptor of phosphorylation driven functions of vimentin in mitosis (Fig. 3E and SI Appendix, Fig. S3H) (28). Furthermore, vimentin filaments were disrupted or “tangled” in WHIM12 shDPYSL3 and Hs578T shDPYSL3, in contrast to WHIM12 shLuc and Hs578T shLuc where vimentin radiated in all directions in a delicate filamentous network (Fig. 3F and SI Appendix, Fig. S3I).
DPYSL3 Controls Migration in CLOW Breast Cancer.
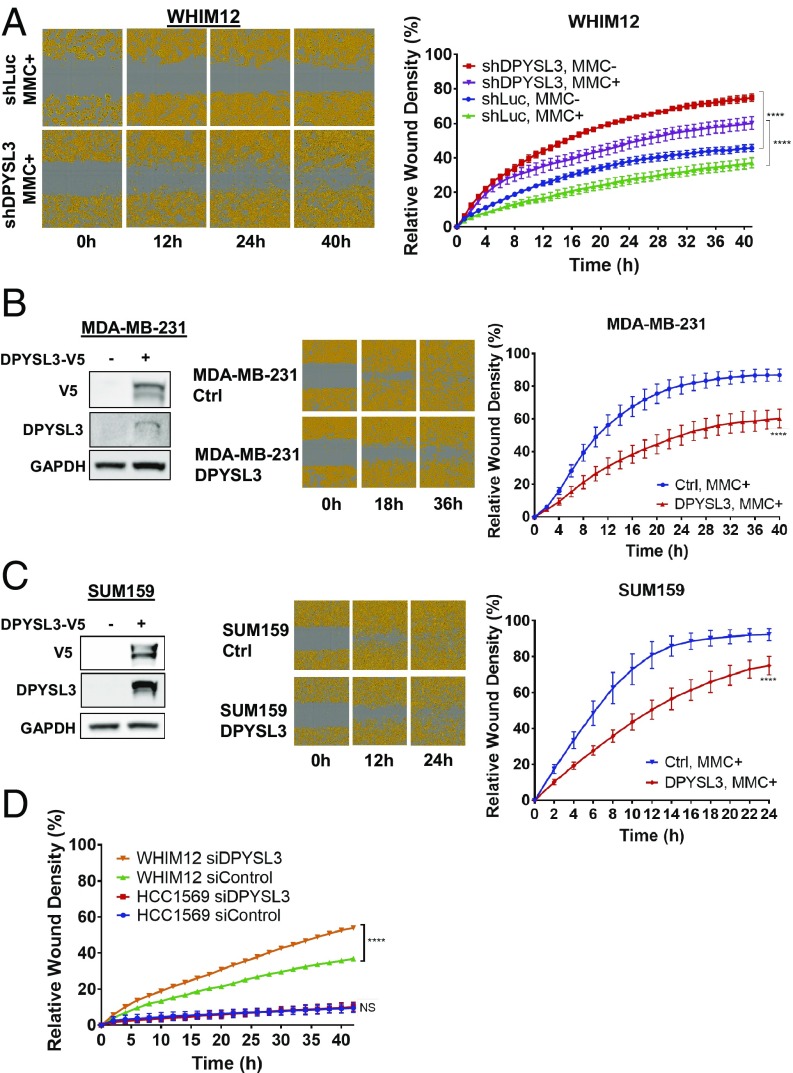
To evaluate the effect of DPYSL3 on motility, a scratch healing assay was performed. Given the effects of DPYSL3 on mitosis, mitomycin C (MMC) was used to prevent proliferation, thereby constraining the experiment to observations on migration alone, since differences in proliferation rates would confound the assay. These experiments demonstrated that shDPYSL3 WHIM12 cells had >20% greater wound healing efficiency at 12 h with an increase to 30% at 40 h compared with shLuc control cells (Fig. 4A). In addition to motility analysis, transwell migration assays and Matrigel invasion assays also indicated increased motility and invasion in shDPYSL3 WHIM12 cells (SI Appendix, Fig. S5 A and B). In addition to the WHIM12 model, the scratch healing assay was also performed with SUM159, MCF7, ZR75.1, and Hs578T after transfection with control siRNA vs. DPYSL3 siRNA. Motility within the SUM159, MCF7, and ZR75.1 siDPYSL3-treated cells were comparable to that of these siControl-treated cells because of low levels of DPYSL3, while the wound width of Hs578T siDPYSL3 cells decreased at a higher rate than control, indicating DPYSL3 siRNA treated cells were more likely to undergo migration (SI Appendix, Fig. S5 C–F). When DPYSL3 was overexpressed using a DPYSL3 expression vector in the CLOW breast cancer cell lines, MDA-MB-231 and SUM159, the relative wound density of DPYSL3 expressed cells was lower in both of MDA-MB-231 cells and SUM159 cells, consistent with a suppressive role for DPYSL3 in cell migration (Fig. 4 B and C). Finally, the migration of HCC1569, a non-CLOW line that expresses DPYSL3, was unaffected by siDPYSL3 treatment, while the relative wound density of WHIM12 siDPYSL3 increased significantly compared with that of siControl (P < 0.0001) (Fig. 4D).
Fig. 4.
DPYSL3 inhibits migration in CLOW breast cancer. (A, Left) Representative cell migration images of stable WHIM12 shLuc and shDPYSL3 cells in the presence of mitomycin C (MMC+) at indicated times postwounding. Cells are pseudocolored orange to aid visualization. (A, Right) Quantification of relative wound density of indicated stable WHIM12 cell lines during the course of the wound recovery assay in the absence (MMC–) or presence of MMC. P value was determined by ANOVA. ****P < 0.0001 (Tukey’s multiple comparisons test). (B and C, Left) Western blots showing V5-tagged DPYSL3 expression in transfected MDA-MB-231 cells (B) and SUM159 cells (C). GAPDH was used as a loading control. (B and C, Center) Representative cell migration images of transfected cells from B and C, Left postwounding at the indicated time points. (B and C, Right) Relative wound density of indicated cell lines during the course of the wound recovery assay in the presence of MMC (MMC+). ****P < 0.001 (derived from Student t test comparing last time point measured). (D) Quantification from scratch wound healing assay of WHIM12 and HCC1569 transfected with siControl or siDPYSL3 in the presence of mitomycin C proliferation blocking agent. Data are averages from three independent experiments ± SEM. ****P < 0.001 (Bonferroni’s multiple comparisons test).
DPYSL3 Modulates Migration via EMT.
To further investigate the consequences of DPYSL3 knockdown on the biology of WHIM12 growth in vivo, WHIM12 shLuc and shDPYSL3 xenograft tumor sections were stained with H&E, CD44, and CD24. WHIM12 shLuc tumors demonstrated a CD44-positive and CD24-negative profile consistent with stemness, but the expression of these markers were unaffected by shDPYSL3 mediated knockdown (Fig. 5A). However, during these experiments, it was noted that shDPYSL3 knockdown markedly increased spindle-cell morphology compared with the shLuc control tumors. By using visual point counting, the percentage of spindle cells in shDPYSL3 was significantly elevated compared with shLuc (P < 0.001) (Fig. 5A).
Fig. 5.
DPYSL3 suppresses EMT in CLOW breast cancer. (A, Left) Representative H&E staining and CD44/CD24 IHC in WHIM12 xenograft tumors. (A, Right) Bar graphs quantifying the number of cells with spindle morphology. More than 3,000 cancer cells from each group were counted, and then the percentage of spindle cancer cells was evaluated. Data are average counts from four tumor sections from each group ± SD. ***P < 0.001 (Student t test). (B, Left) Western blotting in stable WHIM12 shLuc and shDPYSL3 cells showing expression of SNAIL, TWIST, and DPYSL3 proteins. (B, Right) Bar graphs depicting average protein levels quantified from B, Left from three independent experiments ± SEM. *P < 0.05; **P < 0.01 (Student t test). (C) Expression of SNAI1 and TWIST1 by mRNA-qPCR in stable WHIM12 cell lines. (D) Western blotting showing expression of SNAIL, TWIST, and DPYSL3 levels in WHIM12 shLuc and WHIM12 shDPYSL3 xenograft tumors. DNA-PKcs was used as a loading control for human cell content. (E) Bar graphs depicting average expression of SNAI1 and TWIST1 by mRNA-qPCR in stable WHIM12 xenograft tumors ± SEM. (F) Western blotting in SUM159 transfected with control vector (DPYSL3–) or V5-tagged DPYSL3 (DPYSL3+) showing expression of SNAIL, TWIST, and DPYSL3.
Given the spindle cell morphology of DPYSL3 knockdown cells, markers of epithelial-to-mesenchymal transition (EMT) were examined. Interestingly, measurement of SNAIL/SNAI1 and TWIST/TWIST1 expression level revealed both protein and mRNA levels were increased in WHIM12 shDPYSL3 cells compared with WHIM12 shLuc cells (Fig. 5 B and C). SNAIL/SNAI1 and TWIST/TWIST1 were also upregulated in Hs578T DPYSL3 knockdown cells (SI Appendix, Fig. S6 A and B). In addition to in vitro studies, protein and mRNA levels of SNAIL/SNAI1 and TWIST/TWIST1 in WHIM12 shDPYSL3 xenograft tumors was higher than that in shLuc tumors (Fig. 5 D and E). Additionally, when DPYSL3 was overexpressed in CLOW cell line, SUM159, SNAIL and TWIST levels decreased (Fig. 5F).
DPYSL3 Provides Negative Feedback on EMT.
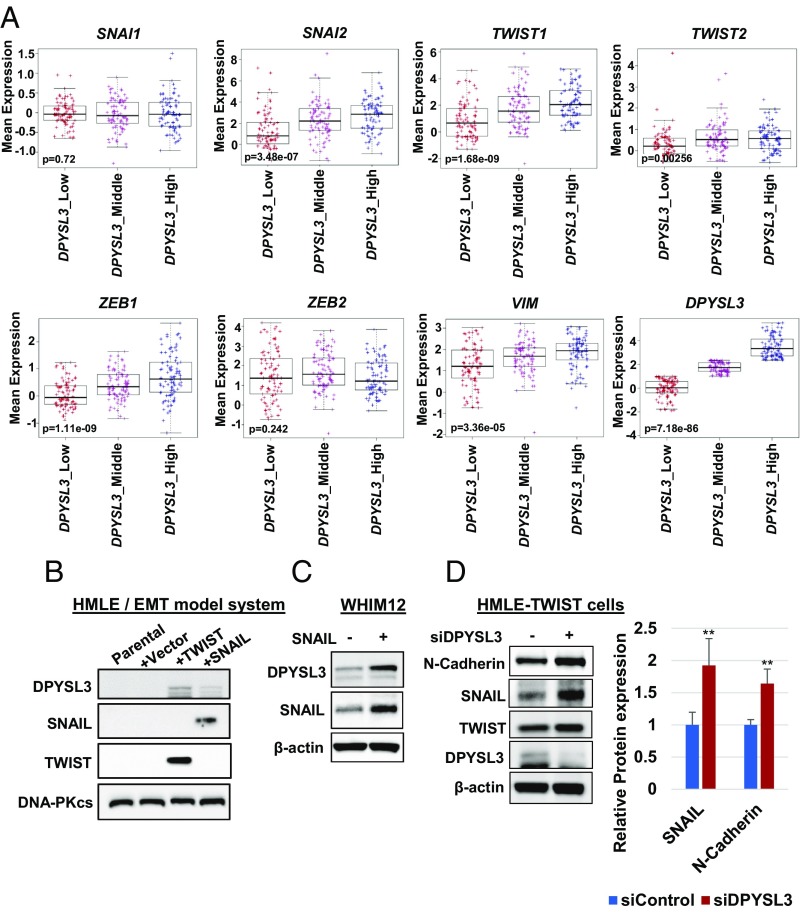
We subsequently examined the relationship between SNAI1/2, TWIST1/2, ZEB1/2, VIM, and DPYSL3 mRNA levels in tumors designated CLOW in the METABRIC database, demonstrating that DPYSL3 expression positively correlated with multiple EMT markers (Fig. 6A). To further investigate the EMT regulation of DPYSL3, Western blotting was performed in the HMLE model of EMT in which HMLE-SNAIL and HMLE-TWIST cells were generated by infecting immortalized human mammary epithelial cells with retroviral vectors expressing SNAI1 or TWIST1. This data demonstrated that TWIST and SNAIL induced DPYSL3 (Fig. 6B). Additionally, SNAIL overexpression in WHIM12 up-regulated DPYSL3, consistent with the result of the HMLE model of EMT, suggesting that EMT induced DPYSL3 (Fig. 6C). On the other hand, transient knock down of DPYSL3 in HMLE-TWIST cells increased SNAIL (Fig. 6D), which was consistent with data of WHIM12 and Hs578T (Fig. 5 and SI Appendix, Fig. S6). These data suggest that EMT and DPYSL3 are regulated in a reciprocal manner.
Fig. 6.
The EMT negative-feedback loop is mediated by DPYSL3. (A) Box and whiskers plots with center line representing mean mRNA expression levels of SNAI1/2, TWIST1/2, ZEB1/2, and VIM of CLOW breast tumors in the METABRIC dataset (25, 26). Box extends to interquartile range (IQR), and whiskers extend to 1.5 × IQR of mean gene expression. Comparison of gene expression signatures among three ranks (low, middle, or high) of DPYSL3 expression is shown. P value was determined by Kruskal–Wallis test. (B) Western blotting of DPYSL3 in HMLEs expressing control vector, TWIST, and SNAIL. DNA-PKcs was used for a loading control. (C) Western blotting of DPYSL3 in WHIM12 expressing control vector and SNAIL. (D, Left) Western blotting of EMT markers in HMLE-TWIST siControl and HMLE-TWIST siDPYSL3 cells. (D, Right) Quantification on the right from three independent experiments ± SEM. **P < 0.01 (Student t test).
PAK2 Is a Critical DPYSL3-Interacting Kinase that Promotes Migration.
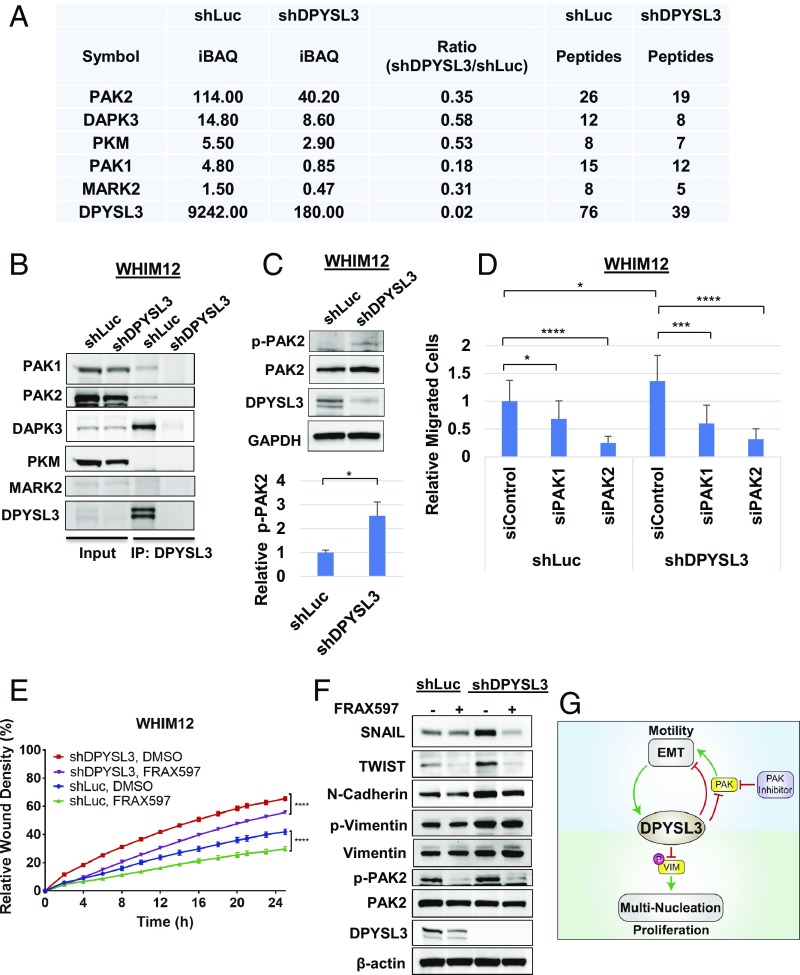
To further investigate effects on migration, the DPYSL3 interactome was determined by immunoprecipitation (IP) of DPYSL3 within WHIM12 shLuc and shDPYSL3 cells followed by mass spectrometry. The mass-spectrometry data revealed five DPYSL3-associated kinases: p21-activated kinase 2 (PAK2), Death-associated protein kinase 3 (DAPK3), pyruvate kinase (PKM), p21-activated kinase 1 (PAK1), and Microtubule Affinity Regulating Kinase 2 (MARK2) (Dataset S3 and Fig. 7A). PAK2 demonstrated the highest intensity-based absolute quantification and peptide values in the WHIM12 shLuc IP proteome, while DPYSL3 knockdown cells had reduced PAK2 precipitation by 65%, suggesting that PAK2 was the most strongly associated kinase with DPYSL3 (Fig. 7A). To confirm these data, IP and Western blotting (IP Western blot) was performed by using WHIM12 shLuc and shDPYSL3 cells and WHIM12 parental cells. PAK1, PAK2, and DAPK3 precipitated with DPYSL3 in the IP Western blot, thus validating DPYSL3 and PAK1/2 interaction (Fig. 7B and SI Appendix, Fig. S7A). In a different CLOW cell line, Hs578T cells, IP Western blots showed that these observations were consistent (SI Appendix, Fig. S7B). DPYSL3 interacted with PAK1, PAK2, and DAPK3, but the expression level of MARK2 was too low to be further evaluated (SI Appendix, Fig. S7B).
Fig. 7.
PAK2 interacts with DPYSL3 to modulate migration. (A) Table depicting DPYSL3 interacting kinases pulled down by DPYSL3 IP/mass spectrometry data from stable WHIM12 shLuc and shDPYSL3 cell lines. Accompanying information is presented in Dataset S3. iBAQ, intensity-based absolute quantification. (B) Western blots from inputs and DPYSL3 immunoprecipitated lysates from stable WHIM12 shLuc and shDPYSL3 cell lines. (C, Upper) Western blots from stable WHIM12 shLuc and shDPYSL3 cells. (C, Lower) Phospho-PAK2 (p-PAK2) from three independent experiments ± SEM. *P < 0.05 (Student t test). (D) Quantification of cell migration from stable WHIM12 cells transfected with siRNA after transwell migration assay relative to shLuc cells transfected with siControl. Data were shown as the averages from three independent experiments ± SEM. *P < 0.05; ***P < 0.001; ****P < 0.0001 (ANOVA with Tukey’s multiple comparisons test). (E) Quantification of cell migration as relative wound densities of stable WHIM12 shLuc and shDPYSL3 cells treated with DMSO or FRAX597 (1 M) at time points postwounding. P value was determined by ANOVA. ****P < 0.0001 (Bonferroni’s multiple comparisons test). (F) WHIM12 shLuc and shDPYSL3 cells were treated with DMSO or FRAX597 (1 M) for 36 h. Western blotting was performed with the indicated antibodies in WHIM12 shLuc and WHIM12 shDPYSL3 with or without FRAX597 treatment. (G) Working model of DPYSL3 as a multifunctional signaling scaffold. DPYSL3 is induced during EMT and provides a negative feedback on EMT. PAK inhibition suppresses cell migration in CLOW breast cancer. DPYLS3 promotes proliferation via positive regulation of the mitotic spindle. Loss of DPYSL3 induces vimentin phosphorylation leading to cytokinetic failure and multinucleation, thus impairing proliferation.
Of note, phosphorylation of serine-20 of PAK2 was up-regulated by DPYSL3 knockdown (Fig. 7C), suggesting that DPYSL3 is indeed a negative regulator of PAK2 phosphorylation status. To determine whether PAK is critical for migration in DPYSL3 expressing CLOW cells, a migration assay was performed with siRNA-based transient knockdowns of PAK1 and PAK2 in WHIM12 shLuc and shDPYSL3 cells. The siPAK2 treatment significantly decreased cellular migration by 75% compared with siControl, and siPAK2 opposed migration more effectively that siPAK1 in both shLuc and shDPYSL3 WHIM12 cells (Fig. 7D and SI Appendix, Fig. S7C). Interestingly, SNAIL and TWIST expression levels in shLuc and shDPYSL3 decreased with depletion of PAK2 (SI Appendix, Fig. S7C). To further support these siRNA-based findings, a migration assay was conducted with or without the PAK family inhibitor FRAX597. FRAX597 significantly inhibited the migration of control shLuc and shDPYSL3 cells (P < 0.0001) (Fig. 7E). Additionally, Western blotting showed that FRAX597 suppressed EMT markers in WHIM12 cells, regardless of DPYSL3 status, although the effect was more marked in the shDPYSL3 cells as SNAIL and TWIST up-regulated in this setting (Fig. 7F). These data are consistent with DPYSL3 functioning as negative regulator of PAK-dependent migration.
Discussion
Proteogenomics—the field of integrating data from mass spectrometry-based proteomic profiling, and phosphoproteomics into next-generation RNA-seq and DNA-sequencing data analysis pipelines promises new insights into cancer biology and therapeutic targeting (29). As well as analyses of clinical samples for disease phenotype association (30), the application of proteogenomics to model systems also has considerable potential. Here, we leveraged a proteogenomic dataset from a series of breast cancer PDXs described in an earlier publication (8). A search for breast cancer subtype-associated multilevel expression (RNA, protein, and phosphoprotein) outliers identified an association between DPYSL3 and CLOW breast cancer that had not been previously explored. In our view, discovery approaches that triangulate multiple tiers of ‘omics data with literature search engines to identify novel and targetable cancer biology should be more widely applied.
The CLOW subtype designation is based on a transcriptional signature indicating lack of expression from claudin family members that are components of tight junctions, where they form an intracellular barrier that regulates the flow of molecules between the cells of an epithelium. The clinical significance of a diagnosis of a CLOW tumor is not established, mostly because there are few reports of a specific therapeutic vulnerability for this subtype, unlike HER2 or ER-expressing subtypes or BRCA mutant triple-negative breast cancer. The role of DPYSL3 outlined in our study is an early report on specific druggable biology for CLOW tumors. DPYSL3 is not an appealing direct therapeutic target, because inhibition of phosphorylation produces a complex phenotype that depends on whether the reduction of phosphorylation promotes or inhibits the function of the DPYSL3-regulated protein. Thus, loss of DPYSL3 promotes vimentin phosphorylation, which inhibits vimetin function due a collapse of the filamentous network and mitotic failure. Interventions that increase or stabilize vimentin phosphorylation is predicted to be effective in CLOW tumors (28). On the other hand, PAK-dependent EMT and motility is promoted by loss of DPYSL3, as increased phosphorylation promotes PAK activity. Therapeutic vulnerability to PAK inhibition is therefore predicted for CLOW tumors, whereby suppression of PAK activity will inhibit EMT and motility and, theoretically, metastasis.
These studies raise the question of whether DPYSL3 has a role in other cancers or in breast cancer subtypes other than CLOW. DPYSL3 expression has been observed in several malignant tumors, including prostate cancer, pancreatic cancer, gastric cancer, and neuroblastoma (19, 20, 23, 31). Gao et al. and Yang et al. demonstrated DPYSL3 was a metastasis suppressor in prostate cancer and lung cancer, respectively (20, 32). In contrast, Hiroshima et al. reported DPYSL3 was a metastasis promoter in pancreatic cancer (23). To account for the opposite effects of DPYSL3 expression on prostate and pancreatic cancer metastasis, Guo et al. investigated a difference in the DPYSL3 splice variant that was predominantly expressed in gastric cancer models and concluded that two different isoforms of DPYSL3 (a long isoform: 75 KDa, a short isoform: 62 KDa) had opposite functions in regulating gastric cancer cell proliferation as well as invasion and metastasis (31). In the present study, WHIM12 cells and tumors are long isoform dominant by quantitative PCR (qPCR), thus, the multiple roles of DPYSL3 in CLOW breast cancer cells seem unlikely to relate to the short isoform (SI Appendix, Fig. S8). The question of subtype specificity within breast cancer was addressed by examining the HCC1569 cell line. Despite DPYSL3 expression, HCC1569 did not respond to siDPYSL3 (Figs. 2F and 4D and SI Appendix, Fig. S3J). While a more extensive screen could still reveal non-CLOW lines that are DPYSL3-regulated, the cell line data presented herein, as well as the elevated expression in CLOW lines remains suggestive that DPYSL3 has specific roles in the CLOW breast cancer subtype.
The role of DPYSL3 in preventing multiple nucleation and polyploidy during mitosis of CLOW cells was anticipated by studies in other cell systems. DPYSL3 binds directly to tubulin, promotes microtubule assembly, and is also known as a microtubule-associated protein (33, 34). In fact IP and mass-spectrometry data from WHIM12 shLuc and WHIM12 shDPYSL3 demonstrates that DPYSL3 interacts with -tubulin (Dataset S3). Additionally, some previous reports demonstrate that DPYSL3 regulates the actin and microtubule growth cone cytoskeleton (18, 19, 33, 35), and other publications demonstrate that GSK3-dependent phosphorylation of DPYSL3 regulates chromosomal alignment and mitotic progression through its effect on spindle microtubules (27). In CLOW breast cancer, depletion of DPYSL3 causes multiple nucleation and then polyploidy due to cytokinetic failure, likely due to disruption of vimentin function through increased phosphorylation. In other words, DPYSL3 is a negative regulator of a vimentin protein kinase(s). While we have yet to define which exact vimentin kinase(s) is/are modulated by DPYSL3, it is unlikely to be a PAK family member because the pan-PAK inhibitor FRAX597 does not phenocopy the effect of DPYSL3 knockdown in mitosis or affect vimentin phosphorylation, despite previous observations that PAK kinases are capable of modulating vimentin phosphorylation (Fig. 7F and SI Appendix, Fig. S7 D–G) (28, 36–38).
In contrast, the role of DPYSL3 in migration and EMT in CLOW cells does appear to be PAK-dependent and specifically mediated by PAK2. Cell migration in cells correlated with increased phosphorylation of PAK2 on Ser20 and was sensitive to PAK2 siRNA and pharmacological PAK inhibition. IP and mass spectrometry-based proteomics and Western blotting suggest that DPYSL3 and PAK2 directly interact. Thus, DPYSL3 can be considered a biomarker for CLOW tumors where migration and EMT can be potentially opposed with a PAK inhibitor.
A role for DPYSL3 in EMT is consistent with the multistudy integration data of Parsana et al. (39). Furthermore WHIM12 was derived from a patient with a metaplastic carcinoma with spindle cell formation, an EMT characteristic. Further analyses of human data (METABRIC) revealed a strong positive correlation between five of seven EMT markers and DPYSL3 expression (Fig. 6A). This finding is consistent with induction of DPYSL3 by TWIST and SNAIL in the HMLE EMT system (Fig. 6B). While it appears somewhat paradoxical that a negative regulator of EMT is positively associated with EMT markers in clinical datasets, a logical synthesis of these data is that DPYSL3 is induced by EMT and subsequently provides negative feedback on the EMT state by inhibiting PAK. Within a tumor where EMT is the dominant and pathological state, DPYSL3-mediated feedback may be impaired, causing DPYSL3 levels to increase, yet EMT persists. An example of this effect are the HMLE cells we studied. DPYSL3 is induced by retroviral expression of SNAI1 and TWIST1 in this model; thus, potential feedback inhibition in the transcription of SNAI1 and TWIST1 by DPYSL3 is disrupted because these EMT transcription factors are not controlled by their normal regulatory elements. Certainly, the kinetics and exact mechanism of how DPYSL3 regulates EMT warrants further detailed study.
Additional roles for DPYSL3 are possible. Mass-spectrometry IP data suggests an interaction with DAPK3, a result that suggests DAPK3 may play a role in apoptosis. Consistent with this suggestion DAPK3 transcript levels also showed a strong positive correlation with DPYSL3 in the METABRIC data (SI Appendix, Fig. S9).
We accept as a limitation of our study that we only identified one example of a CLOW PDX. Future studies should include further analyses of additional triple-negative breast cancer PDXs to identify further examples for in vivo studies.
In conclusion, this study provides insights into DPYSL3 as a negative regulator of kinases that regulate cytokinesis, motility, and EMT. These are critical processes in cancer cells, and DPYSL3 expression could thus be used to stratify patients for clinical trials that target these cancer hallmarks.
Materials and Methods
Bioinformatics Analysis.
Three datasets were used for identification of genes differentially expressed in CLOW breast cancer in the WHIM series of PDX models. Across the WHIM subtypes, RNA-seq data were accessed through the dbGAP under accession no. phs000611 (www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000611.v1.p1), while full proteome profiling and phosphosite expression data were downloaded from https://www.nature.com/articles/ncomms14864 (8). WHIM 02, 04, 06, 08, 09, 11, 12, 13, 14, 16, 17, 18, 20, 21, 24, 25, 27, 30, 35, 43, and 46 were used from the proteomics data; WHIMs 09, 37, and 47 were excluded because during analysis, it was determined that a different pooled reference control was used for these WHIM samples, which limited cross-tumor comparisons. WHIM 17 and 46 were also excluded, as these ultimately proved to be EBV-associated lymphoproliferative lesions, not breast cancer-derived. The RNA-seq data analysis included all available WHIM data, including WHIM 03, 05, 22, 26, 37, and 47 (which were not profiled using protomics) but excluded 17 and 46 (as discussed above). Both proteomics and phosphoproteomics datasets were filtered for observations in at least 10 samples as performed previously and mapped to the gene level. The phosphosite data were rolled up into a single value by averaging all phosphosites per gene and sample as an estimate of the relative phosphorylation status of each protein. For RNA-seq, genes were filtered to those quantified in at least one of the proteomics datasets. Transcripts per million expression data were log-transformed and converted into ratios based on the median value per gene. For all datasets, ratios were converted to z-scores with a mean of zero and a SD of 1. Within each dataset, ANOVA was performed on each gene to identify differential genes based on tumor subtype, and resulting P values were false discovery rate-corrected with the Benjamini–Hochberg procedure.
Quantified genes were mapped to PubMed IDs by using PubMed ELink, and each abstract was downloaded by using PubMed EFetch. PubMed IDs that map to >50 genes were removed to eliminate high-dimensional data studies that did not make specific conclusions at the gene level. The resulting abstracts were filtered by the presence of the keywords “breast cancer” and “claudin-low” to obtain the number of publications corresponding to each gene related to breast cancer. These tasks were performed in an in-house Python script, where the final query for this publication was performed on February 26, 2018.
Cell Culture.
Regular cell lines were obtained from ATCC. MDA-MB-231, MCF7, and MDA-MB-175 were maintained in DMEM supplemented with 10% FBS and antibiotic. The base medium for the Hs578T was DMEM supplemented with 10 g/mL insulin and 10% FBS. MDA-MB-436 was cultured in Leibovitz’s L-15 medium with 10 g/ml insulin, 16 g/ml glutathione, and 10% FBS. The SUM159 was maintained in Ham’s F-12 medium with 5% FBS, 5 g/mL insulin, and 1 g/mL hydrocortisone. The WHIM12 cells which were derived from the WHIM12 CLOW human patient derived xenograft (7, 40) were cultured in Ham’s F-12 medium containing 5% FBS with antibiotic and supplements (50 ng/mL sodium selenite, 50 g/mL 3,3’,5-triiodo-l-thyronine, 5 g/mL transferrin, 5 mM ethanolamine, 1 g/mL hydrocortisone, 5 g/mL insulin, 10 ng/mL epidermal growth factor, and 2 mM l-glutamine). This cell line was a gift from S. Li and the HAMLET core at Washington University in St. Louis, where this cell line can be obtained (icts.wustl.edu/icts-researchers/icts-cores/find-services/by-core-name/human-mouse-linked-evaluation-of-tumors). HCC1569 cells were gifted by Meghana Trivedi, University of Houston, Houston. HMLE cells, HMLE-SNAIL cells, and HMLE-TWIST cells were provided by C. Cheng, Baylor College of Medicine, Houston. HMLE-SNAIL and -TWIST cells were generated by infecting immortalized human mammary epithelial cells (41) with retroviral vectors expressing SNAI1 or TWIST1 or the control vector and selecting with 1 g/mL puromycin.
Mouse Xenografts.
All animal experiments were carried out within the guidelines recommended for care and use of laboratory animals by the National Institutes of Health. All animal procedures were approved by the Institutional Animal Care and Use Committee at Baylor College of Medicine (Houston) (protocol no. AN-6934). Athymic nude female mice were purchased from Jackson Laboratories. A total of 3 × cells were injected s.c. into a mammary fat pad of the mouse. Four mice were injected per group. Tumor volumes were measured by caliper every 3–4 d and calculated by V = (length) × /2. P value was assessed by Student t test using GraphPad Prism7.
IP and Mass-Spectrometry Analysis.
Lyastes were prepared from DPYSL3 knocked down WHIM12 cells and luciferase knocked down WHIM12 cells with NETN lysis buffer [50 mM TrisHCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40 with protease inhibitors]. After incubation with anti-DPYSL3 antibody, DPYSL3 complex was pulled down by Dynabeads Protein A. Beads were washed with lysis buffer four times, and then protein complex was resolved on SDS/PAGE. Each gel lane was split into four regions and digested with trypsin overnight. Digested peptides were analyzed with a Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) coupled to an EASY-nLC 1200 UHPLC (Thermo Fisher Scientific). Sample was loaded onto a 2-cm by 100-m ID C18 precolumn and resolved on a 5-cm by 150-m ID column packed with sub2 m C18 beads (Reprosil-Pur Basic C18, catalog no. r119.b9.0003, Dr. Maisch GmbH) over 75-min linear gradient. The gradient phase consisted of water + 0.1% formic acid (buffer A) and 90% acetonitrile + 0.1% formic acid (buffer B), and was ran from 5% A to 28% B. The resulting raw files were searched by using the Mascot search engine (Version 2.5.1, Matrix Science) (42), validated with Percolator (Version 2.05) (43), and grouped with custom in-house software.
In Vitro Proliferation Assay.
A total of 200 cells were plated into 96-well cell culture plates. At 2 d later, the plates were placed into the IncuCyte Zoom Live-content imaging system (Essen Bioscience). And images of cell movement were taken at regular intervals within a 96-h data collection period. The medium was changed once during the periods. Confluence level was calculated for data analysis.
Scratch Assay.
Cells were plated at 20,000 cells per well for WHIM12, or 30,000–40,000 cells per well for Hs578T, MDA-MB-436, MDA-MB-231, MCF7, ZR75.1, HCC1569, or SUM159 in 96-well image lock plates for 24 h before treating with DMSO, MMC (final concentration was 0.5 g/ml), and/or FRAX597 (1 M). A wound maker was used to scratch and cells were removed from a discrete area of the monolayer to form a cell-free zone into which cells at the edges of the wound can migrate. Cells were imaged at 10× magnification in an IncuCyte Zoom Live-content imaging system (Essen Bioscience) at 37°C, 5% CO2. Images were acquired every 1 or 2 h for >24 h, with two images per well. Data were analyzed by using IncuCyte analysis software and GraphPad Prism7.
Supplementary Material
Acknowledgments
We thank Dr. Dan Cao, Mr. Ari Gao, Dr. Meenakshi Anurag, Dr. Eric Chang, Dr. Kimberly R. Holloway, Dr. Svasti Haricharan, and Mr. Nindo Punturi for helpful discussions; Dr. Alyson Fournier for providing expression vectors of V5-DPYSL3 and reagents; Dr. Sendurai A. Mani for a gift of the SNAI1 and TWIST1 expression vectors; Dr. Meghana Trivedi for gifting HCC1569 cells; and the patients and their families for the provision of samples to generate the WHIM series of breast cancer xenografts. This work was primarily supported by Cancer Prevention Institute of Texas Recruitment of Established Investigators Award RR14033 (to M.J.E.); the McNair Foundation; and the Eads Fund for Metastatic Breast Cancer Research. This work was also supported by Susan G. Komen for the Cure Grants BCTR0707808, KG090422, and PG12220321 (to M.J.E.) for PDX work. The WHIM 12 cells and PDX were provided by the HAMLET core at Washington University in St. Louis which was supported by Clinical and Translational Science Awards Grant UL1 RR024992. Additional support included the Breast Cancer Research Foundation (M.J.E. and C.M.P.); National Cancer Institute Grant R01-CA182467 (to C.C.), Breast SPORE Program Grants P50-CA58223 and R01-CA148761 (to C.M.P.), and Grant U24-CA143848 (to C.M.P.) for support of The Cancer Genome Atlas Breast Cancer Project; Cancer Center Support Grant NCI-P30 CA125123 (to C.K.O.); National Cancer Institute Grant U54-CA233223 (to M.J.E.); CPRIT Proteomics and Metabolomics Core Facility Award RP170005 (to A.M.); Clinical Proteomic Tumor Analysis Consortium funding (M.J.E.) for proteomics analysis of breast cancer CA160035 and U01-CA214125; and National Institutes of Health Grant T32-GM088129 (to J.T.L.). Research fellowships were awarded to R.M. from Hamamatsu Oncology Center, Japan; to A.H. from Yonsei University Wonju College of Medicine in Korea; and to L.R.d.S. from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior–Brasil Finance Code 001.
Footnotes
Conflict of interest statement: M.J.E. received ad hoc consulting fees from Pfizer, AstraZeneca, Sermonix, Celgene, Nanostring, Puma, and Novartis, as well as patents and royalty income for PAM50-based diagnostics to Nanostring/Prosigna. C.M.P. and M.J.E. are equity stockholders, consultants, and board of director members of BioClassifier LLC, are listed as inventors on patent applications on the Breast PAM50 assay, and receive income from the “Prosigna” breast cancer assay. All other authors have no potential conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810598115/-/DCSupplemental.
References
- 1.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herschkowitz JI, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prat A, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weigelt B, et al. Metastatic breast carcinomas display genomic and transcriptomic heterogeneity. Mod Pathol. 2015;28:340–351. doi: 10.1038/modpathol.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabatier R, et al. Claudin-low breast cancers: Clinical, pathological, molecular and prognostic characterization. Mol Cancer. 2014;13:228. doi: 10.1186/1476-4598-13-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4:1116–1130. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang KL, et al. Proteogenomic integration reveals therapeutic targets in breast cancer xenografts. Nat Commun. 2017;8:14864. doi: 10.1038/ncomms14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goshima Y, Nakamura F, Strittmatter P, Strittmatter SM. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature. 1995;376:509–514. doi: 10.1038/376509a0. [DOI] [PubMed] [Google Scholar]
- 10.Gaetano C, Matsuo T, Thiele CJ. Identification and characterization of a retinoic acid-regulated human homologue of the unc-33-like phosphoprotein gene (hUlip) from neuroblastoma cells. J Biol Chem. 1997;272:12195–12201. doi: 10.1074/jbc.272.18.12195. [DOI] [PubMed] [Google Scholar]
- 11.Quach TT, et al. Collapsin response mediator protein-3/unc-33-like protein-4 gene: Organization, chromosomal mapping and expression in the developing mouse brain. Gene. 2000;242:175–182. doi: 10.1016/s0378-1119(99)00528-4. [DOI] [PubMed] [Google Scholar]
- 12.Fukada M, et al. Molecular characterization of CRMP5, a novel member of the collapsin response mediator protein family. J Biol Chem. 2000;275:37957–37965. doi: 10.1074/jbc.M003277200. [DOI] [PubMed] [Google Scholar]
- 13.Minturn JE, Fryer H, Geschwind DH, Hockfield S. TOAD-64, a gene expressed early in neuronal differentiation in the rat, is related to unc-33, a C. elegans gene involved in axon outgrowth. J Neurosci. 1995;15:6757–6766. doi: 10.1523/JNEUROSCI.15-10-06757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuasa-Kawada J, et al. Axonal morphogenesis controlled by antagonistic roles of two CRMP subtypes in microtubule organization. Eur J Neurosci. 2003;17:2329–2343. doi: 10.1046/j.1460-9568.2003.02664.x. [DOI] [PubMed] [Google Scholar]
- 15.Yoshimura T, et al. GSK-3 regulates phosphorylation of CRMP-2 and neuronal polarity. Cell. 2005;120:137–149. doi: 10.1016/j.cell.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 16.Ip JP, Fu AK, Ip NY. CRMP2: Functional roles in neural development and therapeutic potential in neurological diseases. Neuroscientist. 2014;20:589–598. doi: 10.1177/1073858413514278. [DOI] [PubMed] [Google Scholar]
- 17.Charrier E, et al. Collapsin response mediator proteins (CRMPs) Mol Neurobiol. 2003;28:51–63. doi: 10.1385/MN:28:1:51. [DOI] [PubMed] [Google Scholar]
- 18.Alabed YZ, Pool M, Tone SO, Fournier AE. Identification of CRMP4 as a convergent regulator of axon outgrowth inhibition. J Neurosci. 2007;27:1702–1711. doi: 10.1523/JNEUROSCI.5055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosslenbroich V, et al. Collapsin response mediator protein-4 regulates F-actin bundling. Exp Cell Res. 2005;310:434–444. doi: 10.1016/j.yexcr.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Gao X, et al. Expression profiling identifies new function of collapsin response mediator protein 4 as a metastasis-suppressor in prostate cancer. Oncogene. 2010;29:4555–4566. doi: 10.1038/onc.2010.213. [DOI] [PubMed] [Google Scholar]
- 21.Li K, et al. Manipulation of prostate cancer metastasis by locus-specific modification of the CRMP4 promoter region using chimeric TALE DNA methyltransferase and demethylase. Oncotarget. 2015;6:10030. doi: 10.18632/oncotarget.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou W, et al. Upregulation of CRMP4, a new prostate cancer metastasis suppressor gene, inhibits tumor growth in a nude mouse intratibial injection model. Int J Oncol. 2015;46:290–298. doi: 10.3892/ijo.2014.2705. [DOI] [PubMed] [Google Scholar]
- 23.Hiroshima Y, et al. Collapsin response mediator protein 4 expression is associated with liver metastasis and poor survival in pancreatic cancer. Ann Surg Oncol. 2013;20:369–378. doi: 10.1245/s10434-012-2491-3. [DOI] [PubMed] [Google Scholar]
- 24.Li C, et al. Enhancing DPYSL3 gene expression via a promoter-targeted small activating RNA approach suppresses cancer cell motility and metastasis. Oncotarget. 2016;7:22893. doi: 10.18632/oncotarget.8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira B, et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tone SO, Dayanandan B, Fournier AE, Mandato CA. GSK3 regulates mitotic chromosomal alignment through CRMP4. PLoS One. 2010;5:e14345. doi: 10.1371/journal.pone.0014345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bollong MJ, et al. A vimentin binding small molecule leads to mitotic disruption in mesenchymal cancers. Proc Natl Acad Sci USA. 2017;114:E9903–E9912. doi: 10.1073/pnas.1716009114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruggles KV, et al. Methods, tools and current perspectives in proteogenomics. Mol Cell Proteomics. 2017;16:959–981. doi: 10.1074/mcp.MR117.000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mertins P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo H, Xia B. Collapsin response mediator protein 4 isoforms (CRMP4a and CRMP4b) have opposite effects on cell proliferation, migration, and invasion in gastric cancer. BMC Cancer. 2016;16:565. doi: 10.1186/s12885-016-2593-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y, et al. Inhibition of cell-adhesion protein DPYSL3 promotes metastasis of lung cancer. Respir Res. 2018;19:41. doi: 10.1186/s12931-018-0740-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khazaei MR, et al. Collapsin response mediator protein 4 regulates growth cone dynamics through the actin and microtubule cytoskeleton. J Biol Chem. 2014;289:30133–30143. doi: 10.1074/jbc.M114.570440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin PC, Chan PM, Hall C, Manser E. Collapsin response mediator proteins (CRMPs) are a new class of microtubule-associated protein (MAP) that selectively interacts with assembled microtubules via a taxol-sensitive binding interaction. J Biol Chem. 2011;286:41466–41478. doi: 10.1074/jbc.M111.283580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan M, et al. CRMP4 and CRMP2 interact to coordinate cytoskeleton dynamics, regulating growth cone development and axon elongation. Neural Plast. 2015;2015:1–13. doi: 10.1155/2015/947423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li QF, et al. Critical role of vimentin phosphorylation at Ser-56 by p21-activated kinase in vimentin cytoskeleton signaling. J Biol Chem. 2006;281:34716–34724. doi: 10.1074/jbc.M607715200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goto H, et al. Phosphorylation and reorganization of vimentin by p21-activated kinase (PAK) Genes Cells. 2002;7:91–97. doi: 10.1046/j.1356-9597.2001.00504.x. [DOI] [PubMed] [Google Scholar]
- 38.Murray ME, Mendez MG, Janmey PA. Substrate stiffness regulates solubility of cellular vimentin. Mol Biol Cell. 2014;25:87–94. doi: 10.1091/mbc.E13-06-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parsana P, Amend SR, Hernandez J, Pienta KJ, Battle A. Identifying global expression patterns and key regulators in epithelial to mesenchymal transition through multi-study integration. BMC Cancer. 2017;17:447. doi: 10.1186/s12885-017-3413-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toneff M, et al. The Z-cad dual fluorescent sensor detects dynamic changes between the epithelial and mesenchymal cellular states. BMC Biol. 2016;14:47. doi: 10.1186/s12915-016-0269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. ELECTROPHORESIS An Int J. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 43.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.