Abstract
We determined the prevalence and genetic characteristics of methicillin-resistant Staphylococcus aureus (MRSA) isolated from pigs and humans between September 2013 and February 2015 in Kogi State, a central region in Nigeria. A total of 680 nasal swabs were collected and analyzed from pigs (n = 425) and “pig-contact” humans (n = 55) on 35 farms, and “non-pig-contact” humans (n = 200). MRSA was recovered from 20 (4.7%) pigs on 12 farms and 18 (7.0%) humans. Six (2.4%) of the human isolates were recovered from “pig-contact” humans, of which only three work on farms also harboring MRSA positive pigs. All 38 MRSA were resistant to β-lactams only, belonged to spa type t1603, sequence type (ST) 88, and mecA was associated with a SCCmec IVa element. Four isolates from a pig, a pig-contact human from the same farm, a pig-contact human from a pig farm in a different district, and a non-pig-contact human were subjected to whole genome sequencing (WGS). Core genome SNP analysis revealed high genetic similarity between strains (3–11 SNP differences), despite the temporal (2 year gap) and geographic (165 km) differences between isolates. Furthermore, these Nigerian isolates form a distinct clade when compared to other African MRSA ST88 isolates. All but one porcine strain was positive for scn suggesting a possible human origin and that pigs were either transiently contaminated by humans or result of a very recent human-to-pig transmission event. To our knowledge, this is the first report of genetically confirmed MRSA in pigs in Nigeria, which appear to be a typical CA-MRSA clone present in the human population.
Keywords: CA-MRSA, porcine, ST88, staphylococci, zoonotic
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is of great concern in both human and veterinary medicine (Vanderhaeghen et al., 2010). Such concerns are related to difficulty in treating infections, prolonged hospitalization and increased health care costs. Pigs can be asymptomatic carriers of S. aureus including methicillin-resistant strains, although infections with S. aureus are infrequent (van der Wolf et al., 2012). The central concern with MRSA in pigs is the risk of spread to humans (Unnerstad et al., 2017).
MRSA sequence type (ST) 398 is the predominant clone observed in pigs from Europe and North America, whereas MRSA ST9 is predominant in Asia. In Africa, there are only four reports of MRSA in pigs; two in South Africa (Adegoke and Okoh, 2014; Van Lochem et al., 2018), one in Senegal (Fall et al., 2012), and one in Nigeria (Okunlola and Ayandele, 2015). Unfortunately, only two of the four studies used molecular methods to confirm the S. aureus species identification and the presence of mecA in oxacillin or cefoxitin resistant strains, and only one study further typed the MRSA strains. Fall et al. (2012) reported that their porcine MRSA belonged mainly to ST5 and a singleton ST88, which are both major MRSA lineages responsible for human infections in Senegal (Breurec et al., 2011). Pigs have been shown to harbor human-associated MRSA clones e.g., USA300 (Arriola et al., 2011; Baez et al., 2017). People in close contact with pigs (farmers, veterinarians, transporters, and slaughterhouse workers) have been shown to be at a greater risk of being colonized with livestock associated MRSA (LA-MRSA) (Denis et al., 2009; Liu et al., 2015). However, LA-MRSA has been reported in people with no known contact to pigs or pig farms (Larsen et al., 2017). In Nigeria, there are no published data regarding colonization of pigs with MRSA and possible transmission of strains between pigs and humans. Our objective was to evaluate the occurrence of MRSA in pigs, and compare the strains to those isolated from pig-contact and non-contact humans.
Materials and Methods
Ethical Statement
The study and protocol were approved by the institutional review board of Kogi State University Teaching Hospital. Informed oral consent was obtained from each participant and pig farm owners consented to the sampling of their animals.
Sample Collection
The study was conducted between September 2013 and February 2015 and covered 16/21 local government areas of Kogi State (Figure 1). Samples were collected from 35 pig farms housing between 28 and 275 pigs per farm. Between 10 and 15 pigs were randomly sampled per farm. When pigs were housed in pens (i.e., on 5/35 farms), two pigs per pen were sampled. On those farms not housing pigs in pens, pigs were randomly selected. Human samples were collected from 55 pig farms workers (pig contact humans). The number of farm workers per farm ranged from 1 to 6 individuals (mode = 1). Two hundred students attending Kogi State University in Anyigba that had no pig contact were also sampled (non-pig-contact humans). All human participants were provided with a standardized questionnaire to collect demographic data, medical history over the previous 12 months e.g., hospitalization and antimicrobial therapy, and information about contact with pigs.
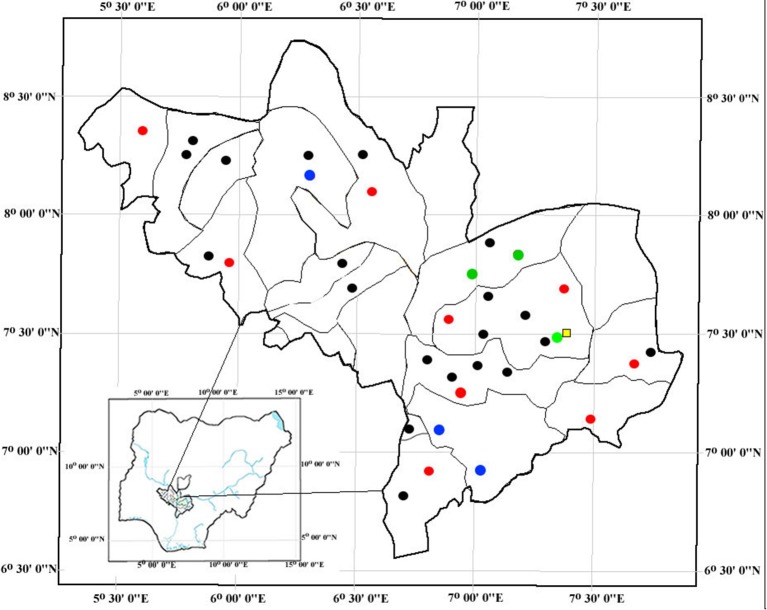
Figure 1.
Geographic location of pig farms in the 21 local government areas and Kogi State University (represented by a yellow square). Black dots indicate MRSA negative pig farms. Red dots indicate MRSA positive pig farm where only pigs were positive. Blue dots indicate pig farms where only humans were positive and no pigs. Green dots indicate pig farms harboring both MRSA positive pigs and humans.
Isolation and Characterization of MRSA
Nasal swabs were collected with a sterile cotton-tip swab inserting into both anterior nares of each pig and human and were placed into 3 ml Mueller Hinton broth (Oxoid, UK) supplemented with 6.5% sodium chloride and incubated at 37°C for 24h. A loopful of the inoculum from the enrichment broth was streaked on Brilliance agar (Oxoid, UK). After incubation at 37°C for 24h, suspected MRSA colonies (denim blue colonies) were sub-cultured on 5% sheep blood agar and incubated at 37°C for 24h. Isolates were speciated by matrix-assisted laser desorption/ionization time-of-flight mass spectrophotometry (MALDI-TOF MS) (BioMérieux, France). Confirmed S. aureus isolates were stored at −20°C for further analysis. Antimicrobial susceptibility testing was done using the Sensititre system and COMPAN1F MIC plate (ThermoFisher Scientific, USA), and susceptibility was interpreted according to CLSI (Clinical and Laboratory Standards Institute (CLSI), 2014). MRSA were characterized by a multiplex PCR to detect spa, mecA, lukF-PV (a marker of the Panton-Valentine leucocidin, PVL), scn and a CC398 specific band (Islam et al., 2017). Direct sequencing of the purified PCR reaction was used for spa typing (Harmsen et al., 2003). Four isolates (one pig, one pig-contact human from the same pig farm, one non-pig-contact human from the same district, and one pig-contact human from a different district) were selected for further SCCmec (Kondo et al., 2007), multi locus sequencing typing (Enright et al., 2000), and whole genome sequencing (WGS).
Whole-Genome Sequencing
Overnight cultures were grown in tryptic soy broth at 37°C with 200-rpm shaking. Genomic DNA was extracted from cultures by using the automated Maxwell DNA instrument using the Maxwell® RSC Cultured Cells DNA Kit (Promega, United Kingdom). Library preparation was carried out using the Nextera XT kit and paired end 2 × 250 bp sequencing on the MiSeq, all following standard Illumina protocols (Illumina, Inc., United States). All raw reads have been deposited in the Sequence Read Archive database in the European Nucleotide Archive under the study accession number PRJEB26533.
Phylogenetics and Comparative Genomics
All analyses were performed in CLC Genomics Workbench v. 11.0 using the tools within the Microbial Genomics Module. First, Kmer analysis was performed against the NCBI RefSeq database and including only complete staphylococcal genomes. This analysis was performed to determine if there was any contamination of the DNA prior to sequencing and as a secondary confirmation of the S. aureus species identification. Secondly, Adapter trimmed, high quality reads were mapped against the reference genome AUS0325, a closed ST88 methicillin-susceptible, penicillin-resistant genome (NCBI accession no. LT615218) to identify single nucleotide polymorphisms (SNPs). A maximum likelihood tree was generated from core genome SNPs. To further investigate the clonal relationship of MRSA ST88 from the African continent, our strains were compared to the 17 publically available ST88 MRSA isolated in Ghana (Kpeli et al., 2017). The Resfinder v.3.0 and Virulence finder v.1.5 web-based pipelines were used to search for the presence of known virulence and antibiotic resistance genes (Zankari et al., 2012; Joensen et al., 2014).
Result
MRSA was recovered from 20/425 (4.7%) pigs, 6/55 (10.9%) pig farms workers, and 12/200 (6.0%) non-pig contact humans (Table 1). The MRSA positive pigs came from 12 farms (Figure 1). Despite sampling multiple pigs per farm, only 4/12 farms had >1 MRSA positive pigs. Six of the human MRSA isolates were recovered from “pig contact” humans on six pig farms. However, only 3/6 individuals came from farms, which also harbored MRSA positive pigs. Moreover, despite sampling more than one pig farm worker on these six farms, only one individual per farm was positive. The remaining 12 MRSA were isolated from University students not having any pig contact. None of the MRSA positive individuals had reported taking antibiotics and only one pig farm worker had reported being hospitalized in the last 12 months prior to sampling.
Table 1.
Characterization of MRSA isolated from pigs, pig farm workers and non-pig contact humans.
| Number of sampled pigs/humans | Number of pig farms | Number of MRSA positive farms | Number of MRSA positive samples | Location of MRSA positive farms/students | Sampling period | Presence of scn amongst MRSA isolates | |
|---|---|---|---|---|---|---|---|
| Pigs | 425 | 35 | 12 | 20 | Ankpa, Anyigba, Echeno, Egbe, Ejule, Itama, Lokoja, Ogidi, Oguma, Okpo, Okura-Offante, Sheria | Nov 2013–Jan 2015 | 19 |
| Pig farm workers | 55 | 35 | 6* | 6 | Ajaka, Anyigba, Elechi, Kabba, Oguma, Sheria | Nov 2013–Jan 2015 | 6 |
| Kogi State University students | 200 | – | – | 12 | Anyigba | May–Nov 2014 | 12 |
3/6 farms harbored MRSA positive pigs. Underlined location of the farms.
All 38 MRSA isolates were resistant to penicillin and cefoxitin only and there was agreement between phenotypic and inferred genotypic resistance. All strains were positive for the CC398 specific band. However, spa sequencing demonstrated that all strains had the same spa type, t1603, which is unrelated to spa types typically associated with CC398. Furthermore, MLST demonstrated that strains belong to ST88. In silico PCR using the CC398 specific primers, which target the C398-specific sau1-hsdS1 variant, (FP2sau1: 5′GAGAATGATTTTGTTTATAACCCTAG3′ and CC398r1: 5′-CAGTATAAAGAGGTGACATGACCCCT-3′) and our de novo assembled contigs, showed that the primers bind 100% and produce the expected 106 bp amplicon. The primers bind to an open reading frame (ORF) with 99% nucleotide identity to hsdS, which is the sequence specificity gene of the Sau1 type I restriction-modification system (data not shown). mecA was harbored on an SCCmec type VIa cassette.
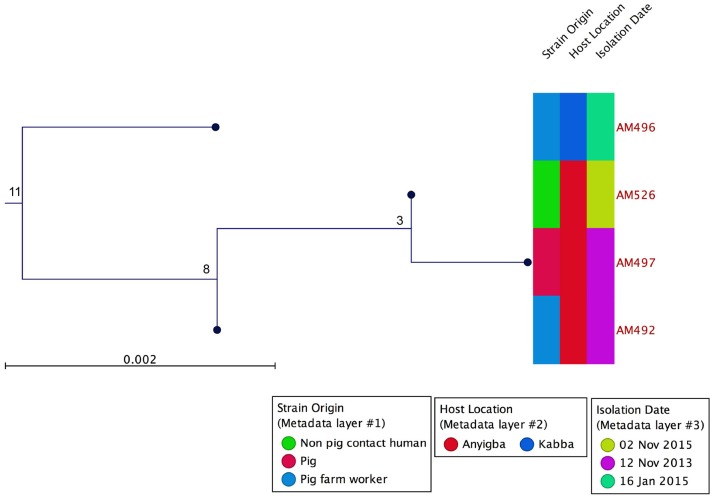
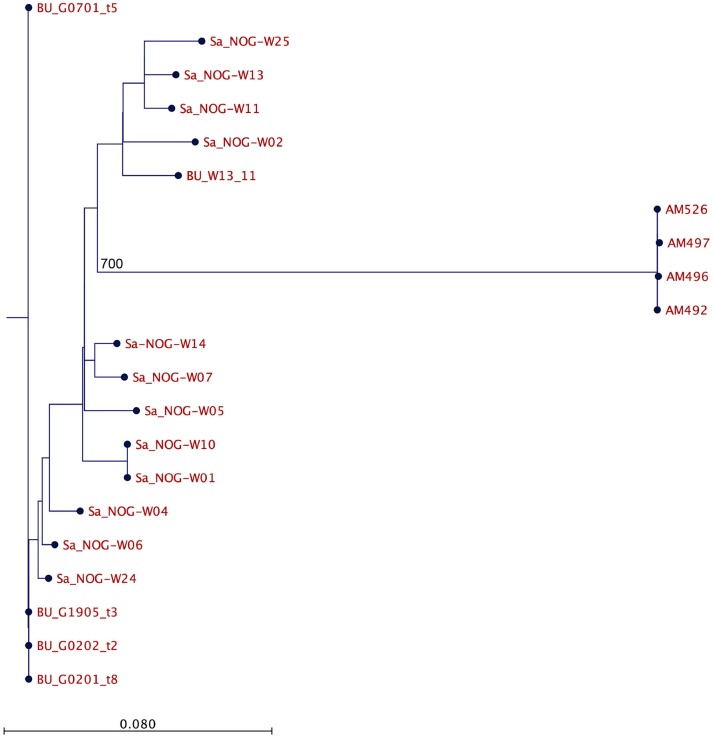
Based on WGS, the core genome consisted of 2407 coding DNA sequences (CDS) with S. aureus AUS0325 used as reference. A core genome phylogeny was inferred by mapping reads from the four sequenced isolates to AUS0325, and a phylogenetic tree was constructed using the core genome SNPs and maximum likelihood estimation. Relative to AUS0325, 1,248 SNPs were identified, but only 11 SNPs were found between the four isolates in this study despite the temporal and geographic differences (Table 2; Figure 2). Isolates from the same region i.e., Angyiba and despite a 2-year gap only differed by 3–8 SNPs. A phylogenetic comparison with the 17 ST88 from Ghana indicates that the Nigerian isolates formed a separate, distinct clade with 700 SNPs differences (Figure 3).
Table 2.
Single nucleotide polymorphisms in AM492, AM496, AM497, and AM526 relative to the ST88 reference genome, AUS0325.
| Nucleotide position | AM526 | AM497 | AM492 | AM496 |
|---|---|---|---|---|
| 853428 | T | T | C | C |
| 853440 | C | Gap | Gap | Gap |
| 853452 | C | Gap | Gap | Gap |
| 1349949 | T | G | T | T |
| 2673340 | G | G | G | A |
| 2673364 | A | A | A | G |
| 2673388 | G | G | Gap | Gap |
| 2673400 | G | G | Gap | Gap |
| 2673412 | Gap | Gap | Gap | G |
| 2673424 | Gap | Gap | G | G |
| 2673436 | Gap | Gap | G | G |
Figure 2.
Core genome phylogeny using AUS0325 as the reference genome. MRSA ST88 from a pig, pig contact human and a non-pig contact human. Phylogeny based on an alignment of 1,248 SNPs. Metadata describing sample origins, sampling location in Kogi State and isolation date is provided. The numbers indicate SNP differences.
Figure 3.
Comparative analysis of the Nigerian and Ghanaian MRSA ST88. Core genome phylogeny based on an alignment of 1492 SNPs. The number in the figure indicates SNP differences.
All MRSA strains were pvl negative and 37/38 strains contained the human immune evasion gene scn. The only strain that was scn-negative was of porcine origin. Furthermore, based on analysis of the de novo contigs in Virulence Finder, all four strains contained the following putative virulence associated genes scn, sak, lukE, lukD, gamma hemolysin, aur, splA, and splB.
Discussion
We describe for the first time the isolation of genetically confirmed MRSA in pigs and pig farm workers in Nigeria. This is only the fifth description of MRSA in healthy pigs on the African continent and appears not to be the typical livestock associated clones found in pigs worldwide namely CC398 and CC9.
All MRSA isolates found in this study belonged to ST88, which has been isolated from both hospital- and community- acquired infections in humans in several Sub-Saharan African countries, including Nigeria (Ghebremedhin et al., 2009; Shittu et al., 2012; Kolawole et al., 2013; Raji et al., 2013; Abdulgader et al., 2015). ST88 appears to be primarily associated with the African continent, but has also been reported in Asia and sporadically in other parts of the world (Schaumburg et al., 2014). The association of ST88 and livestock has only been described in Africa. Specifically, MRSA ST88 has been identified in one pig isolate from Senegal and healthy sheep in the Ivory Coast (Fall et al., 2012; Schaumburg et al., 2015). In China, MRSA ST88 has been found in food of animal origin and recently in livestock workers (Wang et al., 2014; Ye et al., 2016).
MRSA in pigs and pig farmers in Africa was first reported in Senegal (Fall et al., 2012) however, no MRSA was isolated from the farm workers. In Nigeria, there has been only one report of phenotypic oxacillin-resistant S. aureus in pigs from the Oyo State in South Western, Nigeria (Okunlola and Ayandele, 2015). Only 18/200 (9%) pigs were positive for presumptive MRSA on 7 of the 11 farms sampled. MRSA has been isolated from other animals in Nigeria namely; camels, sheep, cattle, goats and recently in poultry (Mai-siyama et al., 2014; Nworie et al., 2017). Similar to (Okunlola and Ayandele, 2015), we observed few MRSA positive animals on each farm. Moreover, all our MRSA isolates in this study belong to the same spa type (t1603) irrespective of the source (pig, pig-contact human or non-pig-contact human) and based on WGS the high genetic similarity, raises questions about the origin of these strains. The immune evasion cluster (IEC), which is carried on a bacteriophage and encodes the secreted proteins staphylococcal complement inhibitor (scn), staphylokinase (sak), and chemotaxis inhibitory protein (chp), are thought to contribute to the immune evasion in humans (van Wamel et al., 2006). Moreover, these genes are less prevalent in livestock adapted S. aureus lineages and are hence considered good genetic markers for identification of human associated S. aureus clones (McCarthy et al., 2011, 2012; Uhlemann et al., 2012). We found a high incidence of scn amongst our porcine isolates (19/20 isolates) and the additional occurrence of sak based on WGS data. The presence of these IEC related genes suggest a possible human origin and that pigs were either transiently contaminated by farm workers or the result of a very recent human-to-pig transmission event. Human associated S. aureus lineages have been previously described in animals including pigs (Arriola et al., 2011; Schaumburg et al., 2012; Nagel et al., 2013; Baez et al., 2017).
The MRSA ST88 in our study was only resistant to β-lactams. Typically, antibiotic resistance amongst animal S. aureus isolates tend be higher compared to human isolates. A study of methicillin resistant coagulase-negative staphylococci (MRCoNS) of porcine origin in Nigeria, found that isolates were multi-drug resistant (MDR), 85% of strains were resistant to fusidic acid, and harbored a number of different antibiotic resistance genes (Ugwu et al., 2015). CoNS are a reservoir of antibiotic resistance genes with the potential to transfer to S. aureus, as has been demonstrated with the plasmid-mediated transfer of mupA, conferring mupirocin resistance, from S. haemolyticus to S. aureus (Rossi et al., 2016). It is possible that these low-level resistant MRSA ST88 strains if allowed to adapt to pigs and occupy the same niche as MDR MRCoNS, provide an opportunity for the transfer of antibiotic resistance genes to potentially create MDR CA-MRSA.
All our strains were presumptively identified as belonging to CC398. However, upon additional strain typing, this was shown to be incorrect. Typing of MRSA is key for surveillance, epidemiological studies and infection control. Rapid, reliable identification methods such as PCR based methods are important tools to identify MRSA and type to the clonal complex level. The CC398 specific PCR is an excellent tool to rapidly identify S. aureus belonging to CC398, which was shown to be 100% specific for CC398 only (Stegger et al., 2011). This PCR is based on sau1-hsdS since it was previously shown that variations in this gene is associated with specific S. aureus clonal lineages but maintained a high sequence homology within the lineage (Waldron and Lindsay, 2006). This approach has also been used for rapid identification of hospital-acquired MRSA lineages (Cockfield et al., 2007). Unfortunately, at the time of the study by Stegger et al. (2011) there were no publically available ST88 genomes and strains belonging to ST88 were not included in the study since ST88 is the not the typical LA-MRSA and it is not a globally disseminated human associated clone. In light of our findings, we recommend that with the current PCR primers the CC398 specific PCR result should be complemented with spa typing or another method to verify the clonal lineage association.
Our study has some limitations. Ideally, all 38 MRSA strains should be whole genome sequenced to better understand ST88 strain diversity in this region and study possible transmission events. However, the latter would be difficult to assess as we do not have information about the movement of pigs e.g., via trade, and the contact and movement of people. Additionally, we could not sequence all strains because of financial limitations.
Livestock are known reservoirs of new pathogenic bacteria with the ability to cross the host species barrier, and evolve in a host-specific manner to become established in the human population. This has been demonstrated for LA-MRSA CC398 (Price et al., 2013). Further research is required in Nigeria to determine whether MRSA carriage amongst pigs was transient or persistent, establish the exact transmission routes on pig farms and explore biosecurity measures for preventing the spread of MRSA in the farm environment. While there are limitations to our study; cross-sectional study design conducted at one time point only, nonetheless we provide valuable information on a continent where there are limited studies describing the occurrence of MRSA in animals and even fewer providing molecular typing data.
Author Contributions
OO, JK, and EO designed of the work. OO, JK, EO, MI, and AM performed the acquisition, the analysis, and interpretation of data. OO, JK, EO, MI, and AM drafted and revised the manuscript. All authors approved the final version of this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Tatjana P. Kristensen and Manja Hanegård for technical assistance.
Footnotes
Funding. Sampling, isolation, and preliminary analysis of MRSA was self financed by OO as part of this PhD project. OO received a travel grant from Kogi State University for a short research stay to perform molecular characterization of MRSA at the University of Copenhagen, Denmark. The molecular characterization was funded by the Danish Council for Independent Research-Technology and Production Sciences (DFF-FTP).
References
- Abdulgader S. M., Shittu A. O., Nicol M. P., Kaba M. (2015). Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Africa: a systematic review. Front. Microbiol. 30:348 10.3389/fmicb.2015.00348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adegoke A. A., Okoh I. A. (2014). Species diversity and antibiotic resistance properties of Staphylococcus of farm animal origin in Nkonkobe Municipality, South Africa. Folia. Microbiol. 59, 133–140. 10.1007/s12223-013-0275-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriola C. S., Güere M. E., Larsen J., Skov R. L., Gilman R. H., Gonzalez A. E., et al. (2011). Presence of methicillin-resistant Staphylococcus aureus in pigs in Peru. PLoS ONE. 6:e28529. 10.1371/journal.pone.0028529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baez M., Collaud A., Espinosa I., Perreten V. (2017). MRSA USA300, USA300-LV and ST5-IV in pigs, Cuba. Int. J. Antimicrob. Agents. 49, 259–261. 10.1016/j.ijantimicag.2016.12.001 [DOI] [PubMed] [Google Scholar]
- Breurec S., Zriouil S. B., Fall C., Boisier P., Brisse S., Djibo S., et al. (2011). Epidemiology of methicillin-resistant Staphylococcus aureus lineages in five major African towns: emergence and spread of atypical clones. Clin. Microbiol. Infect. 17, 160–165. 10.1111/j.1469-0691.2010.03219.x [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) (2014). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Information Suplement, Vol. 34 CLSI document M100-24S. Wayne: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cockfield J. D., Pathak S., Edgeworth J. D., Lindsay J. A. (2007). Rapid determination of hospital-acquired meticillin-resistant Staphylococcus aureus lineages. J. Med. Microbiol. 56, 614–619. 10.1099/jmm.0.47074-0 [DOI] [PubMed] [Google Scholar]
- Denis O., Suetens C., Hallin M., Catry B., Ramboer I., Dispas M., et al. (2009). Methicillin- resistant Staphylococcus aureus ST398 in swine farm personnel, Belgium. Emerg. Infect Dis. 15, 1098–1101. 10.3201/eid1507.080652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright M. C., Day N. P., Davies C. E., Peacock S. J., Spratt B. G. (2000). Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38, 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fall C., Seck A., Richard V., Ndour M., Sembene M., Laurent F., et al. (2012). Epidemiology of Staphylococcus aureus in pigs and farmers in the largest farm in Dakar, Senegal. Foodborne Pathog. Dis. 9, 962–965. 10.1089/fpd.2012.1197 [DOI] [PubMed] [Google Scholar]
- Ghebremedhin B., Olugbosi M. O., Raji A. M. (2009). Emergence of a community associated methicillin resistant Staphylococcus aureus strain with a unique resistant profile in southwest Nigeria. J. Clin. Microbiol. 47, 2975–2980. 10.1128/JCM.00648-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen D., Claus H., Witte W., Rothgänger J., Claus H., Turnwald D., et al. (2003). Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41, 5442–5448. 10.1128/JCM.41.12.5442-5448.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M. Z., Espinosa-Gongora C., Damborg P., Sieber R. N., Munk R., Husted L., et al. (2017). Horses in Denmark are a reservoir of diverse clones of methicillin-resistant and -susceptible Staphylococcus aureus. Front. Microbiol. 8:543. 10.3389/fmicb.2017.00543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensen K. G., Scheutz F., Lund O., Hasman H., Kaas R. S., Nielsen E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Micobiol. 52, 1501–1510. 10.1128/JCM.03617-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolawole O. D., Adeyanju A., Schaumburg F., Akinyoola L. A., Lawal O. O., Amusa Y. B., et al. (2013). Characterization of colonizing Staphylococcus aureus isolated from surgical wards' patients in a Nigerian University Hospital. PLoS ONE. 8:e68721. 10.1371/journal.pone.0068721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y., Ito T., Ma X. X., Watanabe S., Kreiswirth B. N., Etienne J., et al. (2007). Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 51, 264–274. 10.1128/AAC.00165-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kpeli G., Buultjens A. H., Giulieri S., Owusu-Mireku E., Aboagye S. Y., Baines S. L. (2017). Genomic analysis of ST88 community-acquired methicillin resistant Staphylococcus aureus in Ghana. PeerJ. 28:e3047 10.7717/peerj.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J., Petersen A., Larsen A. R., Sieber R. N., Stegger M., Koch A., et al. (2017). Emergence of livestock-associated methicillin-resistant Staphylococcus aureus bloodstream infections in Denmark. Clin. Infect. Dis. 67, 1072–1076. 10.1093/cid/cix504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu Z., Yao Z., Fan Y., Ye X., Chen S. (2015). The prevalence and influencing factors of methicillin-resistant Staphylococcus aureus carriage in people in contact with livestock: a systematic review. Am. J. Infect. Cont. 43:469–475. 10.1016/j.ajic.2014.12.009 [DOI] [PubMed] [Google Scholar]
- Mai-siyama I. B., Okon K. O., Adamu N. B., Askira U. M., Isyaka T. M., Adamu S. G., et al. (2014). Methicillin-resistant Staphylococcus aureus (MRSA) colonization rate among ruminant animals slaughtered for human consumption and contact persons in Maiduguri, Nigeria. Afr. J. Microbiol. Res. 8, 2643–2649. 10.5897/AJMR2014.6855 [DOI] [Google Scholar]
- McCarthy A. J., van Wamel W., Vandendriessche S., Larsen J., Denis O., Garcia-Graells C., et al. (2012). Staphylococcus aureus CC398 clade associated with human-to-human transmission. Appl. Environ. Microbiol. 78, 8845–8848. 10.1128/AEM.02398-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy A. J., Witney A. A., Gould K. A., Moodley A., Guardabassi L., Voss A., et al. (2011). The distribution of mobile genetic elements (MGEs) in MRSA CC398 is associated with both host and country. Genome. Biol. Evol. 3, 1164–1174. 10.1093/gbe/evr092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel M., Dischinger J., Türck M., Verrier D., Oedenkoven M., Ngoubangoye B., et al. (2013). Human-associated Staphylococcus aureus strains within great ape populations in Central Africa (Gabon). Clin. Microbiol. Infect. 19, 1072–1077. 10.1111/1469-0691.12119 [DOI] [PubMed] [Google Scholar]
- Nworie A., Onyema A. S., Okekpa S. I., Elom M. O., Umoh N. O., Usanga V. U., et al. (2017). A novel methicillin-resistant Staphylococcus aureus t11469 and a poultry endemic strain t002 (ST5) are present in chicken in Ebonyi State, Nigeria. Biomed. Res. Int. 2936461. 10.1155/2017/2936461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okunlola I. O., Ayandele A. A. (2015). Prevalence and antimicrobial susceptibility of Methicillin-resistant Staphylococcus aureus (MRSA) among pigs in selected farms in Ilora, South Western Nigeria. Euro. J. Exp. Bio. 5, 50–56. [Google Scholar]
- Price L. B., Stegger M., Hasman H., Aziz M., Larsen J., Andersen P. S., et al. (2013). Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio. 3:e00305–e00311. 10.1128/mBio.00305-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raji A., Ojemhen O., Umejiburu U., Ogunleye A., Blanc D. S., Basset P. (2013). High genetic diversity of Staphylococcus aureus in a tertiary care hospital in Southwest Nigeria. Diagn. Microbiol. Infect. Dis. 77, 367–369. 10.1016/j.diagmicrobio.2013.08.030 [DOI] [PubMed] [Google Scholar]
- Rossi C. C., Ferreira N. C., Coelho M. L., Schuenck R. P., Bastos Mdo C., Giambiagi-deMarval M. (2016). Transfer of mupirocin resistance from Staphylococcus haemolyticus clinical strains to Staphylococcus aureus through conjugative and mobilizable plasmids. FEMS Microbiol Lett. 363:fnw121. 10.1093/femsle/fnw121 [DOI] [PubMed] [Google Scholar]
- Schaumburg F., Alabi A. S., Peters G., Becker K. (2014). New epidemiology of Staphylococcus aureus infection in Africa. Clin. Microbiol. Infect. 20, 589–596. 10.1111/1469-0691.12690 [DOI] [PubMed] [Google Scholar]
- Schaumburg F., Mugisha L., Peck B., Becker K., Gillespie T. R., Peters G., et al. (2012). Drug-resistant human Staphylococcus aureus in sanctuary apes pose a threat to endangered wild ape populations. Am. J. Primatol. 74, 1071–1075. 10.1002/ajp.22067 [DOI] [PubMed] [Google Scholar]
- Schaumburg F., Pauly M., Anoh E., Mossoun A., Wiersma L., Schubert G., et al. (2015). Staphylococcus aureus complex from animals and humans in three remote African regions. Clin. Microbiol. Infect. 21, 345.e1-8. 10.1016/j.cmi.2014.12.001 [DOI] [PubMed] [Google Scholar]
- Shittu A., Oyedara O., Abegunrin F., Okon K., Raji A., Taiwo S., et al. (2012). Characterization of methicillin-susceptible and -resistant staphylococci in the clinical setting: a multicentre study in Nigeria. BMC Infect Dis. 12:286. 10.1186/1471-2334-12-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegger M., Lindsay J. A., Moodley A., Skov R., Broens E. M., Guardabassi L. (2011). Rapid PCR detection of Staphylococcus aureus clonal complex 398 by targeting the restriction-modification system carrying sau1-hsdS1. J. Clin. Microbiol. 49, 732–734. 10.1128/JCM.01970-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugwu C. C., Gomez-Sanz E., Agbo I. C., Torres C., Chah K. F. (2015). Characterization of mannitol-fermenting methicillin-resistant staphylococci isolated from pigs in Nigeria. Braz. J. Microbiol. 46, 885–892. 10.1590/S1517-838246320140644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlemann A. C., Porcella S. F., Trivedi S., Sullivan S. B., Hafer C., Kennedy A. D., et al. (2012). Identification of a highly transmissible animal-independent Staphylococcus aureus ST398 clone with distinct genomic and cell adhesion properties. MBio. 3:e00027–e00012. 10.1128/mBio.00027-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unnerstad E. H., Wahlstrom H., Molander B., Bengtsson B. (2017). Methicillin-resistant Staphylococcus aureus not detected in Swedish nucleus and multiplying pig herds. Infect. Eco Epidemiol. 7:1313068 10.1080/20008686.2017.1313068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wolf P. J., Rothkamp A., Junker K., de Neeling A. J. (2012). Staphylococcus aureus (MSSA) and MRSA (CC398) isolated from post-mortem samples from pigs. Vet. Microbiol. 158, 136–141. 10.1016/j.vetmic.2012.01.025 [DOI] [PubMed] [Google Scholar]
- Van Lochem S., Thompson P. N., Annandale C. H. (2018). Prevalence of methicillin-resistant Staphylococcus aureus among large commercial pig herds in South Africa. Onderstepoort. J. Vet. Res. 85:e1–e4. 10.4102/ojvr.v85i1.1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wamel W. J., Rooijakkers S. H., Ruyken M., van Kessel K. P., van Strijp J. A. (2006). The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 188:1310–1315. 10.1128/JB.188.4.1310-1315.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhaeghen W., Hermans K., Haesebrouck F., Butaye P. (2010). Methicillin-resistant Staphylococcus aureus (MRSA) in food production animals. Epidemiol. Infect. 138, 606–625. 10.1017/S0950268809991567 [DOI] [PubMed] [Google Scholar]
- Waldron D. E., Lindsay J. A. (2006). Sau1: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 188, 5578–5585. 10.1128/JB.00418-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li G., Xia X., Yang B., Xi M., Meng J. (2014). Antimicrobial susceptibility and molecular typing of methicillin-resistant staphylococcus aureus in retail foods in Shaanxi, China. Foodborne Pathog. Dis. 11, 281–286. 10.1089/fpd.2013.1643 [DOI] [PubMed] [Google Scholar]
- Ye X., Wang X., Fan Y., Peng Y., Li L., Li S., et al. (2016). Genotypic and phenotypic markers of livestock-associated methicillin-resistant Staphylococcus aureus CC9 in Humans. Appl. Environ. Microbiol. 82, 3892–3899. 10.1128/AEM.00091-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]