Abstract
Epigenetic modifications, including DNA methylation and histone modifications, determine the way DNA is packaged within the nucleus and regulate cell-specific gene expression. The heritability of these modifications provides a memory of cell identity and function. Common dysregulation of epigenetic modifications in cancer has driven substantial interest in the development of epigenetic modifying drugs. Although these drugs have the potential to be highly beneficial for patients, they act systemically and may have “off-target” effects in other cells such as the patients’ sperm or eggs. This review discusses the potential for epigenomic drugs to impact on the germline epigenome and subsequent offspring and aims to foster further examination into the possible effects of these drugs on gametes. Ultimately, the information gained by further research may improve the clinical guidelines for the use of such drugs in patients of reproductive age.
Keywords: epigenetic, inheritance, development, germline, pharmacology, cancer, reproduction
Introduction
Sperm and oocytes (eggs) occupy a unique position in biology, as they transmit genetic and epigenetic information from parent to offspring in sexually reproducing organisms. An individual’s genes provide the primary genetic information that determines phenotypic outcomes in our children—whether they have blue eyes, are suited to sprinting or long distance running, or will be susceptible to certain diseases, and so on. While the DNA contains the primary genetic sequence, chemical modifications to the DNA and associated histone proteins influence how the DNA is organised within the nucleus and whether specific genes are switched on or off. This epigenetic information is critically important for the interpretation of the DNA during development of the foetus and in adult life, strongly influencing cell specification, phenotypic outcomes, and adult health. Moreover, epigenetic modifications are heritable, ensuring that a memory of cell-specific gene activity is transmitted during cell division, facilitating cell and tissue function. Here, we consider epigenetic modifications to include DNA methylation and histone modifications that are mitotically or meiotically stable (or both) and contribute to cellular memory. As the term “epigenetic” has been more broadly interpreted, more in-depth discussions can be found in recent stimulating reviews from Steven Henikoff and John Greally 1, 2.
In addition to regulating cell-specific gene expression profiles, epigenetic mechanisms provide a potential interface between the environment and genomic function, including in the germline. Changes mediated by environmental influences, such as diet, drugs, or chemicals, are thought to alter epigenetic programming in germ cells, resulting in epigenetic differences in sperm and eggs that may alter outcomes in offspring (reviewed in 3– 9). In this context, examples of environmental factors are provided by epigenetic modifying drugs, which have attracted substantial interest in oncology but have been studied in only very limited detail with respect to their impacts on germline epigenetic programming and epigenetic inheritance. In the context of this discussion, “epigenomic drugs” include pharmaceuticals that specifically alter the activity of enzymes or proteins that mediate DNA methylation and histone modifications. Although these drugs have great potential for improving clinical outcomes in patients, they may also directly alter the germline epigenome and potentially have deleterious outcomes for future offspring.
Despite a substantial number of studies examining the impacts of diet and other environmental effects on the germline epigenome and inheritance, the potential impacts of epigenomic therapies on the germline have been largely “off the radar” when assessing drug impacts on patients. This is likely due to the primary focus of clinical trials and treatment on safety and improving patient health, whereas reproductive and offspring health are usually secondary considerations. Clearly, these primary aspects of therapy are of paramount importance, and effective therapies should be used to ensure the best possible outcomes for patients. The purpose of this review is not to vilify epigenomic drugs or to discourage their use by patients or prescription by clinicians. However, given the potential impacts of the germline epigenome on offspring, it is important that future research aims to understand how these drugs might change the germline epigenome and whether such changes affect offspring development and health. In the long term, this information may facilitate the development of guidelines and pre-treatment advice for patients with respect to future reproduction and, if required, recommendations for fertility preservation.
Germline epigenetics: programming outcomes in future offspring
The potential for environmental agents to alter the germline epigenome and offspring phenotype has driven a range of studies in germline development and epigenetics. The current conceptual framework for epigenetic inheritance in mammals is dominated by our understanding of DNA methylation, particularly genomic imprinting, which has been intensively studied since its discovery 35 years ago 10– 12. Genomic imprinting involves the differential DNA methylation of the paternal or maternal allele of over 120 genes in the developing male or female germlines, resulting in parent-specific epigenetic regulation of these genes in offspring and as a consequence impact on a range of physiological and behavioural outcomes (reviewed in 12– 15). However, recent studies have revealed that other epigenetic modifications in sperm and oocytes can also influence outcomes in offspring, including histone modifications and associated non-coding RNAs 16– 28. Some examples include impacts on histone 3 lysine 4 (H3K4) 22 and H3K27 methylation 20 and DNA methylation-independent imprinting mediated by methylation of H3K27 in the oocyte 21. Furthermore, interactions among histone modifications, DNA methylation, and other interacting molecules, such as non-coding RNAs, add complexity to the mechanisms mediating heritable outcomes in offspring 29, 30. Such interactions are likely to underlie organised retention and patterning of modified histones and DNA methylation in sperm 31– 35 and the potential for environmental challenges, such as diet, chemicals, and drugs, to interact with the germline epigenome and alter paternal inheritance 16, 36. Although these and other studies are making substantial progress in understanding germline epigenetics and inheritance, much remains to be discovered.
Epigenetic changes in the germline potentially lead to intergenerational or transgenerational impacts on offspring 37, 38, and understanding these differences is important for determining the persistence of potential epigenetic changes induced in germ cells. Intergenerational inheritance occurs when the effect of an environmental stressor is transmitted from a parent (the F0 generation) to their offspring (the F1 generation). In the case of in utero exposure, the germ cells of the exposed foetus (F1) ultimately give rise to the F2 generation; therefore, effects transmitted from the F1 foetus to F2 offspring are considered intergenerational, as both the F0 and F1 generations were directly exposed to the environmental agent. Transgenerational inheritance occurs when an effect persists in the absence of direct germline exposure. For example, when the germline of the F0 parent is exposed, effects detected in the F2 generation can be considered transgenerational. Similarly, effects transmitted following in utero exposure of the F1 foetus that are detected in the F3 generation are considered transgenerational 37, 38. Importantly, these effects must survive epigenetic reprogramming in the F1 or F2 germline, respectively, to be transmitted transgenerationally 38.
As with epigenetic inheritance, epigenetic reprogramming in the germline is currently best understood from the perspective of changes in DNA methylation. Two distinct reprogramming events occur in mice: the first occurs in newly specified primordial germ cells (PGCs) during mid-gestation embryo development, and the latter occurs in the preimplantation embryo 39. In mice, PGCs are specified from surrounding epiblast cells at embryonic day 7 (E7) 40– 42 and around week 2 to 3 in humans 43. Initially, the newly specified PGCs carry epigenetic modifications similar to those of their somatic precursors, including epigenetic modifications from both parents. These epigenetic modifications are incompatible with sex-specific epigenetic programming and must be removed from PGCs before new, parent-specific (that is, only maternal or paternal) modifications that are compatible with offspring development can be established ( Figure 1). Reprogramming in the female and male germlines allows the production of oocytes and sperm that contain maternal- and paternal-specific information that is complementary at fertilisation and supports normal offspring development. Moreover, reprogramming in the germline facilitates the resetting of epigenetic errors that may have accumulated in the germline, preventing transmission of these altered epigenetic states to offspring. Fertilisation initiates the second reprogramming event, which establishes developmental competence in the preimplantation embryo, partly by resetting the maternal and paternal genomes to functional equivalence for many developmental genes. However, preimplantation epigenetic reprogramming leaves inherited epigenetic modifications such as parent-specific genomic imprints intact 44– 50, allowing their parent-specific function later in life. This represents a key difference between the two reprogramming events: reprogramming in PGCs occurs in order to remove existing genomic imprints and other epigenetic information, whereas reprogramming in preimplantation embryos occurs to establish equivalence between the paternal and maternal genomes and restore totipotency but with the exception that genomic imprints and possibly other parent-specific epigenetic information are maintained rather than lost.
Figure 1. Primordial germ cells undergo extensive epigenetic reprogramming prior to transmitting epigenetic information to offspring.
Primordial germ cells (PGCs) are the earliest precursors of sperm and oocytes (eggs) and are specified in the mouse around embryonic day 7 (E7). Initially, PGCs carry both paternal (green) and maternal (purple) epigenetic modifications that are similar to the somatic cells from which PGCs are derived. This information is removed by a process of epigenetic erasure before sex-specific epigenomes (paternal in male germ cells and maternal in female germ cells) are established. Epigenetic erasure occurs as PGCs migrate towards and populate the developing gonads. Soon after reaching the developing testis or ovary, germ cells commit to spermatogenesis or oogenesis, respectively. Subsequently, sex-specific epigenetic information is established. This occurs at different stages of development for males and females: during late foetal stages and early post-natal life in males and during oocyte growth in adult females. This results in the production of gametes which are epigenetically non-equivalent but which contain complementary epigenetic information at fertilisation.
Germline epigenetic reprogramming is achieved during PGC migration and final settlement of germ cells in the developing gonads by E10.5 in mice 51, 52 and around week 6 in humans 43. The PGCs undergo extensive global DNA demethylation while migrating to the genital ridge, followed by further demethylation as germ cells colonise the developing gonads (the future testes/ovaries). This results in global DNA methylation levels being reduced from around 70% in germline precursor cells to 14% and 7% in male and female PGCs by the time early testes and ovaries have formed at E13.5 and includes loss of methylation at imprinted regions 53. Global DNA demethylation is likely to occur via both passive and active mechanisms. Whereas passive loss involves the gradual dilution of methylated cytosine during replication in the absence of maintenance DNA methyltransferase 1 (DNMT1) activity 54, active demethylation involves the conversion of 5-methylcytosine to 5-hydroxymethylcytosine by ten-eleven translocase (TET) enzymes TET1 and TET2 and subsequent base excision repair 55, 56. Recent work demonstrated that TET1 alone is not essential for DNA demethylation in PGCs, indicating that TETs may act redundantly in this system or that TET1 is primarily required to maintain, rather than drive, DNA demethylation in PGCs 57. However, TET1 is required for the activation of germline development genes during reprogramming, indicating that DNA demethylation at specific genes is important for sex-specific germline development 57.
After removal, DNA methylation must be re-established in a sex-specific pattern in the male and female germlines. This occurs at quite different times in male and female mice. In male germ cells, new DNA methylation is established at paternally imprinted genes and repetitive sequences by the action of the de novo DNA methyltransferase DNMT3A and the co-factor DNMT3L during foetal life 58, 59. However, substantial remodelling also occurs during spermatogenesis in adult life, including changes in DNA methylation as germ cells enter meiosis 60 (reviewed by 61). In females, new DNA methylation including maternal imprints is established post-natally in oocytes. This occurs in each oocyte after individual primordial follicles have been released from the follicle reserve and progress through an extended growth phase that culminates in oocyte maturation 27, 58, 62– 64.
Extensive chromatin remodelling also occurs on histones during germline reprogramming. When specified, PGCs are enriched with the repressive modification H3K9me2. However, during germ cell migration, H3K9me2 is replaced with an alternative repressive modification, H3K27me3 65, 66. Further removal or reorganisation of repressive histone modifications or both occur once germ cells enter the developing gonad 65– 68. Although the mechanisms and biological significance of these changes are yet to be determined, H3K27me3 is established at developmental genes in germ cells and repetitive sequences during foetal life 33, 69 and is also present at developmental genes in sperm, indicating an important role for this modification in the paternal germline and, potentially, offspring 69, 70. Consistent with this, the complex required for catalysing H3K27me3, PRC2, is required for spermatogenesis and male fertility 71, 72. Moreover, recent work has demonstrated a role for PRC2 in epigenetic programming in foetal male germ cells and in modulating paternal epigenetic inheritance 73. Although H3K27me3 is not essential for female fertility, it is enriched in growing oocytes and PRC2 is required for regulating maternal inheritance 74. Deletion of the PRC2 genes Ezh2 or Eed specifically from the growing oocyte resulted in offspring with altered birth weights 20, 75, bone mineral density, and fat and muscle content and reduced litter size 20. In addition, H3K27me3 is required for regulating a non-coding RNA and consequently genomic imprints in mice 29, and overexpression of the histone demethylase Kdm6b in the zygote revealed a role for H3K27me3 in DNA methylation-independent imprinting 21. Similarly, the H3K4 methylase SETD1B regulates oocyte-specific RNAs 76, and increased levels of H3K4me2 in sperm resulted in paternally transmitted developmental effects in mice 22.
These examples demonstrate the importance of a range of epigenetic mechanisms in the male and female germlines, but they generally do not identify when the germline is most vulnerable to epigenetic change or how specific environmental agents impact on the germline. Given the differences in the timing of sex-specific DNA methylation and establishment of imprints, the periods of greatest sensitivity to environmentally induced epigenetic change may also differ in male and female germ cells. For example, male germ cells may be most vulnerable during foetal life whereas female germ cells may be most vulnerable during oocyte growth in adults. However, this does not exclude changes at other stages, such as during the extensive nuclear remodelling and histone replacement/rearrangement that occur during spermatogenesis or within the follicle reserve that contains the oocytes that underpin the reproductive life of females. Understanding epigenetic programming in both mechanistic and temporal frameworks will help illuminate the stages during which the germline is most sensitive to specific environmental factors and the potential risks of different exposures.
Emerging environmental agents: could epigenomic drugs affect the germline epigenome and future offspring?
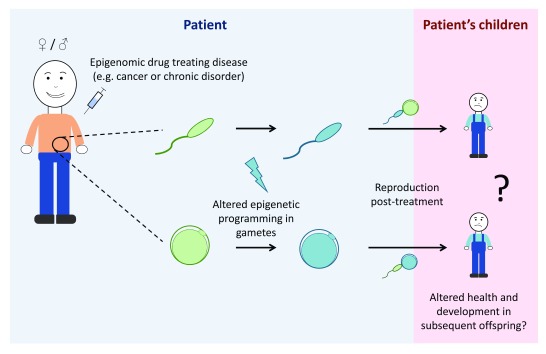
Although many studies indicate that a large range of environmental stimuli may affect the germline epigenome and consequently offspring phenotype, the underlying mechanisms are often poorly understood (reviewed in 3– 9). One relatively obvious way that epigenetic programming in the germline could be altered is through the action of agents that directly inhibit the enzymes that mediate epigenetic change. Indeed, the dynamic nature of epigenetic modifications coupled with the prevalence of dysregulated epigenetic modifying enzymes in tumours has led to the development of an extensive range of pharmacological inhibitors of specific epigenetic modifying enzymes for cancer therapies 77, 78. In the context of oncology, these drugs are being used to either kill cancer cells or drive their differentiation. It has been estimated that approximately half of all tumours involve abnormalities in chromatin modifier proteins (including epigenetic modifiers), and substantial efforts in pharmacological science are directed towards developing therapeutics for as many of these chromatin modifiers as possible (reviewed in 77, 78). This area of pharmacology is rapidly expanding, and these drugs offer highly promising new therapies that are clearly important for patients. However, these drugs act systemically and their potential impacts on the germline and future offspring remain largely unexplored. A number of studies have addressed whether epigenomic drugs alter the germline epigenome but have tended to focus on direct impacts on fertility rather than outcomes in offspring as a result of germline exposure to the drug ( Table 1). Moreover, although some studies have tested clinically relevant drug doses in mice, the clinical relevance of doses used in other studies has not always been clear. As differing drug doses are expected to affect target epigenetic modifications to varying degrees, it is important to test doses that reflect those used in humans as closely as possible when determining germline drug impacts in model organisms. Despite these challenges, it is important to explore the potential impacts of these therapies on the germline and subsequent offspring to generate a greater knowledge base from which clinical guidelines can be developed for the use of epigenomic drugs in children or patients of reproductive age ( Figure 2).
Figure 2. Epigenomic drugs may alter epigenetic programming in the germline and may alter health and development in offspring.
Epigenomic drugs are being used for cancer therapies and other disorders such as epilepsy; however, potential impacts of epigenomic drugs on the germline remain largely unexplored. As germ cells contain substantial epigenetic information, treatment with epigenomic drugs may alter the epigenetic information in sperm and oocytes (eggs). As epigenomic drugs work systemically, changes to the germline epigenome cannot be excluded and may result in altered health and development of subsequent offspring. In this diagram, the pale-blue background represents what is occurring in the patient whereas the pink background represents the patient’s children. Green gametes represent epigenetically normal sperm and oocytes, whereas blue gametes represent sperm and oocytes with altered epigenomes.
Table 1. Studies of intergenerational or transgenerational impacts of epigenomic drugs in utero or in adults.
| Inhibitor type and
mechanism |
Drug | Target | Approval/trial
status |
Studies in germline | Characteristics assessed | Studies in offspring | Characteristics
assessed |
|---|---|---|---|---|---|---|---|
|
DNMTi
Inhibit DNA methyltransferases |
Azacytidine
(Vidaza) |
Pan-
DNMT |
FDA | Zhao
et al. (2013)
79
Doerksen and Trasler (1996) 80 Seifertová et al. (1976) 81 |
Oocyte
in vitro maturation
rate and chromosome condensation 79 Male fertility 80, 81 |
Doerksen and Trasler (1996) 80 | Pre- and post-
implantation development of embryos of treated males (embryo loss, litter size, and gross morphology) 80 Embryo loss following treatment of adult males 81 |
| Decitabine
(Dacogen) |
Pan-
DNMT |
FDA | Kelly
et al. (2003)
82
Oakes et al. (2007) 83 Kläver et al. (2015) 84 Song et al. (2016) 85 Cisneros and Branch (2003) 86 Raman et al. (1995) 87 |
Male fertility and global DNA
methylation in sperm 82, 83. DNA methylation at paternally imprinted regions in sperm 83 Male fertility 84 Male fertility (sperm production only) 85 Fertility of in utero exposed males and females 86 Male fertility (sperm production only) 87 |
Kelly
et al. (2003)
82
Oakes et al. (2007) 83 Kläver et al. (2015) 84 |
Pre- and post-
implantation development of embryos of treated males (embryo loss, litter size, and gross morphology) 82 Progression through pre-implantation development in vitro 83 Fertility in male offspring of treated males 84 |
|
| Guadecitabine | Pan-DNMT | Phase I, II,
and III |
None | None | |||
|
HDACi
Inhibit histone deacetylases |
Belinostat
(Beleodaq) |
HDAC
class I and class II |
FDA | None | None | ||
| Panobinostat
(Farydak) |
HDAC
class I, II, and IV |
FDA | None | None | |||
| Romidepsin
(Istodax) |
HDAC
class I |
FDA | None | None | |||
| Vorinostat
(Zolinza) |
HDAC
class I, II, and IV |
FDA | Wise
et al. (2008)
88
Kläver et al. (2015) 84 |
Male and female fertility and
pregnancy outcome 88 Male fertility 84 |
Kläver et al. (2015) 84 | Fertility in male
offspring of treated males 84 |
|
| Valproic acid | HDAC class I and II | FDA/phase III | Bairy
et al. (2010)
89
Røste et al. (2001a) 90 Røste et al. (2001b) 91 Røste et al. (2003) 92 |
Male fertility (sperm production
only) 89 Effects of valproate on ovarian morphology 90 Male reproductive system and fertility 91 Semen parameters in male epilepsy patients taking valproic acid 92 |
Choi et al. (2016) 93 | Transgenerational
inheritance of autism- like behaviours following in utero exposure 93 |
|
| Tacedinaline
(CI994) |
HDAC
class I, II, III, and VIII |
Phase III | None | None | |||
| HDACi 4b
[N1-(20aminophenyl)-N7- phenoylheltanediamide] |
HDAC
class I, II, and III |
Not in clinic | None | Jia et al. (2015) 94 | Intergenerational
effects in Huntington’s disease 94 |
||
|
BET inhibitors
Inhibit BET domain family proteins |
Apabetalone/RVX000222/RVX-208 | Pan-BET | Phase III | None | None | ||
| JQ1 | Pan-BET | Not in clinic | Matzuk et al. (2012) 95 | Male fertility 95 | Matzuk et al. (2013) 95 | Production of offspring
from treated males 95 |
|
| CPI-0610 | Pan-BET | Phase I | None | None | |||
| TEN-010 | Pan-BET | Phase I | None | None | |||
| BAY1238097 | Pan-BET | Phase I | None | None | |||
| OTX015 | Pan-BET | Phase I | None | None | |||
| INCB054329 | Pan-BET | Phase I and II | None | None | |||
| BMS-986158 | Pan-BET | Phase I and II | None | None | |||
| FT-1101 | Pan-BET | Phase I and II | None | None | |||
| GSK525762 | Pan-BET | Phase I | None | None | |||
|
LSD1 inhibitors
Inhibit histone demethylase LSD1 |
GSK2879552 | LSD1 | Phase I | None | None | ||
| IMG-7289 | LSD1 | Phase I | None | None | |||
| INCB059872 | LSD1 | Phase I | None | None | |||
| Tranylcypromine | LSD1 | Phase I | None | None | |||
|
EZH2 inhibitors
Inhibit histone methyltransferase EZH2 |
Tazemetostat | EZH2 | Phase I and II | Prokopuk et al. (2018) 74 | H3K27me3 levels in oocytes
and foetal germ cells following drug treatment and withdrawal 74 |
None | |
| CP1-1205 | EZH2 | Phase 1 | None | None | |||
| GSK126 | EZH2 | Not in clinic | Prokopuk et al. (2017) 68 | H3K27me3 dynamics during
epigenetic reprogramming in foetal germ cells 68 |
None | ||
| UNC-1999 | EZH2 | Not in clinic | None | None | |||
| EBI-2511 | EZH2 | Not in clinic | None | None |
We have included published articles investigating the effects of post-natal exposure to epigenomic drugs on the germline or subsequent offspring or both. Studies assessing in utero exposure were included if outcomes were assessed in the germline or in the offspring of exposed foetuses. Direct teratogenic effects were excluded. Table modified from Jones et al. 77 to form the basis of the drug list, and additional drugs were added. This drug list is extensive though not exhaustive. BET, bromodomain extra terminal; DNMT, DNA methyltransferase; DNMTi, DNA methyltransferase inhibitor; EZH2, enhancer of zeste 2; FDA, US Food and Drug Administration; H3K27me3, H3K27 trimethylation; HDAC, histone deacetylase; HDACi, histone deacetylase inhibitor.
Impacts of epigenomic drugs may alter reproductive health in a range of ways. From a germline perspective, perhaps the most obvious are the potential impacts on fertility, including zygote (fertilised egg) and early embryo viability. For example, rats treated with the class I/II histone deacetylase (HDAC) inhibitor vorinostat were fertile, but increased peri- and post-implantation embryo loss was observed in treated females crossed with untreated males 88. Furthermore, although some offspring survived, there was no analysis of their developmental outcomes, so it remains unclear whether treatment of adult females resulted in developmental differences in offspring. In another example, increased ovarian cysts and decreased corpora lutea were observed in female rats treated with an alternative HDAC inhibitor, valproic acid (VPA) 90. Mechanistically, extensive HDAC-dependent histone deacetylation occurs during oocyte maturation in mice, which can be blocked by treatment with the HDAC inhibitor trichostatin A 96. Moreover, genetic ablation of both Hdac1 and Hdac2 in growing oocytes resulted in the arrest of follicle development at the secondary follicle stage 97. In addition, VPA affected male fertility in rats and had mild effects in male patients undergoing long-term treatment for epilepsy 91, 92. Whereas these studies largely focussed on fertility and embryo viability, some studies have examined the impacts of HDAC inhibitor treatment in paternal inheritance. In a model of Huntington’s disease (HD), the treatment of adult F0 mice altered DNA methylation patterns in sperm and ameliorated disease phenotype in F1 offspring, apparently through a mechanism involving histone demethylation and DNA methylation 94. Although these effects potentially imparted some beneficial intergenerational effects on behaviour in this HD model, the broader impacts on offspring health were not examined. This is an important point, as impacts of this drug on DNA methylation and histone methylation, as well as histone acetylation, strongly indicate that the effects of treatment were unlikely to be focussed only on HD genes but are likely to have altered other aspects of inheritance. Indeed, the treatment of pregnant female mice with VPA led to the inducement of autism-like behaviours not only in the directly exposed F1 offspring but also in the unexposed F2 and F3 offspring of exposed F1 progeny, demonstrating a detrimental transgenerational effect of VPA in inheritance 93.
While DNA methylation is the best-understood marker of inherited epigenetic modifications, there is limited understanding of the impacts of DNMT inhibitors on the female germline and inheritance. In oocytes matured in vitro in the presence of azacytidine (Vidaza), chromosomes were less condensed and more unstable than in untreated controls 79. Although treatment induced the expression of early apoptotic markers, these oocytes progressed through maturation faster than did untreated controls, and it was concluded that azacytidine treatment imparted a beneficial effect 79. However, the potential for the resulting oocytes to be fertilised or to support normal offspring development was not assessed. In another study, in utero decitabine (Dacagon)-exposed females mated with untreated males had normal fertility and produced offspring of normal weights 86, but other potential impacts on offspring outcomes were not assessed.
A range of studies in males have demonstrated that both azacytidine and decitabine impair spermatogenesis or male fertility or both, and some reported reduced litter size or embryo loss or both 80– 87. Although these treatments reduced male fertility, this effect was reversible when treatment was withdrawn. In one study male mice restored normal testis histology 85, and in another study drug-induced foetal loss was prevented 81 4 weeks after treatment was terminated. Moreover, although subtle effects on reproductive organs and sperm parameters were observed in F1–F3 offspring of treated males, neither decitabine nor vorinostat caused intergenerational or transgenerational effects on male fertility 84. Although these studies demonstrate substantial, though reversible, impacts of DNMT inhibitors on male fertility and litter size, the broader impact of these drugs on developmental outcomes in offspring remains largely unknown.
DNA methyltransferase and HDAC inhibitors are well established in the clinic and have been used in a range of combination therapies, and their actions in relation to cancer treatment are becoming better understood 77, 98– 103. However, more recent developments have focussed on histone methyltransferase (HMT) and bromodomain extra terminal repeat (BET) inhibitors 104– 107 ( Table 1). Very few studies have examined the impacts of BET inhibitors on the germline, although the preclinical BET inhibitor JQ1 has been proposed as a potential male contraceptive because of its ability to reversibly block male fertility 95. As with DNMT inhibitors, withdrawal of JQ1 treatment restored fertility and these males produced apparently normal pups 95, but developmental outcomes in these offspring were not assessed in detail.
Another prominent epigenomic target is the H3K27 histone methyltransferase EZH2, for which drugs include EPZ-6438 (tazemetostat), GSK126, CPI-1205, EBI-2511, and UNC1999 108– 112 ( Table 1). Perhaps the most clinically advanced of these is tazemetostat, which is being assessed in a range of phase I/II clinical trials for human patients presenting with a variety of cancers, including lymphomas, myelomas, mesothelioma, solid tumours, and malignant rhabdoid tumours of the kidney and ovary ( http://clinicaltrials.gov). Patient cohorts include individuals of reproductive age and children as young as 6 months. Recent work demonstrated that the treatment of adult female mice with a clinically relevant dose of tazemetostat for 10 days significantly depleted H3K27me3 in growing oocytes, and H3K27me3 did not recover after a 10-day period of drug withdrawal 74. This is concerning given that oocyte-specific deletion of Ehz2 caused growth restriction in offspring 20, 75 and similar oocyte-specific deletion of the essential Ezh2-interacting gene Eed caused foetal overgrowth, increased bone mineral density, and altered fat and muscle content in offspring 20. Similarly, recent work demonstrated a role for PRC2 in regulating paternal epigenetic inheritance 73. Moreover, de novo germline mutations in the PRC2-encoding genes EZH2, EED, or SUZ12 in humans result in Weaver and Cohen–Gibson syndromes, characterised by a spectrum of abnormalities including over-growth, skeletal defects, and cognitive deficits 113– 119. Although these mutations are genetic, the observed phenotypes may have an epigenetic basis. Therefore, given these outcomes, future work should address whether pharmacological depletion of H3K27me3 in growing oocytes can recapitulate the phenotypic outcomes in subsequent offspring observed in the mouse oocyte deletion models or human genetic conditions 20, 74, 113– 119. Prokopuk et al. also demonstrated depletion of H3K27me3 in the primordial follicle pool of juvenile mouse ovaries cultured with tazemetostat in vitro 74. As human clinical trials currently include children, it is important to examine whether tazemetostat-induced depletion of H3K27me3 is observed in primordial follicles of juvenile mice in vivo, whether H3K27me3 recovers after drug withdrawal, and whether this potential transient loss of H3K27me3 in the oocyte genome affects outcomes in future offspring ( Figure 3).
Figure 3. The potential for epigenomic drugs to impact the germline remains largely unexplored.
Three key areas of investigation for specific drugs are understanding ( A) the capacity of epigenomic drugs to affect the germline epigenome, ( B) the capacity of the germline to recover after drug withdrawal, and ( C) whether changes induced in the germline epigenome impact on the health and development of subsequent offspring. Collectively, this information will aid in refining clinical guidelines for the use of epigenomic drugs in patients of reproductive age and in children/adolescents prior to reproduction. In this diagram, green gametes represent epigenetically normal sperm and oocytes (eggs) and blue gametes represent sperm and oocytes with altered epigenomes.
Epigenomic drugs during pregnancy
Substantial epigenetic reprogramming occurs during foetal development, making this a period of particular interest for germline exposures to environmental factors. However, as epigenomic drugs target proteins that widely influence specification and development, the use of these drugs during pregnancy is contraindicated in most circumstances.
Despite this, the use of epigenomic drugs in pregnancy is not unprecedented, particularly in the case of ongoing chronic illnesses which require continued treatment during pregnancy. VPA has been used extensively since 1974 to treat both epilepsy and bipolar mania, including in women of reproductive capacity and pregnant women 120. However, VPA has been shown to be teratogenic in both animal and human studies, with in utero exposure linked to neural tube, cardiac, limb, kidney, craniofacial, and genitourinary defects 120– 123. Furthermore, a recent study in mice demonstrated autistic-like behaviours in offspring exposed to VPA in utero 93. Remarkably, these behaviours were observed in the two subsequent generations, indicating that these effects were maintained transgenerationally 93.
The continued use of VPA during pregnancy highlights the difficulty that clinicians and patients face under these conditions: how is the use of a drug that is required by the patient balanced with risk to the unborn child? As the risks of epigenomic drug exposures to the unborn child either through the germline or after direct in utero exposure are poorly understood, it is difficult to make informed decisions regarding potential outcomes of such exposures. Animal studies that separate pre-fertilisation from gestational exposures are required to evaluate the underlying mechanisms and relative risks of these two periods.
Conclusions and considerations for future germlines
Though not exhaustive, the examples described in this review provide a snapshot of the germline epigenome and inhibitors specific to a small group of epigenetic complexes. These studies illustrate the broader concept that a range of epigenetic mechanisms act to establish the germline epigenome and indicate that the effects on the germline by many epigenomic drugs currently under development should be explored. The combined use of both pharmaceutical and genetic models in mice or other animals provides opportunities for well-controlled studies of the impacts of epigenomic drugs and the mechanisms involved in epigenetic inheritance. Furthermore, more human epidemiological studies are also required to evaluate drugs that are currently in use. Key outstanding areas include addressing whether epigenomic drugs alter the germline epigenome, how these changes affect the health and development of subsequent children, and whether these changes are reversed following drug withdrawal ( Figure 3). Outcomes from such studies are required to facilitate more informed clinical use of the drugs with regard to fertility and reproduction and determine whether fertility-preserving approaches should be used to decrease germline exposure to specific drugs or maintain fertility for the patient or both.
Abbreviations
BET, bromodomain extra terminal repeat; DNMT, DNA methyltransferase; E, embryonic day; EED, embryonic ectoderm; EZH2, enhancer of zeste 2; H3K4, histone 3 lysine 4; H3K9me2, histone 3 lysine 9 dimethylation; H3K27, histone 3 lysine 27; H3K27me3, H3K27 trimethylation; HD, Huntington’s disease; HDAC, histone deacetylase; PGC, primordial germ cell; PRC2, polycomb repressive complex 2; SUZ12, suppressor of zeste 12; TET, ten-eleven translocase enzyme; VPA, valproic acid.
Acknowledgements
We thank Dr Neil Youngson and the manuscript reviewers for constructive comments on this article.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Douglas T Carrell, Department of Human Genetics, University of Utah School of Medicine, Salt Lake City, Utah, USA
Marisa Bartolomei, Epigenetics Institute, Center for Excellence in Environmental Toxicology, Department of Cell and Developmental Biology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA
David Brocks, Division of Epigenomics and Cancer Risk Factors, German Cancer Research Center, Heidelberg, Germany
Funding Statement
This work was supported by Australian National Health and Medical Research Grants GNT1144966 and GNT1144887 awarded to PSW.
[version 1; referees: 3 approved]
References
- 1. Henikoff S, Greally JM: Epigenetics, cellular memory and gene regulation. Curr Biol. 2016;26(14):R644–8. 10.1016/j.cub.2016.06.011 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 2. Greally JM: A user's guide to the ambiguous word 'epigenetics'. Nat Rev Mol Cell Biol. 2018;19(4):207–208. 10.1038/nrm.2017.135 [DOI] [PubMed] [Google Scholar]
- 3. Radford EJ: Exploring the extent and scope of epigenetic inheritance. Nat Rev Endocrinol. 2018;14(6):345–355. 10.1038/s41574-018-0005-5 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 4. Bohacek J, Mansuy IM: Molecular insights into transgenerational non-genetic inheritance of acquired behaviours. Nat Rev Genet. 2015;16(11):641–52. 10.1038/nrg3964 [DOI] [PubMed] [Google Scholar]
- 5. Xin F, Susiarjo M, Bartolomei MS: Multigenerational and transgenerational effects of endocrine disrupting chemicals: A role for altered epigenetic regulation? Semin Cell Dev Biol. 2015;43:66–75. 10.1016/j.semcdb.2015.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nilsson EE, Sadler-Riggleman I, Skinner MK: Environmentally induced epigenetic transgenerational inheritance of disease. Environ Epigenet. 2018;4(2):dvy016. 10.1093/eep/dvy016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yeshurun S, Hannan AJ: Transgenerational epigenetic influences of paternal environmental exposures on brain function and predisposition to psychiatric disorders. Mol Psychiatry. 2018. 10.1038/s41380-018-0039-z [DOI] [PubMed] [Google Scholar]
- 8. Youngson NA, Morris MJ: What obesity research tells us about epigenetic mechanisms. Philos Trans R Soc Lond B Biol Sci. 2013;368(1609): 20110337. 10.1098/rstb.2011.0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prokopuk L, Western PS, Stringer JM: Transgenerational epigenetic inheritance: adaptation through the germline epigenome? Epigenomics. 2015;7(5):829–846. 10.2217/epi.15.36 [DOI] [PubMed] [Google Scholar]
- 10. Surani MA, Barton SC, Norris ML: Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308(5959):548–50. 10.1038/308548a0 [DOI] [PubMed] [Google Scholar]
- 11. McGrath J, Solter D: Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37(1):179–183. 10.1016/0092-8674(84)90313-1 [DOI] [PubMed] [Google Scholar]
- 12. Ferguson-Smith AC: Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12(8):565–75. 10.1038/nrg3032 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 13. Barlow DP, Bartolomei MS: Genomic imprinting in mammals. Cold Spring Harb Perspect Biol. 2014;6(2): pii: a018382. 10.1101/cshperspect.a018382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stewart KR, Veselovska L, Kelsey G: Establishment and functions of DNA methylation in the germline. Epigenomics. 2016;8(10):1399–1413. 10.2217/epi-2016-0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. John RM: Imprinted genes and the regulation of placental endocrine function: Pregnancy and beyond. Placenta. 2017;56:86–90. 10.1016/j.placenta.2017.01.099 [DOI] [PubMed] [Google Scholar]
- 16. Dalgaard K, Landgraf K, Heyne S, et al. : Trim28 Haploinsufficiency Triggers Bi-stable Epigenetic Obesity. Cell. 2016;164(3):353–64. 10.1016/j.cell.2015.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 17. Sankar A, Kooistra SM, Gonzalez JM, et al. : Maternal expression of the histone demethylase Kdm4a is crucial for pre-implantation development. Development. 2017;144(18):3264–3277. 10.1242/dev.155473 [DOI] [PubMed] [Google Scholar]
- 18. Gapp K, Jawaid A, Sarkies P, et al. : Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 2014;17(5):667–9. 10.1038/nn.3695 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 19. Chen Q, Yan M, Cao Z, et al. : Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351(6271):397–400. 10.1126/science.aad7977 [DOI] [PubMed] [Google Scholar]
- 20. Prokopuk L, Stringer JM, White CR, et al. : Loss of maternal EED results in postnatal overgrowth. Clin Epigenetics. 2018;10(1):95. 10.1186/s13148-018-0526-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Inoue A, Jiang L, Lu F, et al. : Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature. 2017;547(7664):419–424. 10.1038/nature23262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Siklenka K, Erkek S, Godmann M, et al. : Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science. 2015;350(6261):aab2006. 10.1126/science.aab2006 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 23. Posfai E, Kunzmann R, Brochard V, et al. : Polycomb function during oogenesis is required for mouse embryonic development. Genes Dev. 2012;26(9):920–32. 10.1101/gad.188094.112 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 24. Daxinger L, Oey H, Isbel L, et al. : Hypomethylation of ERVs in the sperm of mice haploinsufficient for the histone methyltransferase Setdb1 correlates with a paternal effect on phenotype. Sci Rep. 2016;6: 25004. 10.1038/srep25004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Teperek M, Simeone A, Gaggioli V, et al. : Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Res. 2016;26(8):1034–46. 10.1101/gr.201541.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van de Werken C, van der Heijden GW, Eleveld C, et al. : Paternal heterochromatin formation in human embryos is H3K9/HP1 directed and primed by sperm-derived histone modifications. Nat Commun. 2014;5: 5868. 10.1038/ncomms6868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hanna CW, Taudt A, Huang J, et al. : MLL2 conveys transcription-independent H3K4 trimethylation in oocytes. Nat Struct Mol Biol. 2018;25(1):73–82. 10.1038/s41594-017-0013-5 [DOI] [PubMed] [Google Scholar]
- 28. Hatanaka Y, Tsusaka T, Shimizu N, et al. : Histone H3 Methylated at Arginine 17 Is Essential for Reprogramming the Paternal Genome in Zygotes. Cell Rep. 2017;20(12):2756–2765. 10.1016/j.celrep.2017.08.088 [DOI] [PubMed] [Google Scholar]
- 29. Greenberg MV, Glaser J, Borsos M, et al. : Transient transcription in the early embryo sets an epigenetic state that programs postnatal growth. Nat Genet. 2017;49(1):110–118. 10.1038/ng.3718 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Duffié R, Ajjan S, Greenberg MV, et al. : The Gpr1/Zdbf2 locus provides new paradigms for transient and dynamic genomic imprinting in mammals. Genes Dev. 2014;28(5):463–478. 10.1101/gad.232058.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hammoud SS, Nix NA, Zhang H, et al. : Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460(7254):473–8. 10.1038/nature08162 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Brykczynska U, Hisano M, Erkek S, et al. : Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17(6):679–87. 10.1038/nsmb.1821 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 33. Lesch BJ, Dokshin GA, Young RA, et al. : A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. Proc Natl Acad Sci U S A. 2013;110(40):16061–6. 10.1073/pnas.1315204110 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 34. Denomme MM, McCallie BR, Parks JC, et al. : Alterations in the sperm histone-retained epigenome are associated with unexplained male factor infertility and poor blastocyst development in donor oocyte IVF cycles. Hum Reprod. 2017;32(12):2443–2455. 10.1093/humrep/dex317 [DOI] [PubMed] [Google Scholar]
- 35. Erkek S, Hisano M, Liang CY, et al. : Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat Struct Mol Biol. 2013;20(7):868–75. 10.1038/nsmb.2599 [DOI] [PubMed] [Google Scholar]
- 36. Ben Maamar M, Sadler-Riggleman I, Beck D, et al. : Epigenetic Transgenerational Inheritance of Altered Sperm Histone Retention Sites. Sci Rep. 2018;8(1): 5308. 10.1038/s41598-018-23612-y [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 37. Heard E, Martienssen RA: Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157(1):95–109. 10.1016/j.cell.2014.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stringer JM, Barrand S, Western P: Fine-tuning evolution: germ-line epigenetics and inheritance. Reproduction. 2013;146(1):R37–48. 10.1530/REP-12-0526 [DOI] [PubMed] [Google Scholar]
- 39. Hogg K, Western PS: Refurbishing the germline epigenome: Out with the old, in with the new. Semin Cell Dev Biol. 2015;45:104–113. 10.1016/j.semcdb.2015.09.012 [DOI] [PubMed] [Google Scholar]
- 40. Okamura D, Hayashi K, Matsui Y: Mouse epiblasts change responsiveness to BMP4 signal required for PGC formation through functions of extraembryonic ectoderm. Mol Reprod Dev. 2005;70(1):20–29. 10.1002/mrd.20136 [DOI] [PubMed] [Google Scholar]
- 41. Yamaji M, Seki Y, Kurimoto K, et al. : Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet. 2008;40(8):1016–22. 10.1038/ng.186 [DOI] [PubMed] [Google Scholar]
- 42. Ying Y, Qi X, Zhao GQ: Induction of primordial germ cells from murine epiblasts by synergistic action of BMP4 and BMP8B signaling pathways. Proc Natl Acad Sci U S A. 2001;98(14):7858–62. 10.1073/pnas.151242798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tang WW, Kobayashi T, Irie N, et al. : Specification and epigenetic programming of the human germ line. Nat Rev Genet. 2016;17(10):585–600. 10.1038/nrg.2016.88 [DOI] [PubMed] [Google Scholar]
- 44. Li X, Ito M, Zhou F, et al. : A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15(4):547–57. 10.1016/j.devcel.2008.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 45. Messerschmidt DM, de Vries W, Ito M, et al. : Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335(6075):1499–502. 10.1126/science.1216154 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 46. Nakamura T, Arai Y, Umehara H, et al. : PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9(1):64–71. 10.1038/ncb1519 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 47. Hirasawa R, Chiba H, Kaneda M, et al. : Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22(12):1607–16. 10.1101/gad.1667008 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 48. Howell CY, Bestor TH, Ding F, et al. : Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104(6):829–38. 10.1016/S0092-8674(01)00280-X [DOI] [PubMed] [Google Scholar]
- 49. Cirio MC, Ratnam S, Ding F, et al. : Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC Dev Biol. 2008;8:9. 10.1186/1471-213X-8-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Maenohara S, Unoki M, Toh H, et al. : Role of UHRF1 in de novo DNA methylation in oocytes and maintenance methylation in preimplantation embryos. PLoS Genet. 2017;13(10):e1007042. 10.1371/journal.pgen.1007042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. CHIQUOINE AD: The identification, origin, and migration of the primordial germ cells in the mouse embryo. Anat Rec. 1954;118(2):135–146. 10.1002/ar.1091180202 [DOI] [PubMed] [Google Scholar]
- 52. Molyneaux KA, Stallock J, Schaible K, et al. : Time-lapse analysis of living mouse germ cell migration. Dev Biol. 2001;240(2):488–98. 10.1006/dbio.2001.0436 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 53. Seisenberger S, Andrews S, Krueger F, et al. : The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48(6):849–62. 10.1016/j.molcel.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 54. Kagiwada S, Kurimoto K, Hirota T, et al. : Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J. 2013;32(3):340–353. 10.1038/emboj.2012.331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hackett JA, Sengupta R, Zylicz JJ, et al. : Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339(6118):448–52. 10.1126/science.1229277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hajkova P, Jeffries SJ, Lee C, et al. : Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329(5987):78–82. 10.1126/science.1187945 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 57. Hill PWS, Leitch HG, Requena CE, et al. : Epigenetic reprogramming enables the transition from primordial germ cell to gonocyte. Nature. 2018;555(7696):392–396. 10.1038/nature25964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kaneda M, Okano M, Hata K, et al. : Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429(6994):900–3. 10.1038/nature02633 [DOI] [PubMed] [Google Scholar]
- 59. Bourc'his D, Bestor TH: Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431(7004):96–9. 10.1038/nature02886 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 60. Gaysinskaya V, Miller BF, De Luca C, et al. : Transient reduction of DNA methylation at the onset of meiosis in male mice. Epigenetics Chromatin. 2018;11(1):15. 10.1186/s13072-018-0186-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jenkins TG, Aston K, James ER, et al. : Sperm epigenetics in the study of male fertility, offspring health, and potential clinical applications. Syst Biol Reprod Med. 2017;63(2):69–76. 10.1080/19396368.2016.1274791 [DOI] [PubMed] [Google Scholar]
- 62. Hata K, Okano M, Lei H, et al. : Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129(8):1983–93. [DOI] [PubMed] [Google Scholar]
- 63. Bourc’his D, Xu GL, Lin CS, et al. : Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:(5551)2536–9. 10.1126/science.1065848 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Gahurova L, Tomizawa SI, Smallwood SA, et al. : Transcription and chromatin determinants of de novo DNA methylation timing in oocytes. Epigenetics Chromatin. 2017;10:25. 10.1186/s13072-017-0133-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Seki Y, Hayashi K, Itoh K, et al. : Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278(2):440–458. 10.1016/j.ydbio.2004.11.025 [DOI] [PubMed] [Google Scholar]
- 66. Seki Y, Yamaji M, Yabuta Y, et al. : Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134(14):2627–38. 10.1242/dev.005611 [DOI] [PubMed] [Google Scholar]
- 67. Hajkova P, Ancelin K, Waldmann T, et al. : Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452(7189):877–81. 10.1038/nature06714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Prokopuk L, Stringer JM, Hogg K, et al. : PRC2 is required for extensive reorganization of H3K27me3 during epigenetic reprogramming in mouse fetal germ cells. Epigenetics Chromatin. 2017;10:7. 10.1186/s13072-017-0113-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sachs M, Onodera C, Blaschke K, et al. : Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep. 2013;3(6):1777–1784. 10.1016/j.celrep.2013.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ng JH, Kumar V, Muratani M, et al. : In vivo epigenomic profiling of germ cells reveals germ cell molecular signatures. Dev Cell. 2013;24(3):324–333. 10.1016/j.devcel.2012.12.011 [DOI] [PubMed] [Google Scholar]
- 71. Mu W, Starmer J, Fedoriw AM, et al. : Repression of the soma-specific transcriptome by Polycomb-repressive complex 2 promotes male germ cell development. Genes Dev. 2014;28(18):2056–69. 10.1101/gad.246124.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mu W, Starmer J, Shibata Y, et al. : EZH1 in germ cells safeguards the function of PRC2 during spermatogenesis. Dev Biol. 2017;424(2):198–207. 10.1016/j.ydbio.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Stringer JM, Forster SC, Qu Z, et al. : Reduced PRC2 function alters male germline epigenetic programming and paternal inheritance. BMC Biol. 2018;16(1):104. 10.1186/s12915-018-0569-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Prokopuk L, Hogg K, Western PS: Pharmacological inhibition of EZH2 disrupts the female germline epigenome. Clin Epigenetics. 2018;10:33. 10.1186/s13148-018-0465-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Erhardt S, Su IH, Schneider R, et al. : Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development. 2003;130(18):4235–48. 10.1242/dev.00625 [DOI] [PubMed] [Google Scholar]
- 76. Brici D, Zhang Q, Reinhardt S, et al. : Setd1b, encoding a histone 3 lysine 4 methyltransferase, is a maternal effect gene required for the oogenic gene expression program. Development. 2017;144(14):2606–2617. 10.1242/dev.143347 [DOI] [PubMed] [Google Scholar]
- 77. Jones PA, Issa JP, Baylin S: Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17(10):630–41. 10.1038/nrg.2016.93 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 78. Dawson MA: The cancer epigenome: Concepts, challenges, and therapeutic opportunities. Science. 2017;355(6330):1147–1152. 10.1126/science.aam7304 [DOI] [PubMed] [Google Scholar]
- 79. Zhao FY, Shao CP, Li Y, et al. : 5-Azacytidine induces early stage apoptosis and promotes in vitro maturation by changing chromosomal construction in murine oocytes. Reprod Toxicol. 2013;37:56–61. 10.1016/j.reprotox.2013.01.007 [DOI] [PubMed] [Google Scholar]
- 80. Doerksen T, Trasler JM: Developmental exposure of male germ cells to 5-azacytidine results in abnormal preimplantation development in rats. Biol Reprod. 1996;55(5):1155–62. 10.1095/biolreprod55.5.1155 [DOI] [PubMed] [Google Scholar]
- 81. Seifertová M, Veselý J, Cihák A: Enhanced mortality in offsprings of male mice treated with 5-azacytidine prior to mating. Morphological changes in testes. Neoplasma. 1976;23(1):53–60. [PubMed] [Google Scholar]
- 82. Kelly TL, Li E, Trasler JM: 5-aza-2'-deoxycytidine induces alterations in murine spermatogenesis and pregnancy outcome. J Androl. 2003;24(6):822–30. 10.1002/j.1939-4640.2003.tb03133.x [DOI] [PubMed] [Google Scholar]
- 83. Oakes CC, Kelly TL, Robaire B, et al. : Adverse effects of 5-aza-2'-deoxycytidine on spermatogenesis include reduced sperm function and selective inhibition of de novo DNA methylation. J Pharmacol Exp Ther. 2007;322(3):1171. 10.1124/jpet.107.121699 [DOI] [PubMed] [Google Scholar]
- 84. Kläver R, Sánchez V, Damm OS, et al. : Direct but no transgenerational effects of decitabine and vorinostat on male fertility. PLos One. 2015;10(2):e0117839. 10.1371/journal.pone.0117839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Song N, Endo D, Song B, et al. : 5-aza-2'-deoxycytidine impairs mouse spermatogenesis at multiple stages through different usage of DNA methyltransferases. Toxicology. 2016;361–362:62–72. 10.1016/j.tox.2016.07.005 [DOI] [PubMed] [Google Scholar]
- 86. Cisneros FJ, Branch S: 5-AZA-2'-deoxycytidine (5-AZA-CdR): a demethylating agent affecting development and reproductive capacity. J Appl Toxicol. 2003;23(2):115–20. 10.1002/jat.898 [DOI] [PubMed] [Google Scholar]
- 87. Raman R, Narayan G: 5-Aza deoxyCytidine-induced inhibition of differentiation of spermatogonia into spermatocytes in the mouse. Mol Reprod Dev. 1995;42(3):284–90. 10.1002/mrd.1080420304 [DOI] [PubMed] [Google Scholar]
- 88. Wise LD, Spence S, Saldutti LP, et al. : Assessment of female and male fertility in Sprague-Dawley rats administered vorinostat, a histone deacetylase inhibitor. Birth Defects Res B Dev Reprod Toxicol. 2008;83(1):19–26. 10.1002/bdrb.20139 [DOI] [PubMed] [Google Scholar]
- 89. Bairy L, Paul V, Rao Y: Reproductive toxicity of sodium valproate in male rats. Indian J Pharmacol. 2010;42(2):90–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Røste LS, Taubøll E, Berner A, et al. : Valproate, but not lamotrigine, induces ovarian morphological changes in Wistar rats. Exp Toxicol Pathol. 2001;52(6):545–552. 10.1016/S0940-2993(01)80014-2 [DOI] [PubMed] [Google Scholar]
- 91. Røste LS, Taubøll E, Berner A, et al. : Morphological changes in the testis after long-term valproate treatment in male Wistar rats. Seizure. 2001;10(8):559–565. 10.1053/seiz.2001.0545 [DOI] [PubMed] [Google Scholar]
- 92. Røste LS, Taubøll E, Haugen TB, et al. : Alterations in semen parameters in men with epilepsy treated with valproate or carbamazepine monotherapy. Eur J Neurol. 2003;10(5):501–6. 10.1046/j.1468-1331.2003.00615.x [DOI] [PubMed] [Google Scholar]
- 93. Choi CS, Gonzales EL, Kim KC, et al. : The transgenerational inheritance of autism-like phenotypes in mice exposed to valproic acid during pregnancy. Sci Rep. 2016;6: 36250. 10.1038/srep36250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jia H, Morris CD, Williams RM, et al. : HDAC inhibition imparts beneficial transgenerational effects in Huntington's disease mice via altered DNA and histone methylation. Proc Natl Acad Sci U S A. 2015;112(1):E56–64. 10.1073/pnas.1415195112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Matzuk MM, McKeown MR, Filippakopoulos P, et al. : Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150(4):673–684. 10.1016/j.cell.2012.06.045 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 96. Kim JM, Liu H, Tazaki M, et al. : Changes in histone acetylation during mouse oocyte meiosis. J Cell Biol. 2003;162(1):37–46. 10.1083/jcb.200303047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ma P, Pan H, Montgomery RL, et al. : Compensatory functions of histone deacetylase 1 (HDAC1) and HDAC2 regulate transcription and apoptosis during mouse oocyte development. Proc Natl Acad Sci U S A. 2012;109(8):E481. 10.1073/pnas.1118403109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. McCaw TR, Randall TD, Arend RC: Overcoming immune suppression with epigenetic modification in ovarian cancer. Transl Res. 2018; pii: S1931-5244(18)30099-9. 10.1016/j.trsl.2018.06.003 [DOI] [PubMed] [Google Scholar]
- 99. Bamodu OA, Kuo KT, Yuan LP, et al. : HDAC inhibitor suppresses proliferation and tumorigenicity of drug-resistant chronic myeloid leukemia stem cells through regulation of hsa-miR-196a targeting BCR/ABL1. Exp Cell Res. 2018;370(2):519–530. 10.1016/j.yexcr.2018.07.017 [DOI] [PubMed] [Google Scholar]
- 100. Kiweler N, Brill B, Wirth M, et al. : The histone deacetylases HDAC1 and HDAC2 are required for the growth and survival of renal carcinoma cells. Arch Toxicol. 2018;92(7):2227–2243. 10.1007/s00204-018-2229-5 [DOI] [PubMed] [Google Scholar]
- 101. Muscat A, Popovski D, Jayasekara WS, et al. : Low-Dose Histone Deacetylase Inhibitor Treatment Leads to Tumor Growth Arrest and Multi-Lineage Differentiation of Malignant Rhabdoid Tumors. Clin Cancer Res. 2016;22(14):3560–70. 10.1158/1078-0432.CCR-15-2260 [DOI] [PubMed] [Google Scholar]
- 102. Roulois D, Loo Yau H, Singhania R, et al. : DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell. 2015;162(5):961–73. 10.1016/j.cell.2015.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 103. Liu M, Ohtani H, Zhou W, et al. : Vitamin C increases viral mimicry induced by 5-aza-2'-deoxycytidine. Proc Natl Acad Sci U S A. 2016;113(37):10238–44. 10.1073/pnas.1612262113 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 104. Savickiene J, Treigyte G, Stirblyte I, et al. : Euchromatic histone methyltransferase 2 inhibitor, BIX-01294, sensitizes human promyelocytic leukemia HL-60 and NB4 cells to growth inhibition and differentiation. Leuk Res. 2014;38(7):822–9. 10.1016/j.leukres.2014.04.003 [DOI] [PubMed] [Google Scholar]
- 105. Oh SY, Seok JY, Choi YS, et al. : The Histone Methyltransferase Inhibitor BIX01294 Inhibits HIF-1α Stability and Angiogenesis. Mol Cells. 2015;38(6):528–34. 10.14348/molcells.2015.0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bhadury J, Nilsson LM, Muralidharan SV, et al. : BET and HDAC inhibitors induce similar genes and biological effects and synergize to kill in Myc-induced murine lymphoma. Proc Natl Acad Sci U S A. 2014;111(26):E2721–30. 10.1073/pnas.1406722111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Baker EK, Taylor S, Gupte A, et al. : BET inhibitors induce apoptosis through a MYC independent mechanism and synergise with CDK inhibitors to kill osteosarcoma cells. Sci Rep. 2015;5: 10120. 10.1038/srep10120 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 108. Knutson SK, Kawano S, Minoshima Y, et al. : Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther. 2014;13(4):842–54. 10.1158/1535-7163.MCT-13-0773 [DOI] [PubMed] [Google Scholar]
- 109. McCabe MT, Ott HM, Ganji G, et al. : EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–12. 10.1038/nature11606 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 110. Vaswani RG, Gehling VS, Dakin LA, et al. : Identification of ( R)- N-((4-Methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1 -(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1 H-indole-3-carboxamide (CPI-1205), a Potent and Selective Inhibitor of Histone Methyltransferase EZH2, Suitable for Phase I Clinical Trials for B-Cell Lymphomas. J Med Chem. 2016;59(21):9928–9941. 10.1021/acs.jmedchem.6b01315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lu B, Shen X, Zhang L, et al. : Discovery of EBI-2511: A Highly Potent and Orally Active EZH2 Inhibitor for the Treatment of Non-Hodgkin's Lymphoma. ACS Med Chem Lett. 2018;9(2):98–102. 10.1021/acsmedchemlett.7b00437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Konze KD, Ma A, Li F, et al. : An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem Biol. 2013;8(6):1324–34. 10.1021/cb400133j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Cohen AS, Tuysuz B, Shen Y, et al. : A novel mutation in EED associated with overgrowth. J Hum Genet. 2015;60(6):339–42. 10.1038/jhg.2015.26 [DOI] [PubMed] [Google Scholar]
- 114. Cohen AS, Gibson WT: EED-associated overgrowth in a second male patient. J Hum Genet. 2016;61(9):831–4. 10.1038/jhg.2016.51 [DOI] [PubMed] [Google Scholar]
- 115. Cooney E, Bi W, Schlesinger AE, et al. : Novel EED mutation in patient with Weaver syndrome. Am J Med Genet A. 2017;173(2):541–545. 10.1002/ajmg.a.38055 [DOI] [PubMed] [Google Scholar]
- 116. Imagawa E, Higashimoto K, Sakai Y, et al. : Mutations in genes encoding polycomb repressive complex 2 subunits cause Weaver syndrome. Hum Mutat. 2017;38(6):637–648. 10.1002/humu.23200 [DOI] [PubMed] [Google Scholar]
- 117. Tatton-Brown K, Loveday C, Yost S, et al. : Mutations in Epigenetic Regulation Genes Are a Major Cause of Overgrowth with Intellectual Disability. Am J Hum Genet. 2017;100(5):725–736. 10.1016/j.ajhg.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Gibson WT, Hood RL, Zhan SH, et al. : Mutations in EZH2 cause Weaver syndrome. Am J Hum Genet. 2012;90(1):110–8. 10.1016/j.ajhg.2011.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Tatton-Brown K, Hanks S, Ruark E, et al. : Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget. 2011;2(12):1127–33. 10.18632/oncotarget.385 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 120. Gotlib D, Ramaswamy R, Kurlander JE, et al. : Valproic Acid in Women and Girls of Childbearing Age. Curr Psychiatry Rep. 2017;19(9):58. 10.1007/s11920-017-0809-3 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 121. Jäger-Roman E, Deichl A, Jakob S, et al. : Fetal growth, major malformations, and minor anomalies in infants born to women receiving valproic acid. J Pediatr. 1986;108(6):997–1004. 10.1016/S0022-3476(86)80949-0 [DOI] [PubMed] [Google Scholar]
- 122. Samrén EB, van Duijn CM, Koch S, et al. : Maternal use of antiepileptic drugs and the risk of major congenital malformations: a joint European prospective study of human teratogenesis associated with maternal epilepsy. Epilepsia. 1997;38(9):981–990. 10.1111/j.1528-1157.1997.tb01480.x [DOI] [PubMed] [Google Scholar]
- 123. Epstein RA, Moore KM, Bobo WV: Treatment of nonpsychotic major depression during pregnancy: patient safety and challenges. Drug Healthc Patient Saf. 2014;6:109–129. 10.2147/DHPS.S43308 [DOI] [PMC free article] [PubMed] [Google Scholar]