Abstract
Lentinus tigrinus is a species of wood-decaying fungi (Polyporales) that has an agaricoid form (a gilled mushroom) and a secotioid form (puffball-like, with enclosed spore-bearing structures). Previous studies suggested that the secotioid form is conferred by a recessive allele of a single locus. We sequenced the genomes of one agaricoid (Aga) strain and one secotioid (Sec) strain (39.53–39.88 Mb, with 15,581–15,380 genes, respectively). We mated the Sec and Aga monokaryons, genotyped the progeny, and performed bulked segregant analysis (BSA). We also fruited three Sec/Sec and three Aga/Aga dikaryons, and sampled transcriptomes at four developmental stages. Using BSA, we identified 105 top candidate genes with nonsynonymous SNPs that cosegregate with fruiting body phenotype. Transcriptome analyses of Sec/Sec versus Aga/Aga dikaryons identified 907 differentially expressed genes (DEGs) along four developmental stages. On the basis of BSA and DEGs, the top 25 candidate genes related to fruiting body development span 1.5 Mb (4% of the genome), possibly on a single chromosome, although the precise locus that controls the secotioid phenotype is unresolved. The top candidates include genes encoding a cytochrome P450 and an ATP-dependent RNA helicase, which may play a role in development, based on studies in other fungi.
Keywords: basidiomycetes, bulked segregant analysis, development, fungi, gasteromycetes, transcriptome
Introduction
Mushrooms, the fruiting bodies of Agaricomycetes (Fungi, Basidiomycota), can be divided into two artificial groups based on morphology: hymenomycetes, which produce spores on the surface of the fruiting body, and gasteromycetes, which produce spores internally. Hymenomycetes include gilled mushrooms (“agarics”), toothed fungi, polypores, coral fungi, and crust fungi, whereas gasteromycetes include puffballs, truffles, stinkhorns, earthstars, and bird’s nest fungi. Gasteromycetes have evolved at least 120 times from hymenomycete ancestors (Hibbett et al. 2007; Sánchez-García M, Ryberg M, Hibbett D, unpublished data). Evolution of gasteromycetes involves enclosure of the hymenophore (comprising the spore-bearing structures), and loss of a complex mechanism of spore discharge, called ballistospory, which is presumably irreversible (Hibbett 2004; Liu et al. 2017).
Most gasteromycetes bear scant resemblance to their closest hymenomycete relatives, and their phylogenetic placements have only been resolved with molecular data. Exceptions include the so-called “secotioid” fungi, which are gasteromycetes that retain gross morphological similarities to certain hymenomycetes (Thiers 1984; Bruns et al. 1989). Some secotioid fungi resemble mushrooms in which the cap fails to open (interpreted as a “developmental arrest”), and a few have retained ballistospory (Desjardin et al. 1995). Secotioid fungi may provide clues to the early stages of morphological transformations from hymenomycetes to gasteromycetes, but the genetic bases of such transformations are obscure (Nagy et al. 2018).
Lentinus tigrinus is a white-rot wood-decaying mushroom that has both an agaricoid form, with exposed gills, and a secotioid form (fig. 1). In both forms, the gills originate as ridges of tissue on the surface of the young fruiting body, but in the secotioid form a layer of hyphae later proliferates from the margins of the developing gills and eventually encloses the hymenophore (Hibbett et al. 1994). Both forms are ballistosporic, but only the agaricoid form releases spores into the air; in the secotioid form the spores are trapped within the fruiting body.
Fig. 1.
—Lentinus tigrinus fruiting bodies produced in culture. (A) RLG-9953 (agaricoid); (B) FP-102501 (secotioid); (C) Lenti6/Lenti7.
Analyses of heritability have suggested that the secotioid phenotype is conferred by a recessive allele of a single gene (Rosinski and Faro 1968; Hibbett et al. 1994), but the identity of the locus is unknown. Lentinus tigrinus is widely reported in North America, Eurasia, and North Africa, but the secotioid form only occurs in North America (Pegler 1983). A phylogenetic study of Lentinus with broad geographic sampling suggested that the North American populations of L. tigrinus form a monophyletic group, separate from Eurasian populations (Grand et al. 2011), which is consistent with the view that evolution of the secotioid form is a recent innovation, perhaps the consequence of a single mutation.
We sequenced two strains of L. tigrinus with agaricoid and secotioid forms, and identified genomic regions associated with phenotypes using bulked segregant analysis (BSA). We also examined gene expression between the two phenotypes across four developmental stages. We thus identified 25 top candidate genes, based on both BSA and gene expression criteria, which are likely to be on the same chromosome.
Materials and Methods
Culturing and genetic crosses used methods previously described for L. tigrinus (Hibbett et al. 1993, 1994) and DNA and RNA were obtained from mycelia of Lenti6 (Aga) and Lenti7 (Sec) cultured on liquid malt extract media or from fruiting bodies. Library construction, DNA sequencing, and genome assembly and annotation were conducted at the JGI. Detailed methods including SNP, BSA, and transcriptome analyses are described in Supplementary Material.
Strain Development, Genome Sequencing, Assembly, and Annotation
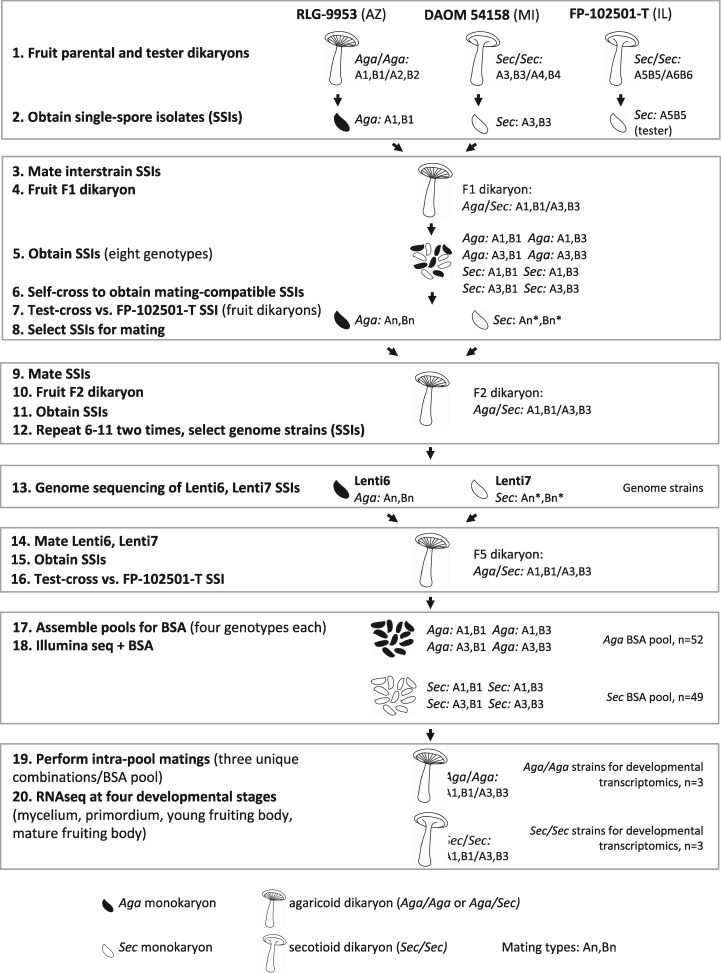
We obtained three wild dikaryons of L. tigrinus from the USDA Forest Products Laboratory, Madison WI, including two derived from secotioid specimens, DAOM-54158 (Michigan) and FP-102501-T (Illinois), and one from an agaricoid specimen, RLG-9953-Sp (Arizona). DAOM-54158 and FP-102501-T produced secotioid fruiting bodies on sawdust-wheat bran medium, and RLG-9953-Sp produced agaricoid fruiting bodies (fig. 1). An F1 cross between single spore isolates (SSIs) derived from DAOM-54158 and RLG-9953-Sp yielded a dikaryon with an agaricoid fruiting body, from which 10 SSIs were obtained, self-crossed to determine mating compatibility, and then mated to a tester strain from FP-102501-T (fig. 2, steps 1–7). Six of the resulting dikaryons produced secotioid fruiting bodies and four produced agaricoid fruiting bodies, which is consistent with prior results suggesting that the secotioid form is conferred by a recessive allele of a single locus. Two additional rounds of self-crossing were performed (intended to purge heterozygosity outside of the mating type and fruiting body morphology loci), yielding monokaryons Lenti6, carrying the genotype of the agaricoid form (Aga), and Lenti7, with the genotype of the secotioid form (Sec) (fig. 2, steps 8–13). A dikaryon obtained by mating Lenti6 and Lenti7 produced an agaricoid fruiting body (fig. 1C).
Fig. 2.
—Strain development for genome sequencing (steps 1–13), BSA (steps 14–18), and developmental transcriptomics (steps 19–20). Mating alleles of single-spore isolates derived from the parental and tester dikaryons were arbitrarily numbered A1, B1 (RLG-9953), A3, B3 (DAOM 54158), and A5, B5 (FP 102501-T). Mating type alleles of single-spore progeny were not determined after step 2. Accordingly, mating types are designated An, An*, Bn, and Bn*, where An ≠ An* and Bn ≠ Bn*. Because Lentinus tigrinus is heterothallic and tetrapolar, all dikaryons produced after step 2 must be A1, B1/A3, B3. See text for further details.
We isolated genomic DNA from cultures in liquid media, conducted Illumina sequencing, and obtained assemblies of 39.88 Mb distributed among 286 scaffolds (97.8 × average coverage) for Lenti6, and 39.53 Mb on 207 scaffolds (96.2 × average coverage) for Lenti7, containing 15,581 and 15,380 protein-coding genes, respectively (supplementary table S1, Supplementary Material online). Subsequent descriptions of genome contents refer to Lenti7 owing to its superior assembly (except as noted).
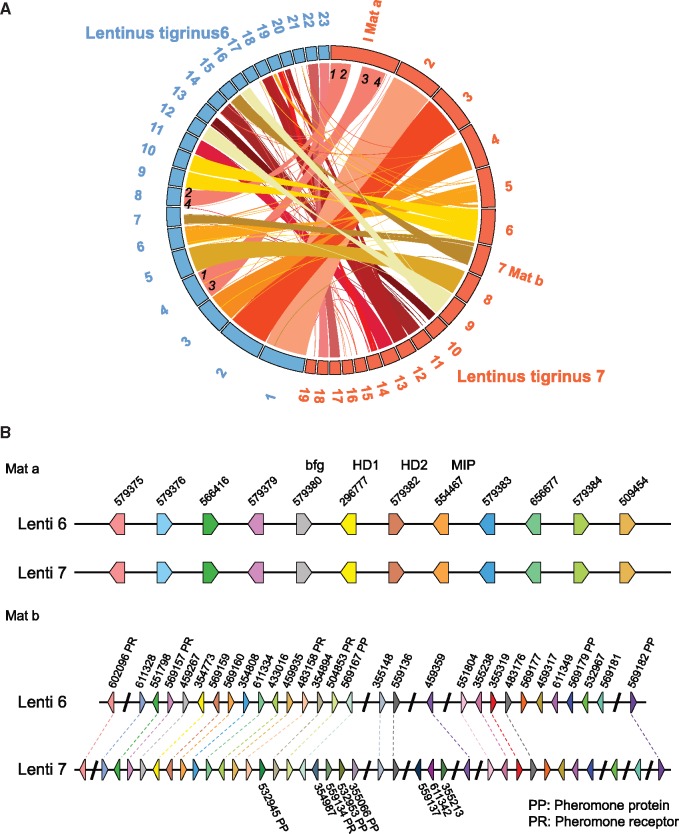
To assess synteny between genomes we used nucmer (Kurtz et al. 2004) to map all scaffolds >0.5 Mb, which comprise 22 Mb (56%) of Lenti6 (23 scaffolds) and 25 Mb (64%) of Lenti7 (19 scaffolds). Lenti6 and Lenti7 share a high degree of conservation, with only a few blocks rearranged, notably a rearrangement between scaffold 1 of Lenti7 and a combination of scaffolds 4 and 8 from Lenti6 (fig. 3A). Because of the length cutoff, some scaffolds do not show their synteny blocks in the plot. The high similarity extends to the whole genome level. Between the two genomes, only 86 rearrangements and 50 inversions were found among 1,404 conserved synteny blocks (96% of the total genome length), with as high as 99% average identity.
Fig. 3.
—Synteny blocks between Lenti6 and Lenti7 (scaffolds >0.5 Mb). The length of each block is relative to its size. Colored bands and lines linking scaffolds represent syntenic blocks (minimum 1 kb) shared by the two strains. Numbers to the inside of Lenti7 scaffold 1 and Lenti6 scaffolds 4 and 8 represent major rearrangements. (A) Scaffold locations of MAT-A HD (homeodomain) and MAT-B PR (pheromone/receptor) loci are indicated on Lenti7. (B) Distribution of genes in the MAT-A HD and MAT-B PR loci of Lenti6 and Lenti7.
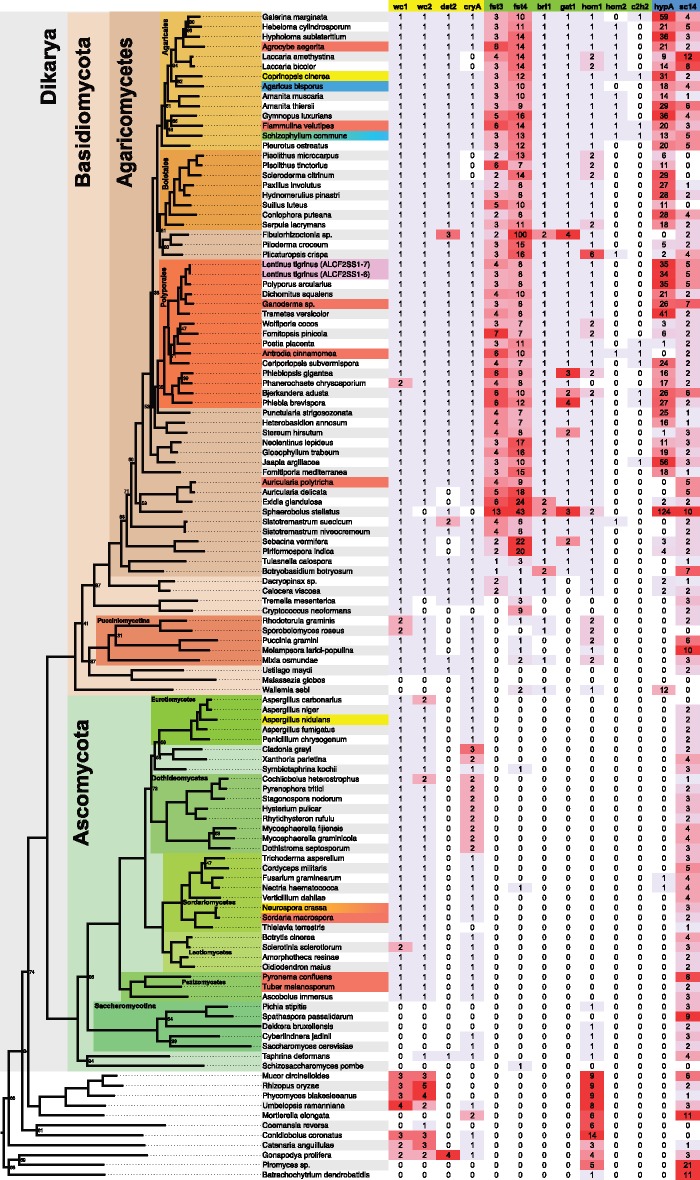
We focused on genes encoding decay enzymes (see supplementary text, figs. S1 and S2, and table S2, Supplementary Material online) and homologs of genes that have previously been suggested to play roles in fruiting body development in Agaricomycetes, including genes encoding transcription factors (TFs) (fst3, fst4, bri1, gat1, hom1, hom2, and c2h2; Ohmet al. 2010), hydrophobins (hypA and sc14; Wessels et al. 1991; Lugones et al. 1998; Santos and Labarere 1999; Banerjeeet al. 2008), and components of light-signaling pathways (wc1, wc2, dst2, and cryA; Terashima et al. 2005; Corrochano 2007; Kuratani et al. 2010; fig. 4). Lenti7 possesses 286 genes encoding TFs, including four copies of fst3, eight copies of fst4, and one copy each of bri1, gat1, and hom1 (supplementary table S3, Supplementary Material online), based on BLAST searches using queries from Coprinopsis cinerea and Schizophyllum commune. Lentinus tigrinus also has one copy each of wc1, wc2, dst2, and cryA, 35 genes homologous to hypA of Agaricus bisporus and five genes homologous to sc14 of S. commune (fig. 4). The complement of putative developmental genes in L. tigrinus is typical for Agaricomycetes, based on a comparison of 118 genomes from across all Fungi (fig. 4; for further details, see Supplementary Material).
Fig. 4.
—Phylogenetic distribution of light-signaling proteins (highlighted in yellow), transcription factors (green), and hydrophobins (blue) implicated in fruiting body morphogenesis, in 118 fungal genomes. Highlighted taxon names indicate species with previously published transcriptomes from fruiting bodies.
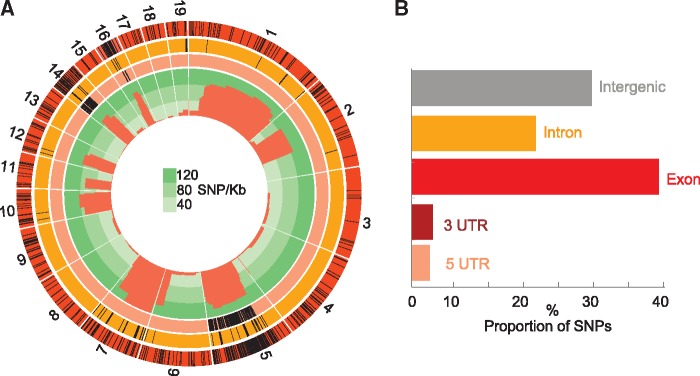
Using stringent search criteria not only relying on the annotation and assembly (see Materials and Methods), we identified 177 (43 expressed: fragments per kilobase of transcript per million mapped reads [FPKM] ≥ 1) and 199 (74 expressed) strain-specific “orphan” genes (vs. “shared” genes) that are unique to Lenti6 and Lenti7, respectively (supplementary table S4, Supplementary Material online). Orphan genes are concentrated on scaffolds 5, 7, and 14 of Lenti7 (fig. 5), and are associated with high average substitution rates of their scaffolds (rho = 0.89, P < 0.0001 based on the 19 largest scaffolds). Over 60% of the orphan genes occur in clusters of two or more genes (supplementary fig. S3, Supplementary Material online). In addition, orphan genes encode significantly shorter proteins than shared genes and are more likely to lack functional domains (supplementary fig. S4, Supplementary Material online).
Fig. 5.
—Genome-wide distribution of SNPs, BSA, DEGs, and orphan genes across the 19 largest scaffolds of Lenti7. (A) Circos-plot of genomic features between Lenti6 and Lenti7. Outer track (red): 605 DEGs; second track (orange): 117 Lenti7-specific orphan genes; third track (peach): 221 BSA candidate genes; inner track (green): SNP density. (B) Distribution of SNPs across intergenic regions, exons, introns, and 3′ and 5′ UTRs in Lenti7.
To determine the evolutionary origin of the orphan genes, we performed BLASTP searches against 19 other species, including 17 Polyporales and two other Agaricomycetes, using the orphan genes as queries. Only seven putative orphan genes in Lenti6 and eleven in Lenti7 have significant hits in at least one of the other species (supplementary fig. S5, Supplementary Material online), suggesting that they may have been lost in one of the two L. tigrinus strains. Thus, 95% of the orphan genes (170 genes in Lenti6 and 188 genes in Lenti7) have probably arisen in the lineage leading to L. tigrinus.
SNP and BSA
To assess the distribution of SNPs between Lenti6 and Lenti7, we performed a genetic diversity scan using a read-mapping approach. Briefly, Illumina reads from Lenti6 were mapped onto the Lenti7 assembly using BWA (Li and Durbin 2009), and then SNPs were called and filtered using SAMtools (Li et al. 2009) and bcftools (Li et al. 2009), respectively (for further details, see the Supplementary Material). The density of 132,014 SNPs among the 19 largest scaffolds of Lenti7 ranges from 0.007/kb (scaffold 13) to 17.8/kb (scaffold 7) (supplementary table S5, Supplementary Material online), which reflects the inbred nature of the strains and possibly the stringency of our SNP detection protocol (only SNPs supported by 90% of the reads were scored) (fig. 5A). SNPs occurred in intergenic regions, introns, exons, and UTRs (fig. 5B). SNPs in exons account for 40% of total SNPs, and occur in 3,073 genes. Among these 3,073 genes, 2,585 genes have diverse essential functions, such as cell wall modification (supplementary fig. S6, Supplementary Material online), and bear nonsynonymous SNPs.
To identify SNPs associated with fruiting body phenotypes, we performed BSA using 101 SSIs that we obtained by fruiting a Lenti6-Lenti7 (Aga/Sec) dikaryon (fig. 2). SSIs were genotyped by mating with an FP-102501-T tester strain (Sec) to screen for Aga and Sec genotypes, and divided into Sec and Aga pools, with 49 and 52 individuals, respectively (fig. 2). We identified 231,797 SNPs (0.5% of the whole genome) between the two phenotype pools, of which 99,993 SNPs (“BSA sites”) have significantly different allele frequencies (FDR < 0.01).
Surprisingly, none of the BSA sites had fixed allelic variants in the two pools, so we repeated the test crosses. Nine of the SSIs that had been scored as Sec produced agaricoid fruiting bodies in the second test crosses, and four putative Aga SSIs produced secotioid fruiting bodies (all test cross progeny are shown in supplementary figs. S7 and S8, Supplementary Material online). On basis of these repeated crosses, the Sec pool contains ∼18.4% Aga SSIs, whereas the Aga pool contains ∼9.2% Sec SSIs. The mixed phenotypes to some extent impair identification of fixed BSA sites. In addition, each pool included “intermediate” forms with varying degrees of fusion in the margins of the gills, which were ultimately coded as agaricoid (9 in the Sec pool and 13 in the Aga pool; e.g., supplementary fig. S7, panels JJ, UU, Supplementary Material online and supplementary fig. S8, panels QQ, UU, Supplementary Material online). It is possible that the intermediate forms are caused by environmental conditions, such as high CO2 concentrations (Moore et al. 2008), although a genetic basis for the intermediate forms cannot be excluded.
We selected the 1,312 BSA sites from 99,993 BSA sites using the overlap of the top 5% lowest P-values based on the Cochran–Mantel–Haenszel test and the top 5% highest different allele frequencies as parameters. Among the selected BSA sites, the median difference in allele frequencies between pools is 0.80 (0.79–0.87), which may reflect the mixture of phenotypes that were inadvertently included in each pool (supplementary fig. S10, Supplementary Material online). The BSA sites are located in coding regions, introns, UTR, and intergenic regions (supplementary table S6, Supplementary Material online). In the coding regions, we found eight start codon gains, one start codon loss, and one stop codon gain (supplementary table S6, Supplementary Material online). BSA sites from exonic regions yield 212 “BSA candidate genes,” of which 105 carry nonsynonymous SNPs and 169 have synonymous SNPs (62 BSA candidate genes have both synonymous and nonsynonymous SNPs; supplementary table S6 and fig. S9, Supplementary Material online).
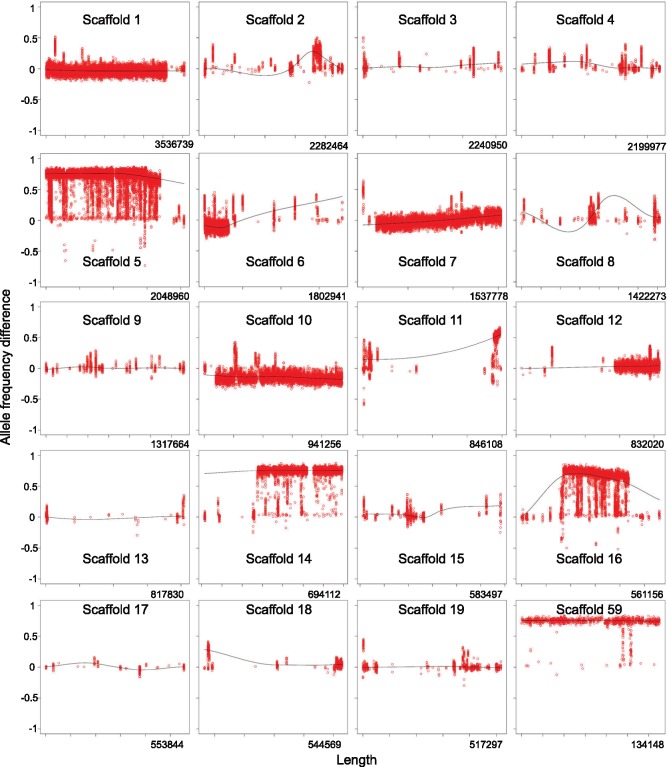
The BSA candidate genes are concentrated on scaffolds 5, 14, and 16 of Lenti7 (corresponding to scaffolds [6, 31], [19, 51], and [62, 63, and 70] of Lenti6), which also have high densities of orphan genes (figs. 5 and 6, supplementary table S4, Supplementary Material online). To assess whether scaffolds 5, 14, and 16 of Lenti7 could be linked, we compared them to the genomes of Polyporus brumalis and Ganoderma lucidum, which are closely related species of Polyporaceae (Justo et al. 2017). Scaffold 5 from P. brumalis and scaffold 3 from Ganoderma sp. contained significant hits to Lenti7 scaffolds 5, 14, and 16, suggesting that they may represent parts of a single chromosome (supplementary fig. S11, Supplementary Material online). Scaffold 59 (which is only 134 kb) shows a similar pattern of differences in allele frequencies as scaffolds 5, 14, and 16 (fig. 6), and it is also likely to be part of the same hypothetical chromosome (supplementary fig. S11, Supplementary Material online).
Fig. 6.
—Differences in allele frequencies between Sec and Aga pools in bulked segregant analyses across the 19 largest scaffolds and scaffold 59 of the Lenti7 genome. Allele frequencies in each pool were averaged over two replicates and plotted relative to their position. Alleles that segregate with fruiting body phenotype are concentrated on scaffolds 5, 14, 16, and 59.
Lentinus tigrinus is heterothallic and tetrapolar (Hibbett et al. 1994) (i.e., mating compatibility among monokaryons requires heteroallelism at each of two MAT loci; Jameset al. 2013). Scaffolds 1 and 7 of Lenti7, which contain the MAT-A HD and MAT-B P/R mating loci (fig. 3B), also carry large numbers of SNPs and orphan genes, but do not contain any of the BSA candidate genes (figs. 5 and 6). The high density of SNPs, but absence of BSA candidate genes on these scaffolds presumably reflects selection in our strain development strategy (fig. 2) for polymorphism at the mating loci, which are unlinked from the regions responsible for the secotioid phenotype.
Differential Gene Expression between Secotioid and Agaricoid Forms
We created three Aga/Aga dikaryons (producing agaricoid fruiting bodies) and three Sec/Sec dikaryons (secotioid) by mating unique combinations of SSIs that were used in BSA, yielding three biological replicates for each phenotype (six samples total) (fig. 2, steps 17–20). We cultured the dikaryons on sawdust-bran medium and sampled RNA at four developmental stages: Vegetative mycelium, fruiting body primordia, young fruiting bodies, and mature fruiting bodies (supplementary fig. S12, Supplementary Material online). 13,584 predicted genes were expressed (FPKM ≥ 1), representing 88% of the total annotated genes from Lenti7 (which was used as the reference genome). Hierarchical clustering of RNA-seq samples from two strains indicates high overall similarity of expression patterns at the same stages between phenotypes (supplementary fig. S13, Supplementary Material online).
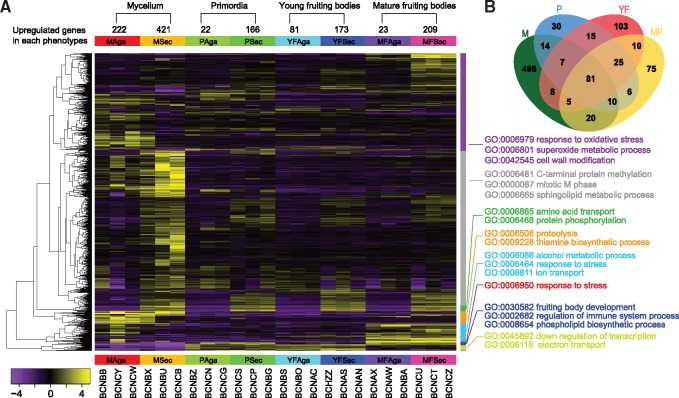
We detected 907 differentially expressed genes (DEGs) that were significantly up or downregulated between secotioid and agaricoid forms (FDR < 0.05 and logFC > 2), with 643, 188, 254, and 232 genes differentially expressed at the mycelium, primordium, young fruiting body, and mature fruiting body stages, respectively (fig. 7A, supplementary table S7, Supplementary Material online). Among the four stages, the DEGs are enriched at the mycelium stage (70% of total DEGs occurred at this stage). Eighty one genes had significantly altered expression during all four stages, and 706 genes were differentially expressed at only one stage (fig. 7B).
Fig. 7.
—Hierarchical clustering and functional classification of 907 genes that are differentially expressed between secotioid (Sec) and agaricoid (Aga) forms. (A) Hierarchical clustering of differentially expressed genes (fold change >4 and FDR < 0.05) in in mycelia (M), primordia (P), young fruiting bodies (YF), and mature fruiting bodies (MF). Enriched GO categories are shown by each cluster. Values of the color key refer to the log base 2 of fragments per kilobase of transcript per million mapped reads (FPKM). (B) Venn diagram of phenotype-related differential expressed genes in four stages.
We compared the distribution of BSA sites in regions upstream of differentially expressed versus nondifferentially expressed genes. 6% (56) of the 907 DEGs and 1% (189) of the 12677 nonDEGs have BSA sites in their upstream 1 kb regions (supplementary fig. S14, Supplementary Material online). Genes that are differentially expressed between phenotypes are significantly more likely to have upstream BSA sites than nonDEGs (Fisher’s exact test P < 0.001), which is consistent with the view that divergence in TF binding sites may alter expression levels. Moreover, we found 12 DEGs having BSA sites in their annotated UTRs (supplementary table S6, Supplementary Material online).
We examined co-expression patterns of the 907 DEGs over all four stages, and grouped them into nine clusters (using a cut-off of 40% of the tree height to delimit clusters; fig. 7A). GO enrichment analysis revealed DEGs from each cluster involved in various biological processes. In one cluster there is an overrepresented function associated with “reproductive fruiting body development” (GO: 0030582), including three genes (523,664, 523,675, and 571,802) encoding aegerolysin-like proteins, which have been suggested to be expressed preferentially in the primordia, and in the early stages of fruiting body development in Pleurotus ostreatus and Agrocybe aegerita (Berne 2002; Vidic et al. 2005; Berne et al. 2009). However, all three aegerolysin genes show differential expression in the mycelium stage, not during fruiting body formation, suggesting that there is greater functional diversity of the aegerolysin family than previously recognized (Nayak et al. 2013).
61 of the 212 BSA candidate genes are differentially expressed between secotioid and agaricoid phenotypes (supplementary fig. S9B, Supplementary Material online). Of these 61 genes, 25 have nonsynonymous BSA sites, and were differentially expressed at the young or mature fruiting body stages. Therefore, these 25 genes, from scaffolds 5 and 14, represent the top candidates for genes that are likely to influence fruiting body form. 21 of these 25 genes (excluding Lenti7_1_546183, Lenti7_1_551037, Lenti7_1_568005, Lenti7_1_610076) are upregulated in secotioid phenotypes relative to agaricoid phenotypes (supplementary fig. S9B, Supplementary Material online, table 1). Two of these genes, one encoding a cytochrome P450 and the other an ATP-dependent RNA helicase, are members of families which include genes that have previously been shown to affect morphology of fruiting bodies in the basidiomycete C. cinerea (Muraguchi and Kamada, 2000) or the ascomycete Cordyceps militaris (Zheng et al. 2015), respectively.
Table 1.
25 Top Candidate Genes Identified with BSA (nonsynonymous) and DEG
| Protein ID (Lenti7/6) | Location | Stages | Gene Description |
|---|---|---|---|
| 85195/ND | scaffold_14: 369055-370013 | YF | — |
| 86465/526915 | scaffold_14: 443879-446907 | YF/MF | ATP-dependent RNA helicase |
| 87620/ND | scaffold_14: 563749-564340 | YF/MF | — |
| 283939/ND | scaffold_5: 195155-196111 | YF | — |
| 456840/496367 | scaffold_5: 918091-920336 | YF/MF | Tripeptidyl-peptidase |
| 457171/556809 | scaffold_5: 1321131-1322690 | YF/MF | — |
| 464033/535943 | scaffold_14: 357810-359372 | YF/MF | Tyrosinase |
| 531647/572283 | scaffold_5: 669180-671138 | YF/MF | Aldo/keto reductase |
| 531677/650807 | scaffold_5: 738255-739089 | YF/MF | — |
| 531846/542591 | scaffold_5: 1163998-1167020 | YF/MF | Sodium/hydrogen exchanger |
| 535696/548680 | scaffold_14: 430337-432896 | YF | Cytochrome P450 |
| 543568/502902 | scaffold_5: 40571-41704 | YF/MF | Protein phosphorylation |
| 543726/650805 | scaffold_5: 747877-749069 | YF/MF | Aldo/keto reductase |
| 546139/118234 | scaffold_14: 424410-425504 | YF/MF | Chromatin assembly factor-I |
| 546183/611691 | scaffold_14: 608506-611139 | MF | Flavin-containing monooxygenase |
| 551037/658517 | scaffold_5: 424249-427854 | YF | Methylase containing SET domain |
| 567942/572310 | scaffold_5: 615457-616764 | YF | — |
| 568005/496312 | scaffold_5: 789648-791747 | MF | Tripeptidyl-peptidase |
| 568067/608283 | scaffold_5: 987789-989330 | MF | — |
| 568092/608272 | scaffold_5: 1055550-1057038 | YF | Leucine Rich Repeat |
| 568118/358653 | scaffold_5: 1122189-1123261 | YF | — |
| 610076/586341 | scaffold_5: 84608-89187 | YF | Triphosphate hydrolase protein |
| 610453/604269 | scaffold_5: 1327281-1329825 | YF/MF | — |
| 610475/581275 | scaffold_5: 1423453-1424456 | YF | — |
| 613650/653772 | scaffold_14: 612169-614204 | MF | Microtubule-associated proteins |
Note.—ND: ortholog of lenti7 gene is not among the annoated genes from lenti6 due to poor assembly or bad annotation.
Seven TF genes were found to show differential expression between phenotypes, mainly at the young fruiting body stage (table 2). One of the differentially expressed TF genes (Lenti7_1_567867) contains a HOX domain similar to that of hom2, which has been shown to affect fruiting body development in S. commune (Ohm et al. 2011). However, none of the TFs previously implicated in fruiting body development that are present in the L. tigrinus genome (fst3, fst4, bri1, gat1, hom1, hom2) show differential expression between phenotypes; they may play roles in development of both phenotypes, but are probably not responsible for the transition to the secotioid form.
Table 2.
Transcription Factor Genes Identified with Bulked Segregant Analysis or with Differential Expression Levels between Phenotypes
| Protein ID (Lenti7/6) | Location | BSAa | Stagesb | Domains |
|---|---|---|---|---|
| 537478/229775 | scaffold_25: 159517-160085 | M | Zn(2)-Cys(6) | |
| 535761/526822 | scaffold_14: 632028-634037 | YES (s) | M, P, YF | Transcription factor Tfb4 |
| 569405/572504 | scaffold_7: 1195929-1196337 | — | M, YF | Zn(2)-Cys(6) |
| 215353/566174 | scaffold_32: 341786-343826 | — | MF | Zn(2)-Cys(6) |
| 465877/613826 | scaffold_19: 280471-283089 | — | YF | Heat shock transcription factor |
| 567867/543015 | scaffold_5: 393051-396002 | — | YF | HOX domain |
| 573265/613178 | scaffold_24: 126363-128314 | — | YF | Serine/threonine protein kinase |
| 464098/502691 | scaffold_14: 412497-415601 | YES (ns) | — | Zn(2)-Cys(6) |
| 503037/584228 | scaffold_5: 1223891-1229088 | YES (s) | — | Zn(2)-Cys(6) |
| 509362/557565 | scaffold_14: 341275-343024 | YES (s) | — | Zn(2)-Cys(6) |
| 548499/586366 | scaffold_59: 106797-116305 | YES (ns) | — | Zinc finger, GATA-type |
| 599750/113648 | scaffold_59: 61101-62947 | YES (s) | — | HOX domain (hom1) |
n, nonsynonymous substitution; s, synonymous substitution.
Stages at which expression varies between phenotypes.
Six TF genes had significantly different allele frequencies between Sec and Aga pools in BSA, and one of these TF genes, Lenti7_1_535761, was also differentially expressed at three stages (table 2). Among these six TF genes, one is a homolog of hom1 (Lenti7_1_599750), which has also been shown to play a role in development in S. commune (Ohm et al. 2010, 2011). The hom1 alleles have only synonymous substitutions in their coding regions, but synonymous substitutions can have phenotypic consequences (Bailey et al. 2014). In addition, the occurrence of noncoding BSA sites upstream (1 kb) of many DEGs also suggest that mutations in TF binding sites could contribute to the secotioid phenotype.
Eleven (32%) hydrophobin genes, another important class of developmental genes, show differential expression between phenotypes, all at the mycelium stage, but none of the genes encoding hydrophobins are among the BSA candidate genes (supplementary table S3, Supplementary Material online).
Conclusions
Lentinus tigrinus provides a unique opportunity to study fruiting body evolution in Agaricomycetes, because it has naturally occurring agaricoid and secotioid forms. Results presented here are consistent with the view that the secotioid phenotype is conferred by a recessive allele at a single locus, but the precise identity of the locus is unresolved. The secotioid form could result from a shift in transcription level without a change in protein sequence, or a nonsynonymous mutation without a change in expression. Our top candidate genes (table 1) were identified with a combination of BSA and differential expression criteria, but it is possible that one of the many genes identified with only one of the two criteria is responsible for the secotioid form. Other potential mechanisms that could underlie the secotioid phenotype include DNA methylation (potentially affecting gene expression) and RNA editing (which could cause amino acid replacements in proteins). However, this study did not include genome-wide analyses of such epigenetic modifications.
The top 25 candidate genes span almost 1.5 Mb (ca. 4% of the genome) on two scaffolds (1,383,885 bp on scaffold 5, and 256,394 bp on scaffold 14). Seven of the candidate TFs, identified with both BSA or differential expression analyses, also occur on scaffolds 5, 14, and 59, with the rest distributed on scaffolds 19–32 (table 2). It is possible that scaffolds 5, 14, and 59 represent parts of a single chromosome, but long-read sequencing (PacBio or nanopore) will be needed to test this hypothesis.
Like many saprotrophic fungi, L. tigrinus grows well in culture, but fruiting body formation is sensitive to environmental conditions, including light, temperature, humidity, and CO2 concentration. Most progeny could be clearly differentiated as secotioid or agaricoid in test crosses, but intermediate forms occurred. Further studies under controlled conditions will be necessary to determine reaction norms for fruiting body development in the secotioid and agaricoid forms of L. tigrinus. Genome-wide association studies using geographically diverse wild strains could help reconstruct the genetic bases and evolutionary history of the secotioid form in L. tigrinus. Other kinds of analyses, such as gene knock-out experiments, will eventually be needed to test specific mechanistic hypotheses, but at present they are constrained by the absence of a transformation protocol for L. tigrinus.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This research was supported by National Science Foundation award DEB-1456588 to D.S.H. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. For permission to present data from unpublished genomes (supplementary fig. S4, Supplementary Material online), we thank Daniele Armaleo and Francois Lutzoni (Cladonia grayi), Joey Spatafora (Ascobolus immersus), Sebastien Duplessis (Melampsora larici-populina), Ken Wolfe (Sporobolomyces roseus), and Paul Dyer (Xanthoria parietina). Amy Yeager produced the mushroom and spore cartoons in figure 2.
Literature Cited
- Bailey SF, Hinz A, Kassen R.. 2014. Adaptive synonymous mutations in an experimentally evolved Pseudomonas fluorescens population. Nat Commun. 5:4076. [DOI] [PubMed] [Google Scholar]
- Banerjee G, Robertson DL, Leonard TJ.. 2008. Hydrophobins Sc3 and Sc4 gene expression in mounds, fruiting bodies and vegetative hyphae of Schizophyllum commune. Fungal Genet Biol. 453:171–179. [DOI] [PubMed] [Google Scholar]
- Berne S, Lah L, Sepcic K.. 2009. Aegerolysins: structure, function, and putative biological role. Protein Sci. 184:694–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berne S. 2002. Pleurotus and Agrocybe hemolysins, new proteins hypothetically involved in fungal fruiting. Biochim Biophys Acta. 15703:153–159. [DOI] [PubMed] [Google Scholar]
- Bruns TD, Fogel R, White TJ, Palmer JD.. 1989. Accelerated evolution of a false-truffle from a mushroom ancestor. Nature 3396220:140–142. [DOI] [PubMed] [Google Scholar]
- Corrochano LM. 2007. Fungal photoreceptors: sensory molecules for fungal development and behaviour. Photochem Photobiol Sci. 67:725–736. [DOI] [PubMed] [Google Scholar]
- Desjardin DE, Martínez-Peck L, Rajchenberg M.. 1995. An unusual psychrophilic aquatic agaric from Argentina. Mycologia 874:547–550. [Google Scholar]
- Grand EA, Hughes KW, Petersen RH.. 2011. Relationships within Lentinus subg. Lentinus (Polyporales, Agaricomycetes) with emphasis on sects. Lentinus and Tigrini. Mycol Progr. 104:399–413. [Google Scholar]
- Hibbett D. 2004. Trends in morphological evolution in homobasidiomycetes inferred using maximum likelihood: a comparison of binary and multistate approaches. Syst Biol. 536:889–903. [DOI] [PubMed] [Google Scholar]
- Hibbett DS, et al. 2007. A higher-level phylogenetic classification of the fungi. Mycol Res. 1115:509–547. [DOI] [PubMed] [Google Scholar]
- Hibbett DS, Murakami S, Tsuneda A.. 1993. Sporocarp ontogeny in Panus (Basidiomycotina): evolution and classification. Am J Bot. 8011:1336–1348. [Google Scholar]
- Hibbett DS, Tsuneda A, Murakami S.. 1994. The secotioid form of Lentinus tigrinus: genetics and development of a fungal morphological innovation. Am J Bot. 814:466–478. [Google Scholar]
- James TY, et al. 2013. Polyporales genomes reveal the genetic architecture underlying tetrapolar and bipolar mating systems. Mycologia 1056:1374–1390. [DOI] [PubMed] [Google Scholar]
- Justo A, et al. 2017. A revised family-level classification of the Polyporales (Basidiomycota). Fungal Biol. 1219:798–824. [DOI] [PubMed] [Google Scholar]
- Kuratani M, et al. 2010. The dst2 gene essential for photomorphogenesis of Coprinopsis cinerea encodes a protein with a putative FAD-binding-4 domain. Fungal Genet Biol. 472:152–158. [DOI] [PubMed] [Google Scholar]
- Kurtz S, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 52:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2514:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2516:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, et al. 2017. Asymmetric drop coalescence launches fungal ballistospores with directionality. J R Soc Interface. 14: 20170083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugones LG, Wosten HA, Wessels JG.. 1998. A hydrophobin (ABH3) specifically secreted by vegetatively growing hyphae of Agaricus bisporus (common white button mushroom). Microbiology 144(Pt 8):2345–2353. [DOI] [PubMed] [Google Scholar]
- Moore D, Gange AC, Gange EG, Boddy L.. 2008. Chapter 5 Fruit bodies: their production and development in relation to environment. Br Mycol Soc Symp Ser. 28:79–103. [Google Scholar]
- Muraguchi H, Kamada T.. 2000. A mutation in the eln2 gene encoding a cytochrome P450 of Coprinus cinereus affects mushroom morphogenesis. Fungal Genet Biol. 29(1):49–59. [DOI] [PubMed] [Google Scholar]
- Nagy LG, Kovacs GM, Krizsan K.. 2018. Complex multicellularity in fungi: evolutionary convergence, single origin, or both? Biol Rev Camb Philos Soc. 93(4):1778–1794. [DOI] [PubMed] [Google Scholar]
- Nayak AP, Green BJ, Beezhold. 2011. Fungal hemolysins Med Mycol.. 51(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm RA, de Jong JF, de Bekker C, Wösten HAB, Lugones LG.. 2011. Transcription factor genes of Schizophyllum commune involved in regulation of mushroom formation. Mol Microbiol. 816:1433–1445. [DOI] [PubMed] [Google Scholar]
- Ohm RA, et al. 2010. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol. 289:957–963. [DOI] [PubMed] [Google Scholar]
- Pegler DN. 1983. The genus Lentinus, a world monograph. Kew Bull Add Ser. 10:1–281. [Google Scholar]
- Rosinski MA, Faro S.. 1968. The genetic basis of hymenophore morphology in Panus tigrinus. (Bull. ex Fr.) Singer. Am J Bot. 55:720. [Google Scholar]
- Santos C, Labarere J.. 1999. Aa-Pri2, a single-copy gene from Agrocybe aegerita, specifically expressed during fruiting initiation, encodes a hydrophobin with a leucine-zipper domain. Curr Genet. 355:564–570. [DOI] [PubMed] [Google Scholar]
- Terashima K, Yuki K, Muraguchi H, Akiyama M, Kamada T.. 2005. The dst1 gene involved in mushroom photomorphogenesis of Coprinus cinereus encodes a putative photoreceptor for blue light. Genetics 1711:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiers HD. 1984. The secotioid syndrome. Mycologia 761:1–8. [Google Scholar]
- Vidic I, et al. 2005. Temporal and spatial expression of ostreolysin during development of the oyster mushroom (Pleurotus ostreatus). Mycol Res. 109(Pt 3):377–382. [PubMed] [Google Scholar]
- Wessels J, De Vries O, Asgeirsdottir SA, Schuren F.. 1991. Hydrophobin genes involved in formation of aerial hyphae and fruit bodies in Schizophyllum. Plant Cell. 38:793–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z-L, Qiu X-H, Han R-C.. 2015. Identification of the genes involved in the fruiting body production and cordycepin formation of Cordyceps militaris Fungus. Mycobiology. 431:37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.