Abstract
Arthrobacter spp. are coryneform Gram-positive aerobic bacteria, belonging to the class Actinobacteria. Representatives of this genus have mainly been isolated from soil, mud, sludge or sewage, and are usually mesophiles. In recent years, the presence of Arthrobacter spp. was also confirmed in various extreme, including permanently cold, environments. In this study, 36 psychrotolerant and metalotolerant Arthrobacter strains isolated from petroleum-contaminated soil from the King George Island (Antarctica), were screened for the presence of plasmids. The identified replicons were thoroughly characterized in order to assess their diversity and role in the adaptation of Arthrobacter spp. to harsh Antarctic conditions. The screening process identified 11 different plasmids, ranging in size from 8.4 to 90.6 kb. A thorough genomic analysis of these replicons detected the presence of numerous genes encoding proteins that potentially perform roles in adaptive processes such as (i) protection against ultraviolet (UV) radiation, (ii) resistance to heavy metals, (iii) transport and metabolism of organic compounds, (iv) sulfur metabolism, and (v) protection against exogenous DNA. Moreover, 10 of the plasmids carry genetic modules enabling conjugal transfer, which may facilitate their spread among bacteria in Antarctic soil. In addition, transposable elements were identified within the analyzed plasmids. Some of these elements carry passenger genes, which suggests that these replicons may be actively changing, and novel genetic modules of adaptive value could be acquired by transposition events. A comparative genomic analysis of plasmids identified in this study and other available Arthrobacter plasmids was performed. This showed only limited similarities between plasmids of Antarctic Arthrobacter strains and replicons of other, mostly mesophilic, isolates. This indicates that the plasmids identified in this study are novel and unique replicons. In addition, a thorough meta-analysis of 247 plasmids of psychrotolerant bacteria was performed, revealing the important role of these replicons in the adaptation of their hosts to extreme environments.
Keywords: Arthrobacter spp., plasmid, Antarctica, psychrotolerant, metalotolerant, adaptation
Introduction
Almost 80% of the Earth's biosphere is permanently cold, i.e., the temperature in these regions stays below 5°C throughout the whole year. The ocean depths constitute the majority of such environments, but there are also vast constantly cold terrestrial areas, such as the polar and alpine regions (De Maayer et al., 2014). Antarctica is known to be one of the coldest regions on Earth, with an average annual temperature ranging between −48°C at the South Pole and −2°C on island areas. It is an extreme habitat for indigenous organisms, not only because of the low temperatures, but also due to the occurrence of other harsh environmental conditions, including increased UV radiation, strong and drying winds, and the low amount of easily accessible nutrients (Martianov and Rakusa-Suszczewski, 1990; D'amico et al., 2006; Kejna et al., 2013; Cowan et al., 2014; Grzesiak et al., 2015; Kosek et al., 2017). Organisms living in Antarctica also have to face the emerging contamination of this region. Despite the fact that most of Antarctica is considered a pristine area with levels of contaminants that are lower than elsewhere in the world (these are mainly the consequence of natural bioweathering of heavy metal-rich rocks), there are areas where human influence is becoming more and more visible. Anthropogenic pollution by sulfur, heavy metals, polycyclic aromatic hydrocarbons and persistent organic pollutants (i.e., toxic organic compounds that are resistant to environmental degradation through chemical, biological, or photolytic processes, e.g., pesticides and pharmaceuticals) has been observed in Antarctica (Boutron and Wolff, 1989; Graf et al., 2010; Kukucka et al., 2010). Interestingly, the anthropogenic contaminants in this region mainly come from indigenous sources (e.g., ships and research stations). Intercontinental atmospheric long-range transport of pollutants to Antarctica seems to be highly limited, mostly due to the strong westerlies at mid-latitudes (40–60° S), which isolate the continent in winter (Kukucka et al., 2010; Stohl and Sodemann, 2010).
The extreme Antarctic environment leads to the selection of well-adapted organisms. These are poly-extremophiles that can withstand a variety of harsh physical and chemical conditions. In this study, we have focused on psychrotolerant, and at the same time metalotolerant, bacteria of the genus Arthrobacter, isolated from contaminated soil collected in the area of the Henryk Arctowski Polish Antarctic Station on King George Island (Romaniuk et al., 2018).
Arthrobacter spp. are Gram-positive, coryneform aerobic bacteria, belonging to the class Actinobacteria. They are frequently found in various extreme environments, including Arctic and Antarctic ice and soil (Dsouza et al., 2015; Singh et al., 2017), and also various contaminated sites (Sun et al., 2015; Ren et al., 2016; Chauhan et al., 2018). This wide distribution of Arthrobacter spp. could be a consequence of their high tolerance to various stress factors, such as permanent cold, long-term nutrient starvation, oxidative stress, osmotic pressure, and toxic compounds (Yao et al., 2015). Arthrobacter spp. can utilize a wide range of organic compounds that are recognized as environmental pollutants, including nitroglycerin, benzene derivatives, polycyclic aromatic hydrocarbons, haloalcohols, haloalkanes, N-heterocyclic compounds, insecticides and herbicides (Sharpee et al., 1973; Cripps, 1975; Pipke et al., 1987; Van Den Wijngaard et al., 1991; Jain et al., 1994; Casellas et al., 1997; Datta et al., 2000; Strong et al., 2002; Seo et al., 2006; Kallimanis et al., 2007; Rong et al., 2009; Husserl et al., 2010; Ren et al., 2016, 2018). In some cases, these abilities are conferred by plasmids, e.g., carbaryl (1-naphthyl N-methylcarbamate) pesticide utilization by Arthrobacter sp. RC1000 was linked with its plasmids pRC1 and pRC2 (Hayatsu et al., 1999), and a nicotine-utilization gene cluster was identified in plasmid pAO1 of Arthrobacter nicotinovorans (Baitsch et al., 2001). The wide distribution of Arthrobacter spp. and their ability to thrive in extreme environments indicates their great potential for environmental biotechnologies (Bjerketorp et al., 2018; Wang et al., 2018).
Currently (Nov. 11, 2018), 20 complete genomic and 33 plasmid sequences of Arthrobacter spp. are available in the GenBank database. The plasmids range in size from 3.7 to 754.2 kb, and they differ significantly in their gene content. Some carry genetic modules of adaptive and biotechnological value, e.g., the 113-kb linear plasmid pAL1 of A. nitroguajacolicus Rue61a, which confers the ability to utilize the N-heteroaromatic compound 2-methylquinoline (Overhage et al., 2005). A number of plasmids of mesophilic Arthrobacter spp. have been described in detail (Baitsch et al., 2001; Eaton, 2001; Igloi and Brandsch, 2003; Sandu et al., 2003; Overhage et al., 2005; Parschat et al., 2007; Jerke et al., 2008; Kolkenbrock and Fetzner, 2010; Kolkenbrock et al., 2010; Wagenknecht and Meinhardt, 2011; Mihasan and Brandsch, 2013, 2016; Mihasan et al., 2013; Mihasan, 2015). However, little is known about the genetic organization, biology and adaptive role of plasmids of Arctic and Antarctic, cold-active Arthrobacter spp. To our best knowledge, only one cryptic plasmid from a psychrotolerant Arthrobacter strain isolated from a Greenland ice core has been thoroughly described (Miteva et al., 2008).
Here, we report the detailed genomic analysis of 11 novel plasmids (named the ANT plasmids) identified within a pool of psychrotolerant and metalotolerant Antarctic Arthrobacter strains. Our analysis have revealed a putative role of the identified replicons in the adaptation of Arthrobacter spp. to the extreme Antarctic environment and gives an insight into the overall Arthrobacter plasmidome. What is more, this analysis complements previous works describing Antarctic plasmidomes, as in the majority they describe the ecological role of plasmids of Gram-negative bacteria (it was revealed that these replicons constitute more than 80% of all already analyzed plasmids of cold-active bacteria) (Dziewit and Bartosik, 2014; Ciok et al., 2018). Therefore, the knowledge on biology, evolution and diversity of plasmids of psychrotolerant Gram-positive bacteria is rather scarce. Therefore, thorough analyses of novel replicons of Arthrobacter spp. representing high GC content, Gram-positive genus give a broader view on the overall role of plasmids in bacteria inhabiting extreme polar environments and tend toward reducing the abovementioned bias in analyses of polar plasmidomes.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
Thirty six Arthrobacter strains isolated from petroleum-contaminated soil collected in Antarctica were analyzed in this study (Romaniuk et al., 2018). Additionally, Escherichia coli DH5α was used (Hanahan, 1983). The strains were grown in LB or minimal salts medium M9 (Sambrook and Russell, 2001) (liquid or solidified with 1.5% agar) at 20°C (Arthrobacter spp.) and 37°C (E. coli). The media were supplemented with kanamycin (50 μg/ml), sodium pyruvate (0.4%) and laminarin (1%). Plasmids used and constructed in this study are listed in Table S1.
All Arthrobacter strains analyzed in this study are deposited at the microbiological repository of Institute of Microbiology, Faculty of Biology, University of Warsaw and are available for further analyses upon request.
Standard Molecular Biology Techniques
Plasmid DNA was isolated using a standard alkaline lysis method (Birnboim and Doly, 1979). Routine DNA manipulations were carried out using standard methods (Sambrook and Russell, 2001). PCRs were performed using a KAPA HiFi PCR Kit and appropriate primer pairs in a Mastercycler (Eppendorf, Hamburg, Germany). The following program was used: a 3-min initial denaturation at 95°C, then 35 cycles of denaturation at 98°C for 20 s, annealing at 46–56°C (depending on the primer pair) for 15 s, and extension at 72°C for 30 s per kb, followed by a final extension at 72°C for 1 min per kb. Primers used in this study are listed in Table S1.
DNA Sequencing
DNA sequencing was performed using an Illumina MiSeq instrument in paired-end mode using a v3 chemistry kit. The obtained sequence reads were filtered for quality and assembled using Newbler v3.0 software (Roche, Basel, Switzerland). Final gap closure was performed by capillary sequencing of PCR products using an ABI3730xl DNA Analyser (Applied Biosystems, Waltham, USA). The summary of the sequencing of particular plasmids was presented in Table S2.
Bioinformatics
Plasmid sequences were manually annotated using Artemis software (Carver et al., 2012). Similarity searches were performed using the BLAST programs (Altschul et al., 1997) and Conserved Domains Database (Marchler-Bauer et al., 2017), provided by the NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Pfam (Finn et al., 2016). The detection of RNA sequences was performed using the ARAGORN v1.2.38 (Laslett and Canback, 2004) and tRNAscan-SE programs (Lowe and Chan, 2016). Helix-turn-helix motifs were identified using the helix-turn-helix DNA-binding motif prediction tool (Dodd and Egan, 1990). EC numbers were assigned using the KEGG database (Kanehisa et al., 2017) and UniProt Knowledgebase (UniProtKB) (Pundir et al., 2017). Restriction-modification systems were analyzed and classified using the REBASE database (Roberts et al., 2015). Insertion sequences were analyzed using ISfinder (Siguier et al., 2006). Comparative genomic analyses were performed using Circoletto (Darzentas, 2010) and BLASTp. The BLASTp search using a manually curated database of proteins encoded within the phenotypic modules of the ANT plasmids, was performed for investigation of the distribution of particular genetic modules in Arthrobacter genomes. For this analysis, the following cutoff values were applied (e-value < 1e-05 and amino acid sequence identity of at least 40%). Phylogenetic analyses were performed using the MEGA6 tool (Tamura et al., 2013) and trees were constructed using the Maximum Likelihood method based on the Tamura-Nei model (for the 16S rDNA tree) and Le and Gascuel model (for the ParA tree).
Growth Kinetics
Growth curves were determined for bacteria cultivated in 2 ml lots of appropriate media in the glass tubes at 20°C (Arthrobacter spp.) or 37°C (E. coli) with shaking (150 rpm). The OD600 of the culture was measured every 24 h using SunriseTM plate reader (Tecan, Männedorf, Switzerland) with Magellan software (Tecan).
Tellurium Resistance Testing
Analytical grade salt (Na2TeO3) was used in a resistance assay performed in 96-well plates, as described previously (Dziewit et al., 2013). Triplicate cultures of each strain were challenged with a range of concentrations of those heavy metal salts. Isolates that grew in the presence of 1 mM Te(IV) were considered resistant (Lewis et al., 2018).
Direct-Plate UV Irradiation Assay
Responses to UV irradiation of various strains were compared by applying the modified direct-plate irradiation protocol (Lin and Wang, 2001). Bacterial cultures were grown overnight to about 1 x 108 cells per ml. These cultures were plated on the LB plates and irradiated with the UVC lamp (Philips TUV−30-W-245 nm Lamp, type No. 57413-P/40) at a distance of 45 cm at exposure times (5, 10, 15, 18, 21, 24 s). Before each exposition, lids of the treatment plates were removed to avoid shielding. After irradiation, the lids were replaced and the plates were immediately placed in the incubator at 37°C for 48 h.
Nucleotide Sequence Accession Numbers
Nucleotide sequences of the Arthrobacter plasmids were deposited in the GenBank (NCBI) database under the accession numbers MH067967-MH067977. Additionally, nucleotide sequences and information concerning insertion sequences and transposons were deposited in ISfinder (Siguier et al., 2006) and transposon registry (https://transposon.lstmed.ac.uk/) databases, respectively.
Results and Discussion
General Features of the ANT Plasmids
In a previous study, we isolated and characterized 90 psychrotolerant bacterial strains from petroleum-contaminated soil collected near the Henryk Arctowski Polish Antarctic Station on King George Island, Antarctica (62°09.601′ S, 58°28.464′ W) (Romaniuk et al., 2018). PCR amplification and sequencing of the 16S rRNA genes of these isolates followed by comparative sequence analysis allowed taxonomic assignment of all strains. The results showed that the main group of bacteria (36 isolates, named the ANT strains in this work) belong to the Arthrobacter genus (Romaniuk et al., 2018). This observation corroborates previous reports that Arthrobacter spp. are among the most frequently isolated cultivable soil bacteria, even in polar regions, e.g., they comprised 14.7% of cultivable bacteria isolated from soil samples collected at the Browning Peninsula, Eastern Antarctica and 22% of isolates originating from postglacial soils of Ecology Glacier, King George Island, Antarctica (Zdanowski et al., 2013; Pudasaini et al., 2017). Also the results of various metagenomic analyses suggest that Arthrobacter spp. are amongst most frequent bacterial genera in soil from polar regions as they were identified in >3 clone-library-based or 454-based studies (as it was revealed from the summarized literature data) (Pearce et al., 2017). Moreover, metagenomic analyses of other soils worldwide also indicate that Arthrobacter spp. often constitute a dominant or subdominant group, e.g., in rhizosphere microbiome of burned holm-oak in the Sierra Nevada Natural and National Park (SE Spain) they comprised more than 21% of the total bacterial community, while in the permafrost from the Qinghai-Tibet Plateau Arthrobacter spp. comprised up to 15% of all bacteria (Hu et al., 2016; Fernandez-Gonzalez et al., 2017).
Phylogenetic analysis using partial 16S rDNA sequences (1,206 nt) of 36 ANT strains and 54 type strains representing various Arthrobacter species retrieved from the NCBI database was performed. The tree topology revealed that the ANT strains form three separate clusters, which strongly suggests their divergence, although they were all isolated from the same soil sample (Figure 1). The levels of identity of 16S rDNA sequences within each cluster are high. Cluster I groups 23 strains, including 15 ANT strains. Within this cluster two subclusters were distinguished: IA grouping 12 ANT strains together with a reference strain Arthrobacter russicus A1-3, isolated from an air sample from the Russian space station Mir (exhibiting reciprocal identity of 16S rDNA sequences between 97.8 and 100%) (Li et al., 2004), and IB clustering three ANT strains showing the highest identity level (99.8%) with the sequence of Arthrobacter psychrochitiniphilus GP3, isolated from Adélie penguin guano from Antarctica (Wang et al., 2009). Cluster II groups seven ANT isolates and two reference strains, i.e., Arthrobacter oryzae KV-651 and Arthrobacter humicola KV-653, isolated from a paddy soil sample collected in Japan (Kageyama et al., 2008). These strains exhibit very high reciprocal identity (between 99.8 and 100%) of their 16S rDNA sequences. The remaining 14 ANT strains form cluster III gathering strains with 16S rRNA genes identical in 98 to 100%. An outlier for this cluster is Arthrobacter arilaitensis Re117, sharing between 97.2 and 97.6% 16S rDNA identity with the ANT strains grouped within cluster III (Irlinger et al., 2005) (Figure 1).
Figure 1.
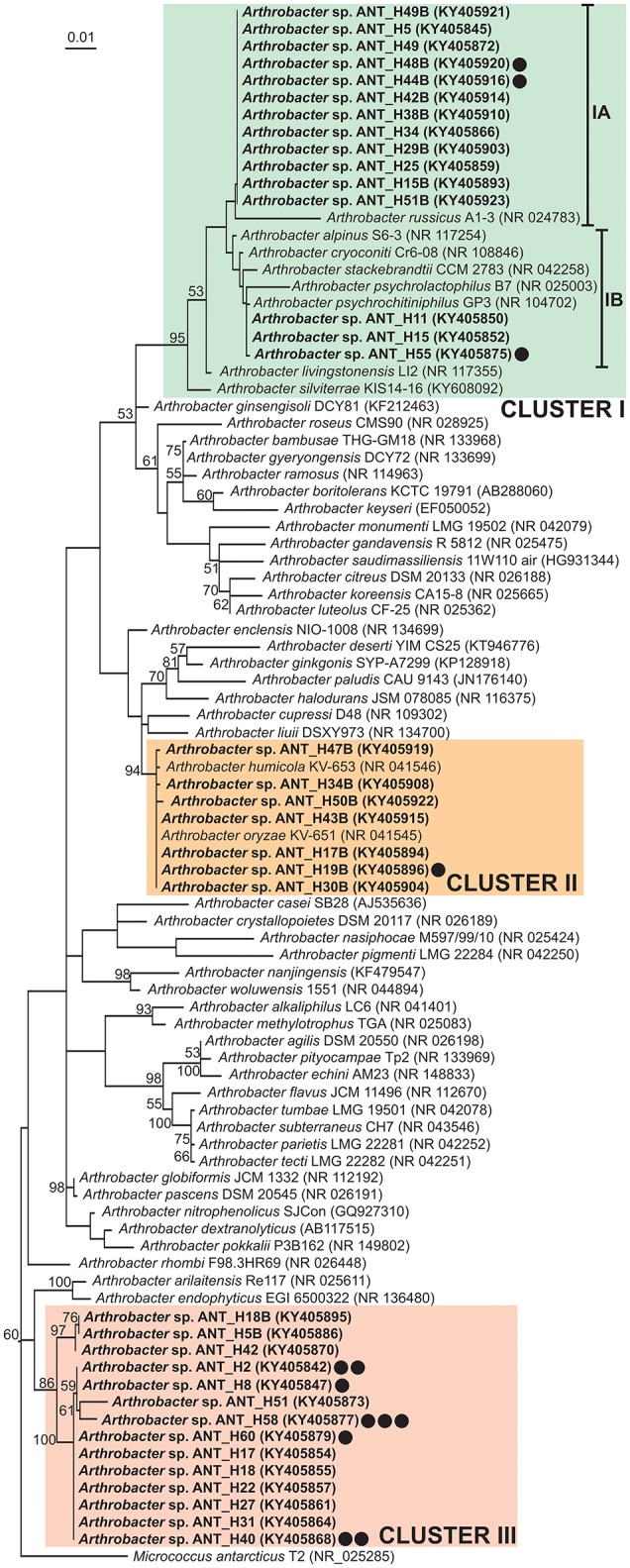
Phylogenetic tree for 16S rDNA sequences of Arthrobacter spp. The tree was constructed by applying the Maximum Likelihood method based on the Tamura-Nei model. Statistical support for the internal nodes was determined by 1,000 bootstrap replicates and values of ≥50% are shown. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 91 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1,121 positions in the final dataset. The 16S rDNA sequence of Micrococcus antarcticus T2 was used as an outgroup. GenBank accession numbers of the 16S rDNA sequences used for the phylogenetic analysis are given in brackets. The strains analyzed in this study are in bold text. Clusters I–III, distinguished by colored backgrounds, contain the ANT strains. The IA and IB subclusters are within the cluster I. Strains containing plasmids were indicated by black dots. Number of dots corresponds to the number of plasmids found within the particular strain.
Plasmids, as mobile extrachromosomal elements, may potentially contribute to bacterial adaptation to various environmental conditions occurring in diverse geographical locations, including Antarctica (Dziewit and Bartosik, 2014). The Arthrobacter strains analyzed in this study were isolated from a multi-extreme environment that is not only permanently cold, but is also highly contaminated with heavy metals and toxic organic compounds (Romaniuk et al., 2018). Therefore, it is possible that plasmids of bacteria originating from this environment carry various genetic modules of adaptive value. To identify and characterize the plasmidome of Antarctic Arthrobacter spp., all of the aforementioned ANT strains were screened for extrachromosomal replicons. This analysis revealed the presence of 13 plasmids (named the ANT plasmids) within nine strains (six carry a single plasmid, two have two plasmids, and one strain has three plasmids). Based on analysis of NCBI database resources, we noticed that plasmid-containing Arthrobacter spp. usually carry one to three extrachromosomal replicons, although a recent report describes Arthrobacter sp. ZXY-2, a strain hosting five plasmids (Zhao et al., 2017).
The complete nucleotide sequences of the 13 ANT plasmids were determined and thoroughly analyzed. All of these plasmids are circular replicons ranging in size from 8.4 to 90.6 kb, with a high GC content of between 57.4 and 63.7%. Our analysis revealed the presence of 11 unique plasmid sequences, since the smallest of the plasmids, pA2H1 (8,447 bp), was found in three different strains: ANT_H2, ANT_H55, and ANT_H60. Interestingly, the second plasmid in the ANT_H2 strain, pA2H2, was not present in either ANT_H55 or ANT_H60 (Table 1). The presence of the same plasmid in three strains suggests that horizontal gene transfer occurs in Antarctic soil. It is also noteworthy that two nearly identical plasmids, pA44BH1 and pA48BH1, differing by only 10 bp, were found in two different ANT strains (Table 1).
Table 1.
General features of the ANT plasmids.
| Plasmid name | GenBank accession number | Strain | Plasmid size (bp) | GC content (%) | Number of genes | % of coding regions | Genetic modulesa |
|---|---|---|---|---|---|---|---|
| pA2H1 | MH067968 | ANT_H2 | 8,447 | 57.4 | 9 | 74.8 | MOB, REP, TNP |
| ANT_H55 | |||||||
| ANT_H60 | |||||||
| pA2H2 | MH067969 | ANT_H2 | 64,347 | 61.8 | 60 | 82.6 | EFX, LIP, PAR, TER, TNP, TRA, UMU, XER |
| pA8H1 | MH067977 | ANT_H8 | 43,280 | 60.8 | 46 | 80.3 | LAM, PAR, RM, SLP, SUL, TER, UMU, XER |
| pA19BH1 | MH067967 | ANT_H19B | 69,439 | 63.7 | 54 | 86.1 | ARM, EFX, PAR, TNP, TRA |
| pA40H1 | MH067970 | ANT_H40 | 78,287 | 59.0 | 71 | 84.7 | COP, PAR, RM, TA, TNP, TRA |
| pA40H2 | MH067971 | ANT_H40 | 90,618 | 59.1 | 89 | 83.0 | CYT, PAR, RM, SLP, SUL, TER, TNP, TRA, UMU, XER |
| pA44BH1 | MH067972 | ANT_H44B | 40,676 | 59.3 | 36 | 86.9 | PAR, RE, TNP, TRA |
| pA48BH1 | MH067973 | ANT_H48B | 40,674 | 59.3 | 37 | 86.9 | PAR, RE, TNP, TRA |
| pA58H1 | MH067974 | ANT_H58 | 33,641 | 57.7 | 31 | 84.9 | PAR, RM, TRA |
| pA58H2 | MH067975 | ANT_H58 | 73,138 | 60.4 | 73 | 87.0 | CYT, PAR, RM, SUL, TNP, TRA |
| pA58H3 | MH067976 | ANT_H58 | 84,207 | 59.8 | 78 | 83.1 | ARM, MFS, PAR, RM, TAU, TCT, TNP, TRA, UMU |
ARM, aromatic compound utilization; COP, copper resistance; CYT, cytochrome biogenesis; EFX, drug-specific efflux system; LAM, laminarinase; LIP, lipoprotein export; MFS, major facilitator transporter; MOB, mobilization for conjugal transfer; PAR, partitioning; RE, type IV restriction enzyme; REP, replication; RM, restriction-modification; TA, toxin-antitoxin; TNP, transposition; SLP, sulfate transport; SUL, sulfur metabolism; TAU, taurine transport; TCT, tricarboxylate transport; TER, tellurium resistance; TRA, conjugal transfer; UMU, UV-damage protection/repair; XER, recombination.
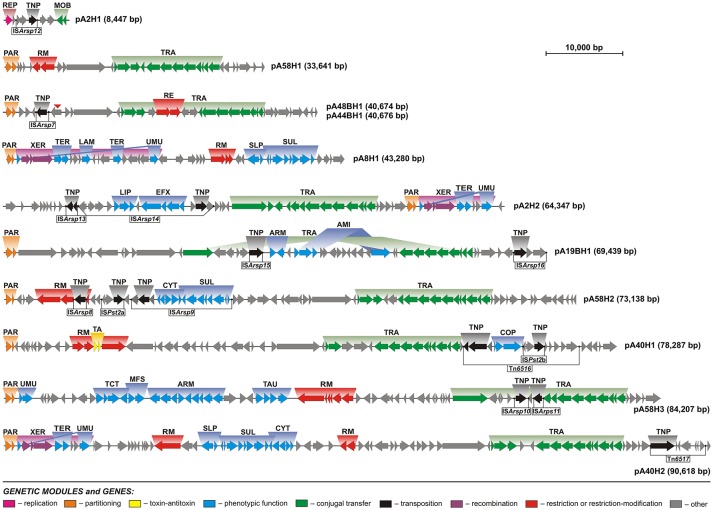
After manual annotation, 582 predicted genes were distinguished in the sequenced plasmid genomes and putative biological functions were assigned to 56% of them (Figure 2, Table S3). The smallest (pA2H1) and largest (pA40H2) ANT plasmids, carry 9 and 89 predicted genes, respectively. In comparison, the smallest Arthrobacter plasmid known until now, pAG001 (3.7 kb) of Arthrobacter sp. IHBB 11108, contains 6 genes (Kiran et al., 2015), while the largest, unnamed2 (754.2 kb), isolated from strain ERGS1:01, carries 675 genes (Kumar et al., 2015).
Figure 2.
Linear maps showing the genetic structure and organization of the circular plasmids of Antarctic Arthrobacter spp. Arrows indicate genes and their transcriptional orientation. Predicted genetic modules are indicated by colored boxes: ARM, aromatic compound utilization; COP, copper resistance; CYT, cytochrome biogenesis; EFX, drug-specific efflux; LAM, laminarinase; LIP, lipoprotein export system; MFS, major facilitator transporter; MOB, mobilization for conjugal transfer; PAR, partitioning; RE, type IV restriction enzyme; REP, replication; RM, restriction-modification; TA, toxin-antitoxin; TNP, transposition; SLP, sulfate transport; SUL, sulfur metabolism; TAU, taurine transport; TCT, tricarboxylate transport; TER, tellurium resistance; TRA, conjugal transfer; UMU, UV-damage protection/repair; XER, recombination. Potentially active transposable elements were distinguished and the location of their IR sequences is marked by black dots. A red triangle indicates the gene that was disrupted in the plasmid pA44BH1, but is intact within pA48BH1.
Genetic Modules Comprising the ANT Plasmid Backbones
Bacterial plasmids have a modular structure and it is possible to dissect them into several functional genetic modules. The backbone is comprised of a set of gene clusters, coding for replication, stabilization and conjugal transfer functions, which are necessary for plasmid maintenance and spread (Toussaint and Merlin, 2002). Comprehensive analyses of the Arthrobacter plasmid sequences allowed us to distinguish and characterize the genetic modules comprising their functional backbones (Figure 2).
Replication and Stabilization Modules
Detailed genomic analyses of the ANT plasmids led to the identification of a recognizable replication system only in the replicon pA2H1. This module is similar to a previously described replication system of the plasmid pPRH of Arthrobacter rhombi PRH1 (Stanislauskiene et al., 2012). It is composed of two overlapping genes, pA2H1_01 and pA2H1_02 (Figure 2, Table S3). Analysis of the putative polypeptide encoded by pA2H1_01 using BLASTp identified the closest homolog as a hypothetical protein of Glutamicibacter sp. BW77 (acc. no. WP_096254262) (91% amino acid sequence identity), containing replicase (pfam03090) and primase C-terminal 1 (PriCT_1) domains characteristic for replication proteins. Analysis of the protein encoded by pA2H1_02 revealed the presence of two helix-turn-helix motifs (RTARELAQKTGLSERTIRAWTA, residues 11–32 and LSMRAISKEVGIAVSAVHYALQ, residues 57–78), which suggests that this protein can interact with DNA.
Attempts to identify replication systems within the sequences of the other ANT plasmids were unsuccessful. A previous study of Arthrobacter plasmids also concluded that the identification of replication systems by in silico analysis is probably not possible based on our current knowledge (Mihasan, 2015).
Plasmids, as extrachromosomal elements that are usually not crucial for cell viability, have to carry their own survival kit, i.e., a stabilization system that helps to maintain the replicon in a bacterial cell. This could be especially important in the meaning of bacteria inhabiting extreme environments, where a general tendency to lose extrachromosomal elements may be even stronger comparing with more favorable environmental conditions. Therefore, analyzing the role of plasmids in adaptation, it is important to understand the mechanisms enabling their stable maintenance in bacterial cell. There are two main groups of plasmid-stabilizing modules: (i) partitioning systems (PAR) that promote the active segregation of plasmids into daughter cells during cell division, and (ii) toxin-antitoxin systems (TA) that cause post-segregational elimination of plasmid-less cells (Thomas, 2000; Salje, 2010; Harms et al., 2018).
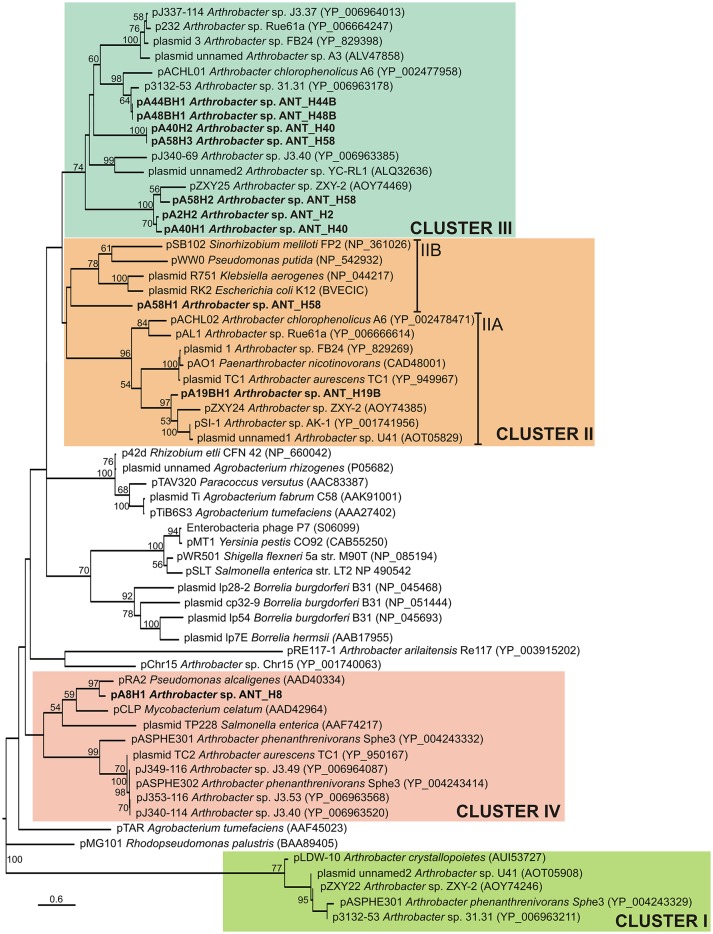
Analyses of the ANT plasmids revealed that 10 of them carry partitioning systems composed of two genes encoding ParA (ATPase) and ParB (DNA-binding protein), that may potentially interact with a predicted cis-located partitioning site, parS (Table S4). The only exception is the smallest identified plasmid pA2H1, which lacks a recognizable stabilization system. Based on a comparative analysis of ParA protein sequences, Mihăşan proposed that Arthrobacter plasmids should be divided into 4 clades (Mihasan, 2015). In the present study, we have extended this classification by adding all currently available Arthrobacter plasmid-encoded ParA sequences.
Phylogenetic analysis was performed using 62 plasmid-encoded ParA protein sequences (122 amino acid positions), including 9 ParAs originating from the ANT plasmids and 53 sequences used previously (Mihasan, 2015), or identified by us in searches of the NCBI database. The resulting phylogenetic tree topology revealed that the ParA proteins encoded by the ANT plasmids group within three separate clusters (Figure 3). The seven ParAs of plasmids pA2H2, pA41H1, pA40H2, pA44BH1, pA48BH1, pA58H2, and pA58H3 were grouped together with nine other Arthrobacter spp. plasmid ParAs previously classified within cluster III (Mihasan, 2015) (Figure 3). The ParA proteins of pA19BH1 and pA58H1 were classified within cluster II (Mihasan, 2015) (Figure 3). However, it was possible to distinguish two subclusters within this grouping. The first (IIA) includes the ParA of pA19BH1 and sequences from Arthrobacter spp. and closely related Paenarthrobacter nicotinovorans ATCC 49919 (Igloi and Brandsch, 2003), while the second (IIB) contains the ParA of pA58H1 and proteins originating from the plasmids RK2 of E. coli (Easter et al., 1998), R751 of Klebsiella aerogenes (Thorsted et al., 1998), pWW0 of Pseudomonas putida (Greated et al., 2002), and pSB102 of Sinorhizobium meliloti FP2 (Schneiker et al., 2001) (Figure 3). The ParA protein of pA8H1 was grouped within cluster IV. Interestingly, this ParA formed a separate subcluster together with three plasmids of diverse origin, i.e., pRA2 of Pseudomonas alcaligenes NCIB 9867 (Kwong et al., 2000), pCLP of Mycobacterium celatum (Le Dantec et al., 2001), and plasmid TP228 of Salmonella enterica subsp. enterica serovar Newport (Hayes, 2000) (Figure 3).
Figure 3.
Phylogenetic tree of partitioning proteins (ParAs) encoded within the Arthrobacter plasmids. The analysis was based on 39 sequences of the ParA proteins (122 amino acid positions) encoded within the Arthrobacter plasmids. Additionally, 23 reference ParA sequences of other bacteria, previously used by Mihasan (2015), were included in this analysis. The unrooted tree was constructed using the Maximum Likelihood method based on the Le and Gascuel model (Le and Gascuel, 2008). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Statistical support for the internal nodes was determined by 1,000 bootstrap replicates and values of ≥50% are shown. Accession numbers of the protein sequences used for the analysis are given in brackets. Clusters I–IV group ParA proteins of Arthrobacter plasmids, with reference to previously proposed clustering (Mihasan, 2015). The names of plasmids analyzed in this work are in bold text.
A stabilizing module of the second type, i.e., a toxin-antitoxin system, was found only within plasmid pA40H1 (Table S3). This is composed of two small non-overlapping genes with the putative antitoxin gene downstream of that encoding the toxin. In silico analyses showed that it is a tad-ata-like, type II TA system (Dziewit et al., 2007). The predicted toxin displays homology to proteins of the RelE/ParE superfamily, while the putative antitoxin belongs to the HTH_XRE family of transcriptional regulators (Dziewit et al., 2007). This observation suggests that TA systems are rare within Arthrobacter plasmids compared to PAR systems, and they probably play a minor role as stabilizing modules.
Conjugal Transfer Modules
Plasmids can be transferred between bacterial cells via conjugation, a complex process involving pilus synthesis, aggregate stabilization, surface exclusion and DNA metabolism (Llosa et al., 2002). Some plasmids carry only a minimal set of genetic elements (i.e., usually an oriT sequence and a gene encoding a relaxase that initiates the conjugal transfer by nicking the DNA at oriT) enabling their conjugal transfer. Such replicons are called mobilizable plasmids, and their transfer depends on the presence of other self-transmissible elements in the bacterial cell (Smillie et al., 2010). The presence of the conjugal transfer systems within plasmids is important from the evolutionary and ecological point of view as it enables dissemination of adaptive features between bacteria inhabiting particular environment. Therefore, because of a central role in prokaryote adaptation and evolution, studying genetic diversity, biology, and biochemistry of plasmid transfer systems in various groups of bacteria (including extremophilic ones) is currently under active investigation (Pistorio et al., 2008).
Genomic analysis of the ANT plasmids revealed that all but one (pA8H1), carry between 2 and 13 genes encoding proteins potentially involved in conjugal transfer (Figure 2, Table S3). Based on comparative analyses we predict that nine ANT plasmids (pA2H2, pA19BH1, pA40H1, pA40H2, pA44BH1, pA48BH1, pA58H1, pA58H2, and pA58H3) are self-transmissible, i.e., conjugative replicons, while pA2H1 is a mobilizable plasmid. Genes encoding putative relaxases were identified and the subsequent analysis of conserved sequence motifs in these enzymes allowed their classification into two families: MOBF and MOBP (Garcillan-Barcia et al., 2009).
Relaxases (TraAs) encoded within plasmids pA2H2, pA58H3, and pA19BH1 belong to the MOBF family. The TraAs of pA2H2 and pA58H3 share 94% amino acid sequence identity, and the conjugal transfer modules of these two plasmids are highly similar. However, this gene cluster is disrupted by the insertion of two transposable elements in pA58H3 (Figure 2), and this may influence the efficiency of the plasmid conjugation process. A homologous conjugal transfer module was previously identified in plasmid pZXY24 (acc. no. CP017425) of Arthrobacter sp. ZXY-2, originating from a pesticide plant in China. The last plasmid, pA19BH1, encodes a relaxase sharing only 39% amino acid sequence identity with the TraAs of pA2H2 and pA58H3. Interestingly, the conjugal transfer module of this replicon seems to be divided (and possibly inactivated) by the insertion of a large (24-kb) DNA fragment.
The relaxases of plasmids pA40H1, pA40H2, pA44BH1, pA48BH1, pA58H1, and pA58H2 were classified into the MOBP family. BLASTn searches revealed that gene clusters found within these plasmids that encode components of a type IV secretion system involved in bacterial conjugation are related to several other conjugal transfer systems found within various Arthrobacter plasmids (Mihasan and Brandsch, 2016). The presence of conserved genes encoding proteins involved in plasmid conjugation, gene synteny and the occurrence of similar DNA repeats (previously identified within conjugal transfer modules of Arthrobacter spp. by Mihăşan and Brandsch) suggests that many Arthrobacter plasmids may be related and could have evolved from a common ancestor (Mihasan and Brandsch, 2016). Interestingly, the predicted relaxase of the mobilizable plasmid pA2H1 also belongs to the MOBP family. This plasmid carries two non-overlapping genes encoding a predicted relaxase (MobA) and a mobilization protein (MobC). BLASTp analysis revealed the presence of a homologous mobilization module within plasmid KLBMP5180 (acc. no. CP012751) of Glutamicibacter halophytocola KLBMP 5180, isolated from the inner tissues of the halophyte plant Limonium sinense in China.
Phenotypic Modules of the ANT Plasmids
Besides the genes comprising replicon backbones, many plasmids also carry auxiliary genes that determine phenotypes that may be beneficial under particular environmental conditions, e.g., antibiotic and heavy metal resistance, and the utilization of complex organic compounds (Toussaint and Merlin, 2002). Analysis of the auxiliary genetic load of the ANT plasmids revealed that seven of these replicons carry gene clusters that may have adaptive value, i.e., they might contribute to the survival of their hosts in the extreme polar environment. Phenotypic modules recognized in these plasmids are possibly involved in (i) protection against ultraviolet (UV) radiation, (ii) resistance to heavy metals, (iii) transport and metabolism of organic compounds, (iv) sulfur metabolism, and (v) protection against exogenous DNA (Table S5).
Protection Against UV Radiation
Due to stratospheric ozone depletion, Antarctica is a region where the level of UV radiation is higher than in other areas (Madronich, 1994). UV-B exposure may cause DNA damage and increase production of reactive oxygen species (ROS), thus inducing mutagenesis and leading to genomic instability (Santos et al., 2012). The umuCD DNA repair system is a component of the global SOS system and its expression is regulated by the transcriptional repressor LexA (Butala et al., 2009). In response to DNA damage (e.g., caused by UV radiation), umuCD-encoded DNA polymerase of the Y family, enables DNA synthesis past lesions in the template strand. Therefore, the umuCD genes play an important role in maintaining genome stability in bacteria (Goodman et al., 2016; Murison et al., 2017). Genomic analysis revealed that these DNA repair systems are often found within plasmids of cold-active bacteria, especially those originating from polar regions (Dziewit and Bartosik, 2014).
UMU modules (comprised of umuCD) were identified within four of the analyzed ANT plasmids: pA2H2, pA8H1, pA40H2, and pA58H3 (Table S5). However, in the first three replicons these modules were disrupted by the insertion of DNA fragments between the umuC and umuD genes. These insertions (XER modules) may in fact represent integrative elements of a novel type. They contain (i) conserved core regions, composed of three genes encoding a predicted tyrosine recombinase XerD, a transcriptional regulator of the XRE family, and site-specific recombinase XerC, as well as (ii) variable regions, composed of 4 or 14 genes. Interestingly, the variable regions contain genetic modules of potential adaptive value, e.g., a tellurium resistance module and a gene encoding a putative laminarinase (Figure 2). We hypothesize that the conserved core regions of these elements are responsible for DNA rearrangements, while the variable regions constitute an auxiliary genetic load. The elements inserted within the umuCD modules of plasmids pA2H2 and pA40H2 are 7.5 kb in size and exhibit 99% nucleotide sequence identity. The element found in pA8H1 is larger (16.6 kb) and contains a DNA region present within two other XER modules plus an additional DNA fragment.
For testing the predicted biological function of the UMU module carried by the pA58H3 plasmid, it was PCR amplified and cloned within the pBBR1 MCS-2 vector. Comparing the UV tolerance of E. coil carrying obtained pBBR-UMU and “empty” vector revealed lack of differences. Based on the obtained results we think that the tested genetic module is inactive in E. coli.
Resistance to Heavy Metals
In a previous study, we performed a chemical analysis of soil samples collected near the Arctowski Polar Station, which revealed high concentrations of chromium, copper, mercury, nickel, and zinc. Moreover, we showed that bacteria isolated from these samples were multi-resistant and carried various heavy metal resistance genes. In particular, the ANT strains were resistant to the following heavy metals: As(V)−34 strains (MIC values between 25 and 700 mM), Cu(II)−34 (2–8 mM), As(III)−32 (3–18 mM), Ni(II)−20 (2–4 mM), Zn(II)−7 (2–3 mM), Cr(VI)−4 (2 mM), and Co(II)−2 (2 mM) (Romaniuk et al., 2018). In the present study, we tested for the presence of heavy metal resistance genes within the Arthrobacter plasmids. Four heavy metal resistance modules were detected by in silico analysis of these replicons (Table S5).
Three putative tellurium resistance modules (TER) were found in plasmids pA2H2, pA8H1, and pA40H2 (Figure 2, Table S5). Tellurium is toxic to most living organisms and its toxicity is related to the generation of ROS and disturbance of the thiol:redox buffering system. Previously, it was shown that tellurium-resistant bacteria are common in Antarctic soils (Arenas et al., 2014; Rodriguez-Rojas et al., 2016a,b; Munoz-Villagran et al., 2018; Valdivia-Gonzalez et al., 2018). Our analysis revealed that the tellurium resistance modules of the ANT plasmids are composed of two overlapping genes encoding tellurium resistance protein TerC and a hypothetical protein that probably operates as a cation-transporting ATPase. This module is present in the three aforementioned plasmids and also within the genome of Paeniglutamicibacter gangotriensis Lz1yT, a bacterium isolated from a penguin rookery soil sample collected in Antarctica (Shivaji et al., 2013).
We tested whether the TER modules carried by the Arthrobacter plasmids confer resistance to Te(IV). The MIC values were determined to establish the level of tellurium resistance of the Arthrobacter plasmid-bearing strains. All analyzed strains were recognized as sensitive and the MIC value for all them was 1 mM. All identified TER modules are nearly identical, thus only one of them was used for further analyses. The TER module of the pA40H2 plasmid was amplified by PCR and cloned within the pBBR1 MCS-2 vector for testing its activity in the heterological host—E. coli. Unfortunately, it was found to be inactive in this host.
The fourth heavy metal resistance module (COP) was found in plasmid pA40H1, as a passenger gene cluster of the transposon Tn6516. COP is predicted to be involved in copper resistance. Cu is an essential trace element for all bacteria, functioning as a cofactor of various enzymes. However, an excess of Cu is toxic. Under aerobic conditions, Cu is thought to catalyze the production of hydroxyl radicals, via the Fenton and Haber-Weiss reactions, that may cause oxidative damage to various cellular macromolecules. Under anaerobic conditions an excess of copper leads to the formation of adventitious Cu(I)-thiolate bonds, which inactivates enzymes whose activity depends on free cysteines or disulfide bonds. In addition, copper in excess can lead to incorrect disulfide bond formation in the periplasm (Ladomersky and Petris, 2015). The analyses performed in our previous study revealed that the concentration (elemental composition) of copper in soil collected near the Arctowski station was 126 μg g−1, i.e., up to 7 times higher than at other sites on King George Island (Romaniuk et al., 2018). These concentration is above the acceptable norms for Cu content in various soils and sediments, including agricultural soils in European Union (Kabata-Pendias and Pendias, 2001; Hooda et al., 2010; Toth et al., 2016). Moreover, we found that 94% of Arthrobacter strains isolated from this region were resistant to Cu(II) (Romaniuk et al., 2018). However, the identification of only one copper resistance module within the ANT plasmids, suggests that there are other chromosomal Cu-resistance genes.
The COP module of the pA40H1 plasmid is composed of three genes encoding (i) copper chaperone CopZ (COG2608) which functions as a heavy metal transporter, (ii) a protein containing a heavy metal-binding domain, and (iii) a copper-translocating P-type ATPase (COG2217) (Lu et al., 2003; Singleton et al., 2009) (Figure 2, Table S5). These proteins exhibit between 97 and 100% amino acid sequence identity with homologous proteins (acc. nos. WP_047117894-WP_047117894) of Arthrobacter sp. YC-RL1, an aromatic compound-degrading bacterium isolated from polluted agricultural soil in China (Ren et al., 2016).
Transport and Metabolism of Organic Compounds
Lichens and bryophytes are abundant in Antarctica (Colesie et al., 2018). These organisms accumulate a storage glucan in their cells called moss starch, which is composed of glucose units linked by β-1,3 and β-1,4 glycosidic bonds. One of the enzymes that enables bacteria to use moss starch as a carbon source is laminarinase. Interestingly, the same enzyme may be used by marine bacteria for the utilization of the algal storage glucan laminarin (Davis, 1992; Alderkamp et al., 2007). In silico analyses of the plasmid pA8H1 identified a predicted gene (pA8H1_p12) encoding a glycoside hydrolase family protein. Detailed analysis of this pA8H1-encoded protein led to the identification of a LamG domain, that is characteristic of laminarinases, responsible for the hydrolysis of 1,3-beta-D-glucosidic bonds in glucans such as laminarins or curdlans. The presence of a predicted laminarinase (LAM) module in pA8H1, suggested that the strain ANT_H8 may be able to utilize moss starch as an alternative carbon and/or energy source. This was tested in a series of growth experiments. It was shown that none of analyzed Arthrobacter strains is able to use laminarin as a sole carbon source for growth. Additionally, the LAM module was tested in a heterological host. This analysis revealed that E. coli carrying pBBR-LAM plasmid was not able to utilize laminarin. This may suggest that the module is not functional or needs some specific (yet not known) environmental factors for its activation. Alternatively, we may also speculate that this enzyme is responsible for utilization of a glucose polimer, other than laminarin.
Analysis of the plasmid pA58H3 revealed the presence of a large gene cluster containing four genetic modules potentially involved in the transport and metabolism of organic compounds. The modules encode (i) a tricarboxylate transport system, (ii) an ABC-type nitrate/sulfonate/bicarbonate transport system, (iii) major facilitator transporters, and (iv) enzymes potentially involved in aromatic compound utilization (Figure 2, Table S5).
The putative tricarboxylate transport system (TCT module) of pA58H3 is composed of 4 genes encoding homologs of the TctA, TctB, and TctC proteins, as well as a LysR family transcriptional regulator. This gene cluster is homologous to a genetic module of Arthrobacter sp. Leaf337 (Bai et al., 2015). It was proposed that this type of module may be involved in citrate transport (Sweet et al., 1979). Also present within pA58H3 are genes encoding an ABC-type nitrate/sulfonate/bicarbonate transport system, which may be involved in taurine transport (TAU module). Taurine is widely distributed in animal tissues. Excess taurine produced by animals is excreted either directly in urine, or as taurocholate in bile (Sekowska et al., 2000; Schuller-Levis and Park, 2003). While plants and animals cannot degrade taurine, various bacteria and fungi are capable of doing this. They can use taurine as the sole source of carbon, nitrogen, sulfur and energy (Shimamoto and Berk, 1979). In Arthrobacter spp. taurine most probably supports growth when utilized as the sole sulfur source, since sulfite formation by the action of taurine dioxygenase was confirmed for Arthrobacter sp. Rue61a (Niewerth et al., 2012). It is also noteworthy that taurine can protect bacteria from osmotic stress, and thus it could be an important protective agent under extreme environmental conditions (Mosier et al., 2013).
A gene encoding a predicted major facilitator transporter (MFS module) was also found in plasmid pA58H3. The pA58H3_p18 transporter shares >50% amino acid sequence identity with several aromatic acid, vanillate and benzoate transporters of Rhodococcus spp. In addition, it contains a conserved TIGR00895 domain that is shared by members of the cl26865 superfamily, grouping transporters of benzoate, carbohydrates, organic alcohols or acids. Therefore, we speculate that the activity of this transporter may be linked with the function of the fourth genetic module within the aforementioned gene cluster of pA58H3, that contains genes potentially involved in the utilization of aromatic compounds such as phenoxybenzoates (ARM module). This module contains genes encoding enzymes involved in the transformation of aromatic compounds: (i) aldehyde dehydrogenase (pA58H3_p20), belonging to a large family of NAD(P)+-dependent enzymes enabling the oxidation of endogenous and exogenous aliphatic and aromatic aldehydes to their corresponding carboxylic acids, and playing an important role in detoxification (Sophos and Vasiliou, 2003); (ii) 3-ketoacyl-(acyl-carrier-protein) reductase (pA58H3_p21), belonging to the SDR superfamily of enzymes catalyzing the metabolism of steroids, cofactors, carbohydrates, lipids, aromatic compounds, amino acids, and acting in redox sensing (Oppermann et al., 2003); (iii) phthalate dioxygenase reductase (pA58H3_p24), an FMN-dependent reductase mediating electron transfer from the two-electron donor reduced nicotinamide adenine nucleotide (NADH), to the one-electron acceptor, [2Fe-2S] (Correll et al., 1992; Gassner et al., 1995); (iv) Zn-dependent alcohol dehydrogenase (pA58H3_p26), that may catalyze the NAD(P)(H)-dependent interconversion of alcohols to aldehydes or ketones (Persson et al., 2008); and (v) phthalate 4,5-dioxygenase (pA58H3_p27), belonging to a large class of aromatic ring-hydroxylating dioxygenases that enable microorganisms to tolerate and utilize aromatic compounds for growth (Tarasev and Ballou, 2005).
Genes encoding proteins potentially involved in the utilization of aromatic compounds were also found in plasmid pA19BH1: (i) pA19BH1_p21 encoding a predicted 3-ketoacyl-(acyl-carrier-protein) reductase belonging to the SDR superfamily; and (ii) pA19BH1_p22 encoding a putative class III extradiol dioxygenase which uses a non-heme Fe(II) to cleave aromatic rings between a hydroxylated carbon and an adjacent non-hydroxylated carbon (Broderick, 1999).
The presence of genes encoding enzymes potentially involved in aromatic compound utilization within plasmids pA53H3 and pA19BH1 may be strictly linked to the presence of pollutants in the petroleum-contaminated soil sample from which the Arthrobacter strains were isolated. This soil sample was collected near the petroleum pumping and storage warehouse at the Henryk Arctowski Polish Antarctic Station (Romaniuk et al., 2018).
It is also important to mention that within the transposable element ISArsp14, carried by plasmid pA2H2, two predicted efflux systems (LIP and EFX) were found. The putative ABC-type lipoprotein export system, namely LIP, is composed of three genes encoding (i) an ATPase (COG1136) homologous to LolD, (ii) a permease of the FtsX-like family (COG0577)—the LolC homolog, and (iii) a transcriptional regulator of the TetR family. This efflux system is similar to LolCDE, found in gram-negative bacteria (Zuckert, 2014). The second module EFX is composed of four genes encoding (i) a predicted transporter of the MMPL family, most probably involved in lipid transport, (ii) a putative drug efflux protein of EmrB/QacA subfamily, and (iii) two TetR family transcriptional regulators. Interestingly, a homologous genetic module was also found within plasmid pA19BH1 (Figure 2). It is possible that the EFX module is a drug-specific efflux system (Lomovskaya and Lewis, 1992; Saier et al., 1998).
Sulfur Metabolism
Sulfur oxidation is an important and well-characterized process of chemolithotrophic bacteria. It may also be used by heterotrophic bacteria for detoxication and probably energy generation (Liu et al., 2014). Glutathione spontaneously reacts with elemental sulfur to generate glutathione persulfide, and this compound may be oxidized by the enzyme sulfur dioxygenase (EC 1.13.11.18) to form sulfite and glutathione. It has been proposed that this reaction is not only a detoxification process, but it may also generate ATP via oxidative phosphorylation (Liu et al., 2014).
Analysis of the ANT plasmids revealed that three of them (pA8H1, pA40H2, and pA58H2) contain gene clusters that are probably linked to sulfur metabolism (SUL module). These clusters are nearly identical (≥97% nucleotide sequence identity) and each of them contains a gene encoding a putative sulfur dioxygenase. Seven other proteins are encoded by these SUL modules: (i) a CsoR family transcriptional regulator, (ii) a small hypothetical protein of unknown function, (iii and iv) two rhodanese-related sulfurtransferases (COG0607), (v) a TauE/SafE sulfite exporter (Weinitschke et al., 2007), (vi) a transporter of the major facilitator superfamily, and (vii) a DUF302 domain-containing hypothetical protein (Figure 2, Table S5). Plasmids pA8H1 and pA40H2 also contain an additional genetic module (named SLP) composed of two genes encoding a MerR family transcriptional regulator and sulfate permease (SulP) involved in sulfate uptake.
Furthermore, plasmids pA40H2 and pA58H2, in close proximity to the aforementioned SUL modules, contain nearly identical (97% nucleotide sequence identity) five-gene clusters (named CYT) encoding proteins required for cytochrome biogenesis or related to sulfur metabolism: (i) a transcriptional regulator (COG3682), (ii and iii) two cytochrome c-type biogenesis proteins CcdA (DsbD analogs, COG0785), (iv) a thiol-disulfide oxireductase (DsbA family protein, COG1651), and (v) intracellular sulfur oxidation protein (DsrE/DsrF family, COG1416) (Figure 2, Table S5). It is hypothesized that the combined activities of the CYT, SUL and SLP modules may be responsible for energy gain from the oxidation of sulfur compounds.
Protection Against Exogenous DNA
Bacteria have developed several mechanisms to protect their cells against invading exogenous DNA (mostly bacteriophages). The most common of these are restriction-modification (RM) systems that allow discrimination between foreign (non-methylated) DNA invading the bacterial cell and endogenous (methylated) genetic material. As a consequence, the restriction endonuclease destroys exogenous DNA. RM systems have been classified into four major types according to subunit composition, recognition site, cofactor requirements and cleavage position (Tock and Dryden, 2005; Vasu and Nagaraja, 2013).
The biological role of the RM systems is to protect the host against (i) infecting bacteriophages, which would otherwise convert the cell into a factory to replicate phage particles and either kill it or compromise its health, or (ii) other selfish DNA (including plasmids), which may consume cellular energy and substrates for their own maintenance, thus affecting cell growth (Tock and Dryden, 2005). The prevention of energy and substrate loss is especially important for bacteria inhabiting nutrient-poor and extreme environments like Antarctica (De Maayer et al., 2014). Moreover, plasmid-borne RM systems may function as a mechanism of post-segregational cell killing that increases the stability of the plasmid in the bacterial population (Dziewit et al., 2011; Mruk and Kobayashi, 2014; Werbowy et al., 2015; Sitaraman, 2016).
Within the analyzed ANT plasmids we found seven putative RM modules (Figure 2, Table 2). Four systems, identified in plasmids pA8H1, pA40H1, pA58H1, and pA58H3, were classified as type I RM modules. They are composed of three genes encoding a restriction endonuclease, DNA methyltransferase and the S subunit determining the specificity of both restriction and methylation (Table 2) (Vasu and Nagaraja, 2013). Two other systems, carried by pA40H2, represent type II RMs (Table 2). One belongs to the IIG subtype, and encodes a single polypeptide with both cleavage (endonuclease) and modification domains (Pingoud et al., 2005). The second represents the IIP subtype and encodes two proteins: a DNA m5C methyltransferase and a restriction endonuclease with the probable recognition sequence 5′-GGCGCC-3′ (predicted on the basis of detailed comparative sequence analysis performed at the REBASE database) (Roberts et al., 2015). The last RM system represents an example of type III and is present within pA58H2. This module is most probably inactive due to the insertion of the transposable element ISArsp8 (Figure 2).
Table 2.
Restriction-modification systems and genes encoding type IV restriction enzymes identified within the ANT plasmids.
| Plasmid name | Genes for restriction or restriction-modification systems (coordinates) | Typea | Best BLASTp hit for the restriction endonuclease (GenBank acc. no.) |
|---|---|---|---|
| pA8H1 | pA8H1_p28-p30 (26,211–29,034) | I | Restriction endonuclease of Paenibacillus sabinae (WP_025334610) |
| pA40H1 | pA40H1_p13-p14 and pA40H1_p17 (8,581–15,359) | I | Restriction endonuclease of Arthrobacter crystallopoietes (WP_005269194) |
| pA58H1 | pA58H1_p07-p08 (3,622–6,419) | I | Restriction endonuclease of Gordonia sp. 852002-10350_SCH5691597 (WP_064861386) |
| pA58H3 | pA58H3_p35-p40 (37,630–44,790) | I | Restriction endonuclease subunit R of Arthrobacter sp. Leaf69 (KQN86732) |
| pA40H2 | pA40H2_p24 (19,320–22,664) | IIG | Hypothetical protein of Arthrobacter sp. RIT-PI-e (WP_082177329) |
| pA40H2 | (pA40H2_p46-p47) (43,092–45,084) | IIP | Nuclease of Arthrobacter sp. AK-YN10 (ERI35594) |
| pA58H2 | (pA58H2_p06-p07 and pA58H2_p09) (4,195–10,824) | III | Type III restriction-modification system endonuclease of Actinotignum timonense (WP_087070271) |
| pA44BH1 | (pA44BH1_p16-p17) (19,883–22,958) | IV | Hypothetical protein of Microbacterium sp. Cr-K29 [WP_029260290] |
| pA48BH2 | (pA48BH1_p17-p18) (19,871–22,946) | IV | Hypothetical protein of Microbacterium sp. Cr-K29 [WP_029260290] |
Classification based on comparison with the REBASE database.
In addition, within plasmids pA44BH1 and pA48BH1, we detected identical pairs of genes (mcrB-like and mcrC-like) encoding a type IV McrBC-like methylation-dependent restriction enzyme. McrB and McrC are both necessary for methyl-directed restriction activity (Raleigh, 1992).
Distribution of the Phenotypic Modules of the ANT Plasmids in Arthrobacter Genomes
To evaluate the distribution of 14 phenotypic modules carried by the ANT plasmids in various Arthrobacter genomes a BLAST search using 20 chromosomes and 33 plasmids of Arthrobacter spp. retrieved from the NCBI database was performed (Table S6). It revealed that within the chromosomes of analyzed Arthrobacter spp. there are between 2 and 10 genetic modules, homologous to the gene clusters of the ANT plasmids described above. It was also shown that such modules are not common within the plasmids of Arthrobacter spp., as they were found only in nine replicons. Amongst these plasmids, the pLDW-10 replicon originating from A. crystallopoietes DSM 20117, isolated from the soil sample collected in USA, contains as many as four modules (ARM, CYT, SUL, and TER), while other plasmids contain only one or two such modules (Table S6).
Further analysis revealed that the LAM and TAU gene clusters are specific for the ANT plasmids, as they were found in any of the analyzed Arthrobacter genomes. Interestingly both modules are involved in metabolism of alternative carbon sources (putatively, laminarin, and taurine), which may be extremely beneficial for the host under nutrient limitation. In contrast, COP, SLP, TER, and UMU modules were found in majority of Arthrobacter chromosomes, and additionally TER and UMU modules were also recognized within analyzed Arthrobacter plasmids (including: unnamed plasmid of A. alpinus A3, pAL1 plasmid of Arthrobacter sp. Rue61a, and unnamed plasmid 2 of Arthrobacter sp. U41) (Table S6).
Transposable Elements Found Within the ANT Plasmids
Transposable elements (TEs) are common in prokaryotic genomes and play an important role in the evolution of bacteria. They are responsible for various DNA rearrangements, including duplications, insertions, inversions, and deletions (Mahillon and Chandler, 1998). A thorough analysis of the ANT plasmids enabled the identification of 14 TEs, including two transposons of the Tn3 family and 12 insertion sequences (IS) of the IS1380, IS3, IS30, IS481, ISAs1, ISNCY, and ISL3 families (Table 3).
Table 3.
Transposable elements found within the ANT plasmids.
| Transposable element | Family/Group | Plasmid | Number of passenger genes | Size (bp) | IR sequencea | DR sequencea |
|---|---|---|---|---|---|---|
| ISArsp7 | ISL3 | pA44BH1 pA48BH1 | 0 | 1,451 | IRL: GGCTCTTCGCGGTTGGTGGTGAAT | CCTTTGCC |
| IRR: GGCTCTTCGCGTTTCGTGGTGAAT | ||||||
| ISArsp8 | IS481 | pA58H2 | 0 | 1,718 | IRL:TGTGATCTCTCAGAACATCGTTTACAACATCACTCAGGACATCGCTTACA | TGCTGCAAATGCCGA |
| IRR:TGTGAACACTCGCTTCTTTGTTTACAGTCGCACTCACTACATCGTTTACA | ||||||
| ISArsp9 | ISNCY | pA58H2 | 11 | 12,978 | IRL:TGTGAGCAACAACGATAAGTGGCCTGTTTTTCAC | AAGGATGCTC |
| IRR:TGTGAATCGCAACGGTAAGTGGCGTGTTTTTCGC | ||||||
| ISArsp10 | IS1380 | pA58H3 | 0 | 1,667 | IRL: CCTTGATCCACCGAT | CTTAG |
| IRR: CCTTGATCCACCGAT | ||||||
| ISArsp11 | ISAs1 | pA58H3 | 0 | 1,306 | IRL: CAGGGCTGATGCAAAGTCG | GGAACAGCAC |
| IRR: CAGGGCTAATGCAAAGTCG | ||||||
| ISArsp12 | IS481 | pA2H1 | 2 | 2,615 | IRL: TGTACTGACCGGACACGTTG | CCTAGA |
| IRR: TGTACTGACCGGACACGTTG | ||||||
| ISArsp13 | IS3/ IS150 | pA2H2 | 0 | 1,442 | IRL: TGAACTGCTCCCCGAAAGTTGGACT | TCC |
| IRR: TGAACTGGTCCCCGAAAGTTGGACT | ||||||
| ISArsp14 | ISNCY | pA2H2 | 12 | 17,535 | IRL: CTGTGAATCGCAGCGGTAAATGGCGTGTTTTTTGCAGCG | GCTTCTT |
| IRR: CTGTGAATCGCAACGGTAAATGGCGTGTTTTTCGCAGCG | ||||||
| ISArsp15 | IS30 | pA19BH1 | 0 | 1,862 | IRL: GGATTCTATTGATCGAAGCAACG | GACCCACGCG |
| IRR: GGATTTCAGTGGTCGACGCAACG | ||||||
| ISArsp16 | ISNCY | pA19BH1 | 2 | 4,338 | IRL: TGAGAATGCTCATTGGTATGTCAGTAA | GTT |
| IRR: TGAGAATGCTCATCGAGATGTCAGTAA | ||||||
| ISPst2a | ISL3 | pA58H2 | 2 | 2,985 | IRL: GGGTATCCGGAATTTCTGGTTGAT | TTTTTAAA |
| IRR: GGGTATACGGATTTAATGGTTGAT | ||||||
| ISPst2b | ISL3 | pA40H1 | 2 | 2,985 | IRL: GGGTATCCGGAATTTCTGGTTGAT | CAAAAAAG |
| IRR: GGGTATACGGATTTAATGGTTGAT | ||||||
| Tn6516 | Tn3 | pA40H1 | 12 | 14,887 | IRL: GGGGTGGCTTGTGAAAAAGTGAAATATCCGACGCTAAG | TCTAT |
| IRR: GGGGTGGCTTGTGAAAAAGTGAAATATCCGACGCTAAG | ||||||
| Tn6517 | Tn3 | pA40H2 | 6 | 7,080 | IRL: GGGGTCTGAGCTGCAGCGGAACAGTGGTTGGCGCTAAG | TGGTA |
| IRR: GGGGTCTGAACTGCAGCGGAACAGTGGTTGGCGCTAAG |
Inverted and direct repeats are shown in the 5′ to 3′ orientation.
Transposons of the Tn3 Family
The characteristic feature of Tn3 family transposons is their modular structure. Three types of functional genetic module can be distinguished within these elements: (i) a transposase gene, (ii) a cointegrate resolution module, and (iii) passenger genes (Toussaint and Merlin, 2002; Nicolas et al., 2015). Analysis of the ANT plasmids identified two novel Tn3-family transposons, Tn6516 and Tn6517, within the plasmids pA40H1 and pA40H2, respectively (Figure 2, Table 3).
The Tn6516 transposon (14,887 bp) contains 15 genes, including the core genetic module encoding transposase and resolvase, as well as passenger genes (Figure 2, Table 3). The auxiliary genes of Tn6516 include the copper resistance module (COP) described above. Interestingly, an additional TE, ISPst2b of the ISL3 family, was detected downstream of the COP module. The ISPst2b element is composed of 3 genes encoding a transposase, putative permease (COG0701) and an ArsR family transcriptional regulator. The remaining passenger genes of Tn6516 encode four hypothetical proteins, an FrmR family transcriptional regulator and a putative alkylhydroperoxidase family enzyme (COG2128).
The second Tn3 family transposon, Tn6517, is of the size 7,080 bp. It carries transposase and serine resolvase genes, and five passenger genes encoding hypothetical proteins (Figure 2, Table 3).
Insertion Sequences
In silico analysis of the ANT plasmids revealed the presence of 12 insertion sequences. Six of them are typical ISs carrying only transposase genes: ISArsp7 (ISL3 family), ISArsp8 (IS481 family), ISArsp10 (IS1380 family), ISArsp11 (ISAs1 family), ISArsp13 (IS3 family/IS150 group), and ISArsp15 (IS30 family). Other ISs found within the ANT plasmids carry between 2 and 12 passenger genes (Figure 2, Table 3).
ISArsp12 (2,615 bp) belonging to the IS481 family, identified within the smallest ANT plasmid pA2H1, carries 2 passenger genes, encoding an ArsR family transcriptional regulator and a predicted permease of unknown specificity (Figure 2, Table 3). This is highly similar (86% nucleotide sequence identity) to ISPfr21 of Propionibacterium freudenreichii (Falentin et al., 2010).
The two 2,985-bp long elements, ISPst2a (of pA58H2) and ISPst2b (of pA40H1) of the ISL3 family, also carry 2 passenger genes encoding an ArsR family transcriptional regulator and putative permease (Figure 2, Table 3). It is important to mention that these auxiliary genes share no similarity with those of ISArsp12. These two ISs differ in only 2 bp and are isoforms of ISPst2 that is present in several copies in the genomes of three Pseudomonas stutzeri strains (Bolognese et al., 1999).
In addition, three ISNCY family ISs were found within the ANT plasmids: ISArsp9 (of pA58H2), ISArsp14 (of pA2H2), and ISArsp16 (of pA19BH1). These elements all carry passenger genes. ISArsp9 (12,978 bp) contains 11 passenger genes, which comprise the CYT and SUL modules described above. ISArsp14 (17,535 bp) encodes a transposase, resolvase, and ATP-binding protein showing a high level of similarity to the equivalent proteins of ISArsp9 and ISBli29 of Brevibacterium linens BS258 (acc. no. CP014869). Seven of the 12 passenger genes of ISArsp14 comprise the predicted transport system modules LIP and EFX, described above. The third identified element of the ISNCY family, ISArsp16, is much smaller (4,338 bp). It encodes a transposase showing 69% amino acid sequence identity to the transposase of ISAau5 found within plasmid TC1 of Arthrobacter aurescens TC1 (Mongodin et al., 2006). ISArsp16 carries two passenger genes encoding hypothetical proteins of unknown function (Figure 2, Table 3).
Comparative Analysis of Arthrobacter Plasmids and Meta-Analysis of Plasmids of Psychrotolerant Bacteria
Looking for tracks of possible horizontal gene transfer and common or distinct origin of the ANT replicons their comparative genomic analysis was performed. It was based on all against all BLASTn comparison. Similar approach was applied previously in several other analyses of bacterial plasmids, e.g., extrachromosomal replicons of Streptococcus macedonicus and Lactococcus lactis (Gorecki et al., 2011; Papadimitriou et al., 2015). Performed analysis of the ANT plasmids revealed that there are several DNA regions shared by these replicons. This is particularly apparent for plasmids pA40H2, pA8H1, and pA2H2, that possess nearly identical XER modules (96% nucleotide sequence identity), and plasmids pA40H2 and pA58H2, which carry almost identical CYT and SUL modules (97% identity) (Figure S1). This may reflect possible horizontal transfer of genetic modules between these replicons. On the other hand, lack of conservation of plasmids' backbones may suggest that the ANT replicons originate from various ancestors.
Comparison of the nucleotide sequences of the ANT plasmids and other Arthrobacter plasmids (mostly from mesophilic strains) revealed their divergence, since only limited similarity (usually confined to the conjugal transfer modules) was found. This indicates that the ANT plasmids are novel and unique replicons. Interestingly, the highest level of similarity detected is between pA8H1 and the unnamed plasmid (acc. no. CP013298) of Arthrobacter sp. YC-RL1, which possess nearly identical modules predicted to participate in sulfur metabolism (97% nucleotide sequence identity) (Figure S1).
The proteomes of Arthrobacter plasmids were then examined by an all against all comparative protein analysis. This confirmed the results of the nucleotide sequence comparisons showing that the ANT plasmids share many more homologous proteins with each other, than with the plasmids of mesophilic Arthrobacter spp. The only exceptions were the nearly identical plasmids pA44BH1 and pA48BH1, that encode as many as 19 (>50%) proteins with homology to those of p3132-53 of Arthrobacter sp. 31-32 (acc. no. JQ418520), isolated from aromatic hydrocarbon-contaminated soil from Indiana, USA. A more detailed analysis of the possible functions of putative proteins common to various Arthrobacter plasmids revealed that they are mostly involved in conjugal transfer or replicon stabilization. Therefore, they are products of the conserved genetic backbones of the plasmids, whereas proteins encoded by auxiliary genes are mostly unique. This finding seems to support the hypothesis that many Arthrobacter plasmids may be related and have a common ancestor (Mihasan and Brandsch, 2016), and in the course of evolution they have gained various auxiliary genes that may confer an adaptive advantage to their hosts in their specific habitats.
The auxiliary genetic information stored in all sequenced so far Arthrobacter plasmids was carefully inspected (Table 4). It was shown that seven (43.75%) out of 16 plasmids carry genetic modules encoding proteins involved in metabolism and transport of organic compounds. Interestingly, four plasmids (all belonging to the group of ANT replicons) carry genes enabling protection against UV radiation and resistance to heavy metals. It was also shown that Arthrobacter plasmids are rich of restriction-modification systems, as they were found within nine (56.25%) plasmids (Table 4).
Table 4.
Functions of auxiliary genetic modules identified within Arthrobacter plasmids.
| Function | Plasmid name/GenBank accession numbera/Strain name | |
|---|---|---|
| Protection against: | Low temperature | Plasmid unnamed1/CP015733/Arthrobacter sp. U41 |
| Oxidative stress | Plasmid unnamed3/CP015735/Arthrobacter sp. U41 | |
| UV radiation | pA2H2/MH067969/Arthrobacter sp. ANT_H2 pA40H2/MH067971/Arthrobacter sp. ANT_H40 pA58H3/MH067976/Arthrobacter sp. ANT_H58 pA8H1/MH067977/Arthrobacter sp. ANT_H8 | |
| Resistance to heavy metals | pA2H2/MH067969/Arthrobacter sp. ANT_H2 pA40H1/MH067970/Arthrobacter sp. ANT_H40 pA40H2/MH067971/Arthrobacter sp. ANT_H40 pA8H1/MH067977/Arthrobacter sp. ANT_H8 | |
| Protection against exogenous DNA—restriction-modification system | Plasmid unnamed1/CP015733 Arthrobacter sp. U41 pA19BH1/MH067967/Arthrobacter sp. ANT_H19B pA40H1/MH067970/Arthrobacter sp. ANT_H40 pA40H2/MH067971/Arthrobacter sp. ANT_H40 pA44BH1/MH067972/Arthrobacter sp. ANT_H44B pA48BH1/MH067973/Arthrobacter sp. ANT_H48B pA58H1/MH067974/Arthrobacter sp. ANT_H58 pA58H3/MH067976/Arthrobacter sp. ANT_H58 pA8H1/MH067977/Arthrobacter sp. ANT_H8 | |
| Metabolism and transport of: | Carbohydrates | pA19BH1/MH067967/Arthrobacter sp. ANT_H19B pA2H2/MH067969/Arthrobacter sp. ANT_H2 pA40H1/MH067970/Arthrobacter sp. ANT_H40 pA40H2/MH067971/Arthrobacter sp. ANT_H40 pA58H2/MH067975/Arthrobacter sp. ANT_H58 pA58H3/MH067976/Arthrobacter sp. ANT_H58 pA8H1/MH067977/Arthrobacter sp. ANT_H8 |
| Amino acids | pA40H1/MH067970/Arthrobacter sp. ANT_H40 pA58H2/MH067975/Arthrobacter sp. ANT_H58 pA58H3/MH067976/Arthrobacter sp. ANT_H58 | |
| Nucleotides | pA58H2/MH067975/Arthrobacter sp. ANT_H58 | |
| Lipids | pA19BH1/MH067967/Arthrobacter sp. ANT_H19B pA2H2/MH067969/Arthrobacter sp. ANT_H2 | |
| Inorganic ions | Plasmid unnamed1/CP015733 Arthrobacter sp. U41 pA40H2/MH067971/Arthrobacter sp. ANT_H40 pA58H2/MH067975/Arthrobacter sp. ANT_H58 pA58H3/MH067976/Arthrobacter sp. ANT_H58 pA8H1/MH067977/Arthrobacter sp. ANT_H8 | |
| Toxic organic compounds | pA19BH1/MH067967/Arthrobacter sp. ANT_H19B pA58H3/MH067976/Arthrobacter sp. ANT_H58 | |
| Energy production and conversion | pA19BH1/MH067967/Arthrobacter sp. ANT_H19B pA40H2/MH067971/Arthrobacter sp. ANT_H40 pA58H2/MH067975/Arthrobacter sp. ANT_H58 pA58H3/MH067976/Arthrobacter sp. ANT_H58 | |
GenBank accession numbers of plasmids identified and analyzed in this study are in bold text.
For a broader view of the possible role of the ANT plasmids in the adaptation of extremophilic Arthrobacter spp., a meta-analysis of 247 plasmids of psychrotolerant bacteria representing 36 genera was performed. This analysis revealed that the most common feature found within plasmids of bacteria from cold regions is the presence of genes conferring resistance to heavy metals (Figure S2, Table S7). It was shown that 11.74% of the analyzed plasmids carry such genes. In the ANT plasmids, heavy metal resistance genes were found in four replicons (Figure S2, Table S7). The abundance of heavy metal resistance genes and their presence within mobile genetic elements (i.e., plasmids) is understandable because these toxic elements are the most commonly occurring bactericidal factors in nature (Nies, 1999). It is important to mention that heavy metals are not only anthropogenic contaminants, they also occur naturally in minerals and are released during (bio)weathering processes (Gadd, 2007; Matlakowska et al., 2012).
Our meta-analysis also revealed that within the plasmids of psychrotolerant bacteria, and especially those isolated from Arctic and Antarctic regions, genes conferring protection against low temperature, oxidative stress and UV radiation are very common. Amongst the analyzed plasmids 18.22% carry such genes (Figure S2, Table S7). In polar regions, low temperature is the predominant life-limiting factor (D'amico et al., 2006). In addition, the increased UV radiation and oxygen solubility favor the formation of ROS, which may cause damage to DNA, RNA, proteins, and lipids (Cabiscol et al., 2000; Casanueva et al., 2010). Therefore, the acquisition (on a plasmid) of genes conferring protection against these stressors could be highly beneficial for the bacterial host.
We also performed a meta-analysis of the distribution (within the plasmids of psychrotolerant bacteria) of genes whose products are involved in the transport and metabolism of carbohydrates, amino acids, nucleotides, lipids, inorganic ions, and toxic organic compounds, as well as energy production and conversion (Figure S3, Table S7). This revealed that 17% of the analyzed plasmids carry such genes (Figure S3, Table S7). The abundance of metabolic genes within plasmids of psychrotolerant bacteria may represent a specific form of adaptation, since cold-active microorganisms usually inhabit nutrient-poor environments, where genes enabling the utilization of poorly-degradable compounds are highly beneficial.
Conclusions
In this study, we explored the plasmidome of Antarctic Arthrobacter strains isolated from soil samples collected at the Henryk Arctowski Polish Antarctic Station on King George Island. In total, 11 various plasmids were identified, sequenced and analyzed. These replicons exhibit great diversity in both size and nucleotide sequence. The plasmids carry from 9 to 89 predicted genes, some forming genetic modules that potentially benefit their bacterial hosts. A comparative analysis of nucleotide sequences allowed us to define the ANT plasmids as novel and unique replicons in the Arthrobacter genus.
The analysis of the ANT plasmid backbones revealed that most of them contain partitioning systems. This allowed ParA-based phylogenetic classification of all Arthrobacter plasmids. In addition, despite previously reported difficulties in identifying (with the use of bioinformatic tools) replication modules in Arthrobacter plasmids, we were able to distinguish one such system in the plasmid pA2H1. Moreover, based on in silico analysis, the majority of the ANT plasmids were recognized as self-transmissible replicons.
The bioinformatic analyses that we performed indicated the presence of genetic modules that may be responsible for adaptation of the bacteria to the extreme Antarctic environment. The following genetic modules of predicted adaptive value were distinguished: (i) umuCD genes (UMU module) conferring protection against UV radiation, (ii) tellurium and copper resistance determinants (TER and COP), (iii) genes encoding laminarinase (LAM), several enzymes involved in aromatic compound utilization (ARM) and various transporters (EFX, LIP, MFS, TAU, and TCT), that may be involved in the transport and utilization of alternative carbon sources, and (iv) sulfur metabolism genes (CYT, SLP, and SUL).
It was also revealed that the ANT plasmids contain a number of restriction or restriction-modification systems whose function could be protection against exogenous DNAs, such as bacteriophages, or alternatively some may act as plasmid stabilization modules. Moreover, 14 novel transposable elements, including two transposons of the Tn3 family and 12 insertion sequences of the IS1380, IS3, IS30, IS481, ISAs1, ISNCY, and ISL3 families, were identified within the ANT plasmids. Interestingly, our analysis also revealed the presence of putative novel integrative elements (XER modules) that contain a conserved core region composed of three genes encoding a predicted tyrosine recombinase XerD, transcriptional regulator of the XRE family and site-specific recombinase XerC, accompanied by 4 or 14 passenger genes.
It is important to mention that the presented work has also several limitations, that one should be aware of. First of all, the biological function of the distinguished genetic modules is based only on bioinformatic analyses, and although the investigations were very complex, these predictions are still speculative. Therefore, there is a need for experimental validation of biological functions of the predicted genetic modules. However, this needs a development of a proper host-vector systems for analyses of Arthrobacter spp. The majority of the Arthrobacter genes characterized so far have been cloned and expressed in E. coli. However, heterologous expression, can lead to production of inactive proteins due to misfolding, a lack of appropriate enzyme cofactors and proper insertion of these cofactors into apoenzymes, or various posttranslational modifications. Such approach was presented in this study and we hypothesize that inability of showing the functionality of selected Arthrobacter modules in the heterological host may be in fact a consequence of abovementioned reasons.
As a limitation of this study we can also recognize lack of full genomic sequences of all analyzed Arthrobacter strain, which forecloses testing if the plasmid modules confer novel and unique biological functions or genes responsible for the particular process are already present within the host's chromosome. Other limitation of this work is that although 36 strains were analyzed this is still a relatively small pool of bacteria. For more complex analyses of a larger pool of bacteria, a joint-collaboration approach involving various scientific centers working with Antarctic prokaryotes could be proposed. An alternative is also a shotgun metagenomic approach for discovering a total metaplasmidome of Antarctic soil samples, and this is an aim of our current works.
In summary, our analysis of the ANT plasmids, supported by a meta-analysis of 247 plasmids of various psychrotolerant bacteria, shows that extrachromosomal replicons may play an important role in the adaptation of Arthrobacter strains (and bacteria in general) to the extreme Antarctic environment.
Author Contributions
LD and KR conceived and designed experiments. KR sequenced and annotated plasmids. KR, PG, and LD analyzed the data. LD contributed reagents, materials, and analysis tools. KR, PG, and LD wrote manuscript. All authors read and approved the submitted version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jan Gawor from the DNA Sequencing and Oligonucleotide Synthesis Laboratory, IBB Polish Academy of Science for his technical assistance during plasmid sequencing. Library construction and plasmid genome assembly were carried out at the DNA Sequencing and Oligonucleotide Synthesis Laboratory IBB PAN using the CePT infrastructure financed by the European Union—the European Regional Development Fund [Innovative economy 2007–13, Agreement POIG.02.02.00-14-024/08-00].
Footnotes
Funding. This work was financed by the National Science Centre, Poland (grants no. 2013/09/D/NZ8/03046 and partially 2016/23/B/NZ9/02909).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03144/full#supplementary-material
References
- Alderkamp A. C., Van Rijssel M., Bolhuis H. (2007). Characterization of marine bacteria and the activity of their enzyme systems involved in degradation of the algal storage glucan laminarin. FEMS Microbiol. Ecol. 59, 108–117. 10.1111/j.1574-6941.2006.00219.x [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas F. A., Pugin B., Henríquez N. A., Arenas-Salinas M. A., Diaz-Vasquez W. A., Pozo M. F., et al. (2014). Isolation, identification and characterization of highly tellurite-resistant, tellurite-reducing bacteria from Antarctica. Polar Sci. 8, 40–52. 10.1016/j.polar.2014.01.001 [DOI] [Google Scholar]
- Bai Y., Muller D. B., Srinivas G., Garrido-Oter R., Potthoff E., Rott M., et al. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369. 10.1038/nature16192 [DOI] [PubMed] [Google Scholar]
- Baitsch D., Sandu C., Brandsch R., Igloi G. L. (2001). Gene cluster on pAO1 of Arthrobacter nicotinovorans involved in degradation of the plant alkaloid nicotine: cloning, purification, and characterization of 2,6-dihydroxypyridine 3-hydroxylase. J. Bacteriol. 183, 5262–5267. 10.1128/JB.183.18.5262-5267.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. (1979). A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7, 1513–1523. 10.1093/nar/7.6.1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerketorp J., Roling W. F. M., Feng X. M., Garcia A. H., Heipieper H. J., Hakansson S. (2018). Formulation and stabilization of an Arthrobacter strain with good storage stability and 4-chlorophenol-degradation activity for bioremediation. Appl. Microbiol. Biotechnol. 102, 2031–2040. 10.1007/s00253-017-8706-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognese F., Di Lecce C., Galli E., Barbieri P. (1999). Activation and inactivation of Pseudomonas stutzeri methylbenzene catabolism pathways mediated by a transposable element. Appl. Environ. Microbiol. 65, 1876–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutron C. F., Wolff E. W. (1989). Heavy-metal and sulfur emissions to the atmosphere from human activities in Antarctica. Atmos. Environ. 23, 1669–1675. 10.1016/0004-6981(89)90051-6 [DOI] [Google Scholar]
- Broderick J. B. (1999). Catechol dioxygenases. Essays Biochem. 34, 173–189. 10.1042/bse0340173 [DOI] [PubMed] [Google Scholar]
- Butala M., Zgur-Bertok D., Busby S. J. (2009). The bacterial LexA transcriptional repressor. Cell. Mol. Life. Sci. 66, 82–93. 10.1007/s00018-008-8378-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabiscol E., Tamarit J., Ros J. (2000). Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 3, 3–8. [PubMed] [Google Scholar]
- Carver T., Harris S. R., Berriman M., Parkhill J., Mcquillan J. A. (2012). Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464–469. 10.1093/bioinformatics/btr703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanueva A., Tuffin M., Cary C., Cowan D. A. (2010). Molecular adaptations to psychrophily: the impact of 'omic' technologies. Trends Microbiol. 18, 374–381. 10.1016/j.tim.2010.05.002 [DOI] [PubMed] [Google Scholar]
- Casellas M., Grifoll M., Bayona J. M., Solanas A. M. (1997). New metabolites in the degradation of fluorene by Arthrobacter sp. strain F101. Appl. Environ. Microbiol. 63, 819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A., Pathak A., Jaswal R., Edwards B., Iii, C.happell D., Ball C., et al. (2018). Physiological and comparative genomic analysis of Arthrobacter sp. SRS-W-1-2016 provides insights on niche adaptation for survival in uraniferous soils. Genes 9:31. 10.3390/genes9010031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciok A., Budzik K., Zdanowski M. K., Gawor J., Grzesiak J., Decewicz P., et al. (2018). Plasmids of psychrotolerant Polaromonas spp. isolated from Arctic and Antarctic glaciers - diversity and role in adaptation to polar environments. Front. Microbiol. 9:1285. 10.3389/fmicb.2018.01285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colesie C., Budel B., Hurry V., Green T. G. A. (2018). Can Antarctic lichens acclimatize to changes in temperature? Glob. Chang. Biol. 24, 1123–1135. 10.1111/gcb.13984 [DOI] [PubMed] [Google Scholar]
- Correll C. C., Batie C. J., Ballou D. P., Ludwig M. L. (1992). Phthalate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleotides to [2Fe-2S]. Science 258, 1604–1610. 10.1126/science.1280857 [DOI] [PubMed] [Google Scholar]
- Cowan D. A., Makhalanyane T. P., Dennis P. G., Hopkins D. W. (2014). Microbial ecology and biogeochemistry of continental Antarctic soils. Front. Microbiol. 5:154. 10.3389/fmicb.2014.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripps R. E. (1975). The microbial metabolism of acetophenone. Metabolism of acetophenone and some chloroacetophenones by an Arthrobacter species. Biochem. J. 152, 233–241. 10.1042/bj1520233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'amico S., Collins T., Marx J. C., Feller G., Gerday C. (2006). Psychrophilic microorganisms: challenges for life. EMBO Rep. 7, 385–389. 10.1038/sj.embor.7400662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzentas N. (2010). Circoletto: visualizing sequence similarity with Circos. Bioinformatics 26, 2620–2621. 10.1093/bioinformatics/btq484 [DOI] [PubMed] [Google Scholar]
- Datta J., Maiti A. K., Modak D. P., Chakrabartty P. K., Bhattacharyya P., Ray P. K. (2000). Metabolism of gamma-hexachlorocyclohexane by Arthrobacter citreus strain BI-100: identification of metabolites. J. Gen. Appl. Microbiol. 46, 59–67. 10.2323/jgam.46.59 [DOI] [PubMed] [Google Scholar]
- Davis C. L. (1992). Production of laminarinase and alginase by marine bacteria after starvation. FEMS Microbiol. Lett. 86, 349–355. 10.1111/j.1574-6968.1992.tb04827.x [DOI] [Google Scholar]
- De Maayer P., Anderson D., Cary C., Cowan D. A. (2014). Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep. 15, 508–517. 10.1002/embr.201338170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. (1990). Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18, 5019–5026. 10.1093/nar/18.17.5019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dsouza M., Taylor M. W., Turner S. J., Aislabie J. (2015). Genomic and phenotypic insights into the ecology of Arthrobacter from Antarctic soils. BMC Genomics 16:36. 10.1186/s12864-015-1220-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziewit L., Bartosik D. (2014). Plasmids of psychrophilic and psychrotolerant bacteria and their role in adaptation to cold environments. Front. Microbiol. 5:596. 10.3389/fmicb.2014.00596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziewit L., Jazurek M., Drewniak L., Baj J., Bartosik D. (2007). The SXT conjugative element and linear prophage N15 encode toxin-antitoxin-stabilizing systems homologous to the tad-ata module of the Paracoccus aminophilus plasmid pAMI2. J. Bacteriol. 189, 1983–1997. 10.1128/JB.01610-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziewit L., Kuczkowska K., Adamczuk M., Radlinska M., Bartosik D. (2011). Functional characterization of the type II PamI restriction-modification system derived from plasmid pAMI7 of Paracoccus aminophilus JCM 7686. FEMS Microbiol. Lett. 324, 56–63. 10.1111/j.1574-6968.2011.02388.x [DOI] [PubMed] [Google Scholar]
- Dziewit L., Pyzik A., Matlakowska R., Baj J., Szuplewska M., Bartosik D. (2013). Characterization of Halomonas sp. ZM3 isolated from the Zelazny most post-flotation waste reservoir, with a special focus on its mobile DNA. BMC Microbiol. 13:59. 10.1186/1471-2180-13-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easter C. L., Schwab H., Helinski D. R. (1998). Role of the parCBA operon of the broad-host-range plasmid RK2 in stable plasmid maintenance. J. Bacteriol. 180, 6023–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton R. W. (2001). Plasmid-encoded phthalate catabolic pathway in Arthrobacter keyseri 12B. J. Bacteriol. 183, 3689–3703. 10.1128/JB.183.12.3689-3703.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falentin H., Deutsch S. M., Jan G., Loux V., Thierry A., Parayre S., et al. (2010). The complete genome of Propionibacterium freudenreichii CIRM-BIA1, a hardy actinobacterium with food and probiotic applications. PLoS ONE 5:e11748. 10.1371/journal.pone.0011748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez A. J., Martinez-Hidalgo P., Cobo-Diaz J. F., Villadas P. J., Martinez-Molina E., Toro N., et al. (2017). The rhizosphere microbiome of burned holm-oak: potential role of the genus Arthrobacter in the recovery of burned soils. Sci. Rep. 7:6008. 10.1038/s41598-017-06112-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic. Acids Res. 44, D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadd G. M. (2007). Geomycology: biogeochemical transformations of rocks, minerals, metals and radionuclides by fungi, bioweathering and bioremediation. Mycol. Res. 111, 3–49. 10.1016/j.mycres.2006.12.001 [DOI] [PubMed] [Google Scholar]
- Garcillan-Barcia M. P., Francia M. V., De La Cruz F. (2009). The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33, 657–687. 10.1111/j.1574-6976.2009.00168.x [DOI] [PubMed] [Google Scholar]
- Gassner G. T., Ludwig M. L., Gatti D. L., Correll C. C., Ballou D. P. (1995). Structure and mechanism of the iron-sulfur flavoprotein phthalate dioxygenase reductase. FASEB J. 9, 1411–1418. 10.1096/fasebj.9.14.7589982 [DOI] [PubMed] [Google Scholar]
- Goodman M. F., McDonald J. P., Jaszczur M. M., Woodgate R. (2016). Insights into the complex levels of regulation imposed on Escherichia coli DNA polymerase V. DNA Repair (Amst) 44, 42–50. 10.1016/j.dnarep.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorecki R. K., Koryszewska-Baginska A., Golebiewski M., Zylinska J., Grynberg M., Bardowski J. K. (2011). Adaptative potential of the Lactococcus lactis IL594 strain encoded in its 7 plasmids. PLoS ONE 6:e22238. 10.1371/journal.pone.0022238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf H. F., Shirsat S. V., Oppenheimer C., Jarvis M. J., Podzun R., Jacob D. (2010). Continental scale Antarctic deposition of sulphur and black carbon from anthropogenic and volcanic sources. Atmos. Chem. Phys. 10, 2457–2465. 10.5194/acp-10-2457-2010 [DOI] [Google Scholar]
- Greated A., Lambertsen L., Williams P. A., Thomas C. M. (2002). Complete sequence of the IncP-9 TOL plasmid pWW0 from Pseudomonas putida. Environ. Microbiol. 4, 856–871. 10.1046/j.1462-2920.2002.00305.x [DOI] [PubMed] [Google Scholar]
- Grzesiak J., Zdanowski M. K., Gorniak D., Swiatecki A., Aleksandrzak-Piekarczyk T., Szatraj K., et al. (2015). Microbial community changes along the ecology glacier ablation zone (King George Island, Antarctica). Polar Biol. 38, 2069–2083. 10.1007/s00300-015-1767-z [DOI] [Google Scholar]
- Hanahan D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. 10.1016/S0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- Harms A., Brodersen D. E., Mitarai N., Gerdes K. (2018). Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol. Cell 70, 768–784. 10.1016/j.molcel.2018.01.003 [DOI] [PubMed] [Google Scholar]
- Hayatsu M., Hirano M., Nagata T. (1999). Involvement of two plasmids in the degradation of carbaryl by Arthrobacter sp. strain RC100. Appl. Environ. Microbiol. 65, 1015–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F. (2000). The partition system of multidrug resistance plasmid TP228 includes a novel protein that epitomizes an evolutionarily distinct subgroup of the ParA superfamily. Mol. Microbiol. 37, 528–541. 10.1046/j.1365-2958.2000.02030.x [DOI] [PubMed] [Google Scholar]
- Hooda P. S., Tack F. M. G., Edwards A. C., Laing G. D., Chen W., Chang A. C., et al. (2010). Trace Elements in Soils. Chichester: John Wiley and Sons, Ltd. [Google Scholar]
- Hu W., Zhang Q., Tian T., Li D., Cheng G., Mu J., et al. (2016). Characterization of the prokaryotic diversity through a stratigraphic permafrost core profile from the Qinghai-Tibet Plateau. Extremophiles 20, 337–349. 10.1007/s00792-016-0825-y [DOI] [PubMed] [Google Scholar]
- Husserl J., Spain J. C., Hughes J. B. (2010). Growth of Arthrobacter sp. strain JBH1 on nitroglycerin as the sole source of carbon and nitrogen. Appl. Environ. Microbiol. 76, 1689–1891. 10.1128/AEM.02400-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igloi G. L., Brandsch R. (2003). Sequence of the 165-kilobase catabolic plasmid pAO1 from Arthrobacter nicotinovorans and identification of a pAO1-dependent nicotine uptake system. J. Bacteriol. 185, 1976–1986. 10.1128/JB.185.6.1976-1986.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irlinger F., Bimet F., Delettre J., Lefevre M., Grimont P. A. (2005). Arthrobacter bergerei sp. nov. and Arthrobacter arilaitensis sp. nov., novel coryneform species isolated from the surfaces of cheeses. Int. J. Syst. Evol. Microbiol. 55, 457–462. 10.1099/ijs.0.63125-0 [DOI] [PubMed] [Google Scholar]
- Jain R. K., Dreisbach J. H., Spain J. C. (1994). Biodegradation of p-nitrophenol via 1,2,4-benzenetriol by an Arthrobacter sp. Appl. Environ. Microbiol. 60, 3030–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerke K., Nakatsu C. H., Beasley F., Konopka A. (2008). Comparative analysis of eight Arthrobacter plasmids. Plasmid 59, 73–85. 10.1016/j.plasmid.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Kabata-Pendias A., Pendias H. (2001). Trace Elements in Soils and Plants. Boca Raton, FL: CRC Press. [Google Scholar]
- Kageyama A., Morisaki K., Omura S., Takahashi Y. (2008). Arthrobacter oryzae sp. nov. and Arthrobacter humicola sp. nov. Int. J. Syst. Evol. Microbiol. 58, 53–56. 10.1099/ijs.0.64875-0 [DOI] [PubMed] [Google Scholar]
- Kallimanis A., Frillingos S., Drainas C., Koukkou A. I. (2007). Taxonomic identification, phenanthrene uptake activity, and membrane lipid alterations of the PAH degrading Arthrobacter sp. strain Sphe3. Appl. Microbiol. Biotechnol. 76, 709–717. 10.1007/s00253-007-1036-3 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361. 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kejna M., Arazny A., Sobota I., Piszczek J., Łabno R. (2013). Warunki meteorologiczne na Stacji Arctowskiego (Wyspa Króla Jerzego, Antarktyka) w 2012 roku. Probl. Klimat. Polar. 23, 43–56. [Google Scholar]
- Kiran S., Swarnkar M. K., Pal M., Thakur R., Tewari R., Singh A. K., et al. (2015). Complete genome sequencing of protease-producing novel Arthrobacter sp. strain IHBB 11108 using PacBio single-molecule real-time sequencing technology. Genome Announc. 3:e00346–15. 10.1128/genomeA.00346-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolkenbrock S., Fetzner S. (2010). Identification and in vitro deoxynucleotidylation of the terminal protein of the linear plasmid pAL1 of Arthrobacter nitroguajacolicus Ru61a. FEMS Microbiol. Lett. 304, 169–176. 10.1111/j.1574-6968.2010.01900.x [DOI] [PubMed] [Google Scholar]
- Kolkenbrock S., Naumann B., Hippler M., Fetzner S. (2010). A novel replicative enzyme encoded by the linear Arthrobacter plasmid pAL1. J. Bacteriol. 192, 4935–4943. 10.1128/JB.00614-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosek K., Jankowska K., Polkowska Z. (2017). Bacterial presence in polar regions associated with environment modification by chemical compounds including contaminants. Environ. Rev. 25, 481–491. 10.1139/er-2017-0007 [DOI] [Google Scholar]
- Kukucka P., Lammel G., Dvorska A., Klanova J., Moller A., Fries E. (2010). Contamination of Antarctic snow by polycyclic aromatic hydrocarbons dominated by combustion sources in the polar region. Environ. Chem. 7, 504–513. 10.1071/EN10066 [DOI] [Google Scholar]
- Kumar R., Singh D., Swarnkar M. K., Singh A. K., Kumar S. (2015). Complete genome sequence of Arthrobacter sp. ERGS1:01, a putative novel bacterium with prospective cold active industrial enzymes, isolated from East Rathong glacier in India. J. Biotechnol. 214, 139–140. 10.1016/j.jbiotec.2015.09.025 [DOI] [PubMed] [Google Scholar]
- Kwong S. M., Yeo C. C., Suwanto A., Poh C. L. (2000). Characterization of the endogenous plasmid from Pseudomonas alcaligenes NCIB 9867: DNA sequence and mechanism of transfer. J. Bacteriol. 182, 81–90. 10.1128/JB.182.1.81-90.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladomersky E., Petris M. J. (2015). Copper tolerance and virulence in bacteria. Metallomics 7, 957–964. 10.1039/C4MT00327F [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D., Canback B. (2004). ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32, 11–16. 10.1093/nar/gkh152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Dantec C., Winter N., Gicquel B., Vincent V., Picardeau M. (2001). Genomic sequence and transcriptional analysis of a 23-kilobase mycobacterial linear plasmid: evidence for horizontal transfer and identification of plasmid maintenance systems. J. Bacteriol. 183, 2157–2164. 10.1128/JB.183.7.2157-2164.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S. Q., Gascuel O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- Lewis G. L., Jorgensen Q. R., Loy J. D., Moxley R. A. (2018). Tellurite resistance in Shiga toxin-producing Escherichia coli. Curr. Microbiol. 75, 752–759. 10.1007/s00284-018-1444-x [DOI] [PubMed] [Google Scholar]
- Li Y., Kawamura Y., Fujiwara N., Naka T., Liu H., Huang X., et al. (2004). Rothia aeria sp. nov., Rhodococcus baikonurensis sp. nov. and Arthrobacter russicus sp. nov., isolated from air in the Russian space laboratory Mir. Int. J. Syst. Evol. Microbiol. 54, 827–835. 10.1099/ijs.0.02828-0 [DOI] [PubMed] [Google Scholar]
- Lin K., Wang A. (2001). UV mutagenesis in Escherichia coli K-12: cell survival and mutation frequency of the chromosomal genes lacZ, rpoB, ompF, and ampA. J. Exp. Microbiol. Immunol. 1, 32–46. [Google Scholar]
- Liu H., Xin Y., Xun L. (2014). Distribution, diversity, and activities of sulfur dioxygenases in heterotrophic bacteria. Appl. Environ. Microbiol. 80, 1799–1806. 10.1128/AEM.03281-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llosa M., Gomis-Ruth F. X., Coll M., De La Cruz Fd F. (2002). Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45, 1–8. 10.1046/j.1365-2958.2002.03014.x [DOI] [PubMed] [Google Scholar]
- Lomovskaya O., Lewis K. (1992). Emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. U.S.A. 89, 8938–8942. 10.1073/pnas.89.19.8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M., Chan P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic. Acids Res. 44, W54–W57. 10.1093/nar/gkw413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z. H., Dameron C. T., Solioz M. (2003). The Enterococcus hirae paradigm of copper homeostasis: copper chaperone turnover, interactions, and transactions. Biometals 16, 137–143. 10.1023/A:1020709307589 [DOI] [PubMed] [Google Scholar]
- Madronich S. (1994). Increases in biologically damaging UV-B radiation due to stratospheric ozone depletion: a brief review. Arch. Hydrobiol. Beih. 43, 17–30. [Google Scholar]
- Mahillon J., Chandler M. (1998). Insertion sequences. Microbiol. Mol. Biol. Rev. 62, 725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C. J., Lu S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic. Acids. Res. 45, D200–D203. 10.1093/nar/gkw1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov V., Rakusa-Suszczewski S. (1990). Ten years of climate observations at the Arctowski and Bellingshausen stations (King George Island, South Shetlands, Antarctia) in Global Change, Regional Research Centres: Scientific Problems and Concept Developments, ed Breymeyer A. I. (Warsaw: Institute of Geography and Spatial Organization; Polish Academy of Sciences; ), 80–87. [Google Scholar]
- Matlakowska R., Sklodowska A., Nejbert K. (2012). Bioweathering of Kupferschiefer black shale (Fore-Sudetic Monocline, SW Poland) by indigenous bacteria: implication for dissolution and precipitation of minerals in deep underground mine. FEMS Microbiol. Ecol. 81, 99–110. 10.1111/j.1574-6941.2012.01326.x [DOI] [PubMed] [Google Scholar]
- Mihasan M. (2015). Bioinformatics-based molecular classification of Arthrobacter plasmids. Cell Mol. Biol. Lett. 20, 612–625. 10.1515/cmble-2015-0036 [DOI] [PubMed] [Google Scholar]
- Mihasan M., Brandsch R. (2013). pAO1 of Arthrobacter nicotinovorans and the spread of catabolic traits by horizontal gene transfer in gram-positive soil bacteria. J. Mol. Evol. 77, 22–30. 10.1007/s00239-013-9576-x [DOI] [PubMed] [Google Scholar]
- Mihasan M., Brandsch R. (2016). A predicted T4 secretion system and conserved DNA-repeats identified in a subset of related Arthrobacter plasmids. Microbiol. Res. 191, 32–37. 10.1016/j.micres.2016.05.008 [DOI] [PubMed] [Google Scholar]
- Mihasan M., Stefan M., Hritcu L., Artenie V., Brandsch R. (2013). Evidence of a plasmid-encoded oxidative xylose-catabolic pathway in Arthrobacter nicotinovorans pAO1. Res. Microbiol. 164, 22–30. 10.1016/j.resmic.2012.10.003 [DOI] [PubMed] [Google Scholar]
- Miteva V., Lantz S., Brenchley J. (2008). Characterization of a cryptic plasmid from a Greenland ice core Arthrobacter isolate and construction of a shuttle vector that replicates in psychrophilic high G+C Gram-positive recipients. Extremophiles 12, 441–449. 10.1007/s00792-008-0149-7 [DOI] [PubMed] [Google Scholar]
- Mongodin E. F., Shapir N., Daugherty S. C., Deboy R. T., Emerson J. B., Shvartzbeyn A., et al. (2006). Secrets of soil survival revealed by the genome sequence of Arthrobacter aurescens TC1. PLoS Genet. 2:e214. 10.1371/journal.pgen.0020214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosier A. C., Justice N. B., Bowen B. P., Baran R., Thomas B. C., Northen T. R., et al. (2013). Metabolites associated with adaptation of microorganisms to an acidophilic, metal-rich environment identified by stable-isotope-enabled metabolomics. mBio 4, e00484–e00412. 10.1128/mBio.00484-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mruk I., Kobayashi I. (2014). To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res. 42, 70–86. 10.1093/nar/gkt711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Villagran C. M., Mendez K. N., Cornejo F., Figueroa M., Undabarrena A., Morales E. H., et al. (2018). Comparative genomic analysis of a new tellurite-resistant Psychrobacter strain isolated from the Antarctic Peninsula. PeerJ 6:e4402. 10.7717/peerj.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murison D. A., Timson R. C., Koleva B. N., Ordazzo M., Beuning P. J. (2017). Identification of the dimer exchange interface of the bacterial DNA damage response protein UmuD. Biochemistry 56, 4773–4785. 10.1021/acs.biochem.7b00560 [DOI] [PubMed] [Google Scholar]
- Nicolas E., Lambin M., Dandoy D., Galloy C., Nguyen N., Oger C. A., et al. (2015). The Tn3-family of replicative transposons. Microbiol Spectr. 3:MDNA3-0060-2014. 10.1128/microbiolspec.MDNA3-0060-2014 [DOI] [PubMed] [Google Scholar]
- Nies D. H. (1999). Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51, 730–750. 10.1007/s002530051457 [DOI] [PubMed] [Google Scholar]
- Niewerth H., Schuldes J., Parschat K., Kiefer P., Vorholt J. A., Daniel R., et al. (2012). Complete genome sequence and metabolic potential of the quinaldine-degrading bacterium Arthrobacter sp. Rue61a. BMC Genomics 13:534. 10.1186/1471-2164-13-534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppermann U., Filling C., Hult M., Shafqat N., Wu X., Lindh M., et al. (2003). Short-chain dehydrogenases/reductases (SDR): the 2002 update. Chem. Biol. Interact. 143–144, 247–253. 10.1016/S0009-2797(02)00164-3 [DOI] [PubMed] [Google Scholar]
- Overhage J., Sielker S., Homburg S., Parschat K., Fetzner S. (2005). Identification of large linear plasmids in Arthrobacter spp. encoding the degradation of quinaldine to anthranilate. Microbiology 151, 491–500. 10.1099/mic.0.27521-0 [DOI] [PubMed] [Google Scholar]
- Papadimitriou K., Anastasiou R., Maistrou E., Plakas T., Papandreou N. C., Hamodrakas S. J., et al. (2015). Acquisition through horizontal gene transfer of plasmid pSMA198 by Streptococcus macedonicus ACA-DC 198 points towards the dairy origin of the species. PLoS ONE 10:e0116337. 10.1371/journal.pone.0116337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parschat K., Overhage J., Strittmatter A. W., Henne A., Gottschalk G., Fetzner S. (2007). Complete nucleotide sequence of the 113-kilobase linear catabolic plasmid pAL1 of Arthrobacter nitroguajacolicus Ru61a and transcriptional analysis of genes involved in quinaldine degradation. J. Bacteriol. 189, 3855–3867. 10.1128/JB.00089-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce D. A., Newsham K. K., Thorne M. A., Calvo-Bado L., Krsek M., Laskaris P., et al. (2017). Metagenomic analysis of a southern maritime antarctic soil. Front. Microbiol. 3:403. 10.3389/fmicb.2012.00403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson B., Hedlund J., Jornvall H. (2008). Medium- and short-chain dehydrogenase/reductase gene and protein families: the MDR superfamily. Cell. Mol. Life. Sci. 65, 3879–3894. 10.1007/s00018-008-8587-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingoud A., Fuxreiter M., Pingoud V., Wende W. (2005). Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 62, 685–707. 10.1007/s00018-004-4513-1 [DOI] [PubMed] [Google Scholar]
- Pipke R., Amrhein N., Jacob G. S., Schaefer J., Kishore G. M. (1987). Metabolism of glyphosate in an Arthrobacter sp. GLP-1. Eur. J. Biochem. 165, 267–273. 10.1111/j.1432-1033.1987.tb11437.x [DOI] [PubMed] [Google Scholar]
- Pistorio M., Giusti M. A., Del Papa M. F., Draghi W. O., Lozano M. J., Tejerizo G. T., et al. (2008). Conjugal properties of the Sinorhizobium meliloti plasmid mobilome. FEMS Microbiol. Ecol. 65, 372–382. 10.1111/j.1574-6941.2008.00509.x [DOI] [PubMed] [Google Scholar]
- Pudasaini S., Wilson J., Ji M., Van Dorst J., Snape I., Palmer A. S., et al. (2017). Microbial diversity of Browning Peninsula, Eastern Antarctica revealed using molecular and cultivation methods. Front. Microbiol. 8:591. 10.3389/fmicb.2017.00591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pundir S., Martin M. J., O'donovan C. (2017). UniProt protein knowledgebase. Methods Mol. Biol. 1558, 41–55. 10.1007/978-1-4939-6783-4_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh E. A. (1992). Organization and function of the mcrBC genes of Escherichia coli K-12. Mol. Microbiol. 6, 1079–1086. 10.1111/j.1365-2958.1992.tb01546.x [DOI] [PubMed] [Google Scholar]
- Ren L., Jia Y., Ruth N., Zhao B., Yan Y. (2016). Complete genome sequence of an aromatic compound degrader Arthrobacter sp. YC-RL1. J. Biotechnol. 219, 34–35. 10.1016/j.jbiotec.2015.12.008 [DOI] [PubMed] [Google Scholar]
- Ren L., Jia Y., Zhang R., Lin Z., Zhen Z., Hu H., et al. (2018). Insight into metabolic versatility of an aromatic compounds-degrading Arthrobacter sp. YC-RL1. Front. Microbiol. 9:2438. 10.3389/fmicb.2018.02438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J., Vincze T., Posfai J., Macelis D. (2015). REBASE - a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 43, D298–D299. 10.1093/nar/gku1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rojas F., Diaz-Vasquez W., Undabarrena A., Munoz-Diaz P., Arenas F., Vasquez C. (2016a). Mercury-mediated cross-resistance to tellurite in Pseudomonas spp. isolated from the Chilean Antarctic territory. Metallomics 8, 108–117. 10.1039/C5MT00256G [DOI] [PubMed] [Google Scholar]
- Rodriguez-Rojas F., Tapia P., Castro-Nallar E., Undabarrena A., Munoz-Diaz P., Arenas-Salinas M., et al. (2016b). Draft genome sequence of a multi-metal resistant bacterium Pseudomonas putida ATH-43 isolated from Greenwich Island, Antarctica. Front. Microbiol. 7:1777. 10.3389/fmicb.2016.01777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk K., Ciok A., Decewicz P., Uhrynowski W., Budzik K., Nieckarz M., et al. (2018). Insight into heavy metal resistome of soil psychrotolerant bacteria originating from King George Island (Antarctica). Polar Biol. 41, 1319–1333. 10.1007/s00300-018-2287-4 [DOI] [Google Scholar]
- Rong L., Guo X., Chen K., Zhu J., Li S., Jiang J. (2009). Isolation of an isocarbophos-degrading strain of Arthrobacter sp. scl-2 and identification of the degradation pathway. J. Microbiol. Biotechnol. 19, 1439–1446. 10.4014/jmb.0811.0626 [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr., Paulsen I. T., Sliwinski M. K., Pao S. S., Skurray R. A., Nikaido H. (1998). Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 12, 265–274. 10.1096/fasebj.12.3.265 [DOI] [PubMed] [Google Scholar]
- Salje J. (2010). Plasmid segregation: how to survive as an extra piece of DNA. Crit. Rev. Biochem. Mol. Biol. 45, 296–317. 10.3109/10409238.2010.494657 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2001). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sandu C., Chiribau C. B., Brandsch R. (2003). Characterization of HdnoR, the transcriptional repressor of the 6-hydroxy-D-nicotine oxidase gene of Arthrobacter nicotinovorans pAO1, and its DNA-binding activity in response to L- and D-nicotine Derivatives. J. Biol. Chem. 278, 51307–51315. 10.1074/jbc.M307797200 [DOI] [PubMed] [Google Scholar]
- Santos A. L., Gomes N. C., Henriques I., Almeida A., Correia A., Cunha A. (2012). Contribution of reactive oxygen species to UV-B-induced damage in bacteria. J. Photochem. Photobiol. B 117, 40–46. 10.1016/j.jphotobiol.2012.08.016 [DOI] [PubMed] [Google Scholar]
- Schneiker S., Keller M., Droge M., Lanka E., Puhler A., Selbitschka W. (2001). The genetic organization and evolution of the broad host range mercury resistance plasmid pSB102 isolated from a microbial population residing in the rhizosphere of alfalfa. Nucleic Acids Res. 29, 5169–5181. 10.1093/nar/29.24.5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller-Levis G. B., Park E. (2003). Taurine: new implications for an old amino acid. FEMS Microbiol. Lett. 226, 195–202. 10.1016/S0378-1097(03)00611-6 [DOI] [PubMed] [Google Scholar]
- Sekowska A., Kung H. F., Danchin A. (2000). Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J. Mol. Microbiol. Biotechnol. 2, 145–177. [PubMed] [Google Scholar]
- Seo J. S., Keum Y. S., Hu Y., Lee S. E., Li Q. X. (2006). Phenanthrene degradation in Arthrobacter sp. P1-1: initial 1,2-, 3,4- and 9,10-dioxygenation, and meta- and ortho-cleavages of naphthalene-1,2-diol after its formation from naphthalene-1,2-dicarboxylic acid and hydroxyl naphthoic acids. Chemosphere 65, 2388–2394. 10.1016/j.chemosphere.2006.04.067 [DOI] [PubMed] [Google Scholar]
- Sharpee K. W., Duxbury J. M., Alexander M. (1973). 2,4-Dichlorophenoxyacetate metabolism by Arthrobacter sp.: accumulation of a chlorobutenolide. Appl. Microbiol. 26, 445–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto G., Berk R. S. (1979). Catabolism of taurine in Pseudomonas aeruginosa. Biochim. Biophys. Acta 569, 287–292. 10.1016/0005-2744(79)90064-0 [DOI] [PubMed] [Google Scholar]
- Shivaji S., Ara S., Bandi S., Singh A., Kumar Pinnaka A. (2013). Draft genome sequence of Arthrobacter gangotriensis strain Lz1yT, isolated from a penguin rookery soil sample collected in Antarctica, near the Indian station Dakshin Gangotri. Genome Announc. 1, e00347–e00313. 10.1128/genomeA.00347-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Singh S. M., Singh R. N., Naik S., Roy U., Srivastava A., et al. (2017). Bacterial communities in ancient permafrost profiles of Svalbard, Arctic. J. Basic Microbiol. 57, 1018–1036. 10.1002/jobm.201700061 [DOI] [PubMed] [Google Scholar]
- Singleton C., Hearnshaw S., Zhou L., Le Brun N. E., Hemmings A. M. (2009). Mechanistic insights into Cu(I) cluster transfer between the chaperone CopZ and its cognate Cu(I)-transporting P-type ATPase, CopA. Biochem. J. 424, 347–356. 10.1042/BJ20091079 [DOI] [PubMed] [Google Scholar]
- Sitaraman R. (2016). The role of DNA restriction-modification systems in the biology of Bacillus anthracis. Front. Microbiol. 7:11. 10.3389/fmicb.2016.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie C., Garcillan-Barcia M. P., Francia M. V., Rocha E. P., De La Cruz F. (2010). Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74, 434–452. 10.1128/MMBR.00020-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sophos N. A., Vasiliou V. (2003). Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem. Biol. Interact. 143-144, 5–22. 10.1016/S0009-2797(02)00163-1 [DOI] [PubMed] [Google Scholar]
- Stanislauskiene R., Gasparaviciute R., Vaitekunas J., Meskiene R., Rutkiene R., Casaite V., et al. (2012). Construction of Escherichia coli-Arthrobacter-Rhodococcus shuttle vectors based on a cryptic plasmid from Arthrobacter rhombi and investigation of their application for functional screening. FEMS Microbiol. Lett. 327, 78–86. 10.1111/j.1574-6968.2011.02462.x [DOI] [PubMed] [Google Scholar]
- Stohl A., Sodemann H. (2010). Characteristics of atmospheric transport into the Antarctic troposphere. J. Geophys. Res. 115:D02305 10.1029/2009JD012536 [DOI] [Google Scholar]
- Strong L. C., Rosendahl C., Johnson G., Sadowsky M. J., Wackett L. P. (2002). Arthrobacter aurescens TC1 metabolizes diverse s-triazine ring compounds. Appl. Environ. Microbiol. 68, 5973–5980. 10.1128/AEM.68.12.5973-5980.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Dong Y., Gao P., Fu M., Ta K., Li J. (2015). Microbial communities inhabiting oil-contaminated soils from two major oilfields in Northern China: implications for active petroleum-degrading capacity. J. Microbiol. 53, 371–378. 10.1007/s12275-015-5023-6 [DOI] [PubMed] [Google Scholar]
- Sweet G. D., Somers J. M., Kay W. W. (1979). Purification and properties of a citrate-binding transport component, the C protein of Salmonella typhimurium. Can. J. Biochem. 57, 710–715. 10.1139/o79-089 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasev M., Ballou D. P. (2005). Chemistry of the catalytic conversion of phthalate into its cis-dihydrodiol during the reaction of oxygen with the reduced form of phthalate dioxygenase. Biochemistry 44, 6197–6207. 10.1021/bi047724y [DOI] [PubMed] [Google Scholar]
- Thomas C. M. (2000). Paradigms of plasmid organization. Mol. Microbiol. 37, 485–491. 10.1046/j.1365-2958.2000.02006.x [DOI] [PubMed] [Google Scholar]
- Thorsted P. B., Macartney D. P., Akhtar P., Haines A. S., Ali N., Davidson P., et al. (1998). Complete sequence of the IncPbeta plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 282, 969–990. 10.1006/jmbi.1998.2060 [DOI] [PubMed] [Google Scholar]
- Tock M. R., Dryden D. T. (2005). The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 8, 466–472. 10.1016/j.mib.2005.06.003 [DOI] [PubMed] [Google Scholar]
- Toth G., Hermann T., Da Silva M. R., Montanarella L. (2016). Heavy metals in agricultural soils of the European Union with implications for food safety. Environ. Int. 88, 299–309. 10.1016/j.envint.2015.12.017 [DOI] [PubMed] [Google Scholar]
- Toussaint A., Merlin C. (2002). Mobile elements as a combination of functional modules. Plasmid. 47, 26–35. 10.1006/plas.2001.1552 [DOI] [PubMed] [Google Scholar]
- Valdivia-Gonzalez M. A., Diaz-Vasquez W. A., Ruiz-Leon D., Becerra A. A., Aguayo D. R., Perez-Donoso J. M., et al. (2018). A comparative analysis of tellurite detoxification by members of the genus Shewanella. Arch. Microbiol. 200, 267–273. 10.1007/s00203-017-1438-2 [DOI] [PubMed] [Google Scholar]
- Van Den Wijngaard A. J., Reuvekamp P. T., Janssen D. B. (1991). Purification and characterization of haloalcohol dehalogenase from Arthrobacter sp. strain AD2. J. Bacteriol. 173, 124–129. 10.1128/jb.173.1.124-129.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu K., Nagaraja V. (2013). Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 77, 53–72. 10.1128/MMBR.00044-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenknecht M., Meinhardt F. (2011). Replication-involved genes of pAL1, the linear plasmid of Arthrobacter nitroguajacolicus Ru61a - phylogenetic and transcriptional analysis. Plasmid 65, 176–184. 10.1016/j.plasmid.2010.12.005 [DOI] [PubMed] [Google Scholar]
- Wang F., Gai Y., Chen M., Xiao X. (2009). Arthrobacter psychrochitiniphilus sp. nov., a psychrotrophic bacterium isolated from Antarctica. Int. J. Syst. Evol. Microbiol. 59, 2759–2762. 10.1099/ijs.0.008912-0 [DOI] [PubMed] [Google Scholar]
- Wang X., Sun L., Wang H., Wu H., Chen S., Zheng X. (2018). Surfactant-enhanced bioremediation of DDTs and PAHs in contaminated farmland soil. Environ. Technol. 39, 1733–1744. 10.1080/09593330.2017.1337235 [DOI] [PubMed] [Google Scholar]
- Weinitschke S., Denger K., Cook A. M., Smits T. H. (2007). The DUF81 protein TauE in Cupriavidus necator H16, a sulfite exporter in the metabolism of C2 sulfonates. Microbiology 153, 3055–3060. 10.1099/mic.0.2007/009845-0 [DOI] [PubMed] [Google Scholar]
- Werbowy O., Boratynski R., Dekowska A., Kaczorowski T. (2015). Genetic analysis of maintenance of pEC156, a naturally occurring Escherichia coli plasmid that carries genes of the EcoVIII restriction-modification system. Plasmid 77, 39–50. 10.1016/j.plasmid.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Yao Y., Tang H., Su F., Xu P. (2015). Comparative genome analysis reveals the molecular basis of nicotine degradation and survival capacities of Arthrobacter. Sci. Rep. 5:8642. 10.1038/srep08642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdanowski M. K., Zmuda-Baranowska M. J., Borsuk P., Swiatecki A., Górniak D., Wolicka D., et al. (2013). Culturable bacteria community development in postglacial soils of Ecology Glacier, King George Island, Antarctica. Polar Biol. 36, 511–527. 10.1007/s00300-012-1278-0 [DOI] [Google Scholar]
- Zhao X., Ma F., Feng C., Bai S., Yang J., Wang L. (2017). Complete genome sequence of Arthrobacter sp. ZXY-2 associated with effective atrazine degradation and salt adaptation. J. Biotechnol. 248, 43–47. 10.1016/j.jbiotec.2017.03.010 [DOI] [PubMed] [Google Scholar]
- Zuckert W. R. (2014). Secretion of bacterial lipoproteins: through the cytoplasmic membrane, the periplasm and beyond. Biochim. Biophys. Acta 1843, 1509–1516. 10.1016/j.bbamcr.2014.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.