Abstract
Female parenting is obligate in mammals, but fathering behavior among mammals is rare. Only 3–5% of mammalian species exhibit biparental care, including humans, and mechanisms of fathering behavior remain sparsely studied. However, in species where it does exist, paternal care is often crucial to the survivorship of offspring. The present study is the first to identify new gene targets linked to the experience of fathering behavior in a biparental species using RNA sequencing. In order to determine the pattern of gene expression within the medial preoptic area that is specifically associated with fathering behavior, we identified genes in male prairie voles (Microtus ochrogaster) that experienced one of three social conditions: virgin males, pair bonded males, and males with fathering experience. A list of genes exhibiting different expression patterns in each comparison (i.e. Virgin vs Paired, Virgin vs Fathers, and Paired vs Fathers) was evaluated using the gene ontology enrichment analysis, and Kyoto Encyclopedia of Genes and Genomes pathways analysis to reveal metabolic pathways associated with specific genes. Using these tools, we generated a filtered list of genes that exhibited altered patterns of expression in voles with different amounts of social experience. Finally, we used NanoString to quantify differences in the expression of these selected genes. These genes are involved in a variety of processes, with enrichment in genes associated with immune function, metabolism, synaptic plasticity, and the remodeling of dendritic spines. The identification of these genes and processes will lead to novel insights into the biological basis of fathering behavior.
Keywords: fathering, dendritic spines, plasticity, gene expression, RNA sequencing, parenting
Introduction
Biparental care, in which both mother and father contribute to the care of the offspring, is displayed by a minority of mammalian species – usually cited as 3–5% [1–3]. Female parenting is obligate because mammalian offspring need to nurse, but the participation of the male is seen only in our own and a limited number of other mammalian species [4–8]. In species where paternal care does exist, including humans, it is often crucial to the survivorship of offspring; or at least has significant long-term impacts on growth as well as neural, reproductive, and social development [9, 10]. However, much is still unknown about the specific hormonal and neurobiological regulation of paternal care [6].
The vast majority of parenting research focuses on the mother, while the role of the father has mostly been considered in the context of paternal absence [9, 11–13]. Considering paternal care through the absence of the father in a biparental species has drawbacks, however, since it confounds the quantitative absence of another caregiving individual with the qualitative absence of the father in particular. Paternal absence is the most extreme situation. Individual variation in fathering can also have long-term effects on offspring [9], and in the context of nonhuman mammals is always carried out in a biparental care situation. In prairie voles, we have shown that natural variation in biparental parenting behavior predicts pup development and juvenile social behavior [14], exploratory behavior and pair-bonding, and adult aggression and stress responses [15–17]. It is not always possible in an intact biparental family to disentangle which outcomes in offspring are due to maternal care and which are due to paternal care. However, some very interesting roles for the father have been observed. For instance, males may compensate for poor maternal care (or allow mothers to expend less energy on non-nutritive tasks like carrying) [14, 18]; or a paternal behavior such as retrievals (carrying pups back to the nest or territory) may be directly linked to offspring display of retrievals and aggression as an adult [19, 20].
The hormonal mechanisms underlying fathering behavior have been much less studied than those underlying maternal behavior, although it has been hypothesized that similar neural circuits are responsible for both maternal and paternal behaviors [21]. While alterations in neural activity with parenthood appear to be hormonally regulated in females, hormonal manipulation in males has often resulted in outcomes that are either ambiguous or species-specific [22]. For instance, testosterone is inversely related to paternal care in most species [9, 23, 24], but is obligate for paternal care in California mice [25]. Prolactin, another leading candidate for the regulation of male parenting, decreases male parenting when administered, as well as when blocked [26]. These inconsistencies have led some to suggest that across species, paternal behavior depends upon nonhomologous neuroendocrine circuits [6], and has raised the question of what factors are involved in the generation of these behaviors.
Although the hormonal regulation of parenting may vary by sex, it is believed that the neural circuit governing parental behavior is similar in mothers and fathers [27]. The medial preoptic area of the hypothalamus (MPOA) is a central node in the neural circuit that regulates both maternal and paternal care and has long been recognized as playing a critical role in the generation and regulation of parental behavior (see [9, 28] for reviews). In biparental California mice, paternal experience increases Fos immunoreactivity in the MPOA [29]. Virgin male prairie voles that were exposed to pups also showed an increase in Fos immunoreactivity within the MPOA [30]. Lesions of the MPOA disrupt both maternal and paternal behavior in California mice [31]. In California mouse males, aromatase levels within the MPOA vary in response to parental status [32], while another study in male California mice showed a decrease in progesterone receptor mRNA expression in the MPOA of fathers compared with virgin males [33]. Studies in mice, which are not parental in the wild but can show induced paternal care in the laboratory [34–36], have bolstered the view of a central role for the MPOA in paternal care.
The goal of this study was to identify novel gene targets and potential mechanisms that may contribute to the production and regulation of paternal behavior. We analyzed gene expression in three groups of adult male prairie voles: virgin males, males who had formed a pair bond with a female, and males who had both pair bonded and gained fathering experience. Samples were taken from the MPOA, a region that is central to the expression of both maternal and paternal behaviors [21, 37–39], and RNA was extracted and sequenced.
Materials and Methods
Subjects
Subjects were 18 adult male prairie voles. Animals were born and housed in the Psychology Department Vivarium at the University of California, Davis. These animals were descendants of a wild stock originally caught near Champaign, IL. The animals were weaned at 20 days of age and pair housed with an animal of the same sex (sibling if available, similarly aged nonsibling if not) in small laboratory cages (27 × 16 × 13 cm) in which food and water were available ad libitum. All animals were maintained at ∼70°F (21°C) on a 14:10 light/dark cycle with the lights on at 6 a.m. All experiments were performed under National Institutes of Health guidelines for the care of animals in research and were approved by the Institutional Animal Care and Use Committee of the University of California, Davis.
At postnatal day (P) 42–45 subjects were placed in one of three groups of age-matched males: (i) virgin males, (ii) sexually experienced, ‘pair-bonded’ males, or (iii) males with fathering experience. This was designed to dissociate alterations in gene expression that were related to pair bonding from alterations related to paternal behavior. Virgin males were housed with a male same-age conspecific for ∼20 days, and they were euthanized without engaging in sexual contact with females. Pair-bonded males were housed with a same-age female conspecific for ∼20 days, after which the males were euthanized. Because mating and pregnancy strengthens pair bonds in prairie voles [40–43], we confirmed that females were pregnant. Pair-bonded males were euthanized before females gave birth, ensuring they had no contact with pups. The third group consisted of males that had 3 days of paternal experience. These males were also housed with female pair-mates with whom they presumably formed a pair-bond. The females gave birth, and the males were permitted three days of contact with pups before they were euthanized. Three days of parental experience was chosen to minimize age differences between subjects. Furthermore, prairie vole fathers already exhibit large amount of paternal care by postnatal day 3 [14, 44].
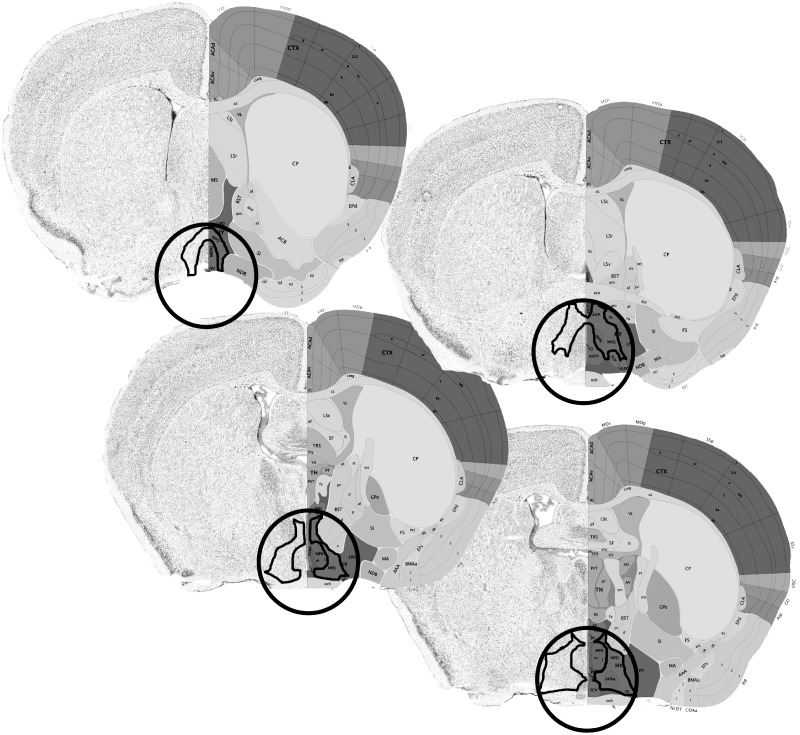
Subjects were anesthetized using isoflurane and euthanized via cervical dislocation. Upon euthanasia, brains were removed and flash frozen. The brains were sliced on a cryostat into 120 µm sections and mounted on slides. Punches were taken from the MPOA using a 15.5-gauge blunt needle (Fig. 1) and were stored in a −80°C freezer until RNA extraction.
Figure 1.
A schematic representing the area from which tissue samples were taken. The circumference of the tissue punch is delineated by a circle, and the MPOA is outlined in black. The tissue punches removed the entirety of the MPOA, as well as small portions of adjacent hypothalamic tissue
RNA Extraction
Total RNA was isolated with Qiazol reagent (Qiagen, Valencia, CA) and purified with an RNeasy® Plus Micro Kit (74004; Qiagen) as well as the optional DNase digestion (Qiagen 129046). A Nanodrop™ Spectrophotometer was used to determine the quality and the quantity of the RNA. All samples had a 260/280 ratio >1.8.
RNA Sequencing
A total of 18 RNA-seq libraries were prepared from the RNA of the 18 male prairie voles (Table 1). RNA sequencing and library preparation was performed by the DNA Technologies and Expression Analysis Core in the Genome Center of the University of California, Davis. A total RNA analysis ng sensitivity (Eukaryotes) of all 18 samples resulted in a mean RIN of 9.2 (range 8.3–9.9). Barcoded RNA-seq libraries were generated from 1 μg total RNA each after poly-A enrichment using the Kapa Stranded RNA-seq kit (Kapa Biosystems, Cape Town, South Africa) following the instructions of the manufacturer. The libraries were generated on a Sciclone G3 liquid handler (Caliper Life Sciences, Alameda, CA). Quality was verified with the Bioanalyzer 2100 instrument (Agilent, Santa Clara, CA) and quantified by fluorometry on a Qubit instrument (Life Technologies, Carlsbad, CA) and pooled in equimolar ratios. The pooled library was then quantified by qPCR with a Kapa Library Quant kit (Kapa Biosystems) and sequenced on one lane of an Illumina HiSeq 4000 (Illumina, San Diego, CA) with paired-end 150 bp reads.
Table 1.
RNA data
| ID | Raw reads | Trimmed reads | % Reads kept | % Aligned | % Aligned to rRNA |
|---|---|---|---|---|---|
| V1 | 25764898 | 25657648 | 99.58373598 | 95.1 | 5.67 |
| V2 | 20747099 | 20665472 | 99.60656186 | 95.99 | 2.63 |
| V3 | 23633483 | 23500072 | 99.43550005 | 96.13 | 2.07 |
| V4 | 17827384 | 17709907 | 99.34103063 | 96.06 | 2.38 |
| V5 | 22459377 | 22360940 | 99.56171091 | 95.59 | 4.09 |
| V6 | 22690769 | 22591612 | 99.56300732 | 96.09 | 2.78 |
| P1 | 22451193 | 22317681 | 99.40532336 | 92.7 | 8.72 |
| P2 | 23931482 | 23837973 | 99.60926365 | 95.84 | 2.26 |
| P3 | 21258757 | 21155922 | 99.51626993 | 94.97 | 5.00 |
| P4 | 20114070 | 20011415 | 99.48963586 | 95.86 | 3.16 |
| P5 | 21770575 | 21677246 | 99.57130668 | 96.11 | 2.06 |
| P6 | 19282593 | 19185302 | 99.49544649 | 95.73 | 3.28 |
| F1 | 20833672 | 20702987 | 99.3727222 | 95.14 | 4.82 |
| F2 | 21680207 | 21568888 | 99.48654088 | 95.99 | 2.53 |
| F3 | 20565896 | 20479195 | 99.57842342 | 96.06 | 1.65 |
| F4 | 19759347 | 19680046 | 99.59866589 | 96.17 | 2.27 |
| F5 | 21472445 | 21367346 | 99.51054014 | 95.95 | 3.06 |
| F6 | 22055401 | 21961628 | 99.57482977 | 96.31 | 2.32 |
Raw sequencing data have been deposited at NCBI’s sequence read archive under study accession number SRP128134.
Bioinformatic Analysis
Bioinformatic analysis was performed by the UC Davis Bioinformatics Core Facility, also in the Genome Center. Briefly, reads were trimmed for adapter contamination and quality using scythe (version c128b19) and sickle (version 7667f147e6), respectively. The reads were then aligned to the prairie vole genome (MicOch1.0) using bwa mem (version 0.7.13), after which featureCounts (version 1.5.0-p1) was used to create the raw gene expressions counts. Finally, R (version 3.3.2) with the edgeR and limma/voom packages were used to filter and transform (voom transformation), and test for statistical significances between groups. Briefly, data were prepared by first choosing to keep genes that achieved at least 0.5 count per million in at least five samples, normalization factors were calculated using trimmed mean of M-value, and the voom transformation was applied. A completely randomized design was implemented, comparisons of interest were extracted using contrasts, and moderated statistics were computed using the empirical Bayes procedure eBayes. Finally, each gene was corrected for multiple testing using the Benjamini–Hochberg false discovery rate correction. Gene expression was directly compared between each pair of groups, resulting in three comparisons: Virgin males vs Paired males (V vs P), Virgin males vs Fathers (V vs F), and Paired males vs Fathers (P vs F). No genes reached a statistical significant threshold (adjusted P-value <0.05) in any of the three pairwise comparisons.
Gene Ontology Analysis
In order to capture the genes that were most likely to show common functional differentiation between comparison groups we performed gene set enrichment analysis with Gene Ontologies (cellular component, molecular function, or biological process). Enrichment testing was conducted using the Kolmogorov–Smirnov test as implemented in the Bioconductor package topGO [45]. We next examined the gene ontology (GO) annotations that were significantly enriched (P-value < 0.05) and filtered the GO annotations in each comparison that were related to the brain or behavior, excluding unrelated annotations (i.e. GO: 0003014, Renal system process or GO: 0008354, Germ cell migration). We then categorized the remaining annotations based on gross function within each comparison group.
Kyoto Encyclopedia of Genes and Genomes Pathways Analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways offer insight into how genes interact within biological and metabolic processes. We identified individual genes associated with significantly enriched GO annotations and ran each individual gene through the KEGG pathways database (http://www.genome.jp/kegg/) to identify molecular signaling pathways associated with that gene. A single gene may be involved in a number of different pathways, so we then identified commonly recurring pathways associated with the individual genes. Pathways that were unrelated to brain function (e.g. those that were involved in kidney, liver, or heart metabolism) were not included in the analysis. When a specific gene was associated with multiple pathways of interest it was identified as a candidate gene for further analysis. For example, Grin2a was associated with six pathways that are involved in neural plasticity.
We identified 49 candidate genes across 9 biologically significant KEGG pathways and examined how their expression changed between conditions of social experience. For each gene, we standardized expression relative to virgin males. This allowed us to identify whether there were coordinated changes between genes that were involved in a specific pathway. We averaged gene expression across animals within each condition and transformed the data into ratios; the values we used for all analyses were the ratios of gene expression in each condition relative to the virgin condition. For each KEGG pathway, we determined the mean relative expression for all genes in each condition. The expression ratio of genes in virgin animals was set at 1, a value >1 indicated that genes were more expressed relative to virgins, and a value <1 indicated that genes were less expressed relative to virgins. Thus, we analyzed whether gene expression for each gene varied between paired animals and fathers. Effect size was measured using Cohen’s d.
Assessment of Gene Interaction Networks
After generating a list of candidate genes, we analyzed the connectivity of the gene network using the STRING Database (string-db.org) [46]. The STRING database identifies protein–protein interactions between members of a gene set, which allows the user to build a network of functional gene interactions. STRING also measures the functional and interaction enrichments of the gene network, calling upon GO annotations, KEGG pathways, and connections between nodes.
NanoString Analysis
Following the identification of candidate genes, we performed a quantitative analysis of the expression of 33 genes (30 target genes and 3 housekeeping genes) using the nCounter SPRINT profiler (NanoString Technologies, Seattle, WA). Genes were chosen to be included in the NanoString analysis based on their log fold change values as determined by the expression data, as well as their functional significance. One additional gene, Bdnf, was chosen due to previous studies indicating that it plays a significant role in plasticity and parenting [47, 48]. The nCounter analysis assay was conducted using RNA that remained after the completion of the sequencing experiment.
Briefly, NanoString is a medium-throughput method that can analyze many genes within a single sample with comparable sensitivity and accuracy to quantitative real-time RT-PCR [49]. NanoString designed and manufactured custom probes corresponding to the 33 genes we identified for quantitative analysis, consisting of 30 target genes and 3 housekeeping genes (Gusb, Pgk1, and Eif4a2). A code set specific to a 100-base region of the target mRNA was designed using a 3’ biotinylated capture probe and a 5’ reporter probe tagged with a specific fluorescent barcode. Data were collected using the nCounter Digital Analyzer by counting the number of individual barcodes.
Each transcript of interest was recognized by a capture probe and a reporter probe, each containing 30–50 bases complementary to the target mRNA. To minimize assay variability, the code sets also included negative and positive control reporter probes that were developed by the External RNA Control Consortium (ERCC). Six positive control reporter probes (ERCC-selected mRNA targets) were pre-mixed with (Spike-Ins) the code set at a concentration range (0.125–128 fM), a range corresponding to the expression levels of most mRNA of interest, to control for overall efficiency of probe hybridization and determine the detection range for transcripts of interest in each assay. A scaling factor was calculated for each sample, and a scaling factor outside the range of 0.3–3 indicated suboptimal hybridization. In our samples, the scaling factor always fell within the optimal range and was thus applied to all counts in the sample.
Quantitative expression data from the nCounter were downloaded and analyzed using the nSolver software package (NanoString Technologies). The raw counts for all transcripts were multiplied by the scaling factor to produce the adjusted counts. The relative expression was determined for each comparison group, and the effect size of the difference between expression values was determined using Cohen’s d. Expression was also compared using t-tests, and P-values were adjusted for multiple comparisons in nSolver.
Results
GO Analysis
Individual genes are associated with GO annotations in order to describe the various functions of a particular gene product. The cellular component analysis describes the locations of gene expression, at the levels of subcellular structures. The molecular function analysis describes the function that each gene product performs within the cell. The biological process analysis describes a recognized series of events or collection of molecular functions associated with a gene or gene product. Each analysis was completed for all genes with differential patterns of expression between the three comparison groups, V vs P, V vs F, and P vs F. Because each GO annotation references many genes, in some instances the same GO annotation was present in multiple comparison groups.
The initial GO enrichment analysis returned 209 GO annotations in the V vs P comparison, 222 annotations in the V vs F comparison, and 264 annotations in the P vs F comparison that were significantly enriched. Upon selecting the GO annotations in each comparison that were related to the brain or behavior, we were left with 47 GO annotations in the V vs P comparison, 47 annotations in the V vs F comparison, and 61 annotations in the P vs F comparison (Tables 2–4). We then categorized these annotations based on gross function (Fig. 2). The functional categories of GO annotations were differentially distributed across the three comparison groups. Annotations related to Neuropeptide activity were only found in the V vs P comparison, whereas immune function annotations were most predominant in the V vs F comparison. The P vs F comparison contained the greatest number of annotations related to Plasticity, DNA/RNA/Transcription, and Axon/Dendrite/Synapse.
Table 3.
virgin vs father GO annotations
| GO ID | GO annotation | # genes | Raw P value |
|---|---|---|---|
| GO: 0001975 | Response to amphetamine | 11 | 0.00033 |
| GO: 0016310 | Phosphorylation | 981 | 0.00097 |
| GO: 0007191 | Adenylate cyclase-activating dopamine receptor signaling pathway | 6 | 0.00172 |
| GO: 1903861 | Positive regulation of dendrite extension | 16 | 0.00206 |
| GO: 0043278 | Response to morphine | 7 | 0.00239 |
| GO: 0060391 | Positive regulation of SMAD protein signal transduction | 8 | 0.00258 |
| GO: 0005254 | Chloride channel activity | 29 | 0.0027 |
| GO: 0000082 | G1/S transition of mitotic cell cycle | 95 | 0.00378 |
| GO: 0005516 | Calmodulin binding | 36 | 0.0038 |
| GO: 0042110 | T-cell activation | 210 | 0.00406 |
| GO: 0008091 | Spectrin | 3 | 0.006 |
| GO: 0043406 | Positive regulation of MAP kinase activity | 98 | 0.00781 |
| GO: 0071277 | Cellular response to calcium ion | 28 | 0.0082 |
| GO: 0019228 | Neuronal action potential | 12 | 0.0093 |
| GO: 0097440 | Apical dendrite | 4 | 0.0098 |
| GO: 0030857 | Negative regulation of epithelial cell differentiation | 23 | 0.01001 |
| GO: 0051098 | Regulation of binding | 214 | 0.01046 |
| GO: 0007626 | Locomotory behavior | 143 | 0.01116 |
| GO: 0014002 | Astrocyte development | 18 | 0.01447 |
| GO: 0010862 | Positive regulation of pathway-restricted SMAD protein phosphorylation | 20 | 0.0151 |
| GO: 0007249 | I-kappaB kinase/NF-kappaB signaling | 104 | 0.01513 |
| GO: 0048681 | Negative regulation of axon regeneration | 9 | 0.01835 |
| GO: 0005815 | Microtubule organizing center | 413 | 0.0185 |
| GO: 0048715 | Negative regulation of oligodendrocyte differentiation | 10 | 0.0188 |
| GO: 0001963 | Synaptic transmission, dopaminergic | 19 | 0.01883 |
| GO: 0001726 | Ruffle | 80 | 0.0191 |
| GO: 0008023 | Transcription elongation factor complex | 29 | 0.0194 |
| GO: 0019233 | Sensory perception of pain | 38 | 0.0198 |
| GO: 0060158 | Phospholipase C-activating dopamine receptor signaling pathway | 4 | 0.02016 |
| GO: 0043235 | Receptor complex | 178 | 0.0212 |
| GO: 0002407 | Dendritic cell chemotaxis | 6 | 0.02176 |
| GO: 0005912 | Adherens junction | 151 | 0.0224 |
| GO: 0019538 | Protein metabolic process | 2271 | 0.02608 |
| GO: 0017146 | NMDA selective glutamate receptor complex | 8 | 0.0274 |
| GO: 0042098 | T-cell proliferation | 88 | 0.03093 |
| GO: 0031340 | Positive regulation of vesicle fusion | 4 | 0.03095 |
| GO: 0003909 | DNA ligase activity | 3 | 0.0332 |
| GO: 0002682 | Regulation of immune system process | 531 | 0.03451 |
| GO: 0005921 | Gap junction | 9 | 0.0375 |
| GO: 0000778 | Condensed nuclear chromosome kinetochore | 4 | 0.0399 |
| GO: 0071144 | Heteromeric SMAD protein complex | 2 | 0.0451 |
| GO: 0035240 | Dopamine binding | 4 | 0.0455 |
| GO: 0099604 | Ligand-gated calcium channel activity | 19 | 0.0469 |
| GO: 0032444 | Activin responsive factor complex | 2 | 0.0471 |
| GO: 0005247 | Voltage-gated chloride channel activity | 2 | 0.0475 |
| GO: 0001591 | Dopamine neurotransmitter receptor activity, coupled via Gi/Go | 2 | 0.0479 |
| GO: 0001042 | RNA polymerase I core binding | 2 | 0.0493 |
Table 4.
paired vs father GO annotations
| GO ID | GO Annotation | # genes | Raw P value |
|---|---|---|---|
| GO: 0005955 | Calcineurin complex | 4 | 0.00104 |
| GO: 0050840 | Extracellular matrix binding | 33 | 0.00172 |
| GO: 0046959 | Habituation | 4 | 0.0031 |
| GO: 0007626 | Locomotory behavior | 144 | 0.0032 |
| GO: 2001223 | Negative regulation of neuron migration | 7 | 0.004 |
| GO: 0046330 | Positive regulation of JNK cascade | 64 | 0.0043 |
| GO: 0060079 | Excitatory postsynaptic potential | 36 | 0.0043 |
| GO: 0060391 | Positive regulation of SMAD protein signal transduction | 8 | 0.0047 |
| GO: 0015116 | Sulfate transmembrane transporter activity | 3 | 0.00473 |
| GO: 0070723 | Response to cholesterol | 10 | 0.0048 |
| GO: 0001696 | Gastric acid secretion | 6 | 0.005 |
| GO: 0051281 | Positive regulation of release of sequestered calcium ion into cytosol | 20 | 0.005 |
| GO: 0060395 | SMAD protein signal transduction | 38 | 0.0051 |
| GO: 0042755 | Eating behavior | 10 | 0.0057 |
| GO: 0033192 | Calmodulin-dependent protein phosphatase activity | 4 | 0.00583 |
| GO: 0000403 | Y-form DNA binding | 4 | 0.00644 |
| GO: 0010001 | Glial cell differentiation | 118 | 0.0065 |
| GO: 0048407 | Platelet-derived growth factor binding | 11 | 0.0071 |
| GO: 0017134 | Fibroblast growth factor binding | 11 | 0.00772 |
| GO: 0016575 | Histone deacetylation | 32 | 0.008 |
| GO: 0045893 | Positive regulation of transcription, DNA-templated | 783 | 0.009 |
| GO: 0005516 | Calmodulin binding | 36 | 0.01032 |
| GO: 0007616 | Long-term memory | 18 | 0.0104 |
| GO: 0005882 | Intermediate filament | 30 | 0.01192 |
| GO: 0061014 | Positive regulation of mRNA catabolic process | 24 | 0.0122 |
| GO: 0060080 | Inhibitory postsynaptic potential | 9 | 0.0124 |
| GO: 0035418 | Protein localization to synapse | 22 | 0.0141 |
| GO: 0008009 | Chemokine activity | 4 | 0.01592 |
| GO: 0007015 | Actin filament organization | 169 | 0.0166 |
| GO: 0070410 | Co-SMAD binding | 8 | 0.01861 |
| GO: 0007212 | Dopamine receptor signaling pathway | 20 | 0.0198 |
| GO: 0001973 | Adenosine receptor signaling pathway | 5 | 0.0199 |
| GO: 0005102 | Signaling receptor binding | 554 | 0.02035 |
| GO: 0000978 | RNA polymerase II proximal promoter sequence-specific DNA binding | 232 | 0.02098 |
| GO: 0005736 | DNA-directed RNA polymerase I complex | 7 | 0.02166 |
| GO: 0004930 | G-protein coupled receptor activity | 110 | 0.02186 |
| GO: 0050882 | Voluntary musculoskeletal movement | 6 | 0.0224 |
| GO: 0000307 | Cyclin-dependent protein kinase holoenzyme complex | 27 | 0.02343 |
| GO: 0030374 | Ligand-dependent nuclear receptor transcription coactivator activity | 24 | 0.02383 |
| GO: 0097110 | Scaffold protein binding | 35 | 0.02393 |
| GO: 0008622 | Epsilon DNA polymerase complex | 3 | 0.02522 |
| GO: 0005881 | Cytoplasmic microtubule | 29 | 0.02579 |
| GO: 0000118 | Histone deacetylase complex | 28 | 0.02583 |
| GO: 0099061 | Integral component of postsynaptic density membrane | 2 | 0.02667 |
| GO: 0044309 | Neuron spine | 40 | 0.02738 |
| GO: 0044295 | Axonal growth cone | 7 | 0.02809 |
| GO: 0071144 | Heteromeric SMAD protein complex | 2 | 0.02824 |
| GO: 0043197 | Dendritic spine | 37 | 0.02853 |
| GO: 0043235 | Receptor complex | 179 | 0.0291 |
| GO: 0015271 | Outward rectifier potassium channel activity | 5 | 0.0317 |
| GO: 0042805 | Actinin binding | 16 | 0.03185 |
| GO: 0003700 | DNA binding transcription factor activity | 385 | 0.03189 |
| GO: 0019905 | Syntaxin binding | 25 | 0.03526 |
| GO: 0098831 | Presynaptic active zone cytoplasmic component | 2 | 0.03658 |
| GO: 0005794 | Golgi apparatus | 562 | 0.04007 |
| GO: 0000976 | Transcription regulatory region sequence-specific DNA binding | 368 | 0.04263 |
| GO: 0017016 | Ras GTPase binding | 161 | 0.04338 |
| GO: 0030864 | cortical actin cytoskeleton | 34 | 0.04449 |
| GO: 0014069 | Postsynaptic density | 77 | 0.04529 |
| GO: 0060053 | Neurofilament cytoskeleton | 2 | 0.04637 |
| GO: 0008076 | Voltage-gated potassium channel complex | 41 | 0.04825 |
Table 2.
virgin vs paired GO annotations
| GO ID | GO Annotation | # genes | Raw P value |
|---|---|---|---|
| GO: 0032286 | Central nervous system myelin maintenance | 4 | 0.003 |
| GO: 0044224 | Juxtaparanode region of axon | 6 | 0.0034 |
| GO: 0019933 | cAMP-mediated signaling | 61 | 0.0056 |
| GO: 0045597 | Positive regulation of cell differentiation | 475 | 0.006 |
| GO: 0050790 | Regulation of catalytic activity | 800 | 0.0063 |
| GO: 0048406 | Nerve growth factor binding | 4 | 0.0076 |
| GO: 0035749 | Myelin sheath adaxonal region | 4 | 0.0078 |
| GO: 0042043 | Neurexin family protein binding | 5 | 0.008 |
| GO: 0008277 | Regulation of G-protein coupled receptor protein signaling pathway | 53 | 0.0081 |
| GO: 0061002 | Negative regulation of dendritic spine morphogenesis | 4 | 0.0083 |
| GO: 0007218 | Neuropeptide signaling pathway | 16 | 0.0092 |
| GO: 0071277 | Cellular response to calcium ion | 28 | 0.0106 |
| GO: 0042102 | Positive regulation of T-cell proliferation | 38 | 0.0117 |
| GO: 0098656 | Anion transmembrane transport | 58 | 0.012 |
| GO: 0042048 | Olfactory behavior | 6 | 0.0129 |
| GO: 0044548 | S100 protein binding | 9 | 0.0132 |
| GO: 0043679 | Axon terminus | 29 | 0.0138 |
| GO: 0048485 | Sympathetic nervous system development | 13 | 0.014 |
| GO: 0061014 | Positive regulation of mRNA catabolic process | 24 | 0.0145 |
| GO: 0006401 | RNA catabolic process | 107 | 0.0147 |
| GO: 0005184 | Neuropeptide hormone activity | 5 | 0.0152 |
| GO: 0043220 | Schmidt–Lanterman incisure | 8 | 0.0158 |
| GO: 0002052 | Positive regulation of neuroblast proliferation | 12 | 0.0158 |
| GO: 1902711 | GABA-A receptor complex | 4 | 0.016 |
| GO: 1900271 | Regulation of long-term synaptic potentiation | 11 | 0.0168 |
| GO: 0051965 | Positive regulation of synapse assembly | 49 | 0.0176 |
| GO: 2000144 | Positive regulation of DNA-templated transcription, initiation | 10 | 0.018 |
| GO: 0022851 | GABA-gated chloride ion channel activity | 3 | 0.018 |
| GO: 0000123 | histone acetyltransferase complex | 56 | 0.0186 |
| GO: 0035976 | Transcription factor AP-1 complex | 5 | 0.019 |
| GO: 0008626 | Granzyme-mediated apoptotic signaling pathway | 3 | 0.0194 |
| GO: 0021879 | Forebrain neuron differentiation | 38 | 0.0198 |
| GO: 0048011 | Neurotrophin TRK receptor signaling pathway | 15 | 0.0202 |
| GO: 2000147 | Positive regulation of cell motility | 235 | 0.0212 |
| GO: 0071933 | Arp2/3 complex binding | 4 | 0.0216 |
| GO: 0005125 | Cytokine activity | 15 | 0.0217 |
| GO: 0007271 | Synaptic transmission, cholinergic | 7 | 0.0228 |
| GO: 0035176 | Social behavior | 31 | 0.0249 |
| GO: 0035198 | miRNA binding | 11 | 0.0255 |
| GO: 0070723 | Response to cholesterol | 10 | 0.0256 |
| GO: 0005856 | Cytoskeleton | 919 | 0.0257 |
| GO: 0005272 | Sodium channel activity | 23 | 0.0263 |
| GO: 0042391 | Regulation of membrane potential | 194 | 0.0276 |
| GO: 0050775 | Positive regulation of dendrite morphogenesis | 13 | 0.0283 |
| GO: 0008188 | Neuropeptide receptor activity | 15 | 0.0299 |
| GO: 0006814 | Sodium ion transport | 69 | 0.0316 |
| GO: 0008021 | Synaptic vesicle | 52 | 0.0355 |
Figure 2.
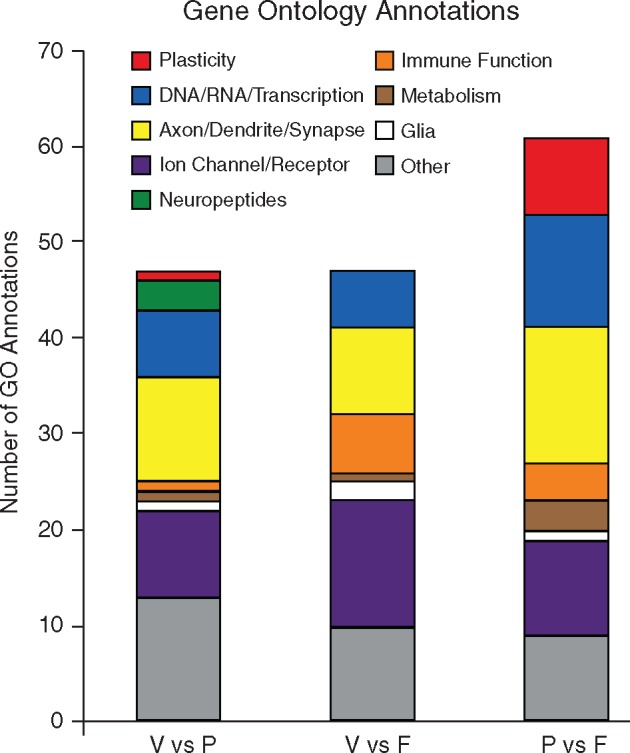
enrichment of GO annotations across comparison groups. The gene enrichment analysis grouped the differentially expressed genes using GO annotations data. We selected significantly enriched GO annotations and identified the annotations that were involved in brain or behavioral processes. Those annotations were then categorized by function within each comparison group. We identified nine functional groups: plasticity (red), DNA/RNA/Transcription (blue), axon/dendrite/synapse (yellow), ion channel/receptor (purple), neuropeptides (green), immune function (orange), metabolism (brown), glia (white), and other (gray). We saw differences in the relative distribution of GO annotation functional groups across the comparison groups. Neuropeptides were only seen in the V vs P group, whereas the V vs F group showed a high number of annotations related to immune function. The P vs F group contained the largest number of annotations related to plasticity, DNA/RNA/transcription, and axon/dendrite/synapse
KEGG Pathways Analysis
In the V vs P comparison group, the commonly recurring pathways included: protein export, protein processing in the endoplasmic reticulum, thyroid hormone synthesis, antigen processing and presentation, Ras signaling, Rap1 signaling, neuroactive ligand–receptor pathway, calcium signaling, and regulation of the actin cytoskeleton. In the V vs F comparison group, the commonly recurring pathways included: protein processing in the endoplasmic reticulum, regulation of the actin cytoskeleton, Ras signaling, metabolic pathways, axon guidance, protein processing in the endoplasmic reticulum, thyroid hormone synthesis, and antigen processing and presentation. In the P vs F comparison group, the commonly recurring pathways included: Ras signaling, Rap1 signaling, neuroactive ligand–receptor pathway, calcium signaling, MAPK signaling, LTP, glutamatergic pathways, dopaminergic pathways, and regulation of the actin cytoskeleton.
Using the list of genes generated from the GO annotations analysis, we next identified genes that were associated with multiple KEGG pathways. By excluding genes that were not associated with any KEGG pathways, or were associated with pathways that were not related to brain function, we further narrowed the range of genes of interest. Ultimately, in each comparison group we identified genes with patterns of expression that differed across social experience and that were linked to biological pathways within the brain (Table 5). We standardized the expression of each gene relative to its expression in virgin males then grouped genes that were associated with nine commonly recurring KEGG pathways and compared the expression of those genes across groups. Since this was an exploratory study, we did not perform statistical tests, and instead used Cohen’s d as a measure of effect size (Table 6). Figure 3 shows the changes in gene expression in paired males (left) and fathers (right) compared with virgin males (dashed line). Overall, we observed only moderate changes in gene expression in paired males, but fathers exhibited an overall decrease in gene expression, especially in genes that were associated with long-term potentiation (LTP) and long-term depression (LTD) (d = 1.072), neurotransmitters (d = 0.911), and Ca2+ signaling (d = 0.877). We found medium effects of differential patterns of expression in genes that were associated with oxytocin signaling (d = 0.787), protein processing in the endoplasmic reticulum (d = 0.599), and Ras/Rap1 signaling (0.578). These results suggest that the genes associated with these KEGG pathways undergo coordinated changes in expression patterns that are related to social experience. Furthermore, different social experiences can result in dramatically different patterns of gene expression, i.e. Ca2+ signaling.
Table 5.
genes with altered patterns of expression
| Comparison | Genes |
|---|---|
| V vs P | Cckar, Dnajc3, Enah, Hspa5, Hyou1, Pak3, Pdia3, Pdia4, Rala, Sorbs1, Th |
| V vs F | Arpc5, Baiap2, Cbl, Chrm1, Chrna1, Cyp2s1, Derl1, Dnajc3, Elovl1, Elovl6, Enah, Epha2, Erp29, Faah, Glra3, Hspa5, Itgb4, Kcnj4, Ksr1, Lamtor3, Nf2, Pdia3, Pdia4, Pdia6, Pigh, Pigo, Pla2g16, Pomgnt2, Prkgc, Pvrl3, Rdx, Tram1, Txndc5 |
| P vs F | Adcy4, Adora2a, Atp2b1, Baiap2, Cacna2d3, Cacnb3, Cckbr, Chrm1, Ddn, Dlg4, Gabrd, Gpr156, Grin2a, Grin2b, Ifngr1, Itpr1, Kcnj2, Kcnj4, Kcnn3, Kdr, Lama2, Ngef, P2rx3, Park2, Prkcg, Ptk2b, Rasgrf2, Rgs14, Rin1, Sipa1l1, Tiam1 |
Table 6.
KEGG pathways associated with differentially expressed genes
| Neurotransmitters | Adcy4, Adora2a, Cacna2d3, Cacnb3, Cckar, Cckbr, Chrm1, Dlg4, Gabrd, Gpr156, Grin2a, Grin2b, Itpr1, Kcnj2, Kcnj4, P2rx3, Prkcg, Rin1 |
| Ca signaling | Adcy4, Adora2a, Cckar, Cckbr, Chrm1, Grin2a, Grin2b, Itpr1, P2rx3, Ptk2b, Rin1 |
| OT signaling | Adcy4, Cacna2d3, Cacnb3, Kcnj2, Kcnj4, Itpr1, Prkcg |
| Regulation of actin cytoskeleton | Arpc5, Baiap2, Chrm1, Enah, Itgb4, Rdx, Tiam1, Enah, Pak3 |
| Ras/Rap1 signaling | Adcy4, Adora2a, Epha2, Grin2a, Grin2b, Kdr, Ksr1, Pak3, Pla2g16, Prkgc, Rala, Rasgrf2, Rgs14, Rin1, Sipa1l1, Tiam1 |
| Protein processing in ER | Derl1, Dnajc3, Erp29, Hspa5, Hyou1, Pdia3, Pdia4, Pdia6, Tram1, Txndc5 |
| Thyroid hormone synthesis | Adcy4, Itpr1, Prkcg, Pdia4, Hspa5 |
| LTP/LTD | Grin2a, Grin2b, Itpr1, Rin1 |
Figure 3.
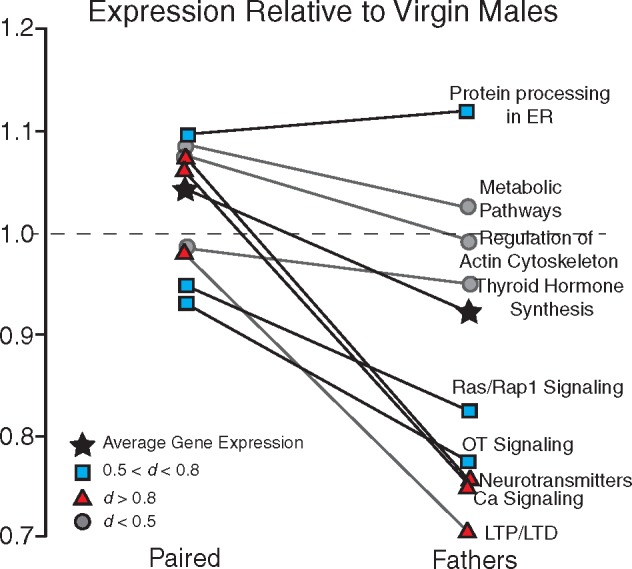
gene expression in paired males and fathers relative to virgin males. Using the KEGG pathways analysis, we identified genes with differential patterns of expression that were linked to nine pathways of biological or behavioral significance. The mean expression of genes associated with each pathway in fathers was averaged and compared against expression in virgin males. On the whole, gene expression was decreased in fathers relative to both virgins and paired males, suggesting that patterns of gene expression undergo coordinated changes in expression relative to social experience. Of the nine pathways, only one showed an increase in gene expression in fathers (protein processing in the endoplasmic reticulum), while five showed decreases in gene expression in fathers (Ras/Rap1 signaling, oxytocin signaling, neurotransmitters, calcium signaling, and LTP/LTD). The overall average gene expression is indicated by black stars. Values that exhibited large effect sizes (Cohen’s d > 0.8) are indicated by red triangles, values that exhibited medium effect sizes (0.5 < Cohen’s d < 0.8) are indicated by blue squares, and values that exhibited small effect sizes (Cohen’s d < 0.5) are indicated by gray circles
STRING Database Analysis
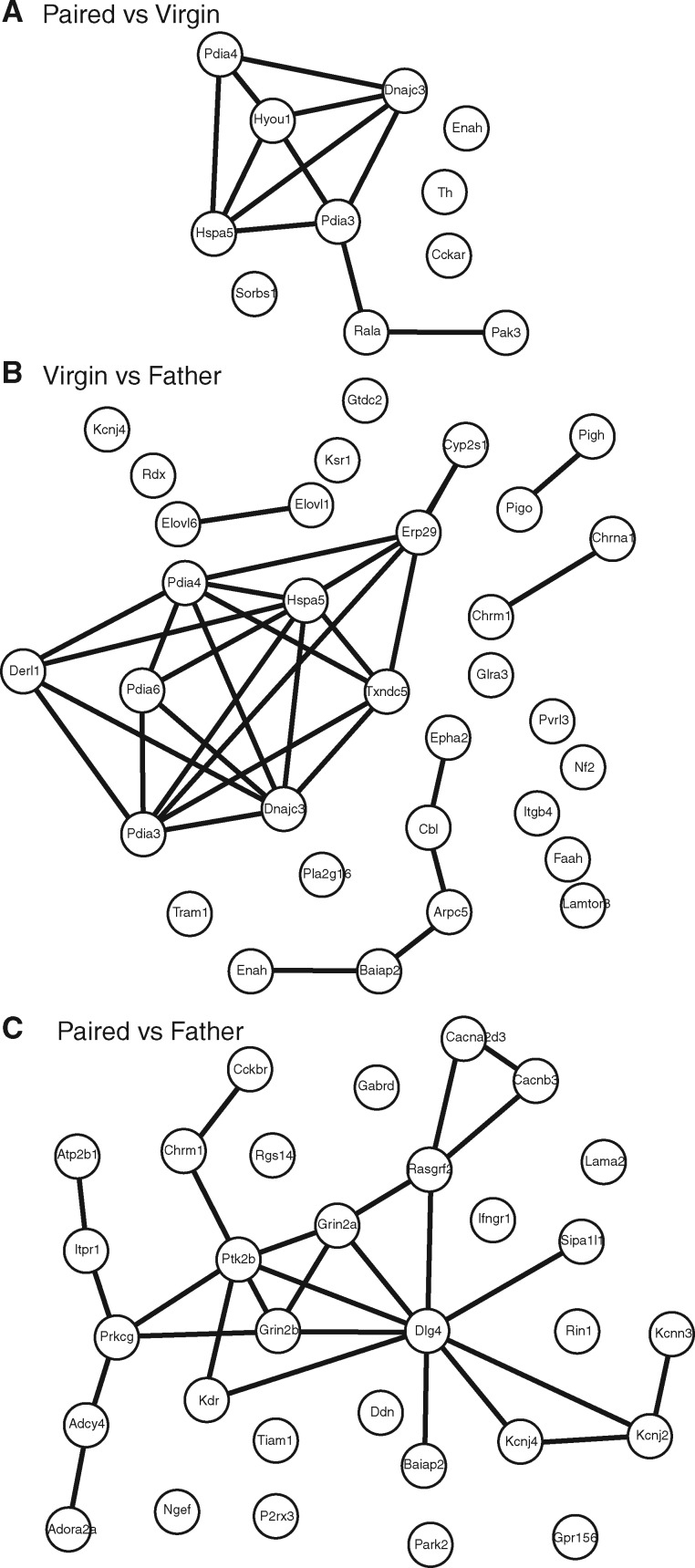
We used the STRING database to assess the network connectivity between the genes in each comparison group that were identified as having both differential patterns of expression and functional significance.
The 11 genes from the V vs P comparison group produced a network with 11 nodes and 11 edges, and a protein-protein interaction (PPI) enrichment P-value of 5.86 × 10−7 (Fig. 4A). Thus, the proteins expressed by these genes have significantly more interactions than would be expected by chance, as defined as a random set of similarly sized proteins selected from the genome. There was one cluster of seven interacting proteins, and the functions of these gene products were primarily related to functions of the endoplasmic reticulum, as well as the cellular response to stimulation.
Figure 4.
STRING database analysis of gene product interaction networks. Selected genes were run through the STRING database of gene product interactions, and networks were generated for each comparison. (A) Paired vs Virgin network; (B) Virgin vs Father; (C) Paired vs Father
The 33 genes from the V vs F comparison group produced a network with 32 nodes and 29 edges, and a PPI enrichment P-value of 6.99 × 10−15 (Fig. 4B), indicating that the proteins expressed by these genes have significantly more interactions than would be expected by chance. These gene products produced one large cluster of nine interacting proteins, one medium cluster of five interacting proteins, and three separate small clusters of two interacting proteins. The large cluster was predominantly involved with the function of the endoplasmic reticulum. The medium cluster was involved with process of neural plasticity, including signaling pathways and modification of the actin cytoskeleton. The three small clusters were involved with the elongation of fatty acid chains, the formation of cholinergic receptors, and GPI-anchor synthesis.
The 31 genes from the P vs F comparison group produced a network with 31 nodes and 27 edges, and a PPI enrichment P-value of 1.36 × 10−12 (Fig. 4C), indicating that the proteins expressed by these genes have significantly more interactions than would be expected by chance. These gene products produced 1 large network consisting of 20 interacting proteins. The genes in this network were involved in a variety of functions, including synaptic plasticity and neural transmission, ion transmembrane transport, the cellular response to stimulus, and the structure of the synapse and dendrite.
NanoString Analysis
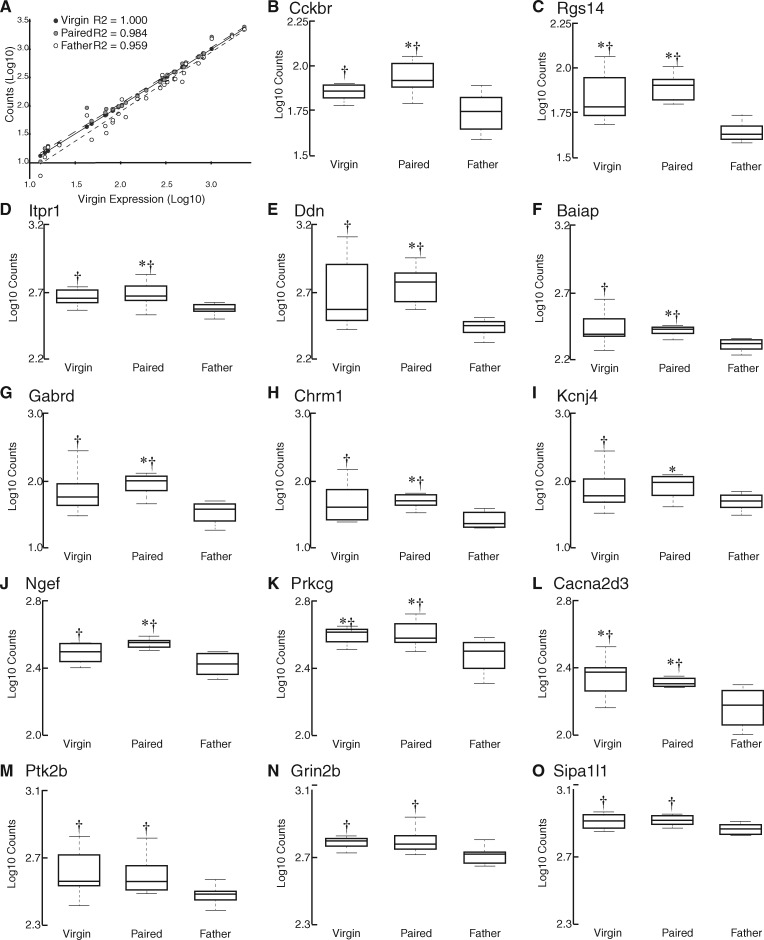
A total of 33 genes (30 target genes and 3 housekeeping genes) were selected for quantitative analysis using NanoString. The housekeeping genes (Gusb, Pgk1, and Eif4a2) did not show different levels of expression across conditions, confirming that these genes can serve as a good baseline in prairie voles. A heat map analysis revealed that 23 of our 30 target genes had lower expression levels in fathers than in either virgins or paired males (Fig. 5). Six genes had lower expression levels in virgins, and no gene in any group appeared to show inordinately high levels of expression. A regression analysis revealed similar levels of gene expression across all experimental conditions (Fig. 6A).
Figure 5.
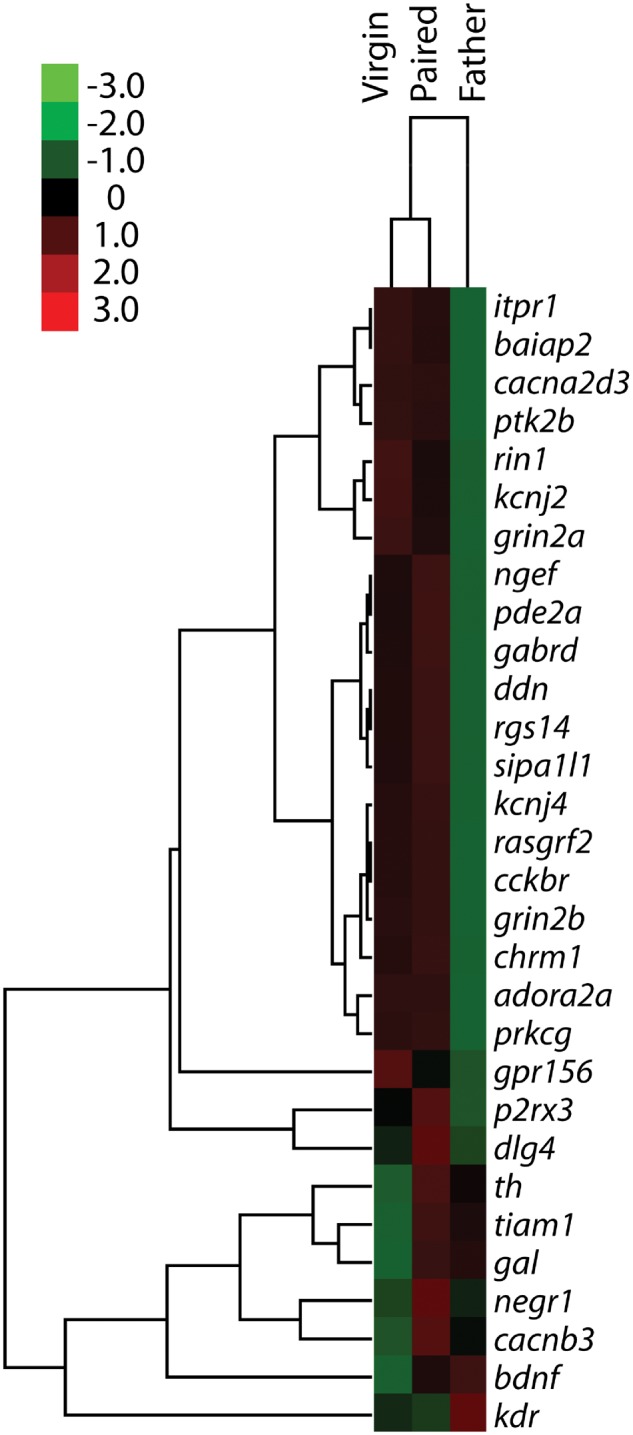
heat map representing the relative expression of individual genes in virgin males, paired males, and males with fathering experience. Gene enrichment is encoded in the heat map ranging from low (green) to high (red). Genes that show similar expression patterns are clustered together, as indicated by the dendrogram to the left of the heat map
Figure 6.
quantitative analysis of gene expression. (A) A scatterplot showing the distribution of gene expression in virgin males (black circles), paired males (gray circles), and fathers (white circles). (B–O) Box and whisker plots showing the expression of genes in virgins, paired males, and fathers. The whiskers represent × 1.5 the interquartile range. While we quantified the expression of 30 genes using NanoString, here we show the 14 genes that exhibited significantly different levels of expression across groups or exhibited large effect size. In each gene of these genes, expression in fathers was lower than expression in paired males, and in three cases (Rgs14, Prkcg, and Cacna2d3) expression in fathers was also significantly lower than in virgin males. *Significantly differs from fathers (P < 0.05). †Large effect size compared with fathers (Cohen’s d > 0.8).
Expression data for each individual gene were compared across groups using t-tests, which were run and P-values were adjusted for multiple comparisons using nSolver software. Of the 30 target genes, 11 genes showed significant differential expression between comparison groups (P < 0.05; Cckbr, Rgs14, Itpr1, Ddn, Baiap, Gabrd, Chrm1, Kcnj4, Ngef, Prkcg, and Cacna2d3; Fig. 6B–O; Table 7). We also calculated the effect sizes using Cohen’s d, examining differential expression of each gene across groups (Fig. 6B–O; Table 7). In the V vs P group, we saw a large effect (defined as 0.8 < d < 1.2) in Tiam1. In the P vs F group, we saw large effects (0.8 < d < 1.2) in Baiap2, Cacna2d3, Cckbr, Chrm1, Ddn, Dlg4, Gabrd, Itpr1, Kdr, P2rx3, Pde2a, Ptk2b, and Rasgrf2. In the P vs F group, we also saw very large effects (defined as d > 1.2) in Grin2b, Ngef, Prkcg, Rgs14, and Sipa1l1. In the V vs F group we saw large effects (0.8 < d < 1.2) in Adora2a, Cacna2d3, Cckbr, Chrm1, Ddn, Gabrd, Grin2a, Grin2b, Itpr1, Kcnj4, Ngef, Prkcg, Ptk2b, and Sipa1l1. In the V vs F group we also saw very large effects (d > 1.2) in Baiap2, Rgs14, and Rin1.
Table 7.
P-values and Cohen’s d values
| Gene name | F vs P |
F vs V |
P vs V |
|||
|---|---|---|---|---|---|---|
| P-value | Cohen’s d | P-value | Cohen’s d | P-value | Cohen’s d | |
| Adora2a | 0.1014 | 0.5380 | 0.2444 | 0.9243 | 0.9891 | 0.3863 |
| Baiap2 | 0.0014 | 0.9039 | 0.0776 | 1.2258 | 0.7946 | 0.3220 |
| Bdnf | 0.6273 | 0.2066 | 0.2702 | 0.6260 | 0.4012 | 0.4194 |
| Cacna2d3 | 0.0460 | 1.0766 | 0.0326 | 1.1004 | 0.9266 | 0.0239 |
| Cacnb3 | 0.4862 | 0.4467 | 0.5735 | 0.3052 | 0.2516 | 0.7520 |
| Cckbr | 0.0120 | 0.9881 | 0.0767 | 0.9884 | 0.7745 | 0.0003 |
| Chrm1 | 0.0050 | 0.9371 | 0.0876 | 1.0050 | 0.7353 | 0.0679 |
| Ddn | 0.0021 | 1.0688 | 0.0829 | 1.0685 | 0.6383 | 0.0003 |
| Dlg4 | 0.2146 | 0.8759 | 0.6877 | 0.1721 | 0.2711 | 0.7037 |
| Gabrd | 0.0018 | 0.9480 | 0.0803 | 0.9667 | 0.5384 | 0.0187 |
| Gal | 0.6783 | 0.2902 | 0.2519 | 0.4687 | 0.1944 | 0.7589 |
| Gpr156 | 0.8590 | 0.0103 | 0.7849 | 0.2844 | 0.8507 | 0.2947 |
| Grin2a | 0.2294 | 0.6531 | 0.1292 | 0.8749 | 0.7381 | 0.2218 |
| Grin2b | 0.0778 | 1.2351 | 0.0877 | 1.0075 | 0.7291 | 0.2276 |
| Itpr1 | 0.0500 | 0.8004 | 0.0906 | 1.0156 | 0.8406 | 0.2152 |
| Kcnj2 | 0.3466 | 0.5576 | 0.1956 | 0.7754 | 0.7144 | 0.2178 |
| Kcnj4 | 0.0474 | 0.6183 | 0.2362 | 0.8591 | 0.8533 | 0.2407 |
| Kdr | 0.2402 | 0.8634 | 0.3649 | 0.6950 | 0.8613 | 0.1683 |
| Negr1 | 0.5673 | 0.3141 | 0.8722 | 0.1085 | 0.4501 | 0.4227 |
| Ngef | 0.0061 | 1.1987 | 0.1282 | 0.9933 | 0.5444 | 0.2054 |
| P2rx3 | 0.0605 | 1.1447 | 0.3671 | 0.4699 | 0.2208 | 0.6748 |
| Pde2a | 0.0677 | 0.9381 | 0.2581 | 0.7403 | 0.6827 | 0.1978 |
| Prkcg | 0.0427 | 1.2933 | 0.0390 | 1.1543 | 0.8554 | 0.1389 |
| Ptk2b | 0.0729 | 0.8524 | 0.0996 | 0.9836 | 0.9024 | 0.1313 |
| Rasgrf2 | 0.1027 | 0.9509 | 0.1289 | 0.7855 | 0.8217 | 0.1654 |
| Rgs14 | 0.0002 | 1.5018 | 0.0237 | 1.2370 | 0.4867 | 0.2648 |
| Rin1 | 0.1646 | 0.7439 | 0.0696 | 1.4011 | 0.2535 | 0.6572 |
| Sipa1l1 | 0.0612 | 1.2207 | 0.1155 | 0.9080 | 0.5573 | 0.3127 |
| Th | 0.4437 | 0.3234 | 0.4343 | 0.4063 | 0.1845 | 0.7297 |
| Tiam1 | 0.7299 | 0.3216 | 0.2976 | 0.5269 | 0.2510 | 0.8484 |
P-values: bold indicates significant values (P ≤ 0.05). Cohen’s d values: bold indicates large effect (0.8 < d < 1.2). Bold italics indicates very large effect (d > 1.2).
Discussion
In this experiment, we compared gene expression in the MPOA of virgin male prairie voles, males that had formed a pair bond, and males with fathering experience. We found that these groups differed in gene expression. Distinct patterns were revealed using a series of analyses, including GO annotation enrichment, KEGG pathways, STRING network analysis, and quantitative assessment using NanoString. Males with fathering experience showed a relative decrease in gene expression compared with virgins and paired males, and many of the genes that exhibited decreased expression were involved in synaptic transmission and plasticity. These results suggest that fathers may exhibit a decreased amount of synaptic plasticity within the MPOA.
The transition to fatherhood is associated with a variety of potential environmental and behavioral changes, such as the presence of infants, changes in energetic requirements and feeding behavior, and stress responsiveness [50–52]. In this experiment, we sought to identify alterations in central nervous system gene expression that are associated with fathering experience. To our knowledge, this is the first time that RNA sequencing has been performed on prairie vole brain tissue. As such, we faced several technical challenges over the course of this study. For instance, while the prairie vole genome has been sequenced [53, 54], its annotation is incomplete, leaving us to rely on the annotated mouse genome (Mus musculus) for many of our analyses. In addition, it is important to consider the consequences associated with working in an outbred rodent-like prairie voles. The individual differences associated with an outbred population may have masked additional target genes associated with the onset of paternity. Furthermore, in this study we concentrated on traditional analyses. By examining protein coding transcripts, and restricting our analysis to pathways and genes that were related to the brain and behavior, it is possible that we overlooked some nontraditional candidate genes. Regardless, we still observed significantly altered expression on both the individual gene and system level. These results suggest that paternity engages similar physiological mechanisms across prairie vole males despite genetic diversity.
Biparental care is rare in mammals, but prairie voles are not the only rodents who exhibit this behavior. The males of several species of Peromyscus, including Peromyscus californicus and Peromyscus polionotus, exhibit paternal care, while other species, including P.maniculatus, do not. This behavioral distinction allowed Bendesky et al. to investigate genetic differences between P. polionotus and P. maniculatus that are linked to parenting behavior [55]. In a series of experiments, they identified several quantitative trait loci that were linked to specific behaviors of interest, including nest building. Further analysis revealed that the gene for arginine vasopression (AVP) was directly related to nest building, and when AVP was administered intracerebroventricularly there was a significant decrease in the quality of nest building [55]. Unlike, the study by Bendesky et al., we did not find changes implicating AVP. However, there are several differences between the two experiments. In this study, we specifically examined gene expression within one hypothalamic nucleus, the MPOA. Our study was in a different species and used males that had very specific social experiences: virgin males, pair bonded males, and males with fathering experience.
RNA sequencing is a powerful technique that allows us to identify alterations in gene expression that are associated with behavioral and other phenotypic changes [56]. The greatest challenge with this technique, however, is the large amount of data it produces. There is no one agreed upon analysis that most effectively identifies specific genes of interest [57, 58]. Thus, in this study we used several techniques to reveal novel gene targets to further our understanding of paternal behavior. We believe that this is a strength rather than a weakness. The ultimate goal of this experiment was to increase our understanding of the alterations that occur within the MPOA following exposure to different social contexts in male prairie voles. As such, we have identified a set of genes and their associated pathways that we can use to further explore male parenting behavior.
Our quantitative assessment of gene expression in the MPOA revealed an overall decrease in the expression of many genes in fathers relative to both virgins and pair-bonded males. The specific genes of interest that we identified were involved in a range of physiological processes, including metabolism, stress responsiveness, and plasticity. However, most of the genes that showed different patterns of expression between groups, and specifically decreased expression in fathers, were associated with synaptic transmission and dendritic spine motility (Table 8). For example, several genes involved in the production and maintenance of receptors (including Cckbr, Chrm1, Gabrd, Grin2b, and Itpr1) and ion channels (including Cacna2d3, Kcnj4, and P2rx3) were significantly downregulated. These results suggest that GABA, glutamate, and cholinergic systems are all affected by fathering experience, as are calcium and potassium channels. Other genes that exhibited significant downregulation in fathers were involved with the actin cytoskeleton, dendritic spine motility, and other components of the physical plasticity of dendrites. We emphasize that this is not an exhaustive list of differentially expressed genes; however, these results suggest that synaptic plasticity may be diminished in the MPOA of male prairie voles with fathering experience.
Table 8.
genes of interest
| Gene ID | Gene name | Function | GO Annotations |
|---|---|---|---|
| Baiap2 | Brain-specific angiogenesis inhibitor 1-associated protein 2 | Insulin receptor tyrisone kinase substrate | Signaling, regulation of biological quality, membrane part, dendrite, dendritic spine, synapse |
| Cacna2d3 | Calcium voltage gated channel, auxiliary subunit alpha 2 delta 3 | Voltage gated calcium channel | Ion channel activity |
| Cckbr | Cholecystokinin B receptor | Multipass transmembrane receptor protein | Signaling, regulation of biological quality |
| Chrm1 | Cholinergic receptor, muscarinic 1 | Muscarinic receptor | Signaling, regulation of biological quality, membrane part, dendrite, synapse |
| Ddn | Dendrin | Plasma membrane surrounding dendritic spine | Membrane part |
| Gabrd | GABA A receptor, subunit delta | GABA receptor | Signaling, dendrite, synapse, ion channel activity |
| Grin2b | Glutamate receptor, ionotropic, NMDA 2b | NMDA receptor | Signaling, regulation of biological quality, membrane part, dendrite, dendritic spine, synapse, ion channel activity |
| Itpr1 | Inositol 1,4,5-triphosphate receptor 1 | Calcium channel | Signaling, regulation of biological quality, membrane part, dendrite, synapse, ion channel activity |
| Kcnj4 | Potassium voltage gated channel subfamily J member 4 | Potassium channels – ion homeostasis | Membrane part, dendrite, synapse, ion channel activity |
| Ngef | Neuronal guanine nucleotide exchange factor | Dendritic spine morphogenesis | Signaling, membrane part |
| P2rx3 | Purinergic receptor p2x, ligand gated ion channel | ATP receptor | Signaling, regulation of biological quality, membrane part, dendrite, dendritic spine, synapse, ion channel activity |
| Pde2a | Phosphodiesterase 2a | 2nd messenger signaling/dendritic spines | Signaling, regulation of biological quality, membrane part, dendrite |
| Prkcg | Protein kinase c gamma | Signaling protein | Signaling, membrane part, dendrite, synapse |
| Ptk2b | Protein tyrosine kinase 2 beta | Ion channel regulation; MapK signaling | Signaling, regulation of biological quality, membrane part, dendrite, dendritic spine, synapse, ion channel activity |
| Rgs14 | Regulator of g-protein signaling 14 | Scaffold protein | Signaling, regulation of biological quality, membrane part, dendrite, dendritic spine, synapse |
| Rin1 | Ras and Rab interactor 1 | Ras effector | Signaling, regulation of biological quality, membrane part, dendrite |
| Sipa1l1 | Signal induced proliferation associate 1 like 1 | Ras effector | Signaling, regulation of biological quality, membrane part, dendrite, dendritic spine, synapse |
We were surprised by the lack of differential expression of oxytocin and vasopressin-related genes; however, this finding is not unique within the literature. In a series of experiments, Kenkel et al. examined the neuroendocrine correlates of pup exposure in male prairie voles that were virgins or had fathering experience [52, 59]. They saw changes in OT immunoreactivity in PVN/BNST, but there were no changes to OT/AVP in the MPOA. Another study examined OT immunoreactive cells in male prairie voles that were virgins, had established pair bonds, or had fathering experience [60]. They saw an increase in the number of OT immunoreactive cells in the MPOA of paired males and fathers compared with virgin males, but there was a greater increase of OT-ir cells in the PVN of fathers compared with paired and virgin males. It is likely that examination of gene expression in the PVN would show alterations in OT gene expression. In future studies we hope to examine patterns of gene expression in additional brain regions.
Fatherhood also seems to be associated with structural alterations in neural plasticity, as measured by changes in the number and density of dendritic spines. Mice with fathering experience show increased survival of newborn neurons and increased dendritic spine density within the hippocampus [61, 62]. Male marmosets show an increase in dendritic spine density in the prefrontal cortex after fathering experience [63]. However, other studies have shown reductions in the survival of adult-generated neurons in the amygdala of the prairie vole and hippocampus of California mice [64, 65]. The effects of fatherhood clearly vary across brain regions, but we do not yet know what is causing these changes in neural plasticity.
The lower gene expression related to dendritic spines, associated with fatherhood in the present study, is evocative of similar changes seen in a recent study of the MPOA of mother rats [66]. Rem2, a gene associated with reduction of dendritic branching but increases in spine density [67, 68] was increased in the MPOA of high licking/grooming rats, but only in lactating mothers (not in virgins). This increase was accompanied by decreased dendritic complexity. Rem2 is involved with GTPase activity and GTP binding. While we did not see alterations in Rem2 expression in this study, we found altered expression in several genes that are involved in Ras and Rap1 signaling. Both Ras and Rap1 are GTPases that work in concert to modulate cellular growth and plasticity [69–71]. Ras relays NMDA receptor signaling that drives the delivery of AMPA receptors during LTP, while Rap1 is involved in the NMDA receptor-dependent removal of AMPA receptors during LTD [69, 72]. The altered pattern of expression of these genes in fathers suggests that this extremely salient social experience triggers a molecular cascade that is involved in neuronal plasticity.
The down-regulation of genes associated with dendritic complexity in the present study, as well as the study by Parent et al., is similar to what one would expect in an animal that had experienced high amounts of stress. It is well established that stress, mediated by corticotropin-releasing hormone, results in a loss of dendritic spines [73–77]. Additionally, rat mothers show a decrease in the number and density of dendritic spines in the amygdala and stria terminalis 4 days after birth [78], and an increase in dendritic spine density in the hippocampus during the postpartum period [79]. This suggests that alterations in dendritic spine density in mothers may be both transient and region specific, perhaps linked to the peak in corticosterone that occurs during parturition [80]. More studies must be done to determine if the same holds true for vole fathers.
In many species, the transition to fatherhood is associated with a suite of behavioral and hormonal changes, including those indicative of stress. In California mice (P.californicus), fathers exhibit attenuated anxiety-like behavior ∼2 weeks after pups are born [61, 62]. Human males show a peak in cortisol levels during the transition to fatherhood [81]. Prairie vole fathers show increased anxiety-like behavior, and chronic pup exposure (in this case, 20 min of unrelated pup exposure per day for 10 days) results in an increase in basal CORT levels [64]. In an open field test, fathers spent more time in corner squares, and in an elevated plus maze, fathers spent less time in open arms. In forced swim tests, fatherhood decreased the latency to immobility, and increased the number and duration of immobility bouts [64]. In the long-term, fatherhood may be beneficial for male health, but the transition to fatherhood is a tremendously stressful period [82].
In male voles with fathering experience, we also see the upregulation of genes related to protein processing in the endoplasmic reticulum. The endoplasmic reticulum is instrumental in managing the protein folding process, including disposing of misfolded proteins [83]. Homeostatic imbalances, including stress, can alter the functioning of the endoplasmic reticulum, leading to the initiation of the unfolded-protein response, which can in turn lead to apoptosis [84, 85]. This may be one mechanism by which physiological stress can result in homeostatic perturbations [86, 87], including some of the changes that are evident in vole fathers, such as weight loss [50, 59].
While many of the changes we saw in gene expression may be partially attributable to stress, there are likely many other additional factors at play. Fathers in many species show systematic endocrine changes [22]. Environmental factors, including changes in the types and amount of sensory stimulation, or the amount of parental care they received, may play a role as well [88, 89]. Much more work must be done to tease apart these many factors.
In this study, we saw the most varied and interesting differences between the paired males and males with fathering experience. This was surprising, as we expected that the greatest differences would be between the virgin males and fathers. However, examination of the quantitative results begins to clarify these findings (Figs 3 and 6). The expression of genes of interest is slightly elevated in paired animals relative to virgins, but the expression in fathers is decreased relative to virgins. Thus, while the expression levels of some genes do not significantly differ between virgins and paired males, and virgins and fathers, we found significant differences between paired males and fathers. This may suggest that the experience of fathering is functionally distinct from any other type of social interactions that these animals have encountered.
Conclusions
The purpose of this study was to explore how gene expression changed across the transition to fatherhood, and to identify novel targets to allow for deeper investigation of male parenting behavior. The use of RNA sequencing confirmed that there are differences in gene expression between voles that had different social experiences, including virgin males, males that had formed a pair bond with a female, and males with parenting experience. The genes identified in this study suggest novel processes that are related to paternal behavior and offer new targets for the further exploration of fathering behavior.
Acknowledgements
The sequencing was carried out at the DNA Technologies and Expression Analysis Cores at the UC Davis Genome Center, supported by NIH Shared Instrumentation Grant 1S10OD010786–01, and was made possible by a pilot grant from the UC Davis Genome Center. Thanks to Chris Harshaw for helpful analytical suggestions. Thanks to Cindy Clayton and Rhonda Oates-O’Brien for husbandry and veterinary care of the prairie vole colony. And special thanks to the anonymous reviewers for their helpful and constructive comments on this manuscript.
Conflict of interest statement: None declared.
References
- 1. Kleiman DG. Monogamy in mammals. Q Rev Biol 1977; 52:39–69. [DOI] [PubMed] [Google Scholar]
- 2. Lukas D, Clutton-Brock TH. The evolution of social monogamy in mammals. Science 2013; 341: 526–30. [DOI] [PubMed] [Google Scholar]
- 3. Opie C, Atkinson QD, Dunbar RI, Shultz S. Male infanticide leads to social monogamy in primates. Proc Natl Acad Sci USA 2013; 110: 13328–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gubernick DJ, Alberts D Jr. The biparental care system of the California mouse, Peromyscus californicus. J Comp Psychol 1987; 101: 169–77. [PubMed] [Google Scholar]
- 5. Runcie MJ. Biparental care and obligate monogamy in the rock-haunting possum, Petropseudes dahli, from tropical Australia. Anim Behav 2000; 59: 1001–8. [DOI] [PubMed] [Google Scholar]
- 6. Wynne-Edwards KE, Timonin ME. Paternal care in rodents: weakening support for hormonal regulation of the transition to behavioral fatherhood in rodent animal models of biparental care. Horm Behav 2007; 52: 114–21. [DOI] [PubMed] [Google Scholar]
- 7. Mendoza SP, Mason WA. Attachment relationships in New World primates. Ann N Y Acad Sci 1997; 807:203–9. [DOI] [PubMed] [Google Scholar]
- 8. Thomas JA, Birney EC. Parental care and mating system of the prairie vole, Microtus ochrogaster. Behav Ecol Sociobiol 1979; 5: 171–86. [Google Scholar]
- 9. Bales KL, Saltzman W. Fathering in rodents: neurobiological substrates and consequences for offspring. Horm Behav 2016; 77:249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cantoni D, Brown RE. Paternal investment and reproductive success in the California mouse, Peromyscus californicus. Anim Behav 1997; 54: 377–86. [DOI] [PubMed] [Google Scholar]
- 11. Ahern TH, Hammock EA, Young LJ. Parental division of labor, coordination, and the effects of family structure on parenting in monogamous prairie voles (Microtus ochrogaster). Dev Psychobiol 2011; 53: 118–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jia R, Tai F, An S, Zhang X, Broders H. Effects of neonatal paternal deprivation or early deprivation on anxiety and social behaviors of the adults in mandarin voles. Behav Processes 2009; 82: 271–8. [DOI] [PubMed] [Google Scholar]
- 13. Ovtscharoff W Jr, Helmeke C, Braun K. Lack of paternal care affects synaptic development in the anterior cingulate cortex. Brain Res 2006; 1116: 58–63. [DOI] [PubMed] [Google Scholar]
- 14. Perkeybile AM, Griffin LL, Bales KL. Natural variation in early parental care correlates with social behaviors in adolescent prairie voles (Microtus ochrogaster). Front Behav Neurosci 2013; 7: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Perkeybile AM, Bales KL. Early rearing experience is related to altered aggression and vasopressin production following chronic social isolation in the prairie vole. Behav Brain Res 2015; 283: 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perkeybile AM, Bales KL. Early rearing experience is associated with vasopressin immunoreactivity but not reactivity to an acute non-social stressor in the prairie vole. Physiol Behav 2015; 147:149–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arias Del Razo R, Bales KL. Exploration in a dispersal task: effects of early experience and correlation with other behaviors in prairie voles (Microtus ochrogaster). Behav Processes 2016; 132:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bales K, French JA, Dietz JM. Explaining variation in maternal care in a cooperatively breeding mammal. Anim Behav 2002; 63: 453–61. [Google Scholar]
- 19. Bester-Meredith JK, Marler CA. Vasopressin and the transmission of paternal behavior across generations in mated, cross-fostered Peromyscus mice. Behav Neurosci 2003; 117: 455–63. [DOI] [PubMed] [Google Scholar]
- 20. Frazier CR, Trainor BC, Cravens CJ, Whitney TK, Marler CA. Paternal behavior influences development of aggression and vasopressin expression in male California mouse offspring. Horm Behav 2006; 50: 699–707. [DOI] [PubMed] [Google Scholar]
- 21. Dulac C, O’Connell LA, Wu Z. Neural control of maternal and paternal behaviors. Science 2014; 345: 765–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saltzman W, Ziegler TE. Functional significance of hormonal changes in mammalian fathers. J Neuroendocrinol 2014; 26: 685–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gray PB, McHale TS, Carre JM. A review of human male field studies of hormones and behavioral reproductive effort. Horm Behav 2017; 91:52–67. [DOI] [PubMed] [Google Scholar]
- 24. Lynn SE. Endocrine and neuroendocrine regulation of fathering behavior in birds. Horm Behav 2016; 77: 237–48. [DOI] [PubMed] [Google Scholar]
- 25. Trainor BC, Marler CA. Testosterone, paternal behavior, and aggression in the monogamous California mouse (Peromyscus californicus). Horm Behav 2001; 40: 32–42. [DOI] [PubMed] [Google Scholar]
- 26. Ziegler TE, Prudom SL, Zahed SR, Parlow AF, Wegner F. Prolactin’s mediative role in male parenting in parentally experienced marmosets (Callithrix jacchus). Horm Behav 2009;56: 436–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kohl J, Babayan BM, Rubinstein ND, Autry AE, Marin-Rodriguez B, Kapoor V, Miyamishi K, Zweifel LS, Luo L, Uchida N et al. . Functional circuit architecture underlying parental behaviour. Nature 2018; 556: 326–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kohl J, Dulac C. Neural control of parental behaviors. Curr Opin Neurobiol 2018; 49:116–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Jong TR, Chauke M, Harris BN, Saltzman W. From here to paternity: neural correlates of the onset of paternal behavior in California mice (Peromyscus californicus). Horm Behav 2009; 56: 220–31. [DOI] [PubMed] [Google Scholar]
- 30. Kirkpatrick B, Kim JW, Insel TR. Limbic system Fos expression associated with paternal behavior. Brain Res 1994; 658: 112–8. [DOI] [PubMed] [Google Scholar]
- 31. Lee AW, Brown RE. Medial preoptic lesions disrupt parental behavior in both male and female California mice (Peromyscus californicus). Behav Neurosci 2002; 116: 968–75. [DOI] [PubMed] [Google Scholar]
- 32. Trainor BC, Bird IM, Alday NA, Schlinger BA, Marler CA. Variation in aromatase activity in the medial preoptic area and plasma progesterone is associated with the onset of paternal behavior. Neuroendocrinology 2003; 78: 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Perea-Rodriguez JP, Takahashi EY, Amador TM, Hao RC, Saltzman W, Trainor BC. Effects of reproductive experience on central expression of progesterone, oestrogen alpha, oxytocin and vasopressin receptor mRNA in male California mice (Peromyscus californicus). J Neuroendocrinol 2015; 27: 245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Akther S, Korshnova N, Zhong J, Liang M, Cherepanov SM, Lopatina O, Komleva YK, Salmina AB, Nishimura T, Fakhrul AA et al. . CD38 in the nucleus accumbens and oxytocin are related to paternal behavior in mice. Mol Brain 2013; 6: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tsuneoka Y, Tokita K, Yoshihara C, Amano T, Esposito G, Huang AJ, Yu LM, Odaka Y, Shinozuka K, McHugh TJ et al. . Distinct preoptic-BST nuclei dissociate paternal and infanticidal behavior in mice. EMBO J 2015; 34: 2652–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu Z, Autry AE, Bergan JF, Watabe-Uchida M, Dulac CG. Galanin neurons in the medial preoptic area govern parental behaviour. Nature 2014; 509: 325–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuroda KO, Numan M. The medial preoptic area and the regulation of parental behavior. Neurosci Bull 2014; 30: 863–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stolzenberg DS, Champagne FA. Hormonal and non-hormonal bases of maternal behavior: the role of experience and epigenetic mechanisms. Horm Behav 2016; 77: 204–10. [DOI] [PubMed] [Google Scholar]
- 39. Rilling JK, Young LJ. The biology of mammalian parenting and its effect on offspring social development. Science 2014; 345: 771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Insel TR, Preston S, Winslow JT. Mating in the monogamous male: behavioral consequences. Physiol Behav 1995; 57: 615–27. [DOI] [PubMed] [Google Scholar]
- 41. Williams JR, Catania KC, Carter CS. Development of partner preferences in female prairie voles (Microtus ochrogaster)—the role of social and sexual experience. Horm Behav 1992; 26: 339–49. [DOI] [PubMed] [Google Scholar]
- 42. Winslow JT, Hastings N, Carter CS, Harbaugh CR, Insel TR. A role for central vasopressin in pair bonding in monogamous prairie voles. Nature 1993; 365: 545–8. [DOI] [PubMed] [Google Scholar]
- 43. Young LJ, Wang Z. The neurobiology of pair bonding. Nat Neurosci 2004; 7: 1048–54. [DOI] [PubMed] [Google Scholar]
- 44. Oliveras D, Novak M. A comparison of paternal behaviour in the meadow vole Microtus pennsylvanicus, the pine voles, M. pinetorum and the prairie vole M. Ochrogaster. Anim Behav 1986; 34: 519–26. [Google Scholar]
- 45. Alexa A, Rahnenfuhrer J. topGo: enrichment analysis for gene ontology. R Package Version 2016; 2: 320. [Google Scholar]
- 46. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P et al. . The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res 2017; 45: D362–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tabbaa M, Lei K, Liu Y, Wang Z. Paternal deprivation affects social behaviors and neurochemical systems in the offspring of socially monogamous prairie voles. Neuroscience 2017; 343: 284–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pereira M. Structural and functional plasticity in the maternal brain circuitry. In: Rutherford HJV, Mayes LC (eds), Maternal brain plasticity: preclinical and human research and implications for intervention: new directions for child and adolescent development. 2016, 23–46. [DOI] [PubMed] [Google Scholar]
- 49. Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T et al. . Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 2008; 26: 317–25. [DOI] [PubMed] [Google Scholar]
- 50. Campbell JC, Laugero KD, Van Westerhuyzen JA, Hostetler CM, Cohen JD, Bales KL. Costs of pair-bonding and paternal care in male prairie voles (Microtus ochrogaster). Physiol Behav 2009; 98: 367–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chauke M, Malisch JL, Robinson C, de Jong TR, Saltzman W. Effects of reproductive status on behavioral and endocrine responses to acute stress in a biparental rodent, the California mouse (Peromyscus californicus). Horm Behav 2011; 60: 128–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kenkel WM, Paredes J, Yee JR, Pournajafi-Nazarloo H, Bales KL, Carter CS. Neuroendocrine and behavioural responses to exposure to an infant in male prairie voles. J Neuroendocrinol 2012; 24: 874–86. [DOI] [PubMed] [Google Scholar]
- 53. McGraw LA, Davis JK, Lowman JJ, ten Hallers BF, Koriabine M, Young LJ, de Jong PJ, Rudd MK, Thomas JW. Development of genomic resources for the prairie vole (Microtus ochrogaster): construction of a BAC library and vole-mouse comparative cytogenetic map. BMC Genomics 2010; 11:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McGraw LA, Davis JK, Young LJ, Thomas JW. A genetic linkage map and comparative mapping of the prairie vole (Microtus ochrogaster) genome. BMC Genet 2011; 12: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bendesky A, Kwon YM, Lassance JM, Lewarch CL, Yao S, Peterson BK, He MX, Dulac C, Hoekstra HE. The genetic basis of parental care evolution in monogamous mice. Nature 2017; 544: 434–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009; 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang ZH, Jhaveri DJ, Marshall VM, Bauer DC, Edson J, Narayanan RK, Robinson GJ, Lundberg AE, Bartlett PF, Wray NR et al. . A comparative study of techniques for differential expression analysis on RNA-Seq data. PLoS One 2014; 9: e103207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szczesniak MW, Gaffney DJ, Elo LL, Zhang X et al. . A survey of best practices for RNA-seq data analysis. Genome Biol 2016; 17: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kenkel WM, Suboc G, Carter CS. Autonomic, behavioral and neuroendocrine correlates of paternal behavior in male prairie voles. Physiol Behav 2014; 128:252–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang B, Li Y, Wu R, Zhang S, Tai F. Behavioral responses to pups in males with different reproductive experiences are associated with changes in central OT, TH and OTR, D1R, D2R mRNA expression in mandarin voles. Horm Behav 2015; 67:73–82. [DOI] [PubMed] [Google Scholar]
- 61. Glasper ER, Hyer MM, Katakam J, Harper R, Ameri C, Wolz T. Fatherhood contributes to increased hippocampal spine density and anxiety regulation in California mice. Brain Behav 2016; 6: e00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hyer MM, Hunter TJ, Katakam J, Wolz T, Glasper ER. Neurogenesis and anxiety-like behavior in male California mice during the mate’s postpartum period. Eur J Neurosci 2016; 43: 703–9. [DOI] [PubMed] [Google Scholar]
- 63. Kozorovitskiy Y, Hughes M, Lee K, Gould E. Fatherhood affects dendritic spines and vasopressin V1a receptors in the primate prefrontal cortex. Nat Neurosci 2006; 9: 1094–5. [DOI] [PubMed] [Google Scholar]
- 64. Lieberwirth C, Wang Y, Jia X, Liu Y, Wang Z. Fatherhood reduces the survival of adult-generated cells and affects various types of behavior in the prairie vole (Microtus ochrogaster). Eur J Neurosci 2013; 38: 3345–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Glasper ER, Kozorovitskiy Y, Pavlic A, Gould E. Paternal experience suppresses adult neurogenesis without altering hippocampal function in Peromyscus californicus. J Comp Neurol 2011; 519: 2271–81. [DOI] [PubMed] [Google Scholar]
- 66. Parent C, Wen X, Dhir SK, Ryan R, Diorio J, Zhang TY. Maternal care associates with differences in morphological complexity in the medial preoptic area. Behav Brain Res 2017; 326:22–32. [DOI] [PubMed] [Google Scholar]
- 67. Ghiretti AE, Paradis S. The GTPase Rem2 regulates synapse development and dendritic morphology. Dev Neurobiol 2011; 71: 374–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ghiretti AE, Moore AR, Brenner RG, Chen LF, West AE, Lau NC, Van Hooser SD, Paradis S. Rem2 is an activity-dependent negative regulator of dendritic complexity in vivo. J Neurosci 2014; 34: 392–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhu JJ, Qin Y, Zhao M, Van Aelst L, Malinow R. Ras and Rap control AMPA receptor trafficking during synaptic plasticity. Cell 2002; 110: 443–55. [DOI] [PubMed] [Google Scholar]
- 70. Cahill ME, Bagot RC, Gancarz AM, Walker DM, Sun H, Wang ZJ, Heller EA, Feng J, Kennedy PJ, Koo JW et al. . Bidirectional synaptic structural plasticity after chronic cocaine administration occurs through Rap1 small GTPase signaling. Neuron 2016; 89: 566–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shah S, Brock EJ, Ji K, Mattingly RR. Ras and Rap1: a tale of two GTPases. Semin Cancer Biol 2018; 10.1016/j.semcancer.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang L, Zhang P, Wang G, Zhang H, Zhang Y, Yu Y, Zhang M, Xiao J, Crespo P, Hell JW et al. . Ras and Rap signal bidirectional synaptic plasticity via distinct subcellular microdomains. Neuron 2018; 98: 783–800.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen Y, Dube CM, Rice CJ, Baram TZ. Rapid loss of dendritic spines after stress involves derangement of spine dynamics by corticotropin-releasing hormone. J Neurosci 2008; 28: 2903–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Radley JJ, Rocher AB, Miller M, Janssen WG, Liston C, Hof PR, McEwen BS, Morrison JH. Repeated stress induces dendritic spine loss in the rat medial prefrontal cortex. Cereb Cortex 2006; 16: 313–20. [DOI] [PubMed] [Google Scholar]
- 75. Leuner B, Shors TJ. Stress, anxiety, and dendritic spines: what are the connections? Neuroscience 2013; 251:108–19. [DOI] [PubMed] [Google Scholar]
- 76. Chen Y, Kramar EA, Chen LY, Babayan AH, Andres AL, Gall CM, Lynch G, Baram TZ. Impairment of synaptic plasticity by the stress mediator CRH involves selective destruction of thin dendritic spines via RhoA signaling. Mol Psychiatry 2013; 18: 485–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Liao XM, Yang XD, Jia J, Li JT, Xie XM, Su YA, Schmidt MV, Si TM, Wang XD. Blockade of corticotropin-releasing hormone receptor 1 attenuates early-life stress-induced synaptic abnormalities in the neonatal hippocampus. Hippocampus 2014; 24: 528–40. [DOI] [PubMed] [Google Scholar]
- 78. Matsuo S, Matsuda KI, Takanami K, Mori T, Tanaka M, Kawata M, Kitawaki J. Decrease in neuronal spine density in the postpartum period in the amygdala and bed nucleus of the stria terminalis in rat. Neurosci Lett 2017; 641:21–5. [DOI] [PubMed] [Google Scholar]
- 79. Kinsley CH, Trainer R, Stafisso-Sandoz G, Quadros P, Marcus LK, Hearon C, Meyer EA, Hester N, Morgan M, Kozub FJ et al. . Motherhood and the hormones of pregnancy modify concentrations of hippocampal neuronal dendritic spines. Horm Behav 2006; 49: 131–42. [DOI] [PubMed] [Google Scholar]
- 80. Pawluski JL, Charlier TD, Lieblich SE, Hammond GL, Galea LA. Reproductive experience alters corticosterone and CBG levels in the rat dam. Physiol Behav 2009; 96: 108–14. [DOI] [PubMed] [Google Scholar]
- 81. Storey AE, Walsh CJ, Quinton RL, Wynne-Edwards KE. Hormonal correlates of paternal responsiveness in new and expectant fathers. Evol Hum Behav 2000; 21: 79–95. [DOI] [PubMed] [Google Scholar]
- 82. Bartlett E. The effects of fatherhood on the health of men: a review of the literature. J Men’s Health Gender 2004; 1: 159–69. [Google Scholar]
- 83. Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature 2008; 454: 455–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Banhegyi G, Baumeister P, Benedetti A, Dong D, Fu Y, As Lee, Li J, Mao C, Margittai E, Ni M et al. . Endoplasmic reticulum stress. Ann N Y Acad Sci 2007; 1113:58–71. [DOI] [PubMed] [Google Scholar]
- 85. Mandl J, Meszaros T, Banhegyi G, Hunyady L, Csala M. Endoplasmic reticulum: nutrient sensor in physiology and pathology. Trends Endocrinol Metab 2009; 20: 194–201. [DOI] [PubMed] [Google Scholar]
- 86. Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 2011; 334: 1081–6. [DOI] [PubMed] [Google Scholar]
- 87. Zhang K, Kaufman RJ. The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology 2006; 66(2 Suppl 1): S102–S109. [DOI] [PubMed] [Google Scholar]
- 88. Champagne FA. Epigenetic legacy of parental experiences: dynamic and interactive pathways to inheritance. Dev Psychopathol 2016; 28: 1219–28. [DOI] [PubMed] [Google Scholar]
- 89. Braun K, Champagne FA. Paternal influences on offspring development: behavioural and epigenetic pathways. J Neuroendocrinol 2014; 26: 697–706. [DOI] [PubMed] [Google Scholar]