Abstract
AIM
To establish a permanent piwi like RNA-mediated gene silencing 1 (PIWIL1) gene knockout in AGP01 gastric cancer cell line using CRISPR-Cas9 system and analyze phenotypic modifications as well as gene expression alterations.
METHODS
CRISPR-Cas9 system used was purchased from Dharmacon GE Life Sciences (Lafayette, CO, United States) and permanent knockout was performed according to manufacturer’s recommendations. Wound-healing assay was performed to investigate the effect of PIWIL1 knockout on migration capability of cells and Boyden chamber invasion assay was performed to investigate the effect on invasion capability. For the gene expression analysis, a one-color microarray-based gene expression analysis kit (Agilent Technologies, Santa Clara, CA, United States) was used according to the protocol provided by the manufacturer.
RESULTS
PIWIL1 gene knockout caused a significant decrease in AGP01 migration capacity as well as a significant decrease in cell invasiveness. Moreover, functional analysis based on grouping of all differentially expressed mRNAs identified a total of 35 genes (5 up-regulated and 30 down-regulated) encoding proteins involved in cellular invasion and migration. According to current literature, 9 of these 35 genes (DOCK2, ZNF503, PDE4D, ABL1, ABL2, LPAR1, SMAD2, WASF3 and DACH1) are possibly related to the mechanisms used by PIWIL1 to promote carcinogenic effects related to migration and invasion, since their functions are consistent with the changes observed (being up- or down-regulated after knockout).
CONCLUSION
Taken together, these data reinforce the idea that PIWIL1 plays a crucial role in the signaling pathway of gastric cancer, regulating several genes involved in migration and invasion processes; therefore, its use as a therapeutic target may generate promising results in the treatment of gastric cancer.
Keywords: Gastric cancer, Piwi like RNA-mediated gene silencing 1, CRISPR-Cas9, Migration, Invasion
Core tip: Piwi like RNA-mediated gene silencing 1 (PIWIL1) gene emerged as an interesting target for gastric cancer, as it is expressed in cancer, stem and germ cells, but it is absent in normal somatic tissue. Our results propose that PIWIL1 plays a crucial role in the signaling pathway of gastric cancer, regulating several genes involved in migration and invasion processes; therefore, its use as a therapeutic target may generate promising results in the treatment of gastric cancer.
INTRODUCTION
Gastric cancer is a major contributor to global cancer burden, being the third leading cause of cancer death worldwide and in both sexes[1]. This type of cancer is thought to be consequence of a multi-step process, resulting from different genetic and epigenetic changes. Specifically, dysfunction of oncogenes and tumor suppressor genes contributes to this malignant disease, and many candidate genes have been implicated to serve as gastric cancer biomarkers[2].
In this context, the piwi like RNA-mediated gene silencing 1 (PIWIL1) gene, located in 12q24.33 and having 22 exons, became an attractive target for gastric cancer treatment. PIWIL1 protein is expressed at increased levels in cancer tissues, stem cells and germ cells, but it has been shown to be absent in normal somatic tissues. This means that it could be a potential target for therapy, since most non-cancer cells would not be affected by cytotoxic effects[3-7].
PIWIL1 plays a key role in tumor cell viability, migration and invasion, and its expression is associated with the maintenance of stem-like characteristics of tumors, which in turn contribute to more severe histological grade, advanced stage and worse clinical outcome[8-10].
Wang et al[11] showed that expression of PIWIL1 in gastric cancer tissue was significantly higher than in adjacent-to-tumor tissue (tumor front). They also demonstrated that patients with a lower expression of PIWIL1 presented a significant better overall survival rate compared to patients with a higher expression levels. Additionally, the 5-year survival rate of patients with a higher expression level of PIWIL1 was significantly lower (36.5% vs 67.6%).
Liu et al[12] reported that expression of PIWIL1 progressively increases during cancer development. The expression ratio in normal gastric tissues, atrophic gastritis, intestinal metaplasia and gastric cancers varied from 10% to 76%.
To further investigate the potential functions of the PIWIL1 gene, Liu et al[12] also silenced PIWIL1 by antisense or short hairpin RNA and noted that suppression of this gene inhibited the growth of gastric cancer cells and induced G2/M arrest. Although relevant information regarding the possible role of PIWIL1 in gastric cancer carcinogenesis is provided by the current literature, the exact molecular mechanisms involved in this carcinogenic process remain unclear.
A recently introduced technology, based on the adaptive immune system of prokaryotes and known as type II clustered, regularly interspaced, short palindromic repeats (CRISPR)/associated protein (Cas), has been demonstrated to cleave double-stranded DNA and has emerged as a relevant genome editing tool[13-15].
This technology can be used both to perform permanent gene knockouts and the site-specific integration of a gene (knock-in)[16-19]. Importantly, it allows for the permanent silencing of the target gene, and it also creates a stable and permanent cell line with the desired modification[14,16].
Here we applied CRISPR/Cas9 technology for the first time to knockout PIWIL1 gene in a gastric cancer cell line and analyzed its phenotypic modifications.
MATERIALS AND METHODS
Cell lines
The human gastric cancer cell line AGP01 was maintained in DMEM-F12 medium supplemented with 10% fetal bovine serum. The cell culture grew attached to a plastic flask in a monolayer in a humidified incubator maintained at 37 °C and 5% CO2.
The AGP01 cell line was established by our research group in 2009[20] from cancer cells present in the ascitic fluid of a female individual with intestinal gastric cancer, located at the antrum and the body region of the stomach, and staged as T3N2M1. The cell line was tested and authenticated by conventional cytogenetics[20]. Recently, the AGP01 cell line was tested by multicolor-fluorescence in situ hybridization (FISH), and results are presented here.
24-color-FISH using all human whole chromosome painting probes
24-color-FISH using simultaneous all human whole chromosome painting (WCP) probes was done as previously reported[21,22]. A total of 20 metaphases was analyzed, using a fluorescence microscope (Axio Imager Z1 mot; Carl Zeiss AG, Oberkochen, Germany) equipped with appropriate filter sets to discriminate between a maximum of five fluorochromes and the counterstain DAPI; the latter was used to induce a GTG-like banding pattern. Image capturing and processing were carried out using ISIS imaging system (MetaSystems, Altlussheim, Germany).
Targeted knockout of PIWIL1 using the CRISPR-Cas9 system
The CRISPR-Cas9 system used was purchased from Dharmacon GE Life Sciences (Lafayette, CO, United States). First, 1 × 104 AGP01 cells/well were seeded in DMEM-F12 medium to a 96-well plate for 24 h. Subsequently, transfection was performed using CR-0046-03-005 (Dharmacon GE Life Sciences) for 48 h. For the transfection procedure, a solution containing 1 μL of the CRISPR RNAs (crRNAs) mixed with the trans-activating small RNA, 2 μL of Cas9 and 7 μL of DMEM-F12 medium (for each well) was prepared first. In another tube, 0.4 μL of DharmafecDUO and 9.6 μL of DMEM-F12 medium (to each well) were added.
The solutions were then incubated for 5 min at room temperature before being combined. After combining, the solution was incubated for 20 min at room temperature, and finally 80 μL of DMEM-F12 medium/10% fetal calf serum (FCS) (per well) was added. At the end of the transfection, all contents (from each well) were transferred to a 24-well plate containing DMEM-F12 medium/10% FCS/1% pen-strep.
After 24 h, samples were treated with 6 μg/mL of puromycin for 72 h to select the resistant clones. Next, 40 cells were plated per well in a 6-well plate to isolate the clones by the filter paper method.
This method consists of using a cut and autoclaved piece of filter paper so that it, after being soaked in trypsin, can be positioned above a single colony of cells that has grown from an isolated cell, allowing for the collection of this clone. Subsequently, each clone grew in a separate well in a 6-well plate, so it could reach the confluence needed to perform DNA extraction and sequencing.
Sequencing
DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega Corporation, Madison, WI, United States) according to the manufacturer’s instructions. For PCR, specific primers targeting the binding region of the purchased crRNA were constructed using the online program Primer3 (Supplementary Table 1).
The quantities of the reagents used in the PCR for a final volume of 12 μL were as follows: 6.25 μL of nuclease-free H2O, 0.5 μL of forward primer (10 ng/μL), 0.5 μL of reverse primer (ng/μL), 4.25 μL of GoTaq Colorless Master Mix 2× (Promega Corporation) and 1 μL of DNA (10 ng/μL).
The conditions using the MasterCycler Gradient thermal cycler (Eppendorf, Hamburg, Germany) were 1 cycle at 95 °C for 3 min for initial denaturation followed by 35 cycles consisting of denaturation at 94 °C for 2 min, primer annealing at 59 °C for 1 min and extension at 70 °C for 2 min, ending with 1 cycle at 70 °C for a final extension for 30 min.
For direct sequencing of the PCR product, the quantities of the reagents used for a final volume of 20 μL were as follows: 15 μL of nuclease-free water, 0.5 μL of forward or reverse primer (10 ng/μL), 0.5 μL of Big Dye, 3 μL of Save Money and 1 μL of the PCR reaction. For this reaction, the ABI PRISM Big Dye Terminator 3.1 Cycle Sequencing Kit (Applied Biosystems, Hercules, CA, United States) was used.
The sequencing was performed using the MasterCycler Gradient (Eppendorf) thermal cycler according to the following 25-cycle thermocycling conditions: denaturation at 96 °C for 50 s, primer annealing at 59 °C for 1 min and extension at 60 °C for 4 min, ending with 1 cycle at 4 °C for 5 min.
After this procedure, a precipitation step was carried out in order to purify the product of the reaction before continuing. For this step, samples were washed with 70% isopropanol and 70% ethanol. Subsequently, sequencing was performed using the ABI Prism 3500 DNA Sequencer (Applied Biosystems). The methodology used was based on the biochemical synthesis of the DNA strand by the Sanger method.
Sequencing analysis
Reference sequence for exon 15 in the PIWIL1 gene was obtained from the National Center for Biotechnology Information (NCBI) and compared with the DNA sequence of the modified cell line as well as the negative control (cell line without the gene knockout). To infer the effect of changes in protein synthesis, we applied Gene Runner v.3.05 (Hastings Software Inc., Hastings, NY, United States; http://www.generunner.com).
Wound-healing assay
Cells were grown in 12-well plates at a density of 2 × 105 cells/well and maintained for 24 h in 5% CO2 at 37 °C. After this period, cells were injured with a 10 μL tip in the center of each well. The medium was then removed to eliminate suspended cells, and wells were washed with 1 × phosphate buffered saline before fresh DMEM-F12 medium/10% FCS/1% pen-strep was added again.
The behavior of cells was observed and photographed immediately after injury and at 6 h, 12 h and 24 h after injury. All experiments were performed in triplicate.
Boyden chamber invasion assay
Boyden inserts (8 μm pores) (BD BiosciencesTM, Franklin Lakes, NJ, United States) were coated with 200 μL of Matrigel (10-13 mg/mL) in 12-well plates. Cells (2 × 105) were seeded in the upper chamber in 1 mL of DMEM without fetal bovine serum. In the lower chamber, DMEM-F12 medium/10% FCS/1% pen-strep was added, functioning as a chemoattractant for the cells present in the upper chamber.
After 48 h, the remaining cells above the filter were removed by scraping with a sterile swab. The cells at the bottom of the filter were fixed with 4% paraformaldehyde and stained with Giemsa. Cells were photographed and analyzed using a light microscope and counted in optical fields (100 ×). All experiments were performed in triplicate.
Total RNA extraction
The mRNA extraction was performed using Promega’s Total RNA Isolation System kit, according to the manufacturer’s specifications. AGP01 and AGP01 PIWIL1 knockout cells were prepared for mRNA extraction. Samples were lysed with lysis buffer containing beta-mercaptoethanol and then diluted in RNA dilution buffer. The samples were centrifuged for 10 min at maximum speed. Then, 95% ethanol was added to ensure adequate membrane binding conditions.
The samples were then transferred to centrifuge columns where the RNA could bind the membrane of the column, facilitating washing to eliminate possible contaminants as well as favoring the extraction of high quality of total RNA.
At the end of the procedure, the RNA was diluted in 60 μL of nuclease-free water. The total RNA was quantified using a Nanodrop spectrophotometer ND-1000 UV-VIS version 3.2.1 (Nanodrop Technologies, Wilmington, DE, United States). The RNA quality was also evaluated by analyzing the A260/A280 ratio according to the manufacturer’s specifications. Purified RNA was stored at -80 °C for the microarray expression assay.
Microarray expression
For the microarray assay, a one-color microarray-based gene expression analysis kit (Agilent Technologies, Santa Clara, CA, United States) was used according to the protocol provided by the manufacturer. The gene expression profile was evaluated in both cell lines (AGP01 with and without PIWIL1 knockout).
The total RNA obtained during the extraction phase was used as the template for the synthesis of the first cDNA strand by reverse transcription using T7 RNA polymerase. Synthesis of the second cDNA strand was used as the template for the in vitro transcription reaction for cRNA production. The cRNA was then incorporated into the fluorochrome 3-cyanine (Cy-3) using the Low Input Quick Amp Labeling kit (Agilent Technologies) according to the protocol provided by the manufacturer. Thereafter, the cRNA purification process was carried out.
The cRNA was quantified by a spectrophotometer (pmol/L), by which it was possible to analyze the absorbance ratio (260 nm/280 nm) and the cRNA (ng/μL) concentration in each sample. After, hybridization was performed for 17 h in a hybridization chamber at 65 °C at 10 rpm. After this period, the slide was washed and immediately scanned in the Agilent G4900DA SureScan Microarray System.
The following setup was used to scan the microarray slides for one color: scan region of 61 mm × 21.6 mm, 5 μm scan resolution, dye channel of green. Next, the images were obtained by using Feature Extraction v10.10 software, and the data were analyzed with GeneSpring GX 9.0 and IPathwayguide (Advaita Bioinformatics Company, Plymouth, MI, United States) programs. Gene identification followed a restriction criterion with a fold-change of > 2.
Differential expression and gene ontology enrichment analysis
To identify differentially expressed (DE) mRNAs, we compared the probes’ expression profiles before and after PIWIL1 knockout. Probes with a mean fold-change < 0.5 and a mean fold-change of > 2 [|Log2(fold-change)| > 1] were selected for differential analysis. Student’s t test was performed in 1222 selected probes, false discovery rate adjustment[23] was performed, and genes with an adjusted P-value of < 0.05 were tagged as DE. For the volcano plot, the fold-change and the P-value of all probes were used, and probes were tagged as DE following the previous criteria. Gene ontology enrichment was performed using org.Hs.eg.db[24] and Gostats[25] R libraries. All graphical and statistical analyses were performed in the R platform (R Core Team, 2017, Vienna, Austria; https://www.R-project.org/).
RESULTS
24-color-FISH using all human WCP probes
24-color-FISH using all human WCP probes revealed in AGP01 cell line a complex karyotype as follows: 63,XX,inv(1)(p12q43),der(1)(1pter->1p12::1q43->1p12::9p12->9pter),+der(1)t(1;12)(q21;q12),+del(1)(p12),+del(2)(p12),+der(2)t(2;8)(p12;q11.2),inv(3)(p21q13)x2,+inv(3)(p21q13),+der(3)t(3;5)(p14;q13),t(4;14)(p12;q11.2),dic(4;12)(p15;q12),+der(4)t(2;4)(p or q?;q12),del(5)(p13)x2,+der(5)x2,del(6)(q12)x2,+del(6)(p21),inv(7)(p12;q11.2),der(7),der(9)t(9;acro)(p21;p12),der(9)t(6;9)(p12;p12),t(13;13)(p10),der(15)(:q13->p11::p11->qter),-15,der(16)t(X;16)(q or p?;q23),+der(16)t(12;16)(q12;q23),21p+,t(22;22)(p10) (Figure 1).
Figure 1.
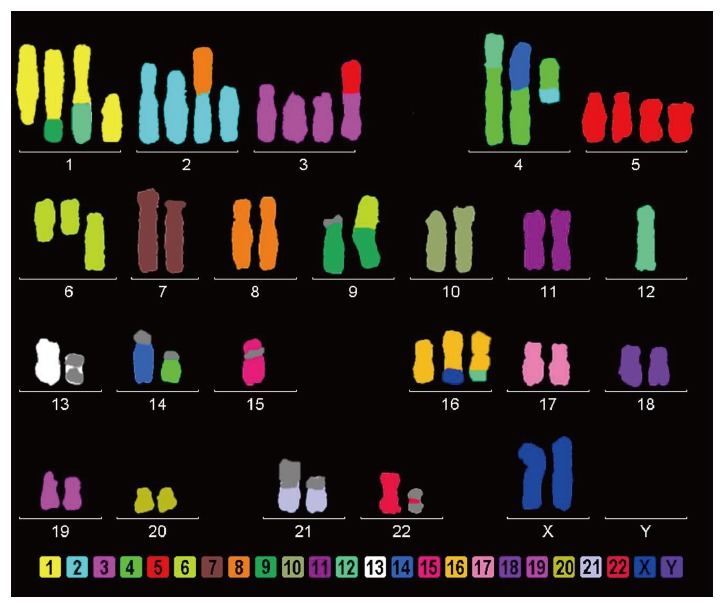
AGP01 cell line multicolor-FISH showing several chromosomal changes, including the monosomy of chromosome 12, where the PIWIL1 gene is located.
From the analysis, we verified that the unique chromosome 12 (where PIWIL1 gene is located) remains intact, without translocations or derivative chromosomes. It is important to note that the monosomy of chromosome 12 agrees with the sequencing result, since the 7 bp insert sequence was observed in hemizygous status.
Targeted knockout of the PIWIL1 gene using the CRISPR-Cas9 system
PIWIL1 gene knockout was successful, as determined by Sanger sequencing. The latter revealed an insertion of 7 adenines in the PIWIL1 gene sequence (Figure 2), which caused a frameshift mutation that impaired protein synthesis.
Figure 2.
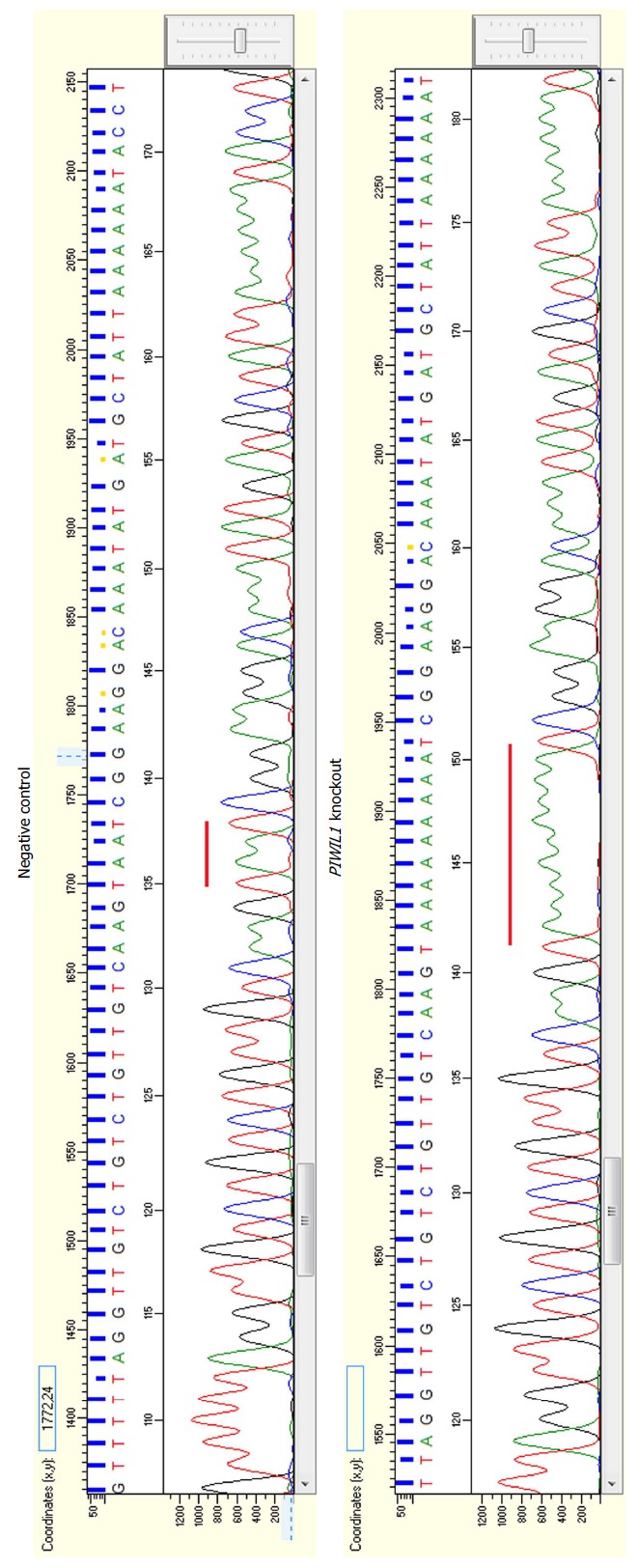
Electropherograms showing the insertion of seven adenines in the PIWIL1 gene sequence after using the CRISPR-Cas9 system (B) in comparison with the reference sequence of the negative control (A).
Prediction of the encoded protein indicated a premature stop codon (Figure 3), suggesting that this insertion generates a truncated protein consisting of 573 amino acids (the wild-type contains 861) with a loss-of-function phenotype, which means that knockout was efficient.
Figure 3.

Amino acid sequences of the wild-type PIWIL1 protein and the PIWIL1 protein after the insertion of seven adenines by the CRISPR-Cas9 system.
Notably, the PIWIL1 knockout cell line remained viable and could be used for further experiments.
Wound-healing assay
The PIWIL1 gene knockout caused a significant decrease in AGP01 migration capacity after 24 h (P < 0.01; Figure 4), which is consistent with the fact that this protein is related to various pathways that regulate cell motility.
Figure 4.
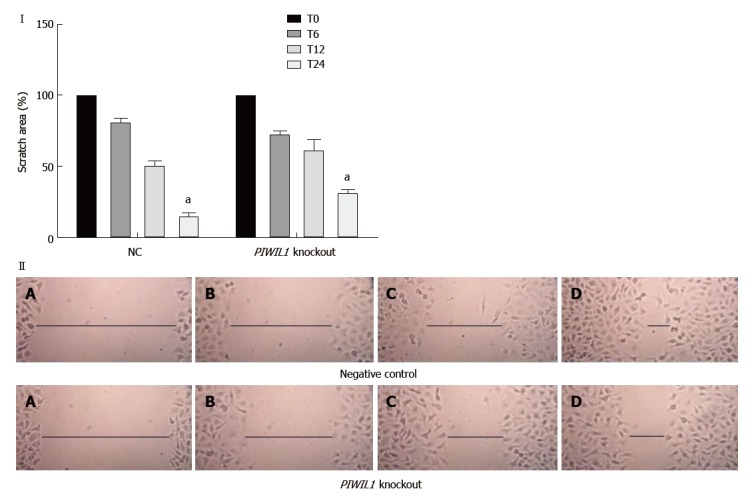
Analysis of the migration capacity of the AGP01 cell line with and without PIWIL1 gene knockout. T0: Immediately after injury; T6: 6 h after injury; T12: 12 h after injury; T24: 24 h after injury. NC: Negative control. aP < 0.01. Two-way ANOVA, Bonferroni post-test. Photomicrography of AGP01 cell migration. A: Immediately after injury; B: 6 h after injury; C: 12 h after injury; D: 24 h after injury. The black lines represent approximation of the edges over time, demonstrating the migration capacity of the cells.
Boyden chamber invasion assay
Also, PIWIL1 gene knockout caused a significant decrease in AGP01 invasiveness (P < 0.001; Figure 5), which is also consistent with the fact that this protein is related to various pathways that regulate cell motility.
Figure 5.
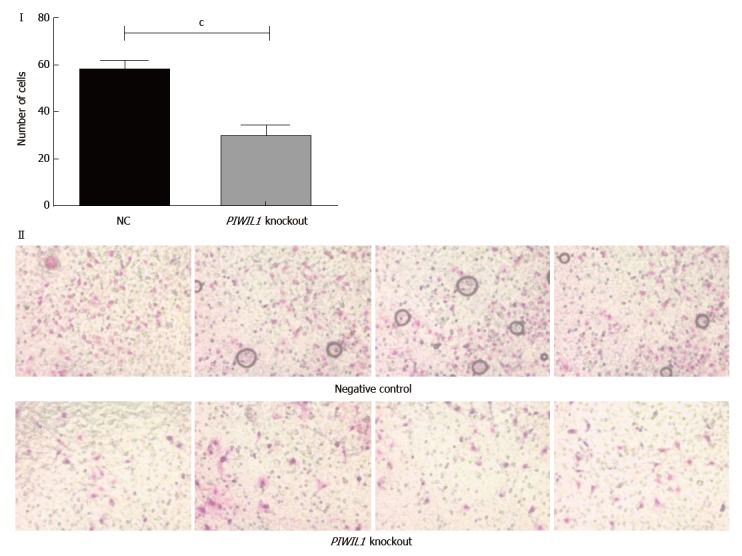
Analysis of the invasion capacity of the AGP01 cell line with and without the PIWIL1 gene knockout. Statistically significant difference between groups was shown by the Student’s t test (cP < 0.001). Photomicrography of the cell invasion assay demonstrating the decrease in the number of cells that invaded when PIWIL1 was knocked out. NC: Negative control.
mRNA array and gene ontology enrichment
Differential analysis: By comparing expression profiles after PIWIL1 permanent knockout in the AGP01 cell line, a total of 251 mRNA were found to be DE [adjusted P-value of < 0.05 and |Log2(Fold-Change)| > 1]. A total of 43 mRNA probes were up-regulated and 208 were down-regulated (Figure 6). The DE genes are described in Supplementary Table 2.
Figure 6.
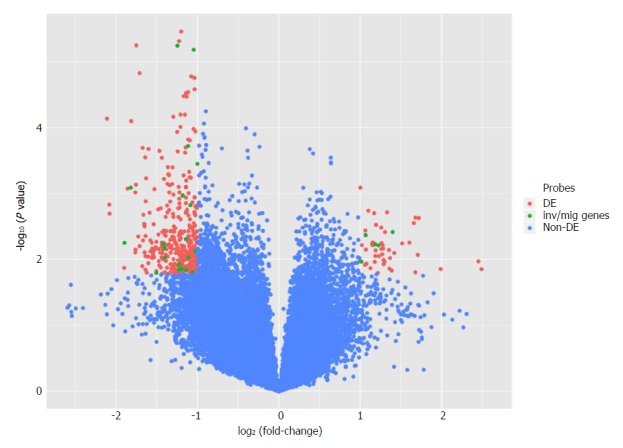
Volcano plot comparing gene expression after PIWIL1 permanent knockout in the gastric cancer AGP01 cell line. Differentially expressed probes [adjusted P-value of < 0.05 and |Log2(Fold-Change)| > 1] are on superior left and right areas (red). mRNAs involved in invasion and migration processes are in green.
We performed a functional analysis by grouping all DE mRNAs. This approach revealed that a total of 35 genes (five up-regulated and 30 down-regulated) encoded proteins involved in invasion and migration cellular processes (Supplementary Figure 1; Supplementary Tables 3 and 4).
According to the current literature, 9 of these 35 genes (DOCK2, ZNF503, PDE4D, ABL1, ABL2, LPAR1, SMAD2, WASF3 and DACH1) are possibly related to the mechanisms used by PIWIL1 to promote carcinogenic effects related to migration and invasion, since their functions are consistent with the changes observed (being up- or down-regulated after knockout).
DISCUSSION
Evidence indicating that PIWIL1 is an oncogene that regulates several cellular mechanisms important for the carcinogenic process of many types of cancers is rising in recent years. PIWIL1 seems to be especially implicated in proliferating activity of cancer cells[8,10,26,27].
Here, we introduce a new gastric cell line, i.e., AGP01, and provide a first description of its karyotype. As is usual for cancer cell lines, there were slight changes from cell to cell concerning number of chromosomes or single cell chromosomal rearrangements[28]. The here-given karyotype was the most frequently observed one. Interestingly, inversions, dicentrics and reciprocal translocations of homologous acrocentric (#13, #15, #22) was observed. Overall, gain of the following regions was present: #1, #3, large parts of #2 and #5, 8q11.2 to 8qter, 12pter to 12q12. Besides, the following regions were under-represented: 2pter to 2p12, 9pter to 9p12, 15q12 to 15qter. These imbalances are in concordance with the literature[29].
In AGP01, we performed for the first time an in vitro knockout experiment of PIWIL1 gene using the CRISPR-Cas9 system. It could be shown that absence of this gene significantly impairs migration and invasion capacity of AGP01 cells. Thus, the AGP01 cell line behaves like that previously reported for gastric cancer cells[12] or lung adenocarcinoma[26]. Together, these studies suggest that PIWIL1 expression is strongly associated with an increased aggressiveness of cancer cells.
According to Wang et al[30], one of the mechanisms by which PIWIL1 regulates the migration and invasion of cancer cells is by promoting the expression of MMP2 and MMP9, two important metalloproteinases involved in the degradation of the extracellular matrix, thereby creating paths for the locomotion of cancer cells[31].
Additionally, Amaar et al[32] demonstrated that the over-expression of PIWIL1 down-regulates the tumor suppressor gene IGFBP5, a member of the insulin-like growth factor binding protein family and whose expression is implicated in suppressing epithelial-mesenchymal transition and reducing the expression of E-cadherin and HIF1α, indicating that is it critically related to cancer progression[33,34].
Data obtained from our gene expression experiments also provided corroborating evidence that the PIWIL1 gene plays a key role in cancer cell migration and invasion because several genes involved in these cellular processes were observed as DE when the cell lines were compared before and after PIWIL1 knockout.
Many studies have demonstrated the oncogenic activities of the DOCK2, ZNF503, PDE4D, ABL1, ABL2, LPAR1, SMAD2 and WASF3 genes and their relation to tumor aggressiveness in several types of cancer, including gastric cancer[35-58]. Interestingly, PIWIL1 knockout led to a decreased expression of these genes as well as an increased expression of the tumor suppressor gene DACH1, demonstrating that PIWIL1 plays a crucial role in the pathway of development and progression of gastric cancer, and is likely a promising candidate for therapeutic intervention.
Zhu et al[53] reported that the over-expression of DACH1 impaired the proliferation and invasion ability of lung adenocarcinoma cells in vitro via the down-regulation of PRX3, an oncoprotein required for the maintenance of mitochondrial function and tumorigenesis[59]. DACH1 expression also inhibited epithelial-mesenchymal transition and metastasis by affecting TGF-β signaling and decreased proliferation of cancer cells by inducing cell cycle arrest at the G2/M phase[60,61].
Regarding the reported oncogenes, Rahrmann et al[37] observed that PDE4D is over-expressed in human prostate cancer and demonstrated that the knockdown of this gene reduced the growth and migration of prostate cancer cells in vitro as well as the growth and proliferation rate of prostate cancer xenografts in vivo[38]. Delyon et al[39] demonstrated that PDE4D is also over-expressed in melanoma cell lines and pinpointed this gene as a regulator of cell invasion by interacting with FAK through RACK1, constituting a signaling pathway that when activated promotes tumor progression and metastasis[43].
Recent studies have determined the role of ABL members from the tyrosine kinase family, ABL1 and ABL2, in the development of many types of solid tumors. These proteins induce the activation of actin polymerization machinery by modulating the expression of several MMPs to promote morphological changes, including the formation of membrane protrusions and altered cell adhesion. Consequently, activation of ABL1 and ABL2 in cancer cells promote enhanced proliferation, migration and invasion, as well as drug resistance[39-42].
DOCK2 (dedicator of cytokinesis) belongs to the DOCK family of proteins and is expressed in hematopoietic cells[44]. According to Kulkarni et al[45], DOCK2 has been reported to activate Rac, which is known to regulate several crucial processes, including lymphocyte migration, activation and differentiation of T cells[46]. Wang et al[47] knocked out DOCK2 in a B-cell lymphoma cell line and observed a decrease in Rac1 expression. Additionally, analysis of the growth curves of both cell lines demonstrated that the DOCK2 knockout grew less than DOCK2, as evidenced by the lower cell proliferation.
ZNF503 is expressed in the mammary gland and other tissues, and there is a high incidence of association between this gene deregulation and tumor aggressiveness in several kinds of tissues, such as lung, kidney and intestine[49]. Shahi et al[48] performed scratch and 3D Matrigel culture assays in two mammary epithelial cell lines to analyze the cell motility and migration. Both assays demonstrated that cell lines with ZNF503 knockout migrated less, did not close the gaps, and inhibited invasiveness when compared to the control cells. These data indicate that ZNF503 promotes cellular invasion and migration, and high levels of this gene are closely related with poor patient survival, breast cancer progression and increased metastasis.
Yu et al[50] demonstrated that the lysophosphatidic acid receptor 1 (LPAR1) gene is related to migration and invasion in ascites from ovarian cancer and is expressed at higher levels in metastatic cell lines, when compared to non-metastatic cell lines. They also observed that the presence of high levels of lysophosphatidic acid are directly connected to cell migration stimulation, and LPAR1 silencing reduced lysophosphatidic acid-induced invasion. Additionally, in breast tumors, a higher expression of LPAR1 is related to a worse lung metastasis-free survival rate[51].
Wiskott-Aldrich syndrome protein family 3 (WASF3) is an important gene, which has C-terminal domains that are responsible for actin polymerization activation, playing a role in cell proliferation and migration. The WASF3 gene is normally over-expressed in several types of tumors, such as breast cancer, osteosarcoma and prostate cancer[52,53]. In gastric cancer, little is known about this gene; however, since micro (mi)RNAs and their targets are considered potential biomarkers for gastric cancer, Wang et al[54] performed a luciferase assay and western blotting to investigate the relationship between miR-218 and WASF3. Their results demonstrated that over-expression of WASF3 harms miR-218 and results in the inhibition of cell proliferation and migration, suggesting that WASF3 is over-expressed in gastric cancer and induces cell proliferation and migration. Additionally, in qRT-PCR, WASF3 mRNA expression levels were higher when compared to normal gastric cell lines.
Smad2 is the first intracellular protein in the signaling cascade of the TGF-β1 signaling pathway, which is involved in the progression of gastric cancer. In advanced stages of cancer, TGF-β1 acts as an oncogene, regulating multiple cellular functions, including stimulation of proliferation, differentiation and the inhibition of apoptosis[55,57].
Interestingly, Lv et al[56] observed that TGF-β1 levels in peritoneal lavage fluid are directly connected to peritoneal metastasis. Corroborating evidence was provided by Shinto et al[58], whose experiments demonstrated that p-Smad2 expression was higher in diffuse-type tumors and in peritoneal metastasis cases. Notably, the AGP01 cell line used to perform the PIWIL1 knockout in our study was obtained from a patient with peritoneal metastasis, and we found SMAD2 was over-expressed.
Taken together, these data reinforce the idea that PIWIL1 plays a crucial role in the signaling pathway of gastric cancer, regulating several genes involved in migration and invasion processes; therefore, its use as a therapeutic target may generate promising results in the treatment of gastric cancer, mainly in patients with peritoneal carcinomatosis, which is a condition associated with poor prognosis and a decreased overall survival.
ARTICLE HIGHLIGHTS
Research background
Gastric cancer (GC) remains a major public health problem, having the third highest incidence of death worldwide. Piwi like RNA-mediated gene silencing 1 (PIWIL1) is involved in regulation of widespread biological processes, including stem cell proliferation, embryogenesis, growth, and development, and has been found to be frequently over-expressed in various tumor types, including GC. Previous studies have demonstrated that PIWIL1 is implicated in improving tumor malignant behavior. PIWIL1 expression has been shown to be absent in normal somatic tissues, making it a very intriguing target for therapy. We attempted to investigate the role of PIWIL1 on the migration and invasion capacity of metastatic GC cells, using the AGP01 cell line, as well as checking the expression status of genes and proteins involved in these cellular processes, in order to elucidate the mechanisms by which PIWIL1 provokes tumorigenic effects and to shed light on potential new strategies to target PIWIL1
Research motivation
Many aspects of gastric carcinogenesis remain elusive, and much effort has been made to improve patient prognosis. The PIWIL1 has been identified as a novel extremely highly expressed gene in many types of cancer and its expression in GC tissue is related to poorer overall survival, suggesting that high expression of PIWIL1 is associated with poor prognosis and that it could be used as a predictive marker or even a target for therapy. Although PIWIL1 has been correlated with worse outcome, the involved mechanisms remain unclear, and many hypotheses are being tested. Once the upstream and downstream signaling pathways of PIWIL1 are elucidated, it will be possible to create new therapeutic strategies for gastric carcinogenesis, in order to improve the overall health of patients affected by this disease.
Research objectives
We performed permanent knockout of the PIWL1 gene to verify phenotypic modifications in the AGP01 metastatic GC cell line, as well as alterations in expression level of mRNA and protein, in an attempt to better understand the mechanisms by which PIWIL1 promotes tumor malignant behavior. This research demonstrates the importance of studying PIWIL1 in GC, since data obtained through the achievement of our objectives showed that this protein has a crucial role in gastric carcinogenesis, promoting molecular and phenotypic alterations compatible with enhanced tumor aggressiveness. The elucidation of the role of PIWIL1 protein in cancer cell invasion and migration will pave the way for developing potential clinical interventions, aiming to control GC dissemination.
Research methods
We applied CRISPR/Cas9 technology to knockout the PIWIL1 gene in a metastatic GC cell line, and analyzed its phenotypic modifications, as well as alterations in gene and protein expression. CRISPR-Cas9 technology was considered in 2015 as one of the most important technological advances of science. Mainly, it allows permanent silencing of the target gene and also creates a stable and permanent cell line with the desired modification. By this way, multiple experiments can be carried on, including long term evaluation of the downstream events caused by the molecular alteration, as well as discovering potential pathways influenced by the studied gene. Therefore, after permanent knockout of PIWIL1 in the AGP01 cell line, we analyzed phenotypic modifications by performing wound-healing and Boyden chamber invasion assays, to assess migration and invasion, respectively. Moreover, aiming to shed light on the molecular mechanisms used by PIWIL1 to make changes in the migration and invasion capability of cells, we carried out proteomic and microarray assays, using multidimensional protein identification technology (commonly known as MudPIT) and a one-color microarray-based gene expression analysis kit, respectively.
Research results
PIWIL1 gene knockout was successfully performed and confirmed by Sanger sequencing, which revealed an insertion of seven adenines in the PIWIL1 gene sequence. In silico prediction of the encoded protein pointed to the appearance of a premature termination codon, suggesting that this insertion generates a truncated protein with a loss-of-function phenotype. PIWIL1 knockout promoted a significant decrease in cell migration and invasion capacity (P < 0.01 and P < 0.001, respectively), which is consistent with data present in the literature demonstrating that this protein is implicated in several signaling pathways that regulate cell motility. By comparing expression profiles after PIWIL1 knockout, a total of 251 mRNA were found to be differentially expressed, with 43 up-regulated and 208 down-regulated mRNA. A functional analysis grouping all differentially expressed mRNAs demonstrated that 35 genes encoded proteins were involved in invasion and migration cellular processes. After extensive review of data presented in the literature, we selected 9 of these 35 genes (DOCK2, ZNF503, PDE4D, ABL1, ABL2, LPAR1, SMAD2, WASF3 and DACH1) as possibly related to the mechanisms used by PIWIL1 to promote carcinogenic effects related to migration and invasion, since their functions are consistent with the changes observed (being up- or down-regulated after knockout). Additionally, the analysis of proteomic data revealed that PIWIL1 knockout caused modification in the expression of 27 proteins involved in epithelial-mesenchymal transition (EMT). Twenty-two oncoproteins related to EMT promotion, including FGFR1, PCNA, ACTN4, GSN and TUBB3, were expressed in the AGP01 cell line before knockout and reduced to a level that were not detectable by the technique after knockout. On the other hand, PIWIL1 knockout caused an increase in the expression of six proteins implicated in EMT suppression, such as ACSM3, ADGRG1 and ANPEP, that were absent in AGP01 before knockout. To the best of our knowledge, this is the first report describing molecular alteration compatible with phenotypic alterations after permanent knockout of PIWL1 in GC. Detailed mechanisms leading to PIWIL1 over-expression in cancer as well as the pathways by which this protein improves the malignant phenotype should be further investigated.
Research conclusions
In the current study, we pioneered the performance of an in vitro knockout of the PIWIL1 gene by using the CRISPR-Cas9 system, and found that absence of this gene significantly impaired the migration and invasion capacity of the AGP01 cell line, besides modifying mRNA and protein expression of potential molecular targets involved in the EMT process. The results of such experiments contributed to understanding of the mechanisms used by PIWIL1 to promote alteration in migration and invasion capacity of gastric cells during tumorigenesis, and also revealed the participation of new players related to PIWIL1 expression, such as FGFR1, PCNA, ACTN4, PDE4D and SMAD2. Our results demonstrated that knockout of PIWIL1 promotes several changes in cell phenotype, suggesting the critical role of the PIWIL1 oncogene in GC, and confirmed the hypothesis that PIWIL1 expression provokes migration, invasiveness and EMT as potential mechanisms of improved tumor aggressiveness. The presented findings open new perspectives for molecular interventions in GC
Research perspectives
Definite silencing of PIWIL1 by the CRISPR-Cas9 system resulted in robust findings favoring the discovery of new mechanisms involved in gastric carcinogenesis. The presented results must be validated by other researchers, and if confirmed, might lead to innovative interventions aiming to treat GC.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Brazil
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
Conflict-of-interest statement: The authors declare that they have no conflicts of interest.
Data sharing statement: No additional data are available.
Peer-review started: August 14, 2018
First decision: August 31, 2018
Article in press: October 5, 2018
P- Reviewer: Chen Z, Kimura A, Matowicka-Karna J, Park WS S- Editor: Wang XJ L- Editor: Filipodia E- Editor: Huang Y
Contributor Information
Taíssa Araújo, Núcleo de Pesquisas em Oncologia, Universidade Federal do Pará, Belém 66073-000, Brazil.
André Khayat, Núcleo de Pesquisas em Oncologia, Universidade Federal do Pará, Belém 66073-000, Brazil.
Luciana Quintana, Núcleo de Pesquisas em Oncologia, Universidade Federal do Pará, Belém 66073-000, Brazil.
Danielle Calcagno, Núcleo de Pesquisas em Oncologia, Universidade Federal do Pará, Belém 66073-000, Brazil.
Ronald Mourão, Núcleo de Pesquisas em Oncologia, Universidade Federal do Pará, Belém 66073-000, Brazil.
Antônio Modesto, Núcleo de Pesquisas em Oncologia, Universidade Federal do Pará, Belém 66073-000, Brazil.
Juliana Paiva, Núcleo de Pesquisas em Oncologia, Universidade Federal do Pará, Belém 66073-000, Brazil.
Adhara Lima, Núcleo de Pesquisas em Oncologia, Universidade Federal do Pará, Belém 66073-000, Brazil.
Fabiano Moreira, Núcleo de Pesquisas em Oncologia, Universidade Federal do Pará, Belém 66073-000, Brazil.
Edivaldo Oliveira, Laboratório de Cultura de Tecidos e Citogenética, Instituto Evandro Chagas, Belém 66087-082, Brazil.
Michel Souza, Laboratório de Cultura de Tecidos e Citogenética, Instituto Evandro Chagas, Belém 66087-082, Brazil.
Moneeb Othman, Institute of Human Genetics, Universitätsklinikum Jena, Jena 07747, Germany.
Thomas Liehr, Institute of Human Genetics, Universitätsklinikum Jena, Jena 07747, Germany.
Eliana Abdelhay, Laboratório de Célula Tronco, Centro de Transplante de Medula Óssea, Instituto Nacional de Câncer José Alencar Gomes da Silva, Rio de Janeiro 20230-130, Brazil.
Renata Gomes, Laboratório de Célula Tronco, Centro de Transplante de Medula Óssea, Instituto Nacional de Câncer José Alencar Gomes da Silva, Rio de Janeiro 20230-130, Brazil.
Sidney Santos, Núcleo de Pesquisas em Oncologia, Universidade Federal do Pará, Belém 66073-000, Brazil.
Paulo Assumpção, Núcleo de Pesquisas em Oncologia, Universidade Federal do Pará, Belém 66073-000, Brazil. assumpcaopp@gmail.com.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Wu HH, Lin WC, Tsai KW. Advances in molecular biomarkers for gastric cancer: miRNAs as emerging novel cancer markers. Expert Rev Mol Med. 2014;16:e1. doi: 10.1017/erm.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Assumpção CB, Calcagno DQ, Araújo TM, Santos SE, Santos ÂK, Riggins GJ, Burbano RR, Assumpção PP. The role of piRNA and its potential clinical implications in cancer. Epigenomics. 2015;7:975–984. doi: 10.2217/epi.15.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navarro A, Tejero R, Viñolas N, Cordeiro A, Marrades RM, Fuster D, Caritg O, Moises J, Muñoz C, Molins L, et al. The significance of PIWI family expression in human lung embryogenesis and non-small cell lung cancer. Oncotarget. 2015;6:31544–31556. doi: 10.18632/oncotarget.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan Y, Liu L, Liao M, Zhang C, Hu S, Zou M, Gu M, Li X. Emerging roles for PIWI proteins in cancer. Acta Biochim Biophys Sin (Shanghai) 2015;47:315–324. doi: 10.1093/abbs/gmv018. [DOI] [PubMed] [Google Scholar]
- 6.Siddiqi S, Matushansky I. Piwis and piwi-interacting RNAs in the epigenetics of cancer. J Cell Biochem. 2012;113:373–380. doi: 10.1002/jcb.23363. [DOI] [PubMed] [Google Scholar]
- 7.Siddiqi S, Terry M, Matushansky I. Hiwi mediated tumorigenesis is associated with DNA hypermethylation. PLoS One. 2012;7:e33711. doi: 10.1371/journal.pone.0033711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litwin M, Szczepańska-Buda A, Piotrowska A, Dzięgiel P, Witkiewicz W. The meaning of PIWI proteins in cancer development. Oncol Lett. 2017;13:3354–3362. doi: 10.3892/ol.2017.5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C, Zhou X, Chen J, Lu Y, Sun Q, Tao D, Hu W, Zheng X, Bian S, Liu Y, et al. PIWIL1 destabilizes microtubule by suppressing phosphorylation at Ser16 and RLIM-mediated degradation of Stathmin1. Oncotarget. 2015;6:27794–27804. doi: 10.18632/oncotarget.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z, Che Q, He X, Wang F, Wang H, Zhu M, Sun J, Wan X. Stem cell protein Piwil1 endowed endometrial cancer cells with stem-like properties via inducing epithelial-mesenchymal transition. BMC Cancer. 2015;15:811. doi: 10.1186/s12885-015-1794-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Liu Y, Shen X, Zhang X, Chen X, Yang C, Gao H. The PIWI protein acts as a predictive marker for human gastric cancer. Int J Clin Exp Pathol. 2012;5:315–325. [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Sun Y, Guo J, Ma H, Li J, Dong B, Jin G, Zhang J, Wu J, Meng L, et al. Expression of hiwi gene in human gastric cancer was associated with proliferation of cancer cells. Int J Cancer. 2006;118:1922–1929. doi: 10.1002/ijc.21575. [DOI] [PubMed] [Google Scholar]
- 13.Riordan SM, Heruth DP, Zhang LQ, Ye SQ. Application of CRISPR/Cas9 for biomedical discoveries. Cell Biosci. 2015;5:33. doi: 10.1186/s13578-015-0027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Sun H, Miao K, Deng CX. CRISPR-Cas9: from Genome Editing to Cancer Research. Int J Biol Sci. 2016;12:1427–1436. doi: 10.7150/ijbs.17421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawahara A, Hisano Y, Ota S, Taimatsu K. Site-Specific Integration of Exogenous Genes Using Genome Editing Technologies in Zebrafish. Int J Mol Sci. 2016:17. doi: 10.3390/ijms17050727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeder ML, Gersbach CA. Genome-editing Technologies for Gene and Cell Therapy. Mol Ther. 2016;24:430–446. doi: 10.1038/mt.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Travis J. Breakthrough of the Year: CRISPR makes the cut. Science Magazine. 2015 Available from: http://www.sciencemag.org/news/2015/12/and-science-s-breakthrough-year. [Google Scholar]
- 20.Leal MF, Martins do Nascimento JL, da Silva CE, Vita Lamarão MF, Calcagno DQ, Khayat AS, Assumpção PP, Cabral IR, de Arruda Cardoso Smith M, Burbano RR. Establishment and conventional cytogenetic characterization of three gastric cancer cell lines. Cancer Genet Cytogenet. 2009;195:85–91. doi: 10.1016/j.cancergencyto.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 21.Liehr T, Heller A, Starke H, Rubtsov N, Trifonov V, Mrasek K, Weise A, Kuechler A, Claussen U. Microdissection based high resolution multicolor banding for all 24 human chromosomes. Int J Mol Med. 2002;9:335–339. [PubMed] [Google Scholar]
- 22.Liehr T. The Standard FISH Procedure. Fluorescence In Situ Hybridization (FISH) - Application Guide. Berlin: Springer. 2017:109–118. [Google Scholar]
- 23.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc. 1995;57:289–300. [Google Scholar]
- 24.Carlson M. org.Hs.eg. db: Genome wide annotation for Human; 2017. p. R package version 3.5.0. [Google Scholar]
- 25.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 26.Xie K, Zhang K, Kong J, Wang C, Gu Y, Liang C, Jiang T, Qin N, Liu J, Guo X, et al. Cancer-testis gene PIWIL1 promotes cell proliferation, migration, and invasion in lung adenocarcinoma. Cancer Med. 2018;7:157–166. doi: 10.1002/cam4.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun R, Gao CL, Li DH, Li BJ, Ding YH. Expression Status of PIWIL1 as a Prognostic Marker of Colorectal Cancer. Dis Markers. 2017;2017:1204937. doi: 10.1155/2017/1204937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bloomfield M, Duesberg P. Karyotype alteration generates the neoplastic phenotypes of SV40-infected human and rodent cells. Mol Cytogenet. 2015;8:79. doi: 10.1186/s13039-015-0183-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gebhart E, Liehr T. Patterns of genomic imbalances in human solid tumors (Review) Int J Oncol. 2000;16:383–399. doi: 10.3892/ijo.16.2.383. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Tong X, Gao H, Yan X, Xu X, Sun S, Wang Q, Wang J. Silencing HIWI suppresses the growth, invasion and migration of glioma cells. Int J Oncol. 2014;45:2385–2392. doi: 10.3892/ijo.2014.2673. [DOI] [PubMed] [Google Scholar]
- 31.Rundhaug JE. Matrix metalloproteinases and angiogenesis. J Cell Mol Med. 2005;9:267–285. doi: 10.1111/j.1582-4934.2005.tb00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amaar YG, Baylink DJ, Mohan S. Ras-association domain family 1 protein, RASSF1C, is an IGFBP-5 binding partner and a potential regulator of osteoblast cell proliferation. J Bone Miner Res. 2005;20:1430–1439. doi: 10.1359/JBMR.050311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Ding N, Li Y, Cheng H, Wang D, Yang Q, Deng Y, Yang Y, Li Y, Ruan X, et al. Insulin-like growth factor binding protein 5 (IGFBP5) functions as a tumor suppressor in human melanoma cells. Oncotarget. 2015;6:20636–20649. doi: 10.18632/oncotarget.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Güllü G, Karabulut S, Akkiprik M. Functional roles and clinical values of insulin-like growth factor-binding protein-5 in different types of cancers. Chin J Cancer. 2012;31:266–280. doi: 10.5732/cjc.011.10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henry C, Llamosas E, Knipprath-Meszaros A, Schoetzau A, Obermann E, Fuenfschilling M, Caduff R, Fink D, Hacker N, Ward R, et al. Targeting the ROR1 and ROR2 receptors in epithelial ovarian cancer inhibits cell migration and invasion. Oncotarget. 2015;6:40310–40326. doi: 10.18632/oncotarget.5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henry CE, Llamosas E, Djordjevic A, Hacker NF, Ford CE. Migration and invasion is inhibited by silencing ROR1 and ROR2 in chemoresistant ovarian cancer. Oncogenesis. 2016;5:e226. doi: 10.1038/oncsis.2016.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahrmann EP, Collier LS, Knutson TP, Doyal ME, Kuslak SL, Green LE, Malinowski RL, Roethe L, Akagi K, Waknitz M, et al. Identification of PDE4D as a proliferation promoting factor in prostate cancer using a Sleeping Beauty transposon-based somatic mutagenesis screen. Cancer Res. 2009;69:4388–4397. doi: 10.1158/0008-5472.CAN-08-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delyon J, Servy A, Laugier F, André J, Ortonne N, Battistella M, Mourah S, Bensussan A, Lebbé C, Dumaz N. PDE4D promotes FAK-mediated cell invasion in BRAF-mutated melanoma. Oncogene. 2017;36:3252–3262. doi: 10.1038/onc.2016.469. [DOI] [PubMed] [Google Scholar]
- 39.Greuber EK, Smith-Pearson P, Wang J, Pendergast AM. Role of ABL family kinases in cancer: from leukaemia to solid tumours. Nat Rev Cancer. 2013;13:559–571. doi: 10.1038/nrc3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Pendergast AM. The Emerging Role of ABL Kinases in Solid Tumors. Trends Cancer. 2015;1:110–123. doi: 10.1016/j.trecan.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu JJ, Rouse C, Xu X, Wang J, Onaitis MW, Pendergast AM. Inactivation of ABL kinases suppresses non-small cell lung cancer metastasis. JCI Insight. 2016;1:e89647. doi: 10.1172/jci.insight.89647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Rouse C, Jasper JS, Pendergast AM. ABL kinases promote breast cancer osteolytic metastasis by modulating tumor-bone interactions through TAZ and STAT5 signaling. Sci Signal. 2016;9:ra12. doi: 10.1126/scisignal.aad3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sulzmaier FJ, Jean C, Schlaepfer DD. FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer. 2014;14:598–610. doi: 10.1038/nrc3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tajiri H, Uruno T, Shirai T, Takaya D, Matsunaga S, Setoyama D, Watanabe M, Kukimoto-Niino M, Oisaki K, Ushijima M, et al. Targeting Ras-Driven Cancer Cell Survival and Invasion through Selective Inhibition of DOCK1. Cell Rep. 2017;19:969–980. doi: 10.1016/j.celrep.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 45.Kulkarni K, Yang J, Zhang Z, Barford D. Multiple factors confer specific Cdc42 and Rac protein activation by dedicator of cytokinesis (DOCK) nucleotide exchange factors. J Biol Chem. 2011;286:25341–25351. doi: 10.1074/jbc.M111.236455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu M, Small D, Duffield AS. DOCK2: A novel FLT3/ITD leukemia drug target. Oncotarget. 2017;8:88253–88254. doi: 10.18632/oncotarget.21390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Nishihara H, Kimura T, Kato Y, Tanino M, Nishio M, Obara M, Endo T, Koike T, Tanaka S. DOCK2 regulates cell proliferation through Rac and ERK activation in B cell lymphoma. Biochem Biophys Res Commun. 2010;395:111–115. doi: 10.1016/j.bbrc.2010.03.148. [DOI] [PubMed] [Google Scholar]
- 48.Shahi P, Slorach EM, Wang CY, Chou J, Lu A, Ruderisch A, Werb Z. The Transcriptional Repressor ZNF503/Zeppo2 Promotes Mammary Epithelial Cell Proliferation and Enhances Cell Invasion. J Biol Chem. 2015;290:3803–3813. doi: 10.1074/jbc.M114.611202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shahi P, Wang CY, Lawson DA, Slorach EM, Lu A, Yu Y, Lai MD, Gonzalez Velozo H, Werb Z. ZNF503/Zpo2 drives aggressive breast cancer progression by down-regulation of GATA3 expression. Proc Natl Acad Sci USA. 2017;114:3169–3174. doi: 10.1073/pnas.1701690114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu X, Zhang Y, Chen H. LPA receptor 1 mediates LPA-induced ovarian cancer metastasis: an in vitro and in vivo study. BMC Cancer. 2016;16:846. doi: 10.1186/s12885-016-2865-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sahay D, Leblanc R, Grunewald TG, Ambatipudi S, Ribeiro J, Clézardin P, Peyruchaud O. The LPA1/ZEB1/miR-21-activation pathway regulates metastasis in basal breast cancer. Oncotarget. 2015;6:20604–20620. doi: 10.18632/oncotarget.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teng Y, Ghoshal P, Ngoka L, Mei Y, Cowell JK. Critical role of the WASF3 gene in JAK2/STAT3 regulation of cancer cell motility. Carcinogenesis. 2013;34:1994–1999. doi: 10.1093/carcin/bgt167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu Z, Chen W, Yin X, Lai J, Wang Q, Liang L, Wang W, Wang A, Zheng C. WAVE3 Induces EMT and Promotes Migration and Invasion in Intrahepatic Cholangiocarcinoma. Dig Dis Sci. 2016;61:1950–1960. doi: 10.1007/s10620-016-4102-9. [DOI] [PubMed] [Google Scholar]
- 54.Wang G, Fu Y, Liu G, Ye Y, Zhang X. miR-218 Inhibits Proliferation, Migration, and EMT of Gastric Cancer Cells by Targeting WASF3. Oncol Res. 2017;25:355–364. doi: 10.3727/096504016X14738114257367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lv ZD, Wang HB, Li FN, Wu L, Liu C, Nie G, Kong B, Qu HL, Li JG. TGF-β1 induces peritoneal fibrosis by activating the Smad2 pathway in mesothelial cells and promotes peritoneal carcinomatosis. Int J Mol Med. 2012;29:373–379. doi: 10.3892/ijmm.2011.852. [DOI] [PubMed] [Google Scholar]
- 56.Lv ZD, Na D, Ma XY, Zhao C, Zhao WJ, Xu HM. Human peritoneal mesothelial cell transformation into myofibroblasts in response to TGF-ß1 in vitro. Int J Mol Med. 2011;27:187–193. doi: 10.3892/ijmm.2010.574. [DOI] [PubMed] [Google Scholar]
- 57.Wu Y, Li Q, Zhou X, Yu J, Mu Y, Munker S, Xu C, Shen Z, Müllenbach R, Liu Y, et al. Decreased levels of active SMAD2 correlate with poor prognosis in gastric cancer. PLoS One. 2012;7:e35684. doi: 10.1371/journal.pone.0035684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shinto O, Yashiro M, Toyokawa T, Nishii T, Kaizaki R, Matsuzaki T, Noda S, Kubo N, Tanaka H, Doi Y, et al. Phosphorylated smad2 in advanced stage gastric carcinoma. BMC Cancer. 2010;10:652. doi: 10.1186/1471-2407-10-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song IS, Jeong YJ, Jeong SH, Heo HJ, Kim HK, Bae KB, Park YH, Kim SU, Kim JM, Kim N, et al. FOXM1-Induced PRX3 Regulates Stemness and Survival of Colon Cancer Cells via Maintenance of Mitochondrial Function. Gastroenterology. 2015;149:1006–1016.e9. doi: 10.1053/j.gastro.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 60.Bu XN, Qiu C, Wang C, Jiang Z. Inhibition of DACH1 activity by short hairpin RNA represses cell proliferation and tumor invasion in pancreatic cancer. Oncol Rep. 2016;36:745–754. doi: 10.3892/or.2016.4843. [DOI] [PubMed] [Google Scholar]
- 61.Yan W, Wu K, Herman JG, Brock MV, Zhou Y, Lu Y, Zhang Z, Yang Y, Guo M. Epigenetic silencing of DACH1 induces the invasion and metastasis of gastric cancer by activating TGF-β signalling. J Cell Mol Med. 2014;18:2499–2511. doi: 10.1111/jcmm.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]


