Abstract
Mildly elevated serum bilirubin levels were reported to be associated with decreased risk of non-alcoholic fatty liver disease (NAFLD). Whether this is a causal relationship remains unclear. We tested the hypothesis that genetically elevated plasma bilirubin levels are causally related to reduce risk of NAFLD. A total of 403 eligible participants were enrolled. NAFLD was determined by liver ultrasonography. The uridine diphosphate glucuronosyltransferase 1A1 (UGT1A1) gene variants (UGT1A1*6 and UGT1A1*28) were genotyped through sequencing. We applied a Mendelian randomization approach to assess the effects of genetically elevated bilirubin levels on NAFLD. NAFLD was diagnosed in 19% of participants in our study (NAFLD = 76; Non-NAFLD = 327). The variants of UGT1A1*28 and UGT1A1*6 were strongly associated with increased total bilirubin (TB), direct bilirubin (DB), and indirect bilirubin (IB) levels (each P < 0.001). These two common variants explain 12.7% (TB), 11.4% (IB), and 10.2% (DB) of the variance in bilirubin levels, respectively. In logistic regression model, after multifactorial adjustment for sex, age, aminotransferase (ALT), white blood count (WBC), and body mass index (BMI), variant UGT1A1*28 (OR = 1.39; 95%CI: 0.614–3.170; P = 0.43) and UGT1A1*6 (OR = 1.64, 95%CI, 0.78–3.44; P = 0.19) genotypes were not significantly associated with the risk of NAFLD. Moreover, the plasma bilirubin level (TB, IB, and DB) were not significantly associated with the risk of NAFLD (P > 0.30). A Mendelian randomization analysis of the UGT1A1 variants suggests that bilirubin is unlikely causally related with the risk of NAFLD.
Keywords: bilirubin, NAFLD, variant, UGT1A1, mendelian randomization
Introduction
Bilirubin, the by-product of hemoglobin catabolism, is generally considered to be a lipid-soluble waste product that needs to be excreted (Fujiwara et al., 2018; Hamoud et al., 2018). However, bilirubin levels may play an important physiological role as antioxidant (Stocker et al., 1987). It has been reported that elevated plasma levels of bilirubin are associated with reduced risk of non-alcoholic fatty liver disease (NAFLD) in case-control studies (Hjelkrem et al., 2012; Salomone et al., 2013) and in retrospective epidemiological studies(Chang et al., 2012; Tian et al., 2016). However, whether these associations reflect a true biological protective effect of bilirubin rather than confounding or reverse causation remains unknown. Whether bilirubin has negative or positive impact on NAFLD has important implication for clinical management. If positive, that means hyperbilirubinemia has a protective effect on NAFLD and UGT1A1 gene can be used as a gene target for the treatment of NAFLD. An artificially increasing plasma unconjugated bilirubin strategy as “Iatrogenic Gilbert syndrome” were proposed for reducing cardiovascular disease and cancer risks (Mccarty, 2007; Schwertner and Vítek, 2008).
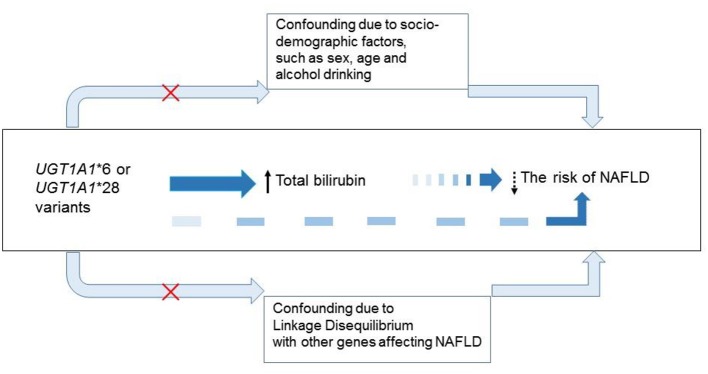
Mendelian randomization is an epidemiological method based on the fact that random assortment of genetic variants during meiosis yields a random distribution of genetic variants among individuals in a population (Emdin et al., 2017; Paternoster et al., 2017). The Mendelian randomization approach is ideal to assess the causal relationship of an intermediate phenotype with a disease phenotype by avoiding confounding by reverse causation, a limitation inherent in observational epidemiological studies (Johansen and Hegele, 2013; Ding et al., 2017; Holmes et al., 2017) (Figure 1). Genetic variation in the uridine diphosphate glucuronosyltransferase 1A1 gene (UGT1A1) is the major cause of hyperbilirubinemia (Johnson et al., 2009; Fujiwara et al., 2018) and is therefore suitable for exploring whether elevated bilirubin levels is a direct cause of reduced risk of NAFLD using a Mendelian randomization approach.
Figure 1.
Application of the Mendelian randomization framework in assessing causal role of bilirubin in NAFLD. According to the Mendelian randomization framework, if a genetic variant (UGT1A1) reliably governs a disease risk factor (Bilirubin), and there is a causal association between the risk factor (Bilirubin) and disease end-point (NAFLD), then the genetic variant (UGT1A1) should itself be associated with the disease end-point (NAFLD). Confounding factors, such as socio-demographic and disease status may affect the total bilirubin levels and the risk of NAFLD by modulate other biological pathways, but do not affect the random distribution of UGT1A1*6 and UGT1A1*28 variants. UGT1A1*6 and UGT1A1*28 were also not in linkage disequilibrium with known genes (e.g., PNPLA3 and TM6SF2) that predispose to NAFLD.
Herein, we tested the hypothesis that UGT1A1 variants (UGT1A1*28 and UGT1A1*6) are associated with higher plasma bilirubin levels and subsequently protect against NAFLD, using a Mendelian randomization approach. We first investigated whether the UGT1A1*6 and UGT1A1*28 variants were associated with elevated plasma bilirubin [total bilirubin (TB), direct bilirubin (DB), and indirect bilirubin (IB)], and explored the effect of genetic variants on bilirubin levels. We next tested whether elevated baseline plasma bilirubin levels (TB, IB, and DB) were association with decreased risk of NAFLD. Finally, we investigated whether these variants were associated with reduced risk of NAFLD.
Patients and Methods
Study Participants
The study population consisted of 446 Han Chinese adults who visited the second affiliated hospital of Luohe Medical College for routine health checkups from June to July in 2017. Among these participants, we excluded 43 participants who met one of the following exclusion criteria; (1) a medical history of CLD [viral hepatitis (n = 8), excessive alcohol consumption or cirrhosis]; (2) abnormal liver functions [Alanine aminotransferase (ALT) or Aspartate aminotransferase (AST) >50 U/L (n = 3)]; (3) Hemolysis signs [Hemoglobin (HB) < 100 g/L (n = 5)]; (4) evidence of infection or biliary obstruction or pregnancy: [White blood count (WBC) >10 × 109/L (n = 8); Total bile acid (TBA) >15 umol/L (n = 6); Glutamyltransferase (γGT) >80 U/L (n = 1), or alkaline phosphatase (ALP) >120 U/L(n = 6); pregnancy (n = 6)]. The eligible sample size for analyses was 403 (NAFLD = 76; Controls = 327) in the present study.
The study was approved by the Ethics Committee of Beijing YouAn hospital, Capital Medical University and a written informed consent form was obtained from all studied participants.
Clinical Data
All individuals underwent an abdominal B-type ultrasonography (Siemens S2000, Germany) performed by a trained sonographer. The diagnosis of NAFLD was based on the guidelines developed by the Fatty Liver Disease and Alcoholic Liver Disease Group, Chinese Society of Hepatology (Zeng et al., 2008). Ultrasonographic diagnosis was based on internationally well-accepted Criteria: liver-to-kidney echo contrast, parenchymal brightness, deep beam attenuation, bright vessel walls, obscure hepatic vessel structures, visibility of the diaphragm, and the neck of the gallbladder (Lin Y. C. et al., 2009; Hernaez et al., 2011; Ballestri et al., 2015); (2) there is no excessive alcohol consumption (defined as >140 g/week for male or 70 g/week for female, respectively) (Zeng et al., 2008); and (3) there are no competing etiologies for HS or chronic liver disease(CLD). Body mass index (BMI) was calculated as weight divided by height squared (kg/m2). All participants underwent plasma biochemistry, hemograms, and virological testing for HBV and HCV. Liver function tests [ALT, AST, γGT, TBA, total protein (TP), albumin(ALB), ALP, total bilirubin (TB), direct bilirubin (DB), and indirect bilirubin (IB)], fasting plasma glucose(FPG), low-density (LDL) lipoprotein, high-density lipoprotein (HDL), total cholesterol(TC), triglyceride(TG), and uric acid(UA) were measured using an ADVIA 2400 Clinical Chemistry System (Siemens, Germany). Total and direct bilirubin were measured by Vanadate oxidation method (Siemens, Germany). WBC, red blood cell (RBC), platelet (PLT), and hemoglobin (HB) were measured using XT-1800i Automated Hematology Analyzer (Sysmex, Japan). All biochemical tests were carried out in the same laboratory with standardized laboratory methods.
DNA Extraction, Primer Design, and PCR Amplification
Genomic DNA was extracted from whole blood using a QIAamp DNA Blood Mini Kit (QIAGEN, Germany) according to the manufacturer's protocol. The UGT1A1 gene promoter and first exon were PCR amplified and sequenced as previously described (Huang et al., 2002). Briefly, PCR conditions were as follows: 35 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The amplified products were purified from agarose gel and sequenced via an ABI3730XL sequencer (Applied Biosystems, Foster City, CA, USA).
Statistical Analysis
Continuous variables were expressed as mean and standard deviation (SD) and then statistically analyzed using Student's t-test or one-way ANOVA. Categorical variables were analyzed using the chi-squared (χ2) test. Stepwise linear regression models were used to detect the effects of SNPs with total, indirect, and direct bilirubin levels. Candidate covariates in stepwise selection included the two UGT1A1 variants (UGT1A1*6 and UGT1A1*28), sex, age, WBC, HB, PLT, ALB, TBA, and BMI. Binary logistic regressions were used to calculate ORs and the 95% confidence interval (CI) for NAFLD of bilirubin layering and UGT1A1 variants after adjustment for the effects of sex, age, WBC, ALT, and BMI. All statistical analyses were performed using SPSS version 23.0 (SPSS, Chicago, IL, USA). A two-tailed P-value < 0.05 was considered statistically significant.
Results
Demographic Characteristics
Clinical characteristics stratified by UGT1A1 genotypes are presented in Table 1. Among 403 enrolled individuals, the mean age was 37.3 ± 10.0 years old and 36.7% are men. Individuals in the study distributed into 4 genotype groups: 162 (40.2%) homozygous for wild-type genotypes, 87 (21.6%) with variant UGT1A1*6 genotypes, 126(31.3%) with variant UGT1A1*28 genotypes, and 28 (6.9%) with compound variant genotypes. The minor allele frequencies (MAF) of the UGT1A1 variants were 15.1% (UGT1A1*28) and 21.5% (UGT1A1*6). UGT1A1*6 was in low degree of linkage disequilibrium with UGT1A1*28 (D′ = 1.000; r2 = 0.049). Except for BMI, there were no obvious difference among the four genotype groups in respect to ALT, AST, TP, ALB, TBA, ALP, γGT, WBC, HB, PLT, FPG, TC, TG, HDL, LDL, UA, and diastolic blood pressure (Table 1).
Table 1.
Basic characteristics of the participants stratified by the UGT1A1 Genotypes.
| Variables | Genotypes | P-value | |||||
|---|---|---|---|---|---|---|---|
| Wild-type | UGT1A1*6 | UGT1A1*28 | UGT1A1*6-*28 | P6/wild | P28/wild | Pcom/wild | |
| (n = 160; 40.2%) | (n = 87; 21.6%) | (n = 126; 31.3%) | (n = 28; 6.9%) | ||||
| Sex, Female (%) | 105 (64.8%) | 56 (64.4%) | 78 (61.9%) | 16 (57.1%) | 0.944 | 0.611 | 0.436 |
| Age (years) | 36.2 ± 9.1 | 38.9 ± 11.7 | 37.4 ± 9.7 | 38.1 ± 10.8 | 0.045 | 0.312 | 0.357 |
| FPG (mmol/L) | 4.75 ± 0.51 | 4.80 ± 0.45 | 4.87 ± 0.81 | 4.87 ± 0.64 | 0.527 | 0.093 | 0.339 |
| TC (mmol/L) | 4.29 ± 0.70 | 4.37 ± 0.82 | 4.37 ± 0.87 | 4.87 ± 0.64 | 0.490 | 0.398 | 0.479 |
| TG (mmol/L) | 1.27 ± 0.72 | 1.30 ± 0.86 | 1.47 ± 1.06 | 1.28 ± 0.80 | 0.770 | 0.060 | 0.956 |
| HDL (mmol/L) | 1.34 ± 0.30 | 1.34 ± 0.29 | 1.32 ± 0.30 | 1.31 ± 0.31 | 0.764 | 0.376 | 0.519 |
| LDL (mmol/L) | 2.51 ± 0.60 | 2.55 ± 0.79 | 2.56 ± 0.72 | 2.44 ± 0.54 | 0.666 | 0.539 | 0.673 |
| UA (umol/L) | 299.9 ± 74.0 | 289.0 ± 76.9 | 310.9 ± 86.3 | 313.2 ± 83.8 | 0.308 | 0.263 | 0.443 |
| BMI (kg/m2) | 22.7 ± 3.1 | 23.6 ± 3.6 | 24.2 ± 3.6 | 23.6 ± 3.1 | 0.055 | < 0.001 | 0.196 |
| Systolic (mmHg) | 116.1 ± 16.8 | 118.5 ± 16.7 | 117.8 ± 16.9 | 122.8 ± 22.3 | 0.321 | 0.425 | 0.074 |
| Diastolic (mmHg) | 72.1 ± 11.0 | 73.7 ± 10.2 | 75.2 ± 11.4 | 73.4 ± 11.6 | 0.290 | 0.023 | 0.575 |
| ALT ± SD (U/L) | 17.6 ± 7.9 | 17.4 ± 7.0 | 19.0 ± 9.1 | 17.5 ± 6.7 | 0.804 | 0.144 | 0.929 |
| AST ± SD (U/L) | 17.7 ± 3.9 | 18.4 ± 4.6 | 18.2 ± 4.8 | 17.6 ± 3.9 | 0.218 | 0.292 | 0.888 |
| TP ± SD (g/L) | 75.9 ± 3.8 | 75.9 ± 4.4 | 75.9 ± 4.1 | 76.4 ± 4.3 | 0.869 | 0.975 | 0.557 |
| Alb ± SD (g/L) | 44.5 ± 2.3 | 44.6 ± 2.7 | 44.4 ± 2.7 | 44.8 ± 2.3 | 0.922 | 0.594 | 0.617 |
| TBA ± SD (umol/L) | 5.73 ± 2.41 | 5.91 ± 2.52 | 5.62 ± 2.37 | 5.43 ± 1.25 | 0.581 | 0.720 | 0.582 |
| ALP ± SD (U/L) | 65.9 ± 18.9 | 65.7 ± 15.1 | 68.0 ± 20.6 | 65.8 ± 15.6 | 0.928 | 0.338 | 0.986 |
| γGT ± SD (U/L) | 18.1 ± 10.2 | 17.9 ± 10.9 | 19.3 ± 11.6 | 18.5 ± 10.8 | 0.923 | 0.345 | 0.832 |
| WBC ± SD(× 109/L) | 6.26 ± 1.34 | 6.22 ± 1.53 | 6.14 ± 1.28 | 5.88 ± 1.25 | 0.830 | 0.479 | 0.177 |
| Hb ± SD (g/L) | 135.2 ± 13.8 | 134.9 ± 14.1 | 136.1 ± 15.1 | 137.0 ± 16.0 | 0.923 | 0.581 | 0.529 |
| PLT ± SD(× 109/L) | 227.1 ± 50.2 | 229.5 ± 47.4 | 235.7 ± 49.5 | 221.6 ± 53.8 | 0.713 | 0.145 | 0.591 |
Wild-type genotypes were homozygous, without either UGT1A1*6 or UGT1A1*28 variants. Variant UGT1A1*6 includes carriage of heterozygous or homozygous UGT1A1*6 variant allele. Variant UGT1A1*28 genotypes included heterozygous or homozygous UGT1A1*28 variants. The UGT1A1*6-*28 compound variant genotypes carry both UGT1A1*6 and UGT1A1*28 variants, either heterozygous or homozygous. P-values were calculated between groups by using analysis of variance for continuous variables and the χ2 test for categorical variables.
The Effects of the UGT1A1 Variants on Bilirubin Levels
We examined the effects of the variants (UGT1A1*28 and UGT1A1*6) on total, direct, and indirect bilirubin levels (Table 2). Total bilirubin levels were correlated with carriage of 1 and 2 copies of the variant alleles: compound heterozygotes (22.1 ± 12.6 umol/L), UGT1A1*6 (17.8 ± 8.6 umol/L), or UGT1A1*28 (16.4 ± 7.4 umol/L), and wild-type genotype (13.0 ± 4.9 umol/L), P < 0.001. The correlation between carriages of variant UGT1A1 was similar for the indirect and direct bilirubin groups (P < 0.001, Table 2). The overall incidence of NAFLD was 18.9%, consisted with the reported incidence in China (Fan et al., 2017), while the distribution of incidence of NAFLD was 21.4, 17.2, 25.4, and 14.2% for compound heterozygotes, UGT1A1*6 carriers, UGT1A1*28 carriers, and non-carriers, respectively. There was a significant difference in incidence of NAFLD between the non-carrier group and UGT1A1*28 carrier group (P = 0.016, Table 2).
Table 2.
The UGT1A1 genotypes and bilirubin levels.
| Variables | Genotypes | P-value | |||||
|---|---|---|---|---|---|---|---|
| Wild-type | UGT1A1*6 | UGT1A1*28 | UGT1A1*6-*28 | P6/wild | P28/wild | Pcom/wild | |
| (n = 160; 40.2%) | (n = 87; 21.6%) | (n = 126; 31.3%) | (n = 28; 6.9%) | ||||
| TB ± SD (μmol/L) | 13.0 ± 4.9 | 17.8 ± 8.6 | 16.4 ± 7.4 | 22.1 ± 12.6 | < 0.001 | < 0.001 | < 0.001 |
| IB ± SD (μmol/L) | 8.1 ± 3.3 | 11.6 ± 6.2 | 10.3 ± 4.2 | 14.3 ± 10.4 | < 0.001 | < 0.001 | < 0.001 |
| DB ± SD (μmol/L) | 4.9 ± 1.7 | 6.2 ± 2.7 | 6.1 ± 3 | 7.7 ± 2.8 | 0.001 | 0.002 | < 0.001 |
| NAFLD (%) | 14.2% | 17.2% | 25.4% | 21.4% | 0.524 | 0.016 | 0.485 |
The UGT1A1*6-*28 compound variant genotypes carry both UGT1A1*6 and UGT1A1*28 variants, either heterozygous or homozygous. P-values were calculated between groups by using ANOVA for continuous variables and the χ2 test for categorical variables.
UGT1A1 variant genotypes explained a substantial portion of the variation in bilirubin levels (Table 3). Using a stepwise regression model selection procedure, bilirubin levels were found to be influenced by the UGT1A1 variants as well as age, hemoglobin levels, platelet count, white blood count albumin, and total bile acid. With these co-factors being adjusted, UGT1A1*28 explained 5.6% and UGT1A1*6 explained 7.1% of the variation of total bilirubin levels. Similar results were observed for correlation with IB and DB (Table 3). In summary, the two common variants combined explain 12.7% of TB, 11.4% of IB, and 10.2% of DB levels.
Table 3.
Genetic, demographic and biochemical factors that affect bilirubin levels.
| Explanatory variables | Total bilirubin(TB) | Indirect bilirubin(IB) | Direct bilirubin (DB) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Coefficient | R2 | P-value | Coefficient | R2 | P-value | Coefficient | R2 | P-value | |
| UGT1A1*28 | 3.743 ± 0.577 | 5.6% | 3.5%10−10 | 2.469 ± 0.383 | 4.8% | 4.5%10−10 | 1.269 ± 0.212 | 4.2% | 6.3%10−9 |
| UGT1A1*6 | 2.908 ± 0.476 | 7.1% | 2.9%10−9 | 1.869 ± 0.316 | 6.6% | 8.8%10−9 | 1.032 ± 0.175 | 6.0% | 9.7%10−9 |
| Sex(Male) | 1.599 ± 0.959 | 0.6% | 0.096 | 1.320 ± 0.637 | 1.1% | 0.039 | 0.277 ± 0353 | 0.1% | 0.433 |
| Age (years) | 0.136 ± 0.032 | 2.9% | 2.2%10−5 | 0.115 ± 0.021 | 10.0% | 8.7%10−8 | 0.021 ± 0.012 | 0.6% | 0.072 |
| WBC (%109/L) | −0.388 ± 0.215 | 3.1% | 0.072 | −0.183 ± 0.143 | 0.3% | 0.201 | −0.204 ± 0.079 | 5.8% | 0.010 |
| HB (g/L) | 0.150 ± 0.033 | 9.4% | 8.0%10−6 | 0.101 ± 0.022 | 6.3% | 6.0%10−6 | 0.049 ± 0.012 | 8.9% | 5.7%10−5 |
| PLT (%109/L) | −0.018 ± 0.006 | 1.8% | 0.002 | −0.010 ± 0.004 | 1.9% | 0.008 | −0.008 ± 0.002 | 2.8% | 3.7%10−5 |
| ALB (g/L) | 0.512 ± 0.127 | 1.6% | 6.8%10−5 | 0.306 ± 0.088 | 2.6% | 3.2%10−4 | 0.206 ± 0.047 | 3.7% | 1.4%10−5 |
| TBA (umol/L) | −0.391 ± 0.115 | 4.8% | 0.001 | −0.282 ± 0.077 | 4.2% | 2.7 × 10−4 | −0.108 ± 0.042 | 1.8% | 0.011 |
| BMI (kg/m2) | −0.144 ± 0.089 | 0.7% | 0.104 | −0.069 ± 0.059 | 0.3% | 0.245 | −0.076 ± 0.033 | 1.0% | 0.021 |
Associated of Bilirubin Level With the Risk of NAFLD
We then analyzed the plasma bilirubin levels (TB, IB, and DB) and the distribution of UGT1A1 genotypes with or without NAFLD (Table 4). Of 403 individuals, 76 (19%) were diagnosed as NAFLD, close to the 25% population prevalence of NAFLD in Asia (Fan et al., 2017). However, there was no significant difference in TB, IB, and DB levels between the NAFLD and non-NAFLD groups, either in the crude or fully adjusted model (P > 0.30, Table 4). Furthermore, there were no significant difference in the incidence of NAFLD among the 4 quartile groups with different levels of TB, IB, and DB, which further support no association between plasma bilirubin level (TB, DB, and IB) and NAFLD (P > 0.15, Supplementary Table 1).
Table 4.
Distribution and multivariate analysis of UGT1A1 genotypes for participants with and without NAFLD.
| Explanatory variables | NAFLD | Non-NAFLD | P-value | OR (95%CI) | P-value | OR (95% CI)adjusted | P-valueadjusted |
|---|---|---|---|---|---|---|---|
| (n = 76) | (n = 327) | ||||||
| TB (μmol/L) | 16.2 ± 8.6 | 15.6 ± 7.5 | 0.548 | 1.009 (0.979–1.041) | 0.547 | 1.028 (0.971–1.088) | 0.349 |
| DB (μmol/L) | 5.9 ± 4.7 | 5.7 ± 2.4 | 0.622 | 1.025(0.949–1.107) | 0.526 | 1.028 (0.861–1.227) | 0.763 |
| IB (μmol/L) | 10.2 ± 4.9 | 9.9 ± 5.5 | 0.523 | 1.011(0.968–1.057) | 0.622 | 1.045 (0.962–1.234) | 0.300 |
| VARIANT UGT1A1*6 | |||||||
| G/G | 38 (50.0%) | 211 (64.5%) | 1.000 | ||||
| G/A or A/A | 38 (50.0%) | 116 (35.5%) | 0.019 | 1.819 (1.099–3.009) | 0.020 | 1.638 (0.780–3.442) | 0.193 |
| VARIANT UGT1A1*28 | |||||||
| 6/6 | 55 (72.4%) | 233 (71.3%) | 1.000 | ||||
| 6/7 or 7/7 | 21 (27.6%) | 94 (28.7%) | 0.846 | 0.946 (0.542–1.652) | 0.846 | 1.395 (0.614–3.170) | 0.427 |
| COMPOUND VARIANT GENOTYPES | |||||||
| 6/6; G/G or G/A; 6/6 or 6/7; G/G | 70 (92.1%) | 305 (93.3%) | 1.000 | ||||
| G/A or A/A and 6/7 or 7/7 | 6 (7.9%) | 22 (6.7%) | 0.719 | 1.118 (0.465–3.040) | 0.719 | 2.131 (0.589–7.704) | 0.249 |
Systolic blood pressure, diastolic blood pressure, fasting plasma glucose, total cholesterol, triglyceride, low-density lipoprotein, high-density lipoprotein, and uric acid were also included in the regression model and were not significant (each P > 0.05). Adjusted for age, sex, ALT, WBC, and BMI.
The Association Between the UGT1A1 Variants and NAFLD
We used Mendelian randomization to determine if UGT1A1 variant alleles differentially distribute between NAFLD cases and Non-NAFLD controls. The frequency of UGT1A1*6 variant was significantly higher in NAFLD group [50.0%; (OR = 1.82; 95% CI, 1.09–3.01; P = 0.02)] than in the non-NAFLD (35.5%), when compare with GG genotype (wild-type). However, in fully adjusted multivariate model, the association was attenuated and no longer significant (OR = 1.64, 95%CI, 0.78–3.44; P = 0.19) (Table 4). The prevalence of UGT1A1*28 genotypes or compound heterozygous genotypes did not differ significantly between the NAFLD and non-NAFLD groups in the crude analysis and in the fully adjusted model.
Discussion
In the present study, we assessed the effects of UGT1A1 variants and bilirubin levels on NAFLD. Two common variants (UGT1A1*6 and UGT1A1*28) were significantly associated with increased plasma bilirubin levels (TB, IB, and DB). However, these genetic variants and genetically elevated bilirubin levels (TB, IB, and DB) were not associated with decreased risk of NAFLD. Based on a Mendelian randomization approach, our data suggest that increased bilirubin levels is unlikely a causal factor for a reduced risk of NAFLD.
In our study, the frequencies of UGT1A1*28 and UGT1A1*6 variant alleles were 15.1 and 21.5%, respectively, similar to previously reported MAF in Asian populations (Huang et al., 2004; Memon et al., 2016). Previous studies (Bosma et al., 1995; Teng et al., 2007) have shown that UGT1A1*28 and UGT1A1*6 decrease the expression levels and activity of UGT1A1, thereby increasing TB levels. In this study, we found that these variants not only raise TB levels but also increase DB and IB levels. This is in agreement with a previous finding that UGT1A1*6 variant can increase urobilinogen levels, which correlates with DB levels (Kataoka et al., 2011). We found that the effect of UGT1A1*6 explains 7.1% of the total variation of TB levels, slightly higher than the previous results from a case-control study (5.2%) (Lin R. et al., 2009) and a GWAS (4.5%) (Dai et al., 2013) in Chinese. Further, we found that UGT1A1*28 had stronger effect on bilirubin levels (TB, IB, and DB) than either rs6742078 or rs887829 reported in the GWAS study (Chen et al., 2012; Dai et al., 2013). As rs6742078 and rs887829 are in high linkage disequilibrium with UGT1A1*28, it is likely that the reported GWAS signal for rs6742078 or rs887829 is tracking the functional UGT1A1*28 variant (Johnson et al., 2009; Yang et al., 2015). Moreover, the UGT1A1*28 and UGT1A1*6 (rs4148323) explain 12.7% of the total variation of TB levels, and UGT1A1 is the only major gene that control total bilirubin variance in different populations (Johnson et al., 2009; Dai et al., 2013). The proportion of total phenotypic variance in most studied traits explained by the collective effects of known, common variants (R2) is rarely >10% but often >1%(Pierce et al., 2011), e.g., C-reactive protein, urate, lipids, triglycerides and FPG, as reviewed by Pierce et al. (2011). Therefore, the two variables are appropriate instruments for Mendelian Randomization analysis.
At present, there are two unresolved questions regarding the relationship between bilirubin levels and NAFLD. First, is there a relationship between the bilirubin levels and NAFLD? Two studies (Hjelkrem et al., 2012; Salomone et al., 2013) based on liver pathological diagnosis reported no significant association of bilirubin levels with NAFLD without non-alcoholic steatohepatitis (NASH), whereas a significantly lower prevalence of unconjugated hyperbilirubinemia in patients with histopathological evidence of NASH. Besides, there is a protective effect of elevated bilirubin level was found in three studies on risk of liver ultrasonography-diagnosed NAFLD including Han Chinese retirees (Tian et al., 2016), middle aged Korean Workers (Chang et al., 2012), and people for routine health checkups in Korean (Kwak et al., 2012). Whether the study on bilirubin and NAFLD were negative or positive, it has important value for clinical, and scientific research.
Furthermore, Variant UGT1A1*6 genotypes were associated with a lower risk of NAFLD in obese children, while the variant UGT1A1*28 genotypes and total bilirubin level were not significantly associated with the occurrence of pediatric NAFLD (Lin Y. C. et al., 2009). In our study, the UGT1A1 variants (UGT1A1*6 and UGT1A1*28) and bilirubin level were not significantly associated with the risk of NAFLD in Han Chinese adult in Mendelian randomization trial. The inconsistent results could be owing to differences in study designs, genetic background, age, sex, severity of the disease and other variables. Second, which type of bilirubin is related to lower risk of NAFLD? Total bilirubin (IB and DB were not measured) was significantly associated with the incidence of NAFLD in some studies (Kwak et al., 2012; Puri et al., 2013), while direct bilirubin was associated with reduced NAFLD risk in other two studies (Chang et al., 2012; Tian et al., 2016). We did not observe an association between genetically bilirubin level (TB, IB, and DB) and risk of NAFLD. Of course, further studies are certainly needed to address the above issues.
Our study meets the three assumptions for Mendelian randomization method (Figure 1) (Johansen and Hegele, 2013; Emdin et al., 2017): (1) the genetic variant is related with the risk factor (bilirubin). In our study, the variants of UGT1A1*28 and UGT1A1*6 were strongly associated with increased TB, DB, and IB levels (each P < 0.001); (2) the genetic variant is not related with confounders. In our study, except for the bilirubin levels, other clinical variables were no significant different among the four UGT1A1 genotype groups; (3) the genetic variant affect the consequence (NAFLD) only through the risk factor (bilirubin). UGT1A1 is the only enzyme catalyzing the generation of water-soluble bilirubin glucuronides in hepatocytes (Memon et al., 2016; Wagner et al., 2018). UGT1A1*6 and UGT1A1*28 were also not in linkage disequilibrium with the known genes [e.g., PNPLA3 and TM6SF2 (Macaluso et al., 2015; Fan et al., 2017)] that predispose to NAFLD. Moreover, in order to enhance statistical power of Mendelian randomization (Emdin et al., 2017), two variants in UGT1A1 gene (UGT1A1*6 and UGT1A1*28) that affect the bilirubin levels the most were included in the analysis. To our knowledge, this is the first Mendelian randomization trial to explore the association of elevated bilirubin levels and NAFLD. Our findings that lifelong elevated bilirubin levels as well as the bilirubin level-controlling genetic variants, which are unconfounded by socio-economic and/or environmental factors, did not support a role of bilirubin in protecting the occurrence of NAFLD.
This study has several limitations. First, the study is a single-center, small-sample study and the results should be validated in large-scale prospective studies; Second, this study only analyzed the two common variants in UGT1A1 gene, and other SNPs related to bilirubin levels were not included in the study, such as rs2417940 in SLCO1B3 gene, which reportedly explains only 0.64% of TB variation (Kang et al., 2010). Nevertheless, the two common variants combined (UGT1A1*6 and UGT1A1*28) explains 12.7% of TB variation and are the major genetic factors controlling bilirubin levels. Third, our study enrolled mainly mid-aged adults that may have lower incidence of NAFLD than elders, hence the lower statistical power. Nevertheless, the incidence of NAFLD in the study is consistent with the prevalence in China (~20%) (Fan et al., 2017) and the common UGT1A1 variants tested strongly predict bilirubin levels as expected. Fourth, the UGT1A1*28 group had higher BMI and incidence of NAFLD than the wild-type group (Tables 1, 2), consistent with the finding that obesity increases incidence of NAFLD (Chang et al., 2016; Targher and Byrne, 2016). We thus adjusted BMI as a covariable for all association tests. We also found that BMI had none or minor effect on bilirubin levels (Table 3), unsupported of an UGT1A1 variants-BMI-bilirubin-NAFLD pathway, a potential confounder for Mendelian randomization trial. Lastly, it is well-known that liver biopsy is the gold standard for diagnosis of NAFLD (Chalasani and Younossi, 2018). The diagnosis of NAFLD was based on liver ultrasonography in this study, which may lead to potential attenuation of the associations between the NAFLD and bilirubin levels due to its insensitivity in detection of mild fatty liver. Whereas, the B-type ultrasonography is reliable and accurate in detection of moderate-severe fatty liver with sensitivity of 85% and specificity of 94% (Hernaez et al., 2011), and has been a cost-effective tool for screening NAFLD in epidemiological studies (Chang et al., 2012; Tian et al., 2016) and clinical practice (Chalasani and Younossi, 2018). Of course, future well-powered studies using biopsy-proven NAFLDs will provide more definitive conclusion.
In conclusion, genetically elevated plasma bilirubin levels were not associated with reduced risk of NAFLD in Mendelian randomization trial of a general adult population. These data suggest that bilirubin is unlikely causally related with the risk of NAFLD.
Author Contributions
LL, PA, and ZD designed this study. LL carried out the experiments and analyses with the help of XJ, XY, SZ, SL, YC, WA, and ZD. LL, PA, CW, and ZD drafted the manuscript. All others contributed to the revision of the manuscript. All the authors read and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Gailing Guo, Lanxia Li (Department of Laboratory Medicine), and Qiaoe Chu (Department of Physical Examination) in the second affiliated hospital of Luohe Medical College for their technical assistance.
Glossary
Abbreviations
- ALB
albumin
- ALP
alkaline phosphatase
- ALT
aminotransferase
- AST
aspartate aminotransferase
- BMI
Body mass index
- DB
direct bilirubin
- FPG
fasting plasma glucose
- HB
hemoglobin
- HDL
high-density lipoprotein
- IB
indirect bilirubin
- LDL
low-density lipoprotein
- PLT
platelet
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- γGT
glutamyltransferase
- TB
total bilirubin
- TC
total cholesterol
- TG
triglyceride
- TBA
total bile acid
- TP
total protein
- UA
uric acid
- UGT1A1
uridine diphosphate glucuronosyltransferase 1A1
- WBC
white blood count.
Footnotes
Funding. This study was supported by Beijing Municipal Science and Technology Project (No. Z171100002217070), National Key R&D Program of China (No. 2017YFA0103000), National Science and Technology Key Project on Major Infectious Diseases HIV/AIDS, Viral Hepatitis Prevention and Treatment (No. 2012ZX10002004-006, No. 2017ZX10203201-005, 2017ZX10201201, No. 2017ZX10202203-006-001, and No. 2017ZX10302201-004-002), Beijing Municipal Administration of Hospitals Ascent Plan (No. DFL20151601), Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (No. ZYLX201806), The Digestive Medical Coordinated Development Center of Beijing Municipal Administration of Hospitals (XXZ0503). This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of health, under contract HHSN26120080001E. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00662/full#supplementary-material
References
- Ballestri S., Romagnoli D., Nascimbeni F., Francica G., Lonardo A. (2015). Role of ultrasound in the diagnosis and treatment of nonalcoholic fatty liver disease and its complications. Exp. Rev. Gastroenterol. Hepatol. 9, 603–627. 10.1586/17474124.2015.1007955 [DOI] [PubMed] [Google Scholar]
- Bosma P. J., Chowdhury J. R., Bakker C., Gantla S., De Boer A., Oostra B. A., et al. (1995). The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N. Engl. J. Med. 333, 1171–1175. 10.1056/NEJM199511023331802 [DOI] [PubMed] [Google Scholar]
- Chalasani N., Younossi Z. (2018). The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the study of liver diseases. Hepatology 67, 328–357. 10.1002/hep.29367 [DOI] [PubMed] [Google Scholar]
- Chang Y., Jung H. S., Cho J., Zhang Y., Yun K. E., Lazo M., et al. (2016). Metabolically healthy obesity and the development of nonalcoholic fatty liver disease. Am. J. Gastroenterol. 111, 1133–1140. 10.1038/ajg.2016.178 [DOI] [PubMed] [Google Scholar]
- Chang Y., Ryu S., Zhang Y., Son H. J., Kim J. Y., Cho J., et al. (2012). A cohort study of serum bilirubin levels and incident non-alcoholic fatty liver disease in middle aged Korean workers. PLoS ONE 7:e37241. 10.1371/journal.pone.0037241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Ramos E., Adeyemo A., Shriner D., Zhou J., Doumatey A. P., et al. (2012). UGT1A1 is a major locus influencing bilirubin levels in African Americans. Eur. J. Hum. Genet. 20, 463–468. 10.1038/ejhg.2011.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Wu C., He Y., Gui L., Zhou L., Guo H., et al. (2013). A genome-wide association study for serum bilirubin levels and gene-environment interaction in a Chinese population. Genet. Epidemiol. 37, 293–300. 10.1002/gepi.21711 [DOI] [PubMed] [Google Scholar]
- Ding M., Huang T., Bergholdt H. K., Nordestgaard B. G., Ellervik C., Qi L. (2017). Dairy consumption, systolic blood pressure, and risk of hypertension: mendelian randomization study. BMJ 356:j1000. 10.1136/bmj.j1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emdin C. A., Khera A. V., Kathiresan S. (2017). Mendelian Randomization. JAMA 318, 1925–1926. 10.1001/jama.2017.17219 [DOI] [PubMed] [Google Scholar]
- Fan J. G., Kim S. U., Wong V. W. (2017). New trends on obesity and NAFLD in Asia. J. Hepatol. 67, 862–873. 10.1016/j.jhep.2017.06.003 [DOI] [PubMed] [Google Scholar]
- Fujiwara R., Haag M., Schaeffeler E., Nies A. T. (2018). Systemic regulation of bilirubin homeostasis: potential benefits of hyperbilirubinemia. Hepatology 67, 1609–1619. 10.1002/hep.29599 [DOI] [PubMed] [Google Scholar]
- Hamoud A. R., Weaver L., Stec D. E., Hinds T. D., Jr. (2018). Bilirubin in the liver-gut signaling axis. Trends Endocrinol. Metab. 29, 140–150. 10.1016/j.tem.2018.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernaez R., Lazo M., Bonekamp S., Kamel I., Brancati F. L., Guallar E., et al. (2011). Diagnostic accuracy and reliability of ultrasonography for the detection of fatty liver: a meta-analysis. Hepatology 54, 1082–1090. 10.1002/hep.24452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelkrem M., Morales A., Williams C. D., Harrison S. A. (2012). Unconjugated hyperbilirubinemia is inversely associated with non-alcoholic steatohepatitis (NASH). Aliment. Pharmacol. Ther. 35, 1416–1423. 10.1111/j.1365-2036.2012.05114.x [DOI] [PubMed] [Google Scholar]
- Holmes M. V., Ala-Korpela M., Smith G. D. (2017). Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat. Rev. Cardiol. 14, 577–590. 10.1038/nrcardio.2017.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. S., Chang P. F., Huang M. J., Chen E. S., Chen W. C. (2002). Glucose-6-phosphate dehydrogenase deficiency, the UDP-glucuronosyl transferase 1A1 gene, and neonatal hyperbilirubinemia. Gastroenterology 123, 127–133. 10.1053/gast.2002.34173 [DOI] [PubMed] [Google Scholar]
- Huang M. J., Kua K. E., Teng H. C., Tang K. S., Weng H. W., Huang C. S. (2004). Risk factors for severe hyperbilirubinemia in neonates. Pediatr. Res. 56, 682–689. 10.1203/01.PDR.0000141846.37253.AF [DOI] [PubMed] [Google Scholar]
- Johansen C. T., Hegele R. A. (2013). Using Mendelian randomization to determine causative factors in cardiovascular disease. J. Intern. Med. 273, 44–47. 10.1111/j.1365-2796.2012.02586.x [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Kavousi M., Smith A. V., Chen M. H., Dehghan A., Aspelund T., et al. (2009). Genome-wide association meta-analysis for total serum bilirubin levels. Hum. Mol. Genet. 18, 2700–2710. 10.1093/hmg/ddp202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T. W., Kim H. J., Ju H., Kim J. H., Jeon Y. J., Lee H. C., et al. (2010). Genome-wide association of serum bilirubin levels in Korean population. Hum. Mol. Genet. 19, 3672–3678. 10.1093/hmg/ddq281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka R., Kimata A., Yamamoto K., Hirosawa N., Ueyama J., Kondo T., et al. (2011). Association of UGT1A1 Gly71Arg with urine urobilinogen. Nagoya J. Med. Sci. 73, 33–40. [PMC free article] [PubMed] [Google Scholar]
- Kwak M. S., Kim D., Chung G. E., Kang S. J., Park M. J., Kim Y. J., et al. (2012). Serum bilirubin levels are inversely associated with nonalcoholic fatty liver disease. Clin. Mol. Hepatol. 18, 383–390. 10.3350/cmh.2012.18.4.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Wang Y., Wang Y., Fu W., Zhang D., Zheng H., et al. (2009). Common variants of four bilirubin metabolism genes and their association with serum bilirubin and coronary artery disease in Chinese Han population. Pharmacogenet. Genom. 19, 310–318. 10.1097/FPC.0b013e328328f818 [DOI] [PubMed] [Google Scholar]
- Lin Y. C., Chang P. F., Hu F. C., Chang M. H., Ni Y. H. (2009). Variants in the UGT1A1 gene and the risk of pediatric nonalcoholic fatty liver disease. Pediatrics 124:e1221–1227. 10.1542/peds.2008-3087 [DOI] [PubMed] [Google Scholar]
- Macaluso F. S., Maida M., Petta S. (2015). Genetic background in nonalcoholic fatty liver disease: a comprehensive review. World J. Gastroenterol. 21, 11088–11111. 10.3748/wjg.v21.i39.11088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccarty M. F. (2007). “Iatrogenic Gilbert syndrome”–a strategy for reducing vascular and cancer risk by increasing plasma unconjugated bilirubin. Med. Hypotheses 69, 974–994. 10.1016/j.mehy.2006.12.069 [DOI] [PubMed] [Google Scholar]
- Memon N., Weinberger B. I., Hegyi T., Aleksunes L. M. (2016). Inherited disorders of bilirubin clearance. Pediatr. Res. 79, 378–386. 10.1038/pr.2015.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paternoster L., Tilling K., Davey Smith G. (2017). Genetic epidemiology and Mendelian randomization for informing disease therapeutics: conceptual and methodological challenges. PLoS Genet. 13:e1006944. 10.1371/journal.pgen.1006944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce B. L., Ahsan H., Vanderweele T. J. (2011). Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int. J. Epidemiol. 40, 740–752. 10.1093/ije/dyq151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri K., Nobili V., Melville K., Corte C. D., Sartorelli M. R., Lopez R., et al. (2013). Serum bilirubin level is inversely associated with nonalcoholic steatohepatitis in children. J. Pediatr. Gastroenterol. Nutr. 57, 114–118. 10.1097/MPG.0b013e318291fefe [DOI] [PubMed] [Google Scholar]
- Salomone F., Li Volti G., Rosso C., Grosso G., Bugianesi E. (2013). Unconjugated bilirubin, a potent endogenous antioxidant, is decreased in patients with non-alcoholic steatohepatitis and advanced fibrosis. J. Gastroenterol. Hepatol. 28, 1202–1208. 10.1111/jgh.12155 [DOI] [PubMed] [Google Scholar]
- Schwertner H. A., Vítek L. (2008). Gilbert syndrome, UGT1A1*28 allele, and cardiovascular disease risk: possible protective effects and therapeutic applications of bilirubin. Atherosclerosis 198, 1–11. 10.1016/j.atherosclerosis.2008.01.001 [DOI] [PubMed] [Google Scholar]
- Stocker R., Yamamoto Y., Mcdonagh A. F., Glazer A. N., Ames B. N. (1987). Bilirubin is an antioxidant of possible physiological importance. Science 235, 1043–1046. 10.1126/science.3029864 [DOI] [PubMed] [Google Scholar]
- Targher G., Byrne C. D. (2016). Obesity: metabolically healthy obesity and NAFLD. Nat. Rev. Gastroenterol. Hepatol. 13, 442–444. 10.1038/nrgastro.2016.104 [DOI] [PubMed] [Google Scholar]
- Teng H. C., Huang M. J., Tang K. S., Yang S. S., Tseng C. S., Huang C. S. (2007). Combined UGT1A1 and UGT1A7 variant alleles are associated with increased risk of Gilbert's syndrome in Taiwanese adults. Clin. Genet. 72, 321–328. 10.1111/j.1399-0004.2007.00873.x [DOI] [PubMed] [Google Scholar]
- Tian J., Zhong R., Liu C., Tang Y., Gong J., Chang J., et al. (2016). Association between bilirubin and risk of Non-Alcoholic Fatty Liver Disease based on a prospective cohort study. Sci. Rep. 6:31006. 10.1038/srep31006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner K. H., Shiels R. G., Lang C. A., Seyed Khoei N., Bulmer A. C. (2018). Diagnostic criteria and contributors to Gilbert's syndrome. Crit Rev Clin Lab Sci. 55:129–39. 10.1080/10408363.2018.1428526 [DOI] [PubMed] [Google Scholar]
- Yang H., Wang Q., Zheng L., Lin M., Zheng X. B., Lin F., et al. (2015). Multiple genetic modifiers of bilirubin metabolism involvement in significant neonatal hyperbilirubinemia in patients of chinese descent. PLoS ONE 10:e0132034. 10.1371/journal.pone.0132034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M. D., Fan J. G., Lu L. G., Li Y. M., Chen C. W., Wang B. Y., et al. (2008). Guidelines for the diagnosis and treatment of nonalcoholic fatty liver diseases. J. Dig. Dis. 9, 108–112. 10.1111/j.1751-2980.2008.00331.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.