Abstract
Several lines of evidence highlight the important application of human spermatogonial stem cells (SSCs) in translational medicine. The fate decisions of SSCs are mainly mediated by genetic and epigenetic factors. We have recently demonstrated that PAK1 regulates the proliferation, DNA synthesis, and early apoptosis of human SSCs through the PDK1/KDR/ZNF367 and ERK1/2 and AKT pathway. However, the underlying epigenetic mechanism of PAK1 in human SSCs remains unknown. In this study, we found that the level of miRNA-31-5p was elevated by PAK1 knockdown. CCK-8, PCNA, and 5-ethynyl-2′-deoxyuridine (EDU) assays revealed that miRNA-31-5p mimics inhibited cell proliferation and DNA synthesis of human SSCs. Annexin V/propidium iodide (PI) staining and flow cytometry showed that miRNA-31-5p increased the early and late apoptosis of human SSCs. Furthermore, JAZF1 was predicted and verified as a target of miRNA-31-5p, and the three-dimensional (3D) structure model of JAZF1 protein was illustrated. JAZF1 silencing led to a reduction of cell proliferation and DNA synthesis as well as an enhancement of the early and late apoptosis of human SSCs. Finally, miRNA-31-5p mimics decreased the level of cyclin A2 rather than cyclin D1 or cyclin E1, and JAZF1 knockdown led to the reduction of cyclin A2 in human SSCs. Collectively, miRNA-31-5p regulates the proliferation, DNA synthesis, and apoptosis of human SSCs by the PAK1-JAZF1-cyclin A2 pathway. This study thus offers a novel insight into the molecular mechanisms underlying the fate determinations of human SSCs and might provide novel targets for molecular therapy of male infertility.
Keywords: miRNA-31-5p, human spermatogonial stem cells, proliferation, apoptosis, JAZF1, cyclin A2
Introduction
Human spermatogonia stem cells (SSCs) are located along the basement of the seminiferous tubules. They could both differentiate to mature sperm and self-renew to maintain the stem cell population.1 There are two kinds of type A spermatogonia in human, namely A dark and A pale spermatogonia,2 which are thought to be the reserve stem cells and renewing stem cells, respectively. Spermatogenesis is a complex and strictly organized process, which comprises spermatogonial mitotic proliferation, two times of meiosis, and spermiogenesis.3 Various kinds of factors are involved in this process, and Sertoli cells can support male germ cells in structural and nutritional aspects.4
Human SSCs are the foundation for male fertility, and their self-renewal is vital for maintaining the spermatogenesis throughout the mammalian lifetime.5 It has been estimated that infertility affects 10%–15% of the couples worldwide, and male infertility accounts for half of these cases.6 Notably, there are obvious differences in cell types and biochemical phenotypes of SSCs between the primates and rodents.7, 8 Besides, it has been demonstrated that human SSCs showed a limited proliferation capacity under culture conditions for mouse SSCs.9 It is difficult to use human primary SSCs for research due to the fact that their number is few and it is hard to expand them in vitro. Alternatively, an SSC line can be utilized for basic research by stably expressing SV40LT (simian virus 40 large antigen)10 or hTERT (human telomerase reverse transcriptase).11 SV40LT is widely used to immortalize primary cells, such as mouse germline stem cell line.12 We have recently established human SSC line by means of transfecting SV40 plasmid to primary human SSCs.13 This SSC line can express a number of spermatogonial stem cell biomarkers, and notably, it has an unlimited proliferation ability without tumor formation. Therefore, this human SSC line could be used as an excellent cell model to explore the molecular mechanisms of self-renewal of human SSCs.13
Nonobstructive azoospermia (NOA) patients have impaired spermatogenesis. The molecular basis of male infertility remains largely unknown,14 and only certain NOA patients could obtain offspring through assisted reproductive technology.15 Accumulating studies have indicated the significance of noncoding RNAs (ncRNAs) in male infertility.16 MicroRNAs (miRNAs) are endogenous ncRNAs of 18∼25 nucleotides in length, and they regulate the mRNA stability at posttranscriptional level mainly through binding their 3′ UTR.17 Drosha and Dicer1 are key enzyme in the biogenesis and maturation of miRNA, and their mutations could lead to abnormal spermatogenesis and male infertility.18, 19 MiR-17-92 cluster and miR-290-295 cluster are highly expressed in primordial germ cells (PGCs), and deletion of Dicer in PGCs and spermatogonia exhibits a decrease of proliferation capacity.20 It has been demonstrated that deletion of Dicer1 in mice could result in infertility, since DICER1 is essential for meiotic progression and its deletion increases the apoptosis of spermatocytes and defect in the maturation of spermatozoa.21, 22 Polymorphism in DICER1 (rs12323635) could increase the risk of abnormal semen parameters and idiopathic male infertility.23 We have recently revealed that PAK1 promotes the proliferation and inhibits the apoptosis of human SSCs via the PDK1-KDR-ZNF367 and ERK1/2 and AKT pathway. We have also found that there is lower level of PAK1 in the NOA population. Here, we further uncover the underlying epigenetic mechanism of PAK1 in human SSCs. We found that the level of miRNA-31-5p was elevated by PAK1 silencing. Utilizing the stable human SSC line, we have demonstrated that miRNA-31-5p mediates the proliferation, DNA synthesis, and apoptosis of human SSCs by targeting JAZF1 and cyclin A2. Thus, this study provides a new epigenetic regulation of human SSCs and offers molecular targets for treating male infertility.
Results
miRNA-31-5p Is Enhanced by PAK1 Silencing in Human SSC Line
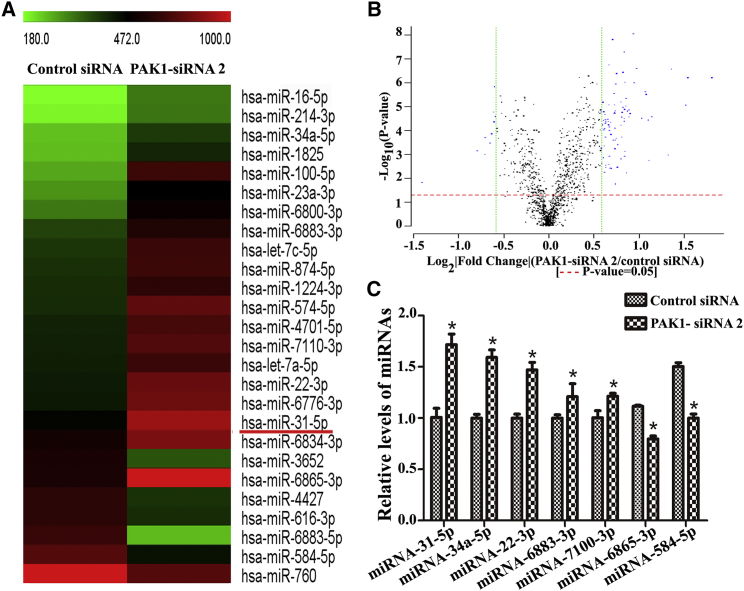
To identify the targets of PAK1, miRNA microarrays were performed to compare the global miRNA-expression patterns between PAK1 knockdown and the control siRNA. We utilized three pairs of small interfering RNAs (siRNAs) targeting different regions of PAK1 mRNA, and we found that the interfering effect of PAK1-siRNA2 is the most prominent.24 Total RNA was extracted from human SSCs at 24 hr after PAK1-siRNA2 and control siRNA treatment. Electropherogram showed the total RNA isolated from human SSC line with control siRNA (Figure S1A) and PAK1-siRNA2 (Figure S1B). The RIN (RNA integrity number) of control siRNA was 7.6, and the RIN of PAK1-siRNA2 was 8.0, which reflects a high quality of RNA used for miRNA microarrays. Histogram plot revealed fold-change distribution of all miRNA probes in human SSC line between PAK1-siRNA2 and control siRNA (Figure S1C).
miRNA microarrays displayed that there were 95 differentially expressed miRNAs with 1.5-fold changes or more in the human SSC line between the PAK1-siRNA2 and control siRNA. Among them, 85 miRNAs were increased significantly, whereas 10 miRNAs were decreased in human SSCs after PAK1-siRNA2 treatment. Hierarchical clustering analysis revealed distinct miRNA expression pattern in human SSCs after PAK1 knockdown (Figure 1A). Notably, the level of miRNA-31-5p was elevated by PAK1 silencing in human SSC line. Scatterplot showed the differential expression of miRNAs between PAK1-siRNA2 and control siRNA (Figure 1B). Real-time qPCR verified a number of the differentially expressed miRNAs identified by miRNA microarrays, including miRNA-31-5p, miRNA-34a-5p, miRNA-22-3p, miRNA-6883-3p, miRNA-7100-3p, miRNA-6865-3p, and miRNA-584-5p, in human SSCs after PAK1 knockdown (Figure 1C), which was consistent with the results of miRNA microarrays.
Figure 1.
Differentially Expressed miRNAs in Human SSC Line between PAK1-siRNA2 and the Control siRNA
(A) Hierarchical clustering illustrated the differentially expressed miRNAs in the human SSC line between PAK1-siRNA2 and the control siRNA. Red dots and green dots represented the upregulated and downregulated miRNAs, respectively. (B) Scatterplot shows the differentially expressed miRNAs in human SSC line between PAK1-siRNA2 and the control siRNA. Note: the selection criteria for the differentially expressed miRNAs (blue dots) was log2|fold change| ≥ 0.585 and p < 0.05. (C) Real-time qPCR showed the levels of miRNA-31-5p, miRNA-34a-5p, miRNA-22-3p, miRNA-6883-3p, miRNA-7100-3p, miRNA-6865-3p, and miRNA-584-5p in the human SSC line between PAK1-siRNA2 and the control siRNA. The asterisk indicates statistically significant differences (p < 0.05) in human SSC line between PAK1-siRNA2 and the control siRNA.
miRNA-31-5p Inhibits the Proliferation and DNA Synthesis of Human SSCs
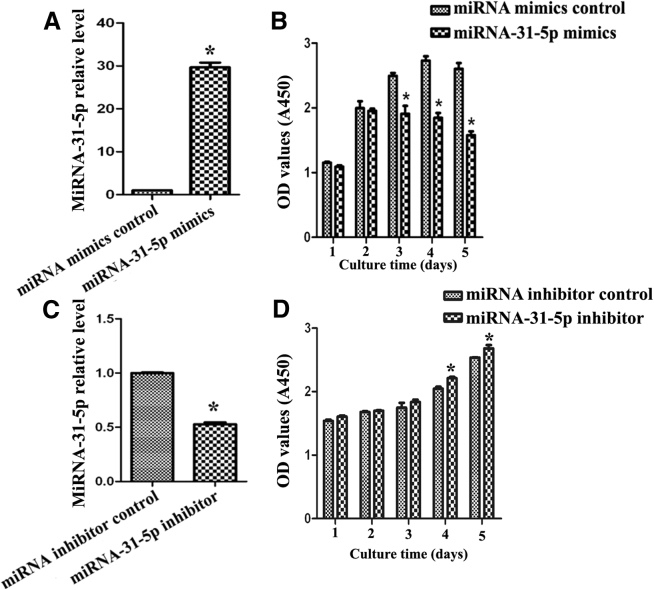
Since miRNA-31-5p level was significantly elevated by PAK1 knockdown, as revealed by miRNA microarrays and real-time qPCR, we further explored the function of miRNA-31-5p mimics and inhibitor in human SSCs. Real-time qPCR revealed that expression level of miRNA-31-5p in human SSC line was significantly increased by miRNA-31-5p mimics when compared with miRNA mimics control (Figure 2A). In contrast, miRNA-31-5p level was statistically decreased by miRNA-31-5p inhibitor compared to miRNA inhibitor control (Figure 2C). After 5 consecutive days of transfection of miRNA-31-5p mimics or inhibitor, cell counting kit-8 (CCK-8) assay was executed to detect the effect of miRNA-31-5p on the proliferation of human SSC line. Notably, miRNA-31-5p mimics suppressed the growth of the human SSC line compared with miRNA mimics control (Figure 2B). Conversely, miRNA-31-5p inhibitor enhanced the proliferation of human SSC line at day 4 and day 5 (Figure 2D).
Figure 2.
The Effect of Overexpression and Knockdown of miRNA-31-5p on the Proliferation of Human SSCs
(A and C) Real-time PCR showed the expression level of miRNA-31-5p in the human SSC line by miRNA-31-5p mimics and miRNA mimics control (A) as well as miRNA-31-5p inhibitor and miRNA inhibitor control (C). (B and D) CCK-8 assay indicated cell growth of the human SSC line by miRNA-31-5p mimics and miRNA mimics control (B) as well as miRNA-31-5p inhibitor and miRNA inhibitor control (D). (A-D) The asterisk indicates statistically significant differences (p < 0.05) between miRNA-31-5p mimics- or inhibitor-treated groups and their controls.
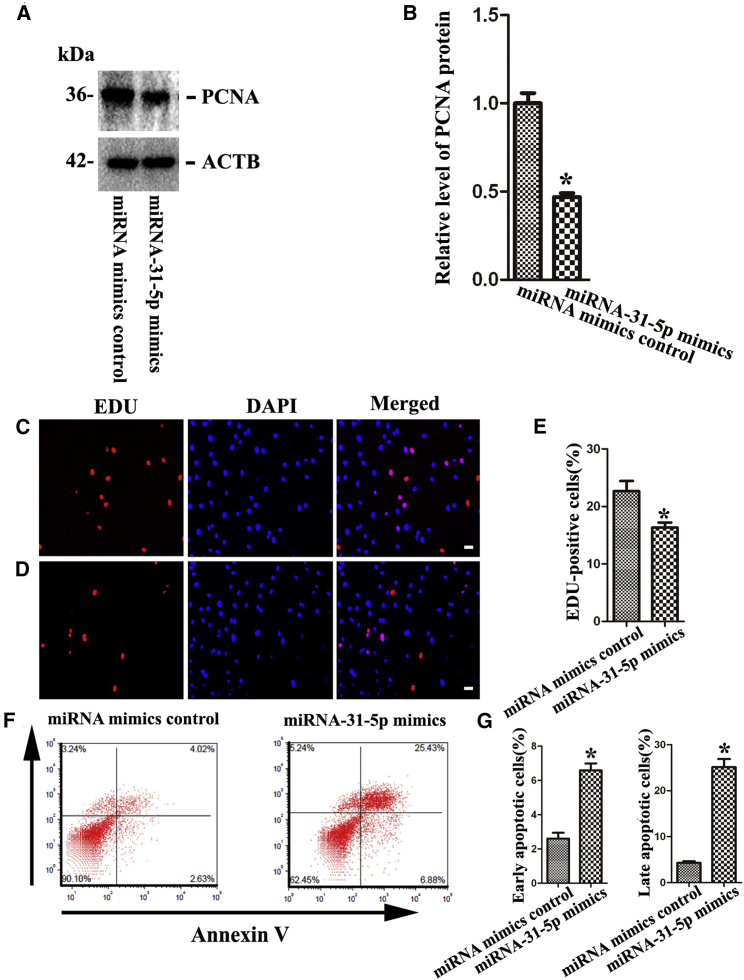
PCNA (proliferating cell nuclear antigen) has been widely recognized as a biomarker for cellular proliferation. Western blots indicated that the expression level of PCNA (Figures 3A and 3B) was obviously decreased by miRNA-31-5p mimics compared with miRNA mimics control in human SSC line. EDU (5-ethynyl-2′-deoxyuridine) incorporation assay demonstrated that the percentage of EDU-positive cells was reduced by miRNA-31-5p mimics compared to miRNA mimics control (Figures 3C–3E). No immunostaining was observed in human SSCs without anti-EDU (Figure S2). Collectively, these results implicate that miRNA-31-5p inhibits the proliferation and DNA synthesis of human SSCs.
Figure 3.
The Influence of miRNA-31-5p Mimics on DNA Synthesis and Apoptosis of Human SSCs
(A and B) Western blots displayed the expression of PCNA (A) and its relative level (B) in the human SSC line by miRNA-31-5p mimics and miRNA mimics control. (C–E) EDU incorporation assay illustrated the percentages of EDU-positive cells in human SSC line by miRNA mimics control (C and E) and miRNA-31-5p mimics (D and E). Scale bars in (C) and (D), 10 μm. (F and G) Annexin V/PI staining and flow cytometry showed the percentages of early apoptosis (F and G, left)and late apoptosis (F and G, right) of the human SSC line by miRNA mimics control and miRNA-31-5p mimics. (B, E, and G) The asterisk indicates the statistically significant differences (p < 0.05) between miRNA-31-5p mimics and miRNA mimics control.
miRNA-31-5p Promotes the Early and Late Apoptosis of Human SSCs
To determine the influence of miRNA-31-5p on the apoptosis of human SSC line, we conducted Annexin V/propidium iodide (PI) staining and flow cytometry. After transfection of miRNA-31-5p mimics, Annexin V/PI staining and flow cytometry revealed that the percentages of early and late apoptosis of human SSC line were obviously increased by miRNA-31-5p mimics compared with miRNA mimics control (Figures 3F and 3G). Considered together, these data indicate that miRNA-31-5p enhances the early and late apoptosis of human SSC cell line.
JAZF1 Is a Target of miRNA-31-5p in Human SSCs
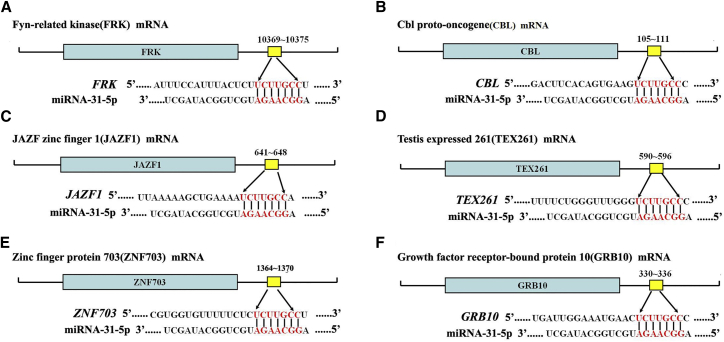
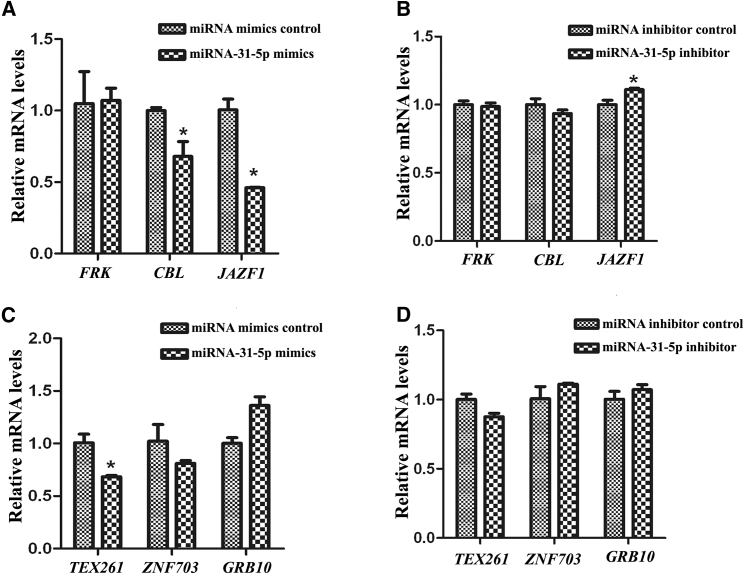
We further uncovered molecular mechanisms underlying the function of miRNA-31-5p in regulating human SSCs. We identified the targets of miRNA-31-5p since miRNAs act as epigenetic regulators by binding with the 3′ UTR of relative mRNAs. Using miRNA predict software Targetscan, we predicted that JAZF1, FRK, CBL, TEX261, ZNF703, and GRB10 were the potential binding targets of miRNA-31-5p. As shown in Figures 4A–4F and Table 1, the seed region (the second to eighth nucleotides) of miRNA-31-5p was base-paired with the 3′ UTR sequence of JAZF1, FRK, CBL, TEX261, ZNF703, and GRB10 mRNA.
Figure 4.
Identification of the Targets of miRNA-31-5p in Human SSCs
(A–F) Schematic diagram illustrated the binding sites of miRNA-31-5p to FRK (A), CBL (B), JAZF1 (C), TEX261 (D), ZNF703 (E), and GRB10 (F) mRNA.
Table 1.
The Predicted Target Genes of miRNA-31-5p Using TargetScan Software
| Gene Symbol | Gene Description | Predicted Target Region |
|---|---|---|
| FRK | fyn-related kinase | position 10,369–10,375 of FRK 3′ UTR |
| CBL | Cbl proto-oncogene | position 105–111 of CBL 3′ UTR |
| JAZF1 | JAZF zinc finger 1 | position 641–648 of JAZF1 3′ UTR |
| TEX261 | testis expressed 261 | position 590–596 of TEX261 3′ UTR |
| ZNF703 | zinc finger protein 703 | position 1,364–1,370 of ZNF703 3′ UTR |
| GRB10 | growth factor receptor-bound protein 10 | position 330–336 of GRB10 3′ UTR |
Furthermore, real-time qPCR revealed the levels of JAZF1 (Figure 5A), CBL (Figure 5A), and TEX261 (Figure 5C) were remarkably reduced by miRNA-31-5p mimics; nevertheless, only JAZF1 level (Figure 5B) was elevated by miRNA-31-5p inhibitor in human SSC line. In addition, there was no significance difference in the levels of FRK (Figures 5A and 5B), CBL (Figure 5B), ZNF703 (Figures 5C and 5D), TEX261 (Figure 5D), and GRB10 (Figures 5C and 5D) transcripts affected by miRNA-31-5p mimics or inhibitor. Together, these data reflect that JAZF1, rather than FRK, CBL, TEX261, ZNF703, or GRB10, is a target of miRNA-31-5p in human SSCs.
Figure 5.
The Effect of miRNA-31-5p Mimics and Inhibitor on the Expression Levels of Potential Targets in Human SSCs
(A–D) Real-time qPCR showed the expression changes of JAZF1 (A and B), CBL (A and B), FRK (A and B), TEX261 (C and D), ZNF703 (C and D), and GRB10 (C and D) in the human SSC line by miRNA-31-5p mimics or miRNA mimics control and miRNA-31-5p inhibitor or miRNA inhibitor control. (A–C) The asterisk indicates statistically significant differences (p < 0.05) between miRNA-31-5p mimics- or inhibitor-treated groups and their controls.
Transcription Factor JAZF1 Silencing Suppresses the Growth and Stimulates the Apoptosis of Human SSCs
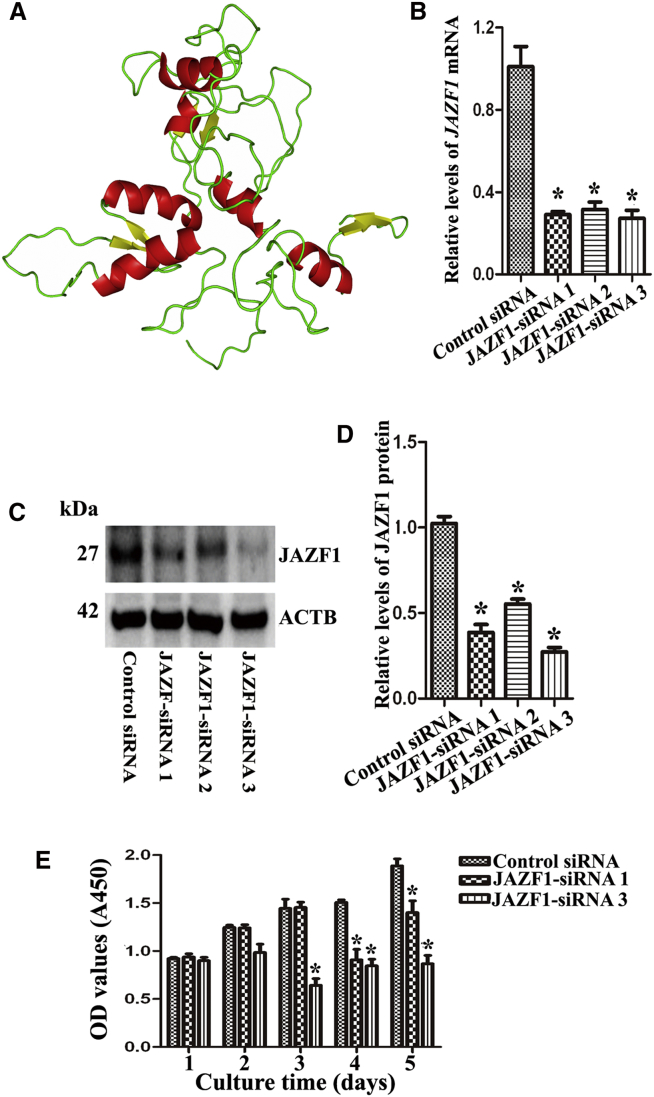
We next uncovered the structure of JAZF1 protein and explored the effect of transcription factor JAZF1 on the proliferation and the apoptosis of human SSC line. The three-dimensional (3D) structure model of JAZF1 protein was drawn and illustrated in Figure 6A. RNAi was utilized to silence JAZF1 in the human SSC line. Three pairs of JAZF1 siRNAs (i.e., JAZF1-siRNA-1, - 2, and -3) with different binding sites were employed to obtain sequence-specific siRNAs of JAZF1. Real-time PCR and Western blots showed that the levels of JAZF1 mRNA (Figure 6B) and JAZF1 protein (Figures 6C and 6D) were diminished by JAZF1-siRNA-1, -2, and -3 in human SSC line, respectively. CCK-8 assay revealed that the cell growth was reduced in the human SSC line by JAZF1-siRNA1 and JAZF1-siRNA3 (Figure 6E).
Figure 6.
The 3D Structure Model of JAZF1 Protein and Influence of JAZF1 Silencing on the Proliferation of Human SSCs
(A) The 3D structure model of JAZF1 protein. Notes: helix, beta-strand, and loop were shown in red, yellow, and green, respectively. This figure was drawn using Pymol (PyMOL Molecular Graphics System). (B) Real time qPCR showed mRNA changes of JAZF1 by JAZF1-siRNA-1, -2, and -3 in the human SSC line. (C and D) Western blots demonstrated protein expression of JAZF1 (C) and its relative levels (D) by JAZF1-siRNA-1, -2, and -3 in human SSC line. (E) CCK-8 assay revealed the growth of human SSC line after transfection of JAZF1-siRNA1 and JAZF1-siRNA3. (B, D, and E) The asterisk indicates statistically significant differences (p < 0.05) in the human SSC line between JAZF1-siRNAs and the control siRNA.
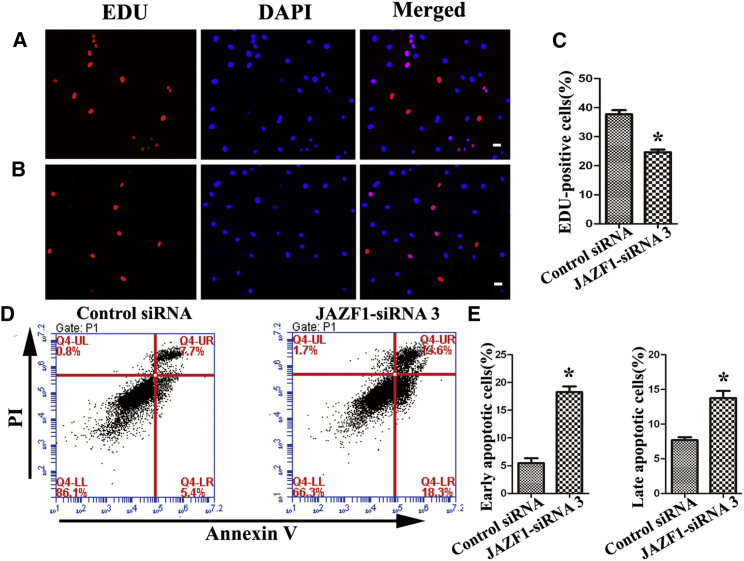
To determine the influence of JAZF1 on the DNA synthesis and apoptosis of human SSC line, an EDU incorporation assay and flow cytometry were performed. The EDU incorporation assay showed that the EDU-positive cells (Figures 7A–7C) were decreased by JAZF1-siRNA3 compared to the control siRNA in the human SSC line. No immunostaining was seen in human SSCs without an antibody to EDU (Figure S2). Furthermore, the early and late apoptotic percentages of the human SSC line were enhanced by JAZF1-siRNA3 (Figures 7D and 7E).
Figure 7.
The Effect of JAZF1 Silencing on DNA Synthesis and Apoptosis of Human SSCs
(A–C) EDU incorporation assay illustrated the percentages of EDU-positive cells affected by control siRNA (A and C) and JAZF1-siRNA3 (B and C) in the human SSC line. Scale bars in (A) and (B), 10 μm. (D and E) Annexin V/PI staining and flow cytometry showed the percentages of early apoptosis (D and E, left) and late apoptosis (D and E, right) in human SSC line treated with JAZF1-siRNA3 and control siRNA. (C and E) The asterisk indicates statistically significant differences (p < 0.05) in human SSC line between JAZF1-siRNA3 and the control siRNA.
miRNA-31-5p Mimics Decreases the Level of Cell Cycle Protein Cyclin A2 but Not Cyclin D1 or Cyclin E1 in Human SSCs
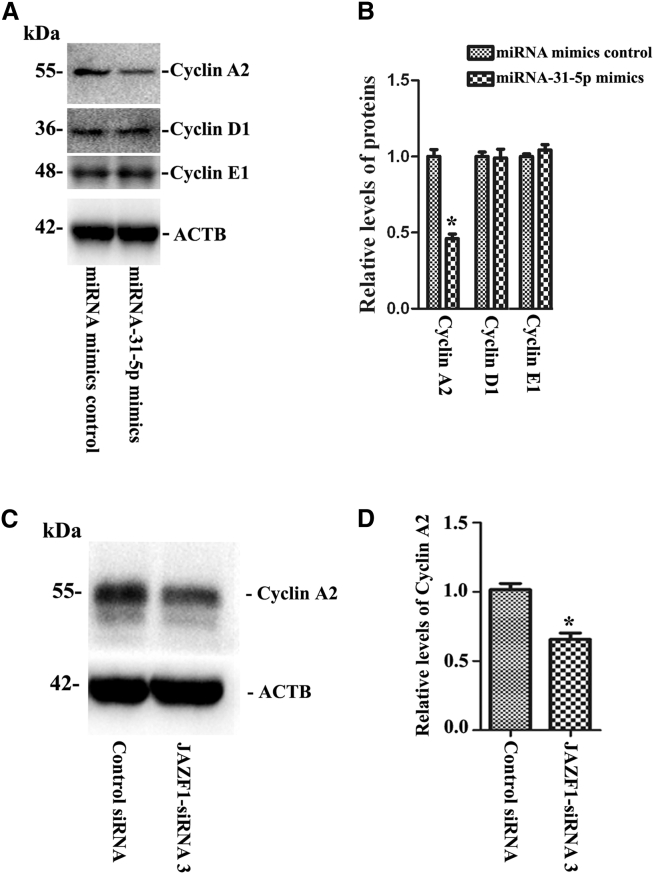
We finally asked whether miRNA-31-5p changed the levels of cell cycle proteins in fate determinations of human SSC line. Here, we examined several cell-cycle regulators, including cyclin A2, cyclin D1, and cyclin E1, in the human SSC line after transfection of miRNA-31-5p mimics. Western blots demonstrated that miRNA-31-5p mimics significantly decreased the expression level of cyclin A2 (Figures 8A and 8B). In addition, no statistical change of cyclin D1 (Figures 8A and 8B) or cyclin E1 (Figures 8A and 8B) was observed in human SSC line with treatment of miRNA-31-5p mimics and the mimics control. Therefore, miRNA-31-5p affects cyclin A2 but not cyclin D1 or cyclin E1 in human SSCs. Furthermore, cyclin A2 protein was significantly reduced by JAZF1-siRNA3 compared to control siRNA in human SSCs (Figures 8C and 8D).
Figure 8.
The Influence of miRNA-31-5p Mimics and JAZF1-siRNA 3 on the Levels of Cell Cycle Regulators in Human SSCs
(A and B) Western blots showed the expression of cyclin A2, cyclin D1, and cyclin E1 proteins (A) and their relative levels (B) in human SSCs by miRNA mimics control and miRNA-31-5p mimics. ACTB served as the control of the loading proteins. (B) The asterisk indicates statistically significant differences (p < 0.05) in the human SSC line between miRNA-31-5p mimics and miRNA mimics control. (C and D) Western blots revealed cyclin A2 protein (C) and its relative levels (D) affected by JAZF1-siRNA3 and control siRNA in human SSCs. ACTB was used as the control of the loading proteins. (D) The asterisk indicates statistically significant differences (p < 0.05) in the human SSC line between JAZF1-siRNA3 and the control siRNA.
Discussion
miRNAs have been recently examined in regulating the physiological process of spermatogenesis and the pathogenesis of male infertility, due to their significant functions and great application potentials. MiR-18 is abundantly expressed in the testis in a cell-type-specific manner during the spermatogenesis, and it controls the transcription factor of HSF2 (heat shock factor 2) in spermatogenesis and is involved in the differentiation and maturation of male germline cells.25 MiR-184 exists in the male germ cells from spermatogonia to round spermatids, and overexpression of miR-184 promotes the proliferation of the GC-1spg germ cell line via by targeting Ncor2 (nuclear receptor corepressor 2).26 Increasing evidence has indicated the effect of miRNAs on the self-renewal, differentiation, and apoptosis of mammalian SSCs.27 It has been reported that miR-21 is enriched in the Thy1+ SSCs,28 and it is essential for maintaining the number of SSCs through the regulation ETV5 (Ets variant gene 5).28 ETV5 is downstream of glial cell line-derived neurotrophic factor (GDNF) signaling, and it is required for SSC self-renewal and critical for fertility.29 We have demonstrated that miR-20 and miR-106a promote mouse SSC self-renewal via inhibiting STAT3 and Ccnd1 at a post-transcriptional level.30 MiR-135a contributes to rat SSC maintenance by targeting FoxO1,31 while miR-224 stimulates the differentiation of mouse SSCs by modulating DMRT1 (doublesex and Mab-3-related transcription factor 1).32 Additionally, miR-544 and miR-204 negatively regulate dairy goat male germline stem cell self-renewal via inhibiting PLZF (promyelocytic leukemia zinc finger gene) and Sirt1.33, 34 Furthermore, miR-34c facilitates the apoptosis of dairy goat male germline stem cells and suppresses their proliferation.35
To our knowledge, very little is known about the role of genetic and epigenetic factors in controlling human spermatogenesis and male infertility. We have recently demonstrated that PAK1 is regulated by epidermal growth factor (EGF) and that PAK1 promotes the proliferation and decreases apoptosis of human SSCs through PDK1-KDR-ZNF367 and ERK1/2-AKT.24 Notably, low levels of PAK1 may be associated with NOA patients.24 To unveil the epigenetic regulation of PAK1 in human SSCs, we performed a miRNA microarray and PAK1 knockdown. We found that 85 miRNAs were downregulated by PAK1 silencing, whereas 10 miRNAs were upregulated by PAK1 knockdown. It is worth noting that the miRNA-31-5p level was significantly increased by PAK1-siRNA2, which was verified by real-time PCR. These data suggest that miRNA-31-5p is negatively regulated by PAK1 in human SSCs.
MiR-31-5p has been reported to play vital roles in numerous human normal cells and malignant cells. It has been demonstrated that miRNA-31 expression level is decreased in the retinoblastoma, and overexpression of miRNA-31-5p represses the proliferation of Y79 cell line.36 Moreover, knockdown of miRNA-31-5p significantly inhibits the formation of a hypertrophic scar by targeting FIH (factor-inhibiting HIF-1) and regulating the HIF-1α pathway.37 However, the roles and molecular mechanism of miRNA-31-5p in regulating fate decisions of human SSCs remain unclear. To investigate the function of miRNA-31-5p in the proliferation and DNA synthesis of human SSCs, we conducted CCK-8, PCNA, and EDU assays. We observed that miRNA-31-5p mimics inhibited the growth of the human SSC line, whereas miRNA-31-5p inhibitors stimulated the proliferation of human SSCs. In addition, miRNA-31-5p mimics reduced the level of PCNA and the percentages of EDU-positive cells, indicating that miRNA-31-5p suppresses proliferation and DNA synthesis of human SSCs. Moreover, Annexin V/PI staining and flow cytometry revealed that miRNA-31-5p mimics enhanced the percentages of early and late apoptosis of the human SSC line, reflecting that miRNA-31-5p promotes the apoptosis of human SSCs.
Using the bioinformatics prediction and verification by real-time PCR, we reveal that JAZF1, rather than FRK, CBL, TEX261, ZNF703, or GRB10, is a target of miRNA-31-5p in human SSCs. Although the levels of CBL and TEX261 were reduced by miRNA-31-5p mimics in human SSCs, there is no significant difference in the transcripts of CBL and TEX261 between miRNA-31-5p inhibitor and miRNA inhibitor control, thus demonstrating that CBL and TEX26 might not be a target of miRNA-31-5p in human SSCs. JAZF1 is a nuclear protein with three C2H2-type zinc fingers, and here we illustrated the 3D structure of JAZF1 protein. CCK-8 and EDU incorporation assays demonstrated that the proliferation and EDU-positive cells were decreased by JAZF1 knockdown in the human SSC line, whereas Annexin V/PI staining and flow cytometry displayed that JAZF1 silencing led to an increase in the percentages of early and late apoptosis of the human SSC line, which was consistent with the function of miRNA-31-5p. Taken together, these data suggest that JAZF1 is a binding target of miRNA-31-5p in human SSCs. Since no information is available for JAZF1 in controlling male germ cells or stem cells, the role and mechanisms of JAZF1 in regulating the fate determinations of SSCs need to be explored further. Cyclin-related proteins change with cell cycle’s progression and thus pattern of cyclins could reflect the specific cell-cycle stage. Here, we investigated the changes of cell-cycle regulators, including cyclin A2, cyclin D1, and cyclin E1, and we observed that miRNA-31-5p mimics could decrease the expression level of cyclin A2 but not cyclin D1 and cyclin E1, suggesting that miRNA-31-5p negatively mediates cyclin A2.
The pathogenesis of NOA patients remains largely unknown. The balance between self-renewal and differentiation of SSCs maintains their number as well as the normal amount of sperm. For example, transgenic mice with one GDNF null allele show significant reduction of undifferentiated spermatogonia, including SSCs, whereas excessive accumulation and improper response to differentiation signals are observed in GDNF-overexpressing mice.38 The molecules produced by Sertoli cells, e.g., GDNF, fibroblast growth factor 2 (FGF2), ETV5, nociceptin, neuregulin 1 (NRG1), and androgen receptor (AR), have been implicated as the most important factors that regulate SSC self-renewal.39 Accumulating evidence indicates the impaired spermatogonial vitality in the NOA population. Both the spermatogonia and SSCs are reduced in the NOA population,40 and SSCs from the NOA population assume a decreased proliferation capacity compared to the cells from the OA population.41 We have recently demonstrated that the level of PAK1 is lower in NOA patients than OA patients.24 In this study, we found that the miRNA-31-5p level was elevated by PAK1 knockdown. These data reflect that high levels of miRNA-31-5p may be associated with male infertility.
In summary, we have found that PAK1 negatively regulates miRNA-31-5p, and we have demonstrated that miRNA-31-5p inhibits the proliferation and DNA synthesis of human SSCs but facilitates the early and late apoptosis of human SSCs via targeting JAZF1 and cyclin A2. Therefore, this study offers a new epigenetic mechanism underlying the fate determinations of human SSCs, and it may provide novel targets for molecular therapy of male infertility.
Materials and Methods
Human SSC Line and Cell Culture
A human SSC line stably expressing human SV40 large T antigen under the control of the EF1α promoter was established previously by our research group.13 This cell line expresses a number of human germ cell and SSC hallmarks, including VASA, DAZL, MAGEA4, GFRA1, RET, UCHL1, GPR125, PLZF, and THY1,13 indicating that these cells are human SSCs phenotypically.
Human SSC line was cultured with DMEM/Nutrient Mixture F12 (DMEM/F12; Gibco Laboratories, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; Gibco Laboratories, Grand Island, NY) and 100 units/mL penicillin and streptomycin (Invitrogen). The cells were passaged every 3–4 days using 0.05% trypsin (Invitrogen) and 0.53 mM EDTA (Invitrogen), and they were maintained at 34°C in a humidified 5% CO2 incubator.
miRNA Microarrays
Total RNA was isolated from the human SSC line treated with PAK1-siRNA2 or control siRNA using RNAiso Plus reagent (Takara, Kusatsu, Japan). DNase I was used to remove potential genomic DNA contamination. The RIN was calculated to evaluate RNA integrity using Agilent Bioanalyzer. miRNA enrichment was performed from total RNA utilizing Phalanx miRNA OneArray system. The enriched RNA was labeled with ULS from KREATECH on miRNA OneArray. The labeled targets were purified with KREApure column. Labeling efficiency and yield of recovered labeled RNA were quantified via NanoDrop ND-1000 Spectrophotometer (Thermo Scientific, Massachusetts, USA).
Pre-hybridization was conducted on microarrays before being hybridized with labeled targets. The microarrays were rinsed with 100% ethanol and deionized water and incubated at 42°C with pre-hybridization solution for 2 hr in dark. After washing with deionized water, miRNA OneArray hybridization was performed and scanned by AXON4000B from Molecular Devices. Raw data (gpr files) were generated from GenePix.4 with the images (tiff files). Data analysis of gene expression profiling, including data filtering, normalization, and statistical calculations, was processed by R version 2.12.1. Arrays (including technical replicates) of any compared sample set were normalized after filtering probes according to flag note from gpr files. The log2(fold change) was calculated by pairwise combination and error-weighted average. The differentially expressed miRNAs were selected according to log2(ratio) and p value with the criteria |log2(fold change)| ≥ 0.585 and p value < 0.05.
Transfection of PAK1-siRNA2, JAZF1 siRNAs, and miRNA-31-5p Mimics and Inhibitor into Human SSC Line
The sequences of JAZF1-siRNA-1, -2, and -3, miRNA-31-5p miRNA mimics and inhibitor, miRNA mimics control, and miRNA inhibitor control are listed in Table 2 and synthesized from GenePharma (Shanghai, China), and PAK1-siRNA2 sequence was indicated previously.24 Control siRNA without targeting any sequence of these genes was used as a negative control.
Table 2.
The Sequences for RNA Oligos
| RNA Oligo Names | RNA Oligo Sequences |
|---|---|
| JAZF1-siRNA1 | sense 5′-CCGACCUCAUCGAGCACAUTT-3′ |
| antisense 5′-AUGUGCUCGAUGAGGUCGGTT-3′ | |
| JAZF1-siRNA2 | sense 5′-GGCACCACACAAUCAAUUUTT-3′ |
| antisense 5′-AAAUUGAUUGUGUGGUGCCTT-3′ | |
| JAZF1-siRNA3 | sense 5′-CCAUCAGCUCCGAAGCCAUTT-3′ |
| antisense 5′-AUGGCUUCGGAGCUGAUGGTT-3′ | |
| miRNA-31-5p mimics | sense 5′-AGGCAAGAUGCUGGCAUAGCU-3′ |
| antisense 5′CUAUGCCAGCAUCUUGCCUUU-3′ | |
| miRNA mimics control | sense 5′-UUCUCCGAACGUGUCACGUTT-3′ |
| antisense 5′-ACGUGACACGUUCGGAGAATT-3′ | |
| miRNA-31-5p inhibitor | 5′-AGCUAUGCCAGCAUCUUGCCU-3′ |
| miRNA inhibitor control | 5′-CAGUACUUUUGUGUAGUACAA-3′ |
Human SSCs were seeded at 1 × 105/cm2 density and cultured in DMEM/F12 supplemented with 10% FBS overnight, and 100 nM PAK1-siRNA2, JAZF1-siRNA-1, -2, and -3, or control siRNA were transfected into human SSC line through lipofectamine 3000 (Life Technologies, Carlsbad, USA) according to manufacturer’s protocol. The efficiency of PAK1-siRNA2 was verified previously.24 Human SSC line was classified into four groups in terms of transfecting different miRNAs: (1) miRNA mimics control, (2) miRNA-31-5p mimics, (3) miRNA inhibitor control, and (4) miRNA-31-5p inhibitor. Transfection of 100 nM miRNA-31-5p miRNA mimics or inhibitor was conducted using lipofectamine 3000 transfection agent according to manufacturer’s protocol.
miRNA RT-PCR and Real-Time PCR
miRNA reverse transcription (RT) was conducted in the human SSC line treated with PAK1-siRNA2 or control siRNA and miRNA-31-5p miRNA mimics or inhibitor using TransScript miRNA First Strand cDNA Synthesis SuperMix (Transgen, China) according to the manufacturer’s protocol. Each RT reaction was composed of 100 ng RNA, 1 μL of miRNA RT Enzyme Mix, 10 μL of 2× TS miRNA Reaction Mix, and RNase-free water in a total volume of 20 μL. Reactions were performed in a Veriti 96-Well Thermal Cycler (Applied Biosystems) for 60 min at 37°C and followed by heat inactivation of RT for 5 s at 85°C. RT reaction mix was diluted by 5 times in nuclease-free water and held at −20°C. The primer sequences of miRNAs, including miR-31-5p, miRNA-34a-5p, miRNA-22-3p, miRNA-6883-3p, miRNA-7100-3p, miRNA-6865-3p, and miRNA-584-5p, were designed and listed in Table 3. Real-time PCR was performed in triplicate using Power SYBR Green PCR Master Mix (Applied Biosystems, Woolston Warrington, UK) and a 7500 Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA) according to the protocol as described previously. To quantify the PCR products, the comparative CT (threshold cycle) method was used as described preciously.42 The CT values of miRNAs were normalized to those of U6 (ΔCT = CT(miRNA) − CT(U6)), and the relative expression of miRNAs to the control was calculated by the formula 2−ΔΔCT(ΔΔCT = ΔCT(miRNA) − ΔCT(control)).
Table 3.
The Sequences of miRNA Primers for Real-Time PCR
| Gene Names | Primer Sequences (5′-3′) | Annealing Temperature (°C) |
|---|---|---|
| miRNA-31-5p | AGGCAAGATGCTGGCATAG | 60 |
| miRNA-34a-5p | TGGCAGTGTCTTAGCTGGTT | 60 |
| miRNA-22-3p | AAGCTGCCAGTTGAAGAACTGT | 60 |
| U6 | GCTCGCTTCGGCAGCACATAT | 60 |
| miRNA-584-5p | TTATGGTTTGCCTGGGACTGAG | 60 |
| primer-reverse | GATCGCCCTTCTACGTCGTAT | 60 |
| miRNA-6865-3p | ACACCCTCTTTCCCTACCG | 60 |
| miRNA-7110-3p | TCTCTCTCCCACTTCCCTG | 60 |
Real-Time qPCR
RNA was extracted from the human SSC line treated with miRNA-31-5p mimics or inhibitor using the RNAiso Plus reagent (Takara, Kusatsu, Japan). The quality and concentrations of total RNA were measured by Nanodrop (Thermo Scientific), and the ratios of A260/A280 of total RNA were set as 1.9–2.0 to ensure good quality. The First Strand cDNA Synthesis Kit (Thermo Scientific) was used to conduct RT of total RNA. The primer sequences of selected genes used for real-time PCR were designed and listed in Table 4. Real-time PCR reactions were conducted using Power SYBR Green PCR Master Mix (Applied Biosystems, Woolston Warrington, UK) and a 7500 Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). To quantify the PCR products, the comparative CT (threshold cycle) method was used as described above. The CT values of genes were normalized to those of housekeeping gene ACTB (ΔCT = CT(GENE) − CT(ACTB)), and the relative expression of genes to the control was calculated by the formula 2−ΔΔCT(ΔΔCT = ΔCT(GENE) − ΔCT(control)).
Table 4.
The Sequences of Gene Primers Used for Real-Time PCR
| Gene Names | Primer Sequences (5′-3′) | PCR Product Size (bp) | Annealing Temperature (°C) |
|---|---|---|---|
| FRK | F: GAGATAGACCGCAACTCCATAC | 81 | 60 |
| R: GTTCCACAGACCTTCCCATAC | |||
| CBL | F: AGGGAGACACATTTCGGATTAC | 91 | 60 |
| R: AAGCTCTTCCAAGGGACTATTG | |||
| JAZF1 | F: CGCCTCCTTCTTCTCCAATAC | 96 | 60 |
| R: ATGTGGTTGTCCTCGATGTG | |||
| TEX261 | F: CTGACCTCGCCTAACTTCATC | 104 | 60 |
| R: AGGACCTCTGAGAAGGGATAAT | |||
| ZNF703 | F: CCTATGGCAAGAGCCACTTAT | 87 | 60 |
| R: GCGCGTATGGCGAATAGTA | |||
| GRB10 | F: CTGTGCCAATTGCTGGTTTAC | 102 | 60 |
| R: GTCTTCCAAGCACCTCTCTAATC | |||
| R: CTTGCTGTAGGTCAGGCACT | |||
| R: CGGGTGGTTTAGGTTCTGTTT | |||
| ACTB | F: CACTCTTCCAGCCTTCCTTC | 104 | 60 |
| R: GTACAGGTCTTTGCGGATGT |
3D Structure Model of JAZF1 Protein
The homology model of JAZF1 (UniProtKB: Q86VZ6.2) was constructed by using the RaptorX server based on amino-acid sequence alignment with zinc finger protein 568 and its structure (PDB: 5V3M). The p value of 2.09e−09 and the uGDT of 77 indicated that the overall quality of the 3D structure model was reliable.
Western Blots
The human SSC line treated with JAZF1-siRNAs or control siRNA and miRNA-31-5p mimics or miRNA mimics control were lysed with RIPA buffer (DingGuoChangSheng Biotech, Bengjing, China) containing protease inhibitor cocktail (MedChemExpress, USA). After 30 min lysis on ice, cell lysates were cleared by centrifugation at 12,000 × g, and the concentrations of proteins were measured by Pierce BCA Protein Assay Kit (Thermo Scientific, USA). Forty micrograms of cell lysate from each sample were used for SDS-PAGE (Bio-Rad Laboratories, Richmond, CA), and Western blots were performed according to the protocol as described previously. The chosen antibodies included PCNA (Abcam), JAZF1 (Sigma-Aldrich, USA), cyclin A2 (Santa Cruz), cyclin D1 (Santa Cruz), cyclin E1 (Santa Cruz), and ACTB (Proteintech). After extensive washes, the membranes were incubated with horseradish peroxidase-conjugated immunoglobulin G (IgG) (Santa Cruz Biotechnology) at 1:2,000 dilution for 1 hr at room temperature. The blots were detected by Chemi-Doc XRS system (Bio-Rad Laboratories, Hercules, CA, USA), and densitometric analyses were processed with ImageJ software. The relative levels of proteins were normalized to the expression of ACTB.
CCK-8 Assay
The human SSC line was plated at a density of 3,000–5,000 cells/well in 96-well plates, and they were tranfected without or with 100 nM of miRNA-31-5p mimics, miRNA-31-5p inhibitor or corresponding control RNA oligos, and JAZF1-siRNA1, JAZF1-siRNA3, or control siRNA. The proliferation capacity of human SSC line was detected by CCK-8 assay (Dojin Laboratories, Kumamoto, Japan) according to the manufacturer’s instruction. In brief, 10 μL of CCK-8 reagents were added into each well of the cells. After 5 days of culture, microplate reader (Thermo Scientific, USA) was used to measure the absorbance at the wavelength of 450 nm.
EDU Incorporation Assay
The human SSC line was seeded at a density of 5,000 cells/well in a 96-well plate containing DMEM/F12 medium, and they were transfected with miRNA-31-5p mimics or miRNA mimics control, and JAZF1-siRNA3 or control siRNA. The culture medium was added with 50 μM EDU (RiboBio, Guangzhou, China). After 12 hr of culture, the cells were washed with DMEM and fixed with 4% paraformaldehyde (PFA). Cells were neutralized with 2 mg/mL glycine and permeabilized in 0.5% Triton X-100 for 10 min at room temperature. EDU antibody was replaced by PBS and served as a negative control. EDU immunostaining was performed with Apollo staining reaction buffer. The nuclei of cells were stained with DAPI, and the EDU-positive cells were counted from at least 500 cells under fluorescence microscopy (Nikon, Tokyo, Japan).
FITC-Annexin V/PI Staining and Flow Cytometry
The apoptosis percentages of the human SSC line affected by miRNA-31-5p mimic and JAZF1-siRNA3 were measured through Annexin V-fluorescein isothiocyanate (FITC)/PI kit by flow cytometry pursuant to the manufacturer’s instruction (Biolegend, London, UK). After transfection of miRNA-31-5p mimics, miRNA mimics control, JAZF1-siRNA3 or control siRNA for 3 days, cells were collected and simultaneously stained with Annexin V-FITC (green fluorescence) and the nonvital dye PI (red fluorescence), which allowed the discrimination of intact cells (FITC−PI−), early apoptotic (FITC+PI−) and late apoptotic or necrotic cells (FITC+PI+). The procedure was carried out according to the protocol as described previously.43
Statistical Analysis
All data were shown as mean (bars) ± SD (error bars) and analyzed by Student’s t test or one-way ANOVA with the appropriate post-hoc tests using GraphPad Prism5.0 (GraphPad Software). Normality and homogeneity of variances were checked prior to carry out Student’s t test or one way ANOVA, and p < 0.05 was considered statistically significant.
Author Contributions
H.F. performed the experiments, wrote the manuscript, and helped with data analysis. F.Z. assisted with the experiments; W.Z. assisted with the experiments; Q.Y. assisted with the experiments and ordered reagents; Q.Q. and X.Y. assisted with the experiments; Z.H. was responsible for the conception and design, supervision of all aspects of the laboratory experiments, data analysis, writing the manuscript, and final approval of the manuscript. All authors approved the manuscript.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
This work was supported by grants from the National Nature Science Foundation of China (31671550, 31872845, 31230048), Chinese Ministry of Science and Technology (2016YFC1000606, 2014CB943101), and Shanghai Hospital Development Center (SHDC12015122).
Footnotes
Supplemental Information includes two figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.11.004.
Supplemental Information
References
- 1.Kanatsu-Shinohara M., Shinohara T. Spermatogonial stem cell self-renewal and development. Annu. Rev. Cell Dev. Biol. 2013;29:163–187. doi: 10.1146/annurev-cellbio-101512-122353. [DOI] [PubMed] [Google Scholar]
- 2.Komeya M., Ogawa T. Spermatogonial stem cells: Progress and prospects. Asian J. Androl. 2015;17:771–775. doi: 10.4103/1008-682X.154995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Rooij D.G. The nature and dynamics of spermatogonial stem cells. Development. 2017;144:3022–3030. doi: 10.1242/dev.146571. [DOI] [PubMed] [Google Scholar]
- 4.Crisóstomo L., Alves M.G., Gorga A., Sousa M., Riera M.F., Galardo M.N., Meroni S.B., Oliveira P.F. Molecular Mechanisms and Signaling Pathways Involved in the Nutritional Support of Spermatogenesis by Sertoli Cells. Methods Mol. Biol. 2018;1748:129–155. doi: 10.1007/978-1-4939-7698-0_11. [DOI] [PubMed] [Google Scholar]
- 5.Garcia T.X., Hofmann M.C. Regulation of germ line stem cell homeostasis. Anim. Reprod. 2015;12:35–45. [PMC free article] [PubMed] [Google Scholar]
- 6.Tuttelmann F., Ruckert C., Ropke A. Disorders of spermatogenesis: Perspectives for novel genetic diagnostics after 20 years of unchanged routine. Med. Genet. 2018;30:12–20. doi: 10.1007/s11825-018-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermann B.P., Sukhwani M., Hansel M.C., Orwig K.E. Spermatogonial stem cells in higher primates: are there differences from those in rodents? Reproduction. 2010;139:479–493. doi: 10.1530/REP-09-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boitani C., Di Persio S., Esposito V., Vicini E. Spermatogonial cells: mouse, monkey and man comparison. Semin. Cell Dev. Biol. 2016;59:79–88. doi: 10.1016/j.semcdb.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Medrano J.V., Rombaut C., Simon C., Pellicer A., Goossens E. Human spermatogonial stem cells display limited proliferation in vitro under mouse spermatogonial stem cell culture conditions. Fertil. Steril. 2016;106:1539–1549.e8. doi: 10.1016/j.fertnstert.2016.07.1065. [DOI] [PubMed] [Google Scholar]
- 10.DenBesten P.K., Gao C., Li W., Mathews C.H., Gruenert D.C. Development and characterization of an SV40 immortalized porcine ameloblast-like cell line. Eur. J. Oral Sci. 1999;107:276–281. doi: 10.1046/j.0909-8836.1999.eos107407.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang H., Wen L., Yuan Q., Sun M., Niu M., He Z. Establishment and applications of male germ cell and Sertoli cell lines. Reproduction. 2016;152:R31–R40. doi: 10.1530/REP-15-0546. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann M.C., Braydich-Stolle L., Dettin L., Johnson E., Dym M. Immortalization of mouse germ line stem cells. Stem Cells. 2005;23:200–210. doi: 10.1634/stemcells.2003-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou J., Niu M., Liu L., Zhu Z., Wang X., Sun M., Yuan Q., Yang S., Zeng W., Liu Y. Establishment and Characterization of Human Germline Stem Cell Line with Unlimited Proliferation Potentials and no Tumor Formation. Sci. Rep. 2015;5:16922. doi: 10.1038/srep16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wosnitzer M., Goldstein M., Hardy M.P. Review of Azoospermia. Spermatogenesis. 2014;4:e28218. doi: 10.4161/spmg.28218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esteves S.C. Clinical management of infertile men with nonobstructive azoospermia. Asian J. Androl. 2015;17:459–470. doi: 10.4103/1008-682X.148719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunes S., Arslan M.A., Hekim G.N.T., Asci R. The role of epigenetics in idiopathic male infertility. J. Assist. Reprod. Genet. 2016;33:553–569. doi: 10.1007/s10815-016-0682-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carthew R.W., Sontheimer E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tahmasbpour E., Balasubramanian D., Agarwal A. A multi-faceted approach to understanding male infertility: gene mutations, molecular defects and assisted reproductive techniques (ART) J. Assist. Reprod. Genet. 2014;31:1115–1137. doi: 10.1007/s10815-014-0280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Procópio M.S., de Avelar G.F., Costa G.M.J., Lacerda S.M.S.N., Resende R.R., de França L.R. MicroRNAs in Sertoli cells: implications for spermatogenesis and fertility. Cell Tissue Res. 2017;370:335–346. doi: 10.1007/s00441-017-2667-z. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi K., Chuva de Sousa Lopes S.M., Kaneda M., Tang F., Hajkova P., Lao K., O’Carroll D., Das P.P., Tarakhovsky A., Miska E.A., Surani M.A. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS ONE. 2008;3:e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romero Y., Meikar O., Papaioannou M.D., Conne B., Grey C., Weier M., Pralong F., De Massy B., Kaessmann H., Vassalli J.D. Dicer1 depletion in male germ cells leads to infertility due to cumulative meiotic and spermiogenic defects. PLoS ONE. 2011;6:e25241. doi: 10.1371/journal.pone.0025241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmermann C., Romero Y., Warnefors M., Bilican A., Borel C., Smith L.B., Kotaja N., Kaessmann H., Nef S. Germ cell-specific targeting of DICER or DGCR8 reveals a novel role for endo-siRNAs in the progression of mammalian spermatogenesis and male fertility. PLoS ONE. 2014;9:e107023. doi: 10.1371/journal.pone.0107023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin Y., Xia Y., Wu W., Han X., Lu C., Ji G., Chen D., Wang H., Song L., Wang S., Wang X. Genetic variants in microRNA biogenesis pathway genes are associated with semen quality in a Han-Chinese population. Reprod. Biomed. Online. 2012;24:454–461. doi: 10.1016/j.rbmo.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Fu H., Zhang W., Yuan Q., Niu M., Zhou F., Qiu Q., Mao G., Wang H., Wen L., Sun M. PAK1 Promotes the Proliferation and Inhibits Apoptosis of Human Spermatogonial Stem Cells via PDK1/KDR/ZNF367 and ERK1/2 and AKT Pathways. Mol. Ther. Nucleic Acids. 2018;12:769–786. doi: 10.1016/j.omtn.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Björk J.K., Sandqvist A., Elsing A.N., Kotaja N., Sistonen L. miR-18, a member of Oncomir-1, targets heat shock transcription factor 2 in spermatogenesis. Development. 2010;137:3177–3184. doi: 10.1242/dev.050955. [DOI] [PubMed] [Google Scholar]
- 26.Wu J., Bao J., Wang L., Hu Y., Xu C. MicroRNA-184 downregulates nuclear receptor corepressor 2 in mouse spermatogenesis. BMC Dev. Biol. 2011;11:64. doi: 10.1186/1471-213X-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harchegani A.B., Shafaghatian H., Tahmasbpour E., Shahriary A. Regulatory Functions of MicroRNAs in Male Reproductive Health: A New Approach to Understanding Male Infertility. Reprod. Sci. 2018;Jan 1 doi: 10.1177/1933719118765972. 1933719118765972. [DOI] [PubMed] [Google Scholar]
- 28.Niu Z., Goodyear S.M., Rao S., Wu X., Tobias J.W., Avarbock M.R., Brinster R.L. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA. 2011;108:12740–12745. doi: 10.1073/pnas.1109987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jamsai D., Clark B.J., Smith S.J., Whittle B., Goodnow C.C., Ormandy C.J., O’Bryan M.K. A missense mutation in the transcription factor ETV5 leads to sterility, increased embryonic and perinatal death, postnatal growth restriction, renal asymmetry and polydactyly in the mouse. PLoS ONE. 2013;8:e77311. doi: 10.1371/journal.pone.0077311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Z., Jiang J., Kokkinaki M., Tang L., Zeng W., Gallicano I., Dobrinski I., Dym M. miRNA-20 and mirna-106a regulate spermatogonial stem cell renewal at the post-transcriptional level via targeting STAT3 and Ccnd1. Stem Cells. 2013;31:2205–2217. doi: 10.1002/stem.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moritoki Y., Hayashi Y., Mizuno K., Kamisawa H., Nishio H., Kurokawa S., Ugawa S., Kojima Y., Kohri K. Expression profiling of microRNA in cryptorchid testes: miR-135a contributes to the maintenance of spermatogonial stem cells by regulating FoxO1. J. Urol. 2014;191:1174–1180. doi: 10.1016/j.juro.2013.10.137. [DOI] [PubMed] [Google Scholar]
- 32.Cui N., Hao G., Zhao Z., Wang F., Cao J., Yang A. MicroRNA-224 regulates self-renewal of mouse spermatogonial stem cells via targeting DMRT1. J. Cell. Mol. Med. 2016;20:1503–1512. doi: 10.1111/jcmm.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song W., Mu H., Wu J., Liao M., Zhu H., Zheng L., He X., Niu B., Zhai Y., Bai C. miR-544 Regulates Dairy Goat Male Germline Stem Cell Self-Renewal via Targeting PLZF. J. Cell. Biochem. 2015;116:2155–2165. doi: 10.1002/jcb.25172. [DOI] [PubMed] [Google Scholar]
- 34.Niu B., Wu J., Mu H., Li B., Wu C., He X., Bai C., Li G., Hua J. miR-204 Regulates the Proliferation of Dairy Goat Spermatogonial Stem Cells via Targeting to Sirt1. Rejuvenation Res. 2016;19:120–130. doi: 10.1089/rej.2015.1719. [DOI] [PubMed] [Google Scholar]
- 35.Li M., Yu M., Liu C., Zhu H., He X., Peng S., Hua J. miR-34c works downstream of p53 leading to dairy goat male germline stem-cell (mGSCs) apoptosis. Cell Prolif. 2013;46:223–231. doi: 10.1111/cpr.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montoya V., Fan H., Bryar P.J., Weinstein J.L., Mets M.B., Feng G., Martin J., Martin A., Jiang H., Laurie N.A. Novel miRNA-31 and miRNA-200a-Mediated Regulation of Retinoblastoma Proliferation. PLoS ONE. 2015;10:e0138366. doi: 10.1371/journal.pone.0138366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X., Zhang Y., Jiang B.H., Zhang Q., Zhou R.P., Zhang L., Wang C. Study on the role of Hsa-miR-31-5p in hypertrophic scar formation and the mechanism. Exp. Cell Res. 2017;361:201–209. doi: 10.1016/j.yexcr.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Meng X., Lindahl M., Hyvönen M.E., Parvinen M., de Rooij D.G., Hess M.W., Raatikainen-Ahokas A., Sainio K., Rauvala H., Lakso M. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 39.Chen S.R., Liu Y.X. Regulation of spermatogonial stem cell self-renewal and spermatocyte meiosis by Sertoli cell signaling. Reproduction. 2015;149:R159–R167. doi: 10.1530/REP-14-0481. [DOI] [PubMed] [Google Scholar]
- 40.Hentrich A., Wolter M., Szardening-Kirchner C., Lüers G.H., Bergmann M., Kliesch S., Konrad L. Reduced numbers of Sertoli, germ, and spermatogonial stem cells in impaired spermatogenesis. Mod. Pathol. 2011;24:1380–1389. doi: 10.1038/modpathol.2011.97. [DOI] [PubMed] [Google Scholar]
- 41.Lim J.J., Sung S.Y., Kim H.J., Song S.H., Hong J.Y., Yoon T.K., Kim J.K., Kim K.S., Lee D.R. Long-term proliferation and characterization of human spermatogonial stem cells obtained from obstructive and non-obstructive azoospermia under exogenous feeder-free culture conditions. Cell Prolif. 2010;43:405–417. doi: 10.1111/j.1365-2184.2010.00691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao C., Sun M., Yuan Q., Niu M., Chen Z., Hou J., Wang H., Wen L., Liu Y., Li Z., He Z. miRNA-133b promotes the proliferation of human Sertoli cells through targeting GLI3. Oncotarget. 2016;7:2201–2219. doi: 10.18632/oncotarget.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H., Yuan Q., Sun M., Niu M., Wen L., Fu H., Zhou F., Chen Z., Yao C., Hou J. BMP6 Regulates Proliferation and Apoptosis of Human Sertoli Cells Via Smad2/3 and Cyclin D1 Pathway and DACH1 and TFAP2A Activation. Sci. Rep. 2017;7:45298. doi: 10.1038/srep45298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.