Abstract
Both common and rare genetic variants of laccase domain-containing 1 (LACC1, previously C13orf31) are associated with inflammatory bowel disease, leprosy, Behcet disease, and systemic juvenile idiopathic arthritis. However, the functional relevance of these variants is unclear. In this study, we use LACC1-deficient mice to gain insight into the role of LACC1 in regulating inflammation. Following oral administration of Citrobacter rodentium, LACC1 knockout (KO) mice had more severe colon lesions compared with wildtype (WT) controls. Immunization with collagen II, a collagen-induced arthritis (CIA) model, resulted in an accelerated onset of arthritis and significantly worse arthritis and inflammation in LACC1 KO mice. Similar results were obtained in a mannan-induced arthritis model. Serum and local TNF in CIA paws and C. rodentium colons were significantly increased in LACC1 KO mice compared with WT controls. The percentage of IL-17A–producing CD4+ T cells was elevated in LACC1 KO mice undergoing CIA as well as aged mice compared with WT controls. Neutralization of IL-17, but not TNF, prevented enhanced mannan-induced arthritis in LACC1 KO mice. These data provide new mechanistic insight into the function of LACC1 in regulating TNF and IL-17 during inflammatory responses. We hypothesize that these effects contribute to immune-driven pathologies observed in individuals carrying LACC1 variants.
Introduction
Genetic variants of laccase domain-containing 1 (LACC1, previously called C13orf31), including the common coding single nucleotide polymorphism Ile254Val (rs3764147), have been associated with inflammatory bowel disease [Crohn (1–3) and ulcerative colitis (4)], the mycobacterial disease leprosy (5, 6), and with Behcet disease (7), a vasculitis of mucosal membranes. In addition, a rare homozygous coding mutation in LACC1 (Cys284Arg) was recently identified in children with a severe form of systemic juvenile idiopathic arthritis (JIA) in consanguineous Saudi Arabian families (8). Systemic JIA is characterized by arthritis and a variety of extra-articular features, including daily fevers, macular rash, enlarged lymph nodes, hepatomegaly, and serositis (9). The children homozygous for LACC1 Cys284Arg had the characteristic quotidian fever and macular rash, symmetrical polyarthritis affecting small and large joints, leukocytosis, thrombocytosis, and elevated markers of inflammation (8).
In a separate Saudi Arabian family, the same Cys284Arg LACC1 mutation resulted in severe pediatric Crohn disease (10). Homozygous truncation mutations in LACC1 (frameshift c.128_129delGT) discovered in a Moroccan family additionally resulted in early onset systemic JIA (11). These human genetic studies suggest that LACC1 is involved in regulation of inflammation in the setting of infection and/or autoimmunity.
LACC1 is named for its sequence similarity to the laccase family of enzymes, also known as multicopper oxidoreductases. In bacteria and other lower organisms, laccases catalyze copper-dependent reduction–oxidation reactions that result in oxidation of a broad range of phenolic substrates (12). Laccases have a type 1 (T1) blue copper and a trinuclear copper core that comprise the catalytic domain of the enzyme (12). The laccase-like domain within LACC1 contains a putative T1 blue copper site but lacks the canonical trinuclear core; thus, it is unclear whether LACC1 functions similarly to other laccases.
LACC1 shows little homology with other mammalian proteins but does have structural similarity to the YfiH protein of Shigella flexneri based on crystal structure (Pfam motif DUF152) (13). Three-dimensional modeling of LACC1 based on its homology to the oxidase YfiH suggests that the rare Cys284Arg mutation may disrupt the catalytic site (at the T1 blue copper site) within the laccase-like domain of LACC1 (10). The common Ile254Val variant of LACC1 may otherwise negatively alter the stability and/or function of LACC1 protein (10).
To better understand the role of LACC1 in inflammation and to gain insight into how genetic variants of LACC1 might contribute to the pathogenesis of disease, we generated and characterized LACC1 knockout (KO) mice. These studies provide new information on the inflammatory networks that LACC1 influences in vivo.
Materials and Methods
Mice
LACC1 KO mice were generated from Lacc1tm1a(KOMP)Wtsi “KO first” sperm purchased from the Knockout Mouse Project on a C57BL/6 background. We additionally treated embryos with HTN-CRE to excise the first translational exon of LACC1. LACC1 KO mice were maintained at Genentech under specific pathogen-free conditions. All wildtype (WT) mice were generated from the same colony and age-matched for all experiments.
Colitis models
For infectious colitis, animals were fasted overnight and the following morning received oral inoculation of 2 × 109 Citrobacter rodentium. Fresh stool samples were collected 3× per wk, weighed, and vortexed in PBS before performing serial dilutions on McConkey plates for fecal bacterial shedding quantification. On day 13, serum was collected, colons’ weight and length were recorded, and half the colon went for histology scoring, whereas the other half went into RNAlater and was frozen at −80°f for RNA analysis. For chemical colitis, mice drinking water was replaced with drinking water containing 3% dextran sulfate sodium (DSS, 35–50 kDa; MP Biomedicals). Mice were weighed daily, and normal water was replaced on day 5. Animals’ colons and serum were harvested on day 8 post–initial DSS treatment.
For colon lesion severity assessment, histology on formalin-fixed samples of the colon (four transverse sections per mouse averaged to give one score per mouse) was scored from 0 to 5. Scoring scheme was as follows: 0, normal; 1, elevation of crypts from lamina muscularis mucosae by inflammatory cells, no or minimal crypt separation, crypt epithelium unaltered; 2, as score 1 but more extensive, may have had slight crypt hyperplasia; 3, crypt elevation but inflammatory cells generally limited to lamina propria, occasional foci of crypt epithelium effacement; 4, marked crypt elevation, some submucosal or transmural inflammation, marked crypt hyperplasia, usually diffuse; 5, as score 4, but submucosal or transmural inflammation marked, could have had crypt abscess and herniation.
Arthritis models
Collagen-induced arthritis (CIA): Mice were immunized intradermally on the side of the back with 200 μg of chick type II collagen (Chondrex catalog no. 20011) in 100 μl of CFA on day 0 and day 21. For the treatment study, starting on day 24 and continuing 3× per wk, mice were injected i.p. with either 400 μg of anti-ragweed IgG or 200 μg of anti–IL-17A/200-μg anti–IL-17F IgG2a. Mannan-induced arthritis (MIA): On day 0, 4, and 8, animals were injected i.p. with 20 mg of mannan (M7504-5G; Sigma) in 200 μl of PBS. For treatment studies, on day 4, 7, and 9, mice were injected i.p. with either 400 μg of anti-ragweed IgG, 200 μg of anti–IL-17A/200-μg anti–IL-17F IgG2a, or 400 μg of anti–TNFR II IgG2a (Genentech). All experimental animals were clinically scored for arthritis 3–5× per wk. Clinical arthritis scoring was as follows: each paw scored 0–4. A score of 0 was assigned for normal joint appearance. A score of 1 was assigned cumulatively to each paw for erythema and/or edema in the tarsal or carpal joints, metatarsal or metacarpal joints, metatarsal phalangeal or metacarpal phalangeal joints, or phalanges. A maximal score of 4 indicated erythema, edema, or both, involving the entire paw. The maximal disease index for each mouse was 16. Additional clinical analysis was carried out by normalizing the arthritis score to onset of animal arthritis (defined as the time at which at least one paw had an arthritis score of 1 or more) or normalizing to the onset of each individual paw’s arthritis onset (independently obtaining an arthritis score of 1 or more). For the “number of paws affected” criteria, an affected paw was defined as having an arthritis score of 1 or more.
At day of harvest, serum was collected, and paws with skin removed were put into either RNAlater or RLT buffer and snap frozen for RNA analysis or put into formalin for histological scoring. For histological scoring, all four paws were analyzed on a 0–5 scale for each inflammation, fibroplasia, cartilage loss, and bone remodeling, and each was equally weighted to give a total paw lesion score (0–5). The animal average lesion score was calculated by obtaining the average of each mouse’s paw lesion scores with or without removal of arthritic paws (classified as having a clinical score of 1 or more).
Isolation of primary immune cells
For the isolation of primary mouse cells, single-cell suspensions were generated from spleen, peritoneal lavage fluid, bone marrow, and lymph nodes and either sorted for specific subsets or negatively enriched using Miltenyi kits. Macrophages (F480hi, CD11bhi, CD19−, CD11c−, SiglecF−) were sorted from the peritoneal fluid. Dendritic cells were negatively enriched from the spleen using Miltenyi’s Pan Dendritic Cell Mouse Isolation Kit (130-100-875) per manufacturer’s instructions (40–70% CD11c+ or BST2+, Fig. 1) or additionally sorted (CD11c+ MHC class IIhi CD11b+ or CD11b−, Supplemental Fig. 1). B cells (CD45+ CD19+), CD4+ T cells (CD45+ CD3+ CD4+ CD25−), regulatory T cells (CD45+ CD3+ CD4+ CD25+), and CD8+ T cells (CD45+ CD3+ CD8+) were sorted from the lymph node. Primary neutrophils were isolated from bone marrow by negative enrichment using Miltenyi’s Neutrophil Isolation Kit (130-097-658) per manufacturer’s instructions (93% Ly6g+ or greater). All sorted populations were ≥90% enriched.
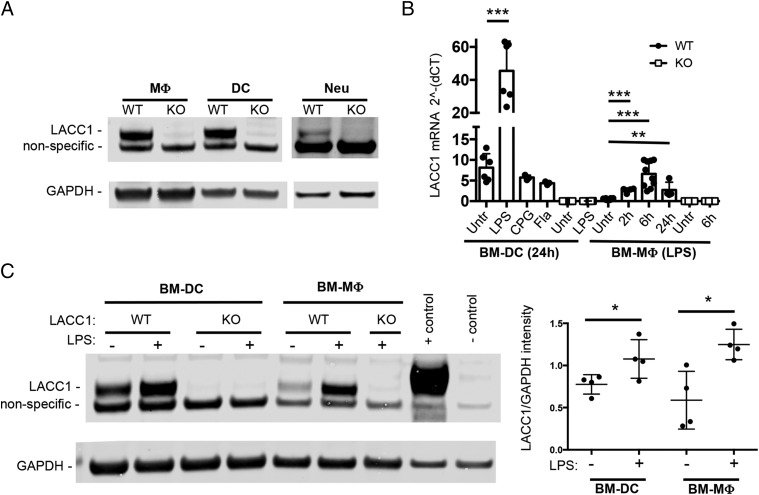
FIGURE 1.
LACC1 is expressed in myeloid cells and is LPS inducible. (A) LACC1 protein levels in primary mouse macrophages (MΦ), dendritic cells (DC), or neutrophils (Neu) isolated from LACC1 KO or WT mice as assessed by Western blotting of total cell lysates (10 μg) with polyclonal Abs to LACC1 or mAbs to GAPDH. (B) LACC1 mRNA expression relative to HPRT levels in BM-DCs in WT or LACC1 KO cells untreated or stimulated with 100 ng/ml LPS, 5 μM CPG (ODN1826), or S. typhimurium flagellin (Fla) for 24 h and a time course for BM-mac. Data from two independent experiments (n = from 6 to 9 mice) or one experiment [BM-DC with CPG/FLA (n = 3) and WT BM-mac with LPS for 2 h/24 h (n = 4)]. Error bars show SD. (C) Protein levels of LACC1 assessed by Western blotting 24 h after LPS or mock treatment, quantification on the right. Positive control, HEK cells transfected with a LACC1 expression construct (Myc-DDK–tagged); negative control, mock-transfected HEK cells. *p < 0.05, **p < 0.01, ***p < 0.00.
Generation of bone marrow–derived cells
Bone marrow was flushed from the femur of mice, 70 μm filtered and cultured in vitro with either conditioned media (DMEM + 10% FBS + 10 U/ml penicillin-streptomycin + 2 mM l-glutamine + 1 mM sodium pyruvate) containing 50 ng/ml M-CSF (PeproTech) for bone marrow–derived macrophages (BM-macs) or conditioned media (RPMI 1640 + 10% FBS + 10 U/ml penicillin-streptomycin +2 mM l-glutamine + 0.01 M HEPES + 50 μM 2-ME) containing 1–10 ng/ml GM-CSF (PeproTech) for BM-derived dendritic cells (BM-DCs). Six days later, BM-macs were harvested, aliquoted, and frozen in FBS with 10% DMSO until needed. For analysis, BM-macs were reseeded for 2 h with M-CSF and then exposed to appropriate treatment. BM-DCs were cultured for 7 d in vitro and assayed fresh. For confocal analysis of BM-DCs, bone marrow cells were additionally cultured with 10 ng/ml IL-4 (R&D Systems), and positive CD11c+ enrichment was performed using Miltenyi’s CD11c+ beads (130-097-059). Macrophages from human blood monocytes were generated by culturing in M-CSF (50 ng/ml) for 7 d before being plated for microscopy. For Western analysis, BM-macs and BM-DCs were exposed to 100 ng/ml ultra-pure LPS (Invivogen Ultra-pure LPS-EK) for 24 h. For TLR agonist stimulation, 0.5 μg/ml Pam3CSK4, 10^8 cells per ml HKLM, 1 μg/ml Poly(I:C) high m.w., 100 ng/ml LPS, 1 μg/ml Salmonella typhimurium flagellin (100 ng/ml for BM-DCs), 100 ng/ml FSL-1, 1 μg/ml ssRNA40, or 5 μM ODN1826 (Invivogen Mouse TLR1-9 Agonist Kit, catalog no. tlrl-kit1mw) were added for 2, 6, or 24 h (only 24 h for BM-DCs) before cells were suspended in RLT supplemented with 2-ME (Sigma) for RNA analysis.
LACC1 polyclonal Ab generation
A polyclonal rabbit anti-mouseLACC1 Ab was generated by inoculating rabbits with full-length mouse LACC1 protein, and serum was affinity purified using protein A, resulting in a total IgG polyclonal Ab fraction (Genentech).
Protein analysis
Protein extracts, separated by SDS-PAGE (NuPage Novex 4–12% Bis-Tris) and transferred onto nitrocellulose membranes, were probed with Abs against LACC1 (in-house polyclonal Ab, 1:500) or GAPDH (1:4000). Proteins of interest were detected with donkey anti-mouse or anti-rabbit IRDye 800CW or 680RD secondary Abs (1:15, 000 Li-Cor). Positive control was HEK 293 cells transfected with LACC1 (c-terminal Myc-DDK–tagged) (Origene MR214267, pCMV6-entry vector), and negative control was mock-transfected HEK 293. Protein was isolated 16 h after transfection.
For serum cytokine detection, serum was collected, and cytokine titers were measured by Luminex using a MILLIPLEX MAP Mouse Ig Cytokine/Chemokine Magnetic Bead Panel (catalog no. MCYTMAG-70K-PX32, MilliporeSigma). Concentration was depicted as an average fold change (KO/WT) for each experiment, with a detection threshold of 1.5 pg/ml and a significance cutoff of p < 0.05. Colored box indicates the degree of fold change, and gray box signifies cytokine was not detected or did not reach significant cutoff.
RNA analysis
At time of RNA extraction, colon was removed from RNAlater, put into RLT buffer supplemented with 2-ME (Sigma-Aldrich), and dissociated using gentleMACS M Tubes. At time of RNA extraction, paws were put into RLT + 2-ME and disrupted with 5 mM stainless steel beads (catalog no. 69989; Qiagen) using a Tissue Lyser II. All RNA was then extracted using an RNAeasy Mini Kit (catalog no. 74104 w) or RNeasy Fibrous Tissue Mini Kit (paws only, catalog no. 74704) with DNase step per the manufacturer’s instructions (Qiagen). cDNA was generated using a qScript cDNA Synthesis Kit per the manufacturer’s instructions (Quanta Biosciences catalog no. 95047-500). For Fluidigm analysis, sequence-specific preamplification was performed for 14 cycles, and the resulting cDNA was analyzed in quadruplicate with TaqMan Universal PCR Master Mix (Thermo Fisher catalog no. 4304437) and TaqMan Gene Expression Assays (Applied Biosystems) in 96.96 Dynamic Arrays on a BioMark System (Fluidigm). Raw cycle threshold (Ct) values were normalized to the average of GAPDH/HPRT/RPL19 (or just GAPDH and RPL19 for colon samples) using the equation 2^−[(gene of interest Ct) − (averaged reference genes Ct)]. Fold change per experiment was determined using the average normalized Ct values, and a heatmap was constructed with the threshold criteria of average normalized Ct value >0.0005 (paws) or 0.0001 (colon). Quantitative PCR of BM-DC and BM-mac LACC1 mRNA was performed using TaqMan Universal PCR Master Mix (Thermo Fisher catalog no. 4304437) and TaqMan Gene Expression Assays and normalized to HPRT levels (Fig. 1B). LACC1 mRNA from Supplemental Fig. 1E was analyzed using Fluidigm.
RNA sequencing and pathway analysis
BM-macs were plated at 250,000 cells/1 ml conditioned media without antibiotics in a 24-well flat-bottom plate and 24 h later infected with log phase Mycobacterium marinum multiplicity of infection (MOI) 1, containing GFP by centrifugation for 10 min (1000 RPM). Cells were then washed two times and put back into conditioned media with 20 μg/ml streptomycin and incubated at 33°C. Bioluminescence, as a readout of the amount of M. marinum in each well, was read out using a Luminometer. Twenty-four hours postinfection, RLT buffer with 2-ME was added and RNA extracted (as above). RNA was quantified using Nanodrop 8000 (Thermo Scientific), and integrity was measured using the Bioanalyzer RNA 6000 Pico Kit (Agilent). Libraries were prepared using the TruSeq RNA Library Prep Kit version 2 (Illumina) with 100–500 ng of input and amplified using 10 cycles of PCR. Libraries were multiplexed and sequenced on a HiSeq 2500 System (Illumina), resulting in 15 M single-end 50-bp reads per library. Alignment, feature counting, normalization, and differential expression analysis were performed similar to as described previously (14). In brief, HTSeqGenie was used to perform filtering, alignment to GRCm38, and feature counting. Normalized reads per kilobase gene model per million total reads (nRPKM) values were computed as a measure of gene expression. Pairwise differential expression analysis was performed using voom and limma. For differential gene expression analysis, significant genes were filtered and identified as p < 0.02, nRPKM >2 and fold change >1.2 or <0.8. Pathway analysis was performed with Ingenuity Pathway Analysis (IPA) software (Qiagen) using the “Diseases and Disorders” pathway module. Heatmap Euclidean clustering of genes was performed by plotting log 2–transformed fold change values for each replicate sample and each gene (log 2 floor set at −3 for heatmap). Colored boxes indicate the degree of fold change.
Confocal microscopy
Fresh BM-DCs and fresh human blood macrophages were added to four-well chambered slides and allowed to adhere for 2 h. Frozen BM-macs were reseeded for 2 h with complete media containing M-CSF and then polarized for 16 h with 50 ng/ml IFN-γ and 20 ng/ml LPS. Slides were then fixed with 2% paraformaldehyde for 15 min. Permeabilization with 0.05% Triton X-100 was conducted for 5 min followed by washing with PBS. Nonspecific binding was blocked with 5% BSA, 0.5% gelatin, and 0.05% Tween 20 for 20 min in PBS followed by incubation with primary Abs in blocking solution at 4°C overnight. The next morning, slides were washed with PBS and covered with Image-IT FX for 15 min before incubation with secondary Ab diluted in blocking solution at room temperature for 30 min. Slides were then washed with PBS and stained with 300 nm of Dapi before being mounted with no. 1.5 coverslips and ProLong Gold. Fluorescence was visualized with a Nikon A1R confocal microscope.
Primary Abs are as follows: in-house rabbit anti-mLACC1 (1.8 mg/ml, 1:500), mouse anti-hLACC1 (sc-374553 Santa Cruz E7, 200 μg/ml 1:50), mouse anti-PMP70 (Sigma SAB4200181, 1 mg/ml 1:100), and rat anti-LAMP1 (0.5 mg/ml 1:200). Secondary Abs: goat anti-rabbit (Fab2) 488 (The Jackson Laboratory 111-545-047, 1:200), goat anti-rat (Fab2) 647 (The Jackson Laboratory 112-605-072, 1:200), and goat anti-mouse (Fab2) 647 (The Jackson Laboratory 115-006-062, 1:200).
Monitoring IL-17 production in T cells and flow cytometry
Lymph nodes and spleen from either 5-mo-old mice (aged mice) or day 20 CIA mice were processed into a single-cell suspension and stimulated with 50 ng/ml PMA (Sigma) and 500 ng/ml ionomycin (aged mice) or stimulated with anti–CD3/28 T cell activator Dynabeads at a ratio of 0 (unstimulated) or 1:1 (Thermo Fisher catalog no. 11452) (day 20 CIA cells) in a 96-well round-bottom plate at 2 × 106 cells per ml. Two hours later, brefeldin A (GolgiPlug; BD Biosciences) was added to the culture, and four additional hours later, cells were surface stained with CD3, CD4, CD8, and CD45. Fixation and permeability, using a Foxp3/Transcription Factor Staining Buffer set (catalog no. 00-5523-00; eBioscience), followed by intracellular staining for IL-17 and IFN-γ, was conducted per the manufacturer’s instructions. For the frequency of IL-17–producing CD4+ T cells in aged mice, fold change was determined by dividing each value by the WT mean value within each experiment. For flow cytometric analysis of immune subsets from the mesenteric lymph node of day 13 infected C. rodentium mice, gating strategies included single, live CD45+ cells. Additional gating included CD4+ cells defined as CD3+CD4+ and subdivided into CD25+ or CD44lo versus CD44hi, CD8+ cells as CD3+CD8+, dendritic cells as CD11b+ CD11c+, neutrophils as CD11b+ Ly6g+, eosinophils as CD11b+ SiglecF+, and macrophages as CD11b+ Ly6g− CD11c− SiglecF− and subdivided based on Ly6c expression.
In all flow cytometry experiments, dead cells were discriminated using the LIVE/DEAD Fixable NIR-IR Dead Cell Stain Kit (Thermo Fisher L10119). Flow cytometry was performed using an LSR II (BD), and data were analyzed using FlowJo (Tree Star) software.
Graphing and statistical analysis
Univariate tests were done using a nonparametric Mann–Whitney U test in Prism or an ordinal mixed-effects regression controlling for mouse when analyzing repeated measures data, with the package ordinal in R version 3.3.3.
For longitudinal models, a mixed-effects model using a random intercept for each animal was fit controlling for time as a fixed effect and testing group as a predictor. Starting timepoints in which mice all had a score of zero were removed. The outcome arthritis score was log transformed prior to fit, and analysis was completed in R using the package nlme using a standard linear mixed-effects model. Ordinal outcomes (such as the number of paws affected) were fit using an ordinal mixed-effects model. The p values were considered significant at *p < 0.05, **p < 0.01, ***p < 0.001.
Results
LACC1 expression in myeloid cells
Previous RNA sequencing (RNAseq) studies in mouse immune cell subsets suggested that LACC1 transcripts are expressed predominantly in myeloid cells [(15) and shown in Supplemental Fig. 1A]. To test this, we raised polyclonal Abs against mouse LACC1 protein and found that LACC1 protein was highly expressed in mouse primary macrophages and dendritic cells sorted from the peritoneum or spleen, respectively (Fig. 1A). Confocal microscopy determined that LACC1 protein was cytoplasmic and nuclear in myeloid cells and did not colocalize with peroxisomes or lysosomes (Supplemental Fig. 1B, 1C). Additionally, highly enriched neutrophils from the blood also expressed detectable LACC1 protein (Fig. 1A).
We next isolated cells from LACC1 KO mice and confirmed the absence of detectable protein or LACC1 mRNA expression (Fig. 1A, 1B). LACC1 protein was not expressed at high levels in several other immune cell subsets analyzed (Supplemental Fig. 1D). A faint band close to the m.w. of LACC1 was identified in LACC1 KO cells (Fig. 1A and Supplemental Fig. 1D). A band with similar intensity was also detected in WT LACC1 T cells and B cells (Supplemental Fig. 1D), cell populations that have no or very little LACC1 RNA expression (Supplemental Fig. 1A), suggesting that this band is nonspecific.
Treatment of BM-macs and BM-DCs with LPS resulted in a robust increase of LACC1 at both the transcript and protein levels (Fig. 1B, 1C). BM-macs exposed to other TLR agonists also showed an increase in LACC1 mRNA, albeit less pronounced than observed with LPS (Supplemental Fig. 1E). The exception to this was Poly(I:C), whose exposure resulted in a similar increase in LACC1 mRNA as LPS (Supplemental Fig. 1E).
Enhanced colonic inflammation in LACC1 KO mice following C. rodentium infection
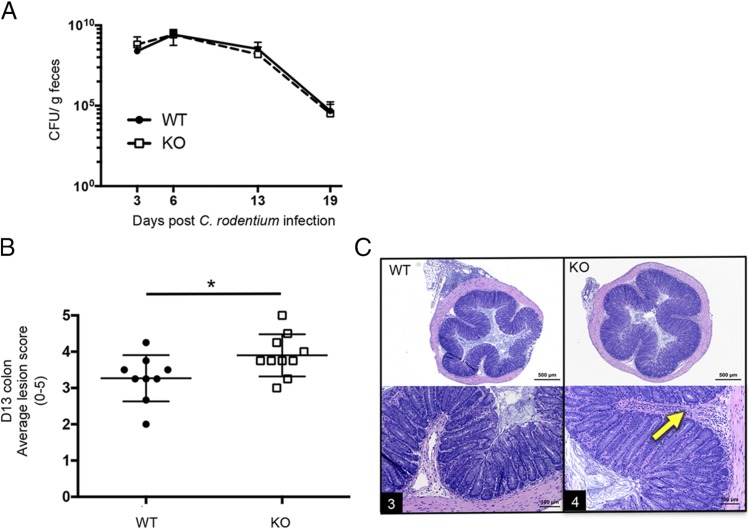
As noted above, single nucleotide polymorphisms within LACC1 have been linked to Crohn disease and ulcerative colitis in humans (1–4); we therefore tested the role of LACC1 in a mouse model of colitis caused by infection with C. rodentium. LACC1 KO or WT controls were orally infected with 2 × 109 CFU C. rodentium, and fecal bacterial clearance was monitored over time. Animals were sacrificed, and colons were analyzed for weight, length, and histologic changes 13 d postinfection.
LACC1 KO mice did not exhibit significant differences in bacterial clearance, colon weight, or colon length as compared with controls (Fig. 2A and data not shown). However, histological analysis of colons showed that inflammatory lesion scores were significantly worse in LACC1 KO colons compared with WT controls (Fig. 2B, 2C). Mesenteric lymph node cells were also analyzed at day 13 postinfection, and no significant differences were identified in immune cell subsets (data not shown). In a separate model of chemical colitis induced by DSS, we found no significant clinical or histologic differences between LACC1 KO and control mice (data not shown).
FIGURE 2.
In vivo infection with C. rodentium. LACC1 KO and age-matched WT control mice were orally inoculated with 2 × 109 C. rodentium. (A) Stools were collected for quantification of bacterial shedding over time. (B and C) On day 13 postinfection, colons were harvested for histologic scoring of colonic lesion severity. (B) Average lesion score (four transverse sections per mouse averaged to give one score per mouse) for each animal. (C) Representative colon histology from LACC1 WT (left) and LACC1 KO (right) colons. Arrow, submucosal infiltration with inflammatory cells. Number in black rectangle is score for the individual section. Data are presented as mean ± SD from two independent experiments (n = 9–19 mice/timepoint). *p < 0.05
LACC1 KO mice exhibit worse CIA
LACC1 KO mice monitored monthly for over 1.5 y did not develop spontaneous arthritis (n = 34 KO mice, n = 23 WT mice, only one animal per group reached an arthritis score of 3 or greater), and serum from 10-mo-old LACC1 KO mice versus WT controls showed no differences in total Ig levels or levels of various autoantibodies including anti-dsDNA or anti–extractable nuclear Ag (Supplemental Fig. 2A, 2B).
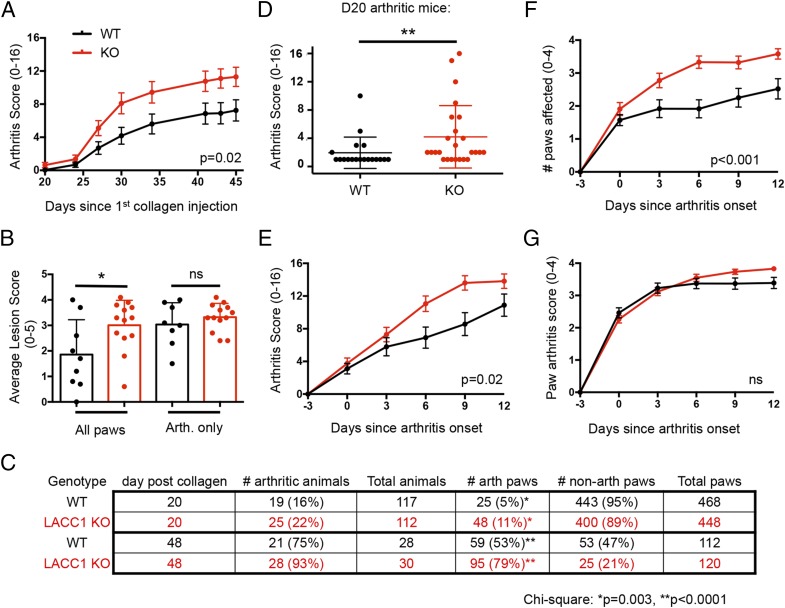
Based on the observation that a rare coding mutation in LACC1 was associated with severe systemic JIA in children (8), we tested the role of LACC1 in the development of CIA in mice. LACC1 KO and WT controls were immunized intradermally with 200 μg of chick collagen II in 100 μl of CFA on day 0 and day 21 and monitored over time for arthritis development. LACC1 KO mice developed significantly more severe arthritis and had a higher arthritis score over time (Fig. 3A). Histological analysis of paws at day 48 revealed an increased average lesion score in LACC1 KO paws as compared with WT controls (Fig. 3B). The percentage of arthritic paws at day 20 and day 48 after the initial collagen injection was significantly higher in the LACC1 KO mice compared with WT (Fig. 3C). From analysis of arthritic mice at day 20, it was clear that LACC1 KO mice exhibited more severe arthritis compared with WT controls (Fig. 3D).
FIGURE 3.
Enhanced CIA in LACC1 KO mice. LACC1 KO (red) and age-matched WT control (black) mice were immunized with 200 μg of chicken type II collagen in CFA intradermally on day 0 and day 21. (A) Paw arthritis scores were monitored over time. Data are presented as mean ± SEM from two independent experiments (n = 29–39 mice per timepoint). (B) At day 40, all paws (left) or arthritic paws (right) were histologically scored for lesion severity from all the mice in one of the experiments (n = 9 WT and 13 LACC1 KO mice). Data are presented as mean ± SD. (C) Number of arthritic animals and paws at day 20 (top two rows) and day 48 (bottom two rows) after initial collagen immunization. (D) Arthritis score from arthritic mice at day 20 CIA, data combined from three independent experiments (n = 19 WT and 25 KO). Data presented as mean ± SD. (E and F) Arthritis scores and numbers of affected paws normalized to arthritis onset. Data represent 18–30 mice in each group per timepoint and are presented as mean ± SEM. (G) The individual arthritis score for each paw normalized to the individual paw arthritis onset over time. Data combined from two independent experiments and presented as mean ± SEM. (n = 55–95 paws per timepoint). *p < 0.05, **p < 0.01.
To decipher the underlying reason for the enhanced arthritis scores in LACC1 KO mice and to overcome variability in disease onset in the CIA model (ranging from 2 to 6 wk from initial collagen immunization), we normalized the arthritis score for each animal based on the day of arthritis onset (Fig. 3E, 3F). Even after excluding animals that never developed arthritis (a trend of increased numbers of arthritic animals was observed in the LACC1 KO group, Fig. 3C), LACC1 KO mice showed worse arthritis compared with WT controls (Fig. 3E). Furthermore, once LACC1 KO mice developed arthritis, a greater number of paws developed arthritis as compared with WT animals (Fig. 3F). After normalizing the arthritis score of each paw based on arthritis onset in each individual paw, arthritis progression was not different between groups (Fig. 3G). Similarly, analysis of each mouse’s average paw lesion score, after excluding nonarthritic paws, showed no difference between WT and LACC1 KO (Fig. 3B). Taken together, these data suggest that LACC1 KO mice show a reduced threshold for developing arthritis after collagen immunization in the CIA model, but the progression of arthritis once initiated within a paw was similar between WT and LACC1 KO.
Exacerbated arthritis and worse psoriasis in LACC1 KO mice in the MIA model
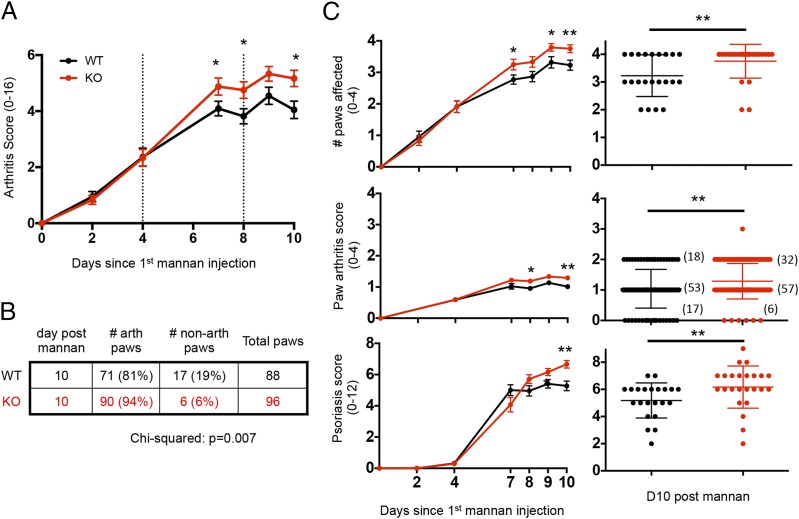
Because of the variability in disease onset in the CIA model, we also wanted to test whether LACC1 KO and WT mice differed in their response to injection with mannan polysaccharide purified from Saccharomyces cerevisiae (15), a more synchronized model of arthritis inflammation. LACC1 KO and WT control mice were injected i.p. with 20 mg of mannan on days 0, 4, and 8, and arthritis scores were monitored over time. Similar to the results obtained in the CIA model, MIA was also significantly worse in LACC1 KO mice compared with WT (Fig. 4A). The increased arthritis score was also mainly driven by the number of paws that developed arthritis in the MIA model (Fig. 4B, 4C).
FIGURE 4.
Worse arthritis and psoriasis in LACC1 KO mice in the mannan-induced model of inflammation. LACC1 KO (red) and age-matched WT control (black) mice were administered 20 mg of mannan i.p. on day 0, 4, and 8. (A) Arthritis scores were assessed over time. (B) Number of arthritic paws at day 10. (C) Number of affected paws per animal (top panel), individual paw arthritis score (middle panel, number of paws in each group are in parentheses), or average psoriasis score (bottom) over time (left) or at day 10 post–initial injection of mannan (right). Data shown in (A)–(C) represent combined results of three independent experiments (n = 22–24 animals per group), including two cohorts of animals receiving 400 μg of anti-ragweed IgG control as shown in Fig. 6A. Time course data presented as mean ± SEM, and day 10 timepoint as mean ± SD. *p < 0.05, **p < 0.01.
Mannan injection also results in psoriatic lesions of the pinnae and paws that can be scored visually (16). Psoriasis scores were significantly higher in LACC1 KO as compared with WT mice following mannan exposure (Fig. 4C).
We also looked for differences between LACC1 KO and WT mice in the K/BxN model of arthritis. Serum from K/BxN mice contains high titers of autoantibodies against the self-antigen glucose-6-phosphate isomerase (GP6PI), and injection of serum into mice leads to immune complex formation and deposition in joints, causing joint inflammation in an Arthus-like reaction (17). Administration of 75 μl of K/BxN serum to LACC1 KO and WT controls showed no differences in arthritis score or other clinical features between groups (data not shown).
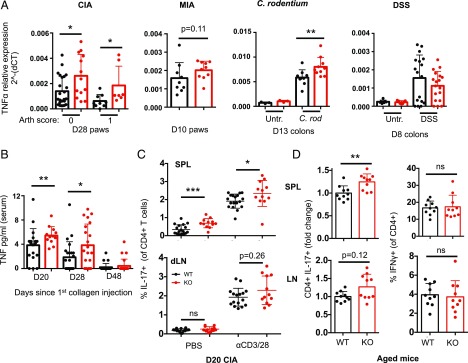
Increased TNF levels in LACC1 KO mice
To identify putative mechanisms contributing to accelerated arthritis and increased colitis in LACC1 KO mice, we assessed mRNA levels for 32 cytokines and chemokines at various timepoints across these models. We isolated RNA from CIA and MIA paws as well as C. rodentium and DSS colons, and specific transcripts were quantified (Supplemental Fig. 2C). Of interest, transcripts for TNF-α were increased in LACC1 KO CIA paws (day 28) and C. rodentium colons (day 8) and showed a similar trend in MIA paws (day 10) as compared with WT controls (Fig. 5A and Supplemental Fig. 3C). TNF-α mRNA was elevated in both arthritic and nonarthritic CIA LACC1 KO paws compared with WT paws 28 d after initial collagen immunization (Fig. 5A).
FIGURE 5.
Dysregulation of TNF and IL-17 in LACC1 KO mice. (A) TNF mRNA expression relative to housekeeping genes in individual WT (black) or LACC1 KO (red) mice from multiple in vivo models. RNA was isolated from day 28 CIA paws with an arthritis score of 0 (left) or 1 (right), MIA paws at day 10, C. rodentium colons at day 13, and DSS colons at day 8. Data represent at least two independent experiments. (B) Serum TNF protein levels measured by Luminex at day 20, 28, or 48 CIA. (C) Day 20 CIA splenocytes (top) or cells from the draining inguinal lymph node (bottom) received no stimulation (left) or were stimulated with anti-CD3 and anti-CD28 Abs for 6 h (right) followed by intracellular staining for IL-17. Data are from one experiment harvested on day 20 or 22 (n = 12–18 mice per condition). (D) Splenocytes (top) and lymphocytes (bottom) from 5-mo-old mice were stimulated with 50 ng/ml PMA and 500 ng/ml ionomycin for 6 h followed by intracellular staining for IL-17 (left) or IFN-γ (right). Data are combined from three independent experiments (n = 10 per group). IL-17 data are shown as fold change relative to WT for each experiment due to experiment-to-experiment variability. Data in all panels in this paper are presented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.
We next measured protein levels of TNF-α and a broader panel of serum cytokines and chemokines (Supplemental Fig. 2D). TNF-α protein was significantly increased in the serum of LACC1 KO mice 20 and 28 d after initial collagen immunization (Fig. 5B). CCL4, CCL5, and G-CSF were also increased in LACC1 KO serum 13 d after C. rodentium infection, and CCL4 was elevated in LACC1 KO DSS serum 8 d after administration (Supplemental Fig. 2D). I.p. LPS challenge (10 mg/kg) of LACC1 KO and WT mice showed no differences in the tested serum cytokines at any time point analyzed postchallenge (data not shown).
Higher numbers of IL-17–producing CD4+ T cells in LACC1 KO mice
CIA is mediated, at least in part, by IL-17 production (18, 19). We therefore assessed the frequency of IL-17–producing CD4+ T cells and observed significantly higher frequency of this cell subset at day 20 in LACC1 KO mice compared with WT controls (Fig. 5C and Supplemental Fig. 3A). This was observed in freshly isolated day 20 CIA splenocytes and following stimulation with anti-CD3/CD28 Abs.
Of interest, although 10-mo-old LACC1 KO mice did not show significant differences in cell numbers across the various lymphoid populations in spleen and lymph node (data not shown), stimulation of splenocytes from these older mice with PMA and ionomycin showed an increased percentage of LACC1 KO CD4+ T cells producing IL-17 (Fig. 5D). This was not observed in the CD8+ T splenocyte population (data not shown).
Of note, cytospin analysis of blood immune cell subsets at day 28 or day 48 in the CIA model showed no differences in the percentage or number of neutrophils, lymphocytes, monocytes, or eosinophils in LACC1 KO versus WT mice (data not shown). Additionally, there were no differences in Ab isotype levels or serum anticollagen Abs between LACC1 KO and WT controls in the CIA model (data not shown). The numbers of CD25+CD4+ regulatory T cells from inguinal lymph nodes were not altered in CIA LACC1 KO mice (data not shown).
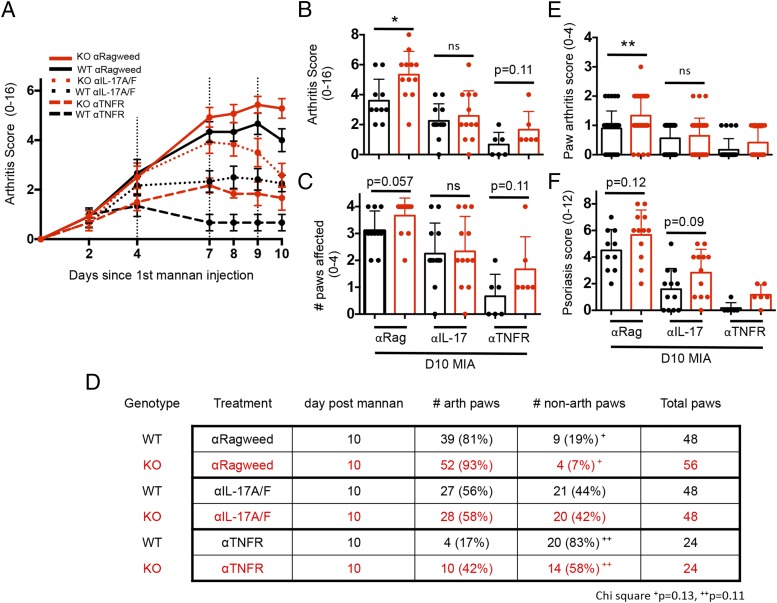
Neutralization of IL-17, but not TNF, prevented the exacerbated MIA initiation in LACC1 KO mice
Following the observation that IL-17A and TNF-α protein levels were elevated in LACC1 KO mice undergoing the arthritis models, we tested whether IL-17 or TNF-α was responsible for the increased MIA arthritis by administering anti–IL-17A/F, anti-TNFR, or control Abs i.p. to LACC1 KO or WT controls on day 4, 7, and 9 following the initial mannan injection.
Both anti–IL-17 and anti-TNFR Abs resulted in reduced arthritis scores compared with the control Ab–treated group in both LACC1 KO and WT mice, indicating that both of these cytokines contribute to arthritis in the MIA model (Fig. 6A), consistent with previous data (16, 20). By day 10 after mannan injection, neutralization of IL-17 diminished the differences in arthritis score observed between LACC1 KO and WT mice receiving the control Abs (Fig. 6A, solid lines versus dotted lines, and Fig. 6B). LACC1 KO and WT mice receiving control Abs did not show statistically significant differences in the number of arthritic paws at day 10 of MIA (Fig. 6C), but neutralization of IL-17 did lessen the difference in independent paw arthritis scores in the control Ab groups (Fig. 6E). However, psoriasis scores of LACC1 KO versus WT mice were not altered following neutralization of IL-17 (Fig. 6F), suggesting that the psoriatic lesions in LACC1 KO mice in this model have a distinct mechanism of pathology.
FIGURE 6.
IL-17 and TNF neutralization in MIA. (A) LACC1 KO (red) and age-matched WT control (black) mice were administered 20 mg of mannan i.p. on day 0, 4, and 8 and administered 400 μg of anti-ragweed IgG control (solid lines), 200 μg of anti–IL-17A/200-μg anti-IL17F IgG2a (dotted lines), or 400 μg of anti–TNFR II IgG2a (dashed lines) i.p. on day 4, 7, and 9. Data are combined from two independent experiments (n = 12–14) or from one experiment (n = 6, anti–TNFR II group only) and shown as mean ± SEM. (B) Arthritis scores and (C) number of affected paws at day 10 MIA. (D) Additional detail on number of affected paws in these experiments. (E) Individual paw arthritis scores and (F) psoriasis scores at day 10 MIA. For (B), (C), (E), and (F), data represent mean ± SD. *p < 0.05, **p < 0.01.
Although neutralization of TNFR diminished the arthritis score in LACC1 KO mice compared with the control Abs, it did not lessen the difference in arthritis score between LACC1 KO and WT mice by day 10 (Fig. 6A, solid lines versus dashed lines). Additionally, although neutralization of IL-17 diminished the observed difference in the percentage of arthritic paws in LACC1 KO versus WT paws, neutralization of TNF did not lessen the percentage of LACC1 KO arthritic paws versus WT (Fig. 6D).
Neutralization of IL-17 in CIA model
We also administered anti–IL-17A/F or anti-ragweed control Abs i.p. to either CIA LACC1 KO or WT controls 3× per wk starting on day 24 of CIA and monitored arthritis development. Neutralization of IL-17 resulted in a decreased arthritis score in WT mice compared with the WT mice receiving anti-ragweed control (Supplemental Fig. 3B). However, neutralization of IL-17 did not diminish the arthritis score in LACC1 KO mice compared with LACC1 KO mice receiving control Abs, although IL-17 neutralization was observed in the day 28 CIA serum of both LACC1 KO and WT animals receiving anti–IL-17A/F Abs (Supplemental Fig. 3B, 3C).
M. marinum infection of LACC1 KO and WT BM-macs
In a final series of experiments, we tested whether the absence of LACC1 altered the control of M. marinum, a bacterium related to M. leprae that causes human leprosy. Live M. marinum bacteria were coincubated with BM-macs at various MOIs and for differing times in vitro, and levels of viable bacteria were measured. No differences in bacterial burden were observed between LACC1 KO and WT BM-macs following infection (Supplemental Fig. 4A).
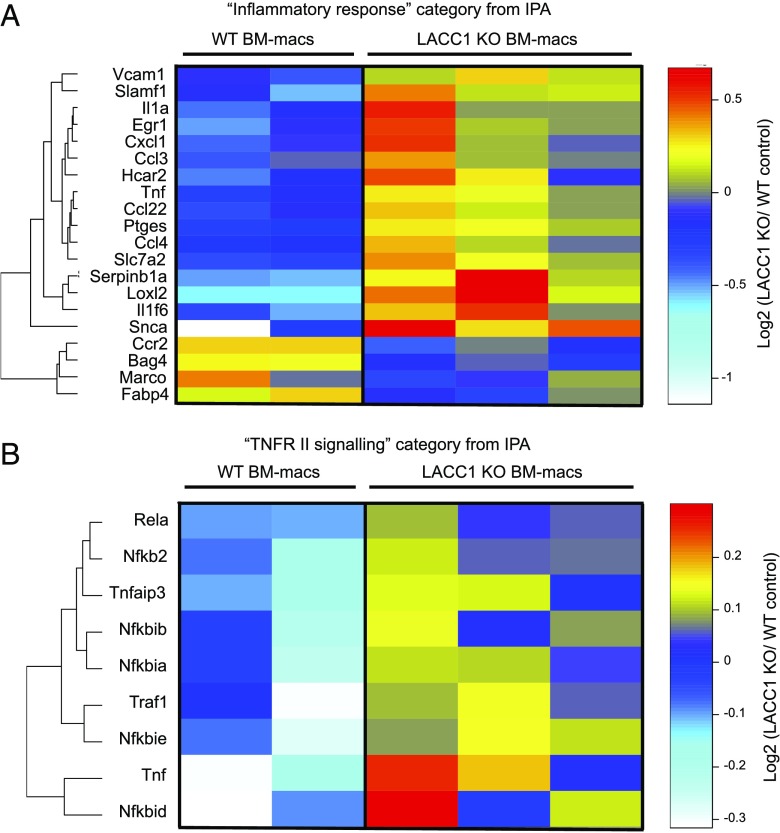
We next performed RNAseq analysis on LACC1 KO versus WT BM-macs infected in vitro (MOI 1) for 24 h with M. marinum. Differentially expressed genes were identified by comparing normalized RPKM between LACC1 KO and WT BM-macs and applying the following set of statistical criteria: fold change >1.2 or <0.8, uncorrected p value <0.02, and mean nRPKM across samples >2.
Following in vitro infection, 139 genes were upregulated, and 140 genes were downregulated in LACC1 KO BM-macs as compared with controls. Formal pathway analysis identified the category “inflammatory response” as the most significantly altered group of genes (56 genes total) within disease and biological functions (Supplemental Fig. 4B). Of the top 20 genes that showed the largest fold change differences between the two groups, several cytokine transcripts were increased in LACC1 KO BM-macs, including TNF, IL-1α, CXCL1, CCL3, and CCL4 (Fig. 7A). Additionally, formal pathway analysis also identified the category “IL-10 signaling” and “TNFR II signaling” as the two top canonical pathways most significantly altered between groups (Fig. 7B, Supplemental Fig. 4C). These data support the hypothesis that LACC1 is involved in repressing inflammatory responses in infected macrophages.
FIGURE 7.
Increased inflammatory gene expression in LACC1 KO macrophages after M. marinum in vitro infection. RNA was isolated from LACC1 KO (n = 3) and WT control (n = 2) BM-macs infected with M. marinum (MOI 1) and then incubated for 24 h followed by RNAseq analysis. (A) Heatmap Euclidean clustering of the 20 genes with the highest fold differences within the inflammatory response classification using IPA profiling. (B) Clustering of differentially expressed genes within the TNFR II signaling IPA classification. For data shown in both panels, expression differences had a p value <0.02, mean nRPKM >2, and a mean fold change ≥ 1.2 or ≤ 0.8.
In conclusion, we have shown that LACC1 KO mice exhibit enhanced arthritis onset in both the CIA and MIA models as well as worse colitis with C. rodentium compared with WT controls. We identified both TNF and IL-17 as increased in LACC1 KO mice and provide mechanistic data that IL-17 is responsible for the enhanced arthritis onset in MIA LACC1 KO animals versus WT controls. In vitro studies of LACC1 KO macrophages suggest that dysregulated myeloid inflammatory responses may contribute to the increased TNF levels observed in LACC1 KO mice compared with controls.
Discussion
To better understand the role for LACC1 in regulating inflammation, we characterized LACC1 KO mice across a series of in vivo models of inflammation and infection. LACC1 KO mice showed worse disease in the C. rodentium model of colitis, in both the CIA and MIA models of inflammatory arthritis, and in the MIA model of psoriasis. No differences between LACC1 KO and WT were observed in the DSS model of colitis or the K/BxN model of arthritis. Thus, these in vivo experiments provide initial evidence that LACC1 functions to downmodulate infectious and inflammatory responses, consistent with the human genetic associations of LACC1 with various autoimmune and inflammatory diseases.
Using mRNA and protein cytokine assays, we found that TNF levels were increased in LACC1 KO mice in both the CIA model and following C. rodentium infection. Although the importance of TNF in driving arthritis in the CIA mouse model is known (21), in our hands, neutralization of TNF, using anti-TNFR II Abs, did not diminish the differences in arthritis scores between LACC1 KO and WT mice in the MIA model. Similarly, although at least three of the diseases that LACC1 variants are associated with (systemic JIA, Behcet vasculitis, and Crohn colitis) are known to be mediated, at least in part, by TNF, the children with systemic JIA carrying two copies of LACC1 Cys284Arg reportedly did not respond well to anti-TNF therapy (8). Based on these data, it is therefore likely that TNF is not the sole driver of increased inflammation in LACC1 KO mice or in humans carrying LACC1 loss of function variants. In contrast, inhibition of IL-17 in the MIA model diminished differences in arthritis inflammation severity between LACC1 KO and WT, suggesting that IL-17 is a primary cytokine driving increased arthritis inflammation in the MIA model in LACC1 KO mice. Notably, this diminishment of differences was not observed fully until day 10 after mannan administration, which suggests that other mechanisms besides an increase in IL-17 production may be driving worse inflammation in LACC1 KO mice.
Significant dysregulation of innate proinflammatory cytokines, including IL-1β, IL-6, and TNF, has previously been shown to contribute to human systemic JIA (22). Patients with systemic JIA also have elevated levels of Th17 cells in their peripheral blood (23), although the precise role of IL-17 in systemic JIA pathology is not well understood. We show in this study that IL-17–producing CD4+ T cells were enriched in LACC1 KO mice in both the CIA model and in aged mice. Therefore, it may be that elevated IL-17 is derived from Th17 cells. TNF is a known regulator of Th17 cell generation (24, 25) and thus may be contributing to the IL-17 dysregulation observed. Other cells types, including δγT cells, neutrophils, and NK T cells, can also produce IL-17 (26). The elevated IL-17 levels in LACC1 KO mice may derive from one of these cell types and not Th17 cells. A recent report suggested that proinflammatory cytokine environments can drive IL-17 overexpression by δγT cells in systemic JIA (27). Future studies are needed to determine the source and mechanism driving elevated IL-17 in LACC1 KO mice.
In contrast to our results in the MIA model, neutralization of IL-17 did not diminish differences in arthritis severity between LACC1 KO and WT mice in the CIA model. The significant variability in disease onset observed in the CIA model makes it challenging to determine the appropriate time when neutralization of IL-17 would be effective. The MIA model provides more synchronized inflammation that minimizes this variable, which could account for the differences observed between the two models.
While this work was underway, Cader and colleagues (28, 29) reported an initial characterization of LACC1 using KO mice and biochemical approaches. Their data showed that LACC1 KO macrophages have diminished levels of reactive oxygen species (ROS) and produce less IL-1β upon various stimuli. Treatment of LACC1 KO mice and controls with 10 mg/kg LPS i.p. resulted in worse sepsis scores in the KO mice and a 2-fold increase in serum IL-1β (29). Cader et al. (29) also presented data suggesting that LACC1 protein controls fatty acid oxidation of endogenously synthesized lipids and colocalizes with peroxisomes, organelles important for lipid metabolism. None of the major findings in the current report overlap with or conflict with the findings of Cader et al. with the following two exceptions. In our hands, we did not observe colocalization of LACC1 protein with peroxisome markers using the same anti-humanLACC1 Ab (Santa Cruz E7) and cell type (human macrophages) as reported by Cader et al. (Supplemental Fig. 1B, 1C). Additionally, in our experiments, in vivo i.p. LPS challenge (10 mg/kg) of LACC1 KO versus WT mice did not result in heightened serum cytokines, including IL-1β, at any time point analyzed (data not shown).
Recently, Lahiri and colleagues (30) provided evidence that human LACC1 increases innate pattern recognition receptor–induced responses. They showed that variants in LACC1 were associated with lower pattern recognition receptor–initiated polyphenol oxidase activity, mitochondrial and cellular ROS production, cytokine secretion, and intracellular bacterial clearance. Lahiri et al. (30) identified LACC1 in both the cytoplasmic and mitochondrial cellular fractions of the cell and report that LACC1 associates with, and is required for, the optimal assembly of NOD2-signaling intermediates following NOD2 stimulation. Given that NOD2 loss of function mutations predispose to Crohn disease (31), these data are consistent with the LACC1 loss of function data reported in this paper.
The MIA, CIA, and C. rodentium infection models used in this study all administer bacterial components to drive inflammation, whereas DSS administration does not. Unlike DSS treatment of LACC1 KO mice, which did not result in worse severity of inflammation, the other three models all showed significantly worse disease severity in LACC1 KO mice compared with WT controls. This suggests that responsiveness to bacterial components is likely what is dysregulated in LACC1 KO versus WT mice and is consistent with the data reported by Lahiri et al. (30).
Currently the connection between LACC1 and dysregulation of TNF and IL-17 remain unclear. Both Cader et al. (29) and Lahiri et al. (30) report reduced ROS in myeloid cells either lacking LACC1 or in small interfering RNA knockdowns. Accumulation of ROS has been shown to limit the generation of Th17 cells (32), and oxidative stress has been reported to inhibit Th17 cell expression of IL-17A (33). Therefore, lowered ROS levels in LACC1 KO cells could lead to increased Th17 cells and IL-17.
Our data show that in vitro infection of LACC1 KO macrophages with M. marinum results in a broad dysregulation of inflammatory cytokines as compared with WT controls, including increased TNF and genes involved in TNFR II signaling. LACC1 transcripts and protein are upregulated by LPS and other TLR ligands in macrophages and dendritic cells (see Fig. 1 and Supplemental Fig. 1). Because similar in vitro infection experiments with dendritic cells did not result in increased inflammatory transcripts (data not shown), we hypothesize that macrophages are the main cell type driving enhanced inflammation in LACC1 KO mice undergoing inflammatory insults.
Taken together, these data indicate that LACC1 regulates inflammation, including production of TNF and IL-17, in a variety of in vivo experimental settings in mice. Additional work using LACC1 KO animals and other genetically engineered LACC1 mutant mice should lead to a better understanding of the role of LACC1 in regulating immunity and the translation of this work into humans with autoimmune and inflammatory disorders.
Supplementary Material
Acknowledgments
We thank Andres Paler Martinez for Luminex, Jovencio Borneo, Terence Ho, Jonathan Paw, and C.K. Poon for cell sorting, Eric Suto and John Liu for assistance with mouse experiments, Scott Stawicki and Jian Payandeh for anti-LACC1 Ab generation, and Raymond Asuncion for mouse colony management. We also thank Jon Sitrin and the rest of the Human Genetics, Immunology, and Diagnostic Discovery departments for scientific discussion. This work benefitted from data assembled by the ImmGen consortium.
This work was supported by Genentech, Inc.
The online version of this article contains supplemental material.
- BM-DC
- BM-derived dendritic cell
- BM-mac
- bone marrow–derived macrophage
- CIA
- collagen-induced arthritis
- Ct
- cycle threshold
- DSS
- dextran sulfate sodium
- IPA
- Ingenuity Pathway Analysis
- JIA
- juvenile idiopathic arthritis
- KO
- knockout
- LACC1
- laccase domain-containing 1
- MIA
- mannan-induced arthritis
- MOI
- multiplicity of infection
- nRPKM
- normalized read per kilobase gene model per million total reads
- RNAseq
- RNA sequencing
- ROS
- reactive oxygen species
- T1
- type 1
- WT
- wildtype.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Jostins L., Ripke S., Weersma R. K., Duerr R. H., McGovern D. P., Hui K. Y., Lee J. C., Schumm L. P., Sharma Y., Anderson C. A., et al. 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franke A., McGovern D. P., Barrett J. C., Wang K., Radford-Smith G. L., Ahmad T., Lees C. W., Balschun T., Lee J., Roberts R., et al. 2010. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 42: 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett J. C., Hansoul S., Nicolae D. L., Cho J. H., Duerr R. H., Rioux J. D., Brant S. R., Silverberg M. S., Taylor K. D., Barmada M. M., et al. 2008. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 40: 955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Assadi G., Saleh R., Hadizadeh F., Vesterlund L., Bonfiglio F., Halfvarson J., Törkvist L., Eriksson A. S., Harris H. E., Sundberg E., D’Amato M. 2016. LACC1 polymorphisms in inflammatory bowel disease and juvenile idiopathic arthritis. Genes Immun. 17: 261–264. [DOI] [PubMed] [Google Scholar]
- 5.Liu H., Irwanto A., Fu X., Yu G., Yu Y., Sun Y., Wang C., Wang Z., Okada Y., Low H., et al. 2015. Discovery of six new susceptibility loci and analysis of pleiotropic effects in leprosy. Nat. Genet. 47: 267–271. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F.-R., Huang W., Chen S.-M., Sun L.-D., Liu H., Li Y., Cui Y., Yan X.-X., Yang H.-T., Yang R.-D., et al. 2009. Genomewide association study of leprosy. N. Engl. J. Med. 361: 2609–2618. [DOI] [PubMed] [Google Scholar]
- 7.Takeuchi M., Mizuki N., Meguro A., Ombrello M. J., Kirino Y., Satorius C., Le J., Blake M., Erer B., Kawagoe T., et al. 2017. Dense genotyping of immune-related loci implicates host responses to microbial exposure in Behcet’s disease susceptibility. Nat. Genet. 49: 438–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wakil S. M., Monies D. M., Abouelhoda M., Al-Tassan N., Al-Dusery H., Naim E. A., Al-Younes B., Shinwari J., Al-Mohanna F. A., Meyer B. F., Al-Mayouf S. 2015. Association of a mutation in LACC1 with a monogenic form of systemic juvenile idiopathic arthritis. Arthritis Rheumatol. 67: 288–295. [DOI] [PubMed] [Google Scholar]
- 9.Petty R. E., Southwood T. R., Manners P., Baum J., Glass D. N., Goldenberg J., He X., Maldonado-Cocco J., Orozco-Alcala J., Prieur A. M., et al. International League of Associations for Rheumatology 2004. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J. Rheumatol. 31: 390–392. [PubMed] [Google Scholar]
- 10.Patel N., El Mouzan M. I., Al-Mayouf S. M., Adly N., Mohamed J. Y., Al Mofarreh M. A., Ibrahim N., Xiong Y., Zhao Q., Al-Saleem K. A., Alkuraya F. S. 2014. Study of Mendelian forms of Crohn’s disease in Saudi Arabia reveals novel risk loci and alleles. Gut 63: 1831–1832. [DOI] [PubMed] [Google Scholar]
- 11.Arostegui J. I., Rabionet R., Remesal A., Mensa-Vilaro A., Murias S., Alcobendas R., Gonzalez-Roca E., Dreschsel O., Ruiz-Ortiz E., Puig A., et al. 2015. A family carrying a homozygous LACC1 truncated mutation expands the clinical phenotype of this disease beyond systemic-onset juvenile idiopathic arthritis. Pediatric Rheumatology 13(Suppl. 1): O76. [Google Scholar]
- 12.Giardina P., Faraco V., Pezzella C., Piscitelli A., Vanhulle S., Sannia G. 2010. Laccases: a never-ending story. Cell. Mol. Life Sci. 67: 369–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim Y., Maltseva N., Dementieva I., Collart F., Holzle D., Joachimiak A. 2006. Crystal structure of hypothetical protein YfiH from Shigella flexneri at 2 A resolution. Proteins 63: 1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sitrin J., Suto E., Wuster A., Eastham-Anderson J., Kim J. M., Austin C. D., Lee W. P., Behrens T. W. 2017. The Ox40/Ox40 ligand pathway promotes pathogenic Th cell responses, plasmablast accumulation, and lupus nephritis in NZB/W F1 mice. J. Immunol. 199: 1238–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heng T. S., Painter M. W., Immunological Genome Project Consortium the. 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 10: 1091–1094. [DOI] [PubMed] [Google Scholar]
- 16.Khmaladze Ia., Kelkka T., Guerard S., Wing K., Pizzolla A., Saxena A., Lundqvist K., Holmdahl M., Nandakumar K. S., Holmdahl R. 2014. Mannan induces ROS-regulated, IL-17A-dependent psoriasis arthritis-like disease in mice. Proc. Natl. Acad. Sci. USA 111: E3669–E3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensen A. D., Haase C., Cook A. D., Hamilton J. A. 2016. K/BxN serum-transfer arthritis as a model for human inflammatory arthritis. Front. Immunol. 7: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lubberts E., Koenders M. I., Oppers-Walgreen B., van den Bersselaar L., Coenen-de Roo C. J., Joosten L. A., van den Berg W. B. 2004. Treatment with a neutralizing anti-murine interleukin-17 antibody after the onset of collagen-induced arthritis reduces joint inflammation, cartilage destruction, and bone erosion. Arthritis Rheum. 50: 650–659. [DOI] [PubMed] [Google Scholar]
- 19.Lubberts E., Joosten L. A., van de Loo F. A., Schwarzenberger P., Kolls J., van den Berg W. B. 2002. Overexpression of IL-17 in the knee joint of collagen type II immunized mice promotes collagen arthritis and aggravates joint destruction. Inflamm. Res. 51: 102–104. [DOI] [PubMed] [Google Scholar]
- 20.Sood S., Brownlie R. J., Garcia C., Cowan G., Salmond R. J., Sakaguchi S., Zamoyska R. 2016. Loss of the protein tyrosine phosphatase PTPN22 reduces mannan-induced autoimmune arthritis in SKG mice. J. Immunol. 197: 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams R. O., Feldmann M., Maini R. N. 1992. Anti-tumor necrosis factor ameliorates joint disease in murine collagen-induced arthritis. Proc. Natl. Acad. Sci. USA 89: 9784–9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruck N., Schnabel A., Hedrich C. M. 2015. Current understanding of the pathophysiology of systemic juvenile idiopathic arthritis (sJIA) and target-directed therapeutic approaches. Clin. Immunol. 159: 72–83. [DOI] [PubMed] [Google Scholar]
- 23.Omoyinmi E., Hamaoui R., Pesenacker A., Nistala K., Moncrieffe H., Ursu S., Wedderburn L. R., Woo P. 2012. Th1 and Th17 cell subpopulations are enriched in the peripheral blood of patients with systemic juvenile idiopathic arthritis. Rheumatology (Oxford) 51: 1881–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tesmer L. A., Lundy S. K., Sarkar S., Fox D. A. 2008. Th17 cells in human disease. Immunol. Rev. 223: 87–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Y., Sun L., Jiang T., Zhang D., He D., Nie H. 2014. TNFα promotes Th17 cell differentiation through IL-6 and IL-1β produced by monocytes in rheumatoid arthritis. J. Immun. Res. 2014: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuwabara T., Ishikawa F., Kondo M., Kakiuchi T. 2017. The role of IL-17 and related cytokines in inflammatory autoimmune diseases. Mediators Inflamm. 2017: 3908061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kessel C., Lippitz K., Weinhage T., Hinze C., Wittkowski H., Holzinger D., Fall N., Grom A. A., Gruen N., Foell D. 2017. Proinflammatory cytokine environments can drive interleukin-17 overexpression by δγT cells in systemic juvenile idiopathic arthritis. Arth. And Rheum. 69: 1480–1494. [DOI] [PubMed] [Google Scholar]
- 28.Assadi G., Vesterlund L., Bonfiglio F., Mazzurana L., Cordeddu L., Schepis D., Mjösberg J., Ruhrmann S., Fabbri A., Vukojevic V., et al. 2016. Functional analyses of the Crohn’s disease risk gene LACC1. PLoS One 11: e0168276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cader M. Z., Boroviak K., Zhang Q., Assadi G., Kempster S. L., Sewell G. W., Saveljeva S., Ashcroft J. W., Clare S., Mukhopadhyay S., et al. 2016. C13orf31 (FAMIN) is a central regulator of immunometabolic function. Nat. Immunol. 17: 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahiri A., Hedl M., Yan J., Abraham C. 2017. Human LACC1 increases innate receptor-induced responses and a LACC1 disease-risk variant modulates these outcomes. Nat. Commun. 8: 15614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sidiq T., Yoshihama S., Downs I., Kobayashi K. S. 2016. Nod2: a critical regulator of ileal microbiota and Crohn’s disease. Front. Immunol. 7: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu G., Xu Q., Qiu Y., Jin X., Xu T., Dong S., Wang J., Ke Y., Hu H., Cao X., et al. 2017. Suppression of Th17 cell differentiation by misshapen/NIK-related kinase MINK1. J. Exp. Med. 214: 1453–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abimannan T., Peroumal D., Parida J. R., Barik P. K., Padhan P., Devadas S. 2016. Oxidative stress modulates the cytokine response of differentiated Th17 and Th1 cells. Free Radic. Biol. Med. 99: 352–363 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.