Abstract
Tissue remodeling of subepithelial mesenchymal cells is a major pathologic condition of chronic obstructive pulmonary disease and asthma. Fibroblasts contribute to fibrotic events and inflammation in both airway diseases. Recent mechanistic studies established a link between mitochondrial dysfunction or aberrant biogenesis leading to tissue remodeling of the airway wall in asthma. Protein arginine methyltransferase-1 (PRMT1) participated in airway wall remodeling in pulmonary inflammation. This study investigated the mechanism by which PRMT1 regulates mitochondrial mass in primary human airway wall fibroblasts. Fibroblasts from control or asthma patients were stimulated with TGF-β for up to 48 h, and the signaling pathways controlling PRMT1 expression and mitochondrial mass were analyzed. PRMT1 activity was suppressed by the pan-PRMT inhibitor AMI-1. The SMAD2/3 pathway was blocked by SB203580 and C/EBPβ by small interference RNA treatment. The data obtained from unstimulated cells showed a significantly higher basal expression of PRMT1 and mitochondrial markers in asthmatic compared with control fibroblasts. In all cells, TGF-β significantly increased the expression of PRMT1 through SMAD2/3 and C/EBPβ. Subsequently, PRMT1 upregulated the expression of the mitochondria regulators PGC-1α and heat shock protein 60. Both the inhibition of the SAMD2/3 pathway or PRMT1 attenuated TGF-β–induced mitochondrial mass and C/EBPβ and α-SMA expression. These findings suggest that the signaling sequence controlling mitochondria in primary human lung fibroblasts is as follows: TGF-β→SMAD2/3→C/EBPβ→PRMT1→PGC-1α. Therefore, PRMT1 and C/EBPβ present a novel therapeutic and diagnostic target for airway wall remodeling in chronic lung diseases.
Introduction
Airway remodeling is the result of increased airway smooth muscle (ASM) mass and of fibroblast dysfunction (1, 2). In the past years, new evidence suggested an essential role of mitochondria in the pathogenesis of chronic inflammatory respiratory diseases (3). Microscopic analysis of lung tissues from patients with chronic inflammatory respiratory diseases, such as pulmonary arterial hypertension, asthma, and chronic obstructive pulmonary disease (COPD), revealed the accumulation of dysmorphic mitochondria and provided the first evidence of mitochondria dysfunction in diseased lungs (4, 5). Recent studies focusing on the mechanistic aspects suggested that mitochondria dysfunction or aberrant biogenesis contributed or caused the pathogenesis of chronic inflammatory respiratory diseases (6). In vitro studies showed that increased proliferation of ASM cells, obtained from patients with severe asthma, was mitochondria controlled, whereas proliferation of control cells was independent from mitochondria (7). However, the mechanism controlling the mitochondria mass in fibroblasts of patients with chronic inflammatory lung diseases was not well studied.
In asthma, the TGF-β is the best studied stimulator of profibrotic processes that drive airway wall remodeling (8). TGF-β is produced by several cell types, including epithelial cells, eosinophils, macrophages, and fibroblasts. It is involved in the damage of the bronchial epithelium, subepithelial fibrosis, ASM remodeling, microvascular changes, and increased mucus production (9, 10). Furthermore, TGF-β may also contribute to the development of glucocorticoid resistance and, thereby, diminish the antiproliferative effect of steroids (11). In an animal model of chronic asthma, TGF-β induced lung tissue remodeling through PRMT1 expression in lung fibroblasts (12), but its function in human cells was not investigated. To treat airway wall remodeling in asthma, the underlying pathologic mechanisms have to be better understood (13).
PRMT1 regulates posttranslational arginine methylation of proteins that play important roles in signal transduction, allowing cells to respond to changes and events in the extracellular milieu. The role of protein acetylation and phosphorylation has been extensively discussed in the context of chronic inflammatory pulmonary diseases (14–16). Arginine methylation of histones and other cellular proteins catalyzed by PRMT1 represents a novel protein modification that was implicated in the following: intracellular signaling, DNA repair, RNA processing, protein–protein interaction, and the regulation of gene expression (17). PRMT1 upregulated extracellular matrix deposition through ERK1/2–STAT1 signaling, both in human fibroblast (18) and ASM cells (19). Therefore, it is not surprising that PRMT1 had been implicated in contributing to mitochondria function.
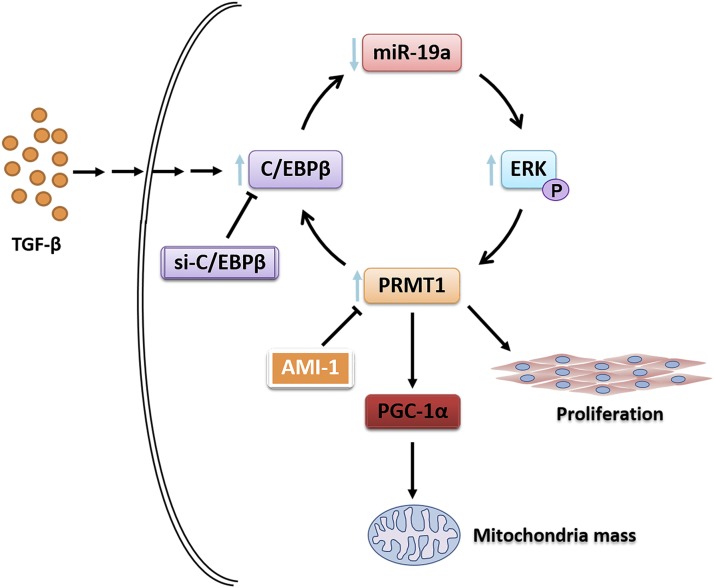
This study further investigated details of the TGF-β–activated signal pathway that augmented PRMT1 expression and caused mitochondria dysfunction in human primary fibroblast. The findings suggested PRMT1 as a novel regulator of mitochondria mass and dysfunction through the following signaling sequence: SMAD2/3→C/EBPβ→PRMT1→PGC-1α.
Materials and Methods
Cell isolation and characterization
Lung tissue specimens were obtained from the clinic of pneumology (University Hospital Basel, Basel, Switzerland), with the approval of the local ethical committee (EK:05/06) and written consent of each tissue donor (n = 6) who underwent endobronchial biopsy for other reasons.
Primary human lung fibroblasts were isolated as previously described, and cells were propagated under standard cell culture conditions (37°C, 100% humidity, 5% CO2, 95% air) (20). Fibroblast growth medium consisted of RPMI 1640 supplemented with 10% FBS, 1 mM sodium pyruvate, 1× nonessential amino acid mix, and 10 mM HEPES (all from Life Technologies, Grand Island, NY). All experiments were performed in fibroblasts between passages 2 and 8.
Cell stimulation and small interfering RNA treatment
Fibroblasts were seeded into six-well plates, allowed to adhere, serum deprived overnight, and then stimulated with 5 ng/ml TGF-β. PRMT1 activity was inhibited by 2 h of preincubating cells with AMI-1 (10 μM) before stimulation, and p-SMAD2/3 was blocked by SB203580. For knockdown experiments, fibroblast cells were pretreated with C/EBPβ small interfering RNAs (siRNA) according to the manufacturer’s protocol (Takara Bio, Kyoto, Japan) for 48 h. The siRNA and the corresponding control sequence were used at a final concentration of 50 nM and were transfected into fibroblasts with HiPerFect Transfection Reagent (Takara Bio) for 48 h.
Plasmid construction and transfection of pcDNA3.1-PRMT1
The full-length cDNA of PRMT1 was amplified from fibroblast cell mRNA by RT-PCR with primers (5′-CGG GAT CCC GAT GGC AGC CGA G-3′ and 5′-CCC TCG AGG GTC AGC GCA TCC GGT AGT CGG-3′) and was cloned into a pcDNA3.1+ vector to construct the pcDNA3.1-PRMT1 recombinant plasmid. The recombinant plasmid pcDNA3.1-PRMT1 was transiently transfected into primary fibroblasts for 24 or 48 h with Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The expression of PRMT1 and PGC-1α in fibroblasts was measured by real-time quantitative PCR (RT-qPCR).
Immunofluorescence and MitoTracker staining
For immunofluorescence analysis, fibroblasts were seeded on 10-mm glass slides and allowed to adhere overnight with or without TGF-β stimulation. Fibroblasts were fixed in 4% paraformaldehyde in PBS for 2 × 5 min. Immunofluorescence staining was performed as described previously (18). Briefly, Abs specific to either PRMT1, C/EBPβ, or PGC-1α (all from Abcam) were diluted 100-fold in 2% BSA–PBS, applied to the slides, incubated at 4°C overnight, followed by three washes with PBS, and subsequently incubated (1 h, room temperature) with PE-labeled secondary Abs (goat anti-rabbit IgG Ab for PRMT1 and PGC-1α, or FITC-labeled goat anti-mouse IgG Ab for C/EBPβ (all from Santa Cruz Biotechnology, Santa Cruz, CA). Nuclei were stained by DAPI.
For MitoTracker staining, 80% confluent cells were incubated for 30 min with serum-free medium containing 100 nM MitoTracker probe (Invitrogen; Thermo Fisher Scientific, Basel, Switzerland) under growth condition. Following three washes with PBS, cells were fixed, and nuclei were stained with DAPI. All images were captured by an Olympus BX61 fluorescence microscope (Olympus Optical, Tokyo, Japan).
Fibroblast proliferation
Direct cell count was performed using an improved Neubauer chamber slide. Fibroblasts were seeded onto six-well plates (0.25 × 105 cells per well) and allowed to adhere overnight, then serum deprived overnight and transiently transfected with C/EBPβ siRNA (48 h) or 10 μM AMI-1 (2 h) before being stimulated with TGF-β. The cell number was counted 24 h after stimulation.
Western blotting
Cells were lysed in RIPA buffer, and the protein concentration was quantified with a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Equal amounts of denatured proteins (20 μg) were separated in 8–16% SDS–PAGE (Thermo Fisher Scientific) and subsequently electrotransferred onto polyvinylidene fluoride membranes. The proteins of interest were detected with Abs specific to the following: PRMT1 or PGC-1α (Abcam), p-SMAD2/3, total SMAD2/3, C/EBPβ, or GAPDH (all from Cell Signaling Technology, Danvers). Protein bands were visualized by incubation of membranes with species-specific secondary HRP-conjugated Abs (Abcam) by chemiluminescence substrate (Thermo Fisher Scientific).
RT-qPCR
RT-qPCR was performed by an Applied Biosystems 7300/7500 Real-Time PCR System (Applied Biosystems, Foster City, CA) with Power SYBR Green PCR Master Mix (Applied Biosystems) for quantification. Gene expression was normalized to β-actin or to U6 snRNA, respectively. Purity of PCR products was confirmed by melting curve analysis, and all data were reanalyzed using the 2-ΔΔCt (relative quantification) method. The information for all primers is shown in Table I.
Table I. Information of primers for RT-qPCR.
| Gene Name | Species | Sequence (5′-3′) | Size (bp) | Annealing Temperature (°C) |
|---|---|---|---|---|
| PRMT1 | Homo sapiens | Forward 5′-TTGACTCCTATGCCCACT-3′ | 126 | 60 |
| Reverse 5′-CCACATCCAGCACCACC-3′ | ||||
| PGC-1α | H. sapiens | Forward 5′-CCTGCATGAGTGTGTGCTCT-3′ | 135 | 62 |
| Reverse 5′-CAGCACACTCGATGTCACTCC-3′ | ||||
| COL1A1 | H. sapiens | Forward 5′-GGACACAGAGGTTTCAGTGGT-3′ | 184 | 60 |
| Reverse 5′-CACCATCATTTCCACGAGCA-3′ | ||||
| Fibronectin | H. sapiens | Forward 5′-ACAAACACTAATGTTAATTGCCCA-3′ | 257 | 60 |
| Reverse 5′-AACTCCCAGGGTGATGCTTG-3′ | ||||
| GAPDH | H. sapiens | Forward 5′-CGGCAAGTTCAACGGCACAG-3′ | 148 | 65 |
| Reverse 5′-GAAGACGCCAGTAGACTCCACGAC-3′ |
Statistical analysis
The null hypothesis was that no treatment had any effect on signaling, PRMT1 expression, or proliferation. A p value <0.05 was regarded as significant. All data are expressed as mean ± SEM. The statistical analysis was performed by Mann–Whitney U test for the comparison between groups. The effect of inhibitors on PRMT1 and remodeling was analyzed by one-way ANOVA.
Results
Constitutive upregulated expression of PRMT1 and mitochondria markers in fibroblasts of asthma patients
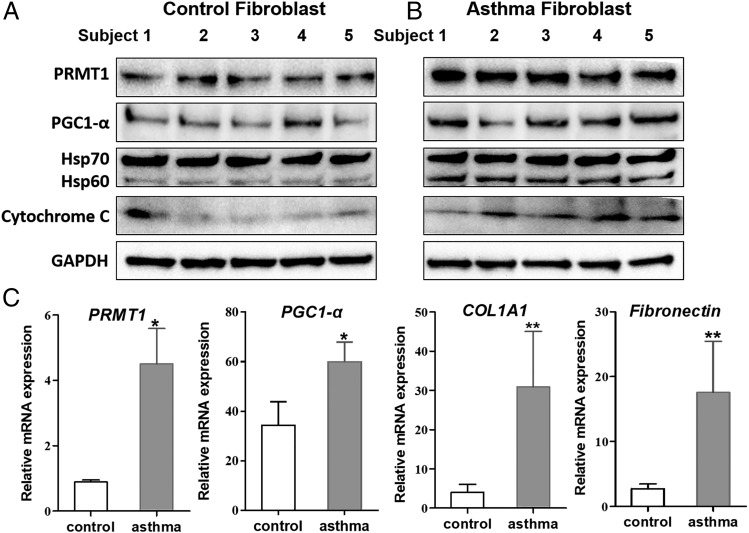
Primary fibroblasts obtained from asthma patients expressed significantly more PRMT1 and mitochondria markers, including PGC-1α, Hsp60, and cytochrome C compared with fibroblasts obtained from nonasthma patients. (Fig. 1A, 1B, Supplemental Fig. 1). The disease-specific upregulated expression of the above-named proteins was confirmed on the mRNA level by RT-qPCR (Table I, Fig. 1C).
FIGURE 1.
PRMT1 and mitochondria markers upregulated in human primary lung asthma fibroblast. (A and B) Primary lung fibroblasts were isolated from human control lungs (n = 5) and asthmatic lungs (n = 5). PRMT1, PGC-1α, Hsp60, Hsp70, and cytochrome C expression was detected by Western blot. GAPDH was used as housekeeping control. (C) The mRNA expression of PRMT1, PGC-1α, COL1A1, and fibronectin were determined by RT-qPCR and normalized to β-actin. The results were expressed as means ± SEM. Comparison of control to asthmatic fibroblast by Mann–Whitney U test (n = 5 for each group): *p < 0.05, **p < 0.01.
TGF-β1 stimulated the expression of PRMT1 and PGC-1α
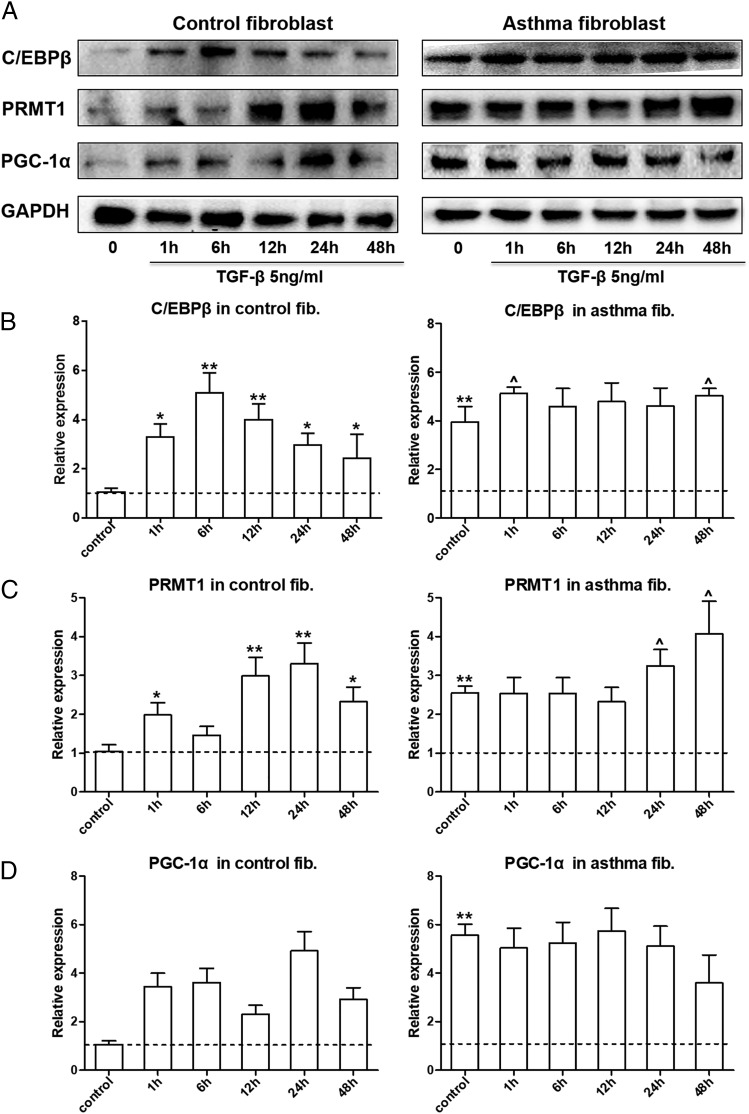
The regulatory mechanism controlling TGF-β1 (5 ng/ml)–induced PRMT1 expression in human lung fibroblasts was further analyzed at various time points (0, 1, 6, 12, 24, and 48 h). The results showed a significant upregulation of C/EBPβ after 1 h, which lasted over 24 h (Fig. 2A, Supplemental Fig. 2). PRMT1 expression significantly increased between 12 to 48 h. PGC-1α showed the slowest response to TGF-β1 but became significant between 24 to 48 h (Fig. 2A).
FIGURE 2.
TGF-β stimulated the expression of PRMT1 and PGC-1α. (A) Representative Western blots of the kinetic for TGF-β–stimulated expression of C/EBPβ, PRMT1, and PGC-1α in primary fibroblast from controls (n = 3) and asthma patients (n = 3). Protein band density of (B) C/EBPβ, (C) PRMT1, and (D) PGC-1α was measured by ImageJ software, and the relative expression was normalized to GAPDH. The results were expressed as mean ± SEM. Comparison of unstimulated control fibroblasts to other groups by Mann–Whitney U test: *p < 0.05, **p < 0.01. Comparison of unstimulated asthmatic fibroblasts to TGF-β1 stimulated cells: ^p < 0.05.
In fibroblasts of asthma patients, the basal expression level of C/EBPβ, PRMT1, and PGC-1α was higher compared with control fibroblasts (Fig. 2A, Supplemental Fig. 2). TGF-β1 stimulation further increased C/EBPβ and PRMT1 expression but had no effect on PGC-1α levels (Fig. 2A). Analysis of protein expression was performed for all Western blots by densitometry (Fig. 2B–D).
TGF-β1 stimulated the expression of PRMT1 and PGC-1α through SMAD2/3
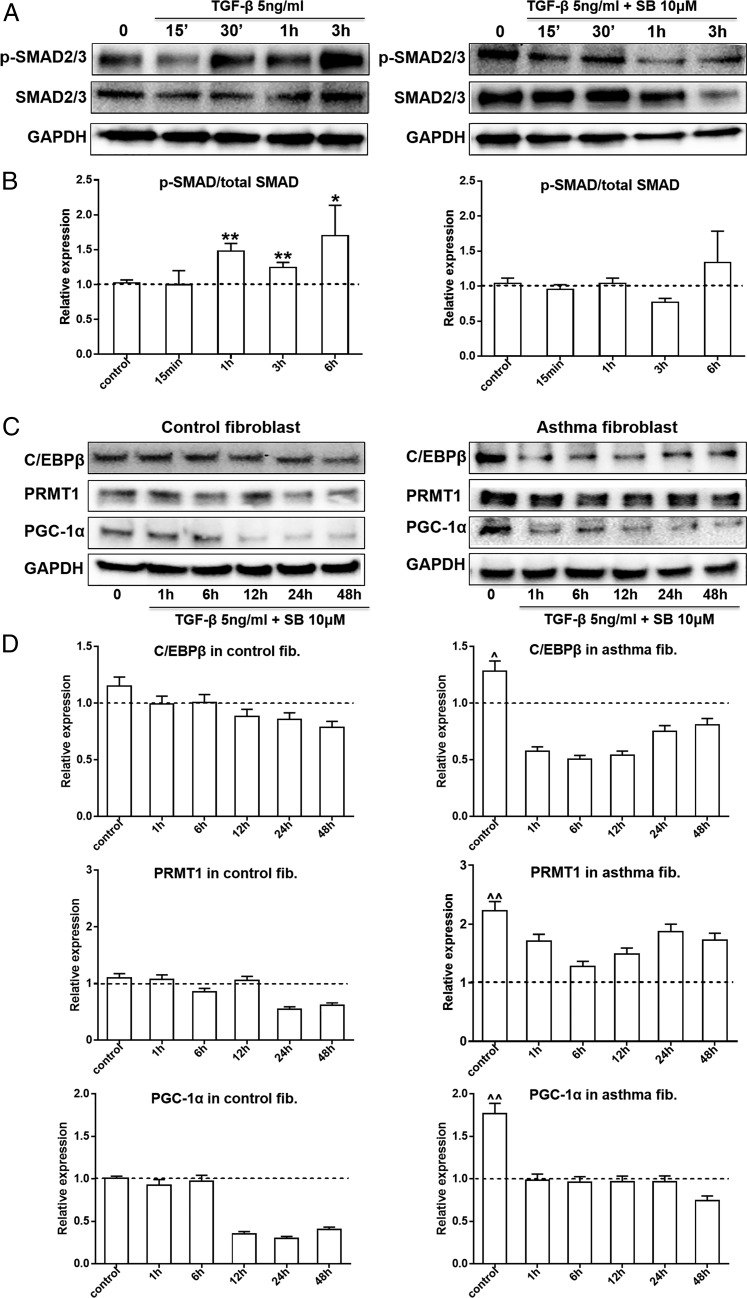
Next, the role of SMAD2/3 in the TGF-β1–stimulated expression of PRMT1 in human lung fibroblasts was analyzed at various time points (15, 30 min, 1 h, and 3 h). TGF-β1 activated SMAD2/3 within 3 h, and this event was suppressed by SB203580 (Fig. 3A). In control and asthmatic fibroblasts, the expression of all TGF-β1–induced proteins, C/EBPβ, PRMT1, and PGC-1α, was suppressed by SB203580 (Fig. 3B). Analysis of protein expression was performed for all Western blots by densitometry (Fig. 3C).
FIGURE 3.
TGF-β stimulated the expression of PRMT1 and PGC-1α through SMAD2/3 pathway. (A) TGF-β–induced phosphorylation of SMAD2/3 was assessed in the presence and absence of the chemical inhibitor SB203580 (SB, 10 μM) for 15, 30 min, 1 h, and 3 h by Western blots in control fibroblasts. (B) Protein band density was measured (ImageJ software), and the expression was normalized to GAPDH. (C) The expression of C/EBPβ, PRMT1, and PGC-1α was determined in the same samples by Western blots (n = 3, each group). (D) Protein band density was measured (ImageJ software), and the expression was normalized to GAPDH. The results were expressed as mean ± SEM and analyzed by Mann–Whitney U test. Comparison of unstimulated control fibroblasts to other groups: *p < 0.05, **p < 0.01. Comparison of asthmatic fibroblasts to unstimulated control cells: ^p < 0.05, ^^p < 0.01.
C/EBPβ knockdown reduced TGF-β–induced expression of PRMT1 and PGC-1α
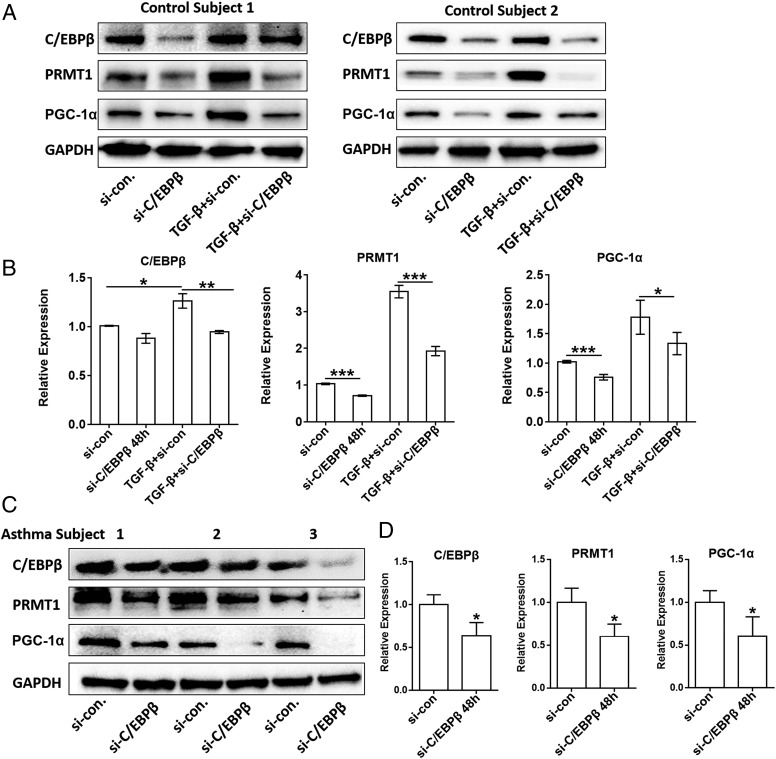
To confirm the role of C/EBPβ in PRMT1 expression, fibroblasts from control or asthma subjects were transfected with C/EBPβ siRNA over 48 h. In control fibroblasts, TGF-β increased PRMT1 and PGC-1α expression, which was prevented in fibroblasts treated with C/EBPβ siRNA (Fig. 4A). Analysis of protein expressions of all Western blots by densitometry was performed in control fibroblasts (Fig. 4B). In nonstimulated fibroblasts of three asthma patients, 48-h transfection with C/EBPβ siRNA significantly reduced the constitutive high expression of C/EBPβ, PRMT1, and PGC-1α (Fig. 4C), and analysis of all Western blots by densitometry is depicted in Fig. 4D.
FIGURE 4.
The knockdown of C/EBPβ reduced PRMT1 expression. (A) Control fibroblasts were transfected with either control siRNA (small interference [si]-Con) or C/EBPβ siRNA (si-C/EBPβ) alone or followed by stimulation with TGF-β (5 ng/ml) for 48 h. The effect of C/EBPβ suppression was verified on the expression of the following: C/EBPβ, PRMT1, and PGC-1α by Western blots. (B) C/EBPβ, PRMT1, or PGC-1α band density was measured (ImageJ software) and normalized to GAPDH (n = 3). (C) Asthmatic fibroblasts (n = 3) were transfected with either control siRNA (si-con) or C/EBPβ siRNA (si-C/EBPβ) for 48 h, and the protein expression for C/EBPβ and PRMT1 was detected by Western blots. (D) C/EBPβ, PRMT1, or PGC-1α band density was measured (ImageJ software) and normalized to GAPDH. Comparison of indicated groups (B) or si-con to si-C/EBPβ group (D) by Mann-Whitney U-test: *p < 0.05, **p < 0.01, ***p < 0.001.
PRMT1 controlled the expression of PGC-1α
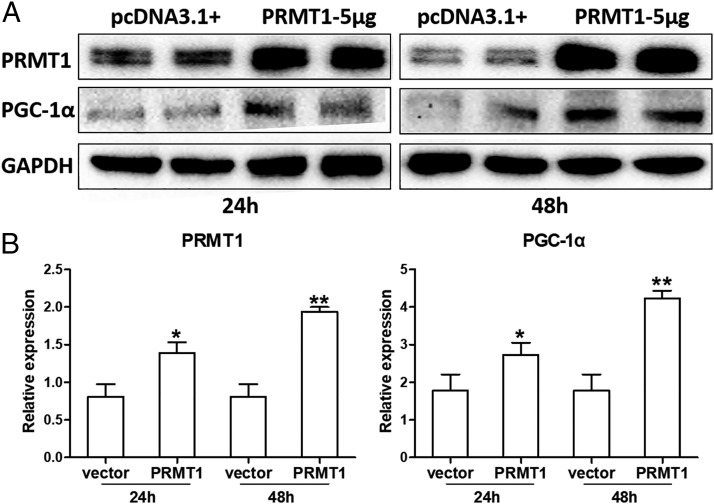
The role of PRMT1 on the expression of PGC-1α was studied by transfecting control fibroblasts with a recombinant plasmid, pcDNA3.1-PRMT1 (empty plasmid pcDNA3.1+ served as control) over 24 or 48 h. The transfection of cells with the plasmid pcDNA3.1-PRMT1 significantly increased the expression of PRMT1 and of PGC-1α (Fig. 5A). The analysis of all Western blots by densitometry is shown in Fig. 5B.
FIGURE 5.
PRMT1 expression is controlled by PGC-1α. (A) Control fibroblasts were transfected with recombinant plasmid pcDNA3.1-PRMT1 (5 μg/well, six-well plate) or plasmid pcDNA3.1+ (mock transfection) for 24 and 48 h. The expression of PRMT1 and PGC-1α was detected by Western blots (n = 4). (B) PRMT1 and PGC-1α band density was measured (ImageJ software) and normalized to GAPDH. The results were expressed as mean ± SEM and analyzed by Mann–Whitney U test. Comparison of pcDNA3.1-PRMT1 to mock transfection: *p < 0.05, **p < 0.01.
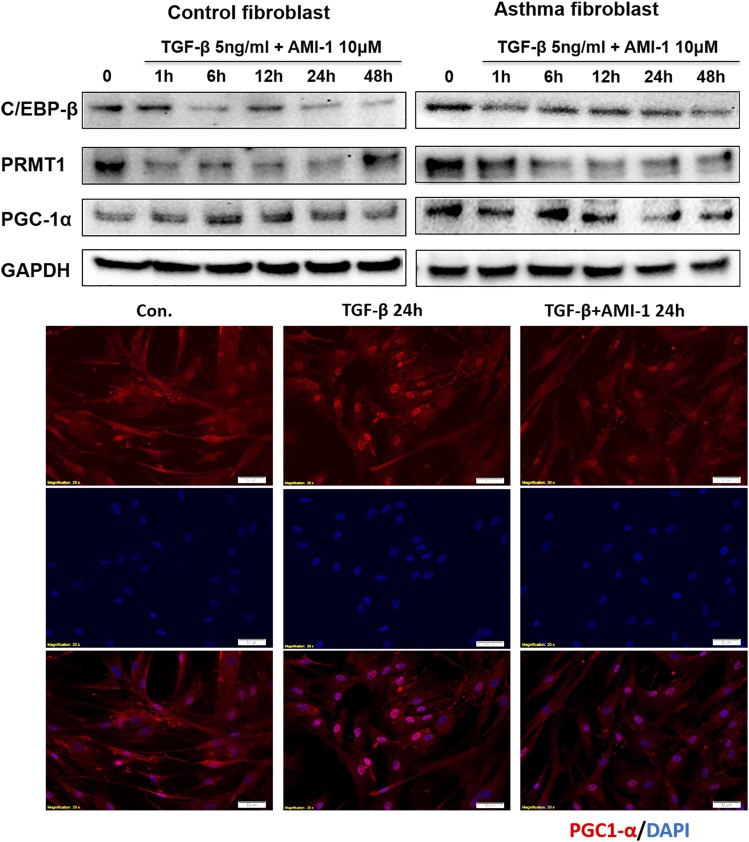
In cells treated with the PRMT enzyme activity inhibitor AMI-1, the expression of C/EBPβ, PRMT1, and PGC-1α was decreased to basal levels (Fig. 6). These data were confirmed by immunofluorescence staining showing that TGF-β1–stimulated PGC-1α expression was prevented by AMI-1 (Fig. 6).
FIGURE 6.
Blocking PRMT1 enzyme activity decreased TGF-β–induced PGC-1α and C/EBPβ expression. Representative Western blots (n = 3) of PRMT1 inhibition by AMI-1 in TGF-β–stimulated asthmatic and control fibroblasts over 48 h. Immunofluorescence: control fibroblasts were stimulated with TGF-β for 24 h in the presence or absence of AMI-1, and PGC-1α expression (red) was detected by immunofluorescence microscopy. Nuclei were stained by DAPI.
TGF-β–stimulated mitochondria mass is regulated by C/EBPβ and PRMT1
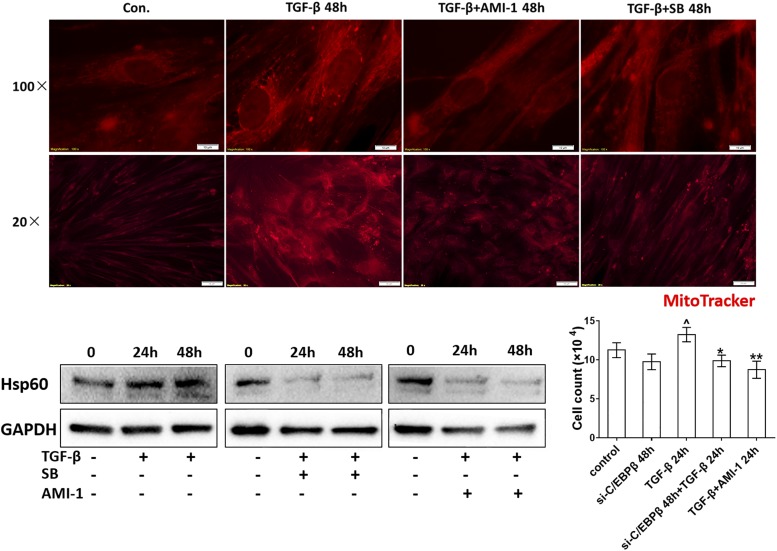
TGF-β significantly increased the mitochondria mass in all fibroblasts. The PRMT1 inhibitor AMI-1 significantly reduced the mitochondria mass in TGF-β–stimulated cells (Fig. 7). A similar reducing effect on TGF-β–stimulated mitochondria mass was observed when the cells had been incubated with SB203580 (Fig. 7).
FIGURE 7.
The inhibition of PRMT1 or SMAD2/3 decreased mitochondria mass and proliferation of fibroblasts. Immunofluorescence microscopy of mitochondria in control fibroblasts stimulated by TGF-β in the absence and presence of AMI-1 or SB203580 (SB) at 48 h. The mitochondrial mass was visualized by MitoTracker staining (red). Representative Western blots of Hsp60 in the presence and absence of either AMI-1 or SB at 24 or 48 h. Incubation of fibroblasts with either C/EBPβ siRNA (small interference [si]-C/EBPβ) or AMI-1 for 48 h reduced TGF-β–induced proliferation (n = 3). Results are expressed as mean ± SEM and were analyzed by Mann–Whitney U test. Comparison of untreated cells to TGF-β stimulation: ^p < 0.05. Comparison of si-C/EBPβ and AMI-1 treatment on TGF-β–stimulated fibroblast proliferation: *p < 0.05, **p < 0.001.
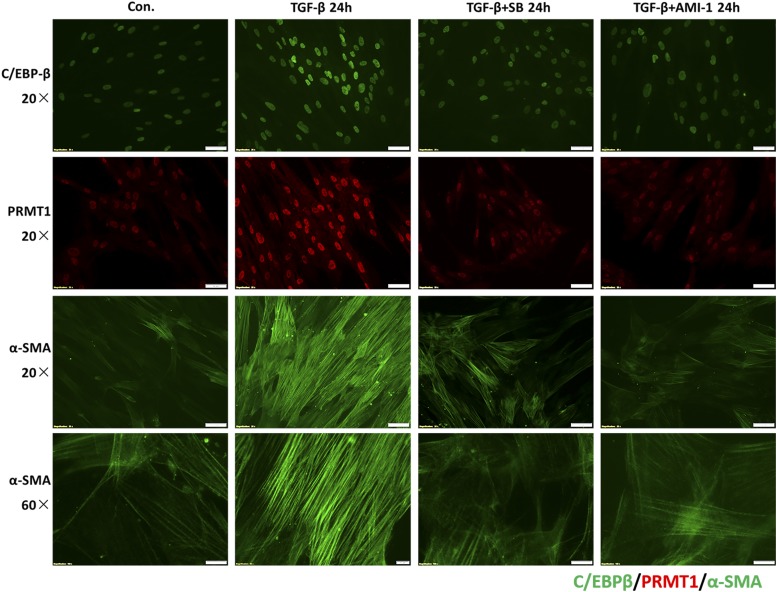
Treatment with either AMI-1 or SB203580 also decreased TGF-β–induced expression of C/EBPβ, PRMT1, and α-SMA immunofluorescence (Fig. 8), suggesting that PRMT1 signaling plays a role for myofibroblast differentiation. In conclusion, the results indicate that in asthma PRMT1 and C/EBPβ play a crucial role in subepithelial fibrosis and inflammation which is summarized in Fig. 9.
FIGURE 8.
PRMT1 inhibition decreased fibroblast differentiation. Immunofluorescence microscopy of the effect of PRMT1 or SMAD2/3 inhibition on the expression of TGF-β–induced C/EBPβ (green), PRMT1 (red), and fibrilar α-SMA (green) proteins. Images are representative for results obtained in cells of controls (n = 3) and asthma patients (n = 3).
FIGURE 9.
The mechanism by which TGF-β controls fibroblast activity in asthma through SMAD2/3→C/EBPβ→PRMT1→PGC-1α signaling. In human lung fibroblasts, TGF-β stimulation activates SMAD2/3 signaling, which induces the expression of C/EBPβ. C/EBPβ is essential for PRMT1 upregulation, which orchestrates PGC-1α, mitochondria mass, and fibroblast proliferation. Therefore, PRMT1 and C/EBPβ may play a crucial role in fibrotic processes and present a novel therapeutic target in chronic inflammatory pulmonary diseases.
Discussion
Fibroblasts contribute to the subepithelial fibrosis and inflammation in chronic inflammatory airway diseases. Recent studies suggested a link between mitochondria dysfunction or aberrant biogenesis to the pathogenesis of asthma, and they were assumed to have a causative role in structural airway wall remodeling. This study demonstrates the sequential signaling cascades that may underlie TGF-β1–induced tissue remodeling in fibroblasts of asthma patients: TGF-β1→SMAD2/3→C/EBPβ→PRMT1→PGC-1α→mitochondria mass. The presented data indicate that TGF-β1 and C/EBPβ are upstream of PRMT1 and PGC-1α expression as summarized in Fig. 9.
Among all cytokines and chemokines that can be produced by bronchial epithelial cells, TGF-β1 was recognized as an important factor inducing proliferation of subepithelial fibroblasts and as a driving force of cell differentiation and activation. Increased levels of TGF-β1 have been reported in bronchoalveolar lavage fluids and biopsy specimens of asthma patients (21, 22), and TGF-β1 expression correlated with the degree of subepithelial fibrosis (23, 24). In healthy humans, the concentration of circulating TGF-β ranges from 0.26 to 20 ng/ml (25–27), whereas in asthma patients, the circulating TGF-β concentration ranges from 38 to 215 ng/ml (28, 29). TGF-β1 induced mitochondria dysfunction in alveolar macrophages and epithelial cells (30, 31), whereas the effect of TGF-β1 on mitochondria dysfunction in subepithelial fibroblasts is less clear.
Mitochondria are regarded as the powerhouse of cells by generating ATP, and their dysfunction was linked to the pathogenesis of various chronic diseases, including diabetes, cancers, asthma, and COPD (32, 33). Mitochondria are the key regulators of cellular processes and function as cellular oxygen sensors, as modulators of intracellular signaling pathways, as a source of intracellular reactive oxygen species, and as elicitors of apoptosis. Abnormal mitochondria function contributed to the pathogenesis of chronic lung remodeling diseases, including asthma (7), COPD (34), and pulmonary arterial hypertension (35). Mitochondria damage and altered function in airway epithelial cells was reported in experimental allergic asthma (33). However, the pathologic mechanism for how mitochondrial abnormalities contribute to or cause asthma was investigated in ASM and in epithelial cells but not in airway wall fibroblasts.
Cytochrome C and heat shock protein-60 (Hsp60) are two typical markers for mitochondria. Cytochrome C is a component of the electron transport chain in mitochondria. The heme group of cytochrome C accepts electrons from the B-C1 complex and transfers electrons to the cytochrome oxidase complex. Extracellular succinate rises when the integrity of cytochromes B and C1 of the electron transport chain is compromised, either by inhibitors or by metabolic reprograming (36).
Hsp60, a mitochondrial stress protein, is a member of the chaperon family that is induced under conditions of cellular stress and injury. Circulating Hsp60 has been documented in asthma (37). The expression of Hsp60 was induced by mitochondrial impairment, and Hsp60 was located in both the mitochondria and cytosol. Hsp60 is essential for the folding and assembly of newly imported proteins into the mitochondria (38, 39). Hsp60 is secreted by a number of cell types, which may contribute to the severity of symptoms in allergic airway inflammation (40). As we showed in Results, cytochrome C and Hsp60 were constitutively expressed in asthmatic fibroblasts, which correlated to the mitochondrial mass. Furthermore, the inhibition of the SMAD2/3 pathway or PRMT1 enzyme activity decreased the expression of cytochrome C and Hsp60.
PGC-1α is a transcriptional coactivator and important metabolic regulator of the cellular energy metabolism, including cholesterol catabolism, fatty acid oxidation, and gluconeogenesis in various organs (41, 42). One of the primary effects of PGC-1α is the activation of mitochondria biogenesis and oxidative phosphorylation (43, 44). In this study, PGC-1α was constitutively increased in fibroblasts of asthma patients and in TGF-β–stimulated control fibroblasts. The methylation of PGC-1α by PRMT1 was reported, which may explain how PRMT1 enhanced the mitochondrial biogenesis (45). In nonalcoholic fatty liver disease, PRMT1 was increased, and PRMT1 siRNA reduced lipogenic markers, including PGC-1α (46). Currently, it is unclear if PGC-1α functions as a coactivator (47) or substrate for PRMT1 (48). In this study, PGC-1α expression was increased in asthmatic fibroblasts and caused the increase of mitochondria mass. PGC-1α expression was induced in control fibroblasts by TGF-β1 or by transfection with a PRMT1 expression vector. Thus, the alteration the PGC-1α affected the content of mitochondria in airway wall fibroblasts.
Subepithelial fibrosis is a consequence of increased TGF-β1 expression, which activates myofibroblasts to produce extracellular matrix components such as collagens and fibronectin. TGF-β1 also enhanced the migration of ASM cells, reduced antiapoptotic signaling, and stimulated proliferation (9, 49). In the context of TGF-β1 signaling, allergen-challenged SMAD3-deficient mice showed reduced numbers of peribronchial myofibroblasts and decreased peribronchial fibrosis (50). Suppressing TGF-β1 by either blocking the SMAD3 pathway or by administration of anti–activin A, significantly reduced peribronchial fibrosis, ASM cell proliferation, and mucus production (51). In this study, TGF-β activated SMAD2/3 signaling and elevated PGC-1α and fibroblast proliferation.
C/EBPβ is a key mediator of TGF-β1–induced gene activity, and its malfunction contributed to the progression of breast cancer (52). C/EBPβ was expressed upon the induction of cell differentiation and it induced the expression of C/EBPα and PPAR-γ (53). It is known that C/EBPs controlled the promoter of mitoribosomal protein S12 and mitochondrial seryl-tRNA ligase, which regulated the mRNA translation of Hsp60, required for protein folding in mitochondria (54, 55). In this study, changes of C/EBPβ expression affected the expression and action of PRMT1 and PGC-1α, and all three factors were constitutively upregulated in fibroblasts of asthma patients. These results suggest that C/EBPβ is an important regulator of the mitochondria mass in fibroblasts.
This study provided additional evidence that TGF-β1–dependent fibroblast remodeling in asthma may be controlled by inhibition of PRMT1 or C/EBPβ. Therefore, both factors present novel diagnostic and therapeutic targets in asthma.
Supplementary Material
Acknowledgments
We are grateful to Xinggang Wang and Hui Wang for expertise, assistance, helpful discussions, and productive critiques. We also thank C.T. S’ng for helping to prepare this manuscript.
This work was supported by National Natural Science Foundation of China 31871314 and 31601033, Swiss National Foundation Grant 31003A_176248/1, and National Natural Science Foundation of Shaanxi Province 2015JM8448.
The online version of this article contains supplemental material.
- ASM
- airway smooth muscle
- COPD
- chronic obstructive pulmonary disease
- Hsp60
- heat shock protein-60
- RT-qPCR
- real-time quantitative PCR
- siRNA
- small interfering RNA.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.James A. 2017. Airway remodeling in asthma: is it fixed or variable? Am. J. Respir. Crit. Care Med. 195: 968–970. [DOI] [PubMed] [Google Scholar]
- 2.Fehrenbach H., Wagner C., Wegmann M. 2017. Airway remodeling in asthma: what really matters. Cell Tissue Res. 367: 551–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schumacker P. T., Gillespie M. N., Nakahira K., Choi A. M., Crouser E. D., Piantadosi C. A., Bhattacharya J. 2014. Mitochondria in lung biology and pathology: more than just a powerhouse. Am. J. Physiol. Lung Cell. Mol. Physiol. 306: L962–L974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravamudan B., Thompson M. A., Pabelick C. M., Prakash Y. S. 2013. Mitochondria in lung diseases. Expert Rev. Respir. Med. 7: 631–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prakash Y. S., Pabelick C. M., Sieck G. C. 2017. Mitochondrial dysfunction in airway disease. Chest 152: 618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy P. H. 2011. Mitochondrial dysfunction and oxidative stress in asthma: implications for mitochondria-targeted antioxidant therapeutics. Pharmaceuticals (Basel) 4: 429–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trian T., Benard G., Begueret H., Rossignol R., Girodet P. O., Ghosh D., Ousova O., Vernejoux J. M., Marthan R., Tunon-de-Lara J. M., Berger P. 2007. Bronchial smooth muscle remodeling involves calcium-dependent enhanced mitochondrial biogenesis in asthma. J. Exp. Med. 204: 3173–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Alawi M., Hassan T., Chotirmall S. H. 2014. Transforming growth factor β and severe asthma: a perfect storm. Respir. Med. 108: 1409–1423. [DOI] [PubMed] [Google Scholar]
- 9.Cohen M. D., Ciocca V., Panettieri R. A., Jr 1997. TGF-beta 1 modulates human airway smooth-muscle cell proliferation induced by mitogens. Am. J. Respir. Cell Mol. Biol. 16: 85–90. [DOI] [PubMed] [Google Scholar]
- 10.Halwani R., Al-Muhsen S., Al-Jahdali H., Hamid Q. 2011. Role of transforming growth factor-β in airway remodeling in asthma. Am. J. Respir. Cell Mol. Biol. 44: 127–133. [DOI] [PubMed] [Google Scholar]
- 11.Breton J. D., Heydet D., Starrs L. M., Veldre T., Ghildyal R. 2018. Molecular changes during TGFβ-mediated lung fibroblast-myofibroblast differentiation: implication for glucocorticoid resistance. Physiol. Rep. 6: e13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Q., Liu L., Roth M., Tian J., He Q., Zhong B., Bao R., Lan X., Jiang C., Sun J., et al. 2015. PRMT1 upregulated by epithelial proinflammatory cytokines participates in COX2 expression in fibroblasts and chronic antigen-induced pulmonary inflammation. J. Immunol. 195: 298–306. [DOI] [PubMed] [Google Scholar]
- 13.Prakash Y. S., Halayko A. J., Gosens R., Panettieri R. A., Jr., Camoretti-Mercado B., Penn R. B., ATS Assembly on Respiratory Structure and Function 2017. An official American thoracic society research statement: current challenges facing research and therapeutic advances in airway remodeling. Am. J. Respir. Crit. Care Med. 195: e4–e19. [DOI] [PubMed] [Google Scholar]
- 14.Brambilla E. 2009. [Epigenetic modifications in lung cancer]. Ann. Pathol. 29: S31–S33. [DOI] [PubMed] [Google Scholar]
- 15.Duru S., Kurt E. B. 2014. [Asthma, environment and epigenetic]. Tuberk. Toraks 62: 165–169. [DOI] [PubMed] [Google Scholar]
- 16.Bégin P., Nadeau K. C. 2014. Epigenetic regulation of asthma and allergic disease. Allergy Asthma Clin. Immunol. 10: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biggar K. K., Li S. S. C. 2015. Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 16: 5–17. [DOI] [PubMed] [Google Scholar]
- 18.Sun Q., Liu L., Mandal J., Molino A., Stolz D., Tamm M., Lu S., Roth M. 2016. PDGF-BB induces PRMT1 expression through ERK1/2 dependent STAT1 activation and regulates remodeling in primary human lung fibroblasts. Cell. Signal. 28: 307–315. [DOI] [PubMed] [Google Scholar]
- 19.Sun Q., Liu L., Wang H., Mandal J., Khan P., Hostettler K. E., Stolz D., Tamm M., Molino A., Lardinois D., et al. 2017. Constitutive high expression of protein arginine methyltransferase 1 in asthmatic airway smooth muscle cells is caused by reduced microRNA-19a expression and leads to enhanced remodeling. J. Allergy Clin. Immunol. 140: 510–524.e3. [DOI] [PubMed] [Google Scholar]
- 20.Tamm M., Roth M., Malouf M., Chhajed P., Johnson P., Black J., Glanville A. 2001. Primary fibroblast cell cultures from transbronchial biopsies of lung transplant recipients. Transplantation 71: 337–339. [DOI] [PubMed] [Google Scholar]
- 21.Redington A. E., Madden J., Frew A. J., Djukanovic R., Roche W. R., Holgate S. T., Howarth P. H. 1997. Transforming growth factor-beta 1 in asthma. Measurement in bronchoalveolar lavage fluid. Am. J. Respir. Crit. Care Med. 156: 642–647. [DOI] [PubMed] [Google Scholar]
- 22.Chakir J., Shannon J., Molet S., Fukakusa M., Elias J., Laviolette M., Boulet L. P., Hamid Q. 2003. Airway remodeling-associated mediators in moderate to severe asthma: effect of steroids on TGF-beta, IL-11, IL-17, and type I and type III collagen expression. J. Allergy Clin. Immunol. 111: 1293–1298. [DOI] [PubMed] [Google Scholar]
- 23.Ram A., Mabalirajan U., Jaiswal A., Rehman R., Singh V. P., Ghosh B. 2015. Parabromophenacyl bromide inhibits subepithelial fibrosis by reducing TGF-β1 in a chronic mouse model of allergic asthma. Int. Arch. Allergy Immunol. 167: 110–118. [DOI] [PubMed] [Google Scholar]
- 24.Mabalirajan U., Aich J., Agrawal A., Ghosh B. 2009. Mepacrine inhibits subepithelial fibrosis by reducing the expression of arginase and TGF-beta1 in an extended subacute mouse model of allergic asthma. Am. J. Physiol. Lung Cell. Mol. Physiol. 297: L411–L419. [DOI] [PubMed] [Google Scholar]
- 25.Cárdaba B., Calzada D., Baos S., Aguerri M., Quiralte J., Lahoz C. 2014. Polymorphisms of tumor necrosis factor-α, transforming growth factor-β, and interleukin-10 in asthma associated with olive pollen sensitization. J. Immunol. Res. 2014: 276345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Czarkowska-Paczek B., Bartlomiejczyk I., Przybylski J. 2006. The serum levels of growth factors: PDGF, TGF-beta and VEGF are increased after strenuous physical exercise. J. Physiol. Pharmacol. 57: 189–197. [PubMed] [Google Scholar]
- 27.Sarahrudi K., Thomas A., Mousavi M., Kaiser G., Köttstorfer J., Kecht M., Hajdu S., Aharinejad S. 2011. Elevated transforming growth factor-beta 1 (TGF-β1) levels in human fracture healing. Injury 42: 833–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manuyakorn W., Kamchaisatian W., Atamasirikul K., Sasisakulporn C., Direkwattanachai C., Benjaponpitak S. 2008. Serum TGF-beta1 in atopic asthma. Asian Pac. J. Allergy Immunol. 26: 185–189. [PubMed] [Google Scholar]
- 29.Jiang K., Chen H. B., Wang Y., Lin J. H., Hu Y., Fang Y. R. 2013. Changes in interleukin-17 and transforming growth factor beta 1 levels in serum and bronchoalveolar lavage fluid and their clinical significance among children with asthma. Transl. Pediatr. 2: 154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grunwell J. R., Yeligar S. M., Stephenson S., Ping X. D., Gauthier T. W., Fitzpatrick A. M., Brown L. A. S. 2018. TGF-β1 suppresses the type I IFN response and induces mitochondrial dysfunction in alveolar macrophages. J. Immunol. 200: 2115–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel A. S., Song J. W., Chu S. G., Mizumura K., Osorio J. C., Shi Y., El-Chemaly S., Lee C. G., Rosas I. O., Elias J. A., et al. 2015. Epithelial cell mitochondrial dysfunction and PINK1 are induced by transforming growth factor-beta1 in pulmonary fibrosis. PLoS One 10: e0121246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harbauer A. B., Zahedi R. P., Sickmann A., Pfanner N., Meisinger C. 2014. The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell Metab. 19: 357–372. [DOI] [PubMed] [Google Scholar]
- 33.Mabalirajan U., Dinda A. K., Kumar S., Roshan R., Gupta P., Sharma S. K., Ghosh B. 2008. Mitochondrial structural changes and dysfunction are associated with experimental allergic asthma. J. Immunol. 181: 3540–3548. [DOI] [PubMed] [Google Scholar]
- 34.Aravamudan B., Kiel A., Freeman M., Delmotte P., Thompson M., Vassallo R., Sieck G. C., Pabelick C. M., Prakash Y. S. 2014. Cigarette smoke-induced mitochondrial fragmentation and dysfunction in human airway smooth muscle. Am. J. Physiol. Lung Cell. Mol. Physiol. 306: L840–L854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomez-Arroyo J., Mizuno S., Szczepanek K., Van Tassell B., Natarajan R., dos Remedios C. G., Drake J. I., Farkas L., Kraskauskas D., Wijesinghe D. S., et al. 2013. Metabolic gene remodeling and mitochondrial dysfunction in failing right ventricular hypertrophy secondary to pulmonary arterial hypertension. Circ Heart Fail 6: 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iyer D., Mishra N., Agrawal A. 2017. Mitochondrial function in allergic disease. Curr. Allergy Asthma Rep. 17: 29. [DOI] [PubMed] [Google Scholar]
- 37.Yang M., Wu T., Cheng L., Wang F., Wei Q., Tanguay R. M. 2005. Plasma antibodies against heat shock protein 70 correlate with the incidence and severity of asthma in a Chinese population. Respir. Res. 6: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deniset J. F., Hedley T. E., Hlaváčková M., Chahine M. N., Dibrov E., O’Hara K., Maddaford G. G., Nelson D., Maddaford T. G., Fandrich R., et al. 2018. Heat shock protein 60 involvement in vascular smooth muscle cell proliferation. Cell. Signal. 47: 44–51. [DOI] [PubMed] [Google Scholar]
- 39.Knowlton A. A., Srivatsa U. 2008. Heat-shock protein 60 and cardiovascular disease: a paradoxical role. Future Cardiol. 4: 151–161. [DOI] [PubMed] [Google Scholar]
- 40.Aguilera-Aguirre L., Bacsi A., Saavedra-Molina A., Kurosky A., Sur S., Boldogh I. 2009. Mitochondrial dysfunction increases allergic airway inflammation. J. Immunol. 183: 5379–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Napolitano G., Barone D., Di Meo S., Venditti P. 2018. Adrenaline induces mitochondrial biogenesis in rat liver. J. Bioenerg. Biomembr. 50: 11–19. [DOI] [PubMed] [Google Scholar]
- 42.Pessayre D. 2007. Role of mitochondria in non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 22(Suppl. 1): S20–S27. [DOI] [PubMed] [Google Scholar]
- 43.Lin J., Wu P. H., Tarr P. T., Lindenberg K. S., St-Pierre J., Zhang C. Y., Mootha V. K., Jäger S., Vianna C. R., Reznick R. M., et al. 2004. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell 119: 121–135. [DOI] [PubMed] [Google Scholar]
- 44.Arany Z., He H., Lin J., Hoyer K., Handschin C., Toka O., Ahmad F., Matsui T., Chin S., Wu P. H., et al. 2005. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab. 1: 259–271. [DOI] [PubMed] [Google Scholar]
- 45.Teyssier C., Ma H., Emter R., Kralli A., Stallcup M. R. 2005. Activation of nuclear receptor coactivator PGC-1alpha by arginine methylation. Genes Dev. 19: 1466–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park M. J., Kim D. I., Lim S. K., Choi J. H., Kim J. C., Yoon K. C., Lee J. B., Lee J. H., Han H. J., Choi I. P., et al. 2014. Thioredoxin-interacting protein mediates hepatic lipogenesis and inflammation via PRMT1 and PGC-1α regulation in vitro and in vivo. J. Hepatol. 61: 1151–1157. [DOI] [PubMed] [Google Scholar]
- 47.Ohno M., Komakine J., Suzuki E., Nishizuka M., Osada S., Imagawa M. 2011. Interleukin enhancer-binding factor 3 functions as a liver receptor homologue-1 co-activator in synergy with the nuclear receptor co-activators PRMT1 and PGC-1α. Biochem. J. 437: 531–540. [DOI] [PubMed] [Google Scholar]
- 48.Garcia M. M., Guéant-Rodriguez R. M., Pooya S., Brachet P., Alberto J. M., Jeannesson E., Maskali F., Gueguen N., Marie P. Y., Lacolley P., et al. 2011. Methyl donor deficiency induces cardiomyopathy through altered methylation/acetylation of PGC-1α by PRMT1 and SIRT1. J. Pathol. 225: 324–335. [DOI] [PubMed] [Google Scholar]
- 49.Ojiaku C. A., Cao G., Zhu W., Yoo E. J., Shumyatcher M., Himes B. E., An S. S., Panettieri R. A., Jr 2018. TGF-β1 evokes human airway smooth muscle cell shortening and hyperresponsiveness via Smad3. Am. J. Respir. Cell Mol. Biol. 58: 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le A. V., Cho J. Y., Miller M., McElwain S., Golgotiu K., Broide D. H. 2007. Inhibition of allergen-induced airway remodeling in Smad 3-deficient mice. J. Immunol. 178: 7310–7316. [DOI] [PubMed] [Google Scholar]
- 51.Gregory L. G., Mathie S. A., Walker S. A., Pegorier S., Jones C. P., Lloyd C. M. 2010. Overexpression of Smad2 drives house dust mite-mediated airway remodeling and airway hyperresponsiveness via activin and IL-25. Am. J. Respir. Crit. Care Med. 182: 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gomis R. R., Alarcón C., Nadal C., Van Poznak C., Massagué J. 2006. C/EBPbeta at the core of the TGFbeta cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell 10: 203–214. [DOI] [PubMed] [Google Scholar]
- 53.Tang Q. Q., Zhang J. W., Daniel Lane M. 2004. Sequential gene promoter interactions of C/EBPbeta, C/EBPalpha, and PPARgamma during adipogenesis. Biochem. Biophys. Res. Commun. 319: 235–239. [DOI] [PubMed] [Google Scholar]
- 54.Roth M., Black J. L. 2009. An imbalance in C/EBPs and increased mitochondrial activity in asthmatic airway smooth muscle cells: novel targets in asthma therapy? Br. J. Pharmacol. 157: 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopez J. M., Hegardt F. G., Haro D. 2001. Differential expression of cytosolic and mitochondrial 3-hydroxy-3-methylglutaryl CoA synthases during adipocyte differentiation. Mol. Cell. Biochem. 217: 57–66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.