Abstract
Targeted delivery of antisense oligonucleotide (AON) drugs is a promising strategy to increase their concentration in the desired tissues and cell types while reducing access to other organs. Conjugation of AONs to N-acetylgalactosamine (GalNAc) has been shown to efficiently shift their biodistribution toward the liver via high-affinity binding to the asialoglycoprotein receptor (ASGPR) expressed at the surface of hepatocytes. Nevertheless, GalNAc conjugation does not prevent accumulation of AONs in the kidney cortex, and GalNAc-conjugated AONs might cause kidney toxicities, for example, under conditions of ASGPR saturation. Here, we investigated the nephrotoxicity potential of GalNAc-conjugated AONs by in vitro profiling of AON libraries in renal proximal tubule epithelial cells (PTECs) and in vivo testing of selected candidates. Whereas GalNAc-conjugated AONs appeared generally innocuous to PTECs, some caused mild-to-moderate nephrotoxicity in rats. Interestingly, the in vivo kidney liabilities could be recapitulated in vitro by treating PTECs with the unconjugated (or naked) parental AONs. An in vitro mechanistic study revealed that GalNAc conjugation attenuated AON-induced renal cell toxicity despite intracellular accumulation similar to that of naked AONs and independent of target knockdown. Overall, our in vitro findings reveal ASGPR-independent properties of GalNAc AONs that confer a favorable safety profile at the cellular level, which may variably translate in vivo due to catabolic transformation of circulating AONs.
Keywords: antisense oligonucleotide, targeted delivery, kidney, ASGPR, toxicity
Introduction
Single-stranded antisense oligonucleotides (AONs) are emerging in clinical development as innovative therapeutics with highly desired properties in respect to target druggability and specificity and to class-related safety.1, 2 Mechanistically, AONs prevent protein synthesis by hybridizing to an intended mRNA, thereby leading to its degradation by the RNase H endonuclease.3 AONs can also act via RNase H-independent mechanisms, for example, by physically blocking the progression of RNA splicing and translation machineries.2, 4 In pre-clinical animal species and in human patients, AONs are currently most commonly formulated in saline solutions, allowing for free uptake (also called gymnosis5) of their naked (unconjugated) form. When administered intravenously or subcutaneously, AONs primarily accumulate in the liver and the kidney, most abundantly in liver Kupffer cells and renal proximal tubular epithelial cells.6, 7 Consequently, liver and kidney are the main tissues where AON-induced toxicities have been observed pre-clinically and clinically.8, 9 The nature of AON-induced organ toxicity is sequence and dose dependent and generally manifests in histopathological changes; namely, single-cell necrosis and apoptosis in the liver and tubular epithelial cell degeneration in the renal cortex.10, 11
Targeted delivery strategies have been used to redistribute AONs from Kupffer cells to hepatocytes for diseases such as hepatitis B, hypercholesterolemia, and transthyretin amyloidosis.12 A well-characterized hepatocyte-targeting approach for small interfering RNAs (siRNAs) and AONs utilizes conjugation to N-acetylgalactosamine (GalNAc), a high-affinity ligand of the asialoglycoprotein receptor (ASGPR), a protein expressed at a high copy number (∼500,000 copies per cell) at the cell surface of hepatocytes.12, 13, 14, 15 GalNAc-mediated liver targeting has been shown to result in robust gene silencing in hepatocytes at a lower effective dose compared to that of naked AONs or siRNA.12, 16 Moreover, by increasing the liver:kidney ratio of AON accumulation, GalNAc conjugation is believed to be associated with a more favorable safety profile.
In drug development, determination of a therapeutic window, i.e., the difference between effective dose and the dose that causes severe side effects, is a necessary step prior to entry into human in order to mitigate the risks associated with new molecules entering clinical testing. In order to detect potential toxicities caused by AON drug candidates pre-clinically, a procedure of repeated high doses in rodents over a 2-week period has been established. At such elevated doses, GalNAc AONs reach the ASGPR saturation on hepatocytes and accumulate in the renal cortex at concentrations similar to that of naked AONs.17 Consequently, nephrotoxicity assessment remains a determinant factor in the preclinical selection of GalNAc AON drug candidates.
We have recently described the implementation of in vitro screening strategies using primary hepatocytes and renal proximal tubular epithelial cells to predict the toxicity risks of naked AONs and select the most favorable AON candidates for further toxicity assessment in mouse and rats. Here, we report that GalNac conjugation attenuates AON-induced tubulotoxicity in vitro, without apparent alterations in intracellular accumulation and efficacy. We found that GalNAc AONs can cause mild-to-moderate kidney lesions in animals, a phenomenon that could be recapitulated in vitro by testing the naked counterparts in renal tubular cells. Our findings highlight the need for in-depth characterization of the biochemical nature of GalNAc AONs and their metabolites in the circulation, tissues, and cells in order to exploit their full potential as therapeutics.
Results
GalNAc-Conjugated AON Libraries Appear Innocuous in an In Vitro Nephrotoxicity Screen
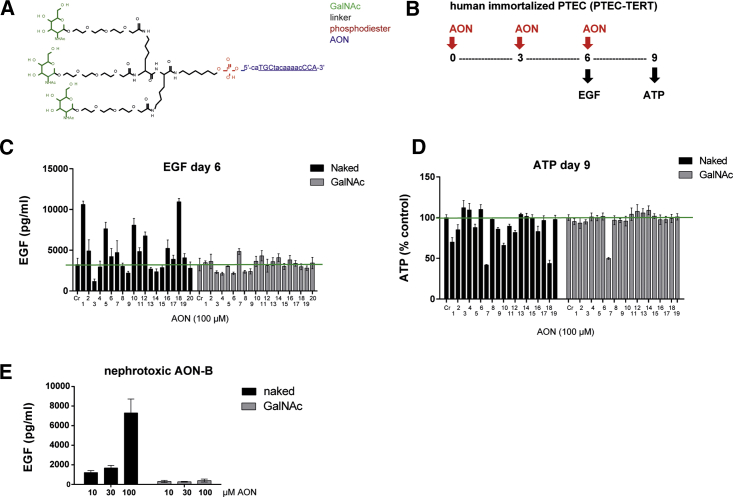
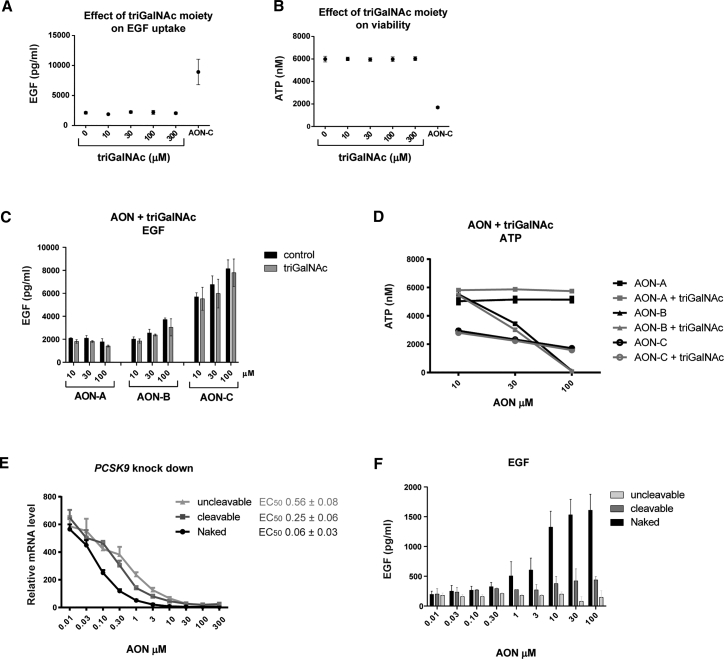
AONs tested in this study consist of locked nucleic acid (LNA) gapmers as described previously18 and in Table S1. In the GalNAc-conjugated form, the 5′ terminus of AONs is linked to three molecules of GalNAc via a triantennary linker (Figure 1A; Table S1). A series of GalNAc-conjugated AONs (hereinafter termed GalNAc AONs) were assessed for potential renal liabilities using a recently described in vitro approach that serves as a primary screen for the selection of AON candidates prior to rodent studies.18 In brief, immortalized human renal proximal tubular epithelial cells (PTECs) are exposed to AONs for up to 9 days, followed by measurement of epidermal growth factor (EGF) consumption at day 6 and intracellular ATP concentrations at day 9 as indicators of cellular functionality and viability, respectively (Figure 1B). Interestingly, GalNAc AONs directed against different targets appeared generally innocuous in the EGF and ATP assays, following an experimental scheme that usually leads to the identification of a significant number of potentially renal toxic AONs, when applied in their naked form at a concentration of 100 μM (Figures 1C, 1D, and S1). Noticeably, a previously characterized in vivo nephrotoxic PCSK9-targeting AON (hereinafter termed AON-B) was included as a control in both the naked and GalNAc-conjugated forms. In striking contrast with the naked AON-B, the GalNAc form of AON-B appeared inert in the EGF assay (Figure 1E), suggesting that GalNAc conjugation may confer novel properties to AONs that alleviate their cytotoxic effects on renal tubular cells.
Figure 1.
GalNAc AONs Are Less Toxic than Naked AONs in Renal Tubular Cells In Vitro
(A) Chemical structure of a GalNAc-AON: 5′-phosphodiester bound (red) of an AON (blue) is coupled to three GalNAc groups (green) via a triantennary linker (black). The 5′-ca (blue) is cleavable. The underlined AON sequence corresponds to the naked AON. (B) Schematic representation of in vitro assay design for AON nephrotoxicity profiling using human immortalized PTECs (PTEC-TERT). Accumulation of EGF in supernatant is detected by ELISA and indicates decreased cell functionality. Reduction of intracellular ATP indicates reduced cell viability. (C and D) Measurement of EGF in the supernatant of PTECs treated for 6 days (C) and measurement of intracellular ATP of PTEC treated for 9 days (D) with a panel of AONs in their Naked or GalNAc form at 100 μM, revealing a generally reduced effect of GalNAc AONs. Data represent means and SD (n = 3). In both assays, statistical significance over control was observed for Naked AONs 1, 2, 5, 7, 9, 10, 16, and 18 (p < 0.005) and GalNAc-AON 7 (p < 0.02); ANOVA with Dunnett’s multiple comparisons test. (E) Concentration-dependent EGF profile of the known nephrotoxic AON-B showing abolition of cytotoxic effect by GalNAc conjugation of AON-B. Data represent means and SD (n = 3).
GalNAc Conjugation Masked the In Vivo Nephrotoxicity Potential of AON Drug Candidates
Abolition of AON-induced cytotoxicity in vitro via GalNAc conjugation did not fully replicate the in vivo profile of GalNac AONs. Indeed, mild histopathological abnormalities were observed in the renal cortex of one rat treated with GalNac AON-B in a 2-week study (Figure 2A). The incidence and degree of severity caused by GalNAc AON-B were significantly reduced compared to naked AON-B (Figure 2A), but the contribution of GalNac conjugation remains unclear, given that GalNac AON-B was administered at half the dose of the naked counterpart. In another rat study, two GalNAc AONs identified as safe candidates in the PTEC screen showed unexpected signs of nephrotoxicity in the standard 2-week rat study (AON-D and -E; Figure 2B). When re-tested in their naked form in PTECs, AON-D and AON-E were cytotoxic, as revealed by elevated EGF level (Figure 2C) and the emergence of morphological alterations (Figure 2D), in stark contrast to the inert profile of GalNAc AON-D and -E (Figures 2C and 2D). It is worth noting that morphological assessment of AON-treated PTECs provided additional insights into the degree of severity of AON-induced tubulotoxicity, allowing for a closer recapitulation of the in vivo degree of toxicity compared to the EGF assay alone. Altogether, the data show that GalNac conjugation of AONs is more protective to renal tubular cells in vitro than in vivo, plausibly due to degradation of the GalNac moiety of circulating AONs; the data also highlight the importance of testing the naked form of AONs when using PTEC safety assays for the selection of candidates prior to animal testing.
Figure 2.
GalNAc AON-Induced Nephrotoxicity Is Recapitulated using Naked AONs In Vitro
(A and B) Histological evaluation of rat kidneys showing histopathological manifestations induced by GalNAc and naked AON-B (20 and 40 mg/kg, respectively) (A) and by GalNAc AON-D and GalNAc AON-E (50 mg/kg dose) (B). Wide, open arrows indicate examples of areas with tubular degeneration and/or regeneration. Fine, filled arrows indicate necrotic or apoptotic cells. 1/4 means that one animal in the group (n = 4) showed minimal tubular degeneration. 4/4 means that all animals in the group (n = 4) showed minimal to moderate lesions. H&E, 20× magnification. Lower panels are enlarged images of sections shown in upper panels, except for vehicle in (B), which is from a different section. (C and D) The toxicity potential of GalNAc AON-D and -E is revealed in PTECs when AONs are applied in their naked form only, as manifested by elevated EGF at day 6 (C) and by morphological abnormalities visible under the bright field (20×, representative of n = 3) at day 9, as shown for the 100-μM concentration (D). Data in (C) represent means and SD (n = 3).
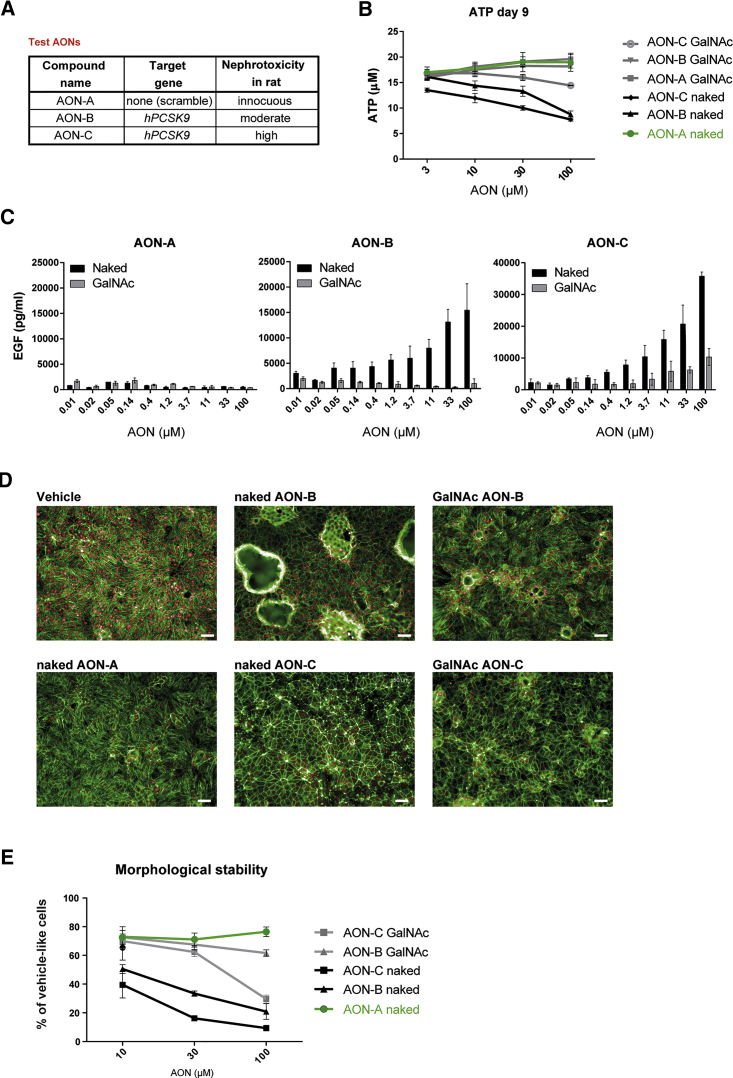
GalNAc Conjugation Attenuates the Cytotoxicity Effect of AONs in Isolated Renal Tubular Cells
We next sought to investigate the mechanisms underlying the differential cytotoxicity of GalNAc versus naked AONs at the cellular level. Tool AONs of known in vivo kidney liabilities (Figure 3A; see also Moisan et al.18)—namely, an innocuous scramble AON (AON-A), a moderate toxic anti-PCSK9 AON (AON-B), and a highly toxic anti-PCSK9 AON (AON-C)—were synthesized as naked and GalNAc conjugates and were tested for their capacity to induce cytotoxicity in PTECs. A dose-response analysis showed a near-complete abolition of the cytotoxic effect of AON-B and a significantly reduced cytotoxic effect of AON-C when tested in the GalNAc-conjugated form in the ATP and EGF assays (Figures 3B and 3C). Morphological changes were visible in PTECs after 9 days of treatment with naked AON-B and AON-C. However, PTECs displayed a more normal appearance when treated with the GalNAc counterparts (Figure 3D). A quantitative analysis of morphological abnormalities in treated versus untreated cells further confirmed the apparent inert or attenuated effect of GalNAc-conjugated AONs on PTECs (Figure 3E). These data further suggest that GalNAc conjugation can mitigate the nephrotoxic potential of AON sequences by direct protection of renal tubular cells.
Figure 3.
GalNAc Conjugation Attenuates the Cytotoxicity Potential of AONs in Renal Tubular Cells
(A) Test AONs and their respective targets and in vivo toxicity scores. (B and C) Measurement of intracellular ATP of PTECs (B) and accumulation of EGF in PTEC supernatants (C) showing concentration-dependent cytotoxicity of nephrotoxic AON-B and AON-C when applied in their naked form for 6 and 9 days, respectively, whereas their GalNAc-conjugated form showed flat EGF and ATP profiles, indicative of reduced cytotoxicity. Innocuous AON-A was used as a control and remained inert in both forms. Data represent means and SD (n = 3). Statistical significance over control was observed at 10, 30, and 100 μM for naked AON-B and AON-C (p < 0.0001) and at 30 and 100 μM for GalNac AON-C (p < 0.001); ANOVA with Dunnett’s multiple comparisons test. (D and E) Morphological alterations induced by 9 days of exposure to 100 μM of AONs are mitigated by GalNAc conjugation. In (D), representative high-content images are shown of AON-treated PTECs stained for actin (phalloidin, green) and nuclei (DRAQ5, red). Scale bars, 50 μm. In (E), quantification of AON-induced changes in RPTEC morphology expressed as cells retaining normal, vehicle-like morphology, nine fields per condition, is shown; data represent mean and SD (n = 3). Statistical significance over control was observed at 10 μM (p < 0.008), 30 μM, and 100 μM (p < 0.0001) for naked AON-B and AON-C and at 100 μM for GalNac AON-C (p < 0.0001); ANOVA with Dunnett’s multiple comparisons test.
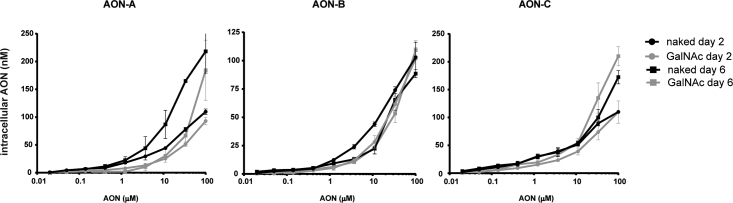
GalNAc Conjugation Does Not Impair Total Intracellular Accumulation of AONs in Renal Tubular Cells
The reduced cytotoxicity potential of GalNAc AONs described earlier could result from a reduction in intracellular accumulation. Indeed, kidney proximal tubular cells do not express the ASGPR, and GalNAc conjugation may hypothetically impair cellular uptake in vitro. This hypothesis was verified by performing a hybridization ELISA (hELISA), using single-stranded oligonucleotide probes that bind to the DNA sequences of AON-A, -B, and C; specifically, on cell lysates from AON-treated PTECs. hELISA quantifies all molecular entities containing the oligonucleotide sequence, whether still conjugated to the intact GalNAc or not.19 The data show a dose-response accumulation of AONs in PTECs with maximum detected concentrations around 100–200 nM in cell lysates after 2 and 6 days of treatment (Figure 4). Interestingly, the intracellular content of AONs was comparable between the GalNAc and naked forms for all three pairs and at the two tested time points (Figure 4). Therefore, the reduced toxicity of GalNAc AONs is unlikely to be the consequence of poor uptake and reduced total intracellular content.
Figure 4.
Similar Intracellular Concentrations of Naked and GalNAc AONs in Renal Tubular Cells
Concentrations of naked and GalNAc-modified AON-A, AON-B, and AON-C in PTEC cell lysates determined by hELISA after 2 days and 6 days of exposure. Data represent means and SD (n = 3).
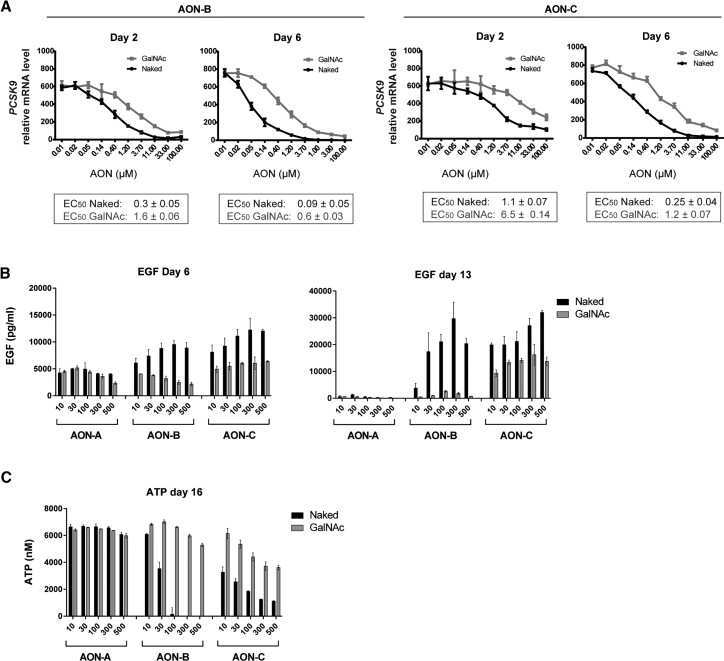
Toxicity and Pharmacology Profiles of GalNAc AONs Do Not Correlate in Renal Tubular Cells
AONs have been reported to accumulate in unproductive cellular compartments after unassisted delivery, implying that a significant fraction of AONs does not reach the target mRNA in the nucleus.20 It is plausible that GalNAc and naked AONs, despite accumulating at equivalent intracellular concentrations in PTECs, differ in their capacity to bind to and knock down on-target as well as off-target sequences. Thus, the potency of GalNAc and naked AONs was compared using a similar experimental scheme as described earlier for intracellular accumulation (Figure 4), except that PTECs were collected in mRNA lysis buffer followed by mRNA analysis of the PCSK9 target gene and PPIB housekeeping gene. Dose-dependent curves of PCSK9 mRNA knockdown were obtained, and half maximal effective concentration (EC50) was calculated for each treatment (Figure 5A). As reported previously, AON-B is more potent in reducing target mRNA than AON-C in PTECs. For both AON-B and AON-C, the naked form was approximately 5 times more potent pharmacologically than the GalNAc as measured after 2 and 6 days of treatment (Figure 5A).
Figure 5.
Reduced Toxicity of GalNac AON Does Not Correlate with Reduced Efficacy
(A) Gene expression analysis showing PCSK9 target gene knockdown by AON-B and AON-C in their GalNAc and naked forms after 2 and 6 days of exposure in PTECs. ECs50 are shown and indicate reduced potency of GalNAc AON-B and GalNAc AON-C compared to the naked counterparts. (B and C) Concentration-dependent effects of GalNAc and Naked AON-A, AON-B, and AON-C on EGF uptake (B) and intracellular ATP (C) in PTECs treated with a concentration of AON up to 500 μM for 16 days. The EGF profile at days 6 and 13 and ATP profile at day 16 showed a reduction of cytotoxic effect by GalNAc conjugation, despite the elevated concentration and extended exposure time. Data represent means and SD (n = 3).
The severity of AON-induced hepatotoxicity and nephrotoxicity of a given oligonucleotide sequence and chemistry is generally dose and time dependent and can be correlated with hybridization-dependent mechanisms such as on- and off-target knockdown. Given the estimated difference in pharmacological potency, we speculated that increasing the concentration of GalNAc AON by 5 times the concentration of the naked form, and for an extended number of treatments, could lead to an equivalent, or more similar, severity of cytotoxicity. An additional set of experiments, in which the maximal AON concentration was increased from 100 to 500 μM and time of treatment was increased from 9 to 16 days, was conducted. Measurement of EGF after 6 and 13 days of treatment (Figure 5B) and ATP at the 16-day endpoint (Figure 5C) revealed that GalNAc AON-B remains generally innocuous and does not show apparent signs of cytotoxicity using these readouts. Noticeably, AON-B showed a dramatic loss of ATP content at day 16, previously unappreciated at the standard 9-day time point but in accordance with the suggested time-dependent effects of cytotoxic AONs on renal cell viability.18 Consistent with Figure 3, GalNAc AON-C did lead to increased EGF and reduced ATP but to a lesser degree than that of the naked AON-C. At the maximal concentration of 500 μM, GalNAc AON-C was better tolerated by PTECs than the naked AON-C at any tested concentrations (Figures 5B and 5C).
Impact of Free GalNAc Moieties and Linker Stability on AON-Mediated Cytotoxicity in Renal Tubular Cells
GalNAc transferases are capable of utilizing GalNAc as a substrate for O-glycosylation of proteins, a post-translational modification that has been shown to occur on epidermal growth factor (EGFR) and enhance its EGF-dependent functions.21, 22 Therefore, subjecting PTECs to GalNAc moieties, via exposure to GalNAc AONs, may have direct effects on EGFR function that could interfere or mask the AON-mediated downregulation of EGF uptake by EGFR. We verified this hypothesis by treating PTECs with free triantennary GalNAc (hereinafter termed triGalNAc) in the absence or presence of naked AONs. Free triGalNAc applied to PTECs at concentrations up to 300 μM had no effect on EGF uptake and intracellular ATP content (Figures 6A and 6B). Moreover, combining free triGalNAc with AONs did not mask the cytotoxic effects of AON-B and AON-C, as measured by EGF accumulation (Figure 6C) and ATP depletion (Figure 6D).
Figure 6.
Effect of Free GalNAc Moiety and Linker Stability on AON-Mediated Cytotoxicity
(A and B) Exposing PTECs to free molecules of triGalNAc at the indicated concentrations did not lead to an apparent effect on EGF uptake after 6 days (A) and cell viability after 9 days (B). AON-C at 100 μM was used as toxic control. (C and D) Concentration-dependent effects of AON-A, AON-B, and AON-C on EGF uptake (C) and intracellular ATP (D) in PTECs in the absence or presence of equimolar concentrations of free GaNAc (triGalNAc), showing that the AON-induced cytotoxicity is not mitigated by exposure to free GalNAc molecules. Data represent means and SD (n = 3). An ANOVA with Sidak’s multiple comparisons test comparing GalNAc to Naked of the same AON and concentration indicates no significance for all conditions. (E) Gene expression analysis showing PCSK9 target gene knockdown in PTECs after 2 days of exposure to AON-B in its naked form or after conjugation to GalNAc via a cleavable versus an uncleavable linker. Data points represent means and SD (n = 3). ECs 50 are shown and indicate a further reduction of GalNAc-AON-B potency when coupled to GalNAc via an uncleavable linker compared to a cleavable linker. (F) Significant EGF accumulation in PTECs is observed after treatment with naked AON-B (p < 0.0001 at 10, 30, and 100 μM) but not with cleavable and uncleavable GalNAc AON-B. Data represent means and SD (n = 3); ANOVA with Dunnett’s multiple comparisons test.
Upon cellular uptake of GalNAc AONs, the GalNAc moiety is degraded, and the linker part is cleaved by intracellular enzymes. By-products of GalNAc-AON biotransformation in hepatocytes have been described,19, 23 but their existence or persistence and effects in kidney cells remain unknown. A non-degradable GalNAc moiety is not available to generate tool compounds, but GalNAc AONs with an uncleavable linker at the 5′-CA position (see Table S1) could be synthesized. Therefore, we investigated the potency and toxicity of such uncleavable linker-baring GalNAc-AON-B, in which the linker is protected from enzymatic cleavage and may prolong the duration of GalNAc conjugation to AON after cellular uptake. The data revealed that the uncleavable GalNAc-AON-B showed more pronounced impairment of target knockdown (Figure 6E) and EGF accumulation (Figure 6F) than the cleavable GalNAc-AON-B in comparison to the naked form. These results suggest that coupling of AON-B to the linker-GalNAc moiety is responsible, at least in part, for the reduced cytotoxicity potential of AON-B.
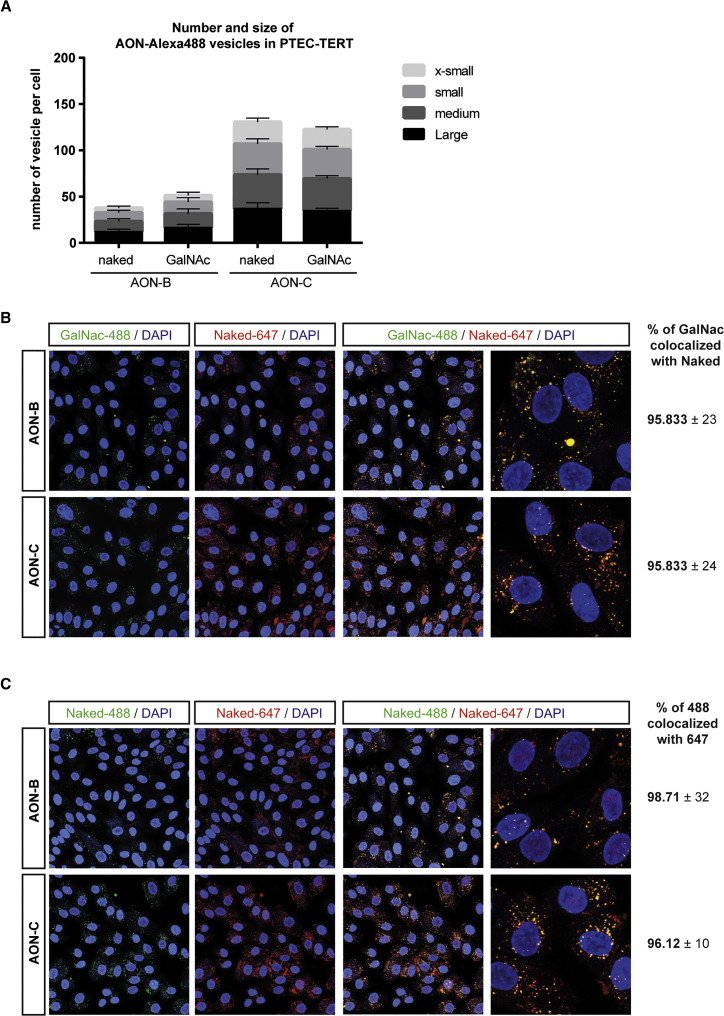
GalNAc and Naked AONs Co-localize in Cytoplasmic Vesicles in Renal Tubular Cells
The fate and effects of GalNAc AONs may differ from naked AONs as a result of non-hybridization-dependent effects, such as protein binding and alternative cell-trafficking routes. We used fluorescently labeled AONs to investigate whether GalNAc conjugation may alter the accumulation patterns of AONs. We first treated sub-confluent PTECs with Alexa 488-labeled AONs in their naked or GalNAc form and captured images of AON accumulation after 60 min of incubation at 37°C. As described previously,20, 24 AONs accumulate mainly in cytoplasmic vesicles. Intracellular vesicles showing accumulation of Alexa 488-GalNAc and Alexa 488-naked AONs were similar in abundance and size (Figure 7A). The higher number of vesicles detected for AON-C compared to AON-B is most likely due to sequence-related properties (Figure 7A). Next, we hypothesized that common trafficking mechanisms will result in co-localization of GalNAc and naked AONs, whereas divergent mechanisms will manifest as distinct intracellular accumulation patterns. Thus, we co-treated PTECs with Alexa Fluor 647-naked AONs and Alexa 488-GalNAc AONs and captured images of AON accumulation after 60 min of incubation at 37°C. The vast majority of vesicles contained both forms of AONs, as indicated by a yellow signal after merging confocal images from the 488-nm and 647-nm channels (Figure 7B). A quantitative analysis revealed that more than 95% of Alexa 488-positive vesicles were also Alexa Fluor 647 positive, suggesting a near-perfect match of intracellular co-localization of GalNAc AONs with naked AONs, for both AON-B and AON-C (Figure 7B). The fluorescence pattern generated by Alexa Fluor 647 AONs was generally more intense, was diffuse, and appeared less specific than that of Alexa 488 and was, therefore, not used to calculate the percentage of co-localization. As positive control of maximal co-localization, PTECs were co-treated with pairs of Alexa Fluor 647-naked/Alexa 488-naked AONs (Figure 7C). This experiment confirmed that the Alexa Fluor 647 labeling generated numerous unspecific small vesicles that were Alexa Fluor 647-positive but Alexa 488-negative vesicles and provided a rationale for the calculation of co-localization used in Figure 7B. Overall, the data suggest that GalNac and naked AONs follow similar intracellular paths in PTECs during the first 60 min of exposure, despite differences in toxicity profile.
Figure 7.
Naked and GalNAc AONs Form Cytoplasmic Vesicles of Similar Abundance, Size, and Localization in Renal Tubular Cells
(A) Abundance and size of AON-containing vesicles: PTECs were treated with Alexa 488 AONs in their naked or GalNAc-conjugated form for 60 min at 37°C prior to confocal imaging. Nuclei were revealed using NucBlue staining. Alexa 488 cytoplasmic vesicles were classified according to size (x-small: 2–3 pixels; small: 3–4 pixels; medium: 4–5 pixels; and large: ≥5 pixels). The number of vesicles of a given size was divided by the number of nuclei counted in the same field of view and reported as “number of vesicles per cell” on the y axis. Data represent mean and SD of 10 fields of view from two independent experiments. ANOVA with Sidak’s multiple comparisons test comparing GalNAc to Naked of the same AON and vesicle size group indicates no significance. (B) Co-localization of naked and GalNAc AONs: PTECs were co-treated with Alexa 488-GalNAc and Alexa Fluor 647-naked AONs for 60 min at 37°C prior to confocal imaging. Nuclei were revealed using NucBlue staining. Selected representative images show that cytoplasmic vesicles formed by GalNAc AONs (green signal, left panels) and naked AONs (red signal, middle panels) co-localize in PTECs (yellow signal, merged channel, right panels). The percentage of Alexa 488-GalNAc AON vesicles that co-localize with Alexa Fluor 647-naked AONs is reported for AON-B and AON-C. (C) Control of co-localization of AONs: Same as in (B), except that PTECs were co-treated with naked AONs only, either Alexa Fluor 647 or Alexa 488 labeled. Selected representative images reveal that Alexa Fluor 647 lead to a more diffuse and less specific signal than Alexa 488 when labeled to AONs. The percentage of Alexa 488-naked AON vesicles that co-localize with Alexa Fluor 647-naked AONs is reported for AON-B and AON-C and represent the maximum co-localization that can be detected for a given sequence and form of AON. Data represent mean and SD of 10 fields of view from two independent experiments.
Discussion
In vitro data presented here revealed properties conferred by GalNAc conjugation that attenuate the toxicity effect of AONs at the cellular level, and that may contribute to their favorable safety profile in vivo, in addition to biodistribution. Studies in human PTECs were performed to address the underlying molecular mechanisms and uncovered novel considerations for cell-based assays aimed at predicting the nephrotoxicity potential of GalNAc AONs prior to animal testing.
Based on in vitro profiling of a broad panel of GalNAc AONs targeting various mRNA targets, we observed that GalNAc conjugation of AONs generally attenuated the cytotoxicity effect of the selected AON libraries. Using the PCSK9-targeting tool AONs of known renal liabilities18 in their naked and GalNAc forms, we confirmed that attachment of a GalNAc moiety to nephrotoxic AONs decreased their cytotoxicity, as measured by functionality, viability, and morphology readouts. At the molecular level, GalNAc AONs were approximately five times less potent pharmacologically than the naked counterparts, despite accumulating at equivalent intracellular concentrations in PTECs. A direct causal effect between reduced gene knockdown and reduced cytotoxicity was not supported by the experimental data overall. Thus, at treatment conditions leading to similar efficacy, i.e., 30–100 μM of AONs for 2 to 6 days, the measured toxicity remained significantly inferior for GalNAc AONs. Moreover, increasing the concentration of GalNAc AONs by a factor of five did not result in equalizing the toxic effect of the matching naked AON, even after additional and prolonged exposure. Remarkably, a nephrotoxic PCSK9-targeting tool AON18 appeared inert in toxicity assays at concentrations up to 500 μM when applied in its GalNAc form, a profile exacerbated by rendering the GalNAc linker uncleavable. These observations suggest that the safer properties of GalNAc AONs are inherent to the biochemical attributes of these new molecular entities, as opposed to the combination of independent effects of GalNAc and AON. Accordingly, the addition of free triGalNAc moieties to vehicle or AON-treated PTEC supernatants did not alter the functionality or viability of PTECs and did not reproduce the effect of GalNAc conjugation to AON-induced toxicity.
We speculated that divergent intracellular trafficking and accumulation patterns may account for the attenuated toxicity of GalNAc AONs. A series of co-localization experiments using fluorescently labeled AONs could not support this hypothesis and rather showed that GalNAc AONs formed cytoplasmic vesicles reminiscent of naked AONs and almost entirely co-localized with naked AONs when co-incubated. In-depth cellular uptake and trafficking studies using unlabeled AONs, and including time resolution and molecular staining of receptors and cell compartments, could confirm or refute the initial attempt presented here.
An alternative explanation to the attenuated cytotoxicity of GalNAc-AON involves the role of metabolite formation in the mechanisms of AON-induced toxicity. Degradation of AONs might be altered by GalNAc conjugation, and the resulting metabolites might be less detrimental to cell functions, either in nature or abundance, than those generated by naked AONs. Experiments dedicated to defining the metabolic profiles of GalNAc and naked AONs in renal tubular cells would be very helpful to verify the correlation between by-products of AON breakdown and emergence of tubulotoxicity in vitro and in vivo.
Whereas a GalNAc AON appeared innocuous in PTECs, administration of the same molecule in rats was associated with tubulotoxicity of a histological nature similar to that observed in animals treated with the naked form, although to a lesser degree of severity. Screening of AONs in their naked rather than GalNAc-conjugated form would have predicted their nephrotoxicity potential. It is plausible that, in vivo, the biotransformation of GalNac AONs releases a pool of naked AONs and metabolic by-products that are potentially toxic to renal tubular cells over time, a phenomenon not reproduced in isolated PTECs in vitro. Therefore, performing early in vitro safety screening assays using the naked version of AONs is likely to provide higher sensitivity in the prediction of the nephrotoxicity potential of GalNac-conjugated AONs. The development of bioanalytical methods capable of discriminating the GalNAc-conjugated form of an AON from the naked counterpart will be key in depicting the pharmacokinetic profile of circulating GalNAc-AON drug candidates and their biodistribution and will certainly contribute to the refinement of targeting approaches and elaboration of risk-mitigation strategies.
Overall, the findings presented here revealed properties conferred by GalNAc conjugation that ameliorate the nephrotoxicity potential of AONs at the cellular level and support a refined in vitro screening strategy toward identification of an optimized AON drug candidate. These may apply broadly to GalNAc-conjugated antisense therapies. In a recent study reporting on the nonclinical safety profile of RNAi, administration of GalNAc-conjugated siRNAs (GalNAc-siRNAs) at doses as high as 300 mg/kg did not result in any histopathologic signs of nephrotoxicity in both rat and monkey.25 A direct comparison to the naked counterparts was not possible, given the instability of naked siRNAs in plasma, but further investigations using various targeting approaches across oligonucleotide-based technologies would be valuable to decipher the full potential of RNAi technologies as medicines.
Materials and Methods
Chemistry and Labeling of AONs
AON-A through AON-E consist of 14- to 16-nt-long gapmers, with gap segments composed of 2′-deoxynucleotides flanked by wing segments comprising nucleotides with LNA sugar modifications. Sequences, backbone modifications, and GalNAc conjugation are provided in Table S1 for AON-A, -B, and -C. AON-D and -E and AONs from libraries shown in Figure 1C and Figure S1 were GalNAc conjugated as described for AON-A through AON-C. Individual AON sequences may be provided upon request.
For labeling of AONs with Alexa 488 and Alexa Fluor 647, AONs were first synthesized with a di-thio linker at the 3′ end. After cleavage from resin and de-protection of the crude AON (according to manufacturer’s instructions; Universal CPG 500A (DMT Off) Resin; Link Technologies, catalog no. 2438-P021), the di-thio bond was reduced in NaHCO3 (pH 8.5) buffer (Fisher Scientific, S/4240/53), with an excess of DTT (Sigma Aldrich, 43815) as a reduction agent. The free thio-AON was purified by preparative high-performance liquid chromatography (HPLC) (according to manufacturer’s instructions; Agilent 1260 with C18 column; Buffer A: NaOAc buffer [pH 8], and Buffer B: acetonitrile [ACN] as mobile phase) and coupled with Alexa Fluor (Thermo Fisher Scientific; Alexa 488 C5 Maleimide, catalog no. A10254, or Alexa Fluor 647 C2 Maleimide, catalog no. A20347) as 2-fold excess in PBS buffer and purified by preparative HPLC to obtain final Alexa Fluor AONs.
Rat Studies
Purpose-bred Wistar Han Crl:WI(Han) male rats obtained from Charles River Laboratories at 7–8 weeks of age were acclimatized for at least 5 days before dosing. The animals were group-housed under standard environmental conditions (22°C ± 2°C; relative humidity, 50% ± 20%; a 12-hr:12-hr light:dark cycle; and pelleted food and water ad libitum), were offered enriched environment in an Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC)-accredited facility, and were regularly and carefully monitored. All procedures were conducted in strict adherence to the Swiss federal ordinance on animal protection and welfare, according to the rules of AAALAC, and with the explicit approval of the local veterinary authority (Kantonales Veterinäramt Basel-Stadt, Basel, Switzerland).
Test compounds were dosed subcutaneously at 40 mg/kg for naked AON-B, 20 mg/kg for GalNAc-AON-B, and 50 mg/kg for Gal-NAc-AON-D and GalNAc-AON-E on days 1 and 8 at 2.5 mL/kg in the interscapular region. Control-group animals received saline as vehicle control. Rats were deeply anesthesized with isoflurane, followed by exsanguination on day 15. Kidney samples were collected and fixed by immersion in 10% neutral buffered formalin. Tissue samples were subsequently dehydrated and embedded in paraffin blocks that were sectioned to 5 μm and stained with H&E. Following histopathological examination, the H&E sections were scanned at 20× magnification with an Aperio ScanScope AT (Leica Biosystems) scanner, and photomicrographs were captured via ImageScope software.
PTEC Culture
PTEC-TERT1 cells (renal PTEC [RPTEC]/TERT1, CHT-003-0002; Evercyte, Wien, Austria) were cultured according to the manufacturer’s instructions in PTEC medium: Mix 1/1 of DMEM (#11966-025) and Ham’s F-12 nutrients (#21765-29) (Thermo Fisher Scientific, Waltham, MA, USA) containing 1% penicillin-streptomycin (GIBCO, #15140-122), 10 mM HEPES (GIBCO, #15630-056), 5.0 μg/mL human insulin (GIBCO, #41400-045), 5.0 μg/mL human transferrin (GIBCO, #41400-045), 8.65 ng/mL sodium selenite (GIBCO, #41400-045), 0.1 μM hydrocortisone (#H6909, Sigma-Aldrich, St. Louis, MO, USA), 10 ng/mL human recombinant EGF (#236-EG-200, R&D Systems, Minneapolis, MN, USA), 3.5 μg/mL ascorbic acid (Sigma-Aldrich, #A4544 powder), 25 ng/mL prostaglandin E1 (Sigma-Aldrich, #P5515), 3.2 pg/mL Triiodo-L-thyronine (Sigma-Aldrich, #T-5516), and 100 μg/mL Geneticin (GIBCO, #10131-027).
AON Treatment
For AON toxicity assessment, PTEC-TERT1 cells were seeded into 96-well plates (#356702; Corning, New York, NY, USA) at a density of 20,000 cells per well in PTEC medium and grown until confluence prior to treatment with AONs. AONs were dissolved in PBS and added to the cell culture at a final concentration of 3, 10, 30, or 100 μM in a final volume of 100 μL for 9 days and at a final concentration of 30, 100, 300, or 500 μM for 16 days as specified in the figure legends. Medium was stored at −20°C for cytokine analysis and refreshed along with AONs every 3 days. PBS served as vehicle control.
For assessment of the effect of GalNAc moieties, PTEC-TERT cells were cultured, seeded, and analyzed as described earlier for AON toxicity. TriGalNAc was synthesized in house, dissolved in PBS, and added to the cell culture at a final concentration of 0.02, 0.05, 0.14, 0.4, 1.2, 3.7, 11, 33, or 100 μM, or together with AONs at a concentration of 10, 30, or 100 μM, in a final volume of 100 μL for 9 days. Medium containing TriGalNAc and AONs was refreshed every 3 days. PBS served as vehicle control.
ATP Assay
Cell viability was determined by measurement of intracellular ATP levels after 9 days or 16 days of AON treatment using the CellTiter-Glo Luminescent Cell Viability Assay (#G7571, Promega, Madison, WI, USA) according to manufacturer’s instructions. For gene expression analysis, cells were collected in Panomics Lysis Mixture (QuantiGene Sample Processing Kit, #QS0101, Affymetrix eBioscience, San Diego, CA, USA) at day 6 of AON treatment, unless otherwise indicated in the figure legends, and stored at −80°C until analysis (described later).
EGF Assay
80 μL of supernatants from PTEC-TERT1 cultures were collected at the indicated time of AON treatment and stored at −20°C until analysis. For EGF measurement, supernatants were thawed on ice, diluted 6× in sample dilution buffer (Bio-Rad, catalog #171-304071), and analyzed by Luminex ELISA using a human EGF singleplex (Bio-Plex Human Cancer Biomarker Panel 2, Bio-Rad, catalog #171-BC603M). The Bio-Plex 200 System (Bio-Rad) was used according to the manufacturer’s instructions. Data are reported as average concentrations and SDs of triplicate wells.
PTEC Morphology
Bright-field images documenting PTEC-TERT1 morphological changes in Figure 1 were taken at day 9 of AON treatment on living cells at 20× magnification.
For high-content imaging of cell morphology, PTEC-TERT1 cells were seeded on CellCarrier-96 collagen-I-coated microplates (PerkinElmer, catalog #6005920). Cell monolayers were fixed with 4% paraformaldehyde in PBS for 20 min at room temperature. Cells were permeabilized and blocked using 0.1% Saponin (Sigma-Aldrich, catalog #47036) and 4% gelatin from cold-water fish skin (Sigma-Aldrich, catalog #G7041) in PBS for 10 min at room temperature. ActinGreen 488 ReadyProbes Reagent (Thermo Fisher Scientific, catalog #R37110) was applied according to the manufacturer’s instructions. DRAQ5 (Thermo Fisher Scientific catalog # 62251) was used for nuclear and cytoplasmic staining according to the vendor’s instructions. Images were acquired on the Operetta High-Content Imaging System (PerkinElmer). Fluorescence images were taken from nine fields per well (central area, 3.1 mm2) at 20× magnification.
To quantify AON-induced changes in cell morphology, high-content image information was extracted and analyzed using the Columbus Image Data Storage and Analysis software (Operetta Application Guide 2013, PerkinElmer). Computational workflow included, in the following order: (1) identification of primary objects (cell nuclei and cytoplasm) based on DRAQ5 staining; (2) exclusion of cropped cells at image borders; and (3) sequential calculation of morphological properties based on two software building blocks—STAR morphology, assessing the outer shape of objects and fluorescence intensity distribution inside the objects, and SER texture properties, representing the regularity of intensities inside the objects. Phenotypes of interest were quantified using the PhenoLOGIC machine learning module of the Columbus Image Analysis software, which classifies the cells based on the pre-calculated STAR and SER features. The algorithm was trained by interactive machine learning on two cell populations: vehicle-treated and AON-C (100 μM)-treated cells. For each population of vehicle- and drug-treated cells, at least 100 training objects were randomly selected over the whole image area. The algorithm identified a linear combination of properties that best separated the training samples. Data are expressed as percentage of cells retaining normal, vehicle-like morphology.
Quantification of Intracellular AON Content by hELISA
Intracellular AON content was determined by hELISA, using a biotinylated capture probe and a digoxigenin-conjugated detection probe as described previously.26 A specific pair of capture and detection probes was designed for AON-A, AON-B, and AON-C, as reported in Table S2.
PTEC-TERT cells were cultured and seeded as described earlier for AON treatment. After 2 and 6 days of incubation with AON, PTEC-TERT cells were washed with 0.1 mg/mL heparin solution (Sigma-Aldrich, #H3393-25KU) and lysed in 75 μL RLT Plus buffer (QIAGEN, #1053393). Cell lysates were diluted and incubated with 35 nM biotinylated capture probe and 30 nM digoxigenin-coupled detection probe for 30 min at room temperature in SSCT buffer—5× saline sodium citrate buffer (SSC Buffer 20× Concentrate, Sigma-Aldrich, #6639) containing 0.05% Tween 20 (Sigma-Aldrich, #P9416)—in a 96-well plate. The assembled complex is then captured on a streptavidine-coated ELISA plate (Nunc 436014) for 1 hr, and after three washing steps with 2× SSCT buffer, each well is incubated with an anti-digoxigenin-alkaline phosphatase (AP)-Fab fragment (Roche, #11093274910) for 1 hr at room temperature. After three additional washing steps, BluePhos substrate (Kirkegaard & Perry Labs [KPL], #50-88-00) was added to the plates, and color development was measured spectrophotometrically at 615 nm after 20 min. The concentration of AON in the cell lysate was calculated according to a standard curve generated with the respective AON stock solution.
PCSK9 Knockdown
PTEC-TERT cells were grown in 96-well plates as described earlier for AON treatment. Cells were collected by replacing culture medium with 100 μL 1× RNA lysis mixture (QuantiGene Sample Processing Kit, Thermo Fisher Scientific, #QS0101) per well. RNA lysis mixtures were kept at −80°C until analysis. 20 μL lysates were mixed with mRNA-capture magnetic bead sets (Panomics QuantiGene Plex Sets, Thermo Fisher Scientific, #12871), incubated overnight, processed for branched DNA amplification, and analyzed according to the manufacturer’s instructions (Panomics QuantiGene Plex Assay Kit, Thermo Fisher Scientific, #QP1015). The PPIB probe was used as a housekeeping gene for normalization. Average fluoresence intensity (FI) and the SD of 3 biological replicates were calculated and normalized on vehicle control.
Visualization of AON-Containing Cytoplasmic Vesicles by Confocal Imaging
PTEC-TERT cells were seeded at a density of 5,000 cells per well on glass-bottom 96-well plates (ibidi, #89626-IBI) in PTEC medium containing 2% fetal bovine serum (FBS). At 70% confluency, cells were FBS starved overnight and then stained with the NucBlue Live ReadyProbes Reagent 1 kit (Thermo Fisher Scientific, #R37605) for 20 min at room temperature and incubated for 60 min at 37°C with medium containing 500 nM Alexa Fluor-labeled AONs. Cells were then fixed with 4% paraformaldehyde for 15 min at 37°C, washed 3 times with PBS 1×, and immediately imaged to avoid artifact due to diffusion of AONs post-fixation. Ten images of randomly selected fields of view per biological duplicates were acquired on a laser-scanning confocal microscope (Leica TCS SP8) using a 40× objective. Settings were adjusted to maximize dynamic range and reduce pixel saturation (pixel size: 120.56 nm × 120.56 nm; image size: 1,024 × 1,024; bit depth: 12 bit; number of z sections: 5). Images were analyzed on FIJI software.
For quantification of abundance and size of AON vesicles, Alexa 488 and Alexa Fluor 647 signals were detected and quantified based on a particle-tracking method introduced by Sbalzarini and Koumoutsakos27 and previously adapted for quantification of cytoplasmic vesicles in PTEC-TERT cells. The same adapted method was applied here, except that four levels of radius were adopted to simultaneously detect all the particles with an extra small radius (2–3 pixels), small radius (3–4 pixels), medium radius (4–5 pixels), and large radius (≥5 pixels). NucBlue nuclear staining was used to report the number of cells per field of detection.
General Statistical Analysis
Statistical analysis was carried out by ANOVA using the Dunnett’s or Sidak’s multiple comparisons test at 95% confidence interval and with an alpha of 0.05. AON treatments were analyzed against the vehicle control of similar conditions, unless otherwise stated in the figure legends; p values were adjusted to account for multiple comparisons. N indicates number of biological replicates, i.e., cells from different well plates and passages.
Author Contributions
A.M. conceived the study, supervised experiments, analyzed the data, and wrote the manuscript. S.S. and M.G. supervised experiments and analyzed the data. R.G., B.A., and Y.M. executed in vitro experiments. A.B.-B. and M.O. performed histopathological evaluation of rat kidney
Conflict of Interests
All authors are employees of F. Hoffmann-La Roche. The funder provided support in the form of salaries for authors but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. A.M. is a non-voting shareholder of F. Hoffmann-La Roche. This does not alter our adherence to policies on sharing data and materials.
Acknowledgments
The authors thank Laurence Hilfiger and Tanja Minz for technical support; Michael Meldgaard and Adrian Schaeublin for synthesis of Alexa Fluor AONs; Christian Weile for design and provision of hELISA probes; Christophe Husser for AON schemes; Matthias Festag and Anne Eichinger for coordinating rat studies; and Yann Tessier, Susanne Mohr, Franz Schuler, Adrian B. Roth, and Thomas Singer for insightful discussions.
Footnotes
Supplemental Information includes one figure and two tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.11.005.
Supplemental Information
References
- 1.Shen X., Corey D.R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018;46:1584–1600. doi: 10.1093/nar/gkx1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett C.F., Swayze E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 3.Wu H., Lima W.F., Crooke S.T. Properties of cloned and expressed human RNase H1. J. Biol. Chem. 1999;274:28270–28278. doi: 10.1074/jbc.274.40.28270. [DOI] [PubMed] [Google Scholar]
- 4.Havens M.A., Hastings M.L. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016;44:6549–6563. doi: 10.1093/nar/gkw533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stein C.A., Hansen J.B., Lai J., Wu S., Voskresenskiy A., Høg A., Worm J., Hedtjärn M., Souleimanian N., Miller P. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 2010;38:e3. doi: 10.1093/nar/gkp841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham M.J., Crooke S.T., Monteith D.K., Cooper S.R., Lemonidis K.M., Stecker K.K., Martin M.J., Crooke R.M. In vivo distribution and metabolism of a phosphorothioate oligonucleotide within rat liver after intravenous administration. J. Pharmacol. Exp. Ther. 1998;286:447–458. [PubMed] [Google Scholar]
- 7.Butler M., Stecker K., Bennett C.F. Cellular distribution of phosphorothioate oligodeoxynucleotides in normal rodent tissues. Lab. Invest. 1997;77:379–388. [PubMed] [Google Scholar]
- 8.Frazier K.S. Antisense oligonucleotide therapies: the promise and the challenges from a toxicologic pathologist’s perspective. Toxicol. Pathol. 2015;43:78–89. doi: 10.1177/0192623314551840. [DOI] [PubMed] [Google Scholar]
- 9.van Poelgeest E.P., Swart R.M., Betjes M.G., Moerland M., Weening J.J., Tessier Y., Hodges M.R., Levin A.A., Burggraaf J. Acute kidney injury during therapy with an antisense oligonucleotide directed against PCSK9. Am. J. Kidney Dis. 2013;62:796–800. doi: 10.1053/j.ajkd.2013.02.359. [DOI] [PubMed] [Google Scholar]
- 10.Monteith D.K., Horner M.J., Gillett N.A., Butler M., Geary R., Burckin T., Ushiro-Watanabe T., Levin A.A. Evaluation of the renal effects of an antisense phosphorothioate oligodeoxynucleotide in monkeys. Toxicol. Pathol. 1999;27:307–317. doi: 10.1177/019262339902700306. [DOI] [PubMed] [Google Scholar]
- 11.Henry S.P., Bolte H., Auletta C., Kornbrust D.J. Evaluation of the toxicity of ISIS 2302, a phosphorothioate oligonucleotide, in a four-week study in cynomolgus monkeys. Toxicology. 1997;120:145–155. doi: 10.1016/s0300-483x(97)03661-5. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y. Preclinical and clinical advances of GalNAc-decorated nucleic acid therapeutics. Mol. Ther. Nucleic Acids. 2017;6:116–132. doi: 10.1016/j.omtn.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanowitz M., Hettrick L., Revenko A., Kinberger G.A., Prakash T.P., Seth P.P. Asialoglycoprotein receptor 1 mediates productive uptake of N-acetylgalactosamine-conjugated and unconjugated phosphorothioate antisense oligonucleotides into liver hepatocytes. Nucleic Acids Res. 2017;45:12388–12400. doi: 10.1093/nar/gkx960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spiess M. The asialoglycoprotein receptor: a model for endocytic transport receptors. Biochemistry. 1990;29:10009–10018. doi: 10.1021/bi00495a001. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz A.L., Rup D., Lodish H.F. Difficulties in the quantification of asialoglycoprotein receptors on the rat hepatocyte. J. Biol. Chem. 1980;255:9033–9036. [PubMed] [Google Scholar]
- 16.Prakash T.P., Yu J., Migawa M.T., Kinberger G.A., Wan W.B., Østergaard M.E., Carty R.L., Vasquez G., Low A., Chappell A. Comprehensive structure-activity relationship of triantennary N-acetylgalactosamine conjugated antisense oligonucleotides for targeted delivery to hepatocytes. J. Med. Chem. 2016;59:2718–2733. doi: 10.1021/acs.jmedchem.5b01948. [DOI] [PubMed] [Google Scholar]
- 17.Javanbakht H., Mueller H., Walther J., Zhou X., Lopez A., Pattupara T., Blaising J., Pedersen L., Albæk N., Jackerott M. Liver-targeted anti-HBV single-stranded oligonucleotides with locked nucleic acid potently reduce HBV gene expression in vivo. Mol. Ther. Nucleic Acids. 2018;11:441–454. doi: 10.1016/j.omtn.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moisan A., Gubler M., Zhang J.D., Tessier Y., Dumong Erichsen K., Sewing S., Gérard R., Avignon B., Huber S., Benmansour F. Inhibition of EGF uptake by nephrotoxic antisense drugs in vitro and implications for preclinical safety profiling. Mol. Ther. Nucleic Acids. 2017;6:89–105. doi: 10.1016/j.omtn.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Husser C., Brink A., Zell M., Müller M.B., Koller E., Schadt S. Identification of GalNAc-conjugated antisense oligonucleotide metabolites using an untargeted and generic approach based on high resolution mass spectrometry. Anal. Chem. 2017;89:6821–6826. doi: 10.1021/acs.analchem.7b01244. [DOI] [PubMed] [Google Scholar]
- 20.Koller E., Vincent T.M., Chappell A., De S., Manoharan M., Bennett C.F. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011;39:4795–4807. doi: 10.1093/nar/gkr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhen Y., Caprioli R.M., Staros J.V. Characterization of glycosylation sites of the epidermal growth factor receptor. Biochemistry. 2003;42:5478–5492. doi: 10.1021/bi027101p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin M.C., Huang M.J., Liu C.H., Yang T.L., Huang M.C. GALNT2 enhances migration and invasion of oral squamous cell carcinoma by regulating EGFR glycosylation and activity. Oral Oncol. 2014;50:478–484. doi: 10.1016/j.oraloncology.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Shemesh C.S., Yu R.Z., Gaus H.J., Greenlee S., Post N., Schmidt K., Migawa M.T., Seth P.P., Zanardi T.A., Prakash T.P. Elucidation of the biotransformation pathways of a GalNAc3-conjugated antisense oligonucleotide in rats and monkeys. Mol. Ther. Nucleic Acids. 2016;5:e319. doi: 10.1038/mtna.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juliano R.L., Ming X., Carver K., Laing B. Cellular uptake and intracellular trafficking of oligonucleotides: implications for oligonucleotide pharmacology. Nucleic Acid Ther. 2014;24:101–113. doi: 10.1089/nat.2013.0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janas M.M., Harbison C.E., Perry V.K., Carito B., Sutherland J.E., Vaishnaw A.K., Keirstead N.D., Warner G. The nonclinical safety profile of GalNAc-conjugated RNAi therapeutics in subacute studies. Toxicol. Pathol. 2018;46:735–745. doi: 10.1177/0192623318792537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Straarup E.M., Fisker N., Hedtjärn M., Lindholm M.W., Rosenbohm C., Aarup V., Hansen H.F., Ørum H., Hansen J.B., Koch T. Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res. 2010;38:7100–7111. doi: 10.1093/nar/gkq457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sbalzarini I.F., Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.