Abstract
Background:
Chinese hamster ovary (CHO) cell line is considered as the most common cell line in the biopharmaceutical industry because of its capability in performing efficient post-translational modifications and producing the recombinant proteins, which are similar to natural human proteins. The optimization of the upstream process via different feed strategies has a great impact on the target molecule expression and yield.
Methods:
To determine and understand the molecular events beneath the feed effects on the CHO cell, a label-free quantitative proteomic analysis was applied. The proteome changes followed by the addition of a designed amino acid feed to the monoclonal antibody producing CHO cell line culture medium were investigated.
Results:
The glutathione synthesis, the negative regulation of the programmed cell death, proteasomal catabolic process, and the endosomal transport pathway were up-regulated in the group fed with a designed amino acid feed compared to the control group.
Conclusion:
Our findings could be helpful to identify new targets for metabolic engineering.
Keywords: CHO cells, Glutathione, Monoclonal antibodies, Proteomics
INTRODUCTION
Chinese hamster ovary (CHO) cells are desired hosts for the production of biopharmaceuticals. Their ability to perform efficient post-translational modifications and to produce the recombinant proteins, which are similar to natural human proteins, making them the most common cell line in the biopharmaceutical industry. Moreover, these cells are simply adapted to grow in a serum-free medium[1,2]. The use of fully chemically defined serum-free media has simplified the downstream processing and minimized the batch-to-batch variations[3]. Over the past two decades, the development of CHO cells with a considerable production yield has mainly been related to the process engineering strategies so as to optimize the media and feeds formulation, new feeding strategies, and the culture conditions[4,5]. Other approaches to achieve a high titer of biopharmaceuticals in CHO cells are genetic engineering through different expression vector design[6], cell engineering via the manipulation of the genes associated with the cell cycle[7], cell metabolism[8], apoptosis[9,10], and the protein secretion pathway[11]. Until now, the achievement rate of the protein titer enhancement through the genetic and cell engineering has been little compared to the bioprocess optimization methods[5].
The full characterization of cellular machinery of the CHO cells will result in more knowledge regarding the improvement of the manufacturing processes[12]. Despite the advances in improving the bio-pharmaceuticals productivity of the CHO cells, the complete characterization of their cellular machinery has still to be achieved[13]. In order to reach the comprehensive understanding of the CHO cell physiology and subsequently, the profound knowledge of the biopharmaceuticals production, systems biology approach has been employed[14,15]. Furthermore, in order to understand the cellular mechanism of the high productivity in CHO cells, different systems biology tools like transcriptomics, proteomics, and metabolomics have been applied[16,17]. Previous investigations that used the systems biology tools have studied differences between the cell line productivity[18-22] and the changes in culture conditions, including the temperature shift[23], sodium butyrate treatment[24,25], and hyperosmolarity[26], which enhance the productivity. Previous proteomic studies have also provided rational strategies for the modifications needed to increase productivity, develop cell growth, improve media consumption, and provide constant glycosylation patterns[27-38].
It is important to comprehend the molecular rationale of the final production increase by the feed compounds. Such knowledge will help bring about the design of new generation feeds to engineer the new cellular processes[17,39]. In this study, by using the label-free quantitative proteomic analysis, protein expression profiles were investigated, followed by the addition of a designed amino acid feed. Through this comparative proteomic study, correlation of the key biological processes influenced by the designed feed, along with the enhancement of the target protein productivity in the CHO cell line were shown. To the best of our knowledge, this is the first proteomic report that considers the effect of feed formulation on the biological processes during the production of the target monoclonal antibody (mAb) in CHO cells.
MATERIALS AND METHODS
Cell line and culture media
Recombinant CHO cell producing Bevasizumab, a mAb against the vascular endothelial growth factor A, was kindly provided by Aryogen Pharmed (Alborz, Iran). The cell line original source was purchased from the Life Technologies (California, USA). The basal culture medium was CDM4CHO (Hyclone Laboratories, Utah, USA), which was supplemented with six mM L-glutamine (Lonza, Verviers, Belgium) at the moment of the preparation according to the manufacturer’s instruction.
Culture conditions
The CHO cells were cultured in a 500-ml shaker flask with an effective volume of 100 ml, incubated at 37 °C with 5% CO2 and agitated at 80 rpm. In the middle of the logarithmic phase, a temperature shift to 32 °C was performed. Each shaker flask was inoculated with an approximate cell density of 5 × 105 cells/ml.
Feeds and their components
The feeds used in this study were a home-made designed amino acid feed (feed A) and a control feed (feed B). The home-made designed amino acid feed containing 75 mM aspartic acid, 15 mM glutamic acid, 2.5 mM glycine, and 12.5 mM arginine (HiMedia Laboratories, Mumbai, India) was prepared based on the design of experiment methods, reported in our previous study[40]. In summary, the critical amino acids for the enhancement of target mAb production were determined employing the Plackett-Burman design. After finding the critical amino acids, their concentration in the feed was optimized by response surface methodology using a Box-Behncken design. The amino acids were dissolved in the basal medium (CDM4CHO), as control feed, and all the feeds were added, as multiple, discrete additions to the culture on days 3, 5, and 7.
Proteomic analysis
Three biological replicate cultures from each group cultured in shaker flasks were examined using the label-free quantitative proteomic analysis. The samples were harvested on day 10. A total of 30 million cells were collected from each replicate of the cell line. The cell suspensions were centrifuged at 180 ×g at 4 °C for 5 min. The cell pellets were washed three times with isotonic sucrose solution (250 mM). The cells were resuspended in lysis buffer (7 M of urea, 2 M of thiourea, 4% CHAPS 40 mM of Tris, 0.2% Biolyte 3/10, and 50 mM of dithiothreitol). The whole-cell extracts were kept at room temperature for 60 min, followed by centrifugation at 14,000 ×g at 4 °C for 15 min. The samples were digested by the filter-aided sample preparation method[41]. The digested peptides were suspended in 10 µl of 0.1% formic acid and subjected to nanoLC-MS/MS analysis.
Reverse-phase ultra-high pressure liquid chromate-graphy (UPLC) separation of the intracellular protein digests was performed on Easy-nLC1000 (Thermo Fisher Scientific, USA) equipped with an in-house made column (100 μm × 10 cm) packed with a reversed-phase ReproSil-Pur C18-AQ resin (3 μm, 120 Å, Dr. Maisch GmbH, Germany). A linear gradient from 0 to 50% mobile phase B (0.1% formic acid in 80% acetonitrile) was achieved against the mobile phase A (0.1% formic acid in water) over 50 min. LTQ-Orbitrap Elite mass spectrometry (Thermo Fisher Scientific) was used along with UPLC to acquire the mass and fragmentation information of the digested peptides.
RAW files were extracted using the Mascot version 2.3.02 (Matrix Science, London, UK) embedded into Proteome Discoverer 2.0 (Thermo Fisher Scientific). Mass spectrometry data were searched against UniProt, Chinese hamster sequence database (http://www.uniprot.org/). Parameters were set as follows: fully tryptic peptides with ≤2 missed cleavages were permitted; carbamidomethylation and oxidization were fixed and variable modifications, respectively: peptide mass tolerance was 10 ppm, and fragment mass tolerance was 20 mmu; the charge state of the peptides were set from +2 to +6. The cut-off of global false discovery rate for peptide and protein identification was set to 0.01.
Chromatographic peak intensity (peak area) was calculated. Protein species with at least two unique peptides were selected for protein species quantitation, and the relative quantitative protein ratios between the two samples were calculated by comparing the average of log2-transformed abundance values (three biological replicates). Student’s t-tests were performed to determine the significance of changes between samples. A fold-change ≥1.5 and p ≤ 0.05 were used as the thresholds to define differentially accumulated protein species.
Bioinformatic analysis
Bioinformatic analysis of proteins was conducted according to Liu et al.[42]. Functional annotation and category analysis was carried out using an online software, Blast2GO (http://www.geneontology.org). A p ≤ 0.05 was used as the threshold to determine the significant enrichments of GO pathways.
Western blot analysis
Western blot analysis was performed as described before in detail[43]. Aliquots of the protein samples (35 µg) were loaded on 12% SDS-PAGE. Subsequently, they were transferred to a nitrocellulose membrane using the Towbin buffer (25 mM of Tris, 192 mM of glycine, and 20% methanol) by a semi-dry Trans-Blot cell (Bio-Rad, USA), and transfer was verified by Ponceau S staining. The membrane was incubated in a blocking buffer (2.5% skim milk, 2.5% glycerol, and 0.05% Tween-20 in TBS) at 4 °C overnight. Furthermore, the membrane was rinsed in TTBS (100 mM of Tris–HCl, 0.9% NaCl, and 0.05% Tween-20, pH 7.5) for 10 min. It was then incubated for 2 h with a blocking solution containing primary antibodies: 1:10,000 rabbit monoclonal to glutathione synthetase (GSS), 1:1000 rabbit polyclonal to glucose-6-phosphate dehydrogenase (G6PDH), 1:1000 rabbit polyclonal to proteasome subunit beta (PSMB), and 1:1000 rabbit polyclonal to beta-actin (all from Abcam, USA). After washing three times for 5 min each with TTBS, the membrane was incubated again for 1 h in 1:500 horseradish peroxidase-conjugated anti-rabbit IgG secondary antibody (RayBiotech, Iran). The immunoreactive bands were then detected by ECL plus kit (GE healthcare, UK) using Kodak Image Station 4000MM Pro.
RESULTS
The label-free quantitative proteomic analysis was incorporated to find the potential pathways and related gene targets to enhance the CHO cell productivity via the appropriate feeds. Comparative proteomics was performed on two groups: control and feed A. Three biological replicates were performed for each group, and the whole cell lysates from six shaker flasks were harvested on day 10 and further processed for the label-free analysis. The feeds were added as multiple discrete additions to the cultures on days 3, 5, and 7. In comparison to the control group, the final mAb titer increased by 70% in the group fed with feed A (Fig. 1A). Moreover, the viable cell density and viability percentage of the designed amino acid feed group improved (Fig. 1B and 1C).
Fig. 1.
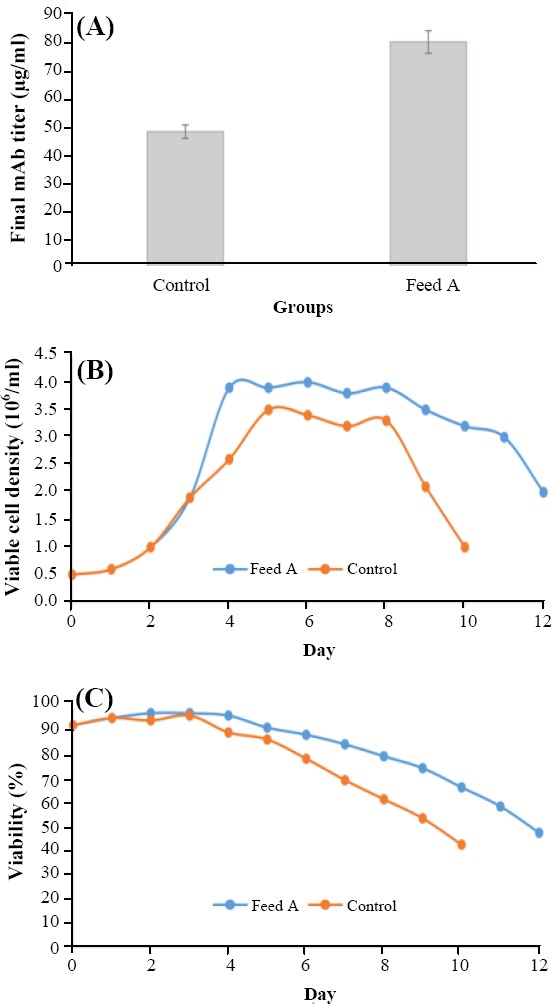
The final mAb titer (A), the viable cell density (B), and the viability percentage (C) in feed A group vs. control group.
The label-free protein identification and the differential expression
The whole cell lysates from the biological replicates were harvested on day 10 and prepared for the label-free quantitative proteomic analysis. Label-free analysis results were provided as supplementary materials. On day 10, 41 proteins in the feed A group were differentially expressed in comparison with the control group. Among these proteins, 30 and 11 proteins were up-regulated and down-regulated, respectively in feed A group in comparison with the control feed group (Table 1).
Table 1.
The list of differentially expressed proteins in feed A group
| No. | Protein name | Protein ID | Fold change | Mus musculus homologous |
|---|---|---|---|---|
| 1 | Cystine/glutamate transporter | A0A098KXA0 | 3.2 | Q9WTR6 |
| 2 | Lactoylglutathionelyase | A0A061 | 2.4 | Q9CPU0 |
| 3 | Carboxylic ester hydrolase | A0A061IAA7 | 2.3 | Q64176 |
| 4 | Hydroxymethylglutaryl-CoA synthase, mitochondrial | G3HP76 | 2.3 | P54869 |
| 5 | Glutathione S-transferase | G3IFJ6 | 2.2 | P30115 |
| 6 | Bifunctionalmethylenetetrahydrofolate dehydrogenase/cyclohydrolase | A0A061HZ57 | 2.0 | P18155 |
| 7 | Plasminogen activator inhibitor 1 | G3HA54 | 2.0 | P22777 |
| 8 | Transgelin | G3H7Z2 | 2.0 | Q9WVA4 |
| 9 | Extracellular matrix protein 1 | G3H2W6 | 1.9 | Q61508 |
| 10 | Low-density lipoprotein receptor | P35950 | 1.9 | P35951 |
| 11 | Alpha-2-macroglobulin receptor-associated protein | G3GWB3 | 1.8 | P55302 |
| 12 | Hemeoxygenase 1 | G3IAI6 | 1.8 | P14901 |
| 13 | 4F2 cell-surface antigen heavy chain | G3IHN6 | 1.8 | P108522 |
| 14 | Macrophage migration inhibitory factor | G3HY08 | 1.8 | P34884 |
| 15 | Glutathione S-transferase | G3I5H1 | 1.8 | P24472 |
| 16 | Phosphoserine aminotransferase | G3IKH9 | 1.7 | Q99K85 |
| 17 | Macrophage metalloelastase | G3GUV3 | 1.7 | Q63341 |
| 18 | 3 beta-hydroxysteroid dehydrogenase/Delta 5-->4-isomerase type 2 | G3I6D1 | 1.7 | P24815 |
| 19 | Calponin | G3I3W0 | 1.7 | Q9DAW9 |
| 20 | von Willebrand factor A domain-containing protein 5A | G3IMX9 | 1.7 | Q99KC8 |
| 21 | Glucose-6-phosphate 1-dehydrogenase | O55044 | 1.6 | Q00612 |
| 22 | Guanine nucleotide-binding protein subunit gamma | G3H7D1 | 1.6 | Q80SZ7 |
| 23 | Prolyl 4-hydroxylase subunit alpha-1 | G3HJ24 | 1.6 | Q60715 |
| 24 | CD44 antigen | P20944 | 1.5 | P15379-2 |
| 25 | Bone marrow stromal antigen 2 | A0A061IGY1 | 1.5 | Q8R2Q8 |
| 26 | Galectin | G3H7B3 | 1.5 | P16110 |
| 27 | Glutathione peroxidase | G3HF60 | 1.5 | O70325 |
| 28 | EH domain-containing protein 1 | G3I6F9 | 1.5 | Q9WVK4 |
| 29 | Glutathione synthetase | G3HAP7 | 1.5 | P51855 |
| 30 | Proteasome subunit beta type | G3H303 | 1.5 | P99026 |
| 31 | Lamin-B receptor | G3GRA0 | -1.5 | Q3U9G9 |
| 32 | UDP-N-acetylhexosaminepyrophosphorylase-like protein 1 | G3IJB9 | -1.5 | Q3TW96 |
| 33 | Lon proteasehomolog, mitochondrial | G3HCJ1 | -1.6 | Q8CGK3 |
| 34 | Translocon-associated protein subunit gamma | G3H223 | -1.6 | Q9DCF9 |
| 35 | Mitochondrial-processing peptidase subunit alpha | G3I6Z3 | -1.6 | Q9DC61 |
| 36 | NAD(P) transhydrogenase, mitochondrial | G3HMX6 | -1.7 | Q61941 |
| 37 | Periaxin | A0A061I2N6 | -1.7 | unreviewed |
| 38 | Protein unc-84-like B | G3I2J5 | -1.7 | Q8BJS4 |
| 39 | Protein AHNAK2-like protein | A0A061I4K9 | -1.9 | O55103-2 |
| 40 | NADH-cytochrome b5 reductase | G3H3L4 | -2.1 | Q9DB73 |
| 41 | CDGSH iron sulfur domain-containing protein 1 | G3H4L8 | -2.8 | Q91WS0 |
The up-regulation of important biological processes
The differentially expressed proteins were identified and clustered by the biological process using Mus musculus homologues and subjected to gene enrichment analysis by the gene ontology consortium. The significant clusters that were up-regulated in feed A group in comparison with the control feed group are presented in Table 2. The pentose-phosphate shunt, the glutathione (GSH) metabolic process, the negative regulation of the programmed cell death, the cellular response to the oxidative stress, the regulation of intracellular transport, and the proteasomal protein catabolic process were up-regulated in the group fed with feed A, in comparison with the control group. There was no significant biological process for the down-regulated proteins in feed A.
Table 2.
Up-regulated significant biological processes in feed A
| No. | Biological process | p value | GO ID |
|---|---|---|---|
| 1 | Pentose-phosphate shunt | 0.01447 | 0006098 |
| 2 | NADPH regeneration | 0.01558 | 0006740 |
| 3 | Glutathione metabolic process, | 0.000001697 | 0006749 |
| 4 | Negative regulation of programmed cell death (apoptotic signaling pathway) | 0.00001436 | 2001234 |
| 5 | Carboxylic acid biosynthetic process | 0.01058 | 0046394 |
| 6 | Cellular response to oxidative stress | 0.02598 | 0034599 |
| 7 | Regulation of vesicle-mediated transport | 0.001639 | 0060627 |
| Regulation of intracellular transport | 0.02796 | 0032386 | |
| 8 | Proteasomal protein catabolic process | 0.02336 | 0010498 |
GO, gene ontology
Western blot analysis result
Western blot analysis of some of the differentially expressed proteins confirmed all the alterations identified by LC-MS/MS (Fig. 2). The selected proteins for Western blot analysis belonged to different biological processes. G6PDH, GSS, and PSMB were from pentose phosphate shunt, GSH metabolic process, and proteasomal protein catabolic process, respectively.
Fig. 2.
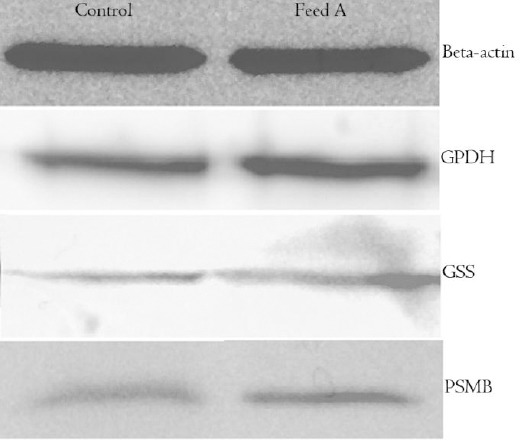
Western blot images of beta-actin, G6PDH, GSS, and PSMB of control group and designed amino acid feed group (feed A). Beta-actin was used as a control protein.
DISCUSSION
The up-regulation of GSH pathway was identified in feed A group as compared to the control feed group. GSH is a significant antioxidant and plays several important roles in cells. GSH synthesis occurs in two steps: (1) glutamatecysteine ligase condensates cysteine and glutamate, and (2) a glycine is added by GSS to form GSH[44]. It was found that GSS was up-regulated in feed A on day 10 and was confirmed by Western blot analysis (Fig. 2). The up-regulation of this protein in the high-producing cell line has also been reported by Orellana et al.[19].
GSH mediates the correct protein disulfide bond formation by reducing the incorrect protein disulfide bonds via the protein disulfide-isomerase in the endoplasmic reticulum (ER)[45,46]. Furthermore, GSH can be oxidized to GSSG in order to detoxify the reactive oxygen species (ROS), which may result in the down-regulation of apoptosis (Fig. 3), and then GSSG can be reduced once more using nicotinamide adenine dinucleotide phosphate (NADPH) to complete the GSH redox cycle[47,48]. NADPH can be provided by the pentose phosphate shunt, which was also up-regulated in feed A group in comparison with the control group (Fig. 3). Moreover, G6PD catalyzes the rate limiting step of the pentose phosphate shunt, and the main function of this protein is to provide NADPH[49]. G6PD was also up-regulated in feed A on day 10 and was confirmed by Western blot analysis (Fig. 2). The up-regulation of GSH in the high mAb producing CHO cells has also been reported in previous studies[19,20]. Two main causes of high oxidative stress in the high-producing CHO cells are: the stress from the protein folding in ER and the stress from the mitochondrial respiration[20]. Recent studies have proposed some strategies to mAb-producing CHO cells for the metabolic engineering of the GSH preservation[50,51].
Fig. 3.
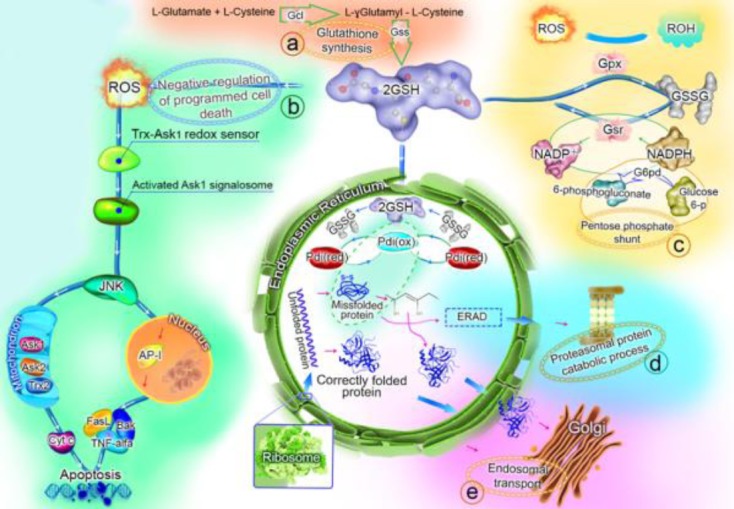
The suggested functions for the up-regulated biological processes in feed A group on day 10. The GSH synthesis (a), the negative regulation of programmed cell death (b), the pentose phosphate shunt (c), the proteasomal protein catabolic process (d), and the endosomal transport (e) were up-regulated processes in feed A group compared with the control feed group. The details and the roles of these processes in the final mAb titer enhancement have been explained in the text. GSH, glutathione (oxidized form); GSSG, glutathione (reduced form); Gcl, glutamatecysteine ligase; GSS, glutathione synthetase; Gpx, glutathione peroxidase; Gsr, glutathione reductase; G6pd, glucose-6-phosphate 1-dehydrogenase; Pdi, protein disulfide-isomerase; ERAD, endoplasmic reticulum-associated protein degradation; ROS, reactive oxygen species.
In the present proteomic analysis, the overexpression of proteins, which were involved in the negative regulation of apoptosis, was found. Furthermore, the batch duration and viability percentage were in line with this event, and the batch duration of feed A group was more than that of the control feed group (Fig. 1B and 1C). The cell apoptosis may occur in a fed-batch culture because of the hyperosmolarity, the by-product accumulation, and the production of ROS[52,53]. The use of a suitably designed feed can delay the apoptosis and extend the culture duration, which leads to an increase in the mAb productivity[54]. The negative regulation of the programmed cell death has been attributed to the role of GSH in the reduction of ROS[47], which also seems to be true in this study (Fig. 3). Based on the results obtained from this study, it seems that increase in productivity is due to increased integral viable cell density and somewhat to increased specific productivity.
In the recombinant protein production process, the protein translation and secretion are considered as bottlenecks[55,56]. There are different transport vesicles in cells, and the up-regulation of the proteins that constitute these vesicles has already been reported, as a result of recombinant protein overexpression[57]. The up-regulation of intracellular protein transport in the high mAb-producing CHO cells has also been reported by Orellana et al.[19]. In line with these reports, the present research demonstrates that the proteins involved in the intracellular transport and the vesicle-mediated transport such as the transitional ER ATPase, filamin A, and AP-2 complex subunit mu are up-regulated after the addition of appropriate feed. AP-2 is involved in the endocytosis and the transport from the trans-Golgi network to the lysosomes[58]. Another report has also showed the up-regulation of AP-2 in the high mAb producing cells[19].
If the protein production in cells increases, the misfolded proteins rate will also increase[59]. In order to prevent the accumulation of misfolded proteins, their retrotranslocation from ER to cytosol, when they undergo the ubiquitination, will occur, and these misfolded proteins will be the substrates for proteasome[60]. In addition, the up-regulation of the proteasomal catabolic process was seen in feed A, and the up-regulation of proteasome subunit beta was confirmed by Western blot analysis. This study shows that the feed composition will affect mAb production by changing the different pathways regulation, which are likely correlated to culture productivity.
ACKNOWLEDGMENTS
This work has financially been supported by the Science and Technology Development division of Aryogen Pharmed, Iran. The authors would like to acknowledge the Creative Proteomics, the proteomics division of the Creative Dynamics Inc, USA.
Footnotes
CONFLICT OF INTEREST. None declared.
REFERENCES
- 1.Kim JY, Kim YG, Lee GM. CHO cells in biotechnology for production of recombinant proteins:current state and further potential. Applied microbiology and biotechnology. 2012;93(3):917–930. doi: 10.1007/s00253-011-3758-5. [DOI] [PubMed] [Google Scholar]
- 2.Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nature biotechnology. 2004;22(11):1393–1398. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- 3.Rodrigues ME, Costa AR, Henriques M, Cunnah P, Melton DW, Azeredo J, Oliveira R. Advances and drawbacks of the adaptation to serum-free culture of CHO-K1 cells for monoclonal antibody production. Applied biochemistry and biotechnology. 2013;169(4):1279–1291. doi: 10.1007/s12010-012-0068-z. [DOI] [PubMed] [Google Scholar]
- 4.Hacker DL, De Jesus M, Wurm FM. 25 years of recombinant proteins from reactor-grown cells - where do we go from here? Biotechnology advances. 2009;27(6):1023–1027. doi: 10.1016/j.biotechadv.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Wuest DM, Harcum SW, Lee KH. Genomics in mammalian cell culture bioprocessing. Biotechnology advances. 2012;30(3):629–638. doi: 10.1016/j.biotechadv.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim Y, Wong NS, Lee YY, Ku Sc, Wong DC, Yap MG. Engineering mammalian cells in bioprocessing - current achievements and future perspectives. Biotechnology and applied biochemistry. 2010;55(4):175–89. doi: 10.1042/BA20090363. [DOI] [PubMed] [Google Scholar]
- 7.Fussenegger M, Schlatter S, Dätwyler D, Mazur X, Bailey JE. Controlled proliferation by multigene metabolic engineering enhances the productivity of Chinese hamster ovary cells. Nature biotechnology. 1998;16(5):468–472. doi: 10.1038/nbt0598-468. [DOI] [PubMed] [Google Scholar]
- 8.Fogolín MB, Wagner R, Etcheverrigaray M, Kratie R. Impact of temperature reduction and expression of yeast pyruvate carboxylase on hGM-CSF-producing CHO cells. Journal of biotechnology. 2004;109(1-2):179–191. doi: 10.1016/j.jbiotec.2003.10.035. [DOI] [PubMed] [Google Scholar]
- 9.Chiang GG, Sisk WP. Bcl-x(L) mediates increased production of humanized monoclonal antibodies in Chinese hamster ovary cells. Biotechnology and bioengineering. 2005;91(7):779–792. doi: 10.1002/bit.20551. [DOI] [PubMed] [Google Scholar]
- 10.Krampe B, Al-Rubeai M. Cell death in mammalian cell culture:molecular mechanisms and cell line engineering strategies. Cytotechnology. 2010;62(3):175–188. doi: 10.1007/s10616-010-9274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohan C, Park SH, Chung JY, Lee GM. Effect of doxycycline-regulated protein disulfide isomerase expression on the specific productivity of recombinant CHO cells:thrombopoietin and antibody. Biotechnology and bioengineering. 2007;98(3):611–615. doi: 10.1002/bit.21453. [DOI] [PubMed] [Google Scholar]
- 12.Meleady P, Hoffrogge R, Henry M, Rupp O, Bort JH, Clarke C, Brinkrolf K, Kelly S, Müller B, Doolan P, Hackl M, Beckmann TF, Noll T, Grillari J, Barron N, Pühler A, Clynes M, Borth N. Utilization and evaluation of CHO-specific sequence databases for mass spectrometry based proteomics. Biotechnology and bioengineering. 2012;109(6):1386–1394. doi: 10.1002/bit.24476. [DOI] [PubMed] [Google Scholar]
- 13.Baycin-Hizal D, Tabb DL, Chaerkady R, Chen L, Lewis NE, Nagarajan H, Sarkaria V, Kumar A, Wolozny D, Colao J, Jacobson E, Tian Y, O'Meally RN, Krag SS, Cole RN, Palsson BO, Zhang H, Betenbaugh M. Proteomic analysis of Chinese hamster ovary cells. Journal of protome research. 2012;11(11):5265–5276. doi: 10.1021/pr300476w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao S, Shiloach J, Betenbaugh MJ. Engineering cells to improve protein expression. Current opinion in structural biology. 2014;26:32, 8. doi: 10.1016/j.sbi.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis AM, Abu-Absi NR, Borys MC, Li ZJ. The use of 'Omics technology to rationally improve industrial mammalian cell line performance. Biotechnology and bioengineering. 2016;113(1):26–38. doi: 10.1002/bit.25673. [DOI] [PubMed] [Google Scholar]
- 16.Dietmair S, Nielsen LK, Timmins NE. Mammalian cells as biopharmaceutical production hosts in the age of omics. Biotechnology journal. 2012;7(1):75–89. doi: 10.1002/biot.201100369. [DOI] [PubMed] [Google Scholar]
- 17.Farrell A, McLoughlin N, Milne JJ, Marison IW, Bones J. Application of multi-omics techniques for bioprocess design and optimization in chinese hamster ovary cells. Journal of proteome research. 2014;13(7):3144–3159. doi: 10.1021/pr500219b. [DOI] [PubMed] [Google Scholar]
- 18.Kang S, Ren D, Xiao G, Daris K, Buck L, Enyenihi AA, Zubarev R, Bondarenko PV, Deshpande R. Cell line profiling to improve monoclonal antibody production. Biotechnology and bioengineering. 2014;111(4):748–760. doi: 10.1002/bit.25141. [DOI] [PubMed] [Google Scholar]
- 19.Orellana CA, Marcellin E, Schulz BL, Nouwens AS, Gray PP, Nielsen LK. High-antibody-producing Chinese hamster ovary cells up-regulate intracellular protein transport and glutathione synthesis. Journal of proteome research. 2015;14(2):609–618. doi: 10.1021/pr501027c. [DOI] [PubMed] [Google Scholar]
- 20.Chong WP, Thng SH, Hiu AP, Lee DY, Chan EC, Ho YS. LC-MS-based metabolic characterization of high monoclonal antibody-producing Chinese hamster ovary cells. Biotechnology and bioengineering. 2012;109(12):3103–3111. doi: 10.1002/bit.24580. [DOI] [PubMed] [Google Scholar]
- 21.Nissom PM, Sanny A, Kok YJ, Hiang YT, Chuah SH, Shing TK, Lee YY, Wong KT, Hu WS, Sim MY, Philp R. Transcriptome and proteome profiling to understanding the biology of high productivity CHO cells. Molecular biotechnology. 2006;34(2):125–140. doi: 10.1385/mb:34:2:125. [DOI] [PubMed] [Google Scholar]
- 22.Gao Y, Ray S, Dai S, Ivanov AR, Abu-Absi NR, Lewis AM, Huang Z, Xing Z, Borys MC, Li ZJ, Karger BL. Combined metabolomics and proteomics reveals hypoxia as a cause of lower productivity on scale-up to a 5000-liter CHO bioprocess. Biotechnology journal. 2016;11(9):1190–1200. doi: 10.1002/biot.201600030. [DOI] [PubMed] [Google Scholar]
- 23.Yee JC, Gerdtzen ZP, Hu WS. Comparative transcriptome analysis to unveil genes affecting recombinant protein productivity in mammalian cells. Biotechnology and bioengineering. 2009;102(1):246–263. doi: 10.1002/bit.22039. [DOI] [PubMed] [Google Scholar]
- 24.Yee JC, de Leon Gatti M, Philp RJ, Yap M, Hu WS. Genomic and proteomic exploration of CHO and hybridoma cells under sodium butyrate treatment. Biotechnology and bioengineering. 2008;99(5):1186–1204. doi: 10.1002/bit.21665. [DOI] [PubMed] [Google Scholar]
- 25.Kantardjieff A, Jacob NM, Yee JC, Epstein E, Kok YJ, Philp R, Betenbaugh M, Hu WS. Transcriptome and proteome analysis of Chinese hamster ovary cells under low temperature and butyrate treatment. Journal of biotechnology. 2010;145(2):143–159. doi: 10.1016/j.jbiotec.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Lee MS, Kim KW, Kim YH, Lee GM. Proteome analysis of antibody-expressing CHO cells in response to hyperosmotic pressure. Biotechnology and progress. 2003;19(6):1734–1741. doi: 10.1021/bp034093a. [DOI] [PubMed] [Google Scholar]
- 27.Van Dyk DD, Misztal DR, Wilkins MR, Mackintosh JA, Poljak A, Varnai JC, Teber E, Walsh BJ, Gray PP. Identification of cellular changes associated with increased production of human growth hormone in a recombinant Chinese hamster ovary cell line. Proteomics. 2003;3(2):147–156. doi: 10.1002/pmic.200390023. [DOI] [PubMed] [Google Scholar]
- 28.Carlage T, Hincapie M, Zang L, Lyubarskaya Y, Madden H, Mhatre R, Hancock WS. Proteomic profiling of a high-producing Chinese hamster ovary cell culture. Analytical chemistry. 2009;81(17):7357–7362. doi: 10.1021/ac900792z. [DOI] [PubMed] [Google Scholar]
- 29.Carlage T, Kshirsagar R, Zang L, Janakiraman V, Hincapie M, Lyubarskaya Y, Weiskopf A, Hancock WS. Analysis of dynamic changes in the proteome of a Bcl-XL overexpressing Chinese hamster ovary cell culture during exponential and stationary phases. Biotechnology progress. 2012;28(3):814–823. doi: 10.1002/btpr.1534. [DOI] [PubMed] [Google Scholar]
- 30.Kuystermans D, Dunn MJ, Al-Rubeai M. A proteomic study of cMyc improvement of CHO culture. BMC biotechnology. 2010;10:25. doi: 10.1186/1472-6750-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JS, Park HJ, Kim YH, Lee GM. Protein reference mapping of dihydrofolate reductase-deficient CHO DG44 cell lines using 2-dimensional electrophoresis. Proteomics. 2010;10(12):2292–2302. doi: 10.1002/pmic.200900430. [DOI] [PubMed] [Google Scholar]
- 32.Baik JY, Ha TK, Kim YH, Lee GM. Proteomic understanding of intracellular responses of recombinant chinese hamster ovary cells adapted to grow in serum-free suspension culture. Biotechnology progress. 2011;27:1680, 1688. [Google Scholar]
- 33.Meleady P, Doolan P, Henry M, Keenan J, O'Sullivan F, Clarke C, Gammell P, Melville MW, Leonard M, Clynes M. Sustained productivity in recombinant Chinese hamster ovary (CHO) cell lines:proteome analysis of the molecular basis for a process-related phenotype. BMC biotechnology. 2011;11:78. doi: 10.1186/1472-6750-11-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei YYC, Naderi S, Meshram M, Budman H, Scharer JM, Ingalls BP, McConkey BJ. Proteomics analysis of chinese hamster ovary cells undergoing apoptosis during prolonged cultivation. Cytotechnology. 2011;63(6):663–677. doi: 10.1007/s10616-011-9385-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stolfa G, Smonskey MT, Boniface R, Hachmann AB, Gulde P, Joshi AD, Pierce AP, Jacobia SJ, Campbell A. CHO-Omics Review:The impact of current and emerging technologies on Chinese hamster ovary based bioproduction. Biotechnology journal. 2018;13(3):e1700227. doi: 10.1002/biot.201700227. [DOI] [PubMed] [Google Scholar]
- 36.Dorai H, Liu S, Yao X, Wang Y, Tekindemir U, Lewis MJ, Wu SL, Hancock W. Proteomic analysis of bioreactor cultures of an antibody expressing CHO-GS cell line that promotes high productivity. Journal of proteomics and bioinformatics. 2013;6:98, 108. [Google Scholar]
- 37.Liu Z, Dai S, Bones J, Ray S, Cha S, Karger BL, Li JJ, Wilson L, Hinckle G, Rossomando A. A quantitative proteomic analysis of cellular responses to high glucose media in Chinese hamster ovary cells. Biotechnology progress. 2015;31(4):1026–1038. doi: 10.1002/btpr.2090. [DOI] [PubMed] [Google Scholar]
- 38.Xu N, Ma C, Ou J, Sun WW, Zhou L, Hu H, Liu XM. Comparative proteomic analysis of three Chinese hamster ovary (CHO) host cells. Biochemical engineering journal. 2017;124:122, 129. doi: 10.1016/j.bej.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neilson KA, Ali NA, Muralidharan S, Mirzaei M, Mariani M, Assadourian G, Lee A, van Sluyter SC, Haynes PA. Less label, more free:approaches in label-free quantitative mass spectrometry. Proteomics. 2011;11(4):535–553. doi: 10.1002/pmic.201000553. [DOI] [PubMed] [Google Scholar]
- 40.Torkashvand F, Vaziri B, Maleknia S, Heydari A, Vossoughi M, Davami F, Mahboudi F. Designed amino acid feed in improvement of production and quality targets of a therapeutic monoclonal antibody. PLoS one. 2015;10(10):e0140597. doi: 10.1371/journal.pone.0140597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nature methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, Pang C, Wei H, Song M, Meng Y, Ma J, Fan S, Yu S. iTRAQ-facilitated proteomic profiling of anthers from a photosensitive male sterile mutant and wild-type cotton (Gossypium hirsutumL.) Journal of proteomics. 2015;126:68, 81. doi: 10.1016/j.jprot.2015.05.031. [DOI] [PubMed] [Google Scholar]
- 43.Vaziri B, Torkashvand F, Eslami N, Fayaz A. Comparative proteomics analysis of mice lymphocytes in early stages of infection by different strains of rabies virus. Indian journal of virology. 2012;23(3):311–316. doi: 10.1007/s13337-012-0093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meister A. Glutathione, ascorbate, and cellular protection. Cancer research. 1994;54(7 Suppl):1969s–1975s. [PubMed] [Google Scholar]
- 45.Kojer K, Riemer J. Balancing oxidative protein folding:the influences of reducing pathways on disulfide bond formation. Biochimica et biophysica acta. 2014;1844(8):1383–1390. doi: 10.1016/j.bbapap.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Lappi AK, Ruddock LW. Reexamination of the role of interplay between glutathione and protein disulfide isomerase. Journal of molecular biology. 2011;409(2):238–249. doi: 10.1016/j.jmb.2011.03.024. [DOI] [PubMed] [Google Scholar]
- 47.Circu ML, Aw TY. Glutathione and modulation of cell apoptosis. Biochimica et biophysica acta. 2012;182(10):1767–1777. doi: 10.1016/j.bbamcr.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McIlwain CC, Townsend DM, Tew KD. Glutathione S-transferase polymorphisms:cancer incidence and therapy. Oncogene. 2006;25(11):1639–1648. doi: 10.1038/sj.onc.1209373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tuttle S, Stamato T, Perez ML, Biaglow J. Glucose-6-phosphate dehydrogenase and the oxidative pentose phosphate cycle protect cells against apoptosis induced by low doses of ionizing radiation. Radiation research. 2000;153(6):781–787. doi: 10.1667/0033-7587(2000)153[0781:gpdato]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 50.Selvarasu S, Ho YS, Chong WP, Wong NS, Yusufi FN, Lee YY, Yap MG, Lee DY. Combined in silico modeling and metabolomics analysis to characterize fed-batch CHO cell culture. Biotechnology and bioengineering. 2012;109(6):1415–1429. doi: 10.1002/bit.24445. [DOI] [PubMed] [Google Scholar]
- 51.Chong WP, Yusufi FN, Lee DY, Reddy SG, Wong NS, Heng CK, Yap MG, Ho YS. Metabolomics-based identification of apoptosis-inducing metabolites in recombinant fed-batch CHO culture media. Journal of biotechnology. 2011;151(2):218–224. doi: 10.1016/j.jbiotec.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 52.Butler M. Animal cell cultures:recent achievements and perspectives in the production of biopharmaceuticals. Applied microbiology and biotechnology. 2005;68(3):283–291. doi: 10.1007/s00253-005-1980-8. [DOI] [PubMed] [Google Scholar]
- 53.Han YK, Kim YG, Kim JY, Lee GM. Hyperosmotic stress induces autophagy and apoptosis in recombinant Chinese hamster ovary cell culture. Biotechnology and bioengineering. 2010;105(6):1187–1192. doi: 10.1002/bit.22643. [DOI] [PubMed] [Google Scholar]
- 54.Han YK, Ha TK, Lee SJ, Lee JS, Lee GM. Autophagy and apoptosis of recombinant Chinese hamster ovary cells during fed-batch culture:effect of nutrient supplementation. Biotechnology and bioengineering. 2011;108(9):2182–2192. doi: 10.1002/bit.23165. [DOI] [PubMed] [Google Scholar]
- 55.Schröder M. Engineering eukaryotic protein factories. Biotechnology letters. 2008;30(2):187–196. doi: 10.1007/s10529-007-9524-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dinnis DM, James DC. Engineering mammalian cell factories for improved recombinant monoclonal antibody production:lessons from nature? Biotechnology and bioengineering. 2005;91(2):180–189. doi: 10.1002/bit.20499. [DOI] [PubMed] [Google Scholar]
- 57.Kuge O, Dascher C, Orci L, Rowe T, Amherdt M, Plutner H, Ravazzola M, Tanigawa G, Rothman JE, Balch WE. Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. The journal of cell biology. 1994;125(1):51–65. doi: 10.1083/jcb.125.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Motley AM, Berg N, Taylor MJ, Sahlender DA, Hirst J, Owen DJ, Robinson MS. Functional analysis of AP-2 alpha and mu2 subunits. Molecular biology of the cell. 2006;17(12):5298–5308. doi: 10.1091/mbc.E06-05-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walter P, Ron D. The unfolded protein response:from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 60.Lemus L, Goder V. Regulation of Endoplasmic Reticulum-Associated Protein Degradation (ERAD) by Ubiquitin. Cells. 2014;3(3):824–847. doi: 10.3390/cells3030824. [DOI] [PMC free article] [PubMed] [Google Scholar]


