Abstract
Background:
Differential expression profile of microRNAs (miRNAs) could be a diagnosis signature for monitoring gastric cancer (GC) progression. In this study, we focus on the comparison of expression levels of miR-21, miR-25, miR-93, miR-106b, and miR-375 during the sequential pattern of GC development, including normal gastric, gastric dysplasia, and GC sample.
Methods:
We used SYBR Green-based quantitative-PCR to quantify miRNAs expression.
Results:
Our analysis revealed the increased expression levels of miR-21 (p = 0.034), miR-25 (p = 0.0003), miR-93 (p = 0.0406), and miR-106b (p = 0.023) in GC samples. In addition, GC patients with positive lymph node metastasis showed the up-regulation of miR-25, miR-93, and miR-106b (p < 0.05).
Conclusion:
Our findings suggested that the expression of miR-21, miR-25, miR-93, and miR-106b altered in GC, and some of them may be further investigated as biomarkers for GC early detection and prognosis prediction.
Keywords: Biomarkers, microRNAs, Stomach cancer
INTRODUCTION
Gastric cancer (GC) is the fourth most prevalent cancer in the world, accounting for the second leading cause of cancer-related mortality[1,2]. Approximately, one million new cases of GC are estimated to occur per year; however, the incidence of GC is relatively higher in Asian than Western countries[2-4]. Risk factors for GC include sex (male to female ratio 2:1), environmental factors (Helicobacter pylori infection, nutrition, smoking, and obesity), blood group A, genetic factors (mutation in P53, MCC, and APC) and epigenetic factors (dysregulation of gene expression in GC)[5]. GC diagnosis in its early stages helps to achieve more successful treatment, but its diagnosis is very difficult for patients with no apparent clinical features. The lack of specific diagnostic biomarkers in the early stages of GC is another problem for the diagnosis and treatment of GC[6]. Unfortunately, GC is diagnosed mostly at advanced stages accompanied by extensive invasion and lymphatic metastasis. Patients with advanced GC tend to develop recurrent diseases, exhibiting poor survival rates. Thus, early detection of GC is crucial to reduce mortality rates and improve the prognosis. In addition, there is a need to develop accurate and versatile diagnostic biomarkers capable of diagnosing GC at various stages[7]. Typically, tumor markers, such as CEA, CA19-9, CA125, and CA 72-4, are commonly used to detect GC; however, such markers suffer from low sensitivity and specificity, highlighting their deficiency in the early diagnosis of GC[8,9].
microRNAs, also known as miRNAs, are small, endogenous non-coding RNA molecules consisting of approximately 22 nucleotides. miRNAs, found in plants, animals, and some viruses, control gene expression through RNA silencing and post-transcriptional regulation[10]. Importantly, miRNAs regulate gene expression via either translational suppression or mRNA degradation[11-14]. miRNAs play an important role in regulating gene expression in tumorigenesis, suggesting their promising potential as diagnostic markers for malignancies. Over the past few years, a variety of studies have demonstrated miRNA expression deregulation in GC development, especially in normal versus tumor samples. Of note, hsa-miR-21, hsa-miR-25, hsa-miR-93, hsa-miR-106b, and hsa-miR-375 are commonly expressed in GC, which are deregulated in GC compared with normal tissues[10,15-18].
We hypothesized that there are unique miRNA expression profiles capable of distinguishing normal gastric tissue (NG), gastric dysplasia (GD), and GC. Furthermore, because miRNA expression is associated with cell differentiation, specific miRNAs may become deregulated in transition from NG tissue to dysplasia and GC. Identification of such differences in miRNA expression can discriminate patients who have a high risk of GC or who need to be followed carefully or treated quickly. The present study set out to identify an ideal biomarker abnormally expressed in the pre-cancerous lesion, normal tissue, and GC.
The selection of miRNAs was based on previous studies[10,15-18] on miRNA profiling of cancerous tissues. Accordingly, in this study, we examined the expression of miR-21, miR-25, miR-93, miR-106b, and miR-375 in the Iranian population.
MATERIALS AND METHODS
Clinical samples
Tissue biopsy samples included in this study were retrospectively selected from cases at the Research Institute for Gastroenterology and Liver Diseases (RIGLD) tissue bank (Tehran, Iran) between 2015 and 2017, with ethical approvals from the organization Ethics Committee (IR.SBMU.MSP.REC.1395.590). Formalin-fixed, paraffin-embedded (FFPE) blocks were available for 29, 33, and 39 cases of NG, GD, and GC, respectively. Demographic data and information on the clinical history and pathologic findings were documented.
RNA extraction
Four to five thick (10 μm) sections of FFPE tissue were used to extract total RNA. For the analysis of GD and GC samples, a pathologist assessed the slides to ensure the appropriate selection of tumor tissue and blocks with more than 50% tumor content per block. Deparaffinization solution (Qiagen, Hilden, Germany) was used to remove paraffin from FFPE tissue. For RNA extraction, we used the Qiagen miRNeasy FFPE kit (Qiagen) according to the manufacturer’s protocol. Total RNA concentration was determined by measuring the ratio of the absorbance at 260 and 280 nm using a NanoDrop™ 2000c Spectrophotometer (Thermo Fisher Scientific, USA). Total extracted RNA was stored at -70 °C until use.
miRNA quantitation
Quantitative real-time PCR (qRT-PCR) was applied to analyze miRNA expression. Briefly, the poly (A) tail was added to the extracted RNA by using the Poly (A) Tailing kit (New England Biolabs GmbH, Germany) at 37 °C for 30 min as per manufacturer’s protocol. cDNA was synthesized using miScript II RT kit (Qiagen) and then diluted 1:5. The diluted cDNA sample (two µl) was used as a template for qRT-PCR by SYBR Premix Ex TaqTM II (TaKaRa, Japan). Two µl of template cDNA was mixed with 12.5 µl of 2× SYBR Green PCR master mix and 1 µl of miR-specific forward and universal reverse primer (Table 1) in a final volume of 25 µl. The PCR was performed in duplicate, according to the standard program on Rotor-Gene Q instrument (Qiagen) as follows: 10 min at 95 °C, followed by three cycles of amplification (15 s at 94 °C and 30 s at 60 °C) and finally a dissociation curve step (ramp from 60 to 95 °C) to verify amplification specificity. All the melting curves contained single peaks, indicating specific PCR amplification. Moreover, the PCR product size was tested by agarose gel electrophoresis. PCR efficiency was evaluated by LinRegPCR software (http://www.hartfaalcentrum.nl). Afterwards, the expression level of miRNA was determined using 2−ΔΔCt and normalized to U6 small nuclear RNA[19].
Table 1.
List of the candidates and primers used in this study
| Names | Sequence |
|---|---|
| Universal reverse primer | GCGAGCACAGAATTAATACGACTC |
| miR-375 | TTTATTCGTTCGGCTCGCGT |
| miR-21 | GGGGTAGCTTATCAGACTGATGTT |
| miR-25 | ACATTGCACTTGTCTCGGTCT |
| miR-106b | CGGTAAAGTGCTGACATTGCA |
| miR-93 | CAAAGTGCTGTTCGTGCAGGT |
| U6 | CGCAAGGATGACACGCAAATTC |
Statistical analysis
Student’s t-test and ANOVA were used to analyze the expression levels of miRs (miR-21, miR-25, miR-93, miR-106b, and miR-375) between two and three groups. In case of non-parametric data distribution, non-parametric tests (Mann-Whitney U and Kruskal-Wallis) were applied. Statistical analysis was performed using SPSS 16 (SPSS, Inc., Chicago, IL, USA). p values less than 0.05 were considered to be statistically significant. The sensitivity and specificity of each miRNA between normal and GC tissue as well as area under curve (AUC) and Youden index were calculated using the MedCalc statistical software 15 (MedCalc Software, Ostend, Belgium).
RESULTS
Our result evidenced no significant differences for the mean of age in NG, GD, and GC (52.03 ± 15.9, 64.32 ± 14.35, and 62.38 ± 13.47, respectively). The male to female sex ratios were found to be 1.23, 3.12, and 1.43 in NG, GD, and GC, respectively.
Identification of elevated miRNAs among normal, dysplastic, and cancerous tissues
Figure 1 shows a significant up-regulation in miR-21 (fold change: 2.53) and miR-25 (fold chage: 2.94) in GC compared to NG (p < 0.00). In addition, miR-25 expression showed a significant up-regulation in GC compared with NG (p = 0.03). As shown in Figure 1A-1D, the expression levels of miR-93, miR-106b, and miR-25 were significantly up-regulated in GC compared to normal tissue (p < 0.05). We found no significant dysregulation of miR-375 in GC compared to NG and GD (Fig. 1E and Table 2).
Fig. 1.
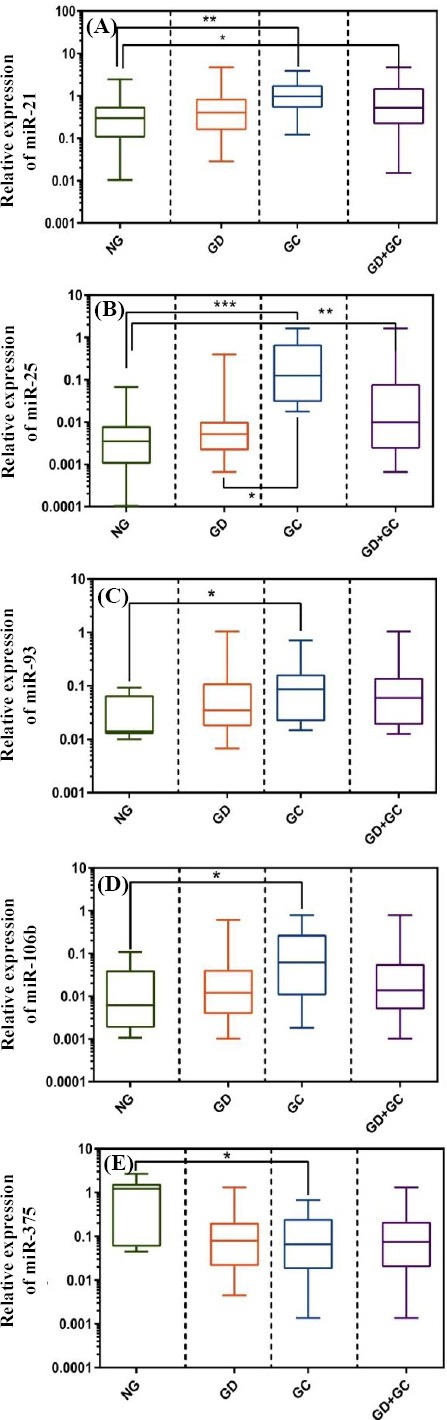
Relative expression (2-ΔΔCt) of miR-21 (A), miR-25 (B), miR-93 (C), miR-106b (D), and miR-375 (E) in the studied samples. *, **, *** indicate p ≤ 0.05, p ≤ 0.01, and p ≤ 0.001, respectively.
Table 2.
ΔCT of each miRNA in three groups (GC, GD, and NG) and between GC and GD groups
| miRNA | GC (n = 39) | GD (n = 33) | NG (n = 29) | p value | GC + GD (n = 72) | NG | p value |
|---|---|---|---|---|---|---|---|
| miR-21 | 2.75±1.068 | 3.3±2.26 | 3.78±1.78 | 0.0048 | 2.65±1.21 | 3.78±1.78 | 0.0175 |
| miR-25 | 5.11±4.24 | 5.96±2.54 | 4.25±3.29 | 0.0003 | 6.08±4.38 | 4.25±3.29 | 0.035 |
| miR-93 | 5.083±2.23 | 6.52±2.34 | 3.47±2.75 | 0.0518 | 6.23±3.28 | 3.47±2.75 | 0.124 |
| miR-106b | 6.127±4.123 | 9.02±4.22 | 9.76±5.43 | 0.0035 | 5.05±4.08 | 9.76±5.43 | 0.0528 |
| miR-375 | 4.63±4.06 | 3.70±3.28 | 3.68±1.57 | 0.3093 | 4.15±3.66 | 3.68±1.57 | 0.72 |
One-way ANOVA test was used to calculate the mean ± SD of miRNA in each group by multiple-comparison, and also the t-test was conducted for analysis between two (GC + NG and NG) groups.
Clinical characteristics of the study subjects
The demographic and clinicopathological data for various miRNAs were retrospectively obtained from patients’ medical records, as depicted in Table 3. As evident from the results of Mann-Whitney test, the expression profile of candidate miRNAs was significantly correlated with several factors, including TNM Classification of Malignant Tumors (TNM) stage, lymph node metastases, and H. pylori infection (p < 0.05).
Table 3.
Comparison between candidate miRNA expression rates (mean of ΔCt ± SD) according to the clinicopathological characteristics of GC patients
| Variable | N | miR-21 | miR-25 | miR-93 | miR-106b | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD | P value | Mean ± SD | P value | Mean ± SD | P value | Mean ± SD | P value | ||
| Age | |||||||||
| >50 | 64 | 3.20 ±2.76 | 0.0893 | 4.175 ± 3.166 | 0.6344 | 6.36 ± 5.176 | 0.0581 | 6.35 ±5.08 | 0.175 |
| <50 | 50 | 5.79 ±4.25 | 5.46 ± 5.038 | 6.32 ± 5.32 | 6.32 ± 5.34 | ||||
| Sex | |||||||||
| Male | 65 | 4.750 ± 3.166 | 0.6654 | 5.33 ± 4.38 | 0.270 | 6.45 ± 5.031 | 0.270 | 6.886 ± 6.36 | 0.270 |
| Female | 49 | 5.038 ± 5.46 | 5.91 ± 4.16 | 5.991± 4.754 | 3.195 ±2.98 | ||||
| TNM | |||||||||
| I, II | 14 | 1.048 ± 1.009 | 0.0309 | 5.59 ± 3.38 | 0.0253 | 4.171 ± 3.521 | 0.5830 | 8.137 ± 7.93 | 0.0138 |
| III, IV | 20 | 5.036 ± 4.070 | 3.280 ±2.524 | 4.843 ± 3.524 | 4.62 ±3.905 | ||||
| LNM | |||||||||
| Positive | 18 | 4.62 ± 4.43 | 0.0259 | 4.62 ±4.43 | 0.0439 | 6.848 ±5.094 | 0.0402 | 3.21 ± 1.32 | 0.0070 |
| Negative | 20 | 4.38 ± 2.635 | 4.38 ±2.63 | 3.427 ±2.327 | 3.169± 2.138 | ||||
| H. Pylori | |||||||||
| Positive | 64 | 4.835 ± 2.935 | 0.0014 | 3.624 ± 3.28 | 0.0128 | 4.865 ± 4.524 | 0.2313 | 7.34 ±6.92 | 0.0023 |
| Negative | 32 | 1.635 ± 1.122 | 6.134 ± 5.35 | 3.427 ± 2.124 | 6.21 ± 4.724 | ||||
miRNAs expression profile for discrimination of GC from normal tissues
The ROC curve analysis was performed on all the miRNAs to investigate the performance of the five mentioned miRNAs as a discriminatory tool to classify tissues in GC, GD, and normal groups. As shown in Figure 2 and Table 4, the ROC curves of the candidate miRNAs demonstrate that these miRNAs could discriminate between cancerous and non-cancer tissue with a relatively high sensitivity and specificity. Besides, the sensitivity and specificity for miR-21 were detected to be 77.08% and 68%, respectively, with an AUC of 0.74 (95% CI = 0.61–0.84, p < 0.00), based on the data presented in Figure 2A. The data presented in Figure 2B demonstrates that miR-25 alone could achieve a suitable diagnosis accuracy to distinguish GC tissue from non-cancerous tissue, with sensitivity of 72.2%, specificity of 100%, and AUC of 0.91 (95% CI = 0.81–0.97; p < 0.00). Moreover, we found that the expression profiles of miR-21, miR-25, miR-93, and miR-106b together have a specificity of 80.08% and sensitivity of 48.30% to distinguish between GC tissue from non-cancer normal, with an AUC of 0.674 (95% CI = 0.611-0.733, p < 0.0001.
Fig. 2.
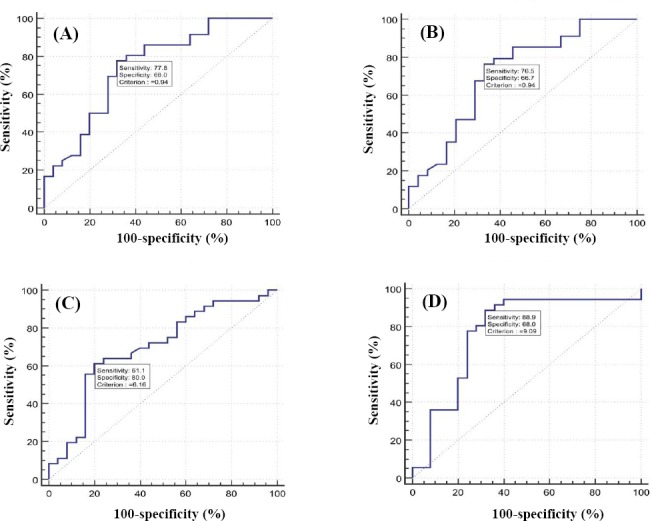
ROC curve analysis using different miRNAs for discriminating GC from normal tissue. ROC curves were constructed to show the specificity and sensitivity of miR-21 (A), miR-25 (B), miR-93 (C), and miR-106b (D).
Table 4.
ROC curves test for determination of specificity, sensitivity, and AUC for candidates miRNA
| miRNA | AUC | Specificity | Sensitivity | P value | Youden index | Cut-off (Δct) |
|---|---|---|---|---|---|---|
| miR-21 | 0.742 | 68 | 77.8 | 0.0003 | 0.4578 | 0.94 |
| miR-25 | 0.726 | 66.7 | 76.5 | 0.0125 | 0.4314 | 0.94 |
| miR-93 | 0.698 | 80 | 61 | 0.0049 | 0.4111 | 6.16 |
| miR-106b | 0.771 | 68 | 88.9 | 0.0001 | 0.5689 | 9.09 |
DISCUSSION
In recent years, a wide variety of studies have focused on the role of miRNAs, as promising biomarkers, in the diagnosis and screening of cancers at early stage[20-23]. Unlike classical biomarkers, miRNAs exhibit a lot of advantages, including high stability, low degradation, and tissue-specific properties[24]. miRNAs are aberrantly expressed in malignant in comparison with normal tissues, indicating the importance of miRNA roles in tumor formation. Importantly, the differential expression patterns of miRNAs may be a useful alternative for both tumor classification and early diagnosis. Additionally, miRNA expression has been exhibited to correlate not only with different cancer types but also with cancer progression[25]. In the present study, we assessed five miRNAs, including miR-21, miR-25, miR-93, miR-106b, and miR-375, followed by qRT-PCR analysis. Our findings revealed that these miRNAs can be used as potentially suitable biomarkers for detecting and distinguishing cancer from non-cancer tissues. Our data also indicated that miR-21, miR-25, miR-93, and miR-106b had a higher expression in the GC tissue when compared with normal tissue. Moreover, pre-cancer tissue (dysplasia) and cancer tissue showed a significant difference in the expression levels of miR-25 (p < 0.05). This implies that miR-25 expression levels can be used to diagnose GC patients at early stage because of the multi-step processes of the gastric carcinogenesis[26]. In parallel, recent studies have suggested that miR-21 can be used as a biomarker for early diagnosis of GC. Shiotani et al.[27] have introduced miR-21 as a potential biomarker and cancer tissue showed a significant difference in the expression levels of miR-25 (p < 0.05). More importantly, miR-25 can be used to diagnose GC for the diagnosis of H. pylori-infected GC patients. Cui et al.[28] have discovered that miR-21 expression levels in GC patients are significantly higher than those in normal individuals. In fact, miR-21 up-regulation can change the biological process of cancer cells, including proliferation, apoptosis, and cellular invasion, probably via regulating RECK and PTEN as the main target genes. Zhang et al.[29] have found a reverse correlation between the expression level of miR-21 and RECK gene, so that the increased expression of miR-21 leads to the decreased expression of RECK. It has been suggested that the down-regulation of RECK plays a role in the progression of GC. Previous investigations have revealed that FOXO3, one of the key target genes of miR-25, critically act as a transcription factor in autophagy processes, cell cycle progression, and apoptosis[30,31].
miR-106b and miR-93 belong to the miR-106b-25 cluster, which resides in the 13th intron of the DNA replication gene MCM7 on chromosome 7 in human beings[32]. These two miRNAs have been found to be highly expressed in many cancers, particularly in GC[33,34]. A study carried out by Petrocca et al.[35] have suggested the oncogenic role of the miR-106b/miR-93 cluster in the progression of GC through the up-regulation effect of these two miRNAs on tumor-suppressor target genes such as P21 and BIM.
Our results indicated that the overexpression of miR-21, miR-25, and miR-106b are closely correlated with the TNM stage (p < 0.05), H. pylori infection (p < 0.05), and lymph node metastases (p < 0.05), which are the main prognostic factors for GC.
In a study conducted in 2011, Matsushima et al.[36] demonstrated that there were significant differences in the miRNA expression signature between H. pylori-infected and H. pylori-uninfected gastric mucosa. Considering the role of H. pylori in chronic inflammation in gastric mucosa, this difference in miRNA expression can be applied as a prognostic factor. Based on recent reports, the expression of miRNAs has a specific relationship with lymph node metastasis, which is a prognostic factor for GC patients[7,23].
In conclusion, given the significant differences in the expression of candidate miRNAs, especially miR-25, in cancer samples compared with normal tissue, as well as their correlation with clinicopathological characteristics that play an important role in the development and progression of GC, these miRNAs can be considered as a suitable biomarker for GC early diagnosis. However, further studies are required to determine their certain roles in GC diagnosis.
ACKNOWLEDGEMENTS
The financial support of this work by Shahid Beheshti University of Medical Sciences is acknowledged. The authors would like to thank Miss Reyhaneh Alaie, laboratory assistant at Central Lab, School of Allied Medical Sciences, Shahid Beheshti University of Medical Sciences, Tehran, Iran for her cooperative attitudes.
Footnotes
CONFLICT OF INTEREST. None declared.
REFERENCES
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA cancer journal for clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Isomoto H, Shikuwa S, Yamaguchi N, Fukuda E, Ikeda K, Nishiyama H, Ohnita K, Mizuta Y, Shiozawa J, Kohno S. Endoscopic submucosal dissection for early gastric cancer:a large-scale feasibility study. Gut. 2009;58(3):331–336. doi: 10.1136/gut.2008.165381. [DOI] [PubMed] [Google Scholar]
- 3.Espinel J, Pinedo E, Ojeda V, del Rio MG. Treatment modalities for early gastric cancer. World journal of gastrointestinal endoscopy. 2015;7(12):1062–1069. doi: 10.4253/wjge.v7.i12.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008:GLOBOCAN 2008. International journal of cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 5.Dhalla F, da Silva S, Lucas M, Travis S, Chapel H. Review of gastric cancer risk factors in patients with common variable immunodeficiency disorders, resulting in a proposal for a surveillance programme. Clinical and experimental immunology. 2011;165(1):1–7. doi: 10.1111/j.1365-2249.2011.04384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooke CL, Torres J, Solnick JV. Biomarkers of Helicobacter pylori-associated gastric cancer. Gut microbes. 2013;4(6):532–540. doi: 10.4161/gmic.25720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng Q, Chen C, Guan H, Kang W, Yu C. Prognostic role of microRNAs in human gastrointestinal cancer:A systematic review and meta-analysis. Oncotarget. 2017;8(28):46611–46623. doi: 10.18632/oncotarget.16679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Leva G, Garofalo M, Croce CM. microRNAs in cancer. Annual review of pathology. 2014;9:287, 314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang S, Chung HC. Novel biomarker candidates for gastric cancer. Oncology reports. 2008;19(3):675–680. [PubMed] [Google Scholar]
- 10.Bartels CL, Tsongalis GJ. MicroRNAs:novel biomarkers for human cancer. Clinical chemistry. 2009;55(4):623–631. doi: 10.1373/clinchem.2008.112805. [DOI] [PubMed] [Google Scholar]
- 11.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. New England journal of medicine. 2005;353(17):1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 12.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 13.Cai H, Yuan Y, Hao YF, Guo TK, Wei X, Zhang YM. Plasma microRNAs serve as novel potential biomarkers for early detection of gastric cancer. Medical oncology. 2013;30(1):452. doi: 10.1007/s12032-012-0452-0. [DOI] [PubMed] [Google Scholar]
- 14.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy BL, Obad S, Bihannic L, Ayrault O, Zindy F, Kauppinen S, Roussel MF. Silencing of the miR-17~92 cluster family inhibits medulloblastoma progression. Cancer research. 2013;73(23):7068–7078. doi: 10.1158/0008-5472.CAN-13-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortés-Sempere M, de Cáceres II. microRNAs as novel epigenetic biomarkers for human cancer. Clinical and translational oncology. 2011;13(6):357–362. doi: 10.1007/s12094-011-0668-z. [DOI] [PubMed] [Google Scholar]
- 18.Shenouda SK, Alahari SK. MicroRNA function in cancer:oncogene or a tumor suppressor? Cancer and metastasis reviews. 2009;28(3-4):369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCTmethod. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Ebert MP, Röcken C. Molecular screening of gastric cancer by proteome analysis. European journal of gastroenterology and hepatology. 2006;18(8):847–853. doi: 10.1097/00042737-200608000-00007. [DOI] [PubMed] [Google Scholar]
- 21.Jinawath N, Furukawa Y, Hasegawa S, Li M, Tsunoda T, Satoh S, Yamaguchi T, Imamura H, Inoue M, Shiozaki H, Nakamura Y. Comparison of gene-expression profiles between diffuse-and intestinal-type gastric cancers using a genome-wide cDNA microarray. Oncogene. 2004;23(40):6830–6844. doi: 10.1038/sj.onc.1207886. [DOI] [PubMed] [Google Scholar]
- 22.Stock M, Otto F. Gene deregulation in gastric cancer. Gene. 2005;360(1):1–19. doi: 10.1016/j.gene.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Cai H, Wang Y. Prognostic significance of tumour markers in Chinese patients with gastric cancer. ANZ journal of surgery. 2014;84(6):448–453. doi: 10.1111/j.1445-2197.2012.06287.x. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. 30. Vol. 105. USA: Proceedings of the national academy of sciences; 2008. pp. 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Correa P. Human gastric carcinogenesis:a multistep and multifactorial process—first American Cancer Society award lecture on cancer epidemiology and prevention. Cancer research. 1992;52(24):6735–6740. [PubMed] [Google Scholar]
- 27.Shiotani A, Murao T, Kimura Y, Matsumoto H, Kamada T, Kusunoki H, Inoue K, Uedo N, Lishi H, Haruma K. Identification of serum miRNAs as novel non-invasive biomarkers for detection of high risk for early gastric cancer. British journal of cancer. 2013;109(9):2323–2330. doi: 10.1038/bjc.2013.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui L, Zhang X, Ye G, Zheng T, Song H, Deng H, Xiao B, Xia T, Yu X, Le Y, Guo J. Gastric juice MicroRNAs as potential biomarkers for the screening of gastric cancer. Cancer. 2013;119(9):1618–1626. doi: 10.1002/cncr.27903. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Laboratory investigation. 2008;88(12):1358–1366. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- 30.Nho RS, Hergert P. FoxO3a and disease progression. World journal of biological chemistry. 2014;5(3):346–354. doi: 10.4331/wjbc.v5.i3.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan C, Wang L, Zhou L, Fu Z. The function of FOXO1 in the late phases of the cell cycle is suppressed by PLK1-mediated phosphorylation. Cell cycle. 2014;13(5):807–819. doi: 10.4161/cc.27727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Lliopoulos D, Pilozzi E, Liu CG, Negrini M, Cavazzini L, Volinia S, Alder H, Ruco LP, Baldassarre G, Croce CM, Vecchione A. E2F1-regulated microRNAs impair TGFβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer cell. 2008;13(3):272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, Tan TM. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer science. 2009;100(7):1234–1242. doi: 10.1111/j.1349-7006.2009.01164.x. [DOI] [PubMed] [Google Scholar]
- 34.Kan T, Sato F, Ito T, Matsumura N, David S, Cheng Y, Agarwal R, Paun BC, Jin Z, Olaru AV, Selaru FM, Hamilton JP, Yang J, Abraham JM, Mori Y, Meltzer SJ. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136(5):1689–1700. doi: 10.1053/j.gastro.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, Cavazzini L, Volinia S, Alder H, Ruco LP, Baldassarre G, Croce CM, Vecchione A. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer cell. 2008;13(3):272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 36.Matsushima K, Isomoto H, Inoue N, Nakayama T, Hayashi T, Nakayama M, Nakao K, Hirayama T, Kohno S. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. International journal of cancer. 2011;128(2):361–370. doi: 10.1002/ijc.25348. [DOI] [PubMed] [Google Scholar]


