miR172b regulates the transcription of FLS2 through TARGET OF EAT1 (TOE1) and TOE2, which governs the ontogeny of plant innate immunity.
Abstract
Innate immunity plays a vital role in protecting plants and animals from pathogen infections. Immunity varies with age in both animals and plants. However, little is known about the ontogeny of plant innate immunity during seedling development. We report here that the Arabidopsis (Arabidopsis thaliana) microRNA miR172b regulates the transcription of the immune receptor gene FLAGELLIN-SENSING2 (FLS2) through TARGET OF EAT1 (TOE1) and TOE2, which directly bind to the FLS2 promoter and inhibit its activity. The level of miR172b is very low in the early stage of seedling development but increases over time, which results in decreased TOE1/2 protein accumulation and, consequently, increased FLS2 transcription and the ontogeny of FLS2-mediated immunity during seedling development. Our study reveals a role for the miR172b-TOE1/2 module in regulating plant innate immunity and elucidates a regulatory mechanism underlying the ontogeny of plant innate immunity.
INTRODUCTION
Plant innate immunity represents the first line of inducible host defense against pathogens (Jones and Dangl, 2006; Takeuchi and Akira, 2010). FLAGELLIN-SENSING2 (FLS2), a well-studied plasma membrane-localized leucine-rich repeat receptor-like kinase (LRR-RLK), recognizes the pathogen-associated molecular pattern (PAMP) flagellin or its derived peptide flg22 to mount PAMP-triggered immunity (PTI) (Gómez-Gómez and Boller, 2000; Jones and Dangl, 2006). The perception of flg22 triggers the rapid association of FLS2 with another LRR-RLK, BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1) (Chinchilla et al., 2007; Heese et al., 2007). A receptor-like cytoplasmic kinase, BOTRYTIS-INDUCED KINASE1 (BIK1), associates with the FLS2 receptor complex and directly phosphorylates the NADPH oxidase RbohD, resulting in a calcium burst and reactive oxygen species production (Lu et al., 2010; Zhang et al., 2010; Kadota et al., 2014; Li et al., 2014). In addition, flagellin perception triggers the activation of mitogen-activated protein kinases (MAPKs), induction of immune-responsive genes, callose deposition to reinforce the cell wall, and immunity to a broad spectrum of pathogens (Jones and Dangl, 2006; Dodds and Rathjen, 2010; Tang et al., 2017).
The immune responses must be tightly controlled to ensure optimal magnitude and appropriate duration. Direct ubiquitination of FLS2 by two closely related U-box E3 ubiquitin ligases, PUB12 and PUB13, leads to the attenuation of immune signaling (Lu et al., 2011). A BAK1-interacting kinase, BAK1-INTERACTING RECEPTOR-LIKE KINASE2 (BIR2), negatively regulates PTI by preventing BAK1-receptor complex formation in the absence of PAMPs. Upon flg22 perception, BIR2 is released from BAK1 and enables the association of BAK1 with FLS2 (Halter et al., 2014). PTI also can be fine-tuned by the rapid release of small peptides like rapid alkalinization factor23, which binds to its receptor FERONIA to inhibit immunity. Otherwise, FERONIA serves as a scaffold to promote the complex assembly of the immune receptor and its coreceptor BAK1 (Stegmann et al., 2017). In addition, protein phosphatase2A (PP2A) associates with BAK1 and modulates its phosphostatus, thereby negatively regulating plant innate immunity (Segonzac et al., 2014). Immune signaling also can be regulated by controlling the turnover of BIK1. Nonactivated BIK1 is ubiquitinated by PUB25 and PUB26 for degradation (Wang et al., 2018). The heterotrimeric G proteins inhibit the E3 ligase activity of PUB25/26, thereby stabilizing BIK1, whereas CALCIUM-DEPENDENT PROTEIN KINASE28 specifically phosphorylates PUB25/26 to boost their activity and promote BIK1 degradation (Monaghan et al., 2014; Liang et al., 2016; Wang et al., 2018). FLS2-mediated immunity also can be positively regulated by the transcriptional control of FLS2 through the transcription factors ETHYLENE INSENSITIVE3 (EIN3) and EIN3-LIKE1 (Boutrot et al., 2010). Overall, relative to the growing number of regulatory mechanisms functioning at the protein level, much less is known about those working through transcriptional regulation of the components of the immune receptor complex.
Immunity varies with age. Age-dependent immune responses have been observed in plants (Zhao et al., 2009; Saur et al., 2016). For example, the immune responses mediated by RECEPTOR-LIKE PROTEIN REQUIRED FOR CSP22 RESPONSIVENESS (NbCSPR) were greater in 6-week-old than in 4-week-old Nicotiana benthamiana plants (Saur et al., 2016). However, to date, little is known about plant innate immunity in the course of seedling development. Here, we show that the microRNA miR172b regulates the transcription of FLS2 through TARGET OF EAT1 (TOE1) and TOE2, which governs the ontogeny of plant innate immunity.
RESULTS
Transcription of FLS2 Governs the Ontogeny of flg22-Triggered Immunity during Seedling Development
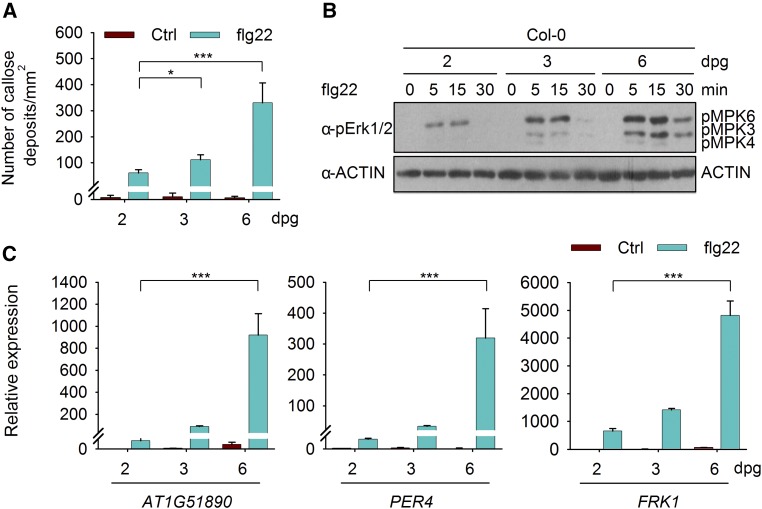
To characterize innate immunity during seedling development, we analyzed flg22-triggered immune responses in Arabidopsis (Arabidopsis thaliana) Col-0 plants 2, 3, and 6 d after germination (Supplemental Figure 1). Flg22-induced callose deposition, MAPK activation, and expression of immune-responsive genes, such as At1g51890, PEROXIDASE4 (PER4) (Malinovsky et al., 2014), and FLG22-INDUCED RECEPTOR-LIKE KINASE1 (FRK1), all were increased progressively during the first week after germination (Figures 1A–1C). This suggests that the flg22-triggered immunity in the course of seedling development may involve an ontogenetic process.
Figure 1.
Flg22-Triggered Immunity Was Increased during Seedling Development.
(A) Flg22-induced callose deposition. Col-0 seedlings at the indicated ages (days post germination [dpg]) were treated with 1 μM flg22 for 12 h. Data are shown as means ± sd (n = 6 leaves from different seedlings). Statistical significance compared with 2-d-old plants was assessed using one-way ANOVA followed by Student-Newman-Keuls tests: *P < 0.05 and ***P < 0.001. Three independent replicates were performed, and similar results were obtained.
(B) Flg22-triggered MAPK activation. Col-0 seedlings were treated with 100 nM flg22. MAPK activation was detected by immunoblotting with anti-pErk1/2 antibodies. ACTIN was detected as a loading control. This experiment was repeated three times with similar results.
(C) Flg22-induced gene induction. Col-0 seedlings were treated with 100 nM flg22 for 3 h. The expression of At1g51890, PER4, and FRK1 was analyzed by RT-qPCR. Expression levels were normalized to those of GAPC. Values are means ± sd (n = 3). Statistical significance compared with 2-d-old plants was assessed using one-way ANOVA followed by Student-Newman-Keuls tests: ***P < 0.001. Three independent replicates were performed, and similar results were obtained.
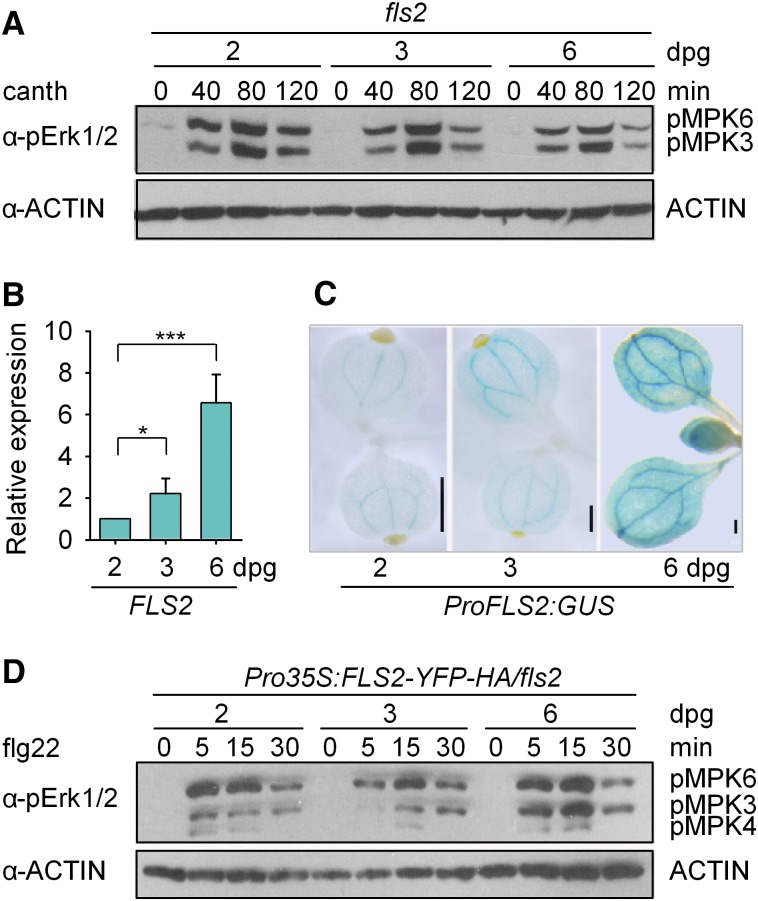
We then monitored MAPK activation in fls2 plants induced by a PP2A inhibitor, cantharidin, which removes the negative regulation of BAK1 imposed by PP2A (Segonzac et al., 2014). During the first week after germination, MAPK activation induced by cantharidin in fls2 seedlings was not increased with time (Figure 2A), suggesting that the ontogeny of immunity during seedling development may not be controlled by the MAPK cascade or BAK1 but may be governed by other upstream components in the PTI signaling pathway. Notably, the expression of FLS2 increases significantly during seedling development (Figure 2B). Furthermore, transgenic plants with the FLS2 promoter (ProFLS2) fused to a reporter gene encoding GUS showed increased GUS activity between 2 and 6 d (Figure 2C). These results suggest that the ontogeny of flg22-triggered immunity during seedling development may be controlled through the transcriptional regulation of FLS2. To confirm this inference, we monitored flg22-induced MAPK activation in Pro35S:FLS2-YFP-HA/fls2 transgenic plants (Shi et al., 2013) and found that MAPK activation did not increase during seedling development when FLS2 was driven by a constitutive promoter (Figure 2D).
Figure 2.
Transcription of FLS2 Controls the Ontogeny of flg22-Triggered Immunity during Seedling Development.
(A) MAPK activation induced by cantharidin is not increased during seedling development. Two-, 3-, or 6-d-old fls2 seedlings were treated with 80 μM cantharidin (canth) for the indicated times. ACTIN was examined as a loading control. This experiment was repeated three times with similar results.
(B) Measurement of FLS2 mRNA levels by RT-qPCR. The levels were normalized to those of GAPC. Values are means ± sd of three biological replicates using independent pools of seedlings grown under the same conditions. Statistical significance compared with 2-d-old Col-0 plants was assessed using one-way ANOVA followed by Student-Newman-Keuls tests: *P < 0.05 and ***P < 0.001.
(C) GUS activity analysis for ProFLS2:GUS transgenic plants. Bars = 250 μm.
(D) Flg22-triggered MAPK activation in Pro35S:FLS2-YFP-HA/fls2 transgenic plants. Seedlings were treated with 100 nM flg22. ACTIN was detected as a loading control. This experiment was repeated three times with similar results.
TOE1/2 Suppress FLS2 Promoter Activity
We hypothesized that the transcriptional activation of FLS2 in the course of seedling development could be due to the absence of transcriptional activators at 2 dpg that are progressively upregulated or to the presence of potential transcriptional suppressors of FLS2 at 2 dpg that are downregulated as plants grow. It has been found that EIN3 serves as a transcriptional activator of FLS2 (Boutrot et al., 2010). Therefore, we focused on screening transcriptional suppressors of FLS2. We performed RNA sequencing (RNA-seq) to identify downregulated transcription factor genes using RNA isolated from 2- and 6-d-old seedlings (Supplemental Figure 2A). We found that the expression of EIN3 was increased during seedling development (Supplemental Figures 2B and 2C). Importantly, a total of 135 transcription factor genes were downregulated in the course of seedling development (Supplemental Figures 2D and 2E, Supplemental Data Set 1). Meanwhile, two brassinosteroid (BR)-repressed genes are expressed at lower levels in 2-d-old than in 6-d-old plants, and a cytokinin (CK)-responsive gene, ARABIDOPSIS RESPONSE REGULATOR3 (ARR3), is expressed at a higher level in 2-d-old than in 6-d-old plants, suggesting that BR and CK signaling may be more active during the early stage of plant growth after germination (Supplemental Figures 2B and 2C).
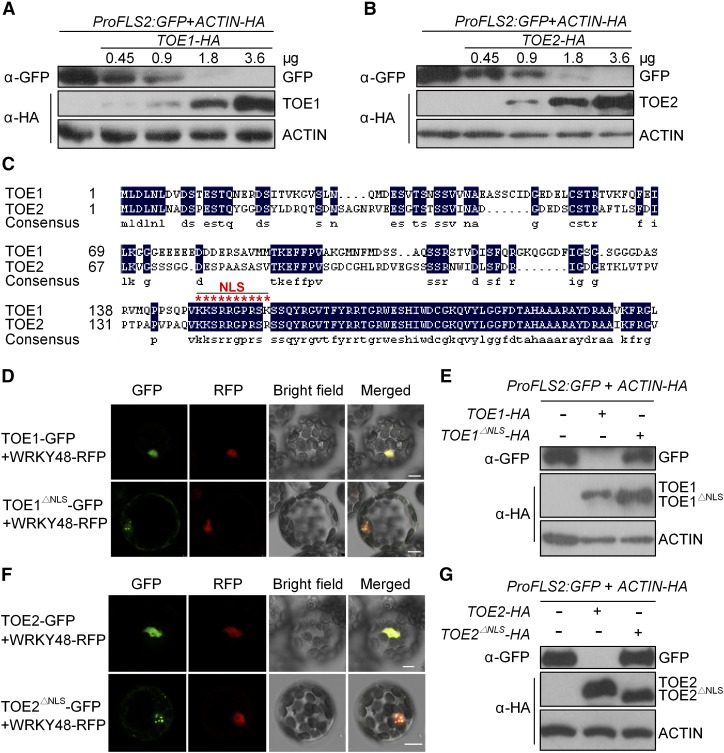
To examine which transcription factors could suppress FLS2 promoter activity, we performed a leaf protoplast cell-based screen by transiently expressing either the firefly luciferase (LUC) reporter gene or the GFP gene driven by the FLS2 promoter along with one of the candidate transcription factor genes (Supplemental Figure 3). To validate the functionality of this screening system, we first transiently expressed EIN3-HA together with ProFLS2:LUC or ProFLS2:GFP. Consistent with the previous report (Boutrot et al., 2010), EIN3 enhanced ProFLS2 activity in protoplasts (Supplemental Figures 4A and 4B). We then screened more than 100 of the 135 transcription factors identified above, using this protoplast transient expression system, and found that TOE1 and its closest homolog, TOE2, significantly suppressed ProFLS2 activity when they were expressed in protoplasts (Supplemental Figures 5 and 6A–6C). TOE1 and TOE2 belong to a subfamily of AP2-like transcription factors (Aukerman and Sakai, 2003; Schmid et al., 2003). However, AP2 did not suppress ProFLS2 activity (Supplemental Figure 6D). Moreover, the suppression of ProFLS2 activity by TOE1/2 was dose dependent (Figures 3A and 3B).
Figure 3.
TOE1/2 Suppress FLS2 Promoter Activity.
(A) and (B) The suppression of ProFLS2 activity by TOE1 (A) or TOE2 (B). TOE1/2-HA was coexpressed with ProFLS2:GFP in protoplasts. ACTIN-HA was coexpressed as an internal transfection control. The expression of ProFLS2:GFP was detected by immunoblotting. The experiments were repeated three times with similar results.
(C) Alignment of the N termini of TOE1 and TOE2. The NLS is highlighted with red asterisks.
(D) and (F) Subcellular localization of TOE1/2 NLS deletion mutants, TOE1/2△NLS. TOE1/2-GFP or TOE1/2△NLS-GFP was coexpressed with WRKY48-RFP in protoplasts. The protoplasts then were subjected to laser confocal imaging. Bars = 7.5 μm.
(E) and (G) TOE1/2△NLS failed to suppress FLS2 promoter activity. These experiments were repeated three times with similar results.
To further confirm these results, we deleted the nuclear localization signals (NLSs) of TOE1/2 and generated the TOE1/2△NLS mutants (Jofuku et al., 1994) (Figure 3C). Then, we transiently expressed TOE1/2-GFP or TOE1/2△NLS-GFP together with WRKY48-RFP, a nucleus-localized protein (Xing et al., 2008), in Arabidopsis protoplasts. We found that the fluorescence patterns of TOE1/2-GFP overlapped with that of RFP-tagged WRKY48 while those of TOE1/2△NLS-GFP did not, although a significant proportion of the mutant proteins could be localized predominantly to nuclear foci. Consequently, TOE1/2△NLS could not suppress ProFLS2 activity (Figures 3D–3G).
To examine whether TOE1 could regulate other PRRs, like the elongation factor EF-Tu receptor (EFR) (Zipfel et al., 2006), we transiently expressed TOE1-HA together with ProEFR:GFP (GFP gene driven by the EFR promoter) in Arabidopsis protoplasts. We found that TOE1 could suppress ProEFR activity (Supplemental Figure 7), suggesting that TOE1 also may regulate EFR-mediated immunity.
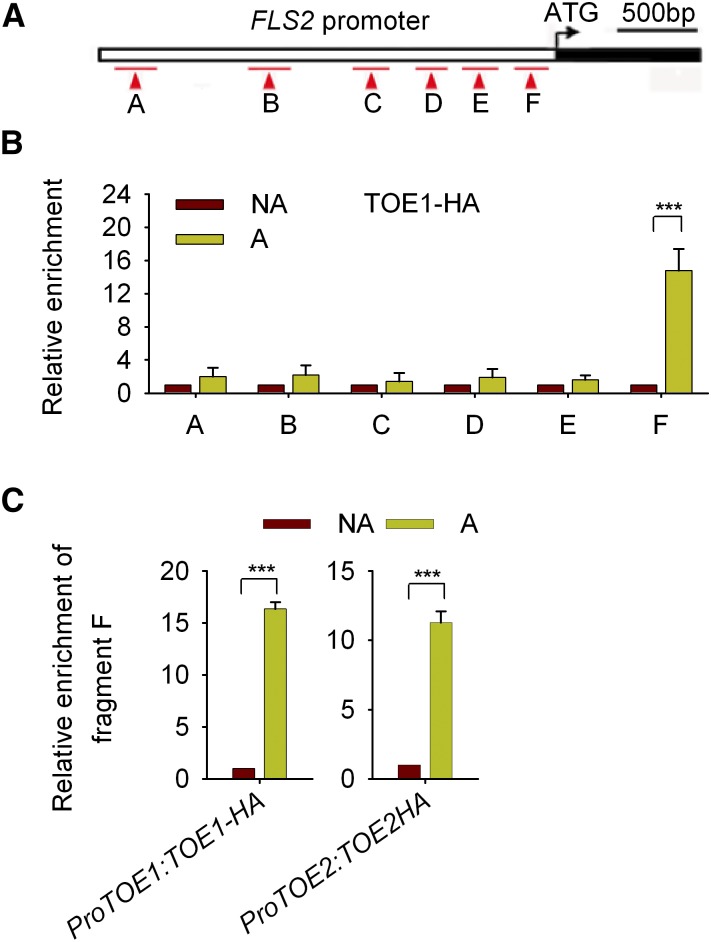
We then performed chromatin immunoprecipitation (ChIP) assays using transiently expressed TOE1-HA in Arabidopsis protoplasts. Region F of the FLS2 promoter was enriched in TOE1-HA-immunoprecipitated chromatin at a markedly higher level than other regions (Figures 4A and 4B). The association between TOE1/2 and region F was confirmed by ChIP assays using ProTOE1:TOE1-HA and ProTOE2:TOE2-HA transgenic plants at 2 dpg, as TOE1 and TOE2 are expressed at relatively high levels at this developmental stage (Figure 4C, Supplemental Data Set 1).
Figure 4.
TOE1/2 Are Associated with Region F of the FLS2 Promoter.
(A) The partial structure of the FLS2 gene and regions amplified by ChIP-PCR.
(B) Enrichment of the indicated TOE1-associated DNA regions in protoplasts expressing TOE1-HA as determined by ChIP-PCR. TOE1-HA was transfected in protoplasts. Chromatin from protoplasts expressing TOE1-HA was immunoprecipitated with anti-HA antibodies, enrichment levels were normalized to those of protoplasts transfected with empty vectors, and no-antibody (NA) immunoprecipitates served as a control. Values are means ± sd of three biological replicates. Statistical significance compared with the NA control was determined by Student’s t tests: ***P < 0.001.
(C) Enrichment of the F region DNA of the FLS2 promoter in ProTOE1:TOE1-HA and ProTOE2:TOE2-HA transgenic plants. Chromatin from 2-d-old ProTOE1/2:TOE1/2-HA transgenic seedlings was immunoprecipitated with anti-HA antibodies, enrichment levels were normalized to those of Col-0 seedlings, and NA immunoprecipitates served as a control. Values are means ± sd (n = 3). Statistical significance compared with the NA control was determined by Student’s t tests: ***P < 0.001. Three independent replicates were performed, and similar results were obtained.
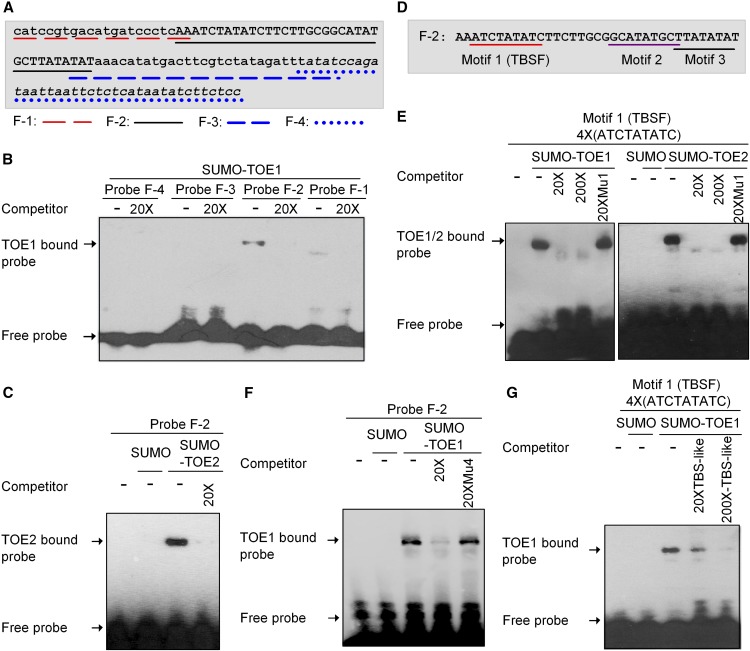
To determine whether TOE1/2 is capable of binding directly to the FLS2 promoter, we conducted electrophoretic mobility shift assays (EMSAs) with purified recombinant SUMO-TOE1/2. Region F of the FLS2 promoter was divided into four fragments (F-1 to F-4) with some overlap (Figure 5A). The SUMO-TOE1 fusion protein was only able to bind to the DNA probes corresponding to F-2. The addition of an excess of unlabeled F-2 probes effectively reduced the binding of SUMO-TOE1 to the biotin-labeled F-2 probes. Similar results also were observed for SUMO-TOE2 (Figures 5B and 5C).
Figure 5.
TOE1/2 Bind Directly to the FLS2 Promoter.
(A) Region F of the FLS2 promoter was divided into four fragments, F-1 to F-4.
(B) TOE1 binds to the F-2 DNA fragment. EMSA was performed with purified recombinant SUMO-TOE1 and each fragment (F-1 to F-4) as a probe. An excess of unlabeled DNA probes was added to compete with the biotin-labeled DNA probes. This experiment was repeated three times with similar results.
(C) TOE2 binds to the F-2 DNA fragment. EMSA was performed with purified recombinant SUMO-TOE2 and the F-2 DNA fragment as a probe. SUMO proteins were used as a negative control. This experiment was repeated three times with similar results.
(D) The F-2 fragment in the FLS2 promoter contains three motifs rich in A/T (Motifs 1–3).
(E) Both TOE1 and TOE2 can bind to Motif 1. EMSA was performed with purified recombinant SUMO-TOE1/2 and Motif 1 (repeated four times) as a probe. An excess of unlabeled wild-type Motif 1 or its mutant (Mu1) DNA probes were added to compete with the biotin-labeled wild-type Motif 1 probes. Motif 1 was named as TBSF. These experiments were repeated three times with similar results.
(F) TOE1 binds to the TBSF motif in the F-2 DNA fragment. EMSA was performed with the F-2 DNA fragment as a probe. SUMO proteins were used as a negative control. Mu4 is a mutant form of the F-2 DNA fragment in which only TBSF was mutated. An excess of unlabeled wild-type F-2 or Mu4 DNA probes were added to compete with the biotin-labeled wild-type F-2 DNA probes. This experiment was repeated three times with similar results.
(G) EMSA competitions with purified recombinant SUMO-TOE1 and the TBS-like motif as a competitor. EMSA was performed with the TBSF motif (repeated four times) as a probe, and an excess of unlabeled probes of the TBS-like motif (from ProFT, repeated four times) was added. This experiment was repeated three times with similar results.
It was shown recently that TOE1 can associate with AT-rich elements (ATREs), TOE binding site (TBS)-like motifs, and other unknown DNA sequences in the promoter of FLOWERING LOCUS T (FT) (Zhai et al., 2015; Zhang et al., 2015). Sequence analysis failed to identify any ATRE, TBS, or TBS-like motif in the F-2 fragment of the FLS2 promoter. However, F-2 contains three motifs (Motifs 1–3) rich in A/T (Figure 5D). To determine whether these motifs can bind to TOE1, we performed EMSAs with each motif repeated four times as a probe. The results showed that only Motif 1 could bind to TOE1, and a mutated form of Motif 1, Mu1, could not compete with wild-type Motif 1 for binding to TOE1 (Figure 5E, Supplemental Figure 8). Motif 1 (ATCTATATC), therefore, is named as TOE binding site in the FLS2 promoter (TBSF). Similarly, TOE2 also was able to bind to the TBSF motif (Figure 5E). This result was further confirmed when only TBSF was mutated in F-2 (Mu4). As demonstrated in Figure 5F, Mu4 failed to compete with wild-type F-2 for binding to SUMO-TOE1. EMSA competition assays also were performed with the previously identified TBS-like motif from the ProFT directly as a competitor (Zhai et al., 2015). The results showed that the addition of an excess of unlabeled probes of TBS-like motifs (repeated four times as a probe) effectively reduced the binding of SUMO-TOE1 to the biotin-labeled TBSF probes (repeated four times as a probe) (Figure 5G). Moreover, a region from the ProFT containing a TBS-like motif can successfully compete with F-2 from the ProFLS2 for binding to SUMO-TOE1 (Supplemental Figure 9).
TOE1/2 Negatively Regulate FLS2-Mediated Immunity during Seedling Development
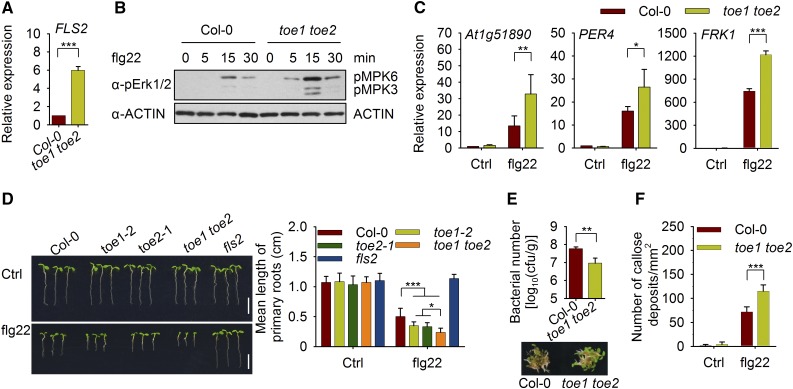
TOE1/2 can suppress FLS2 promoter activity, so we tested the expression of FLS2 in 2-d-old toe1 toe2 mutant plants. As expected, the level of FLS2 transcripts in 2-d-old toe1 toe2 plants was remarkably higher than that of wild-type plants (Figure 6A). Flg22-induced MAPK activation in 2-d-old toe1 toe2 plants was much stronger than that in wild-type plants (Figure 6B). Moreover, the induction of the immune-responsive genes, At1g51890, PER4 (Malinovsky et al., 2014), and FRK1, in 2-d-old toe1 toe2 mutant plants was much greater than that in wild-type plants (Figure 6C).
Figure 6.
TOE1/2 Negatively Regulate FLS2-Mediated Immunity during Seedling Development.
(A) RT-qPCR analysis of FLS2 expression in 2-d-old Col-0 and toe1 toe2 seedlings. Expression levels were normalized to those of GAPC. Values are means ± sd of three biological replicates using independent pools of seedlings grown under the same conditions. Statistical significance compared with Col-0 was determined by Student’s t tests: ***P < 0.001.
(B) Flg22-induced MAPK activation. Two-day-old Col-0 and toe1 toe2 mutant plants were treated with 100 nM flg22 for the indicated times. MAPK activation was detected by immunoblotting with anti-pErk1/2 antibodies. ACTIN was detected as a loading control. This experiment was repeated three times with similar results.
(C) Flg22-induced gene induction in 2-d-old plants. Col-0 and toe1 toe2 mutant seedlings were treated with 100 nM flg22 for 3 h. The expression of At1g51890, PER4, and FRK1 was analyzed by RT-qPCR, and the expression levels were normalized to those of GAPC. Values are means ± sd of three biological replicates using independent pools of seedlings (45 seedlings per pool) grown under the same conditions. Statistical significance compared with Col-0 was assessed using one-way ANOVA followed by Student-Newman-Keuls tests: *P < 0.05; **P < 0.01; and ***P < 0.001.
(D) Flg22-triggered seedling growth inhibition assay for Col-0, toe1-2, toe2-1, and toe1 toe2 double mutant plants. One-day-old plants were transferred to medium containing 1 μM flg22 and grown for another 5 d. The quantification of growth inhibition is shown in the right panel. Values are means ± sd (n = 12 seedlings). Statistical significance was assessed using one-way ANOVA followed by Student-Newman-Keuls tests: *P < 0.05 and ***P < 0.001. Three independent replicates were performed, and similar results were obtained. Bars = 5 mm.
(E) Growth of Pst DC3000 on Col-0 and toe1 toe2 plants. Two-day-old plants were inoculated with bacteria. Values are means ± sd of three biological replicates using independent seedling samples grown and inoculated under the same conditions. Statistical significance compared with Col-0 was determined by Student’s t tests: **P < 0.01. Representative photographs of Col-0 and toe1 toe2 seedlings infected with Pst DC3000 are shown in the lower panel. cfu, colony-forming units.
(F) Flg22-induced callose deposition in Col-0 and toe1 toe2 plants. Two-day-old plants were treated with 1 μM flg22 for 12 h, and the number of callose deposits was counted using ImageJ. Values are means ± sd (n = 6 leaves from different seedlings). Statistical significance compared with Col-0 was assessed using one-way ANOVA followed by Student-Newman-Keuls tests: ***P < 0.001. Three independent replicates were performed, and similar results were obtained.
Then, we examined the sensitivity of toe1-2, toe2-1, and toe1 toe2 mutant plants (Aukerman and Sakai, 2003; Wu et al., 2009) to flg22 by seedling growth inhibition assays. Because the seedling growth inhibition assay involves a relatively long duration of flg22 treatment, we transferred 1-d-old plants to medium containing flg22. After 5 d of growth, toe1-2, toe2-1, and toe1 toe2 plants showed enhanced sensitivity to flg22 compared with wild-type plants; the phenotype was stronger in toe1 toe2 than in either single mutant (Figure 6D), suggesting that TOE1 and TOE2 have overlapping functions in regulating immunity. However, in the absence of flg22, there was no obvious difference among these plants during seedling development (Figure 6D). Moreover, Pro35S:FLS2-YFP-HA/fls2 plants were phenotypically indistinguishable from fls2 and wild-type plants when grown on one-half-strength Murashige and Skoog (1/2 MS) medium or in soil, and the difference among these plants was detected only in the presence of flg22 (Supplemental Figures 10A and 10B).
We also performed pathogen infection assays with 2-d-old toe1 toe2 mutant and wild-type plants. toe1 toe2 plants were more resistant than wild-type plants to Pseudomonas syringae pv tomato (Pst) DC3000 (Figure 6E). Additionally, when 2-d-old plants were treated with flg22 for 12 h, the induction of callose deposition by flg22 was stronger in toe1 toe2 mutant plants than in wild-type plants (Figure 6F). These results suggest that the flg22-triggered immune responses in toe1 toe2 plants are much stronger than those in wild-type plants and that TOE1/2 negatively regulate FLS2-mediated immunity during seedling development.
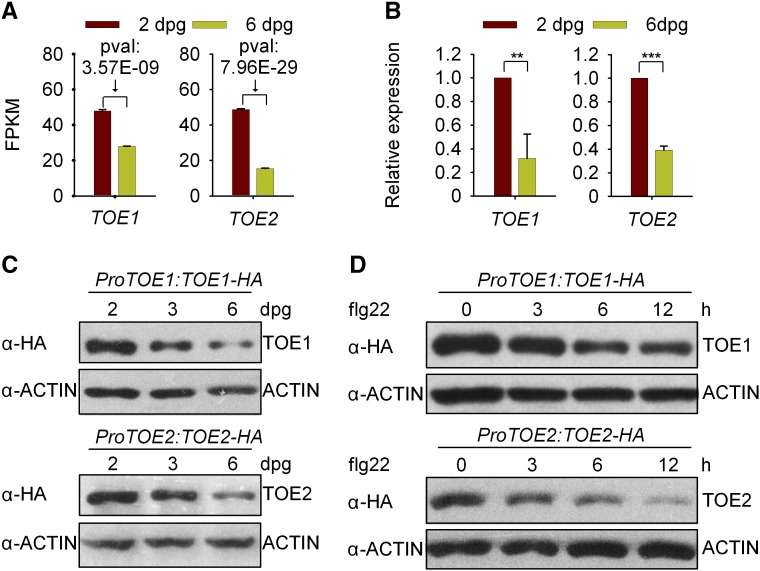
TOE1 and TOE2 are targets of miR172 (Aukerman and Sakai, 2003; Schwab et al., 2005; Jung et al., 2007). It has been found that the expression of miR172 was regulated temporally, and miR172 abundance was increased progressively as plants grew until flowering, which leads to a gradual reduction in TOE1/2 transcript levels (Aukerman and Sakai, 2003; Jung et al., 2007; Wu et al., 2009). Consistently, we found that TOE1/2 transcript levels and the accumulation of TOE1/2 proteins decrease during seedling development (Figures 7A–7C). Furthermore, the TOE1/2 protein levels in 2-d-old plants were decreased upon flg22 treatment (Figure 7D). Moreover, levels of TOE1/2 transcripts and TOE1/2 proteins also decreased upon flg22 treatment at adult plant growth stages (Supplemental Figure 11). These results suggest that the flg22-triggered immunity during seedling development is suppressed by TOE1/2 but that this suppression is gradually removed with time. Moreover, the repression of FLS2 by TOE1/2 also could be relieved upon flg22 treatment.
Figure 7.
Expression Analysis of TOE1 and TOE2.
(A) The fragments per kilobase of transcript sequence per million base pairs sequenced (FPKM) of TOE1/2 during seedling development as determined by RNA-seq analysis. Values are means ± sd of two biological replicates. pval indicates the P value, which was adjusted using the Benjamini and Hochberg approach.
(B) Measurement of TOE1/2 transcript levels during seedling development by RT-qPCR. The transcript levels were normalized to those of GAPC. Values are means ± sd of three biological replicates using independent pools of seedlings grown under the same conditions. Statistical significance compared with 2-d-old Col-0 was determined by Student’s t tests: **P < 0.01 and ***P < 0.001.
(C) Immunoblotting analysis of TOE1/2 protein accumulation during seedling development. Total proteins were isolated from 2-, 3-, or 6-d-old ProTOE1:TOE1-HA or ProTOE2:TOE2-HA transgenic seedlings. ACTIN was examined as a loading control. These experiments were repeated three times with similar results.
(D) TOE1/2 protein levels were decreased upon flg22 treatment. Two-day-old transgenic seedlings were treated with 1 μM flg22 for the indicated times. These experiments were repeated three times with similar results.
In addition, we found that the transgenic plants with the TOE1 promoter (ProTOE1) fused to a GUS reporter gene did not show decreased GUS activity between 2 and 6 d (Supplemental Figure 12) or in response to flg22 treatment (Supplemental Figure 13). These results suggest that the observed decrease of TOE1 transcripts during seedling development, or upon flg22 treatment, is controlled mainly at the posttranscriptional level rather than the transcriptional level.
miR172b Positively Regulates FLS2-Mediated Immunity during Seedling Development
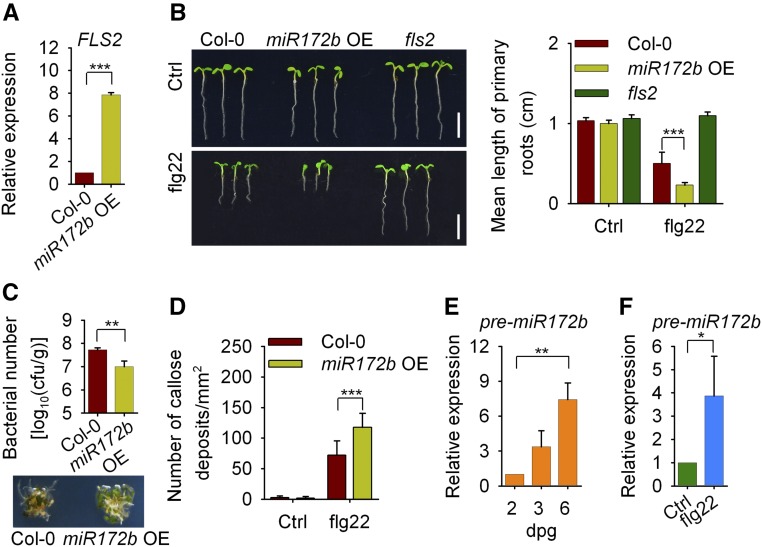
TOE1 and TOE2 are targets of miR172 (Aukerman and Sakai, 2003; Schwab et al., 2005; Jung et al., 2007; Wu et al., 2009). Therefore, we tried to elucidate the role of miR172 in regulating plant innate immunity through TOE1/2. Consistently, the expression of FLS2, flg22-triggered seedling growth inhibition, resistance to Pst DC3000, flg22-induced callose deposition, and the induction of the immune-responsive genes in transgenic plants overexpressing miR172b (OE) during seedling development were very similar to the observations in toe1 toe2 plants (Figures 8A–8D, Supplemental Figure 14).
Figure 8.
miR172b Positively Regulates FLS2-Mediated Immunity during Seedling Development.
(A) RT-qPCR analysis of FLS2 expression in 2-d-old Col-0 and miR172b OE seedlings. Expression levels were normalized to those of GAPC. Values are means ± sd of three biological replicates using independent pools of seedlings (45 seedlings per pool) grown under the same conditions. Statistical significance compared with Col-0 was determined by Student’s t tests: ***P < 0.001.
(B) Flg22-triggered seedling growth inhibition assay. One-day-old wild-type and miR172b OE plants were treated with 1 μM flg22 for 5 d. Quantification of growth inhibition is shown in the right panel. Values are means ± sd (n = 12 seedlings). Statistical significance compared with Col-0 was assessed using one-way ANOVA followed by Student-Newman-Keuls tests: ***P < 0.001. Three independent replicates were performed, and similar results were obtained. Bars = 5 mm.
(C) Growth of Pst DC3000 on Col-0 and miR172b OE plants. Two-day-old plants were inoculated with bacteria (OD600 = 5 × 10−4). Bacterial growth was determined at 3 d post inoculation. Values are means ± sd of three biological replicates using independent seedling samples grown and inoculated under the same conditions. Statistical significance compared with Col-0 was determined by Student’s t tests: **P < 0.01. Representative photographs of Col-0 and miR172b OE seedlings infected with Pst DC3000 are shown in the lower panel. cfu, colony-forming units.
(D) Flg22-induced callose deposition in Col-0 and miR172b OE plants. Two-day-old plants were treated with 1 μM flg22 for 12 h. Values are means ± sd (n = 6 leaves from different seedlings). Statistical significance compared with Col-0 was assessed using one-way ANOVA followed by Student-Newman-Keuls tests: ***P < 0.001. Three independent replicates were performed, and similar results were obtained.
(E) The expression of miR172b during seedling development. Total RNA was isolated from 2-, 3-, or 6-d-old seedlings. The pre-miR172b level was analyzed by RT-qPCR and was normalized to that of GAPC. Values are means ± sd of three biological replicates using independent pools of seedlings grown under the same conditions. Statistical significance compared with 2-d-old plants was assessed using one-way ANOVA followed by Student-Newman-Keuls tests: **P < 0.01.
(F) The expression of miR172b was induced by flg22 treatment. Two-day-old seedlings were treated with or without 100 nM flg22 for 3 h. Values are means ± sd of three biological replicates using independent pools of seedlings grown under the same conditions. Statistical significance compared with the control was determined by Student’s t tests: *P < 0.05.
Consistent with the previous report (Aukerman and Sakai, 2003), levels of pre-miR172b and mature miR172b increased during seedling development or upon flg22 treatment in 2-d-old and 8-week-old plants (Figures 8E and 8F, Supplemental Figures 15 and 16). In addition, we found that ProFLS2 activity was higher in protoplasts isolated from miR172b OE plants than that in protoplasts from Col-0 plants (Figure 9A). Notably, the large increase of FLS2 transcripts observed in the course of seedling development was significantly compromised in MIM172 transgenic plants, a stable knockdown line of miR172 (Todesco et al., 2010), although the level of FLS2 transcripts in 6-d-old MIM172 plants was still higher than that in 2-d-old plants (Supplemental Figure 17). These results suggest that miR172 positively regulates the expression of FLS2 during seedling development, but which also may be regulated by other factors.
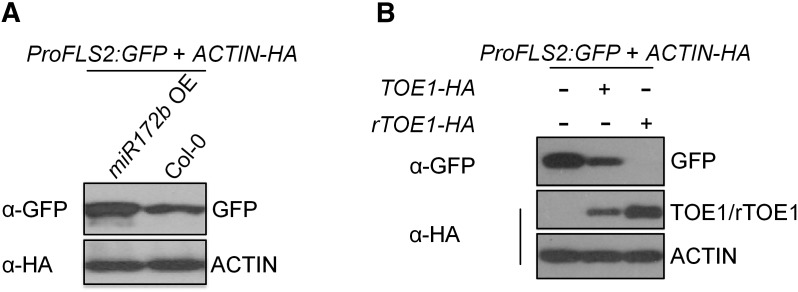
Figure 9.
Regulation of ProFLS2 Activity by miR172b Is Mediated through TOE1/2.
(A) ProFLS2:GFP was expressed in protoplasts isolated from Col-0 or miR172b OE plants.
(B) ProFLS2:GFP was coexpressed with TOE1-HA or rTOE1-HA in protoplasts.
For (A) and (B), ACTIN-HA was coexpressed as an internal transfection control, and the expression of ProFLS2:GFP was detected by immunoblotting with anti-GFP antibodies.
To verify that the regulation of ProFLS2 activity by miR172b is mediated through TOE1/2, we first expressed a miR172-resistant form of TOE1 (rTOE1) (Chen, 2004) in Arabidopsis protoplasts together with ProFLS2:GFP. The results showed that rTOE1 accumulated at a higher level than TOE1, leading to a stronger suppression of ProFLS2 activity (Figure 9B). Moreover, we measured the TOE1/2 mRNA levels in MIM172 transgenic plants (Todesco et al., 2010) and found that the downregulation of TOE1 and TOE2 between 2 and 6 dpg no longer occurred in MIM172 transgenic plants (Figure 7B, Supplemental Figure 18).
Then, we examined the levels of TOE1/2 transcripts and their cleavage products in miR172b OE and wild-type plants. We found that the cleavage products of both TOE1 and TOE2 increased in miR172b OE compared with wild-type plants. The TOE2 mRNA level decreased substantially in miR172b OE, whereas the steady-state level of TOE1 mRNA declined only slightly (Supplemental Figures 19A and 19B), suggesting that, in miR172b OE plants, the steady-state level of at least TOE1 transcripts might be modulated by feedback regulation, as reported previously (Schwab et al., 2005). Furthermore, TOE1 proteins were barely detected in ProTOE1:TOE1-HA/miR172b OE plants (Supplemental Figure 19C). These results confirmed that both TOE1 and TOE2 were downregulated in miR172b OE plants.
DISCUSSION
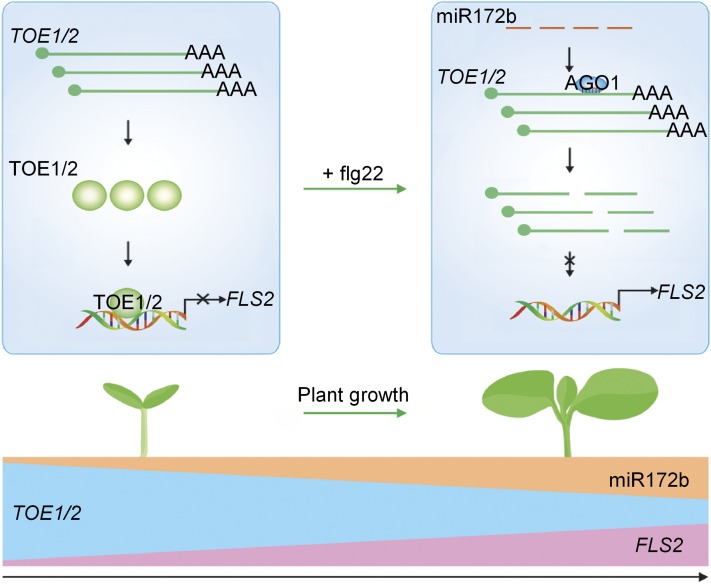
It has been reported that age affects immune responses in plants. The immunity mediated by NbCSPR was greater in 6-week-old than in 4-week-old N. benthamiana plants, which may be related to NbCSPR upregulation in 6-week-old relative to 4-week-old plants (Saur et al., 2016). In another report, the progressively enhanced rice resistance to Xanthomonas oryzae pv oryzae during plant growth from seedling stage to adult stage could be the consequence of the gradually increased expression of the LRR-RLK-type resistance genes Xa3/Xa26 and Xa21 (Zhao et al., 2009). Here, we found that Arabidopsis FLS2 transcription increases progressively during seedling development, which regulates the ontogeny of plant innate immunity. Hence, transcriptional regulation of the immune receptor or related genes plays an important role in regulating age-dependent immunity. The transcription of FLS2 is regulated by miR172b through TOE1/2 that bind directly to the FLS2 promoter and suppress its expression (Figure 10). However, the transcription factors modulating the expression of NbCSPR and resistance genes such as Xa3/Xa26 and Xa21 remain to be determined.
Figure 10.
A Proposed Model of miR172b-Mediated Transcriptional Regulation of FLS2 in Controlling the Ontogeny of Innate Immunity during Seedling Development.
TOE1 and TOE2 bind directly to the FLS2 promoter and inhibit its activity. miR172b targets TOE1 and TOE2 and downregulates their expression. The level of miR172b is very low in the early stage of seedling development but increases over time, which results in decreased TOE1/2 protein accumulation and, consequently, the ontogeny of FLS2-mediated immunity. Additionally, the level of miR172b also is increased in plants challenged with flg22, leading to decreased TOE1/2 accumulation.
In this work, we found that TOE1/2 can bind directly to the TBSF motif in the FLS2 promoter and inhibit its activity, although in vitro DNA affinity purification sequencing data do not provide evidence for the binding of TOE1/2 to this region in the FLS2 promoter (O’Malley et al., 2016). In addition to TBSF, other TOE1 binding sites have been identified in the FT promoter. With its cofactor JASMONATE-ZIM DOMAIN1, TOE1 binds to a TBS-like motif in the FT promoter (Zhai et al., 2015). TOE1 also could interact with CONSTANS (CO) and bind to an ATRE motif in the FT promoter near the CO binding site (Zhang et al., 2015). These results implied that different cofactors may affect the binding of TOE1 to various DNA sequences. However, whether there are cofactors involved in the binding of TOE1 to the FLS2 promoter remains to be determined.
It has been reported that EIN3 also binds directly to the FLS2 promoter and positively controls FLS2 expression (Boutrot et al., 2010). Terrestrial seed plants start growth under the soil. Notably, EIN3 protein levels are increased in response to soil overlay (Zhong et al., 2014). Consistently, our results showed that the expression of EIN3 was increased in the course of seedling development (Supplemental Figures 2B and 2C). Therefore, the transcription of FLS2 is regulated not only by the repression of TOE1/2 but also most likely by the upregulation of EIN3 or other unidentified factors to control the innate immunity ontogeny.
The suppression of FLS2-mediated immune responses during seedling development may be the result of a tradeoff between development and immunity. We found that a number of BR-repressed genes are expressed at lower levels in 2-d-old than in 6-d-old plants, suggesting that BR signaling may be more active during the early stage of seedling development (Supplemental Figures 2B and 2C). However, in the absence of flg22, even Pro35S:FLS2-YFP-HA/fls2 seedlings were phenotypically indistinguishable from fls2 and wild-type seedlings (Supplemental Figure 10). It has been found that the interaction between BR and PTI signaling is unidirectional: activation of BR signaling results in impaired PTI responses in Arabidopsis but not vice versa (Albrecht et al., 2012; Belkhadir et al., 2012; Lozano-Durán et al., 2013). Therefore, the active BR signaling in the course of seedling development may limit PTI responses. Additionally, the plant growth hormone CK modulates various aspects of plant growth (Naseem et al., 2015). We found that a CK-responsive gene, ARR3, is downregulated between 2 and 6 d, suggesting that CK signaling may be more active at the early stage of seedling development. It was reported that moderate activation of CK signaling could lead to the suppression of FLS2 expression and, consequently, the impairment of immune responses like MAPK activation (Hann et al., 2014). However, whether CK signaling is related directly to miR172-mediated FLS2 expression during seedling development remains to be investigated.
miR172 targets AP2-like transcription factors, such as TOE1 and TOE2, and downregulates their expression by transcript cleavage or translation repression (Aukerman and Sakai, 2003; Schwab et al., 2005; Jung et al., 2007; Wu et al., 2009). It was found that the levels of miR172 and TOE1 mRNA display a complementarity in tissues such as flowers, stems, and roots, suggesting that miR172 may regulate TOE1 mainly through mRNA cleavage in these plant tissues (Jung et al., 2007). In addition, miR172 abundance increases as plants grow until flowering, while the levels of TOE1/2 mRNA decrease progressively (Aukerman and Sakai, 2003; Jung et al., 2007). In miR172 overexpression lines, the TOE2 transcript level was reduced significantly but the steady-state levels of TOE1 and AP2 transcripts were not, whereas AP2 protein was decreased dramatically relative to wild-type plants. Therefore, it was suggested that miR172 may negatively regulate its targets through both mRNA cleavage and translational inhibition, and the steady-state transcript levels of miR172 targets also are modulated by feedback regulation (Aukerman and Sakai, 2003; Chen, 2004; Schwab et al., 2005; Jung et al., 2007). However, to date, the detection of TOE1/2 proteins in plants overexpressing miR172 has not been reported.
We found that miR172b abundance increases during seedling development, while the transcript and protein levels of TOE1/2 decrease during the same period (Figures 7B, 7C, and 8E, Supplemental Figure 15). Furthermore, the reduction of TOE1/2 transcript levels between 2 and 6 d was impaired in MIM172 transgenic plants (Supplemental Figure 18). Therefore, miR172-guided transcript cleavage contributes to the downregulation of TOE1 and TOE2 during seedling development.
In miR172 OE transgenic plants, both TOE1 and TOE2 are downregulated, but the underlying mechanisms appear to be more complicated. Consistent with the previous report (Schwab et al., 2005), TOE2 mRNA decreased substantially in miR172b OE transgenic plants compared with wild-type plants, while the steady-state level of TOE1 mRNA only declined slightly, although the cleavage products of both TOE1 and TOE2 were increased significantly in miR172b OE plants. Notably, we found that the TOE1 protein level was reduced dramatically in miR172b OE compared with wild-type plants (Supplemental Figure 19). Taken together, in miR172b OE plants, miR172b may downregulate TOE2 primarily by mRNA cleavage. As for TOE1, feedback regulation may mask the effects of miR172b on TOE1 mRNA level in miR172b OE plants, as has been proposed previously (Schwab et al., 2005; Jung et al., 2007), and TOE1 may be regulated by miR172b through both transcript cleavage and translation repression in miR172b OE plants.
In this study, we demonstrate a function of the miR172-TOE1/2 module. miR172b-mediated transcriptional regulation of FLS2 during seedling development controls the ontogeny of plant innate immunity (Figure 10). The role of the miR172-TOE1/2 module in controlling flowering timing has been studied extensively (Aukerman and Sakai, 2003; Jung et al., 2007; Wu et al., 2009). miR172 abundance was very low in young seedlings but increased progressively as plants grew until flowering, which leads to a gradual reduction of TOE1/2 (Aukerman and Sakai, 2003; Jung et al., 2007; Wu et al., 2009). Once TOE1/2 proteins decrease below a critical threshold, flowering is triggered. It is likely that TOE1/2 prevents precocious flowering by repressing the expression of FT, one of the best characterized floral integrators (Kardailsky et al., 1999). TOE1 and TOE2 can bind directly to FT chromatin or interact with and repress CO, a positive transcriptional regulator of FT (Zhai et al., 2015; Zhang et al., 2015).
The temporal expression of miR172 causes the temporal downregulation of TOE1/2, which presumably results in the increased transcription of FLS2 beyond seedling development and throughout the life cycle. Furthermore, PAMP treatment also could cause the increased expression of miR172 and the decreased expression of TOE1/2 at both the seedling and adult plant stages (Figures 7D and 8F, Supplemental Figures 11 and 16). To ensure survival for any plant species, the most important thing is successful reproduction. Plants have evolved sophisticated mechanisms to coordinate their flowering timing in response to the ever-changing environment. Upon pathogen infections, plants either spend more resources for immunity, which is costly to development, or accelerate flowering to ensure that plants accomplish reproduction before succumbing to disease (Lyons et al., 2015). It has been found that infections by the bacteria Pseudomonas syringae and Xanthomonas campestris accelerate flowering in Arabidopsis (Korves and Bergelson, 2003). Therefore, the miR172-TOE1/2 module not only regulates the ontogeny of innate immunity and the flowering timing during the reproductive phase but also most likely serves as a major integrator to coordinate plant immunity, development, and flowering timing.
METHODS
Plant Material and Growth Conditions
For protoplast isolation and analysis of gene expression at adult plant growth stages, Arabidopsis (Arabidopsis thaliana) plants were grown in soil under 70 μE m−2 s−1 light (white fluorescent bulbs) with a 12-h photoperiod in a growth room at 22°C with 60% relative humidity for 30 d. The mutants fls2, toe1-2 (SALK_069677), toe2-1 (SALK_065370), and toe1 toe2 and the transgenic plants miR172b OE, Pro35S:FLS2-YFP-HA/fls2, and MIM172 were described previously (Shan et al., 2008; Wu et al., 2009; Todesco et al., 2010; Shi et al., 2013). Primers used for genotyping are listed in Supplemental Data Set 2. Seeds were surface sterilized with 50% (v/v) bleach for 5 min and washed at least four times with sterile water. Sterile seeds were germinated and grown on 1/2 MS plates or liquid 1/2 MS medium supplemented with 0.5% (w/v) sucrose. The plates were kept for 2 d in the dark at 4°C to break dormancy (stratification) and transferred thereafter to a growth room at 22°C with a 12-h photoperiod. When comparing plants at different dpg, samples were collected at the same time of day to avoid the effects of photoperiod on the expression of TOE1/2 and other genes (Zhang et al., 2015).
Plasmid Construction, Generation of Transgenic Plants, and Crosses
Arabidopsis TOE1, TOE2, AP2, EIN3, and other transcription factor genes were amplified by PCR from Col-0 cDNA and introduced into a plant expression vector. TOE1/2△NLS mutations were generated by overlap extension PCR as described (Xiao et al., 2007). For ProFLS2:LUC, ProFLS2:GFP, and ProFLS2:GUS, 2.7 kb of genomic DNA sequence upstream of the FLS2 start codon was amplified and fused to LUC, GFP, or GUS in a plant expression vector or pCAMBIA1391 vector. Full-length TOE1/2 was subcloned into a protein expression vector, pET28a-SUMO, using BamHI and HindIII sites for subcloning TOE1 or BamHI and XhoI sites for subcloning TOE2. TOE1/2 transgenic plants were generated in Col-0 by Agrobacterium tumefaciens-mediated transformation with TOE1/2 fused to an HA tag in the pTF101 vector under the control of its native promoter (∼2 kb upstream of its start codon). For ProTOE1:GUS, the TOE1 promoter was fused to a GUS reporter gene in the pCAMBIA1391 vector. The TOE1/2 promoter was amplified from Col-0 genomic DNA. All primers are listed in Supplemental Data Set 2. The ProTOE1:TOE1-HA/miR172b OE plants were generated by crossing ProTOE1:TOE1-HA/Col-0 into the miR172b OE plants.
RT-qPCR, Stem-Loop RT-PCR, and 5′ RACE
To collect RNA of young Arabidopsis seedlings, 0.02 g of 2-, 3-, or 6-d-old plants grown in liquid medium was treated with 100 nM flg22 for 3 h, and total RNA was isolated from whole seedlings. To collect RNA of older plants, rosette leaves from 8-week-old plants were infiltrated with 100 nM flg22 or water, then the samples were harvested at 3 h post infiltration and total RNA was isolated from infiltrated leaves using TRIzol (Invitrogen) according to the manufacturer’s instructions. cDNA was synthesized in 20-μL reactions using 1 µg of DNase I-treated total RNA with a reverse transcription system (Invitrogen). Real-time PCR was performed on a Bio-Rad CFX-96 Real-Time PCR system using a SYBR Green RT-PCR kit (Takara). Expression levels were normalized to the expression of GAPC, a stably expressed reference gene.
Stem-loop-specific reverse transcription was performed as described previously (Chen et al., 2005; Kulcheski et al., 2010). The level of mature microRNA was normalized to that of snoR101. To map the 5′ ends of the cleavage products of TOE1/2 transcripts, 5′ RACE was performed using the 5′-full RACE Kit (Takara). Primers are listed in Supplemental Table 1.
Callose Deposition
Plants were treated with 1 μM flg22 for 12 h, and then callose deposition was analyzed as described previously (Gómez-Gómez and Boller, 2000; Lu et al., 2011). Callose deposits were counted using the analyze particles function of ImageJ 1.42q software (http://rsb.info.nih.gov/ij/). Six different leaves were analyzed for each genotype.
MAPK Assays
MAPK assays were performed using 0.015 g of 2-, 3-, or 6-d-old seedlings grown in liquid medium. Seedlings then were stimulated with 100 nM flg22 or 80 µM cantharidin (Sigma-Aldrich). Total proteins were isolated from whole seedlings. MAPK activation was monitored by immunoblotting with anti-pErk1/2 antibodies (Cell Signaling Technology). ACTIN was examined as a loading control by immunoblotting with anti-β-ACTIN antibodies (Cell Signal Pathway Research Tools Supplier).
GUS Staining
Histochemical staining for GUS activity was performed as described previously (Sun et al., 2009). Plant tissues were incubated in a solution (100 mM sodium phosphate [pH 7.0], 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, 1 mg/mL 5-bromo-4-chrolo-3-indolyl-β-d-glucuronide [Sigma-Aldrich], and 0.1% [v/v] Triton X-100) at 37°C in the dark for 6 h. The GUS-stained samples were dehydrated with 75% (v/v) ethanol three times and then subjected to imaging using a Leica laser scanning microscope.
Transcriptome Sequencing
Total RNA for transcriptome sequencing was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNA concentrations were measured using a NanoDrop 2000 spectrophotometer (ND-2000, ThermoFisher Scientific). Library preparation, sequencing, and data analysis were performed by Novogene. Raw reads in FASTA format were first processed through in-house Perl scripts. At the same time, Q20, Q30, and GC content were calculated for the clean data. Then, the high-quality clean reads were aligned to the reference genome using TopHat v2.0.12. Next, HTSeq v0.6.1 (Trapnell et al., 2009) was used to count the number of fragments per kilobase of transcript sequence per million base pairs sequenced. Differential expression analysis of seedlings at 2 and 6 dpg for two biological replicates was performed using the DESeq R package (1.18.0). P values for differential expression were calculated using a model based on the negative binomial distribution and were adjusted using the Benjamini and Hochberg approach. Genes with P < 0.05 were defined as differentially expressed. The R package pheatmap was used to generate a clustered heat map for differentially expressed genes.
Transient Gene Expression in Protoplasts
Protoplast isolation and gene transient expression were performed as described (Yoo et al., 2007). Briefly, protoplasts were collected 12 h post transfection for promoter activity or gene expression assays. For reporter assays, ProUBQ10:GUS was cotransfected as an internal transfection control, and the promoter activity was calculated as the LUC/GUS ratio. Protein expression was detected by immunoblotting with anti-GFP, anti-FLAG, or anti-HA antibodies (Sigma-Aldrich).
Recombinant Protein Isolation and EMSA
Recombinant SUMO proteins and SUMO-TOE1/2 fusion proteins were purified from Escherichia coli by affinity chromatography using Ni2+-affinity matrices (Qiagen) according to the manufacturer’s instructions. For EMSAs, biotin-labeled DNA probes were prepared by annealing pairs of complementary oligonucleotides with their corresponding sequences. EMSAs were performed using a LightShift Chemiluminescent EMSA Kit (Thermo Scientific). Binding reactions contained 4 μL of protein extract, 2 μL of 10 pM probe, 2 μL of 10× binding buffer, 100 mM MgCl2, 1 μL of 5% (v/v) glycerol, 1 μL of 1 µg/μL poly(dI-dC), and double-distilled water to a total volume of 20 µL. Each reaction was incubated at 25°C for 30 min. All probe sequences are listed in Supplemental Table 2.
ChIP Assays
For ChIP assays using transiently expressed TOE1 proteins, 5 mL of protoplasts was transfected with TOE1-HA or control plasmids and incubated for 8 h, and cells were collected for subsequent use. For ChIP assays using transgenic plants, 2-d-old ProTOE1:TOE1-HA, ProTOE2:TOE2-HA, and wild-type plants were harvested. Cells and seedlings were cross-linked with 1% (v/v) formaldehyde for 10 min in ice and quenched by 0.125 M Gly for 5 min. The remaining steps of ChIP assays were performed as described previously with some modifications (Gendrel et al., 2005; Gao et al., 2013). In both cases, TOE1-HA was immunoprecipitated with anti-HA antibodies (Abcam). Chromatin precipitated without antibodies was used as a negative control, and the chromatin isolated before precipitation was used as an input control. Three independent biological repeats were performed. Primers used in this study are listed in Supplemental Table 3.
Seedling Growth Inhibition Assay
Arabidopsis plants were grown on 1/2 MS plates for 1 d, transferred to 1/2 MS plates containing 1 μM flg22, and then grown for another 5 d. All experiments were repeated three to four times with reproducible results.
Pathogen Assay
The pathogen assay was performed as described previously with modifications (Schreiber et al., 2008). Pseudomonas syringae pv tomato strain DC3000 was grown overnight at 28°C in King’s B medium with 50 μg/mL rifampicin. Bacteria were collected, washed, and diluted to the desired density with 1/2 MS liquid medium (OD600 = 5 × 10−4). Plants were grown in 1/2 MS liquid medium for 2 d on a 12-well tissue culture plate, and then the liquid medium was replaced with the bacterial solution. Three days after inoculation, plants were ground in water, and serial dilutions were plated on King’s B medium with 50 μg/mL rifamycin. The plates were incubated at 28°C for 2 d, and then bacterial colony-forming units were counted.
Replicates of Experiments and Statistical Analysis
The replicates of immunoblotting are shown in Supplemental Data Set 3. All data were analyzed using SigmaPlot 10.0 (Systat Software). The averages and sd of all results were calculated, and one-way ANOVA and Student’s t tests were performed using GraphPad Prism 5 to generate P values. Statistically significant differences are indicated with asterisks as follows: *P < 0.05; **P < 0.01; and ***P < 0.001. The statistical analysis of qPCR and other data is shown in Supplemental Tables 4 and 5.
Accession Numbers
Sequence data from this article can be found in GenBank (https://www.ncbi.nlm.nih.gov/gene/) and TAIR (http://www.arabidopsis.org/) under the following accession numbers: FLS2, NP_199445.1, At5g46330; TOE1, NP_565674.1, At2g28550; TOE2, NP_200820, At5g60120; MIR172B, NR_142746, At5g04275; AP2, NP_195410.1, At4g36920; PER4, NP_172906.1, At1g14540; CPD, NP_196188.1, At5g05690; BR6OX2, NP_566852.1, At3g30180; EIN3, NP_188713.1, At3g20770; FRK1, NP_179509.1, At2G19190; and ARR3, NP_176202.1, At1g59940. The RNA-seq data were deposited in the Gene Expression Omnibus database (GSE119325) at the National Center for Biotechnology Information. Mutants used in this article can be obtained from the ABRC under the following accession numbers: toe1-2 (SALK_069677) and toe2-1 (SALK_065370).
Supplemental Data
Supplemental Figure 1. The Growth of Arabidopsis Plants during the First Week after Germination.
Supplemental Figure 2. Transcriptomic Analyses of Transcription Factor Genes Differentially Expressed during Seedling Development.
Supplemental Figure 3. Schematic Representation of the Constructs ProFLS2:LUC, ProFLS2:GFP, TF-HA, ACTIN-HA, and ACTIN-FLAG.
Supplemental Figure 4. Validation of the Functionality of the Protoplast Cell-Based Screening System.
Supplemental Figure 5. Screening of Transcription Factors That Can Suppress FLS2 Promoter Activity.
Supplemental Figure 6. TOE1/2 Suppressed FLS2 Promoter Activity.
Supplemental Figure 7. TOE1 Suppressed EFR Promoter Activity.
Supplemental Figure 8. TOE1 Cannot Bind to Motif 2/3 in the F-2 Fragment of the FLS2 Promoter.
Supplemental Figure 9. EMSA Competitions with the TBS-Like Motif as a Competitor.
Supplemental Figure 10. The Growth of Pro35S:FLS2-YFP-HA/fls2 Plants during the First Week after Germination.
Supplemental Figure 11. The Expression of TOE1/2 upon PAMP Treatment at Adult Plant Stages.
Supplemental Figure 12. Analysis of TOE1 Promoter Activity during Seedling Development.
Supplemental Figure 13. Analysis of TOE1 Promoter Activity upon flg22 Treatment.
Supplemental Figure 14. Flg22-Induced Gene Induction in 2-d-old miR172b OE Plants.
Supplemental Figure 15. Level of Mature miRNA172a/b Was Increased during Seedling Development.
Supplemental Figure 16. Levels of pre-miR172b and Mature miR172 Were Increased upon flg22 Treatment.
Supplemental Figure 17. Analysis of FLS2 Expression in MIM172 Transgenic Plants.
Supplemental Figure 18. Analysis of TOE1/2 Expression in MIM172 Transgenic Plants.
Supplemental Figure 19. TOE1/2 are Downregulated in miR172b OE Plants.
Supplemental Table 1. Primers Used for RT-qPCR.
Supplemental Table 2. Probes Used for EMSA.
Supplemental Table 3. Primers Used for ChIP-PCR.
Supplemental Table 4. ANOVA Tables.
Supplemental Table 5. Student’s t Test Tables.
Supplemental Data Set 1. Differentially Expressed Transcription Factor Genes during Early Plant Growth as Determined by RNA-seq.
Supplemental Data Set 2. Primers Used for Construction and Genotyping.
Supplemental Data Set 3. Original Blot Images and Replicates of Immunoblotting.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Ping He and Libo Shan (Texas A&M University) for critical reading of the manuscript, R. Scott Poethig (University of Pennsylvania) for the 35S::miR172b seeds, Xia Li (Huazhong Agricultural University) for toe1-2 and toe2-1 seeds, and Jiawei Wang (Chinese Academy of Sciences) and Detlef Weigel (Max Planck Institute for Developmental Biology) for MIM172 seeds. This work was supported by the Chinese Ministry of Science and Technology (2015CB910200), the National Natural Science Foundation of China (31500990 and 31322009), and the State Key Laboratory of Plant Genomics of China.
AUTHOR CONTRIBUTIONS
D.L. conceived the project. Y.Zo., S.W., D.T., and D.L. designed research. Y.Zo., S.W., Y.Zho., J.B., G.H., X.L., and Y.Zha. performed research. Y.Zo., S.W., D.T., and D.L. analyzed data. D.L., Y.Zo., and S.W. wrote the article.
References
- Albrecht C., Boutrot F., Segonzac C., Schwessinger B., Gimenez-Ibanez S., Chinchilla D., Rathjen J.P., de Vries S.C., Zipfel C. (2012). Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. USA 109: 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y., Jaillais Y., Epple P., Balsemão-Pires E., Dangl J.L., Chory J. (2012). Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. USA 109: 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F., Segonzac C., Chang K.N., Qiao H., Ecker J.R., Zipfel C., Rathjen J.P. (2010). Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc. Natl. Acad. Sci. USA 107: 14502–14507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Lao K.Q., Livak K.J., et al. (2005). Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D.G., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500. [DOI] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. (2010). Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Gao X., Chen X., Lin W., Chen S., Lu D., Niu Y., Li L., Cheng C., McCormack M., Sheen J., Shan L., He P. (2013). Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 9: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011. [DOI] [PubMed] [Google Scholar]
- Halter T., Imkampe J., Mazzotta S., Wierzba M., Postel S., Bücherl C., Kiefer C., Stahl M., Chinchilla D., Wang X., Nürnberger T., Zipfel C., et al. (2014). The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 24: 134–143. [DOI] [PubMed] [Google Scholar]
- Hann D.R., Domínguez-Ferreras A., Motyka V., Dobrev P.I., Schornack S., Jehle A., Felix G., Chinchilla D., Rathjen J.P., Boller T. (2014). The Pseudomonas type III effector HopQ1 activates cytokinin signaling and interferes with plant innate immunity. New Phytol. 201: 585–598. [DOI] [PubMed] [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M.E., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku K.D., den Boer B.G.W., Van Montagu M., Okamuro J.K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Jung J.H., Seo Y.H., Seo P.J., Reyes J.L., Yun J., Chua N.H., Park C.M. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19: 2736–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J.D.G., Shirasu K., Menke F., Jones A., Zipfel C. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54: 43–55. [DOI] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D. (1999). Activation tagging of the floral inducer FT. Science 286: 1962–1965. [DOI] [PubMed] [Google Scholar]
- Korves T.M., Bergelson J. (2003). A developmental response to pathogen infection in Arabidopsis. Plant Physiol. 133: 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulcheski F.R., Marcelino-Guimaraes F.C., Nepomuceno A.L., Abdelnoor R.V., Margis R. (2010). The use of microRNAs as reference genes for quantitative polymerase chain reaction in soybean. Anal. Biochem. 406: 185–192. [DOI] [PubMed] [Google Scholar]
- Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., Cai G., Gao L., Zhang X., Wang Y., Chen S., Zhou J.M. (2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338. [DOI] [PubMed] [Google Scholar]
- Liang X., Ding P., Lian K., Wang J., Ma M., Li L., Li L., Li M., Zhang X., Chen S., Zhang Y., Zhou J.M. (2016). Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. eLife 5: e13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R., Macho A.P., Boutrot F., Segonzac C., Somssich I.E., Zipfel C. (2013). The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. eLife 2: e00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Wu S., Gao X., Zhang Y., Shan L., He P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons R., Rusu A., Stiller J., Powell J., Manners J.M., Kazan K. (2015). Investigating the association between flowering time and defense in the Arabidopsis thaliana-Fusarium oxysporum interaction. PLoS ONE 10: e0127699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovsky F.G., Batoux M., Schwessinger B., Youn J.H., Stransfeld L., Win J., Kim S.K., Zipfel C. (2014). Antagonistic regulation of growth and immunity by the Arabidopsis basic helix-loop-helix transcription factor homolog of brassinosteroid enhanced expression2 interacting with increased leaf inclination1 binding bHLH1. Plant Physiol. 164: 1443–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J., Matschi S., Shorinola O., Rovenich H., Matei A., Segonzac C., Malinovsky F.G., Rathjen J.P., MacLean D., Romeis T., Zipfel C. (2014). The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16: 605–615. [DOI] [PubMed] [Google Scholar]
- Naseem M., Kaltdorf M., Dandekar T. (2015). The nexus between growth and defence signalling: Auxin and cytokinin modulate plant immune response pathways. J. Exp. Bot. 66: 4885–4896. [DOI] [PubMed] [Google Scholar]
- O’Malley R.C., Huang S.C., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saur I.M.L., Kadota Y., Sklenar J., Holton N.J., Smakowska E., Belkhadir Y., Zipfel C., Rathjen J.P. (2016). NbCSPR underlies age-dependent immune responses to bacterial cold shock protein in Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 113: 3389–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Uhlenhaut N.H., Godard F., Demar M., Bressan R., Weigel D., Lohmann J.U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012. [DOI] [PubMed] [Google Scholar]
- Schreiber K., Ckurshumova W., Peek J., Desveaux D. (2008). A high-throughput chemical screen for resistance to Pseudomonas syringae in Arabidopsis. Plant J. 54: 522–531. [DOI] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527. [DOI] [PubMed] [Google Scholar]
- Segonzac C., Macho A.P., Sanmartín M., Ntoukakis V., Sánchez-Serrano J.J., Zipfel C. (2014). Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J. 33: 2069–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L., He P., Li J., Heese A., Peck S.C., Nürnberger T., Martin G.B., Sheen J. (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Shen Q., Qi Y., Yan H., Nie H., Chen Y., Zhao T., Katagiri F., Tang D. (2013). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25: 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M., Monaghan J., Smakowska-Luzan E., Rovenich H., Lehner A., Holton N., Belkhadir Y., Zipfel C. (2017). The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289. [DOI] [PubMed] [Google Scholar]
- Sun J., Xu Y., Ye S., Jiang H., Chen Q., Liu F., Zhou W., Chen R., Li X., Tietz O., Wu X., Cohen J.D., et al. (2009). Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21: 1495–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. (2010). Pattern recognition receptors and inflammation. Cell 140: 805–820. [DOI] [PubMed] [Google Scholar]
- Tang D., Wang G., Zhou J.M. (2017). Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 29: 618–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. (2010). A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Grubb L.E., Wang J., Liang X., Li L., Gao C., Ma M., Feng F., Li M., Li L., Zhang X., Yu F., et al. (2018). A regulatory module controlling homeostasis of a plant immune kinase. Mol. Cell 69: 493–504.e6. [DOI] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.H., Yin M.H., Hou L., Luo M., Pei Y. (2007). Asymmetric overlap extension PCR method bypassing intermediate purification and the amplification of wild-type template in site-directed mutagenesis. Biotechnol. Lett. 29: 925–930. [DOI] [PubMed] [Google Scholar]
- Xing D.H., Lai Z.B., Zheng Z.Y., Vinod K.M., Fan B.F., Chen Z.X. (2008). Stress- and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol. Plant 1: 459–470. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhai Q., Zhang X., Wu F., Feng H., Deng L., Xu L., Zhang M., Wang Q., Li C. (2015). Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell 27: 2814–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Wang L., Zeng L., Zhang C., Ma H. (2015). Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev. 29: 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Li W., Xiang T., Liu Z., Laluk K., Ding X., Zou Y., Gao M., Zhang X., Chen S., Mengiste T., Zhang Y., et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301. [DOI] [PubMed] [Google Scholar]
- Zhao J., Fu J., Li X., Xu C., Wang S. (2009). Dissection of the factors affecting development-controlled and race-specific disease resistance conferred by leucine-rich repeat receptor kinase-type R genes in rice. Theor. Appl. Genet. 119: 231–239. [DOI] [PubMed] [Google Scholar]
- Zhong S., Shi H., Xue C., Wei N., Guo H., Deng X.W. (2014). Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc. Natl. Acad. Sci. USA 111: 3913–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760. [DOI] [PubMed] [Google Scholar]