A novel cesa7 mutant revealed that patterned deposition of non-cellulosic secondary wall polymers, xylan and lignin, is not dependent on cellulose patterning during xylem vessel differentiation.
Abstract
The secondary cell wall (SCW) of xylem vessel cells provides rigidity and strength that enables efficient water conduction throughout the plant. To gain insight into SCW deposition, we mutagenized Arabidopsis thaliana VASCULAR-RELATED NAC-DOMAIN7-inducible plant lines, in which ectopic protoxylem vessel cell differentiation is synchronously induced. The baculites mutant was isolated based on the absence of helical SCW patterns in ectopically-induced protoxylem vessel cells, and mature baculites plants exhibited an irregular xylem (irx) mutant phenotype in mature plants. A single nucleic acid substitution in the CELLULOSE SYNTHASE SUBUNIT 7 (CESA7) gene in baculites was identified: while the mutation was predicted to produce a C-terminal truncated protein, immunoblot analysis revealed that cesa7bac mutation results in loss of production of CESA7 proteins, indicating that baculites is a novel cesa7 loss-of-function mutant. In cesa7bac, despite a lack of patterned cellulose deposition, the helically-patterned deposition of other SCW components, such as the hemicellulose xylan and the phenolic polymer lignin, was not affected. Similar phenotypes were found in another point mutation mutant cesa7mur10-2, and an established knock-out mutant, cesa7irx3-4. Taken together, we propose that the spatio-temporal deposition of different SCW components, such as xylan and lignin, is not dependent on cellulose patterning.
INTRODUCTION
The secondary cell wall (SCW) of plants is a thick and rigid composite that is deposited following primary growth cessation in distinct plant cell types, such as xylem vessel cells and fibers. The SCW is composed of cellulose microfibrils embedded in a matrix of hemicellulose and lignin. The most abundant component of SCWs is cellulose, a polymer consisting of linear β-1,4-glucan chains that are synthesized by the cellulose synthase complex (CSC) in the plasma membrane (reviewed in Bashline et al., 2014; McFarlane et al., 2014). In SCWs, cellulose is thought to interact with hemicellulosic polysaccharides such as glucuronoxylan, in which the linear xylan backbone associates with the cellulose microfibrils (Busse-Wicher et al., 2014). The interaction between cellulose microfibrils and xylan is considered to be critical for strength and elasticity of cell walls (reviewed in Scheller and Ulvskov, 2010). In addition, lignin, a phenolic biopolymer, provides rigidity and hydrophobicity to the SCW (reviewed in Vanholme et al., 2010). The lignin monomers (monolignols) are produced inside the cells and exported to the cell walls for polymerization into lignin (reviewed in Mottiar et al., 2016) via random radical coupling. NMR-based structural analysis suggests that lignin is likely associated with hemicellulosic carbohydrates through benzyl ether, γ-ester and phenyl glycoside linkages in SCWs (Balakshin et al., 2011; Yuan et al., 2011).
Accumulated electron microscopy and autoradiography studies on the SCW polymers have suggested that cellulose is deposited at the cell surface, while lignin is deposited within the preexisting wall structure (Ray, 1967; Hepler et al., 1970). Hemicelluloses, such as xylan, are produced by the Golgi and deposited by secretory vesicles throughout SCW synthesis (Northcote et al., 1989; Awano et al., 1998). Moreover, lignin deposition is not observed without prior deposition of polysaccharides (Terashima and Fukushima, 1988; Taylor et al., 1992). Considering that the plasma membrane-localized CSC is responsible for cellulose synthesis, and microtubule orientation is a critical determinant of CSC localization (Paredez et al., 2006; Watanabe et al., 2015), the currently-accepted view of SCW formation can be summarized as 1) SCW formation is initiated with rearrangement of the cortical microtubule network to delimit areas of SCW formation, 2) cellulose synthases are delivered to the MT-enriched plasma membrane domains and cellulose biosynthesis occurs at the cell surface, 3) hemicellulose is simultaneously or subsequently deposited via secretory vesicles and may form hydrogen-bond interactions with cellulose, and 4) monolignols are exported to the cell wall and polymerized into lignin by SCW-localized oxidative enzymes (reviewed in Meents et al., 2018).
Besides detailed microscopy observations, molecular biological approaches continue to improve our understanding of the biosynthetic pathways of these SCW biopolymers. Specifically, Arabidopsis thaliana (Arabidopsis) SCW-specific CELLULOSE SYNTHASE (CESA) genes were identified in irregular xylem (irx) mutants, in which the structural integrity of xylem cells, especially vessel cells, is disrupted (Turner and Somerville, 1997). The irx mutant screen identified alleles of cesa4irx5, cesa8irx1, and cesa7irx3, indicating that all three gene products, CESA4/IRX5, CESA7/IRX3, and CESA8/IRX1, are required for normal cellulose synthesis in the SCW (Taylor et al., 1999, 2000, 2003). The irx screen also revealed SCW-specific hemicellulose biosynthetic genes (IRX7, IRX8, IRX9, IRX10, IRX14, and IRX15) and lignin biosynthetic genes (IRX4 and IRX12) (Turner and Somerville, 1997; Jones et al., 2001; Brown et al., 2005, 2009, 2011; Lee et al., 2007; Peña et al., 2007; Wu et al., 2009; Jensen et al., 2011).
Additional tools to investigate SCW biosynthesis are model plant cell cultures from Zinnia elegans and Arabidopsis, in which the differentiation of tracheary elements can be induced (Fukuda and Komamine, 1980; Demura et al., 2002; Kubo et al., 2005; Pesquet et al., 2010). Early works by Taylor et al. (1992) and Taylor and Haigler (1993) using the Zinnia elegans system showed that treatment with a cellulose synthesis inhibitor, isoxaben or 2,6-dichlorobenzonitrile, during SCW formation disrupts the patterned accumulation of cellulose, as well as xylan and lignin deposition. The authors thus proposed “a self-perpetuating cascade model” for the patterned deposition of SCW polymers, in which localized cellulose deposition mediates the patterning of other SCW polymers (Taylor and Haigler, 1993).
Another important achievement of using these induction systems was the identification of key transcriptional regulators of SCW biosynthesis. Kubo et al. (2005) successfully identified the plant-specific NAM, ATAF1,2, and CUC2 (NAC) transcription factors VASCULAR-RELATED NAC-DOMAIN6 (VND6) and VND7 as master regulators of metaxylem and protoxylem vessel cell fates, respectively. VND6 and VND7 are conserved among a wide range of plant species, including non-vascular land plants such as bryophytes (Zhu et al., 2012; Xu et al., 2014; Nakano et al., 2015). These transcription factors have been exploited experimentally to create transgenic plants and cell cultures that overexpress or activate VND6 or VND7 function. For instance, VND6/VND7 have been fused with the viral VP16 transcription activation domain and the glucocorticoid receptor (GR) and expressed under the control of the 35S promoter (35Spro::VND6/VND7-VP16-GR) and similarly VND6/VND7 were expressed under the control of the inducible XVE system (Zuo et al., 2000) (XVE-VND6/VND7). In these cases, the application of glucocorticoid (35Spro::VND6/VND7-VP16-GR) or estradiol (XVE-VND6/VND7) triggers the activation or the expression of VND6/VND7, respectively, and subsequent transdifferentiation into vessel cells in a synchronous manner (Oda et al., 2010; Yamaguchi et al., 2010). This has created important tools to elucidate the cellular and molecular mechanisms involved in SCW formation (Oda et al., 2010; Ohtani et al., 2011, 2016; Oda and Fukuda, 2012, 2013; Goué et al., 2013; Schuetz et al., 2014; Endo et al., 2015; Watanabe et al., 2015, 2018; Li et al., 2016; Kawabe et al., 2018).
In an attempt to obtain insights into the process of SCW deposition, we performed a forward genetic screen on the inducible VND7-VP16-GR system, screening for plants impaired in the patterned deposition of SCW components in ectopic protoxylem vessel cells. As a result, we successfully isolated the baculites mutant, and identified that the gene responsible for the baculites phenotype was CESA7, encoding a subunit of SCW-specific CSC (cesa7bac). Surprisingly, contrary to the previous observation of cellulose synthesis inhibitor experiments using the Zinnia elegans tracheary element induction system (Taylor and Haigler, 1993; Taylor et al., 1992), while the typical helical pattern of SCW cellulose deposition was abolished in baculites, deposition of xylan and lignin into spiral or annular SCW domains was still observed in ectopic protoxylem vessel cells. Detailed observation during vessel cell differentiation demonstrated that xylan deposition is directed by the microtubule orientation. This observation was also found in an independent cesa7 point mutant allele, cesa7mur10-2, as well as in a known cesa7irx3-4 T-DNA knock-out mutant. Immunoblot analysis with anti-CESA antibodies showed that CESA7 was not detected in the baculites mutant, indicating that cesa7bac is a novel cesa7 null mutant allele. The results of this study indicate that the patterned deposition of xylan/lignin occurs independently of cellulose deposition during differentiation of protoxylem vessel cells.
RESULTS
Forward Genetic Screen for Defects in Secondary Cell Wall Pattern Identifies baculites
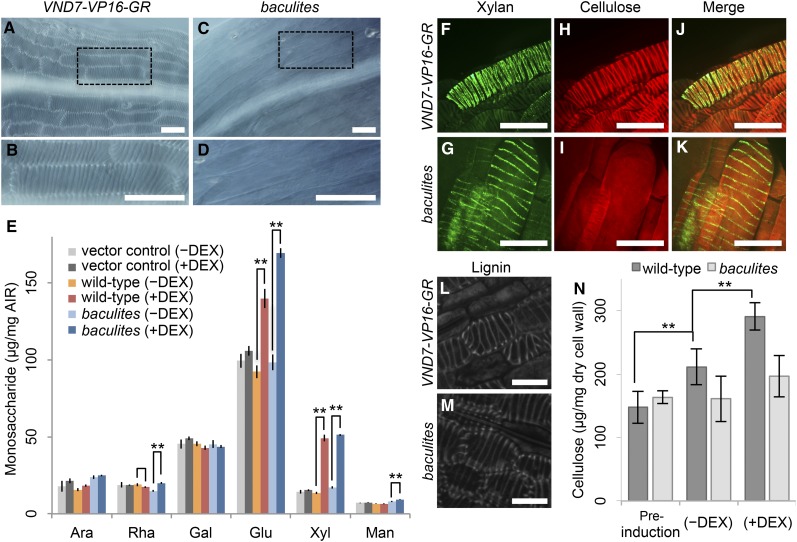
Seeds of 35Spro::VND7-VP16-GR (wild-type VND7-VP16-GR) (Yamaguchi et al., 2010) were mutagenized by ethyl methanesulfonate to obtain M1 plants, which were subsequently individually selfed and grown to produce independent M2 seeds. To identify mutants with defective SCW deposition, we examined transdifferentiated cells from the M2 seedlings treated with 10 μM dexamethasone (DEX) for 3 days via microscopy by the optical microscope equipped with Nomarski optics. After screening more than 2000 lines, we successfully identified the recessive mutant line that we named baculites (Figures 1A to 1D), after a smooth ammonite that lacks the helical pattern characteristic of these extinct molluscs. While the parental wild-type VND7-VP16-GR consistently differentiated ectopic vessel cells with the clear helical SCW characteristic of protoxylem vessel cells (Figures 1A and 1B), plant lines expressing VND7-VP16-GR in the baculites background (baculites VND7-VP16-GR) did not show the helically-patterned SCW of ectopic vessel cells (Figures 1C and 1D). Monosaccharide analysis of the DEX-treated seedlings showed that induction of protoxylem vessel cell differentiation resulted in nearly identical increases in glucose and xylose in both wild-type VND7-VP16-GR and baculites VND7-VP16-GR, while the vector control VP16-GR showed no change in monosaccharide composition (Figure 1E). The data are consistent with previous observation on cell wall fractions of VND7-VP16-GR, which showed glucose and xylose accumulation during xylem vessel cell differentiation induced by DEX treatment (Yamaguchi et al., 2010), suggesting that in baculites VND7-VP16-GR, SCW formation can be initiated, but the deposition of cell wall components may be perturbed.
Figure 1.
baculites Is a Mutant with Impaired SCW Deposition Patterning.
(A) to (D) Typical differential interference contrast (DIC) images of VND7-VP16-GR-induced SCW in hypocotyls of wild-type ([A] and [B]) and baculites ([C] and [D]) cells in 6-day-old seedlings. Dashed area in (A) and (C) represents close-up in (B) and (D), respectively.
(E) Monosaccharide composition of cell walls from seedlings with vector control VP16-GR (vector control), wild-type VND7-VP16-GR (wild type) and baculites VND7-VP16-GR (baculites), with (+) or without (−) DEX treatment, as determined by HPLC. Results are means ±sd (n = 4). Ara, arabinose; Rha, rhamnose; Gal, galactose; Glu, glucose; Xyl, xylose; Man, mannose. Asterisks indicate statistically significant differences (Welch’s t test; *P < 0.05 and **P < 0.01) between the presence and absence of DEX treatment for each genotype.
(F) to (M) Visualization of cell wall components in VND7-VP16-GR-induced SCW of wild-type ([F], [H], and [J]) and baculites ([G], [I], and [K]) cotyledon cells. Xylan was detected by immunostaining using LM10 antibody ([F] and [G]), and cellulose was stained with S4B ([H] and [I]). Merged views are shown in ([J] and [K]).
(L) and (M) Lignin autofluorescence signals detected with multi-photon microscopy in VND7-VP16-GR-induced SCW of wild type (L) and baculites (M).
(N) Measurement of cellulose content of 6-day-old seedlings of wild-type and baculites before VND7-VP16-GR induction, and 3 days after treatment with (+) or without (−) DEX. Results are means ±sd (n = 5). Asterisks indicate statistically significant differences (Welch’s t test, **P < 0.01).
Bars = 100 µm ([A] to [D]), 30 µm ([F] to [K]) and 10 µm ([L] and [M]).
In baculites, Xylan and Lignin, Unlike Cellulose, Form Normal SCW Patterning
For a detailed phenotypic analysis, we next visualized the cell wall components of VND7-mediated ectopic SCWs in wild-type VND7-VP16-GR and baculites VND7-VP16-GR (Figure 1G to 1L). Using confocal scanning laser microscopy, cellulose was visualized with Pontamine Fast Scarlet 4B (S4B), which is a cellulose-specific fluorescent stain (Anderson et al., 2010), while xylan was visualized by immunostaining with the anti-xylan antibody LM10 (McCartney et al., 2005). Both signals for cellulose and xylan were found at the SCW domain formed in wild-type VND7-VP16-GR plants (Figures 1F, 1H and 1J). In contrast to wild-type VND7-VP16-GR, the ectopic vessel cells formed in baculites VND7-VP16-GR lacked the patterned cellulose signals, and an even pattern most likely originating from primary cell walls was observed (Figure 1I). Surprisingly, we found that the pattern of xylan deposition was in the helical SCW domains in the baculites VND7-VP16-GR (Figures 1G and 1K).
To evaluate if lignin deposition was also altered in the baculites VND7-VP16-GR line, we employed multiphoton microscopy to document lignin autofluorescence excited by UV light. A helical pattern of lignin autofluorescence was observed in induced vessel cells in both wild-type VND7-VP16-GR and the baculites VND7-VP16-GR cells (Figures 1L and 1M), thereby indicating that patterned deposition of lignin is unaffected in baculites. Consistent with the non-patterned signal of cellulose (Figure 1I), the cellulose content was not increased in baculites VND7-VP16-GR after the DEX treatment (Figure 1F), indicating that the increased glucose in baculites VND7-VP16-GR (Figure 1E) was not derived from crystalline cellulose. These findings indicate that baculites is a unique mutant that shows that deposition of xylan and lignin into bands of SCW domains can occur without patterned cellulose deposition.
baculites Shows irregular xylem Mutant Phenotypes
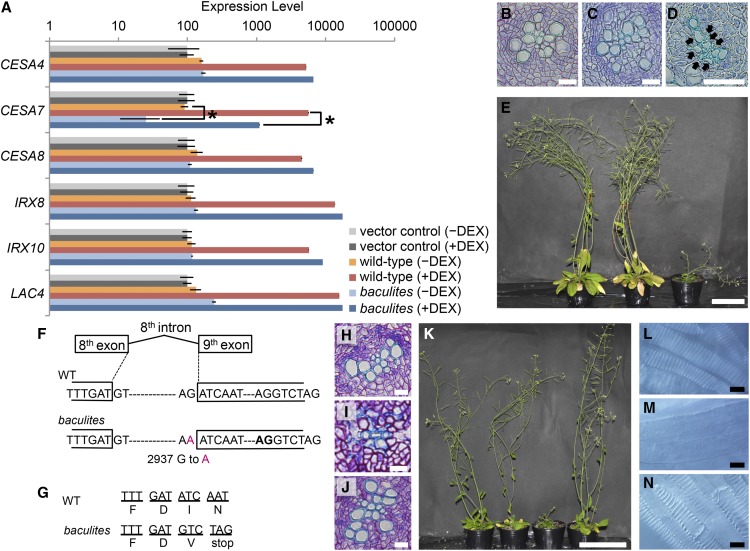
To further examine the abnormalities of SCW formation in baculites VND7-VP16-GR, we performed RT-qPCR analysis on SCW-related genes, including genes integral to cellulose biosynthesis (CESA4/IRX5, CESA7/IRX3, and CESA8/IRX1; Pear et al., 1996; Turner and Somerville, 1997; Taylor et al., 1999), hemicellulose biosynthesis (IRX8 and IRX10; Brown et al., 2005), and lignin biosynthesis (LAC4/IRX12; Brown et al., 2005) using total RNA extracted from 6-day-old seedlings treated with or without DEX for 24 h. In wild-type VND7-VP16-GR, all the genes examined were upregulated by the DEX treatment, as previously reported (Figure 2A; Yamaguchi et al., 2010). However, among the genes tested, expression of only CESA7 was significantly decreased in baculites VND7-VP16-GR compared with wild-type VND7-VP16-GR (Figure 2A), suggesting that the lack of cellulose deposition apparent in baculites VND7-VP16-GR could be attributed to a defect in CESA7.
Figure 2.
CESA7 Is Responsible for the baculites Phenotype.
(A) RT-qPCR analysis of secondary cell wall-related genes. Total RNA was extracted from the 6-day-old seedlings of the vector control VP16-GR (vector control), wild type VND7-VP16-GR (wild type), and baculites VND7-VP16-GR (baculites), which were treated with (+) or without (−) DEX. UBQ10 was used for the internal control, and the results shown are relative values to the expression level of VP16-GR without DEX treatment. Results are mean ±sd (n = 3). Statistically significant differences were found in CESA7 expression between VND7-VP16-GR and baculites (*P<0.05; Student's t test).
(B) to (D) Transverse sections of roots of 20-day-old plants of the vector control VP16-GR (B), wild-type VND7-VP16-GR (C), and baculites VND7-VP16-GR (D). Black arrows indicate collapsed xylem vessels. Bars = 20 μm.
(E) Two-month-old plants of vector control VP16-GR (left), wild-type VND7-VP16-GR (middle), and baculites VND7-VP16-GR (right).
(F) and (G) Single nucleic acid substitution (G2937 to A transition; indicated in red) was found at the acceptor site of the 8th intron of CESA7 in the genome of baculites, resulting in a shift on the acceptor site 13 bp downstream to the next AG (indicated in bold in [F]). This change was presumed to produce the premature stop codon in baculites (G).
(H) to (N) Introduction of CESA7pro::YFP-CESA7 rescued the irx phenotype of baculites. (H) to (J) Transverse sections of roots of VND7-VP16-GR in wild-type (H), baculites (I), and baculites/CESA7pro::YFP-CESA7 backgrounds (J). (K) Growth phenotype of 2-month-old plants with either vector control VP16-GR (left) or VND7-VP16-GR in (left to right) wild-type, baculites, baculites/CESA7pro::YFP-CESA7 backgrounds. (L) to (N) DIC images of ectopic VND7-VP16-GR-induced SCW of wild type (L), baculites (M), and baculites/CESA7pro::YFP-CESA7 (N).
Bars = 20 µm ([B] to [D]), 10 cm ([E], [K], and [L] to [N]), 20 µm ([H] to [J]).
Next, we evaluated if baculites VND7-VP16-GR plants also had defects in the formation of endogenous xylem vessel cells. Histological observations indicated that, compared with control roots (VP16-GR plants; Figure 2B) or roots from un-induced wild-type VND7-VP16-GR lines (Figure 2C), the xylem vessels of baculites VND7-VP16-GR roots were irregularly shaped and frequently collapsed (Figure 2D). Furthermore, two-month-old baculites plants showed a severely dwarfed phenotype with pendent inflorescence stems (Figure 2E). These phenotypes are similar to the irx mutants (Turner and Somerville, 1997; Taylor et al., 1999) including previously reported mutants of SCW-related CESAs (Brown et al., 2005; Bosca et al., 2006), strengthening the hypothesis that baculites is indeed an additional irx mutant.
Mutation in CESA7 Is Responsible for the Phenotype of baculites
Given the results described above, we sequenced the genomic region of CESA7, and found a single nucleic acid substitution (G2937 to A transition) at the splice acceptor site of the 8th intron (Figure 2F). RT-PCR analysis, followed by sequence analysis, on the CESA7 cDNA revealed that in baculites VND7-VP16-GR, the splice acceptor site of the 8th intron was changed to the next AG, 13 bp downstream (boldfaced AG located in the original 9th exon, Figure 2F). The altered splicing is predicted to result in a frame shift to generate a substitution from isoleucine to valine at position 579, followed by a premature stop codon at position 580 (Figure 2G). Introduction of CESA7pro::YFP-CESA7 (Watanabe et al., 2015) into the baculites cell line resulted in the restoration of all observed defects to normal phenotypes, including rescue of the irregular-shaped and collapsed xylem vessel cells (Figures 2H to 2J), and the dwarf phenotype (Figure 2K). In addition, the formation of a helically-patterned SCW was clearly rescued in the ectopic vessel cells induced after DEX treatment in baculites carrying both 35Spro::VND7-VP16-GR and CESA7pro::YFP-CESA7 (Figures 2L to 2N). These results indicate that the phenotypes of baculites VND7-VP16-GR can be attributed to a mutation in CESA7.
baculites Is a Novel Null Mutant Allele of CESA7
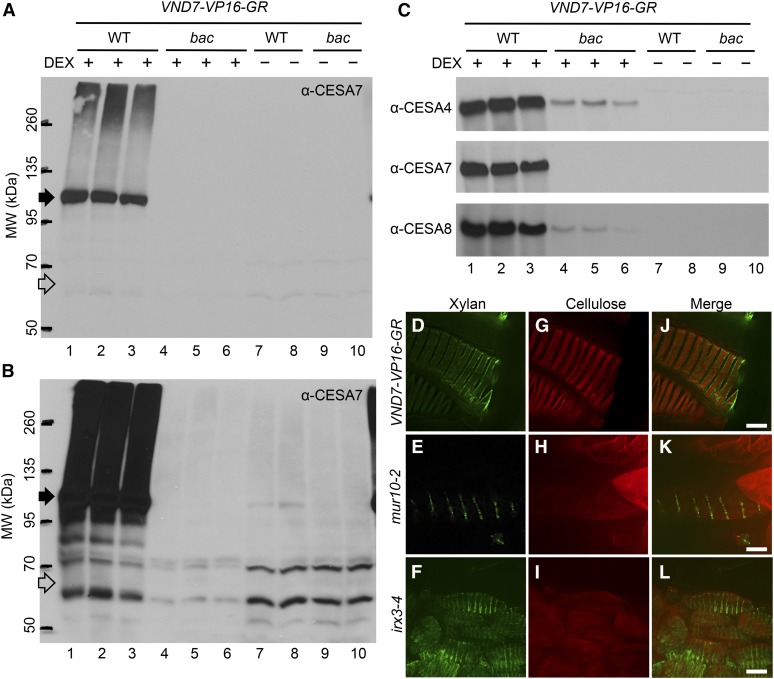
As described above, the mutation in baculites VND7-VP16-GR caused a frame shift and the introduction of a premature stop codon because of the change in the splice site of the 8th intron (Figures 2F and 2G). It was predicted that CESA7 in baculites VND7-VP16-GR is composed of 578 N-terminal amino acids of the native CESA7 with one additional Val residue. Therefore, the ensuing protein would be made up of the N-terminal region, two transmembrane domains, and a truncated central cytoplasmic region (McFarlane et al., 2014). The irx3-1 mutant, containing a stop codon that truncates the CESA7 protein by 168 amino acids (Taylor et al., 1999), was found to be a null mutation as shown by immunoblotting using an anti-CESA7 antibody, where no CESA7 bands were recognized in the irx3-1 extracts (Taylor et al., 2000). This finding suggested that the baculites mutant might also be a null allele of cesa7. To test this hypothesis, we performed immunoblot analysis of baculites VND7-VP16-GR with the anti-CESA7 antibody as per Hill et al. (2014) (Figures 3A to 3C). In the wild-type VND7-VP16-GR lines, the induced accumulation of all SCW-type CESA proteins, CESA4, CESA7, and CESA8, were observed after DEX treatment (Figures 3A and 3C). By contrast, no band was detected by anti-CESA7 antibody in the baculites VND7-VP16-GR after the DEX treatment (Figures 3A and 3C). Even extended exposure did not detect any Anti-CESA7-reacting bands unique to cesa7bac (Figure 3B), clearly indicating that there is no truncated form of CESA7 present in cesa7bac. Thus, we concluded that baculites is a null allele of the cesa7 mutant, similar to irx3-1 (Hill et al., 2014). Severe reductions of CESA4 and CESA8 proteins were also detected in protein extracts from baculites VND7-VP16-GR plants (Figure 3C), consistent with previous observations of decreased levels of all SCW CESAs in the knock-out cesa7 mutant (Taylor et al., 2003; Hill et al., 2014).
Figure 3.
baculites Is a Null Mutant of CESA7 and the Effects of other cesa7 Mutations on SCW Deposition during VND7-VP16-GR-Induced Ectopic Xylem Vessel Cell Differentiation.
(A) to (C) Equal amounts (30 µg) of protein from the wild type VND7-VP16-GR (WT) (lane 1, 2, 3, 7, and 8) and baculites VND7-VP16-GR (bac) (lane 4, 5, 6, 9, and 10) with DEX (lane 1 to 6) or mock (lane 7 to 10) treatments were separated by SDS-PAGE and immunoblotted to detect the SCW-type CESA proteins. Each lane showed an independent biological replicate sample. (A) and (B) The immunodetection of CESA7 proteins by anti-CESA7 antibody. View of extended exposure of (A) was shown in (B). Black and white arrows ([A] and [B]) indicate the bands corresponding to full-length CESA7 proteins and the expected size (∼66 kD) of truncated CESA7 in baculites by nonsense mutation, respectively. (C) Immunodetection of CESA4, CESA7, and CESA8 using specific antibodies.
(D) to (L) Hypocotyl cells of 6-day-old seedlings with VND7-VP16-GR in wild-type ([D], [G] and [L]), mur10-2 ([E], [H], and [K]), and irx3-4 ([F], [I], and [L]) backgrounds treated with DEX for 3 days. Xylan ([D], [E], and [F]) and cellulose ([G], [H], and [I]) were visualized by immunolabeling using LM10 antibody and S4B staining, respectively. Merged view of xylan and cellulose were shown in ([J], [K], and [L]). Bars = 10 µm.
Other Mutants of CESA7 Showed Similar Defects in SCW Deposition to baculites
To examine if the protoxylem vessel cell phenotype of the baculites mutant cesa7 allele (cesa7bac) is also found in other alleles of cesa7, we examined both the cesa7mur10-2 allele in which the histidine at position 734 is replaced with a tyrosine (Bosca et al., 2006) and the cesa7irx3-4 (T-DNA insertion knock-out null mutant; Brown et al., 2005). We crossed VND7-VP16-GR with cesa7mur10-2 or cesa7irx3-4, and established cesa7mur10-2/VND7-VP16-GR and cesa7irx3-4/VND7-VP16-GR plants that were homozygous for each mutation. As with baculites (Figure 1G), the xylan deposition patterns in cesa7mur10-2 and cesa7irx3-4 mutants were in helical SCW patterns (Figures 3E and 3F), although they were less well organized than wild-type VND7-VP16-GR (Figure 3D). Staining of cellulose with S4B revealed patterns of cellulose deposition in cesa7mur10-2 and cesa7irx3-4 mutants that were similar to cesa7bac mutants; i.e. cellulose was not seen in a helical wall pattern as in wild-type VND7-VP16-GR (Figures 3G to 3I). These results clearly indicated that mutations in CESA7 led to similar pattern defects in SCW deposition in protoxylem vessel cells induced by the VND7-VP16-GR system.
Xylan Arrangement, not Helical Patterning, in the SCW Depends on Intact Cellulose
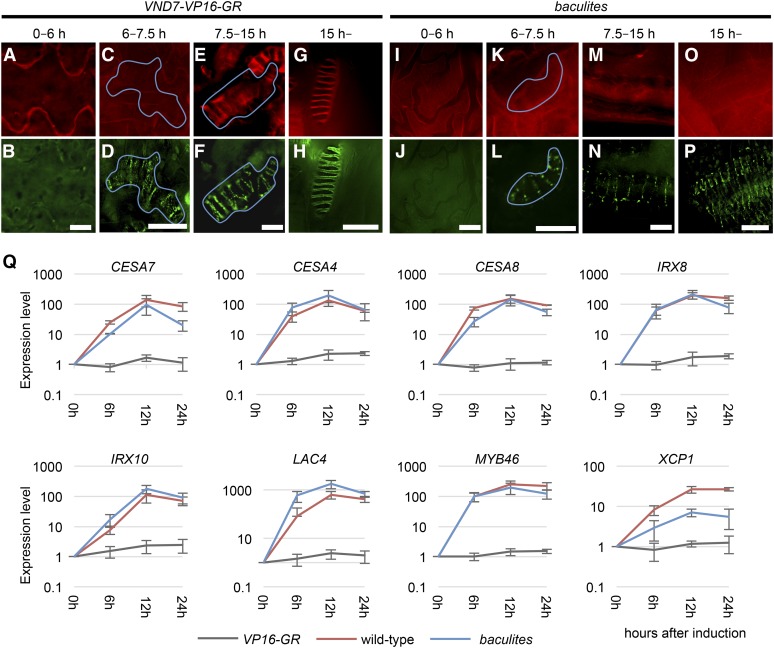
In cesa7bac, despite the lack of SCW cellulose deposition, xylan was still deposited in the helical patterns (Figures 1G and 1M). To further probe cellulose and xylan deposition, we performed time-sequential observations of wild-type VND7-VP16-GR and baculites VND7-VP16-GR. Six-day-old seedlings of wild-type VND7-VP16-GR were treated with DEX, and then subjected to time-course fixation after the DEX treatment (Figure 4). No obvious differences between wild-type (Figures 4A to 4D) and baculites VND7-VP16-GR (Figures 4I to 4L) were apparent early in the differentiation of the protoxylem vessel cells. However, in contrast to wild-type (Figures 4E to 4H), baculites VND7-VP16-GR vessel cells did not exhibit any patterned cellulose signal even as differentiation progressed, i.e. after 7.5 h of DEX treatment (Figures 4M to 4P). In induced baculites VND7-VP16-GR vessel cells, the xylan signals were patchy throughout differentiation, and did not form smooth banded SCW domains, as observed in wild-type VND7-VP16-GR (Figure 4H). To test if the overall timing of differentiation was altered in baculites vessel elements, expression analysis of xylem cell differentiation-related genes was performed, including MYB46 transcription factor (Zhong et al., 2007), CESA4/IRX5, CESA7/IRX3, and CESA8/IRX1 for cellulose biosynthesis, IRX8 and IRX10 for hemicellulose biosynthesis, LAC4/IRX12 for lignin biosynthesis, and XCP1 for programmed cell death (Avci et al., 2008). The timing of expression of these xylem cell differentiation-related genes was similar in wild-type and baculites VND7-VP16-GR plants (Figure 4Q), as 6 h after DEX treatment, all tested genes were upregulated in both wild-type VND7-VP16-GR and baculites VND7-VP16-GR (Figure 4Q). No upregulation of these genes was found in the vector control VP16-GR. Thus, baculites VND7-VP16-GR underwent similar molecular processes of xylem vessel cell differentiation, other than the cellulose deposition, within the observed time window. These results suggest that baculites mutants undergo a typical vessel element developmental program, including xylan being secreted into the bands of SCW domains, despite loss of cellulose deposition. However, the patchy xylan pattern within the bands of SCW demonstrates that the continuous distribution of xylan within the SCW domain relies on the presence of SCW cellulose.
Figure 4.
Time-Course Analysis of Cellulose and Xylan Deposition during Ectopic Xylem Vessel Cell Formation.
Six-day-old VND7-VP16-GR seedlings in wild type ([A] to [H]) and baculites ([I] to [P]) were treated with DEX, and then sampled between 0 and 6 h ([A], [B], [I], and [J]), between 6 and 7.5 h ([C], [D], [K], and [L]), between 7.5 and 15 h ([E], [F], [M], and [N]), and between 15 and 24 h ([G], [H], [O], and [P]) after DEX treatment. Cellulose ([A], [C], [E], [G], [I], [K], [M], and [O]) and xylan ([B], [D], [F], [H], [J], [L], [N], and [P]) were visualized by S4B staining and immunolabeling using LM10 antibody, respectively. Cells in cotyledons are shown. Bars = 10 µm.
(Q) Time course RT-qPCR analysis of secondary cell wall-related genes. Total RNA was extracted every 6 h from the 6-day-old seedlings of the vector control VP16-GR (vector control), wild-type VND7-VP16-GR (wild-type), and baculites VND7-VP16-GR (baculites) after treatment with DEX. UBQ10 was used for the internal control. Results are means ±sd (n = 3).
The Helical Pattern of Xylan Deposition Is Dependent on Cortical Microtubules
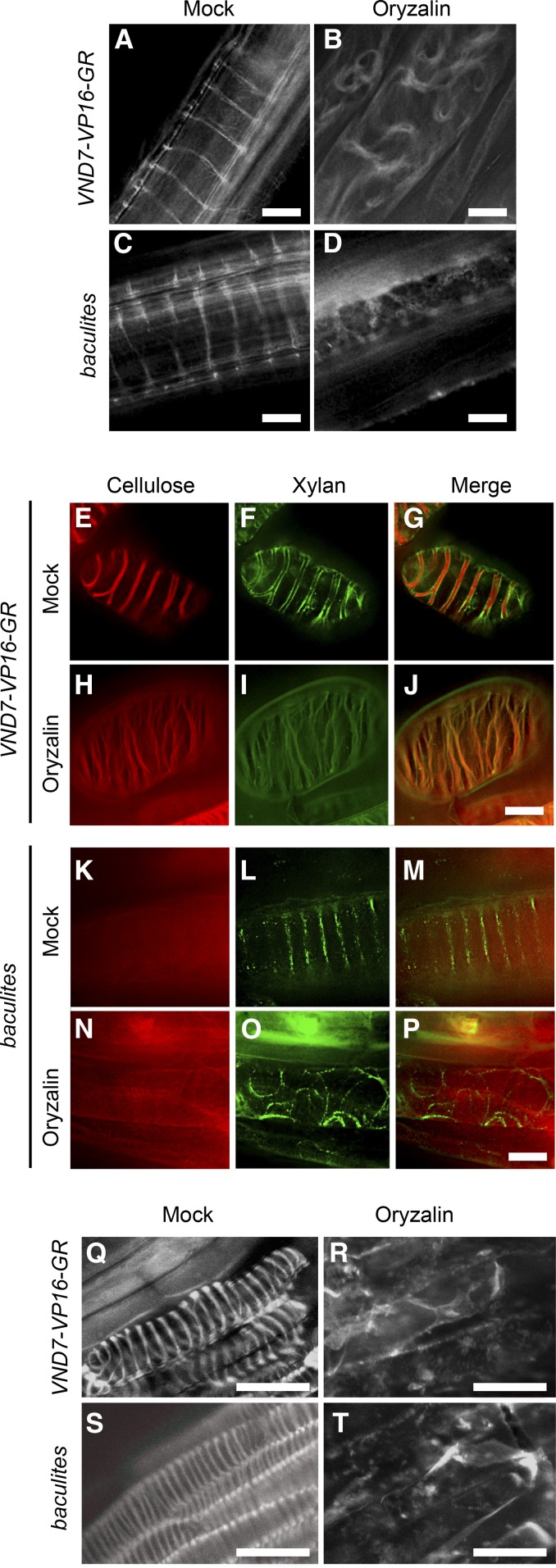
Previous studies have shown that SCW formation is initiated with the rearrangement of the cortical microtubule array, which plays critical roles in determining the pattern of SCW deposition (Oda et al., 2005). Therefore, we visualized cortical microtubules using the UBQ10pro::GFP-TUB6 reporter (Nakamura et al., 2004). Six-day-old seedlings of wild-type and baculites VND7-VP16-GR, both carrying UBQ10pro::GFP-TUB6, were induced with DEX treatment, and observed with confocal microscopy. In the wild-type VND7-VP16-GR, the GFP-TUB6 signals showed clear helical patterns (Figure 5A), which were disrupted by the addition of the microtubule-depolymerizing drug oryzalin (Figure 5B). The GFP-TUB6 pattern in baculites VND7-VP16-GR showed strong helical banding, indicating that the microtubule rearrangement is normal during ectopic protoxylem vessel cell differentiation in baculites VND7-VP16-GR (Figure 5C). This microtubule array was also disrupted by oryzalin in baculites VND7-VP16-GR (Figure 5D).
Figure 5.
Patterned Deposition of Xylan and Lignin Is Directed by the Cortical Microtubule Array.
(A) to (D) Cortical microtubules were visualized during VND7-VP16-GR-induced protoxylem differentiation using the UBQ10pro::GFP-TUB6 reporter line in wild-type ([A] and [B]) and baculites ([C] and [D]) backgrounds, treated with only DEX (Mock; [A] and [C]) or both DEX and the microtubule-depolymerizing drug, oryzalin (Oryzalin; [B] and [D]). Cells in cotyledons are shown.
(E) to (P) Six-day-old seedlings with VND7-VP16-GR in wild type ([E] to [J]) or baculites ([K] to [P]) were treated with only DEX (Mock; [E] to [G] and [K] to [M]) or both DEX and oryzalin (Oryzalin; [H] to [J] and [N] to [P]). Cellulose ([E], [H], [K], and [N]) and xylan ([F], [I], [L], and [O]) were visualized by S4B staining and immunolabeling using LM10 antibody, respectively. Merged view of cellulose and xylan are shown in (G), (J), (M), and (P). Cells in cotyledons are shown.
(Q) to (T) Lignin was labeled by the fluorescently tagged monolignol analogs, γ-nitrobenzofuran (NBD)-tagged coniferyl alcohol (CA) (Tobimatsu et al., 2013; Schuetz et al., 2014). Cells in hypocotyls are shown.
Bars = 10 µm ([A] to [P]), 30 µm ([Q] to [T]).
Next, we examined cellulose, xylan, and lignin in ectopic vessel cells from wild-type and baculites VND7-VP16-GR plants treated with oryzalin. Although the helically-patterned deposition of cellulose and xylan was affected by the oryzalin treatment in wild-type VND7-VP16-GR, the signals of cellulose and xylan were colocalized (Figures 5E to 5J). In baculites, the cellulose signals did not show signs of patterning, regardless of the oryzalin treatment (Figures 5K and 5N), while the helical xylan deposition was disordered by the oryzalin treatment (Figures 5L to 5P). The disorganized xylan patterns (Figure 5O) were similar to the disordered cortical microtubule arrays observed after the oryzalin treatment (Figures 5B and 5D). To test the effect of loss of microtubules on lignin patterning, induced protoxylem vessel elements were treated with oryzalin, and similarly disorganized lignin autofluorescence patterns were observed in both wild-type and baculites VND7-VP16-GR lines. To ensure lignin detection was sensitive enough to detect subtle changes, we used a fluorescently tagged monolignol analog, γ-nitrobenzofuran (NBD)-tagged coniferyl alcohol (CA) (Tobimatsu et al., 2013; Schuetz et al., 2014). This confirmed that the lignin deposition in both wild-type and baculites VND7-VP16-GR lines was perturbed by oryzalin treatment (Figures 5Q to 5T). These observations indicate that the cellulose-independent depositions of xylan and lignin are directed by the cortical microtubules during SCW formation within differentiating protoxylem vessel cells.
DISCUSSION
baculites Is a Novel irx Mutant Allele of CESA7
In the current study, we identified baculites as a novel cesa7 mutant in a genetic screen using VND7-VP16-GR, in which ectopic protoxylem vessel cells can be induced (Figure 1). Surprisingly, the baculites mutants showed SCW formation without the helical pattern typical of protoxylem cells, as well as an irx phenotype (i.e. collapsed xylem vessels and stunted growth). The single nucleic acid substitution in the 8th splice acceptor site of CESA7 was found in the genome of baculites (Figure 2F), and the subsequent introduction of CESA7pro::YFP-CESA7 rescued the phenotype of baculites (Figures 2H to 2N), confirming that baculites is a novel allele of cesa7 (cesa7bac).
The model that CESA7 is one of the essential components of the CSC responsible for normal cellulose biosynthesis in SCWs, and that the CESA7-containing CSC are guided by microtubules for patterned SCW formation, is widely accepted (Wightman and Turner, 2008; Bashline et al., 2014; McFarlane et al., 2014; Li et al., 2015). The irregular xylem phenotype of irx mutants has been attributed to the reduction of cellulose contents of stems (Turner and Somerville, 1997). In cesa7bac, despite the increased amounts of glucose in response to the DEX treatment (Figure 1E), cellulose content did not increase after the DEX treatment (Figure 1O), suggesting that the increased glucose in baculites VND7-VP16-GR may be derived from unidentified cell wall components that are synthesized upon VND7 activation. Nonetheless, these defects in SCW cellulose are in line with previous reports on irx3 mutants, in which cellulose biosynthesis in SCWs is severely affected by the loss of CESA7/IRX3 (Turner and Somerville, 1997).
Xylan Secretion to Helical SCW Domains Is Dependent on Microtubules, not Cellulose
Our immunolabeling observations revealed that xylan was still deposited in a helical pattern in the cesa7bac, cesa7mur10-2, and cesa7irx3-4 mutants that lacked cellulose-rich SCW domains (Figures 1G to 1K, 3E to 3L, and 4I to 4P). Previous monosaccharide analysis has shown that the xylose content of cesa7 mutants is comparable to that of the wild type in SCW-rich stem samples (Zhong et al., 2003; Brown et al., 2005), suggesting that the xylan biosynthetic pathway is independent of cellulose biosynthesis. Interestingly, it is also possible that the cellulose biosynthetic pathway occurs independently of xylan, as a recent transcriptome analysis of xylan-deficient mutants has shown that the expression of many SCW related genes are not affected, including SCW CESA genes (Faria-Blanc et al., 2018). These data support the model in which the cells are committed to forming SCWs with little or no compensatory mechanisms for the loss of one of the SCW components (Faria-Blanc et al., 2018). Here, our data demonstrate that the spatial distribution of xylan into banded SCW domains is independent from cellulose deposition in the induced protoxylem cells. Since the pattern of cortical microtubules is unaffected in cesa7bac mutants, the secretion of CSCs, xylan, and the enzymes required for lignification such as laccases is apparently unchanged at the microtubule-enriched plasma membrane domains. Treatment with microtubule-depolymerizing drug oryzalin disrupted the helical deposition of xylan (Figure 5), indicating that the helically-arranged cortical microtubules are required for helically-patterned xylan deposition. This association between xylan and microtubules is consistent with previous observations in woody plants (Zhu et al., 2015).
On first glance, our observations seem to contradict those of Taylor et al. (Taylor et al., 1992), in which they reported that the treatment of tracheary elements differentiating from Zinnia elegans mesophyll cells with cellulose synthesis inhibitors not only abolished the deposition of cellulose but also the normal patterning of xylan and lignin (Taylor et al., 1992; Taylor and Haigler, 1993). By contrast, we detected normally patterned xylan and lignin in cesa7bac mutants (Figure 1), in which SCW cellulose was not synthesized (Figure 3). As discussed in these earlier articles, it is possible that xylan and monolignol are secreted into culture medium and diffused in the case of the Zinnia elegans system. By contrast, in our system, the synthesized xylan, monolignol, and enzymes are retained within cell walls after secretion, leading to detectable helically-patterned xylan and lignin (Figure 1). Additionally, the different methods used to detect xylan and lignin deposition may be another reason for the differences in results. Nonetheless, our data show that with the VND7-inducible system in planta, proper deposition of both xylan and lignin can occur independently from cellulose deposition. However, it remains to be seen whether this is also true for endogenous vessel elements and further work is needed to determine the effects of xylan patterning and organization in these cells.
Why is xylan deposited independently of cellulose during vessel differentiation? Studies using in vitro cellulose biosynthesis in Acetobacter xylinum showed the addition of xylan greatly altered the cellulose features, likely influencing cellulose aggregation (Atalla et al., 1993). In addition, the suppression of xylan endotransglycosylase activity was reported to affect angles of cellulose microfibrils in Populus tremula×tremuloides (Derba-Maceluch et al., 2015), suggesting that xylan is one of the determinants of cellulose structure during SCW formation. Thus, xylan deposition might enhance and/or moderate the formation of SCW-type cellulose microfibrils, i.e. highly crystalline cellulose with a high degree of polymerization. irx mutants with defects in xylan biosynthesis clearly demonstrate the importance of xylan in SCW integrity (Zhong et al., 2005; Lee et al., 2007; Persson et al., 2007; Brown et al., 2009, 2011; Wu et al., 2010; Jensen et al., 2011). Further analysis of the deposition of cellulose and xylan in endogenous xylem vessels of irx mutants including baculites will provide additional clues into the roles and mechanism of xylan during SCW formation.
Xylan Organization in the Cell Wall Requires Wild-Type SCW Cellulose
We also found that the signals for xylan deposition were strong and linear along the SCW domains that accompany intense cellulose deposition in wild-type VND7-VP16-GR seedlings (Figures 4A to 4H). By contrast, xylan signals at the SCW were uneven along the remnant bands of SCW domains in cesa7bac, cesa7mur10-2 and cesa7irx3-4 (Figures 1G to 1K, 3E to 3L, and 4I to 4P). This indicates that though xylan was secreted to the correct domains, its integration into the cellulose-deficient SCW was disrupted. Thus, the presence of wild-type cellulose appears to be required for the proper organization of xylan with the SCW. Ha et al. (2002)) and Busse-Wicher et al. (2014) proposed a model describing the interaction between cellulose microfibrils and xylan chains in Arabidopsis using mass spectrometry and NMR, in which xylan aligns with vacancies on cellulose microfibril hydrophilic surfaces. The linear and smooth deposition of xylan along cellulose shown in our data may reflect such structural interaction, suggesting that cellulose nanoscale architecture at the cell surface influences the arrangement of xylan. Taken together, our work highlights the interdependence of the xylan-rich matrix and cellulose microfibrils during SCW scaffold formation, likely due to the molecular packing of xylan on microfibrils, as suggested by Ha et al. (2002)) and Busse-Wicher et al. (2014). This scaffold may have a positive effect in anchoring proteins for lignification as well. We aim to further examine SCW deposition and associated properties in our mutant as well as the other types of irx mutants, including xylan-deficient mutants, to elucidate the biological meaning of cellulose-dependent xylan deposition for the structural organization of plant SCWs.
METHODS
Plant Materials and Plant Growth Condition
Arabidopsis (Arabidopsis thaliana) VASCULAR-RELATED NAC-DOMAIN7-inducible line VND7-VP16-GR and the vector control VP16-GR were described in Yamaguchi et al. (2010). The Arabidopsis cesa7 mutants, irx3-4 (Brown et al., 2005) and mur10-2 (Reiter et al., 1997; Bosca et al., 2006), were kindly provided from the Arabidopsis Biological Research Center. Arabidopsis seeds were sown on Murashige and Skoog (MS) medium containing 1% (w/v) sucrose solidified with 0.6% (w/v) gellan gum, and grown under continuous light (45∼85 µmol/m2/s, sunshine color fluorescent tube, NEC FHF32EX-N-HX-S) at 22°C after the cold treatment at 4°C under dark condition for 1 to 2 days. For long-term culture of plants, 3-week-old plants grown on the medium were transferred in the soils and further grown at 22°C under long-day (16-h light/8-h dark) conditions.
DEX and Oryzalin Treatment
For the induction of the VND7-VP16-GR activity, 6-day-old seedlings were immersed in one-half-strength MS medium with 10 μM DEX under continuous light at 22°C (Yamaguchi et al., 2010). The treated samples were collected 3 to 4 days following incubation in the DEX solution, or at the time points indicated in each experiment. For microtubule polymerization inhibition, 6-day-old seedlings were soaked in one-half-strength MS medium containing both 10 µM DEX and 10 µM oryzalin. To monitor the UBQ10pro::GFP-TUB6 signals, the incubation with DEX and oryzalin was performed under dark conditions at 22°C for 16 h.
Histological Observations
For observations of SCW, samples were first incubated in clearing solution (8:1:2 (w/v/v) mixture of chloral hydrate, glycerin, and water) for at least one night. Observations were made with a microscope equipped with Nomarski optics and digital camera system (BX53-DIC and DP72, Olympus). More than 10 seedlings were sampled and tested for each genotype, and the sampling and observation was repeated at least 3 times independently.
For sectioning, 6-day-old seedlings were fixed in FAA solution (3.7% (v/v) formaldehyde, 50% (v/v) ethanol, 5% (v/v) acetic acid) for 1 hour, and dehydrated with a sequential ethanol series of 50%, 60%, 70%, 80%, 90%, and 100% for 30 min at room temperature. Then, the samples were incubated in 100% ethanol for one night at 4°C. The dehydrated samples were embedded in Technovit 7100, according to manufacturer’s protocols (Heraeus-Kulzer). Two or 6 µm-sections were made on a microtome (PR-50, YAMATO KOHKI), followed by staining with toluidine blue-O or by cell wall labeling with immunofluorescence. Sections were prepared from more than 5 seedlings for each genotype, and more than 10 sections per seedling were observed. This experiment was repeated two times independently.
EMS Mutagenesis and Mutant Screening
Seeds of VND7-VP16-GR were incubated with a 0.3% (v/v) ethyl methanesulfonate (EMS, Sigma) solution, according to the methods in Kawabe et al. (2018). After the EMS treatment, M1 seeds were repeatedly washed with 80-mL distilled water 12 times (30-s incubation ×5, 30-min incubation ×2, and then 30-s ×5). Washed M1 seeds were sown on MS medium to grow for 3 weeks, and then separately transferred into the soils for further growth. M2 seeds were harvested after the self-reproduction of each M1 plant independently. The M2 lines were subjected to the DEX treatment, and observed to isolate mutants with abnormal SCW formation in hypocotyl cells.
Cell Wall Labeling
For xylan immunostaining, seedlings were fixed with FAA for 1 hour, and washed with 50% and 20% ethanol for 10 min, and stored in TBST solution (10 mM Tris-HCl (pH 7.0), 0.25 M NaCl, 0.1% (w/v) Tween 20). Samples were then incubated in 5% (w/v) BSA in TBST for 1 h at room temperature, followed by the incubation with anti-xylan antibody LM10 (Plant Probes; McCartney et al., 2005) at 1:36 dilution overnight at 4°C. Samples were washed with TBST three times, and incubated with goat anti-rat IgG-CFL 488 (Santa Cruz) or Alexa488 (Invitrogen) at 1:100 dilution at room temperature for 3 h with gentle shaking. After washing three times with TBST samples were stained with 10 mg/mL Pontamine Fast Scarlet 4B (S4B) (Sigma) in one-half-strength MS medium for 10 min and washed twice in one-half-strength MS medium. Samples were then mounted in one-half-strength MS medium and then observed on a fluorescence microscope (DeltaVision Elite, GE Healthcare) equipped with photometrics cool snap HQ2 camera, and/or a Perkin-Elmer UltraView Vox spinning disk confocal mounted on a Leica DMI6000 inverted microscope equipped with Hamamatsu 9100-02 CCD camera. Excitation and emission filters were set to FITC (475 and 525 nm) and TRITC (542 and 593 nm) or GFP (488 and 525 nm) and RFP (561 and 595 nm).
To detect endogenous deposition of lignin in baculites VND7-VP16-GR, lignin autofluorescence was observed using FV1000 Multiphoton Laser Scanning Microscope (Olympus) fit with a tunable MaiTai BB DeepSee laser adjusted to 730 nm. For lignin localization experiments with oryzalin treatment, the fluorescently tagged monolignol analog, γ-nitrobenzofuran (NBD)-tagged coniferyl alcohol (CA) (NBD-CA; Tobimatsu et al., 2013; Schuetz et al., 2014) was used at a mixture of 0.5 mM NBD-CA and 5 mM CA (Sigma) in one-half-strength MS liquid medium to a final concentration of 1 and 10 μM, respectively, together with 10 μM dexamethasone (Sigma) for VND7 induction. The fluorescence of NBD-CA was detected by confocal laser scanning microscope (FV10i, Olympus).
More than 20 seedlings for each genotype were collected after the DEX treatment, to subject to cell wall labeling experiment. Three to four cells per one seedling were observed, and totally 5 to 10 seedlings for each genotype were tested. We repeated this scheme three times for cell wall labeling shown in Figure 1 and Figure 5, and two times for time-course experiments shown in Figure 4. We made our conclusions of the baculites phenotypes based on ours results of hypocotyl and cotyledon cells, and typical images were selected and shown in figures.
RT-qPCR
Six-day-old seedlings of VP16-GR, VND7-VP16-GR, and baculites VND7-VP16-GR were incubated with or without 10 μM DEX, and 10 seedlings for each genotype were collected after 0, 6, 12, and 24 h. We independently repeated this sampling three times as biological replicates. Total RNA was extracted using RNeasy Plant Mini Kit (Qiagen), and then treated with RQ1 RNase-Free DNase (Promega). One µg of DNase-treated total RNA was reverse-transcribed by Transcriptor Reverse transcriptase (Roche) with oligo (dT) 18 primers. Quantitative PCR from the generated cDNA was performed with the SCW-related gene-specific primers described in Yamaguchi et al. (2010)), except for CESA7 (forward primer 5′-AGCTTCCTCGGCTTGTGTAC-3′ and reverse primer 5′-ATTTGTGAGTACGCCTGCCA-3′) and for LAC4 (forward primer 5′-CGACACCGTCCTTATAGCCC-3′ and reverse primer 5′-GGGCGTCCATGAAAGGAGAA-3′) using LightCycler® 480 system (Roche) according to the manufacturer’s protocols. UBQ10 was used as an internal control.
Monosaccharide Analysis
Six-day-old seedlings of VP16-GR, VND7-VP16-GR, and baculites VND7-VP16-GR were incubated with or without 10 μM DEX for 3 days. We independently repeated this sampling three times as biological replicates. Samples were dried in a 45°C oven overnight and subsequently ground in a Wiley mill to pass a 40 µm mesh screen and extracted with hot acetone overnight using a Soxhlet apparatus, and then dried for 48 h at 50°C. Approximately 10 mg (4 replicates per sample) was cooled in a desiccator and weighed into a 2 mL autoclavable screw-cap tube (Fischer Scientific, Canada). 100 μL of 72% H2SO4 (VWR, Canada) were added to the tube and vortexed to get all the sample in suspension. The slurry was incubated for 1 h on a thermomixer at 30°C and 500 rpm. Following the pre-hydrolysis, 2 mL of nano-pure water were added to the slurry and the tube was vortexed thoroughly for 30 s on maximum speed to re-suspend the sample. Subsequently, the samples and the sugar standards were autoclaved for 60 min at 121°C. After cooling down to room temperature the tubes were spun in a rotor centrifuge at 20,000 rpm for 5 min or until a firm pellet was formed. The supernatant was transferred into a new 2 mL safe-lock Eppendorf tube (Eppendorf Co, USA) for the determination neutral sugars.
The concentration of neutral cell-wall associated carbohydrates (arabinose, rhamnose, galactose, glucose, xylose, and mannose) was quantified by HPLC using Dionex (IC5000, Sunnyvale, CA, USA) equipped with an ion-exchange CarboPac PA-1 (Thermo-Scientific) column, a pulsed amperometric detector (ED 50) with a golden electrode and Dionex AS 100 autosampler (Dionex, Sunnyvale, CA, USA), equipped with a second injection tower to dilute (1:10) and mix the samples. The sample was eluted with nanopure water at a flow rate of 0.850 mL min−1. Aliquots (25 uL) of the diluted sample were injected after being passed through a 0.45-µm nylon syringe filter (Chromatographic Specialties INC, Canada). Optimization of baseline stability and detector sensitivity was achieved by post-column addition of 100 µM mL min−1 NaOH. Cell wall carbohydrates content was determined as a percentage of dry wood, and calculated from standard curves created from external standards.
Cellulose Analysis
Six-day-old seedlings of VND7-VP16-GR and baculities VND7-VP16-GR were incubated with or without 10 μM DEX for 3 days. Seedlings were then flash frozen in liquid nitrogen and then ball milled on SPEX SamplePrep GenoGrinder (Metuchi, NJ, USA). The cell-wall powder was washed with 70% (v/v) ethanol and centrifuged at 16,000 × g for 10 min at 20°C. Subsequently, the pellet was resuspended in a 1:1 methanol:chloroform mixture and was centrifuged again at 16,000 × g for 10 min at 20°C. Finally, the pellet was resuspended in pure acetone and centrifuged at 16,000 × g for 10 min at 20°C. Then the cell-wall pellet was dried overnight at 50°C. Approximately 2.5 mg of the dry, insoluble part of the cell wall material was used for further analysis in 2-mL screw-cap tubes. Subsequently, 750 μL of 2 M trifluoroacetic acid was added to the tubes, and the tubes were incubated for 1 h at 121°C. Afterward, the tubes were supplemented with 600 μL of 2-Propanol (Thermo Fisher Scientific) and left to evaporate under a steady airflow at 40°C. This step was repeated twice; then the tubes were supplemented with 600 μL distilled water, thoroughly vortexed, and centrifuged at 16,000 × g for 15 min at 20°C. The pellet was further used to determine the amount of crystalline cellulose using the Updegraff method (Updegraff, 1969). Briefly, 350 μL of 72% (w/w) sulphuric acid was added to the pellet for 30 min at 20°C. Then 850 μL of water was added to the sample, thoroughly vortexed, centrifuged at 1500 × g for 5 min at 20°C, and the supernatant was kept at 4°C until use. Anthrone solution was used to determine the crystalline cellulose content using a standard curve created with D-glucose. Absorbance was measured at 600 nm in a plate reader.
Plant Transformation
The CESA7pro::YFP-CESA7 and UBQ10pro::GFP-TUB6 plasmids were described in Watanabe et al. (2015) and Nakamura et al. (2004), respectively. The plasmids were electroporated into Agrobacterium tumefaciens strain GV3101/pMP90. A simplified version of the floral dip method was used for plant transformation (Clough and Bent, 1998).
Immunoblot Analysis
For the immunological detection of the CESA4, CESA7, and CESA8 proteins, 4-day-old seedlings treated with or without DEX for 1 day were snap frozen in liquid nitrogen, ground by a drill-driven pestle in a solution of 10% (v/v) trichloroacetic acid in acetone, and protein was extracted as described previously (Wang et al., 2003, 2006). This procedure was independently repeated three times for DEX-treated samples, and two times for mock-treated samples. Protein concentration was measured by a modified Lowry procedure (Peterson, 1977) and equal amounts of protein were separated by SDS-PAGE. Immunoblot analyses were performed with Anti-CESA4.1, Anti-CESA7.1, and Anti-CESA8.2, as described in Hill et al. (2014) and Watanabe et al. (2018).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AF088917 (CESA7/IRX3, AT5G17420), AF458083 (CESA4/IRX5, AT5G44030), AF267742 (CESA8/IRX1, AT4G18780), BT023729 (IRX8, AT5G54690), BX814611 (IRX10, AT1G27440), BX820645 (LAC4/IRX12, AT2G38080), AY519621 (MYB46, AT5G12870), and AF191027 (XCP1, AT4G35350).
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Arabidopsis Biological Research Center for providing Arabidopsis seeds; Dr. Takashi Hashimoto (Nara Institute of Science and Technology, Japan) for providing the UBQ10pro::GFP-TUB6 plasmid; Dr. John Ralph (University of Wisconsin, USA) and Dr. Yuki Tobimatsu (Kyoto University, Japan) for providing NBD-CA; Dr. Minoru Kubo, Dr. Ko Kato, Dr. Ryosuke Sano, Ms. Shizuka Nishida, and Ms. Eriko Tanaka (Nara Institute of Science and Technology, Japan) for critical discussions and technical support. This work was supported in part by Japan Society for the Promotion of Science (KAKENHI Grant Number 25291062 and 18H02466 to T.D.), the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant-in-Aid for Scientific Research on Innovative Areas “The Plant Cell Wall as Information Processing System” Grant Number 25114520 and 15H01235 to M.O., 24114002 to T.D.,“Plant-Structure Optimization Strategy” Grant Number 18H05484 and 18H05489 to M.O. and T.D., and Grants-in-Aid from the NC-CARP project to T.D.), and the Exploratory Research for Advanced Technology (ERATO) from Japan Science and Technology Agency (JST) (Grant Number JPMJER1602 to M.O.). The technical support of UBC Bioimaging is gratefully acknowledged. Canadian funding was from the Natural Sciences and Engineering Research Council (NSERC) Discovery and CREATE ‘Working on Walls’ grants to A.L.S. and S.D.M and Postgraduate Scholarship-Doctoral to Y.W. Work by J.L.H. was supported by The Center for LignoCellulose Structure and Formation, an Energy Frontier Research Center funded by DOE, Office of Science, BES under award no. DE-SC0001090.
AUTHOR CONTRIBUTIONS
Y.T., M.S., L.S., M.O, and T.D. designed the research. Y.T., Y.W., M.S., M.O, P.P., A.Y., F.U., S.D.M., and, J.L.H. performed the experiments. Y.T., Y.W., M.S., L.S., S.D.M, J.L.H., M.O, and T.D. wrote the article. All authors read and approved the article.
Footnotes
Articles can be viewed without a subscription.
References
- Anderson C.T., Carroll A., Akhmetova L., Somerville C. (2010). Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol. 152: 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atalla R.H., Hackney J.M., Uhlin I., Thompson N.S. (1993). Hemicelluloses as structure regulators in the aggregation of native cellulose. Int. J. Biol. Macromol. 15: 109–112. [DOI] [PubMed] [Google Scholar]
- Avci U., Earl Petzold H., Ismail I.O., Beers E.P., Haigler C.H. (2008). Cysteine proteases XCP1 and XCP2 aid micro-autolysis within the intact central vacuole during xylogenesis in Arabidopsis roots. Plant J. 56: 303–315. [DOI] [PubMed] [Google Scholar]
- Awano T., Takabe K., Fujita M. (1998). Localization of glucuronoxylans in Japanese beech visualized by immunogold labelling. Protoplasma 202: 213–222. [Google Scholar]
- Balakshin M., Capanema E., Gracz H., Chang H.M., Jameel H. (2011). Quantification of lignin-carbohydrate linkages with high-resolution NMR spectroscopy. Planta 233: 1097–1110. [DOI] [PubMed] [Google Scholar]
- Bashline L., Li S., Gu Y. (2014). The trafficking of the cellulose synthase complex in higher plants. Ann. Bot. 114: 1059–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosca S., Barton C.J., Taylor N.G., Ryden P., Neumetzler L., Pauly M., Roberts K., Seifert G.J. (2006). Interactions between MUR10/CesA7-dependent secondary cellulose biosynthesis and primary cell wall structure. Plant Physiol. 142: 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.M., Zeef L.A.H., Ellis J., Goodacre R., Turner S.R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.M., Zhang Z., Stephens E., Dupree P., Turner S.R. (2009). Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J. 57: 732–746. [DOI] [PubMed] [Google Scholar]
- Brown D., Wightman R., Zhang Z., Gomez L.D., Atanassov I., Bukowski J.P., Tryfona T., McQueen-Mason S.J., Dupree P., Turner S. (2011). Arabidopsis genes IRREGULAR XYLEM (IRX15) and IRX15L encode DUF579-containing proteins that are essential for normal xylan deposition in the secondary cell wall. Plant J. 66: 401–413. [DOI] [PubMed] [Google Scholar]
- Busse-Wicher M., Gomes T.C.F., Tryfona T., Nikolovski N., Stott K., Grantham N.J., Bolam D.N., Skaf M.S., Dupree P. (2014). The pattern of xylan acetylation suggests xylan may interact with cellulose microfibrils as a twofold helical screw in the secondary plant cell wall of Arabidopsis thaliana. Plant J. 79: 492–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Demura T., Tashiro G., Horiguchi G., Kishimoto N., Kubo M., Matsuoka N., Minami A., Nagata-Hiwatashi M., Nakamura K., Okamura Y., Sassa N., Suzuki S., et al. (2002). Visualization by comprehensive microarray analysis of gene expression programs during transdifferentiation of mesophyll cells into xylem cells. Proc. Natl. Acad. Sci. USA 99: 15794–15799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derba-Maceluch M., Awano T., Takahashi J., Lucenius J., Ratke C., Kontro I., Busse-Wicher M., Kosik O., Tanaka R., Winzéll A., Kallas Å., Leśniewska J., et al. (2015). Suppression of xylan endotransglycosylase PtxtXyn10A affects cellulose microfibril angle in secondary wall in aspen wood. New Phytol. 205: 666–681. [DOI] [PubMed] [Google Scholar]
- Endo H., Yamaguchi M., Tamura T., Nakano Y., Nishikubo N., Yoneda A., Kato K., Kubo M., Kajita S., Katayama Y., Ohtani M., Demura T. (2015). Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 56: 242–254. [DOI] [PubMed] [Google Scholar]
- Faria-Blanc N., Mortimer J.C., Dupree P. (2018). A trascriptomic analysis of xylan mutants does not support the existence of a secondary cell wall integrity system in Arabidopsis. Front. Plant Sci. 9: 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H., Komamine A. (1980). Establishment of an experimental system for the study of tracheary element differentiation from single cells isolated from the mesophyll of Zinnia elegans. Plant Physiol. 65: 57–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goué N., Mortimer J.C., Nakano Y., Zhang Z., Josserand M., Ohtani M., Dupree P., Kakegawa K., Demura T. (2013). Secondary cell wall characterization in a BY-2 inductive system. Plant Cell Tissue Organ Cult. 115: 223–232. [Google Scholar]
- Ha M.A., MacKinnon I.M., Sturcová A., Apperley D.C., McCann M.C., Turner S.R., Jarvis M.C. (2002). Structure of cellulose-deficient secondary cell walls from the irx3 mutant of Arabidopsis thaliana. Phytochemistry 61: 7–14. [DOI] [PubMed] [Google Scholar]
- Hepler P., Fosket D., Newcomb E. (1970). Lignification during secondary wall formation in Coleus: An electron microscopic study. Am. J. Bot. 57: 85–96. [Google Scholar]
- Hill J.L. Jr., Hammudi M.B., Tien M. (2014). The Arabidopsis cellulose synthase complex: a proposed hexamer of CESA trimers in an equimolar stoichiometry. Plant Cell 26: 4834–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J.K., Kim H., Cocuron J.C., Orler R., Ralph J., Wilkerson C.G. (2011). The DUF579 domain containing proteins IRX15 and IRX15-L affect xylan synthesis in Arabidopsis. Plant J. 66: 387–400. [DOI] [PubMed] [Google Scholar]
- Jones L., Ennos A.R., Turner S.R. (2001). Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J. 26: 205–216. [DOI] [PubMed] [Google Scholar]
- Kawabe H., Ohtani M., Kurata T., Sakamoto T., Demura T. (2018). Protein S-nitrosylation regulates xylem vessel cell differentiation in Arabidopsis. Plant Cell Physiol. 59: 17–29. [DOI] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., Mimura T., Fukuda H., Demura T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19: 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Zhong R., Richardson E.A., Himmelsbach D.S., McPhail B.T., Ye Z.-H. (2007). The PARVUS gene is expressed in cells undergoing secondary wall thickening and is essential for glucuronoxylan biosynthesis. Plant Cell Physiol. 48: 1659–1672. [DOI] [PubMed] [Google Scholar]
- Li S., Lei L., Yingling Y.G., Gu Y. (2015). Microtubules and cellulose biosynthesis: the emergence of new players. Curr. Opin. Plant Biol. 28: 76–82. [DOI] [PubMed] [Google Scholar]
- Li Z., Omranian N., Neumetzler L., Wang T., Herter T., Usadel B., Demura T., Giavalisco P., Nikoloski Z., Persson S. (2016). A transcriptional and metabolic framework for secondary wall formation in Arabidopsis. Plant Physiol. 172: 1334–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney L., Marcus S.E., Knox J.P. (2005). Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J. Histochem. Cytochem. 53: 543–546. [DOI] [PubMed] [Google Scholar]
- McFarlane H.E., Döring A., Persson S. (2014). The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 65: 69–94. [DOI] [PubMed] [Google Scholar]
- Meents M.J., Watanabe Y., Samuels A.L. (2018). The cell biology of secondary cell wall biosynthesis. Ann. Bot. 121: 1107–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottiar Y., Vanholme R., Boerjan W., Ralph J., Mansfield S.D. (2016). Designer lignins: harnessing the plasticity of lignification. Curr. Opin. Biotechnol. 37: 190–200. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Naoi K., Shoji T., Hashimoto T. (2004). Low concentrations of propyzamide and oryzalin alter microtubule dynamics in Arabidopsis epidermal cells. Plant Cell Physiol. 45: 1330–1334. [DOI] [PubMed] [Google Scholar]
- Nakano Y., Yamaguchi M., Endo H., Rejab N.A., Ohtani M. (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcote D.H., Davey R., Lay J. (1989). Use of antisera to localize callose, xylan and arabinogalactan in the cell-plate, primary and secondary walls of plant cells. Planta 178: 353–366. [DOI] [PubMed] [Google Scholar]
- Oda Y., Fukuda H. (2012). Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science 337: 1333–1336. [DOI] [PubMed] [Google Scholar]
- Oda Y., Fukuda H. (2013). Rho of plant GTPase signaling regulates the behavior of Arabidopsis kinesin-13A to establish secondary cell wall patterns. Plant Cell 25: 4439–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Mimura T., Hasezawa S. (2005). Regulation of secondary cell wall development by cortical microtubules during tracheary element differentiation in Arabidopsis cell suspensions. Plant Physiol. 137: 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Iida Y., Kondo Y., Fukuda H. (2010). Wood cell-wall structure requires local 2D-microtubule disassembly by a novel plasma membrane-anchored protein. Curr. Biol. 20: 1197–1202. [DOI] [PubMed] [Google Scholar]
- Ohtani M., Nishikubo N., Xu B., Yamaguchi M., Mitsuda N., Goué N., Shi F., Ohme-Takagi M., Demura T. (2011). A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J. 67: 499–512. [DOI] [PubMed] [Google Scholar]
- Ohtani M., Morisaki K., Sawada Y., Sano R., Uy A.L.T., Yamamoto A., Kurata T., Nakano Y., Suzuki S., Matsuda M., Hasunuma T., Hirai M.Y., et al. (2016). Primary metabolism during biosynthesis of secondary wall polymers of protoxylem vessel elements. Plant Physiol. 172: 1612–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez A.R., Somerville C.R., Ehrhardt D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495. [DOI] [PubMed] [Google Scholar]
- Pear J.R., Kawagoe Y., Schreckengost W.E., Delmer D.P., Stalker D.M. (1996). Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. USA 93: 12637–12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña M.J., Zhong R., Zhou G.K., Richardson E.A., O’Neill M.A., Darvill A.G., York W.S., Ye Z.-H. (2007). Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19: 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S., Caffall K.H., Freshour G., Hilley M.T., Bauer S., Poindexter P., Hahn M.G., Mohnen D., Somerville C. (2007). The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 19: 237–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesquet E., Korolev A.V., Calder G., Lloyd C.W. (2010). The microtubule-associated protein AtMAP70-5 regulates secondary wall patterning in Arabidopsis wood cells. Curr. Biol. 20: 744–749. [DOI] [PubMed] [Google Scholar]
- Peterson G.L. (1977). A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 83: 346–356. [DOI] [PubMed] [Google Scholar]
- Ray P.M. (1967). Radioautographic study of cell wall deposition in growing plant cells. J. Cell Biol. 35: 659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter W.D., Chapple C., Somerville C.R. (1997). Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J. 12: 335–345. [DOI] [PubMed] [Google Scholar]
- Scheller H.V., Ulvskov P. (2010). Hemicelluloses. Annu. Rev. Plant Biol. 61: 263–289. [DOI] [PubMed] [Google Scholar]
- Schuetz M., Benske A., Smith R.A., Watanabe Y., Tobimatsu Y., Ralph J., Demura T., Ellis B., Samuels A.L. (2014). Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 166: 798–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.G., Haigler C.H. (1993). Patterned secondary cell wall assembly in tracheary elements occurs in self-perpetuating cascade. Acta Bot. Neerl. 42: 153–163. [Google Scholar]
- Taylor J.G., Owen T.P. Jr., Koonce L.T., Haigler C.H. (1992). Dispersed lignin in tracheary elements treated with cellulose synthesis inhibitors provides evidence that molecules of the secondary cell wall mediate wall patterning. Plant J. 2: 959–970. [Google Scholar]
- Taylor N.G., Scheible W.R., Cutler S., Somerville C.R., Turner S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11: 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N.G., Laurie S., Turner S.R. (2000). Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12: 2529–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N.G., Howells R.M., Huttly A.K., Vickers K., Turner S.R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 100: 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima N., Fukushima K. (1988). Heterogeneity in formation of lignin-XI: An autoradiographic study of the heterogeneous formation and structure of pine lignin. Wood Sci. Technol. 22: 259–270. [Google Scholar]
- Tobimatsu Y., Wagner A., Donaldson L., Mitra P., Niculaes C., Dima O., Kim J.I., Anderson N., Loque D., Boerjan W., Chapple C., Ralph J. (2013). Visualization of plant cell wall lignification using fluorescence-tagged monolignols. Plant J. 76: 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S.R., Somerville C.R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff D.M. (1969). Semimicro determination of cellulose in biological materials. Anal. Biochem. 32: 420–424. [DOI] [PubMed] [Google Scholar]
- Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153: 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Scali M., Vignani R., Spadafora A., Sensi E., Mazzuca S., Cresti M. (2003). Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis 24: 2369–2375. [DOI] [PubMed] [Google Scholar]
- Wang W., Vignani R., Scali M., Cresti M. (2006). A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 27: 2782–2786. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Meents M.J., McDonnell L.M., Barkwill S., Sampathkumar A., Cartwright H.N., Demura T., Ehrhardt D.W., Samuels A.L., Mansfield S.D. (2015). Visualization of cellulose synthases in Arabidopsis secondary cell walls. Science 350: 198–203. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Schneider R., Barkwill S., Gonzales-Vigil E., Hill J.L. Jr., Samuels A.L., Persson S., Mansfield S.D. (2018). Cellulose synthase complexes display distinct dynamic behaviors during xylem transdifferentiation. Proc. Natl. Acad. Sci. USA 115: E6366–E6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman R., Turner S.R. (2008). The roles of the cytoskeleton during cellulose deposition at the secondary cell wall. Plant J. 54: 794–805. [DOI] [PubMed] [Google Scholar]
- Wu A.M., Rihouey C., Seveno M., Hörnblad E., Singh S.K., Matsunaga T., Ishii T., Lerouge P., Marchant A. (2009). The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J. 57: 718–731. [DOI] [PubMed] [Google Scholar]
- Wu A.M., Hörnblad E., Voxeur A., Gerber L., Rihouey C., Lerouge P., Marchant A. (2010). Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol. 153: 542–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Ohtani M., Yamaguchi M., Toyooka K., Wakazaki M., Sato M., Kubo M., Nakano Y., Sano R., Hiwatashi Y., Murata T., Kurata T., et al. (2014). Contribution of NAC transcription factors to plant adaptation to land. Science 343: 1505–1508. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Goué N., Igarashi H., Ohtani M., Nakano Y., Mortimer J.C., Nishikubo N., Kubo M., Katayama Y., Kakegawa K., Dupree P., Demura T. (2010). VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 153: 906–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T.Q., Sun S.N., Xu F., Sun R.C. (2011). Characterization of lignin structures and lignin-carbohydrate complex (LCC) linkages by quantitative 13C and 2D HSQC NMR spectroscopy. J. Agric. Food Chem. 59: 10604–10614. [DOI] [PubMed] [Google Scholar]
- Zhong R., Morrison W.H. III, Freshour G.D., Hahn M.G., Ye Z.-H. (2003). Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant Physiol. 132: 786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Peña M.J., Zhou G.K., Nairn C.J., Wood-Jones A., Richardson E.A., Morrison W.H. III, Darvill A.G., York W.S., Ye Z.-H. (2005). Arabidopsis fragile fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17: 3390–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Richardson E.A., Ye Z.-H. (2007). The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19: 2776–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Ganguly A., Baskin T.I., McClosky D.D., Anderson C.T., Foster C., Meunier K.A., Okamoto R., Berg H., Dixit R. (2015). The fragile Fiber1 kinesin contributes to cortical microtubule-mediated trafficking of cell wall components. Plant Physiol. 167: 780–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T., Nevo E., Sun D., Peng J. (2012). Phylogenetic analyses unravel the evolutionary history of NAC proteins in plants. Evolution 66: 1833–1848. [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]