Altered lignification mediated by microRNA-directed laccase gene silencing is a major contributor to domestication-associated phenotypes of cultivated indica rice.
Abstract
Domestication of rice (Oryza sativa) included conversion of perennial wild species with few seeds to short plants that produced abundant seeds. Most domestication-associated changes were due to variations in transcription factors and other key proteins such as enzymes. Here, we show that multiple yield-related traits associated with indica rice domestication are linked to micro (mi) RNA-mediated regulation. Analysis of small (s) RNA data sets from cultivated indica rice lines, a few landraces, and two wild relatives of rice revealed the presence of abundant 22-nucleotide (nt) reads in wild relatives that mapped to miR397 precursors. miR397 was expressed at very high levels in wild relatives and at negligible levels in high-yielding cultivated lines. In its genera-specific form of 22-nt, miR397 targeted mRNAs encoding laccases that decayed and induced robust secondary cascade silencing in wild species that required RNA-dependent RNA polymerase 6. In wild species of rice, reduced expression of laccases resulted in low lignification. As expected, overexpression of miR397 induced de-domestication phenotypes. At least 26 uncharacterized QTLs previously implicated in rice yield overlapped with laccases and miR397 genes. These results suggest that miRNAs contribute to rice domestication-associated phenotypes.
Introduction
Indica rice was domesticated from Oryza nivara and O. rufipogon in South and East Asia (Sweeney and McCouch, 2007; Wing et al., 2018). Both these wild relatives are still found in many rice-growing areas, often as wild plants next to paddy fields and at the edges of lakes (Sweeney and McCouch, 2007). Some of these wild accessions are perennial. The rice domestication process is reflected in the early history of artificial selection of key agronomically important traits, such as seed shattering, plant architecture, yield, stress tolerance, and grain color (Izawa et al., 2009). In the Oryza complex, four different lines of domestication have been observed. In all these independent domestication events, seed shattering is a common change, mapped to transcription factors now referred to as SHATTERPROOF (SH) genes. Other lines of domestication were linked to other phenotypes such as plant height, panicle structure and grain yield (Sweeney and McCouch, 2007; Wing et al., 2018).
Domestication was accompanied by other changes in genomes in addition to the SH locus, mediated mostly through changes in either sequences or expression levels of transcription factors and their cofactors. Several other genes related mostly to japonica rice domestication, including qSH1, PLOG1, qSW5, GIF1, and Rc, have been cloned and functionally characterized (Kovach et al., 2007; Izawa, 2008). In such instances, simple changes in the promoter sequences of these genes altered their expression (Huo et al., 2017), while in some cases insertion of a transposon was responsible (Li et al., 2017). There were also SNPs that likely alter the key amino acids in important proteins such as enzymes (Kharabian-Masouleh et al., 2012; Ma et al., 2015).
However, it is well established that phenotypic diversity of domesticated crops is not proportional to genomic changes (Meyer and Purugganan, 2013). This has prompted several lines of inquiry into possibilities of other causes that might have contributed to phenotypic diversity and variations associated with domestication. Since the genetic basis for many QTLs that influence the majority of yield-related phenotypes in rice are still unknown, it was speculated that small RNA and epigenetic variations might contribute toward domestication-associated phenotypes (Campo et al., 2013; Qin et al., 2014). Recently, it has been found that heritable phenotypes of hybrids are likely due to epigenetics and small regulatory RNAs (sRNAs) (Shivaprasad et al., 2012b; Bond and Baulcombe, 2014). sRNAs of the micro(mi)RNA class have been implicated in heritable changes in diverse crops (Groszmann et al., 2011; Li et al., 2012; Shivaprasad et al., 2012b; Houston et al., 2013). Epigenetic regulation was also implicated in anthocyanin biosynthetic pathways in maize (Zea mays) (Hollick and Chandler, 2001). Several epialleles have strong genetic linkage to changes in phenotypes of crops and the DNA methylation status in the regulatory sequences of the genes (Manning et al., 2006; Niederhuth and Schmitz, 2014).
sRNAs are key regulators of gene expression across eukaryotes. sRNAs can either be processed from completely complementary dsRNA molecules derived from endogenous or exogenous sources, or from structured RNAs that fold back into dsRNA-like structures (Baulcombe, 2004). In the former case, host RNA-dependent RNA polymerases (RDR) play a crucial role in synthesizing completely complementary dsRNA. The resulting dsRNA molecule is cleaved by the action of multiple Dicer-like proteins (DCL) in plants to generate 20-22-nt miRNAs or small interfering (si)RNAs of 21–24-nt length. The fate of these sRNAs depends on their association with specific Argonaute (AGO) proteins (Baulcombe, 2004).
For example, miRNAs associate with AGO1 to induce targeting of mRNA that shares high complementarity (Baumberger and Baulcombe, 2005). The degree of targeting depends on the nature of the targeted mRNA as well as the extent of complementarity with miRNA. Usually, the target mRNA is cleaved to result in reduced protein accumulation if there is near-complete complementarity. There are ample examples of plant miRNAs participating in feedback loops and subtle spatio-temporal regulations (Willmann and Poethig, 2007). However, most striking among the functions of plant miRNAs is their ability to initiate regulatory cascades to target a large number of mRNAs. This requires conversion of the miRNA-targeted mRNA fragment into a dsRNA molecule by RDR6 (Howell et al., 2007). Most miRNA-mediated targeting does not result in RDR6-dependent biogenesis of secondary siRNAs. Among the reasons known for miRNA-induced RDR6-dependent biogenesis of secondary siRNAs (Felippes and Weigel, 2009; Manavella et al., 2012), length of miRNA, i.e., 22-nt instead of the common 21-nt, is a strong determinant. Once initiated, the cascade silencing generates abundant secondary siRNAs of usually 21-nt in length (Johnson et al., 2009). These siRNAs further associate with specific AGO proteins. The most conserved secondary siRNA loci among plants are derived from the TAS3 locus (Chen et al., 2007) that regulates auxin response factor (ARF) mRNAs, playing crucial roles in development by regulating auxin signaling. The best characterized secondary siRNAs are known as transacting siRNAs (tasi-RNAs) or phased siRNAs (phasi-RNAs), and their targets include a pentatricopeptide and ARF gene mRNAs (Axtell et al., 2006). Although it is not entirely clear why cascade silencing targets are targeted by multiple small RNA species, it is possible to speculate that the secondary siRNAs play a role in either reinforcing or coordinating the action of primary sRNAs or miRNAs.
In this study, using hundreds of indica rice lines, we explored if domestication-related traits are associated with sRNA-mediated regulation. We profiled sRNAs from two wild species of cultivated indica rice, a few landraces and a few high yielding cultivated lines, selected based on maximum phenotypic variability. We observed an unusual abundance of 22-nt sRNAs in wild species that was mapped to miR397 precursor in chromosome (Chr.) 2. miR397 expression in wild species resulted in selective degradation of laccases, thereby reducing lignification in stem tissues. Complementation of high expressing miR397 variant in cultivated lines resulted in phenotypes that match de-domestication. Surprisingly, miR397 was previously implicated in yield when it was overexpressed in japonica rice through regulation of the brassinosteroid pathway (Zhang et al., 2013). Our results suggest the presence of a previously unknown regulator of indica rice domestication.
Results
Wild Relatives of Cultivated Rice Have an Unusual Abundance of 22-nt sRNAs
To explore if domestication-related traits are associated with small RNA-mediated regulation, we initially performed phenotypic analysis of 290 indica rice landraces grown in the Indian sub-continent along with two wild species of rice that are naturally grown (O. nivara and O. rufipogon) and 12 high-yielding cultivated rice lines that are the results of modern breeding. These lines exhibited tremendous variation in phenotypes. Among these lines, we selected two high-yielding lines (O. sativa indica Pusa basmati 1, henceforth PB-1, and whiteponni, hereafter WP), six landraces with diverse phenotypes, along with the two wild species of rice for molecular analysis based on maximum phenotypic variations. O. nivara is an annual diploid grass with an AA genome and O. rufipogon is a perennial diploid grass also with an AA genome (Sweeney and McCouch, 2007). Landraces chosen for analysis generally had variations in vegetative growth, development and panicles, phenotypes strongly linked to domestication-associated phenotypes (Supplemental Figure 1A).
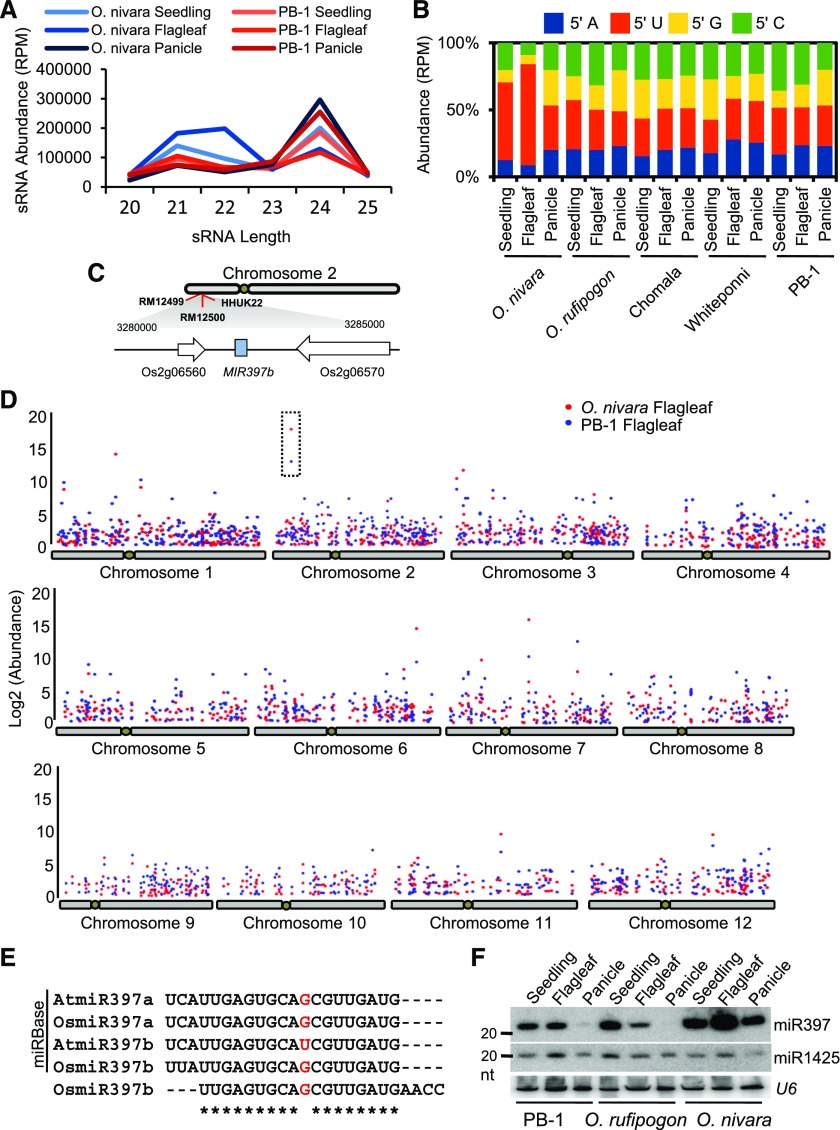
We profiled sRNAs from three different tissues from each of the six rice lines in biological duplicates (Supplemental Table 1). All vegetative tissues had a strong peak for 21-nt, and this class in plants generally includes miRNAs and siRNAs. There was also a strong peak for 24-nt sRNAs (Supplemental Figures 1B and 1C). In flagleaf tissues, as expected for a reproductive tissue (Chávez Montes et al., 2014), there was a higher abundance of 24-nt reads (Supplemental Figure 1B). Most unusually, profiles of sRNAs from O. nivara, and to some extent, O. rufipogon, showed higher accumulation of 22-nt sRNA reads (Figure 1A and Supplemental Figure 1D). This is unusual for multiple reasons. Most plant sRNA profiles lack a strong peak for 22-nt reads since this is not a major sRNA size class (Chávez Montes et al., 2014). Furthermore, 22-nt reads are generally produced from a DCL that is not very active (DCL2) during normal plant development. In addition, a higher percentage of 22-nt reads was prominently observed only in flagleaves, indicating tissue-specific expression of these sRNAs. Most importantly, this unusual abundance was restricted to wild species of rice and was not observed among other rice lines. Among the 22-nt reads, there was a strong bias for 5′ U in data sets derived from wild species of rice (Figure 1B and Supplemental Figure 1E), indicating that the majority of these 22-nt sRNAs were likely associated with AGO1 or its close homologs.
Figure 1.
Variation in sRNA Profiles Across Wild and Cultivated Rice Lines.
(A) Comparison between sRNA profiles of O. nivara and PB-1 indicating higher abundance of 22-nt reads in wild species. Profiles of additional rice lines have been provided in Supplemental Figure 1. Two biological replicates of each sample were used for sRNA sequencing from various rice lines.
(B) The 5′ nucleotide bias of 22-nt sRNA reads across rice lines, represented as the average of two biological replicates.
(C) Location of miR397b on Chr. 2.
(D) Differentially expressed sRNA loci between rice lines, represented as the average of two biological replicates. Box indicates the most differentially expressed loci on Chr. 2.
(E) Alignment of miR397 sequence variants from Arabidopsis, rice (miRBase) and from our data sets. * represents conserved positions and nucleotides in red indicate mis-match.
(F) RNA gel blot confirming the of miR397 across selected rice lines in different tissues. Decade marker size is indicated. The 22-nt miR397b was used as probe, and U6 and miR1425 were used as controls.
Mapping of an Abundant 22-nt sRNA-Producing Locus
We performed differential expression analysis of sRNA loci between rice lines to investigate the unusual abundance of 22-nt sRNAs between the rice lines. This analysis identified 358 differentially expressing sRNA loci (Supplemental Data Set 1). These loci were mapped to all 12 chromosomes of rice. Differentially expressing loci were mapped to many short and long genomic regions; however, one specific locus on chromosome 2, between SSR marker RM12499 and RFLP marker HHUK22, accounted for the most difference between O. nivara and the other rice lines (Figures 1C and 1D). This region alone accounted for nearly 120 thousand reads per million (RPM) of difference between O. nivara and other rice lines such as PB-1 (Supplemental Figure 1F).
The mapped region of chromosome 2 encoded a single non-coding RNA expressing a miRNA precursor. This precursor was the single source for the abundant 22-nt reads in this region of chromosome 2. This precursor generates miR397 in both dicots and monocots (Supplemental Figure 2A); however, the mature form of 22-nt miRNA that we observed is shifted by 3 residues when compared with A. thaliana (Figure 1E) and other plants (Kozomara and Griffiths-Jones, 2014). miR397 is usually 21-nt in length in most plants. The mature miR397 identified here has, in addition to a shift of 3 nucleotides, an extension by 1-nt when compared with other miR397 precursors of other miRBase entries (Supplemental Figure 2B). The 22-nt miR397 is incorporated into the AGO1 isoform (Wu et al., 2009). We also identified a 21-nt version of miR397 in all our rice data sets; however, accumulation of the 21-nt form is at least 20-fold lower than the 22-nt form. miR397 expressed in O. nivara is 22-nt in length (Supplemental Figure 2C). To confirm the high accumulation of miR397 in O. nivara, we performed an RNA gel blot analysis for three different rice lines (Figure 1F). The tissues derived from O. nivara accumulated very high levels of the 22-nt form of miR397.
Since the presence of 22-nt miR397 has been reported only from rice and maize (Jeong et al., 2011), we performed a miRProf analysis to assess the diversity of miR397 sequences in different plant species (Shivaprasad et al., 2012a). The 22-nt version was abundant only among members of the Oryza genera (Supplemental Figure 2D). The presence of genera-specific isoforms indicates that this miRNA has an evolutionarily recent origin. We also used miRProf analysis to confirm differential expression of miRNAs across wild species and other lines (Supplemental Figure 3, Supplemental Data Set 2). Few miRNAs had differential accumulation between rice lines irrespective of the tissues, among which miR397 exhibited maximum variation between lines across three tissues analyzed. Other miRNAs that had variable expression included miR408, miR398 and miR528 across three tissues, and miR1568 and miR168 in flagleaf and panicle tissues. Their targets include genes involved in development and various stress responses. Considering the spectrum of modifications that were introduced through domestication, it is possible that these miRNAs might have contributed to important phenotypes.
miR397 Targets Rice Laccases
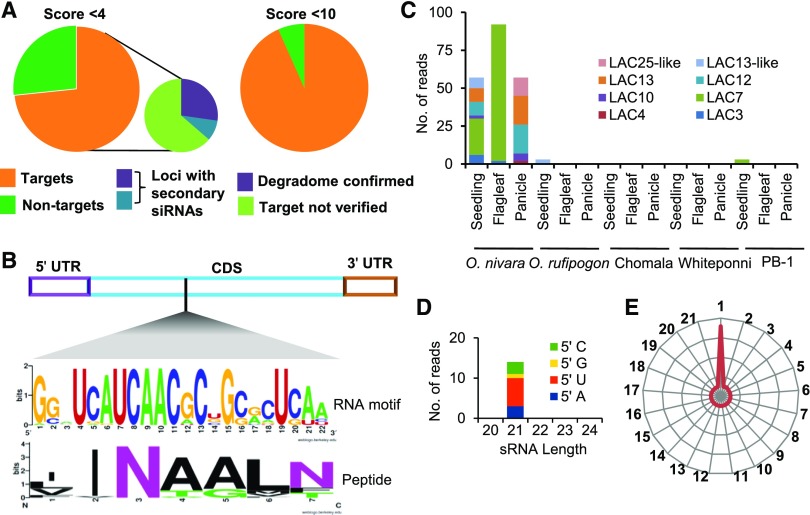
In Arabidopsis, maize, poplar (Populus) and Medicago (Medicago truncatula), miR397 targets a family of laccases (Lu et al., 2013; Wang et al., 2014). Laccases are also the major targets of miR397 in rice, even though the rice miR397 isoform had an extra nucleotide, this made no difference to its targeting ability. Rice has 30 laccases encoded in its genome. To identify the ability of miR397 in targeting rice laccases and other possible previously unidentified targets, we performed psRNA target (Dai and Zhao, 2011) prediction and tapir (Bonnet et al., 2010) tool analysis. We identified 15 laccases as potential targets of miR397 under stringent conditions (tapir score >4), and when the score was relaxed, the number of potentially targeted laccases increased to 28 (Figure 2A). These miRNA targets belonged to different clades of laccases (Supplemental Figure 4), confirming that laccases of diverse sequences were likely targeted by the miRNA. We predicted that miR397 might be able to target multiple clades of laccases due to its ability to target a specific RNA motif encoding a specific domain. Degradome analysis derived from pooled tissues of O. nivara confirmed targeting of at least six laccases by miR397. However, in a publicly available O. rufipogon degradome data set (Wang et al., 2012), reads of OsLAC7 but not of other laccases mapped at the expected slicing site. Analysis of target regions of these laccases identified an RNA motif targeted by miR397 (Figure 2B). This specific RNA motif codes for the INLAAN peptide (Figure 2B). Such a peptide sequence is observed only among laccases in plants. Laccases in other plants were also targeted by miR397 family members in the same region.
Figure 2.
miR397 Targeted Rice Laccases and Induced a Silencing Cascade.
(A) Pie-charts representing potential laccases targeted by miR397b with tapir scores of <4 and <10, respectively.
(B) Conserved RNA and peptide sequences in potential miR397-targeted regions of laccases.
(C) Abundance of laccase-derived PHAS-loci between rice lines, represented as the average of two biological replicates.
(D) The 5′ nucleotide bias of phased siRNAs derived from OsLAC7 in an O. nivara flagleaf sample.
(E) Phasing Register of Secondary siRNAs Arising from OsLAC7 in an O. nivara Flagleaf Sample.
miR397 Induces RDR6-Dependant Cascade Secondary Silencing in Target Loci
Generation of secondary siRNAs from miRNA-targeted RNAs is unusual among plants. These secondary siRNAs, also known as tasi-RNAs or phasi-RNAs generate a cascade silencing effect in miRNA target regions. Since miR397 is 22-nt long, we hypothesized that miRNA-targeted laccase regions might be sources of such secondary siRNAs. We performed tasi-RNA analysis prediction to look for rice regions that have signatures of secondary siRNA generation. As expected, the well-conserved TAS3 locus indeed produced abundant secondary siRNAs in all lines at similar levels (Table 1). Higher numbers of TAS3-derived siRNAs were observed in seedling tissues. However, there were also eight laccase loci acting as PHAS loci in O. nivara, but not in other lines including those from cultivated rice lines such as PB-1 (Figure 2C, Table 1). The presence of secondary siRNAs derived from these laccases, in phase with the miR397 target region, is direct evidence that miR397 targets laccases in O. nivara. This is significant because the presence of phasi-RNAs derived from laccases have not been reported previously, although targeting of laccases by miR397 has been well documented in multiple species where miR397 is expressed in the 21-, but not 22-nt, form. Laccase loci that produced abundant secondary siRNAs in O. nivara included LAC3, LAC7, LAC12 and LAC13 (Figures 2D and 2E and Supplemental Figures 5A to 5C).
Table 1. Abundance of Tasi- and Laccase-Derived Phasi-RNAs among Different Tissues of Rice Lines in Two Biological Replicates.
| Seedling | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| O. nivara | O. rufipogon | Chomala | Whiteponni | PB-1 | ||||||
| Locus/ Gene | Repl1 | Repl2 | Repl1 | Repl2 | Repl1 | Repl2 | Repl1 | Repl2 | Repl1 | Repl2 |
| OsTAS3a | 149 | 169 | 29 | 32 | 262 | 154 | 112 | 97 | 94 | 118 |
| OsTAS3b | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 |
| OsTAS3c | 39 | 44 | 9 | 6 | 45 | 26 | 11 | 15 | 33 | 51 |
| OsTAS3d | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| OsTAS3e | 2891 | 2929 | 655 | 730 | 2587 | 1845 | 1058 | 1205 | 363 | 489 |
| LAC3 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LAC7 | 30 | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| LAC10 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LAC12 | 8 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LAC13 | 8 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LAC13-like | 13 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| Flagleaf | ||||||||||
| O. nivara | O. rufipogon | Chomala | Whiteponni | PB-1 | ||||||
| Locus/ Gene | Repl1 | Repl2 | Repl1 | Repl2 | Repl1 | Repl2 | Repl1 | Repl2 | Repl1 | Repl2 |
| OsTAS3a | 26 | 21 | 43 | 28 | 29 | 26 | 0 | 14 | 27 | 34 |
| OsTAS3b | 0 | 6 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OsTAS3c | 0 | 0 | 27 | 11 | 0 | 0 | 5 | 3 | 4 | 8 |
| OsTAS3d | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OsTAS3e | 404 | 379 | 1343 | 809 | 210 | 395 | 195 | 104 | 56 | 70 |
| LAC3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LAC7 | 80 | 99 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Panicle | ||||||||||
| O. nivara | O. rufipogon | Chomala | Whiteponni | PB-1 | ||||||
| Locus/ Gene | Repl1 | Repl2 | Repl1 | Repl2 | Repl1 | Repl2 | Repl1 | Repl2 | Repl1 | Repl2 |
| OsTAS3a | 0 | 8 | 25 | 44 | 41 | 34 | 74 | 62 | 20 | 21 |
| OsTAS3b | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| OsTAS3c | 34 | 38 | 182 | 318 | 192 | 252 | 276 | 250 | 76 | 70 |
| OsTAS3d | 0 | 0 | 0 | 0 | 0 | 13 | 0 | 7 | 0 | 0 |
| OsTAS3e | 323 | 159 | 1448 | 2615 | 735 | 829 | 1349 | 1376 | 254 | 265 |
| LAC4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LAC10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LAC12 | 29 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LAC13 | 29 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LAC25-like | 18 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
miRNAs of 22-nt initiate secondary silencing in plants that require RDR6 to generate dsRNA substrates from miRNA target regions (Cuperus et al., 2010). To check if laccase-derived secondary siRNAs are indeed dependent on RDR6, we introduced laccase from rice into tobacco plants silenced in RDR6 (Schwach et al., 2005). Tobacco miR397 abundance was extremely low in publicly available tobacco sRNA data sets as well in our RNA gel blot analysis (Supplemental Figure 5D). In wild-type and RDR6i plants, introduced laccases and miRNAs were expressed at high levels. Secondary siRNAs (D10-) were abundantly expressed in wild-type tobacco plants, but not in RDR6i plants (Supplemental Figures 5D and 5E). These results indicate that laccase-derived siRNAs required RDR6 to generate dsRNA intermediates that can be cleaved by DCL4. Multiple secondary siRNAs derived from laccases potentially target parent and other laccases, likely resulting in robust silencing (Supplemental Figure 5F).
The evolution of the 22-nt form in Oryza genera members might have resulted in robust silencing of laccases that is reinforced due to the formation of a dsRNA and subsequent production of secondary cascade siRNAs. This is confirmed with the requirement of RDR6, an enzyme that is generally associated with silencing signal amplification. It is however unclear why this reinforced regulation is required in Oryza genera members where miR397 is a 22-nt miRNA, but not in other plants where miR397 is expressed in a 21-nt form. One possible explanation for the proposed mechanism could be that the precursor of miR397 in Oryza speciation accumulated genetic changes that altered the precursor structure to accommodate robust silencing of laccases (Supplemental Figure 2A).
Differential Expression of Laccases among Rice Lines
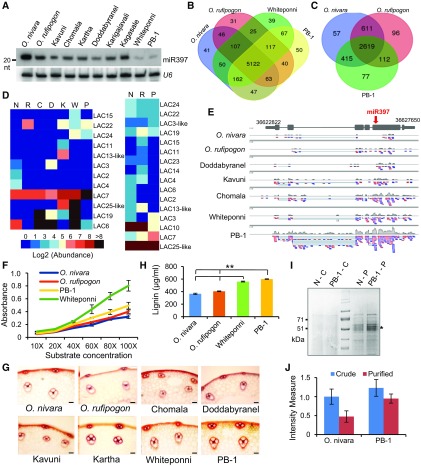
Domestication-associated phenotypes are often complex, involving massive changes in genomes and expression of their contents. To identify transcripts that accumulate differently between rice lines, as well as to identify the outcome of miR397 targeting on laccases and other pathway genes, we performed a transcriptome analysis using two tissues of rice lines with variable expression of miR397. In addition to cultivated rice lines (WP and PB-1) expressing miR397 at lower levels, and wild species that express miR397 at high levels, we also used landraces that have intermediate levels of miR397 (Figure 3A). Up to 90% of all reads matched to the rice genome, almost to similar levels between wild species, landraces and cultivated high yielding lines (Supplemental Table 2). Many transcription factors, disease/pathogen resistance and starch biosynthesis genes had variable expression between rice lines (Figures 3B and 3C and Supplemental Data Sets 3 and 4).
Figure 3.
Abundant miR397 Expression in the Wild Species O. nivara Results in Reduced Lignification.
(A) Abundance of miR397 in flagleaf tissues among rice lines, including landraces. U6 served as the control.
(B) Venn diagram showing differentially expressed genes between rice lines in flagleaf samples, and (C) panicles.
(D) Heatmap showing differential expression of laccases in flagleaf and panicle samples, respectively of O. nivara (N), O. rufipogon (R), Chomala (C), Doddabyranel (D), Kavuni (K), Whiteponni (W) and PB-1 (P).
(E) Coverage graphs showing abundance of OsLAC7 on Chr. 1 in flagleaf RNA-Seq data sets. miR397b target regions have been marked with red arrows.
(F) Laccase activity measurements in flagleaf samples of selected rice lines, using 4-Hydroxyindole as substrate. The assay was performed with three biological replicates and error bars indicate sd.
(G) Stem sections of selected rice lines stained with phloroglucinol. Scale bar = 50 µm.
(H) Quantification of lignin in stems of selected rice varieties. The assay was performed with three biological replicates and error bars indicate sd, ** indicates P value <0.005 (Student’s t test).
(I) SDS-PAGE gel showing reduced accumulation of laccases (*) in crude (C) and concanavalin-bound purified (P) samples derived from stems of O. nivara (N) and PB-1.
(J) Quantification of in-gel activity of laccases from crude and purified protein extracts of stem from O. nivara and PB-1 with 4-Hydroxyindole. The intensity of bands was quantified using ImageJ. The assay was performed with three biological replicates and error bars indicate sd.
Among laccases, we observed expression of only 16 laccases out of 30 in rice tissues analyzed in this study. Flagleaves had expression of 12 laccases, while panicle tissues expressed 16 laccases at variable levels (Figure 3D, Supplemental Figure 6 and Supplemental Data Set 5). Strikingly, almost all laccases were expressed at different levels between rice lines and were among the most differentially expressed genes between wild and cultivated rice lines. miR397-targeted laccase such as LAC7 was abundantly expressed in cultivated rice lines such as PB-1, while it accumulated to negligible levels in O. nivara (Figure 3E). These results, in addition to strongly confirming differential targeting of laccases that depends on expression levels of miR397, also indicate natural variation in the expression of laccases between rice landraces, further suggesting their roles in domestication-associated phenotypes.
Wild Rice Species Accumulate Reduced Levels of Lignin in Stem Tissues
Since laccase RNAs had lower expression in O. nivara due to miR397 targeting, we hypothesized that the lignification process in this species might be affected. We performed laccase assays for total tissues and observed that cultivated rice lines such as PB-1 and WP had very high laccase activity when compared with wild species (Figure 3F). We performed phloroglucinol staining in leaf and stem tissues. While there were many morphological differences between leaf samples, such as the vascular bundle density and arrangement, there was little difference in lignin staining (Supplemental Figures 7A and 7B). However, in stem sections, we observed that wild species accumulated much lower levels of lignin when compared with landraces and PB-1 (Figure 3G). This was also well correlated with total lignin content measured by derivatization with thioglycolic acid (Figure 3H). Similarly, total laccase protein levels, purified using a concanavalin column, also had higher accumulation in cultivated rice line PB-1 when compared with O. nivara (Figure 3I and Supplemental Figure 7C). We separated laccases on a SDS gel and identified several laccases that are expressed only in the cultivated line PB-1 and not in O. nivara. In addition, we performed in-gel staining with 4-hydroxyindole and found that wild species had low accumulation of laccases when compared with PB-1 and WP (Figure 3J, Supplemental Figure 7D).
These results strongly indicate that miRNA-mediated laccase targeting reduced lignification in wild rice lines due to reduced accumulation of laccase RNAs. Together, these results indicate how robust silencing of laccases between rice wild species, landraces and high-yielding cultivated lines is linked to natural variation in expression of a single miRNA that might be the result of selection of phenotypes requiring high lignification.
Overexpression of miR397 in Cultivated Rice Results in Morphological Abnormalities Associated with Reduced Lignification
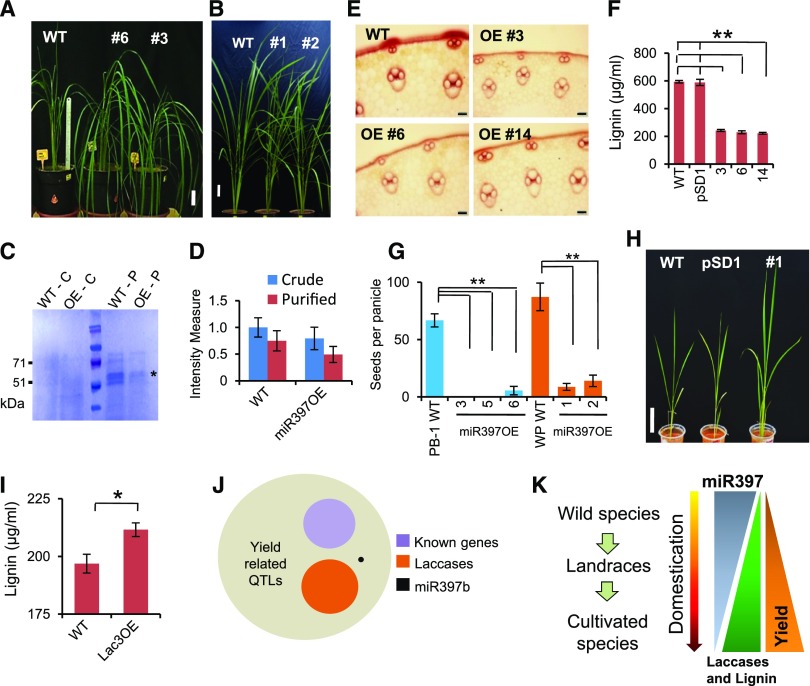
To confirm the presence of phenotypes associated with expression of one miRNA in wild species, we generated overexpression (OE) lines in cultivated rice background. We anticipated that OE of miR397 might alter the phenotype of high-yielding lines through reduced lignification and associated changes. We generated several single-copy T-DNA transformants using Agrobacterium-mediated transformation that expressed miR397 under the maize ubiquitin promoter in PB-1 and WP backgrounds.
These transgenic lines expressed miR397 to very high levels, comparable to O. nivara (Supplemental Figures 8A to 8C). In line with our hypothesis, most miR397 overexpressing transgenic plants in PB-1 as well as WP backgrounds had adverse phenotypes when compared with controls (Figure 4A and 4B). Some of the transgenic plants were feeble, often with reduced stem thickness and exhibited a tendency to lodging. Leaves and stems of few transgenics were narrower with reduced length when compared with controls (Supplemental Figures 8D to 8G, and Supplemental Table 4). Similar to O. nivara plants, transgenic miR397 overexpressing lines had reduced laccases (Figures 4C and 4D and Supplemental Figure 8H and 8I) and lignins in their stems (Figures 4E and 4F). Strikingly, the miR397 OE transgenic plants in PB-1 and WP backgrounds produced very few panicles (most of the plants never flowered) with hardly any seeds (Figure 4G and Supplemental Figures 8J). The phenotypes observed in miR397 OE plants are identical to O. nivara in several key aspects such as reduced lignification, panicle architecture and reduced yield. On the other hand, we also observed stronger and healthier plants when OsLAC3 was overexpressed in the background of cultivated rice line PB-1 (Figures 4H and 4I and, Supplemental Figures 8K to 8O). These results confirm the role of miR397-mediated regulation of laccases in indica rice domestication-associated phenotypes. Association of lignification with yield-related parameters is not limited to rice but was also observed among other monocots such as maize (Sun et al., 2018) and other plants (Pinçon et al., 2001). Our results strongly suggest that miR397 expression might have been a strong selective force in indica rice domestication (Figure 4K).
Figure 4.
OE of miR397b and OsLac3 Induced Lignin-Associated Phenotypes in Cultivated Lines.
(A) Phenotypes of miR397b OE transgenic lines in the PB-1 background. Size bar 5 cm.
(B) Phenotypes of miR397b OE in the WP background.
(C) SDS-PAGE gel showing reduced accumulation of laccases (*) in crude (C) and concanavalin-bound purified (P) protein samples derived from the stems of the wild type (WT) and miR397b OE transgenic line (OE).
(D) Quantification of in-gel activity of laccases in protein extracts of stems from WT and miR397 OE plants with 4-Hydroxyindole. The intensity of bands was quantified using ImageJ. The assay was performed with three biological replicates and error bars indicate sd.
(E) Stem sections of miR397b OE lines stained with phloroglucinol. Scale bar = 50 µm.
(F) Lignin quantification of stems from the WT, vector control (pSD1) and miR397b OE line. The assay was performed with three biological replicates and error bars indicate sd (n = 3), ** indicates P value <0.005 (Student’s t test).
(G) Yield comparison between WT and miR397OE lines in PB-1 and WP backgrounds. Calculations were made using 8 panicles per plant and error bars indicate sd, ** indicates P value <0.005 (Student’s t test).
(H) Phenotypes of LAC3 OE transgenic lines. Size bar = 5 cm.
(I) Lignin quantification from stems of the WT and LAC3 OE line. The assay was performed with three biological replicates and error bars indicate sd (n = 3), * indicates P value <0.05 (Student’s t test).
(J) Overlap of known QTLs for rice yield with finely mapped known domestication-associated genes and laccases.
(K) Proposed model indicating role of miR397:laccase regulation during indica rice domestication.
If miR397:laccase interaction was a key feature of indica rice domestication, then it is possible that these genes are part of QTLs identified for rice yield. We overlapped all uncharacterized QTLs previously mapped for rice yield (from Gramene QTL database) and found that more than 26 previously uncharacterized QTLs overlap with laccases and miR397 (Figure 4J, Supplemental Figure 9, Supplemental Data Set 6).
Discussion
The origin and heritability of agronomic traits during crop domestication are complex events in evolution. Domestication of rice, especially that of the japonica subtype, has been extensively studied and the outcome of such findings was duly used in various breeding and genetic modification efforts to improve agronomic traits (Zuo and Li, 2014). In this study, we identified a novel miRNA-mediated regulation of a large family of genes coding for laccase enzymes as a major sRNA variation between wild species of rice, few selected phenotypically diverse landraces and cultivated rice lines. We propose that this regulation is involved in the evolution of various agronomic traits in indica rice.
In our comparative miRNA analysis, few plants showed high abundance of miR397. Expression levels of miR397 might be a direct proxy for laccase expression between plants, since many trees that accumulate high levels of lignins have low levels of miR397 expression (Lu et al., 2013). Surprisingly, among the 60 different species analyzed in this study, wild species of rice had the highest accumulation of miR397. Wild species of rice originated in semi-aquatic ecosystems and strong mechanical support might not be a requirement for these plants. This could explain why wild species of rice had stronger miR397-mediated silencing of laccases when compared with other plants. Also, it is possible that laccases that cannot be targeted by miR397 might have evolved in some plants where mechanical strength was a prerequisite for changing environment. Alternatively, miR397-mediated regulation might have been a result of co-evolution that was altered during the artificial selection of rice. The altered expression of miR397 might have been due to the incorporation of mutations in miR397 promoter sequences since there is an absence of any change in the precursors, and indeed we do see changes in the promoter sequence between wild and cultivated rice species (Supplemental Figure 2E).
The presence of a 22-nt miRNA form in Oryza genera is extremely interesting since it is not found among other monocots where sequence information is available. Based on the findings in other plants (Chen et al., 2010), it is possible that the evolution of the 22-nt form might have resulted in robust silencing of laccases that is reinforced due to the formation of a dsRNA and subsequent production of secondary cascade siRNAs. This is also confirmed with the requirement of RDR6, an enzyme that is generally associated with silencing signal amplification. It is, however, hard to speculate why this reinforced regulation is required in Oryza genera members where miR397 is a 22-nt miRNA, but not other plants where miR397 is expressed in a 21-nt form. One possible explanation for the proposed mechanism could be that the precursor of miR397 in Oryza speciation accumulated a genetic change that altered the precursor structure to accommodate robust silencing of laccases (Supplemental Figure 2A). It is well-established that precursor structure is a key requirement for 22-nt miRNA biogenesis (Cuperus et al., 2010).
Surprisingly, little sequence variation in nucleotides and amino acids are seen among individual laccase family members, both within and between rice species and landraces. Interestingly, there are natural variations in miR397-targeted RNA motifs among laccases that specify the extent of miR397 targeting. Due to these variations, closely related laccases might not be targets of miR397. For example, miR397 targets OsLAC13 but not OsLAC5 because of five mismatches in the target region, although OsLAC13 and OsLAC5 belong to the same clade (according to the phylogenetic analysis). OsLAC5 escaped from the control of miR397 likely after the whole genome duplication (Guo et al., 2008). Since the functions of individual laccases of rice are not fully understood, it is not possible to interpret the importance of these interactions; however, variations in the expression of individual laccases between landraces studied here indicate a functional importance for these changes.
Based on our analysis, both through sRNA sequencing, phenotyping and genome modifications, we can define functions for rice laccases that were not well-known previously. Our observations indicate that laccases regulate lignification and the associated phenotypes and the architecture of rice plants, and this is in agreement with findings in other plants. For example, disruption of AtLAC4 and AtLAC17 resulted in tissue-specific alterations to lignification of A. thaliana stems specifically in plants grown under continuous light (Berthet et al., 2011). On the other hand, AtLAC15 was involved in two different phenotypes, seed coat coloration and root elongation, both linked to lignin (Liang et al., 2006). OsLAC13 regulated the seed setting rate in rice by suppressing the transport of sugars to the anthers and promoting H2O2 production through unknown mechanisms (Yu et al., 2017). However, heterologous expression of OsLAC10 in Arabidopsis plants increased lignification around vasculature and increased Cu stress tolerance (Liu et al., 2017). Triple mutant of atlac4, atlac11 and atlac17 had very much reduced lignin deposition in the vasculature and the plants were dwarf (Zhao et al., 2013). Cotton GhLAC1 overexpression increased lignin deposition and led to resistance against Verticillium dahliae (Hu et al., 2018). LAC3 from Populus trichocarpa was required for normal cell wall structure and integrity of xylem fibers. Suppression of LAC3 led to increased soluble phenolic content in poplar plants (Ranocha et al., 2002). Lignification also affected seed shattering in rice. For example, OsSH5 enhanced seed shattering in japonica rice by altering lignin deposition at the abscission zone (Yoon et al., 2014).
What are the possibilities that a similar strategy to regulate lignification and mechanical strength is also true for other crops? Among the wild species of crops, few are submerged or prostrate plants with weak stems. In S. pimpinellifolium, a wild relative of cultivated tomato, we observed a higher abundance of miR397 when compared with cultivated tomato lines (21 RPM when compared with 1 in cultivated tomato M82). It is also important to note that S. pimpinellifolium accessions are prostrate. However, in japonica rice that was domesticated from O. rufipogon species, OE of miR397 led to increased yield (Zhang et al., 2013) through an altered brassinosteroid pathway. Since OE of miR397 led to reduced yield in two indica cultivated lines (PB-1 and WP) through altered lignification, we propose the presence of a different regulatory interaction that might be responsible for high yield in the japonica cultivar with miR397 OE. The morphology of japonica lines is very different from those of indica lines, possibly due to their domestication from O. rufipogon instead of O. nivara. The lignification requirement might be different for these two subspecies. At the molecular level, it is possible that a 21-nt form of miR397 was overexpressed in japonica by Zhang et al. (2013) that led to changes in hormonal pathways in addition to changes in lignification. Also, there are differences in the abundance of individual laccase genes between japonica and indica lines, the ones expressed at higher levels in indica lines have a lower abundance in japonica lines. Such variations might alter metabolic and other regulatory pathways, resulting in different outcomes between japonica and indica lines. Further studies are also required to understand the role of miR397 in domestication-associated phenotypes of other crops. It is likely that crops domesticated from wild plants that were prostrate or those from aquatic/semi-aquatic conditions might have undergone similar changes in lignification. Such an analysis can be extremely interesting; however, this requires sRNA data for wild relatives of crops.
De-domestication phenotypes observed in miR397 OE plants in cultivated lines is extremely striking for several reasons. The OE plants (both miR397 and OsLAC3) confirm the role of miR397:laccase interaction in such phenotypes. Variation in expression of individual laccases between landraces that are known for various intermediate phenotypes (domestication is ongoing in these lines) indicates that laccases are likely key regulators of these phenotypes. Furthermore, increased robustness and higher yield of OsLAC3 OE plants suggests that these genes and miR397 offer new tools for genetic engineering and as biomarkers for breeding programs to introgress yield-related traits in rice.
These observations demonstrate a previously unknown genetic and molecular basis for the domestication-associated phenotypes in indica rice. These new tools and markers for further improvement of yield-related characters in indica rice are timely to meet the global food security challenges.
Methods
Plant Genetic Material
Two wild species of rice (O. nivara and O. rufipogon), two cultivated high-yielding lines of indica rice (whiteponni and Pusa basmati-1) and, six indica landraces from South India, namely kavuni, chomala, kartha, doddabyranel, karigajavali and kagasale, were used in this study.
Plant Growth
Rice plants were grown in the greenhouse under natural light conditions and 70% relative humidity (RH). N. benthamiana wild type and RDR6i plants and N. tabacum (Wisconsin 35) were grown in a growth chamber (Panasonic) at 24°C under a light dark cycle of 16/8 h, intensity of 390 µmol/m2/s (fluorescent lamp 40 W) and 70% RH. Transgenic rice plants were initially maintained in a tissue culture facility at 24°C, under 16 h/8 h light/dark cycle and 70% RH. Embryogenic rice calli were induced and maintained in darkness.
Total RNA Extraction and Library Preparation for RNA-Seq and sRNA-Seq
Isolation of total RNA from various tissues of different rice lines was performed using TRIzol Reagent (Ambion) as per manufacturer’s instructions. The concentration of RNA and purity of the samples were estimated using a Nanodrop Spectrophotometer (Thermo Fisher Scientific) and Qubit Fluorometer (Thermo Fisher Scientific), respectively. An aliquot of the samples was also separated on an Agilent RNA Bio-analyzer chip to check for integrity. RNA sequencing libraries were constructed according to the NEXT-flex Rapid directional mRNA-Seq bundle library protocol at Genotypic Technologies, India, and sequenced on an Illumina NextSeq500 platform. sRNA sequencing libraries were prepared with a TruSeq Small RNA Sample Preparation Guide (Illumina, USA) at Genotypic Technologies, India, and sequenced on an Illumina NextSeq500 platform.
Processing of Illumina sRNA- and RNA-Seq Reads and Expression Analysis
The sRNA-Seq reads were processed for adapter removal and filtered for length range of 16–35-nt using UEA small RNA Workbench Version-3 (Stocks et al., 2012). miRNA expression analysis and the identification of the loci producing phased siRNAs and sRNA generating loci were performed using miRProf, the TASI-RNA prediction tool (P value cut-off of 0.01), and SiLoCo (sRNA length 19 to 25 and minimum abundance of 1), respectively, using the UEA small RNA Workbench. RNA-Seq reads were processed to remove adaptors and low-quality bases. The reads obtained after preprocessing were aligned to rice genome (IRGSP-1) and analyzed further as mentioned in Trapnell et al. (2012).
miRNA Target Prediction, Sequence Alignments, and Phased sRNA Analysis
The targets of candidate miRNAs were predicted using the psRNAtarget (Dai and Zhao, 2011) tool and tapir (Bonnet et al., 2010). The sequences in the target regions were extracted and the LOGOs of miRNA target regions were generated using WebLogo (Schneider and Stephens, 1990). Phylogenetic analysis was performed using MEGA6 with the maximum likelihood method and 10,000 bootstrap replicates.
RT-PCR
cDNA was prepared using Invitrogen SuperScript-III kit with 2 µg of RNA from desired samples. Hot start RT-PCR was performed using BioRad thermo cycler. The PCR mix for 20 μL reaction included 1 μL of crude cDNA, 4 μL of RT-Buffer (Promega), 0.4 μL of forward and reverse primers (1:10 dilution of 100µM stock), 0.4 μL of 10µM dNTP mix, 0.8 μL of 50 mM MgCl2, 0.4 μL of DMSO, 0.2 μL of Taq polymerase (BRIT) and 12 μL of water.
Plasmid Clones and Constructs
To generate overexpression constructs, cDNA was prepared from O. nivara flagleaf samples using Superscript III (Invitrogen), according to manufacturer's’ protocol. Amplified cDNAs were cloned into the pBIN19 vector (Bevan, 1984) under the 35S promoter and a binary vector derived from pCAMBIA1300 (pSD1), between the maize ubiquitin promoter and 35S terminator, for transient and stable transformation experiments, respectively (Supplemental Figures 8A and 9A). The binary vectors were then transformed into A. tumefaciens strain LBA4404 with pSB1 (pSB1 harbors extra copies of vir genes for better efficiency). Primers used for cloning are listed in Supplemental Table 3.
RNA Gel Blot Analysis
The RNA gel blots were performed as described by Shivaprasad et al. (2012b). Briefly, ∼12 µg of total RNA extracted as mentioned earlier was dried and resuspended in 8 μL loading buffer (0.10% w/v bromophenol blue, 0.10% w/v xylene cyanol in 100% de-ionized formamide), heated at 95°C for 1 min and loaded onto a 15% denaturing polyacrylamide gel (a 19:1 ratio of acrylamide to bis-acrylamide and 8 M urea). The gel was run at 100 V for 3 h and then transferred to a Hybond-N+ membrane (GE Healthcare) by electro-blotting (Bio Rad) at 10 V overnight at 4°C. The RNA hybridization was performed at 35°C for 12 h using Ultra Hyb-oligo buffer (Ambion) containing appropriate radiolabeled short DNA oligo probes (Supplemental Table 3), end-labeled with 32P-ATP (BRIT, India) by polynucleotide kinase (NEB) and purified through MicroSpin G-25 columns (GE Healthcare). The blot was washed twice with 2X SSC, 0.5% w/v SDS for 30 min at 35°C. The signal was detected after exposure on a phosphor screen using a Molecular Imager (GE Healthcare). For repeated hybridization, the membrane was stripped and re-probed.
Transformation of Rice
miR397 and OsLAC3 overexpressing transgenic rice plants were generated using the constructs PUbi:miR397 and PUbi:OsLAC3 using the A. tumefaciens-mediated transformation method described for PB-1 and WP (Sridevi et al., 2005, 2008). Transgenic plants were regenerated from hygromycin-resistant calli and maintained in a growth chamber (70% RH at 24°C). Regenerated plantlets were transferred to a greenhouse after molecular validation.
Laccase Assays
Laccase assays were performed using 4-hydroxy indole (Sigma) as substrate. About 500 mg of tissue was ground using liquid nitrogen and the total crude protein was extracted using 100 mM sodium acetate buffer (pH 4.5). The 300 μL reaction mixture contained 10 μL of crude enzyme and 100 μL 5 mM 4-hydroxy indole. The reaction was set up at 30°C for 30 min and the absorbance was measured at 620 nm.
Lignin Quantification
Cell wall components were enriched by grinding ∼300 mg of rice stem tissues in liquid nitrogen. The resulting powder was suspended in 1.5 mL of methanol and stirred vigorously for 1 h. The sample was centrifuged at 12,000 rpm for 5 min at 25°C. The pellet was consecutively treated with 1.5 mL of the following solvents: methanol (twice), 1 M NaCl, 1% w/v SDS, water (twice), ethanol, chloroform:methanol (1:1), and tert-butyl methyl ether, with 15 min stirring followed by 5 min centrifugation as previously mentioned. The pellet thus obtained was dried at 80°C for 12 h to obtain alcohol insoluble residue (AIR).
The lignin was quantified by derivatization with thioglycolic acid. To 20 mg of AIR, 1 mL of 2 M HCl and 0.2 mL of thioglycolic acid were added and incubated for 4 h at 95°C with continuous stirring. The samples were rapidly cooled on ice and centrifuged at 12,000 rpm for 10 min at 25°C. Supernatant was discarded and the pellet was washed with 1.5 mL of water thrice by disturbing the pellet, followed by centrifugation and discarding the supernatant. The resulting pellet was resuspended in 1 mL of 0.5 M NaOH and was vigorously shaken overnight at 25°C to extract the lignin thioglycolate. Following centrifugation at 12,000 rpm for 10 min at 25°C, the supernatant was collected and the pellet was resuspended in 0.5 mL of 0.5 M NaOH and centrifuged again. The supernatant obtained was pooled with the supernatant obtained in the previous step. To the combined alkali extract, 0.3 mL of concentrated HCl was added and the sample was incubated at 4°C for 4 h to facilitate lignin thioglycolic acid (LTGA) precipitation. The samples were centrifuged at 12,000 rpm for 10 min and a brown pellet was obtained. The supernatant was discarded and the pellet was dissolved in 1 mL of 0.5 M NaOH. The absorbance was measured at 340 nm and the standard curve was plotted using alkali lignin (Sigma).
Phloroglucinol Staining
Stem and leaf sections were stained with phloroglucinol to visualize lignin deposited in the secondary cell walls. The staining solution used was 2% (w/v) phloroglucinol in 95% (v/v) ethanol. The sections were immersed in the stain for 2 min. The sections were then transferred to 10 N HCl and mounted in 5 N HCl and observed under a bright field microscope (Olympus, BX43).
Laccase Extraction and Purification
Around 20 g of tissue was crushed in liquid nitrogen and incubated in 200 mL of extraction buffer (50 mM Tris-HCl, pH 8, 1.5 M CaCl2 and 1 M NaCl) for 2 h at 4°C on a rocker. The samples were centrifuged at 5000 rpm for 30 min at 4°C and supernatant was collected and subjected to dialysis in 100 mM sodium acetate buffer (pH 4.5) overnight at 4°C. Laccase purification was performed using concanavalin-A (Sigma-Aldrich) beads at 4°C. The column was washed for 4 column volumes with wash buffer (1 M NaCl, 5 mM MgCl2, 5 mM MnCl2, 5 mM CaCl2) and the column was regenerated with regeneration buffers (0.1 M Tris-HCl buffer pH 8.5 with 0.5 M NaCl and 0.1 M sodium acetate buffer pH 4.5 with 1 M NaCl) passed through the column alternatively 4-5 times (started and ended with Tris-HCl buffer). The column was equilibrated for 4 column volumes with equilibration buffer (50 mM Tris-HCl buffer pH 7.5 with 0.5 M NaCl, 1 mM MgCl2, 1 mM CaCl2 and 1 mM MnCl2). The dialyzed sample was passed through the column and following 4 washes in equilibration buffer to wash out the unbound protein, the bound protein was eluted with 50 mM mannose. The presence of laccases was confirmed through LC-MS analysis. In-gel staining of laccases was performed for 2 h, in a 10% (w/v) native gel with separated laccases using 5 mM 4-hydroxy indole (Sigma, a substrate for laccase).
Accession Numbers
High throughput sRNA sequencing from seedling, flagleaf and panicle samples, as well as RNA-seq data has been deposited in Gene Expression Omnibus (GEO). The super series containing both small RNA-Seq (GSE111440) and RNA-Seq (GSE111438) can be accessed through GSE111472.
Supplemental Data
Supplemental Figure 1. Small RNA profiling across selected wild and cultivated rice lines.
Supplemental Figure 2. Phylogenetic analysis of miR397 precursor across plant species and alignment of miR397 sequences from miRBase.
Supplemental Figure 3. Heatmap for differentially expressing miRNAs across three samples derived from rice lines.
Supplemental Figure 4. Phylogenetic relationship between laccases from rice and Arabidopsis.
Supplemental Figure 5. Laccase cascade silencing loci in wild species of rice.
Supplemental Figure 6. Genome browser screen shots of OsLAC3, OsLAC6 and OsLAC19 expression across rice lines.
Supplemental Figure 7. Habit and lignin accumulation in diverse rice lines.
Supplemental Figure 8. Molecular and phenotypic analysis of miR397 OE and LAC3 OE transgenic plants.
Supplemental Figure 9. Overlapping of yield-related QTLs with laccases and miR397.
Supplemental Table 1. Library statistics of sRNA data sets.
Supplemental Table 2. Library statistics of RNA-Seq data sets.
Supplemental Table 3. Primers and oligonucleotide probes used in the study.
Supplemental Table 4. Phenotypic analysis of miR397 OE transgenic plants.
Supplemental Data Set 1. Differentially expressed sRNA loci across selected rice lines.
Supplemental Data Set 2. Differentially expressed miRNAs across rice samples.
Supplemental Data Set 3. Differentially expressed genes across flagleaf samples of rice lines.
Supplemental Data Set 4. Differentially expressed genes across panicle samples of rice lines.
Supplemental Data Set 5. Expression of laccases in flagleaf and panicle samples across rice lines.
Supplemental Data Set 6. Overlapping between rice yield-related QTLs, laccases and miR397.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
The authors acknowledge access to proteomics, radiation lab, central imaging, greenhouse, and sequencing facilities from the host Institute. sRNA and RNA-seq was performed at Genotypic Technologies, Bangalore. Thanks to Prof. David Baulcombe for RDR6i seeds; Prof. K. Veluthambi for pBIN19 vector, Agrobacterium strain LBA4404 (pSB1), PB-1 and WP seeds and suggestions; and Dr. Dhandapani and many farmers for rice seeds. This work is dedicated to Prof. K.S. Krishnan. P.V.S. acknowledges support from Ramanujan Fellowship (SR/S2/RJN-109/2012; Department of Science and Technology, Government of India). P.V.S.’s lab is supported by NCBS-TIFR core funding and a grant from the Department of Biotechnology, Government of India (BT/PR12394/AGIII/103/891/2014). S.C. acknowledges a fellowship from DBT, India.
AUTHOR CONTRIBUTIONS
P.V.S. designed all experiments, discussed results, and wrote the manuscript. S.C. performed most of the bioinformatic analysis, most of the experiments, and wrote the manuscript with P.V.S. V.T. performed infiltration experiments. D.B. made transgenic rice lines and performed their molecular analysis. A.N. and M.P. performed protein analysis. K.P. performed sections and laccase assays.
Footnotes
Articles can be viewed without a subscription.
References
- Axtell M.J., Jan C., Rajagopalan R., Bartel D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. (2004). RNA silencing in plants. Nature 431: 356–363. [DOI] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102: 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet S., Demont-Caulet N., Pollet B., Bidzinski P., Cézard L., Le Bris P., Borrega N., Hervé J., Blondet E., Balzergue S., Lapierre C., Jouanin L. (2011). Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12: 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond D.M., Baulcombe D.C. (2014). Small RNAs and heritable epigenetic variation in plants. Trends Cell Biol. 24: 100–107. [DOI] [PubMed] [Google Scholar]
- Bonnet E., He Y., Billiau K., Van de Peer Y. (2010). TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 26: 1566–1568. [DOI] [PubMed] [Google Scholar]
- Campo S., Peris-Peris C., Siré C., Moreno A.B., Donaire L., Zytnicki M., Notredame C., Llave C., San Segundo B. (2013). Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 199: 212–227. [DOI] [PubMed] [Google Scholar]
- Chávez Montes R.A., de Fátima Rosas-Cárdenas F., De Paoli E., Accerbi M., Rymarquis L.A., Mahalingam G., Marsch-Martínez N., Meyers B.C., Green P.J., de Folter S. (2014). Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat. Commun. 5: 3722. [DOI] [PubMed] [Google Scholar]
- Chen H.-M., Li Y.-H., Wu S.-H. (2007). Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 3318–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-M., Chen L.-T., Patel K., Li Y.-H., Baulcombe D.C., Wu S.-H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 107: 15269–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C. (2010). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17: 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Zhao P.X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39: W155-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippes F.F., Weigel D. (2009). Triggering the formation of tasiRNAs in Arabidopsis thaliana: the role of microRNA miR173. EMBO Rep. 10: 264–270 (Groszmann, M., Greaves, I.K., Albertyn, Z.I., Scofield, G.N., Peacock, W.J., and Dennis). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M., Greaves I.K., Albertyn Z.I., Scofield G.N., Peacock W.J., Dennis E.S.; E.S. (2011). Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc. Natl. Acad. Sci. USA 108: 2617–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Gui Y., Wang Y., Zhu Q.-H., Helliwell C., Fan L. (2008). Selection and mutation on microRNA target sequences during rice evolution. BMC Genomics 9: 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick J.B., Chandler V.L. (2001). Genetic factors required to maintain repression of a paramutagenic maize pl1 allele. Genetics 157: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston K., McKim S.M., Comadran J., Bonar N., Druka I., Uzrek N., Cirillo E., Guzy-Wrobelska J., Collins N.C., Halpin C., Hansson M., Dockter C., et al. (2013). Variation in the interaction between alleles of HvAPETALA2 and microRNA172 determines the density of grains on the barley inflorescence. Proc. Natl. Acad. Sci. USA 110: 16675–16680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M.D., Fahlgren N., Chapman E.J., Cumbie J.S., Sullivan C.M., Givan S.A., Kasschau K.D., Carrington J.C. (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q., Min L., Yang X., Jin S., Zhang L., Li Y., Ma Y., Qi X., Li D., Liu H., Lindsey K., Zhu L., et al. (2018). Laccase GhLac1 Modulates Broad-Spectrum Biotic Stress Tolerance via Manipulating Phenylpropanoid Pathway and Jasmonic Acid Synthesis. Plant Physiol. 176: 1808–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo X., Wu S., Zhu Z., Liu F., Fu Y., Cai H., Sun X., Gu P., Xie D., Tan L., Sun C. (2017). NOG1 increases grain production in rice. Nat. Commun. 8: 1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T. (2008). The Process of Rice Domestication: A New Model Based on Recent Data. Rice (N. Y.) 1: 127–134. [Google Scholar]
- Izawa T., Konishi S., Shomura A., Yano M. (2009). DNA changes tell us about rice domestication. Curr. Opin. Plant Biol. 12: 185–192. [DOI] [PubMed] [Google Scholar]
- Jeong D.-H., Park S., Zhai J., Gurazada S.G.R., De Paoli E., Meyers B.C., Green P.J. (2011). Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23: 4185–4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C., Kasprzewska A., Tennessen K., Fernandes J., Nan G.-L., Walbot V., Sundaresan V., Vance V., Bowman L.H. (2009). Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 19: 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharabian-Masouleh A., Waters D.L.E., Reinke R.F., Ward R., Henry R.J. (2012). SNP in starch biosynthesis genes associated with nutritional and functional properties of rice. Sci. Rep. 2: 557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach M.J., Sweeney M.T., McCouch S.R. (2007). New insights into the history of rice domestication. Trends Genet. 23: 578–587. [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Chen J., Qiu S., Zhang Y., Wang P., Yang L., Lu Y., Shi J. (2012). Deep sequencing and microarray hybridization identify conserved and species-specific microRNAs during somatic embryogenesis in hybrid yellow poplar. PLoS One 7: e43451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Guo K., Zhu X., Chen P., Li Y., Xie G., Wang L., Wang Y., Persson S., Peng L. (2017). Domestication of rice has reduced the occurrence of transposable elements within gene coding regions. BMC Genomics 18: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M., Davis E., Gardner D., Cai X., Wu Y. (2006). Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 224: 1185–1196. [DOI] [PubMed] [Google Scholar]
- Liu Q., Luo L., Wang X., Shen Z., Zheng L. (2017). Comprehensive analysis of rice laccase gene (OsLAC) family and ectopic expression of OsLAC10 enhances tolerance to copper stress in Arabidopsis. Int. J. Mol. Sci. 18: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Li Q., Wei H., Chang M.-J., Tunlaya-Anukit S., Kim H., Liu J., Song J., Sun Y.H., Yuan L., Yeh T.F., Peszlen I., et al. (2013). Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. USA 110: 10848–10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Dai X., Xu Y., Luo W., Zheng X., Zeng D., Pan Y., Lin X., Liu H., Zhang D., Xiao J., Guo X., et al. (2015). COLD1 confers chilling tolerance in rice. Cell 160: 1209–1221. [DOI] [PubMed] [Google Scholar]
- Manavella P.A., Koenig D., Weigel D. (2012). Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. USA 109: 2461–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K., Tör M., Poole M., Hong Y., Thompson A.J., King G.J., Giovannoni J.J., Seymour G.B. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38: 948–952. [DOI] [PubMed] [Google Scholar]
- Meyer R.S., Purugganan M.D. (2013). Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14: 840–852. [DOI] [PubMed] [Google Scholar]
- Niederhuth C.E., Schmitz R.J. (2014). Covering your bases: inheritance of DNA methylation in plant genomes. Mol. Plant 7: 472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinçon G., Maury S., Hoffmann L., Geoffroy P., Lapierre C., Pollet B., Legrand M. (2001). Repression of O-methyltransferase genes in transgenic tobacco affects lignin synthesis and plant growth. Phytochemistry 57: 1167–1176. [DOI] [PubMed] [Google Scholar]
- Qin C., Yu C., Shen Y., Fang X., Chen L., Min J., Cheng J., Zhao S., Xu M., Luo Y., Yang Y., Wu Z., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 111: 5135–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranocha P., Chabannes M., Chamayou S., Danoun S., Jauneau A., Boudet A.-M., Goffner D. (2002). Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar. Plant Physiol. 129: 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T.D., Stephens R.M. (1990). Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18: 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach F., Vaistij F.E., Jones L., Baulcombe D.C. (2005). An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138: 1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad P.V., Chen H.-M., Patel K., Bond D.M., Santos B.A.C.M., Baulcombe D.C. (2012a). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad P.V., Dunn R.M., Santos B.A., Bassett A., Baulcombe D.C. (2012b). Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J. 31: 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridevi G., Dhandapani M., Veluthambi K. (2005). Agrobacterium-mediated transformation of White Ponni, a non-basmati variety of indica rice (Oryza sativa L.). Curr. Sci. 88: 128–132. [Google Scholar]
- Sridevi G., Parameswari C., Sabapathi N., Raghupathy V., Veluthambi K. (2008). Combined expression of chitinase and β-1,3-glucanase genes in indica rice (Oryza sativa L.) enhances resistance against Rhizoctonia solani. Plant Sci. 175: 283–290. [Google Scholar]
- Stocks M.B., Moxon S., Mapleson D., Woolfenden H.C., Mohorianu I., Folkes L., Schwach F., Dalmay T., Moulton V. (2012). The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 28: 2059–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Liu X., Yang J., Liu W., Du Q., Wang H., Fu C. Li W.-X. (2018). microRNA528 affects lodging resistance of maize by Rrgulating lignin biosynthesis under nitrogen-luxury conditions. Mol. Plant. 11: 806–814. [DOI] [PubMed]
- Sweeney M., McCouch S. (2007). The complex history of the domestication of rice. Ann. Bot. 100: 951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-Y., Zhang S., Yu Y., Luo Y.-C., Liu Q., Ju C., Zhang Y.-C., Qu L.-H., Lucas W.J., Wang X., Chen Y.-Q. (2014). MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol. J. 12: 1132–1142. [DOI] [PubMed] [Google Scholar]
- Wang Y., Bai X., Yan C., Gui Y., Wei X., Zhu Q.-H., Guo L., Fan L. (2012). Genomic dissection of small RNAs in wild rice (Oryza rufipogon): lessons for rice domestication. New Phytol. 196: 914–925. [DOI] [PubMed] [Google Scholar]
- Willmann M.R., Poethig R.S. (2007). Conservation and evolution of miRNA regulatory programs in plant development. Curr. Opin. Plant Biol. 10: 503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing R.A., Purugganan M.D., Zhang Q. (2018). The rice genome revolution: from an ancient grain to Green Super Rice. Nat. Rev. Genet. 19: 505–517. [DOI] [PubMed] [Google Scholar]
- Wu L., Zhang Q., Zhou H., Ni F., Wu X., Qi Y. (2009). Rice MicroRNA effector complexes and targets. Plant Cell 21: 3421–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J., Cho L.-H., Kim S.L., Choi H., Koh H.-J., An G. (2014). The BEL1-type homeobox gene SH5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J. 79: 717–728. [DOI] [PubMed] [Google Scholar]
- Yu Y., Li Q.-F., Zhang J.-P., Zhang F., Zhou Y.-F., Feng Y.-Z., Chen Y.-Q., Zhang Y.-C. (2017). Laccase-13 Regulates Seed Setting Rate by Affecting Hydrogen Peroxide Dynamics and Mitochondrial Integrity in Rice. Front. Plant Sci. 8: 1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-C., Yu Y., Wang C.Y., Li Z.Y., Liu Q., Xu J., Liao J.Y., Wang X.J., Qu L.H., Chen F., Xin P., Yan C., et al. (2013). Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 31: 848–852. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Nakashima J., Chen F., Yin Y., Fu C., Yun J., Shao H., Wang X., Wang Z.-Y., Dixon R.A. (2013). Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25: 3976–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Li J. (2014). Molecular dissection of complex agronomic traits of rice: a team effort by Chinese scientists in recent years. Natl. Sci. Rev. 1: 253–276. [Google Scholar]