GHR1 is a receptor-like pseudokinase that activates SLOW ANION CHANNEL1 and acts in stomatal closure through its scaffolding functions rather than by directly phosphorylating its target proteins.
Abstract
Guard cells control the aperture of stomatal pores to balance photosynthetic carbon dioxide uptake with evaporative water loss. Stomatal closure is triggered by several stimuli that initiate complex signaling networks to govern the activity of ion channels. Activation of SLOW ANION CHANNEL1 (SLAC1) is central to the process of stomatal closure and requires the leucine-rich repeat receptor-like kinase (LRR-RLK) GUARD CELL HYDROGEN PEROXIDE-RESISTANT1 (GHR1), among other signaling components. Here, based on functional analysis of nine Arabidopsis thaliana ghr1 mutant alleles identified in two independent forward-genetic ozone-sensitivity screens, we found that GHR1 is required for stomatal responses to apoplastic reactive oxygen species, abscisic acid, high CO2 concentrations, and diurnal light/dark transitions. Furthermore, we show that the amino acid residues of GHR1 involved in ATP binding are not required for stomatal closure in Arabidopsis or the activation of SLAC1 anion currents in Xenopus laevis oocytes and present supporting in silico and in vitro evidence suggesting that GHR1 is an inactive pseudokinase. Biochemical analyses suggested that GHR1-mediated activation of SLAC1 occurs via interacting proteins and that CALCIUM-DEPENDENT PROTEIN KINASE3 interacts with GHR1. We propose that GHR1 acts in stomatal closure as a scaffolding component.
INTRODUCTION
Stomata optimize photosynthetic carbon dioxide uptake with minimal water loss. Guard cells, which form the stomatal pores, allow plants to sense and respond to diverse environmental and endogenous stimuli by adjusting the aperture of stomatal pores. Stomatal aperture decreases in response to drought, low light intensity, low air humidity, elevated intercellular CO2 concentration, pathogens, and the air pollutant ozone (O3). Adjustment of stomatal aperture is achieved by turgor changes resulting from ion transport across the guard cell plasma- and vacuole membranes (Hedrich, 2012). Ion channels and transporters are the primary targets of guard cell signaling networks, and their activity determines the aperture of stomatal pores (Kim et al., 2010; Roelfsema et al., 2012; Kollist et al., 2014; Song et al., 2014). Upstream signaling events involve a complex network of interactions involving the phytohormone abscisic acid (ABA), cytoplasmic calcium (Ca2+), and reactive oxygen species (ROS; Sierla et al., 2016).
During the past decades, several molecular components involved in regulating stomatal movement have been identified in the model plant Arabidopsis thaliana. These components include the guard cell slow-type anion channel, SLOW ANION CHANNEL1 (SLAC1; Negi et al., 2008; Vahisalu et al., 2008) and the core ABA signalosome, composed of the ABA receptors PYRABACTIN RESISTANCE1 (PYR)/REGULATORY COMPONENTS OF ABA RECEPTORS (RCAR; Ma et al., 2009; Park et al., 2009), a group of type 2C protein phosphatases (PP2Cs; Umezawa et al., 2009; Vlad et al., 2009), and the SUCROSE NON-FERMENTING1 (SNF1)-related protein kinase OPEN STOMATA1 (OST1; Mustilli et al., 2002). ABA binding-induced conformational changes in the PYR/RCAR receptors lead to the inhibition of PP2Cs, which allows the activation of the protein kinase OST1 (Ma et al., 2009; Park et al., 2009; Soon et al., 2012). OST1 is then thought to activate SLAC1 via phosphorylation to initiate stomatal closure (Geiger et al., 2009; Lee et al., 2009; Vahisalu et al., 2010; Brandt et al., 2012). Several other kinases, including Ca2+-dependent protein kinases (CPK3, CPK5, CPK6, CPK21, and CPK23), the leucine-rich repeat receptor-like kinase GUARD CELL HYDROGEN PEROXIDE-RESISTANT1 (GHR1), and Calcineurin B-like (CBL)/CBL-interacting protein kinase (CIPK) complexes, have also been implicated in SLAC1 activation (Geiger et al., 2010; Brandt et al., 2012, 2015; Hua et al., 2012; Scherzer et al., 2012; Maierhofer et al., 2014). Although it is clear that phospho-regulation of SLAC1 channel activity is central to stomatal movements, in planta evidence for these events is still mostly lacking.
Genetic studies have established that SLAC1 and OST1 define the core pathway regulating rapid stomatal responses to most closure-inducing cues (Mustilli et al., 2002; Negi et al., 2008; Vahisalu et al., 2008, 2010; Xue et al., 2011; Hedrich and Geiger, 2017). Other components, including ROS produced by the NADPH oxidases RBOHD and RBOHF (Sierla et al., 2016), PYR/RCAR receptors and PP2Cs of the ABA signalosome (Merilo et al., 2013; Chater et al., 2015), and GHR1 (Hua et al., 2012) are also involved in multiple stomatal responses, while others are pathway-specific. For example, the Raf-like protein kinase HIGH LEAF TEMPERATURE1 (HT1) is a CO2-specific guard cell regulator (Hashimoto et al., 2006; Hõrak et al., 2016). Functional redundancy within the 34-member CPK family has hampered their genetic analysis; however, analysis of single and higher order mutants has confirmed a role for certain CPKs in ABA-mediated activation of S-type anion channels in guard cells, which is also reflected in impaired ABA-induced stomatal closure (Mori et al., 2006; Brandt et al., 2015).
Cell surface receptors, such as the plant receptor-like kinases (RLKs), are common components in signal perception and transduction systems. The leucine-rich repeat (LRR) RLKs represent the largest subclass of RLKs (Shiu and Bleecker, 2003; Gou et al., 2010). Typically, the LRR-RLKs consist of an extracellular LRR receptor domain, a transmembrane region, and a cytoplasmic kinase domain. Activation of LRR-RLKs usually involves ligand-induced heterodimerization with co-receptors, followed by transphosphorylation and further activation of the intracellular kinase modules. Activated receptor kinase complexes initiate further signaling events via interaction with downstream signaling components (Couto and Zipfel, 2016; Hohmann et al., 2017). Such complexes also function in stomatal signaling, e.g., flg22-induced stomatal closure involves the pattern recognition receptor FLAGELLIN-SENSITIVE2 (FLS2), the co-receptor BRASSINOSTEROID INSENSITIVE1-ASSOCIATED RECEPTOR KINASE1 (BAK1) and the cytoplasmic kinase BOTRYTIS-INDUCED KINASE1 (BIK1) (Kadota et al., 2014; Li et al., 2014).
At least 10% of all human and Arabidopsis protein kinases and around 20% within the Arabidopsis RLK family lack key residues essential for catalytic kinase activity and are therefore termed pseudokinases (Manning et al., 2002; Castells and Casacuberta, 2007). Accumulating evidence demonstrates that pseudokinases are integral components of signaling pathways and function primarily as kinase scaffolds (either directly or indirectly, through other scaffold proteins) or as allosteric modulators of other signaling components (Langeberg and Scott, 2015). Although the mode of action of plant receptor-like pseudokinases remains mostly unknown, it is clear that they control various biological processes. For example, pseudokinases BAK1-INTERACTING RECEPTOR-LIKE KINASE2 (BIR2) and CORYNE (CRN) mediate receptor complex formation during immune responses and stem cell homeostasis, respectively (Blaum et al., 2014; Halter et al., 2014a, 2014b; Somssich et al., 2015, 2016). POLLEN-SPECIFIC RECEPTOR-LIKE KINASE5 (PRK5) acts as a receptor for a peptide ligand (Wrzaczek et al., 2015), and RECEPTOR DEAD KINASE1 acts as a positive regulator in plant responses to ABA during seedling development through the recruitment of ABA INSENSITIVE1 (ABI1) to the plasma membrane (Kumar et al., 2017).
Apoplastic ROS are signaling molecules involved in both intra- and intercellular signaling during various inducible and developmental processes (Suzuki et al., 2011; Sierla et al., 2013; Waszczak et al., 2018). The air pollutant O3 can be used as a convenient tool to generate apoplastic ROS to study their function. Stomatal conductance determines the effective O3-dose and critical flux rate into the leaf intercellular airspace. Therefore, impaired stomatal regulation can result in altered responses to O3 (Kangasjärvi et al., 2005). A link between O3-sensitivity and stomatal function has been recognized for more than four decades (Mansfield, 1973), but the molecular mechanisms involved remain mostly undefined. Mutant screens for O3 sensitivity have revealed components involved in ROS-related signaling cascades, antioxidant biosynthetic pathways (Conklin et al., 2000; Overmyer et al., 2008), and components in stomatal regulation (Overmyer et al., 2008; Vahisalu et al., 2008, 2010). Here we describe the isolation of Arabidopsis mutants for the LRR-RLK GHR1 from two independent forward genetic screens for O3 sensitivity. We show that GHR1 is an indispensable component required for stomatal movements. Our results show that GHR1 is a pseudokinase and suggest that CPK3 is an interaction partner of GHR1. We propose that GHR1 acts in stomatal closure via SLAC1 activation mediated by the scaffolding functions of GHR1 involving interacting kinases rather than by direct phosphorylation.
RESULTS
Isolation and Mapping of an Ozone-Sensitive Mutant
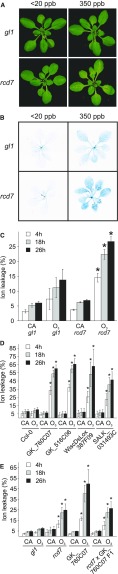
In a previous mutant screen of ethyl methanesulfonate (EMS) mutagenized Arabidopsis of the O3-tolerant Col-0 glabrous1 accession, a set of apoplastic ROS-sensitive radical-induced cell death (rcd) mutants was identified (Overmyer et al., 2000, 2008). Analyses of these mutants have revealed components important for cellular functions, including the transcription factor-interacting protein RCD1 that functions as a hub for hormonal and stress signaling (Jaspers et al., 2010; Vainonen et al., 2012) and the guard cell anion channel SLAC1 (Vahisalu et al., 2008), originally identified as rcd3. From the same screen another mutant, rcd7, was isolated. Upon treatment with 350 ppb O3 for 6 h (Figure 1), rcd7 displayed increased tissue damage compared to the wild-type Col-0 glabrous1 (gl1; Figure 1A). Increased cell death was visualized by trypan blue staining for dead and dying cells and quantified by electrolyte leakage (Figures 1B and 1C; Supplemental Data Set 1). In whole-rosette (Kollist et al., 2007) gas exchange measurements, rcd7 appeared to have slightly higher steady state stomatal conductance compared to wild type (Supplemental Figure 1A), prompting further studies of stomatal function in this mutant.
Figure 1.
Phenotypes of the rcd7 Mutant and Candidate Insertion Mutants and Allelism Tests.
(A) Representative photographs of 3-week-old O3-treated (350 ppb) and clean air (CA) control (<20 ppb) Col-0 gl1 and rcd7 plants taken 18 h after the end of a 6 h exposure to O3.
(B) Trypan blue staining for dead and dying cells performed 18 h after the end of O3 exposure.
(C) to (E) Electrolyte leakage measured 4, 18, and 26 h after the end of O3 exposure. Values are plotted as % of total ion content. At least three independent experiments consisting of four plants per line for each treatment were performed with similar results. Data from a representative experiment is shown. Data are presented as mean ± sd (n = 4 plants). Asterisks indicate statistically significant differences to O3-treated Col-0 gl1 (C and E) or Col-0 (D) (ANOVA with Tukey’s honestly significant difference [HSD] test, P < 0.05).
Segregation analysis of an F2 backcross population of rcd7 (rcd7 gl1 x Col-0) revealed recessive inheritance (Supplemental Table 1A). To identify the rcd7 locus, we generated a mapping population by outcrossing with the C24 accession and established genetic linkage of rcd7 to markers N482S and ciw7 in the lower arm of chromosome 4. Whole genome resequencing revealed possible causal mutations in ten genes within this region (Supplemental Table 1, B and C). Analysis of the insertion mutants of the candidate genes for O3 sensitivity revealed that the line SALK_031493c carrying a T-DNA insertion within At4g20940, a gene encoding a leucine-rich repeat receptor-like kinase (LRR-RLK), exhibited severe O3-damage (Supplemental Table 1C and Supplemental Figure 1B).
During the mapping, a second independent mutation conferring O3 sensitivity and high stomatal conductance was identified segregating in the rcd7 mutant background. The phenotypes of the second mutant were caused by the dominant Ala109Val mutation in the protein kinase HT1, ht1-8D, as described by Hõrak et al. (2016). The mutations were genetically separated and analyzed independently, and the rcd7 lines analyzed here (Figure 1; Supplemental Figure 1) were found to lack this second mutation.
Three additional T-DNA lines for At4g20940 exhibited lesion formation and increased electrolyte leakage in response to O3 (Figure 1D; Supplemental Figure 1C). Allelism tests between rcd7 and a T-DNA allele (GK_760C07) revealed a lack of complementation, confirming that the Ala618Thr mutation in At4g20940 in rcd7 conferred its O3-sensitivity (Figure 1E; Supplemental Figure 1D). As expected, the O3-sensitivity phenotype of the F1 generation was similar to that of rcd7, whereas the T-DNA lines for At4g20940 exhibited severe O3-sensitivity phenotypes (Figure 1E; Supplemental Figure 1D).
Therefore, the loss of the LRR-RLK III 45 (RCD7; AT4G20940) resulted in O3-sensitivity, likely mediated by the associated disruption of stomatal function. At the time of these findings, Hua et al. (2012) reported the identification and characterization of the plasma membrane receptor-like kinase GUARD CELL HYDROGEN PEROXIDE-RESISTANT1 (GHR1). Both RCD7 and GHR1 correspond to At4g20940; therefore, RCD7 will be hereafter referred to as GHR1 (for allele nomenclature, see Supplemental Table 2).
Loss of RCD7/GHR1 Impairs Stomatal Closure
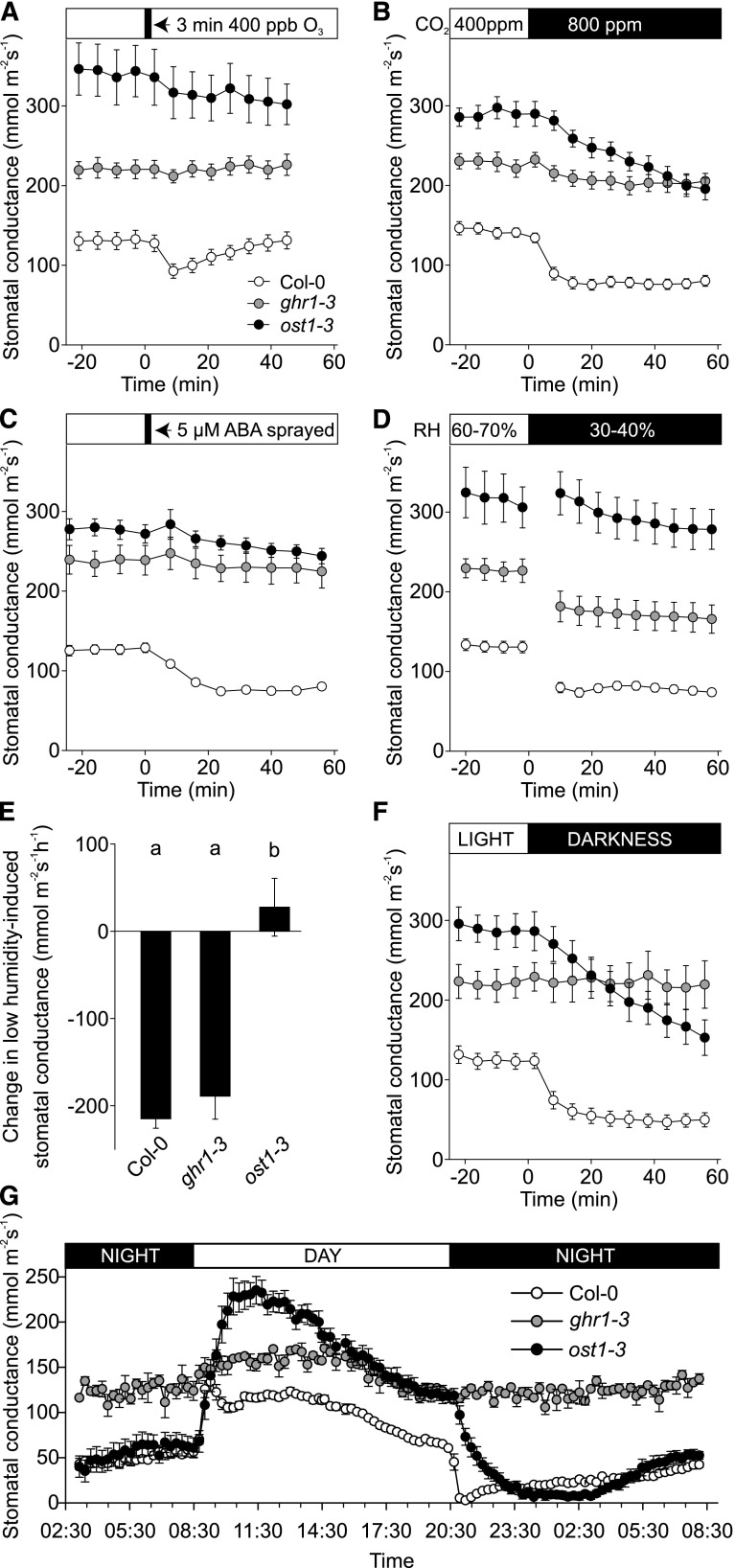
One critical factor that determines the extent of O3 damage in plants is stomatal function, and hence, we assayed the responses of the ghr1 mutants to several stomatal closure-inducing signals. The loss-of-function allele ost1-3 (Yoshida et al., 2002) of the protein kinase OST1, a key regulator of the guard cell anion channel SLAC1, was used as a control in all stomatal measurements. Whole-rosette gas exchange measurements were performed for the deletion mutant ghr1-1 (Hua et al., 2012) and for the T-DNA insertion mutants ghr1-3, ghr1-4, and ghr1-6. Since the phenotypes of all lines were consistent, representative results of the detailed analysis of stomatal function are shown only for ghr1-3 (Figure 2). Daytime steady state stomatal conductance in ghr1-3 was higher than that of Col-0 wild type, but lower than in ost1-3 (Figures 2A to 2D, 2F, and 2G). Rapid OST1-dependent stomatal closure in response to O3 reflects the fast regulation of wild-type Col-0 guard cells by apoplastic ROS (Figure 2A; Kollist et al., 2007; Vahisalu et al., 2010). In the ghr1-3, as in ost1-3 plants (Figure 2A), a three-minute O3 pulse was ineffective at eliciting this stomatal response, indicating that GHR1 is required for the induction of stomatal closure by apoplastic ROS.
Figure 2.
Characterization of ghr1 Stomatal Phenotypes.
Time course of stomatal conductance of Col-0, ghr1-3, and ost1-3 plants in response to (A) O3 pulse, (B) elevated CO2, (C) ABA spray, (D) reduced air humidity, and (F) darkness. Stomatal conductance of 3- to 4-week-old plants was recorded; the indicated treatments were applied at time point zero. Data points represent means ± sem of at least three experiments (n = 8-21 plants). (E) Change in stomatal conductance 16 min after decrease in relative air humidity, calculated based on the data presented in (D). Significant differences (ANOVA with unequal N HSD as post hoc, P < 0.01) between groups are denoted with different letters. (G) Diurnal cycle in stomatal conductance of Col-0, ghr1-3 and ost1-3 plants. Stomatal conductance was recorded for two consecutive days, and diurnal stomatal conductance patterns for the second day are shown. Data points represent mean ± sem (n = 3–8 plants).
In addition, ghr1-3 stomata exhibited impaired responses to elevated CO2, ABA, and light-dark transitions. When the CO2 concentration was elevated from 400 to 800 ppm, ghr1-3 stomata exhibited no response, while the response of ost1-3 stomata was delayed in comparison to wild type (Figure 2B). ABA treatment caused a pronounced decline in stomatal conductance in wild-type plants, whereas the ghr1-3 and ost1-3 mutants were ABA-insensitive (Figure 2C). Stomata of the ghr1-3 plants responded to a reduction in relative air humidity, whereas ost1-3 plants had a strongly impaired response (Figures 2D and 2E). Thus, signaling through GHR1 is more important than that through OST1 for CO2-induced stomatal closure, both proteins are essential for the ABA response, and stomatal closure induced by reduced air humidity appears to be less dependent on GHR1 than on OST1.
Darkness during the light period induced a rapid decline in stomatal conductance in wild-type plants. This process was delayed in ost1-3, whereas the ghr1-3 mutant was non-responsive (Figure 2F). In plants undergoing diurnal light/dark transitions, the onset of the dark period triggered a rapid drop in conductance in wild type and a delayed response in ost1-3 plants, whereas stomatal conductance did not decrease in ghr1-3 plants. Moreover, throughout the night period, stomatal conductance in ghr1-3 remained similar to that observed during the day, and it was ∼2.5-fold higher compared with that of wild type and ost1-3 (Figure 2G).
Thus, in addition to acting in apoplastic ROS- and ABA-induced stomatal closure (Figures 2A and 2C; Hua et al., 2012), GHR1 is an essential component required for stomatal closure provoked by ozone-induced apoplastic ROS, CO2 and light/dark transitions.
Is the Function of GHR1 in Stomatal Responses Based on Its Kinase Activity?
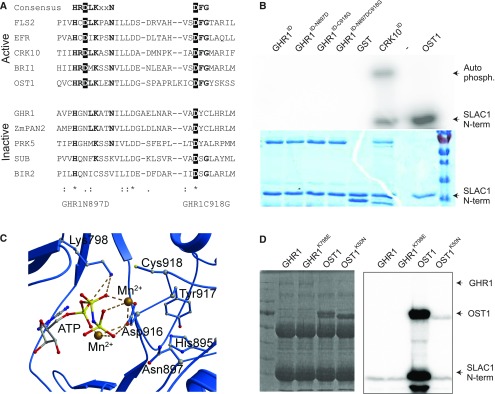
GHR1 consists of 1053 amino acids with a calculated molecular weight of 113.89 kD. Based on cDNA sequencing, GHR1 has a different gene structure from that predicted in TAIR (release 10). Based on the cDNA sequences (Gou et al., 2010; Hua et al., 2012), the correct gene model has different splice sites for the second intron and translation start site 48 base pairs (bp) upstream of the predicted model (TAIR release 10). The N-terminal extracellular domain of GHR1 harbors a signal peptide and 19 predicted LRRs, while the C-terminal intracellular domain contains a kinase domain. However, two of the most critical residues required for kinase function (Figure 3), the aspartate (D) of the HRD motif in subdomain VI B and the glycine (G) of the DFG motif in subdomain VII (Hanks et al., 1988; Stone and Walker, 1995; Hanks, 2003; Kornev et al., 2006), are not conserved (Figure 3A). The lack of these two amino acid residues suggests that GHR1 may not be able to perform a phosphorylation function via classically understood mechanisms.
Figure 3.
GHR1 Kinase Domain and Activity.
(A) Alignment of subdomains VIb and VII of the catalytic core of the kinase domains of active and inactive RLKs and OST1. Residues highlighted in black are considered indispensable for kinase activity. Bold residues are highly conserved in active kinases.
(B) Phosphorylation activity assays of GHR1 intracellular domain (GHR1ID) and the indicated mutant proteins in in vitro kinase assays using γ32P-ATP and SLAC1 N-terminal (N-term) fragment (residues 1–186) as a substrate. GST-CRK10ID and 6xHIS-OST1 were used as positive controls. Protein amount used for the assay was 2 µg apart from 6xHis-OST1, for which 0.04 µg was used. Autoradiograph and Coomassie Brilliant Blue G 250 staining are shown.
(C) Structural prediction of the catalytic core of the intracellular kinase domain of GHR1. Predicted interactions of Lys798 and Asp916 of GHR1 with ATP and Mn2+ ions are shown.
(D) Phosphorylation activity assays of full-length GHR1-TAP, OST1-TAP and ATP binding site mutants GHR1K798E-TAP and OST1K50N-TAP immunoprecipitated from yeast cell extract toward SLAC1 N-terminal fragment. Coomassie Brilliant Blue G 250 staining and autoradiograph are shown.
To determine whether GHR1 is an active kinase, we expressed the full-length GHR1 protein and its intracellular domain (GHR1ID; residues 653 to 1053) as N-terminal GLUTATHIONE-S-TRANSFERASE (GST) or polyhistidine (6xHis)-tagged constructs in E. coli and subjected them to in vitro kinase assays in laboratories at two different universities, under several conditions (see Methods). GST-GHR1ID, GST-GHR1 and 6xHis-GHR1 did not show in vitro kinase activity toward the artificial substrate myelin-basic protein (MBP) or toward the physiologically relevant (Hua et al., 2012) SLAC1 N-terminal domain (Figure 3B; Supplemental Figures 2A to 2D). Under the same conditions, 6xHis-OST1, GST-OST1 (Geiger et al., 2010) and the GST-tagged intracellular domain of CYSTEINE-RICH RLK10 (CRK10ID; Bourdais et al., 2015) were active in both auto- and substrate phosphorylation (Figure 3B; Supplemental Figures 2A to 2D). Introduction of an aspartate (present in the consensus HRD motif of active kinases) into GHR1ID (GHR1ID-N897D) by site-directed mutagenesis did not reconstitute its kinase activity (Figure 3B; Supplemental Figures 2A and 2B). We also generated constructs harboring the GHR1ID-C918G mutation alone or in combination with GHR1ID-N897D. Such mutations have been previously shown to restore the kinase activity of the pseudokinase PRK5 (Wrzaczek et al., 2015), an LRR-RLK of the same subclass as GHR1. However, the introduction of these mutations, either separately or in combination, did not result in the gain of autophosphorylation activity or activity toward SLAC1 N terminus or MBP (Figure 3B; Supplemental Figures 2A and B).
We then performed homology modeling to probe the structure of the GHR1 kinase domain for the signatures of an active kinase. Querying the Structural Bioinformatics Protein Data Bank (RCSB PDB; Bluhm et al., 2011) with the sequence of the GHR1 kinase domain (residues 776 to 1053) produced two major groups of structures, including (1) structures of the plant receptor-like kinase BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1 (BAK1, PDB accession nos. 3UIM, 3ULZ, and 3TL8) and (2) structures of the BRASSINOSTEROID INSENSITIVE 1 (BRI1) kinase domain (PDB accession nos. 4OA2, 4OH4, 4Q5J, 4OA6, 4OA9, 4OAB and 4OAC). The BRI1 structure 4OA9 (Bojar et al., 2014) contained not only AMP, but also two Mn2+ cations and was chosen as a template for modeling of the kinase domain of GHR1. Structurally, the GHR1 kinase domain resembles that of a standard kinase, with smaller N-terminal lobes, larger C-terminal lobes, and no obvious structural abnormalities (Supplemental Figure 3B). According to the predicted structure of the catalytic core of GHR1, the key residues for ATP binding (K798 and D916) are present in the active site and could potentially interact with ATP (Figure 3C). The HGN motif of GHR1 (variation of the conserved HRD motif) also lies in the active site. However, whether the lack of the conserved aspartate of this motif in GHR1 disrupts substrate binding, phosphotransfer, or otherwise interferes with the structure-function of the protein was not apparent from the model.
We then tested whether the posttranslational modifications (PTM) occurring only in eukaryotic cells may be necessary for GHR1 kinase activity. Full-length GHR1 and control constructs were expressed and immunoprecipitated from yeast cell extract prior to in vitro kinase assays. GHR1K798E, where the ATP binding lysine was substituted by glutamate to prevent association of ATP with the N-lobe of GHR1, was used as a negative control. OST1 displayed autophosphorylation and transphosphorylation of the SLAC1 N terminus, whereas GHR1 displayed weak residual activity comparable to that of the kinase-dead OST1K50N and GHR1K798E (Figure 3D). Immunoblot analysis of yeast cell lysates confirmed the expression of all proteins (Supplemental Figure 3C).
In conclusion, although the kinase fold of GHR1 resembles that of active kinases, the absence of critical residues in the active site and the lack of in vitro kinase activity displayed by GHR1 suggest that GHR1 might not be a functionally active kinase.
GHR1 Induces SLAC1 Anion Currents in Xenopus Oocytes
Based on the lack of key residues in its kinase domain, like its maize (Zea mays) counterpart PANGLOSS2 (PAN2; Zhang et al., 2012), GHR1 can be classified as a pseudokinase (Castells and Casacuberta, 2007; Zeqiraj and van Aalten, 2010; Eyers and Murphy, 2013). The lack of activity observed in the in vitro kinase assays also supports this classification. However, GHR1 has been shown to activate the SLAC1 anion channel in African clawed frog (Xenopus laevis) oocyte assays (Hua et al., 2012). To investigate whether kinase activity in GHR1 is required for SLAC1 activation, we co-expressed wild-type and mutant versions of GHR1 together with SLAC1 in the Xenopus oocyte system and monitored anion channel currents (Figure 4).
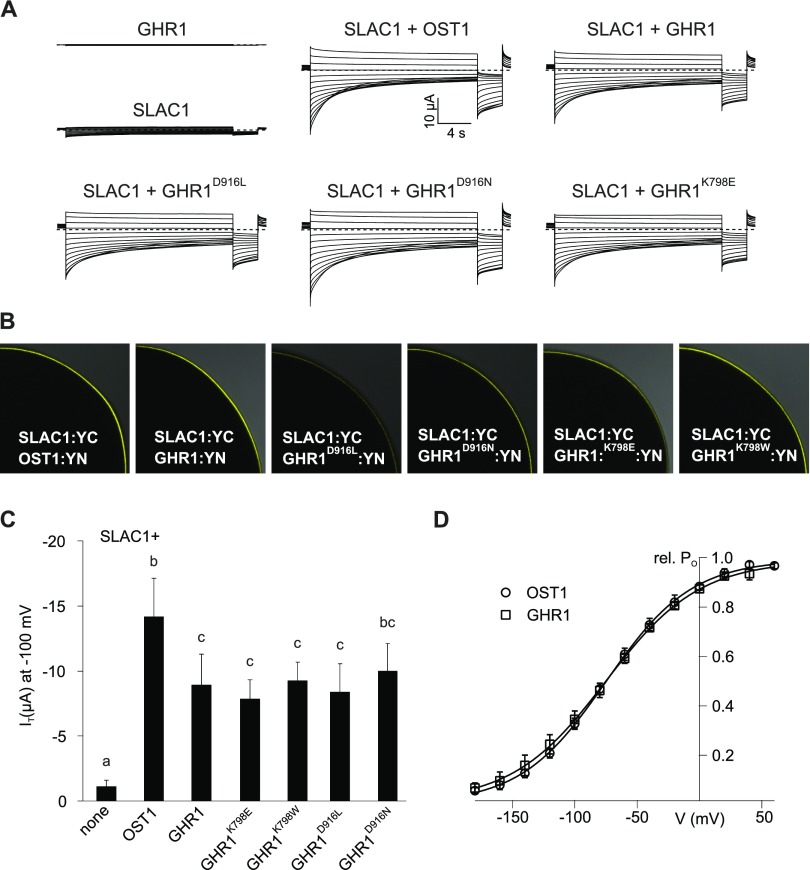
Figure 4.
Activation of SLAC1 Anion Currents by GHR1 Variants That Contain Mutations in Residues Essential for ATP Binding in Xenopus laevis Oocytes.
(A) Representative whole oocyte current traces from cells expressing GHR1 or SLAC1 alone or SLAC1 together with OST1, wild-type GHR1 or GHR1D916L, GHR1D916N and GHR1K798E.
(B) BiFC analysis following co-expression of SLAC1:YC with OST1:YN, GHR1:YN or GHR1D916L:YN, GHR1D916N:YN, GHR1K798E:YN and GHR1K798W:YN in oocytes. One-quarter of a representative oocyte is shown.
(C) Instantaneous currents (IT) recorded at -100 mV in standard buffer of oocytes expressing SLAC1 or GHR1 WT alone or co-expressing SLAC1 with OST1, GHR1 WT or one of the GHR1 mutants GHR1D916L, GHR1D916N, GHR1K798E, or GHR1K798W:YN. Four biological repeats, each performed with oocytes from different batches, were performed. Data of a representative experiment are shown. All data points are mean ± sd (n ≥ 4 oocytes). Significant differences (ANOVA with Tukey’s HSD test, P < 0.05) between groups are denoted with different letters.
(D) Relative voltage-dependent open probability (rel. PO) of oocytes co-expressing SLAC1 with either OST1 or GHR1. Oocytes were perfused with buffers containing 30 mM nitrate. Data points were fitted with a Boltzmann equation (continuous line). All data points are mean ± sd (n = 4 oocytes). Data presented in Figure 4 and Figure 9 derive from the same series of experiments. The control data presented for GHR1:YN and SLAC1:YC (expressed alone or co-expressed) are the same between the figures.
Two different substitutions, GHR1D916L and GHR1D916N, were introduced to replace the aspartate, which is the only conserved amino acid residue of the GHR1 DFG-domain and one of the most important residues for catalysis, as it forms polar contacts with the phosphates of ATP (Figure 3C). In active kinases, these mutations prevent phosphotransfer. In other constructs, the ATP binding lysine was substituted by glutamate or tryptophan, thereby preventing association of ATP with the N-lobe of GHR1 (GHR1K798E, GHR1K798W; Figure 3C). Based on the commonly used approach, SLAC1 and GHR1 variants were fused to split halves of YFP (Hua et al., 2012; Hõrak et al., 2016; in relation to OST1 activation of SLAC1 e.g., Geiger et al., 2009; Brandt et al., 2012) and cRNA combinations were injected into Xenopus oocytes (Figures 4A and 4B). No anion currents were measurable using oocytes expressing SLAC1:YC or GHR1:YN alone (Figures 4A and 4C). However, typical SLAC1-like anion currents were detected (Figures 4A and 4C) upon co-expression of SLAC1:YC with either OST1:YN (positive control), wild-type GHR1:YN or any of the four GHR1 mutants where amino acids required for ATP binding were mutated. Just like OST1:YN, GHR1:YN shifted the voltage-dependent open probability of SLAC1:YC to hyperpolarized membrane potentials (Figure 4D, Maierhofer et al., 2014), thereby activating SLAC1-derived anion currents. These results show that GHR1 is able to activate SLAC1 in oocytes even when the critical ATP binding amino acids required for the phosphorylation reaction had been mutated.
Therefore, in agreement with evidence presented before, the results from in vitro kinase assays support the classification of GHR1 as a pseudokinase. Furthermore, based on functional analysis in Xenopus oocytes, any potential residual kinase activity that might not be detectable in in vitro kinase assays is not required for the function of the protein as an activator of SLAC1.
GHR1 Binds ATP In Vitro, but ATP Binding Is Not Required for Its Function In Planta
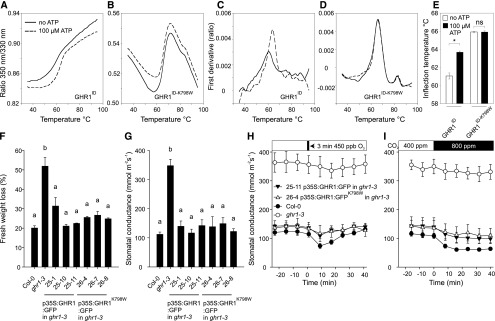
Despite the catalytically inactive nature of most pseudokinases, ∼40% of them, as also predicted for GHR1 (Figure 3C), retain the capacity to bind nucleotides (Hámmaren et al., 2015b; Murphy et al., 2014). Therefore, we studied the ability of the intracellular domain of GHR1 to bind ATP using thermal shift analysis with nano Differential Scanning Fluorimetry (nanoDSF; Figure 5). The presence of ATP significantly shifted the inflection temperature for GST-GHR1ID during the nanoDSF analysis, indicating that GHR1ID can bind ATP and that this interaction stabilizes the protein (Figures 5A, 5C, and 5E). However, the mutant variant of GHR1ID lacking the conserved lysine required for ATP binding, GHR1ID-K798W, did not show any changes in inflection temperature, indicating that GHR1ID-K798W does not bind ATP (Figures 5B, 5D, and 5E).
Figure 5.
Analysis of ATP Binding of GHR1ID and GHR1ID-K798W and Characterization of Stomatal Phenotypes in Independent Transgenic Lines Expressing 35S:GHR1:GFP or 35S:GHR1K798W:GFP in ghr1-3.
(A) and (B) Ratio of 350 nm/330 nm, which represents an unfolding transition of the proteins due to thermal treatment in the absence (solid lines) or presence (dashed lines) of ATP.
(C) and (D) The first derivative of the data presented in (A) and (B) showing a shift in inflection temperature upon ATP binding in the case of GHR1 intracellular domain (GHR1ID) (C), whereas in the case of GHR1ID-K798W, inflection temperatures are the same (D).
All experiments were repeated 6 times; representative curves are shown.
(E) Inflection temperatures for GHR1ID and GHR1ID-K798W in the absence or presence of ATP. The bars represent average value and standard deviations (n = 6). Asterisk denotes statistically significant difference between treatments (independent-samples t test, P < 0.05).
(F) Leaf fresh weight loss in 2 h and (G) stomatal conductance of intact plants. Data are presented as mean ± sem (n = 3-5 plants). Significant differences (ANOVA with Tukey’s HSD test, P < 0.05) between lines are denoted with different letters.
(H) and (I) Time course of stomatal conductance in response to (H) O3 pulse and (I) elevated CO2 in representative transgenic lines. Stomatal conductance of 3- to 4-week-old plants was recorded; the indicated treatments were applied at time point zero. Data points represent means ± sem (n = 3–5 plants).
Furthermore, we used a transgenic approach to test whether ATP binding, and thus any putative kinase activity, is required for the function of GHR1 in planta. We transformed the ghr1-3 mutant with 35S-GHR1-GFP and the active site mutant 35S-GHR1K798W-GFP. Three independent transgenic lines for each construct were analyzed, and all lines showed full complementation of the increased water loss and high steady state stomatal conductance of ghr1-3 (Figures 5F and 5G). All lines also showed complementation of the O3 and CO2 responses of ghr1-3; however, the responses of the transgenic lines appeared to be slightly weaker than those of the wild-type (Figures 5H and 5I; Supplemental Figures 4A to 4D). GFP fluorescence was apparent in the periphery of the guard cells in all transgenic lines (Supplemental Figure 4E), indicating native localization of the fusion proteins.
Thus, although the wild-type GHR1 can bind ATP, this is not required for its function in planta. Therefore, we conclude that GHR1 does not require kinase activity for SLAC1 activation (Figure 4) or for its function in stomatal closure in Arabidopsis.
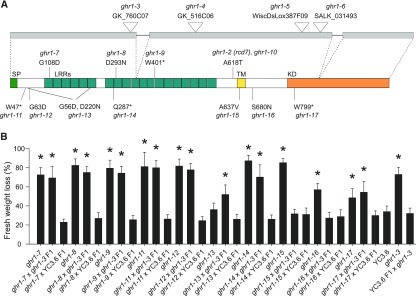
A Screen for Stomatal Regulators Identifies New GHR1 Alleles
The identification of GHR1 variants containing single amino acid changes or protein truncations could potentially give insight into the structure-function relationship of the protein. In a second independent screen for O3 sensitive mutants aimed at identifying novel stomatal regulators, numerous new mutants were retrieved. Sequencing of the sequences encoding proteins related to stomatal responses in this collection revealed 10 additional alleles of GHR1 (Figure 6A). In addition, a second independent double mutant, ghr1-10 ht1-9D, carrying mutations identical to the original ghr1-2 ht1-8D was identified. The water loss phenotypes in all ten lines were recessive, and eight of them did not complement the phenotype of ghr1-3 (Figure 6B), showing that these eight mutations were responsible for the mutant phenotype.
Figure 6.
Additional ghr1 Alleles.
(A) Structure of the GHR1 protein (bottom) and its encoding gene (top). Position of T-DNA insertions, EMS point mutations, and predicted functional domains of the protein are depicted. Gray boxes, exons; SP, signal peptide; LRR, leucine-rich repeat; TM, transmembrane domain; KD, kinase domain.
(B) Leaf fresh weight loss in 2 h in the indicated ghr1 mutants and the F1 progenies from their respective crosses to ghr1-3 (GK_760C07) and Col-0 with GC1:YC3.6. Four independent experiments consisting of at least ten plants for each line were performed. Data from a representative experiment is shown. All data points are mean ± sd (n = 11–16 plants). Significant differences (ANOVA with Tukey’s HSD test, P < 0.01) from YC3.6 are denoted with asterisks.
Although ATP Binding Is Dispensable, the Kinase Domain Is Required but Not Sufficient for GHR1 Function In Planta
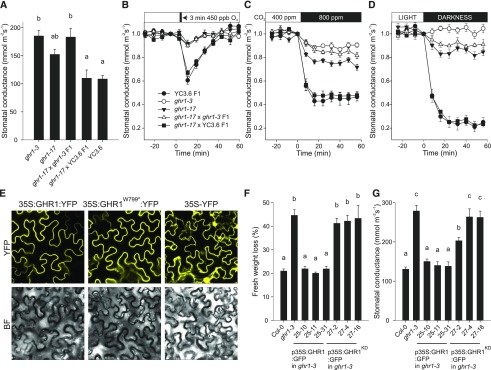
The mutation GHR1W799* in the ghr1-17 allele (Figure 6A) introduces a stop codon in the kinase domain of GHR1 immediately after the ATP binding lysine (GHR1K798), leading to a truncated protein lacking most of the kinase domain. The loss-of-function phenotype of this allele indicates that the kinase domain of GHR1 is required for the function of the protein in planta. Similar to the water loss phenotype of the ghr1-17 allele (Figure 6B), its steady state stomatal conductance (Figure 7A) and stomatal responses to O3, CO2, and darkness were recessive and allelic to ghr1-3 (Figures 7B to 7D). However, apart from the lack of O3-response, the phenotypes displayed by ghr1-17 were not as severe as those of ghr1-3. To verify the expression and correct subcellular targeting of the truncated protein in planta, GHR1 and GHR1W799* were expressed as YFP fusions in stable transgenic Arabidopsis plants and transiently expressed in Nicotiana benthamiana leaf tissue. Immunoblot analysis confirmed the expression of the transgenes (Supplemental Figure 5C), and the strength and localization of the YFP fluorescence signal at the cell periphery were similar between GHR1-YFP and GHR1W799*-YFP (Figure 7E; Supplemental Figure 5D). RT-qPCR analysis revealed that GHR1 transcript levels were lower in ghr1-17 compared to the wild type (Supplemental Figures 5A and 5B). It is thus likely that the phenotype of ghr1-17 is mainly due to the lack of the kinase domain; however, the slightly reduced transcript levels could also contribute to the phenotype in a minor way, even though the abundance of the truncated protein is similar to that of the wild type (Supplemental Figure 5C).
Figure 7.
Functional Domains of GHR1.
(A) to (D) Characterization of stomatal phenotypes of ghr1-17 and the F1 progeny from its cross to ghr1-3 (GK_760C07) and Col-0 with GC1:YC3.6. (A) Stomatal conductance of intact plants. Data are presented as mean ± sem (n = 5–7 plants) and derive from two independent batches of plants. Significant differences (ANOVA with Tukey’s HSD test, P < 0.05) between lines are denoted with different letters. (B) to (D) Stomatal response of intact plants to (B) O3 pulse, (C) elevated CO2, (D) darkness. Stomatal conductance (expressed in relative units) of 3- to 4-week-old plants was recorded; at time point zero, the indicated treatments were applied. Data points represent means ± sem (n = 5–7 plants).
(E) Subcellular localization of 35S:GHR1:YFP, 35S:GHR1W799*:YFP and 35S:YFP fusion proteins in Nicotiana benthamiana epidermal cells. Representative images for each construct taken with identical confocal microscopy acquisition settings are shown.
(F) Leaf fresh weight loss in 2 h and (G) stomatal conductance of intact plants in independent transgenic lines expressing 35S:GHR1:GFP or 35S:GHR1KD:GFP in ghr1-3. Data are pooled from two independent experiments and are presented as mean ± sem (n = 6–10 plants in [F], n = 6-8 plants in [G]). Significant differences (ANOVA with Tukey’s HSD test, P < 0.05) between lines are denoted with different letters.
To address whether the GHR1 kinase domain alone is sufficient for functional complementation in planta, we generated transgenic ghr1-3 lines expressing GFP fusions of full-length GHR1 or the GHR1 kinase domain only (GHR1KD; residues 764-1053) and analyzed their water loss and gas exchange phenotypes. GHR1KD-GFP fusion protein appeared to accumulate in multiple locations within the cell, including the nucleus, but also at the cell periphery (Supplemental Figure 5G). Unlike the wild-type GHR1-GFP protein, GHR1KD -GFP did not complement the ghr1-3 mutant (Figures 7F and 7G; Supplemental Figures 5E and 5F), indicating that the extracellular part, the transmembrane domain, and the intracellular juxtamembrane domain (adjacent to one side of the membrane) are necessary for the function of GHR1. However, we cannot rule out the possibility that the change in localization could contribute to the lack of complementation, although the signal from the cell periphery was clearly visible (Supplemental Figure 5G). Taken together, these data indicate that the kinase domain of GHR1 is necessary but not sufficient for its function in planta.
GHR1 Interacts with SLAC1 and CPK3 Independent of Its Kinase Domain
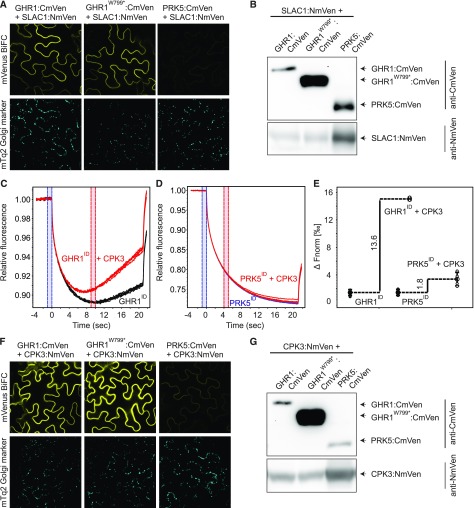
We performed bimolecular fluorescence complementation (BiFC; Gookin and Assmann, 2014) analysis to investigate whether the loss-of-function phenotype of ghr1-17 was due to loss of interaction between GHR1 and SLAC1. The closely related LRR-RLK PRK5 was used as negative control. Strong reconstituted mVenus signal was apparent in the cell periphery upon expression of the constructs bearing SLAC1 and wild-type GHR1 or GHR1W799* (Figure 8A) indicating that the interaction of GHR1 with SLAC1 in planta did not require the kinase domain of GHR1. Little to no signal was detected when constructs carrying SLAC1 and PRK5 coding sequences were expressed (Figure 8A). Immunoblot analysis confirmed expression of GHR1, GHR1W799*, PRK5, and SLAC1 (Figure 8B).
Figure 8.
Interaction of GHR1 with SLAC1 and CPK3.
(A) and (F) Bimolecular fluorescence complementation assays were performed with N. benthamiana leaves infiltrated with 35S:GHR1:CmVen, 35S:GHR1W799*:CmVen or 35S:PRK5:CmVen with 35S:SLAC1:NmVen or 35S:CPK3:NmVen. mVenus (top row) and mTq2 (bottom row) signals are shown. Expression of the mTq2 Golgi marker (bottom row) confirms that transformation of all constructs was successful. N. benthamiana leaf tissue was imaged 48 h after transformation. Identical settings were used for all constructs allowing direct comparison. All experiments were repeated at least three times, and representative BiFC images are shown.
(B) and (G) Immunoblots showing expression of split-YFP fusion proteins in BiFC samples used for confocal imaging.
(C), (D), and (E) Analysis of in vitro binding of GHR1ID and PRK5ID with CPK3. Interaction between intracellular domain of GHR1 (GHR1ID) and PRK5 (PRK5ID) fused to GST with CPK3 was monitored by microscale thermophoresis (MST). (C) MST traces of 66 nM fluorescently labeled GHR1ID in the absence (black) and presence (red) of 5 µM CPK3. (D) MST traces of 40 nM fluorescently labeled PRK5ID in the absence (blue) and presence (red) of 5 µM CPK3. (E) Difference in normalized fluorescence between GHR1ID or PRK5ID alone and in the presence of CPK3 as indicated. All experiments were performed four times.
As shown above (Figure 3B; Supplemental Figure 2), the in vitro kinase assays did not show detectable activity for GHR1, whereas Hua et al. (2012) demonstrated phosphorylation of the SLAC1 N terminus when GHR1-Myc was expressed in Arabidopsis protoplasts and αMyc-immunoprecipitate was used for the kinase assays. This finding suggests that there may possibly be additional components, such as active protein kinases, that interact and copurify with GHR1. To test this hypothesis, we searched the Membrane-based Interactome Network Database (MIND; Jones et al., 2014) for GHR1 interactors. CALCIUM-DEPENDENT PROTEIN KINASE3 (CPK3), which has previously been shown to phosphorylate SLAC1 (Scherzer et al., 2012; Maierhofer et al., 2014), was identified among the GHR1 interactors. We tested the in vitro interaction between the GHR1 intracellular domain and CPK3 by microscale thermophoresis (MST). We detected a clear increase in normalized fluorescence in the presence of CPK3 for GHR1ID, but not for PRK5ID (Figures 8C to 8E). This result indicates that a complex formed between GHR1ID and CPK3 in vitro. The MST measurements revealed a clear concentration-dependent interaction between GHR1ID and CPK3 (Supplemental Figures 6A to 6C); however, aggregation of the sample at high ligand concentrations prevented precise measurement of the Kd; estimates ranged between 58.9 nM and 89.4 nM (Supplemental Figures 6A to 6C). BiFC analysis confirmed that GHR1 and CPK3 interacted in planta and that the interaction was not dependent on the kinase domain of GHR1 and was absent for PRK5 (Figure 8F). Immunoblot analysis confirmed the expression of GHR1, GHR1W799*, PRK5, and CPK3 (Figure 8G).
Thus, GHR1 interacted with SLAC1 and CPK3 in planta. However, the GHR1 kinase domain was not required for these interactions.
Electrophysiology Provides Structural Insights into GHR1 Function
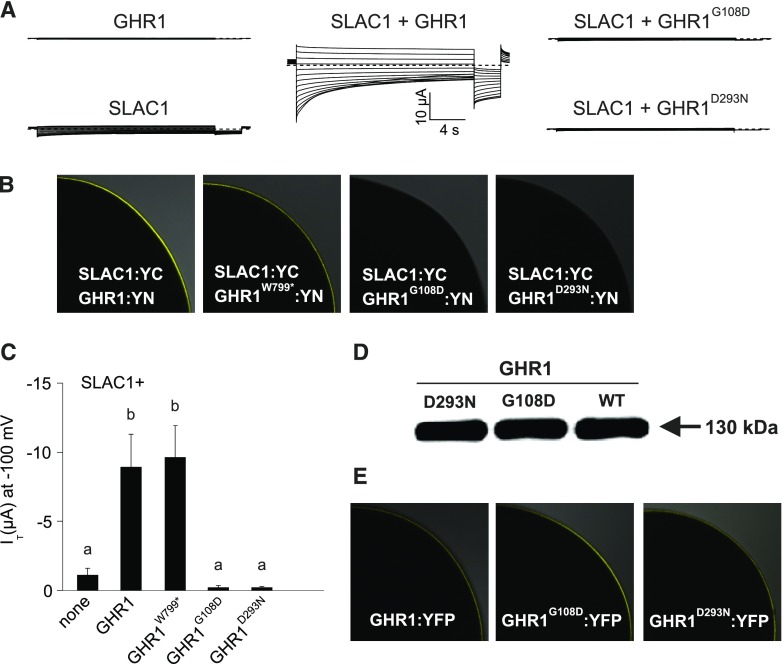
To further address the importance of specific residues and structural elements of GHR1 required for SLAC1 activation, we expressed SLAC1 fused with the C-terminal half of YFP (SLAC1-YC) and GHR1 variants fused with the N-terminal half of YFP (GHR1-YN) in Xenopus oocytes (Figure 9). We addressed the role of the GHR1 N-terminal region (including the ectodomain, transmembrane domain, and intracellular juxtamembrane domain) in SLAC1 activation using a construct bearing the GHR1 truncation lacking the kinase domain (GHR1W799*). GHR1W799*:YN induced SLAC1 currents and interacted with SLAC1:YC on the cell periphery in a manner similar to that of wild-type GHR1 (Figures 9B and 9C).
Figure 9.
Activation of SLAC1 Anion Currents by GHR1 Truncation and Ectodomain Mutants in Xenopus laevis Oocytes.
(A) Representative whole oocyte current traces from cells expressing GHR1 or SLAC1 alone or SLAC1 together with wild-type GHR1 or GHR1G108D and GHR1D293N.
(B) BiFC analysis following co-expression of SLAC1:YC with GHR1:YN or GHR1W799*:YN, GHR1G108D:YN and GHR1D293N:YN in oocytes. One-quarter of a representative oocyte is shown.
(C) Instantaneous currents (IT) recorded at −100 mV in standard buffer of oocytes expressing SLAC1 alone or co-expressing SLAC1 with GHR1 WT or GHR1W799*:YN, GHR1G108D:YN and GHR1D293N:YN. Four biological repeats, each performed with oocytes from different batches, were performed. Data of a representative experiment is shown. All data points are mean ± sd (n ≥ 6 oocytes). Significant differences (ANOVA with Tukey’s HSD test, P < 0.05) between groups are denoted with different letters.
(D) Immunoblot analysis of GHR1 WT:YN, GHR1G108D:YN and GHR1D293N:YN protein levels in oocytes.
(E) Subcellular localization of GHR1 WT:YFP, GHR1G108D:YFP and GHR1D293N:YFP fusion proteins in oocytes. One-quarter of a representative oocyte is shown. Data presented in Figure 4 and Figure 9 derive from the same series of experiments. The control data presented for GHR1:YN and SLAC1:YC (expressed alone or co-expressed) are the same between the figures.
Contrary to the in planta expression experiments conducted in N. benthamiana via Agrobacterium infiltration (Supplemental Figure 7), when expressed in oocytes, the GHR1 ectodomain mutants GHR1G108D:YN and GHR1D293N:YN showed similar protein abundance to the wild-type GHR1:YN (Figure 9D; Supplemental Figure 8). Thus, the Xenopus oocyte studies addressed the importance of the G108 and D293 amino acid residues of GHR1 for SLAC1 activation. No interaction between GHR1G108D:YN or GHR1D293N:YN with SLAC1:YC was detected in oocytes (Figure 9B). In addition, neither mutant was able to induce SLAC1 currents (Figures 9A and 9C). To confirm that the lack of interaction with and activation of SLAC1 by these two ectodomain mutants of GHR1 was not due to mislocalization of the mutant proteins, we analyzed the localization of GHR1G108D:YFP and GHR1D293N:YFP fusion proteins. In oocytes, both mutants localized to the cell periphery, as did wild-type GHR1-YFP fusion protein (Figure 9E).
Thus, in oocytes, GHR1W799* interacted with SLAC1 and activated currents. Moreover, the GHR1 ectodomain residues GHR1G108 and GHR1D293 were required for the interaction and activation of SLAC1.
HT1 Phosphorylates GHR1 at Multiple Sites
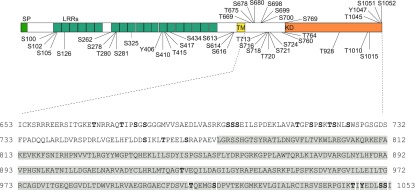
The protein kinase HT1 is one of the few CO2 signaling-specific regulators in guard cells. Thus, an intriguing result from this study is the identification of two independent double mutants from different genetic backgrounds, ghr1-2 ht1-8D and ghr1-10 ht1-9D, containing identical substitutions in GHR1 A618T and in HT1 A109V. We previously showed that GHR1 is a direct phosphorylation substrate for HT1 and that HT1 inhibits the activation of SLAC1 by GHR1 in oocytes (Hõrak et al., 2016). Here, we identified the GHR1 amino acid residues targeted by HT1 in vitro. Purified recombinant GST-GHR1 was incubated with 6xHis-HT1, and the phosphorylation sites were determined by mass spectrometry. A total of 40 GHR1 Ser, Thr, or Tyr residues were phosphorylated by HT1 in vitro (Figure 10; Supplemental Data Set 2). Notably, 16 of the phosphosites were located on the intracellular juxtamembrane domain of GHR1, and seven were located in its pseudokinase domain. In summary, in vitro proteomics analysis demonstrated that HT1, which inhibits the activation of SLAC1 by GHR1 in the oocyte system, phosphorylates GHR1 directly in vitro, and the analysis identified potential sites through which HT1-mediated regulation of GHR1 may occur.
Figure 10.
In Vitro Phosphorylation Sites of GST-GHR1 by 6xHis-HT1 Identified by Mass Spectrometry
Purified recombinant GST-GHR1 was phosphorylated by 6xHis-HT1 in vitro and the phosphorylation sites were subsequently identified by mass spectrometry. The positions of all identified phosphorylation sites are depicted on the GHR1 protein structure (upper panel). Phosphorylation sites located on the intracellular juxtamembrane domain and pseudokinase domain (gray highlighting) are additionally marked (bold letters) on the GHR1 protein sequence (lower panel). SP, signal peptide; LRR, leucine-rich repeat; TM, transmembrane domain; KD, kinase domain. See Supplemental Data Set 2 for MaxQuant Phospho(STY)Sites table.
DISCUSSION
GHR1 Is an Integral Component Required for Stomatal Function
Guard cells sense and respond to a multitude of signals through the activation of various signaling pathways. These pathways often have unique early signaling components, but all ultimately target common elements, such as the anion- and K+ channels that drive stomatal closure (Hedrich, 2012; Hedrich and Geiger, 2017). Large gaps remain in our understanding of the specificity and overlap between stomatal signaling pathways. Here, we identified the LRR-RLK GHR1 as an integral component required for stomatal closure. Our results support a role for GHR1 as an activator of SLAC1, as previously reported (Hua et al., 2012). However, we propose that GHR1 is a pseudokinase that exerts its effect on SLAC1 by acting as a scaffold for additional regulators rather than by direct phosphorylation activity.
The SnRK protein kinase OST1 has long been recognized as a key regulator of guard cell responses that integrates signals from most closure-inducing cues tested to date (Melotto et al., 2006; Vahisalu et al., 2010; Xue et al., 2011; Merilo et al., 2013; Guzel Deger et al., 2015). A role for GHR1 as an integral component required for stomatal function is now emerging, as we demonstrate here the requirement for GHR1 in the control of steady state stomatal conductance and stomatal closure induced by elevated CO2 levels, darkness, diurnal light/dark transitions, and the air pollutant O3, which mimics apoplastic ROS production by NADPH oxidases. Importantly, the total lack of response to CO2 and darkness observed in the ghr1 mutant, as opposed to the delayed response in ost1 (Figure 2), indicates that GHR1 has a relatively larger impact on these responses. Using in vivo gas exchange measurements of intact rosettes (Kollist et al., 2007), we recorded a total absence of responsiveness to diurnal light/dark transitions and to darkness applied during the light period in ghr1-3. These responses have previously been reported to be mostly functional in ghr1-1 (Hua et al., 2012). These diverse results could be due to differences in the experimental setups and methodology used for the measurements or to the different ghr1 alleles used.
Stomatal closure induced by reduced air humidity likely relies on a combination of passive hydraulic movements and active (possibly ABA-mediated) movements (Xie et al., 2006; Bauer et al., 2013; McAdam and Brodribb, 2016; Wang et al., 2017; Merilo et al., 2018). Although ABA-induced stomatal closure was fully impaired in both ost1 and ghr1 plants, the response to reduced air humidity was detected in ghr1, but not in ost1. These data suggest that GHR1 is indispensable for ABA signal transduction leading to stomatal closure, but the same process is dispensable for stomatal closure triggered by a reduction in air humidity. This lends further support to the hypothesis that stomatal responses to reduced air humidity are not solely controlled by ABA signaling in guard cells but may also involve hydropassive components (Wang et al., 2017; Merilo et al., 2018). It thus appears that the signaling cascades in guard cells involving GHR1 and OST1 are at least partially independent of each other and may have other targets in addition to SLAC1.
A strong connection between GHR1 and apoplastic ROS has been suggested (Hua et al., 2012; Devireddy et al., 2018), and genetic analysis indicated that GHR1 acts downstream of ROS production but upstream of Ca2+ channel activation in the Ca2+-dependent ABA signaling pathway (Hua et al., 2012). O3 is degraded to H2O2 in the apoplast, mimicking the activity of RESPIRATORY BURST OXIDASE HOMOLOGs (RBOHs). Thus, the importance of GHR1 in apoplastic ROS signaling was highlighted by the identification of ten EMS mutants representing nine novel alleles of GHR1 from two independent screens for O3 sensitivity. The additional GHR1-mediated stomatal response pathways identified here, which are initiated by elevated CO2 and darkness, both require RBOH-generated ROS (Chater et al., 2015; Zhang et al., 2017) and thus provide further evidence for the role of GHR1 in signaling processes involving apoplastic ROS. How does GHR1 integrate the diverse apoplastic ROS-mediated closure-inducing signals into a functional response? The severity and pervasive nature of stomatal phenotypes in ghr1 mutant plants suggest that GHR1 might play a structural role in connecting diverse components required for proper stomatal function, allowing their regulation in a coordinated manner.
GHR1 Is a Pseudokinase
The in silico, in vitro, and in vivo analyses presented here suggest that GHR1 is a pseudokinase lacking kinase activity. Despite this, the central role of GHR1 in stomatal function is evident from the phenotypes of loss-of-function mutants. Several plant receptor-like pseudokinases, which lack detectable kinase activity, have been shown to act in various biological processes. Mutations in all of these pseudokinase RLKs cause clear phenotypes (Vaddepalli et al., 2011; Zhang et al., 2012; Blaum et al., 2014; Halter et al., 2014a, 2014b; Somssich et al., 2015, 2016; Wrzaczek et al., 2015; Kumar et al., 2017). These findings indicate that, even without kinase activity, pseudokinase RLKs play important roles in specific developmental or inducible processes in plants. Similarly, our results present evidence supporting the notion that GHR1 function does not require kinase activity.
Some pseudokinases retain ATP binding activity independently of phosphotransfer. This ATP binding activity is thought to act as an allosteric switch between active and inactive conformational states (Murphy et al., 2014; Hammarén et al., 2015a, 2015b). Our results show that although wild-type GHR1 can bind ATP, neither phosphotransfer nor ATP binding are necessary for its function, as GHR1 mutants lacking the specific amino acid residues required for ATP binding retained their ability to interact with and activate SLAC1 currents in oocytes. Similarly, a mutation removing the conserved ATP binding lysine (GHR1K798W) and thus the ability of GHR1 to bind ATP (Figures 5A to 5E) did not affect the in planta function of GHR1 in stomatal closure. This finding suggests that GHR1 plays a structural role related to SLAC1 activation in the regulation of stomatal function. This role may be related to signal convergence, as mutations in GHR1 caused severe defects in stomatal movements triggered by almost all internal and external stimuli that function through the activation of SLAC1.
A model for the direct regulation of SLAC1 by GHR1 in planta was proposed by Hua et al. (2012) based on the phosphorylation of the SLAC1 N terminus by a GHR1-Myc fusion protein immuno-purified from Arabidopsis protoplasts. The experimental procedures used to purify GHR1-Myc (Hua et al., 2012) were similar to those commonly used for coimmunoprecipitation and, therefore, it cannot be excluded that the observed effect could have been caused by the copurification of kinases, which could have been responsible for GHR1-mediated activation of SLAC1 in planta. GHR1K798E may interfere with some of these putative interactions, as GHR1K798E-αMyc immunoprecipitate was reported to not phosphorylate SLAC1. CPK3 could be an example of a candidate kinase(s) that relays the signal from GHR1 to SLAC1, as CPK3 was shown to interact with GHR1 in yeast (Jones et al., 2014), in vitro (Figures 8C to 8E) and in planta (Figure 8F), and CPK3 was shown to interact with- and activate SLAC1 by phosphorylating its N terminus (Scherzer et al., 2012). Furthermore, the loss of CPK3 leads to impaired stomatal closure (Mori et al., 2006). The identification of both GHR1 and CPK3 together with the SLAC1 homolog SLAH3 in detergent-resistant membrane fractions (Demir et al., 2013) suggests their co-localization in lipid nanodomains and possible physical and functional connections similar to those demonstrated for other ABA signaling components.
Does GHR1 Function as a Scaffold?
A scaffolding role for the kinase domains of both animal and plant pseudokinases has previously been demonstrated, and the interaction partners are often active kinases (Halter et al., 2014a, 2014b; Langeberg and Scott, 2015). Identification and analysis of the ghr1-17 mutant, which lacks the kinase domain, indicated that this domain is required for GHR1 function in planta. Although this deficiency did not affect the interaction of GHR1 with SLAC1 or CPK3, the loss-of-function phenotype of ghr1-17 could be due to the loss of interaction with thus far unidentified downstream effectors or a potential co-receptor. Analysis of kinase domain mutants for the plant pseudokinases SCRAMBLED (SCM; Kwak et al., 2014) and CRN (Somssich et al., 2016) revealed that while the entire kinase domains or certain residues within them were required for their function in particular pathways, they are dispensable for others, which is likely mediated via interaction with pathway-specific factors. The kinase domains of several canonical kinases and pseudokinases are known to function via their interaction with a co-receptor (Halter et al., 2014a, 2014b; Hohmann et al., 2017). For example, the kinase domain of the pseudokinase BIR2 is an in vitro substrate for BAK1, and the interaction between the proteins is dependent on BAK1 kinase activity (Halter et al., 2014b).
Some of the signaling specificity of GHR1 is likely derived from negative regulation. The protein phosphatase ABI2 (Hua et al., 2012) and the protein kinase HT1 (Hõrak et al., 2016) are thought to negatively regulate GHR1 during ABA- and CO2-induced signaling, respectively. The isolation of two independent double mutants carrying identical mutations in GHR1 and HT1 during this study also points to a functional connection between these proteins. The differential modulation of affinities to interacting proteins through phosphorylation is an established regulatory mechanism of RLKs. The identification of amino acid residues in GHR1 that are phosphorylated by HT1 suggests that HT1 might play a role in regulating these putative interactions. As is typical for plant RLKs (Wang et al., 2005, 2008; Kim and Wang, 2010; Blaum et al., 2014), the intracellular juxtamembrane domain of GHR1 contains multiple potential phosphosites, some of which are targeted by HT1 (Figure 10; Supplemental Data Set 2; Hõrak et al., 2016). The phosphorylation of the juxtamembrane domain in receptor-like pseudokinases could result in the formation of docking platforms for cytosolic factors or modulate the affinities for additional RLKs (Blaum et al., 2014). The negative regulation of GHR1 by HT1 (Hõrak et al., 2016) could be executed through such mechanisms.
What do the experiments in oocytes tell us about the role of GHR1 in the activation of SLAC1? The induction of anion currents by the interaction of GHR1 and SLAC1 suggests that the interaction itself could trigger a conformational change in SLAC1 that modulates the properties of the channel. A kinase-independent activation mechanism for the SLAC1 homolog SLAH3 has been demonstrated involving the electrically silent SLAH1 subunit (Cubero-Font et al., 2016). Similarly, GHR1 could be an auxiliary protein that can gate SLAC1 open by allosteric regulation. Alternatively, GHR1 could function as a scaffold that positions a host kinase in the proximity of SLAC1 to trigger SLAC1 activation via phosphorylation.
When we studied GHR1-mediated activation of SLAC1 in a heterologous system in the absence of other plant-specific factors, it was apparent that the N-terminal region excluding the kinase domain (GHR1W799*) was sufficient for the interaction with and induction of SLAC1 currents (Figure 9). GHR1W799* also retained its ability to interact with SLAC1 in planta. Although the loss-of-function phenotype of ghr1-17 indicated that the kinase domain was required for proper stomatal function, the less severe nature of the phenotypes compared with ghr1-3 suggested that GHR1W799* retained a partial ability for in planta function. This is in line with the observations in oocytes and could be explained by the ability of the truncated protein to modulate the properties of SLAC1 structurally and/or through interaction with a subset of the putative effectors in planta. The interaction with CPK3 is likely mediated via the juxtamembrane domain of GHR1 (Figure 8F) and is thus likely unaffected in ghr1-17. Thus CPK3-mediated activation of SLAC1 could be responsible for some of the residual stomatal function of ghr1-17.
As the SLAH3 anion channel has been shown to be a target of kinase-dependent and -independent activation (Cubero-Font et al., 2016), it is tempting to speculate that SLAC1 could also be activated by two independent mechanisms and that GHR1 could play a role in both. The severity of the phenotypes of ghr1-17 suggests that the kinase domain is required for full GHR1 function in planta, and here we propose that the major role of GHR1 in stomatal closure is based on scaffolding mediated by the kinase domain. This is in line with reports suggesting that while the ectodomains of LRR-RLKs, including pseudokinases, allow their binding to other transmembrane proteins, the signaling specificity may be determined by their intracellular domains (Hohmann et al., 2018a, 2018b).
The two mutations within the extracellular LRR domain (GHR1G108D and GHR1D293N) prevented the interaction with and activation of SLAC1 in oocytes. This observation further highlights the functional importance of the GHR1 ectodomain, but it also serves as an important control verifying the specificity of the oocyte assays. The importance of the extracellular domain for ligand binding and protein-protein interactions has been described for a number of LRR-RLKs, with BRI1, FLS2, and CLAVATA1 (CLV1) being the best-characterized examples (De Smet et al., 2009; Gish and Clark, 2011; Greeff et al., 2012) and BIR3 recently arising as the paradigm for pseudokinases (Hohmann et al., 2018a). The putative role of GHR1D293 and GHR1G108 in protein-protein interactions in planta remains to be addressed under native conditions in Arabidopsis guard cells. The concept of GHR1 as a candidate protein that integrates extra- and intracellular signals is consistent with a recent report demonstrating that structural modifications or signaling events on both sides of the plasma membrane are involved in SLAC1 regulation (Yamamoto et al., 2016).
To fully understand the molecular basis of the role of GHR1 in stomatal function, additional components of the receptor complex will need to be identified: these will likely include co-receptor/s, hitherto unidentified pathway-specific cytosolic factors, and possibly extracellular ligands. It is rather likely that in addition to SLAC1, the complex would also include other channels and transporters that are active in guard cells to allow coordinated regulation of these proteins in membrane-bound protein microdomains during stomatal movements.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana mutants were obtained from the Nottingham Arabidopsis Stock Centre (NASC; http://arabidopsis.info/) unless otherwise noted. A transgenic line expressing the pGC1:YC3.6 (Yang et al., 2008) transgene was obtained from Prof. Julian Schroeder (University of California, San Diego). The ost1-3 (srk2e) seeds were obtained from Prof. Kazuo Shinozaki. Seeds were sown on a 1:1 mixture of peat and vermiculite, vernalized in the dark for two days at 4°C, and germinated for one week, after which the plants were transferred to fresh pots. Plants were grown in controlled growth rooms or chambers (Model SGC120; http://weiss-uk.com) under 12 h light (200 µmol m−2 s−1; OSRAM Lumilux Plus Daylight bulbs)/12 h dark cycle, 22°C/18°C (day/night), 60%/70% relative humidity. Mutant lines (Figures 1D and 1E; Supplemental Figure 1) were genotyped to confirm homozygous plants by PCR using gene-specific primers (Supplemental Data Set 3).
Mutant Screening and Isolation of rcd7 and ghr1 EMS Mutants
Mutagenesis and mutant screening leading to the isolation of rcd7 (ghr1-2) were described previously (Overmyer et al., 2000). The Col-0 glabrous1 (gl1) line, which lacks trichomes (Oppenheimer et al., 1991), was used for mutagenesis to produce plants with a morphological marker to avoid seed contamination and to monitor the success of crosses. All crosses used rcd7/gl1 plants as the pollen acceptor, and the F1 progeny were monitored for the presence of trichomes. The rcd7 mutant was backcrossed to Col-0 and segregation scored in three independent O3-exposures using populations from two independent crosses (418 plants in total). The segregation data were pooled from all experiments. For mapping, rcd7 was outcrossed to the C24 accession; O3 sensitive F2 individuals were selected for linkage analysis with the genetic markers listed in Supplemental Data Set 3. Aseptically grown 7-d-old M3 rcd7/gl1 seedlings were used for DNA isolation using a kit (Qiagen DNeasy Plant Maxi kit; www.qiagen.com), and the DNA was sequenced using the Solid platform at DNA sequencing and Genomics Laboratory, Institute of Biotechnology, University of Helsinki. MAQ software (Li et al., 2008) was used to map the Solid reads to the reference genome (TAIR10 version) and for SNP calling. SNP analysis was performed by developing in-house scripts in R and using the Biostrings and BiomaRt packages (R version 3.0.3). In brief, the SNP list was filtered for the coordinates of the given window, selected for a quality score >20, mapped to exons and introns using gene coordinates from TAIR10 (www.arabidopsis.org), and filtered for homozygosity and consistency with EMS mutagenesis. The mutations causing an amino acid change are reported in Supplemental Table 1C.
The mutants ghr1-7 to ghr1-17 were isolated from a second, separate O3-sensitivity and stomatal function mutant screen. Mutagenesis of Arabidopsis pGC1:YC3.6 (Yang et al., 2008) seeds was performed using 0.4% ethyl methanesulfonate (EMS) for 8 h, as described (Kim et al., 2006). Two-week-old high-density-grown M2 plants were treated with O3 (6 h × 275-350 ppb). Sensitive plants were transplanted and one to two weeks later, rosettes were imaged using a thermal camera (Thermal Imager Optris PI450 with the analysis software, Optris PI Connect, Optris GmbH, Berlin, Germany), and water loss after 2 h was measured from detached leaves. Phenotypes were reconfirmed in the M3 generation before selecting lines for gas exchange measurements.
Lines with gas exchange phenotypes were subjected to candidate gene sequencing by PCR amplification with gene-specific primers (Supplemental Data Set 3) with Phusion DNA polymerase (ThermoFisher Scientific). For each line, amplified genes were pooled and sheared (Covaris S2 ultrasonicator, Covaris) into 800–1000 bp fragments, end repaired, and ligated to barcoded adaptors according to the manufacturer’s protocol (Roche). The final library was size selected (800–1000 bp) as described in (Borgström et al., 2011) and quantified using a Qubit 3.0 fluorometer (ThermoFisher Scientific). Subsequently, the library integrity was analyzed with a 2100 Bioanalyzer (Agilent). Emulsion PCR was performed according to the manufacturer’s instructions and run on a GS FLX+ system (Roche) at DNA Sequencing and Genomics Laboratory, Institute of Biotechnology, University of Helsinki. Data processing and assembly were performed with GS Assembler (Roche). The presence of mutations was assessed manually using Tablet (Milne et al., 2013) and GAP4 (Bonfield et al., 1995) software.
O3 Treatment and Analysis of Leaf Damage
Three-week-old plants were exposed to O3 (350 ppb × 6 h) unless otherwise stated, and damage was visually scored and photographed 24-48 h post exposure. For electrolyte leakage, rosettes were collected into 15 mL of MilliQ water 2 h after the end of exposure, and conductance of the O3 treated and control samples was measured 2 h, 16 h, and 24 h following sample collection with a Mettler conductivity meter (Model FE30; www.mt.com) and analyzed as described (Overmyer et al., 2000). Lactophenol trypan blue staining was performed as described (Koch and Slusarenko, 1990). Rosettes were collected one day post exposure, boiled in staining solution for five minutes, and cleared in chloral hydrate solution. The samples were stored in 60% glycerol before image acquisition with an Epson V750 scanner.
Gas Exchange Measurements
Plants for gas exchange experiments were sown into a 4:2:3 peat:vermiculite:water mixture and grown in custom-made pots (Kollist et al., 2007) in growth cabinets (AR-66LX and AR-22L, Percival Scientific, IA, USA) with 12 h day (23°C) and 12 h night (18°C) cycle at 70% relative humidity and 100–150 μmol m−2 s−1 light. Stomatal conductance measurements were performed with three- to four-week-old plants using a custom-built gas exchange device as described (Kollist et al., 2007). In brief, the plants were inserted into measurement cuvettes where their stomatal conductance was recorded at ∼100 µmol m−2 s−1 light and ∼70% relative air humidity. After stomatal conductance had stabilized, the plants were subject to O3 (400 ppb × 3 min), CO2 elevation (from 400 ppm to 800 ppm), darkness, reduction in relative air humidity (to ∼35%) or 5 µM ABA treatment. ABA was applied by spraying as described previously (Merilo et al., 2015). For diurnal stomatal conductance measurements, the cuvette light was switched on and off so that it mimicked the plant growth day/night regime.
Preparation of Recombinant Proteins
To express the GHR1 intracellular domain (GHR1ID; residues 653 to 1053) as a GST-fusion protein, the coding sequence for GHR1ID, as predicted by a Simple Modular Architecture Research Tool (SMART; http://smart.embl-heidelberg.de/), was cloned with primers introducing EcoRI/NotI sites (Supplemental Data Set 3) into the pGEX-4T-1 vector (GE Healthcare Life Science). This construct was used as template for site-directed mutagenesis with a Phusion Site-Directed Mutagenesis Kit (Thermo Scientific) to generate constructs carrying point mutations in the kinase domain. BL21 cells transformed with expression constructs were induced at OD600 of 0.9 with 500 µM IPTG for 8 h at 28°C. Proteins were purified with Glutathione Sepharose 4B (GE Healthcare) according to the manufacturer’s instructions.
To express full-length GHR1 as a His- or GST-fusion protein, the coding sequence of GHR1 was cloned into either the pET28a vector (Novagen, Merck Millipore) or the pGEX-4T-1 vector (GE Healthcare Life Science). 6xHis-GHR1, 6xHis-SLAC1(1-186), 6xHis-HT1, GST-GHR1, and GST-OST1 were expressed in E. coli BL21(DE3) cells. The cultures were grown in 2xYT medium at 37°C to OD600 ∼0.6. Recombinant protein expression was induced by 0.3 mM IPTG at 16°C for 16 h. 6xHis-tagged proteins were purified by nickel-affinity chromatography. GST-tagged proteins were purified by glutathione-affinity chromatography. For full-length GHR1 and OST1 expression in Saccharomyces cerevisiae W303 (MATa/MATα {leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15} [phi+]), the coding sequences of GHR1, GHR1K798E, OST1, and OST1K50N were cloned into the 2 micron vector pRS426 containing a GAL1 promoter and the C-terminal TAP-tag by exploiting homologous recombination in yeast. Yeast cells were grown in one liter of CSM-Ura medium (Formedium) containing 2% raffinose (Sigma) at 30°C. Recombinant protein expression was induced by 2% galactose (Sigma) for 3 h. The frozen yeast cell pellet was ground in liquid nitrogen with a Retsch MM 400 homogenizer. Lysis buffer (25 mM HEPES, 1 M NaCl, 0.1% NP-40, 1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin, 50 mM NaF, 80 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM EDTA, 30 mM EGTA) was added to the ground yeast cells.
Kinase Analysis and Assays
The kinase domains of GHR1 and a set of well-characterized active and inactive kinases, as predicted by a Simple Modular Architecture Research Tool (SMART; http://smart.embl-heidelberg.de/) were aligned using Clustal Omega (Sievers et al., 2011).
For experiments with the GHR1 intracellular domain, in vitro kinase assays were performed according to Idänheimo et al. (2014), with small modifications. Purified protein (2 µg) was incubated with [γ- 32P]-ATP (6000 Ci/mmol, Perkin Elmer) for 1 h at 30°C in the presence of phosphorylation buffer (50 mM HEPES, pH 7.4, 1 mM DTT, 50 mM NaCl) supplemented with 10 mM Mg2+ and/or 10 mM Mn2+. Additionally, kinase buffer described by Hua et al. (2012) was used. CRK10 (Idänheimo et al., 2014) and OST1 (Vahisalu et al., 2010) were used as positive controls and MBP and the N terminus of SLAC1 (Vahisalu et al., 2010) as substrates. The proteins were separated by 14% SDS-PAGE and exposed on phosphor screens (Fujifilm).
For experiments with full-length GHR1, GHR1, and OST1, kinase activity assays were performed by incubating purified recombinant 6xHis-GHR1 (0.1 μM), GST-GHR1 (0.1 μM), GST-OST1 (0.1 μM), and 6xHis-SLAC1(1-186; 2.5 μM) in reaction buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM MgCl2, 5 mM MnCl2, 500 μM ATP, 1 mM DTT, 0.2 mg/ml insulin and 100 μCi/ml 32P-γ-ATP). C-terminally TAP-tagged GHR1, GHR1K798E, OST1, and OST1K50N were immunoprecipitated from yeast cell extract using 20 μL of IgG beads (IgG Sepharose 6 Fast Flow, GE Healthcare, based on the Sepharose 6 Fast Flow matrix, with human IgG covalently coupled to it). The beads were washed with lysis buffer (25 mM HEPES, 1 M NaCl, 0.1% NP-40, 1mM PMSF, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin, 50 mM NaF, 80 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM EDTA, 30 mM EGTA) and kinase assay buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM MgCl2, 5 mM MnCl2, 500 μM ATP, 1 mM DTT, 0.2 mg/ml insulin, and 100 μCi/ml 32P-γ-ATP). GHR1 and OST1 kinase activity assays were performed by adding washed beads into kinase assay buffer containing purified 6xHis-SLAC1(1-186) (2.5 μM). Reactions were performed at room temperature for 30 min and then stopped by the addition of SDS loading buffer. Proteins were separated by 10% SDS-PAGE and visualized by Coomassie brilliant blue G-250 (Sigma) staining. GHR1 and OST1 activity was determined by autoradiography.
Immunoblot Analysis
For immunodetection of TAP-tagged GHR1 and OST1 or 6xHis-tagged GHR1, 1 μL of yeast cell extract or 1 μL of purified recombinant 6xHis-GHR1 was separated on a 10% SDS polyacrylamide gel. After SDS-PAGE, the proteins were transferred onto a nitrocellulose membrane. Immunodetection of TAP-tagged or 6xHis-tagged proteins was performed with rabbit IgG (1:1000; I5006 Sigma-Aldrich) or anti-6xHis tag antibody [HIS.H8] (ab18184 Abcam) in a 1:1300 dilution, followed by a goat anti-rabbit or mouse HRP-conjugated secondary antibody (1:7500 dilutions; LabAs, Estonia) and chemiluminescent detection.
Mapping of HT1 Phosphorylation Sites in GHR1 by Mass Spectrometry
Purified recombinant GST-GHR1 was phosphorylated by 6xHis-HT1. An aliquot from the kinase reaction was pooled together with SDS-PAGE loading buffer (1:1). Proteins were separated by 10% SDS-PAGE, the gel was stained with Coomassie brilliant blue G-250 (Sigma), and the GHR1 protein band was excised from gel. In-gel digestion of GHR1 protein was performed using Trypsin/P (10 ng μl−1; Sigma). Peptides were purified with C18 StageTips and loaded onto a fused silica emitter (75 μm × 150 mm; Proxeon) packed in house with Reprosil-Pur C18-AQ 3 μm particles (Dr. Maisch HPLC GmbH). Peptides were separated on the Agilent 1200 series nanoflow system (Agilent Technologies) with a 30 min 3%–40% buffer B (0.5% acetic acid/80% acetonitrile) gradient at a flow rate of 200 nl/min, and eluted peptides were sprayed directly into an LTQ Orbitrap XL classic mass-spectrometer (Thermo Electron) with a spray voltage of 2.2 kV. The MS scan range was m/z 300–1,800, and the top five precursor ions were selected for subsequent MS/MS scans. A lock mass of 445.0003 was used for the LTQ Orbitrap to obtain constant mass accuracy during the gradient analysis.
Peptides were identified with the Mascot (http://www.matrixscience.com/) search engine. Peptide mass tolerance of 7 ppm and fragment ion mass tolerance of 0.5 D were used. Two missed cleavage sites for Trypsin/P were allowed. The carbamidomethylation of cysteine was set as a fixed modification. The oxidation of methionine and phosphorylation of serine and threonine were set as variable modifications. Two independent phosphorylation experiments were performed.
Thermal Shift Analysis
Measurements of the unfolding profiles of GST-GHR1 intracellular domain and GST-GHR1ID-K798W kinase domain mutant in the absence or presence of ATP were performed on a Tycho NT.6 instrument. Thermal stability was analyzed by monitoring intrinsic fluorescence of aromatic residues. Upon protein unfolding, aromatic residues buried in the core of the protein are being exposed to the buffer, resulting in a shift of their fluorescence maxima from 330 nm to 350 nm. This shift was presented as 350/330 nm ratio and monitored in a temperature range from 35°C to 95°C. This allowed the inflection point of protein unfolding to be determined. The capillaries were filled with a mixture of 20 μL proteins (0.1 mg/ml) in 50 mM HEPES, pH 8.0, 150 mM NaCl, 10 mM MgCl2 with or without 100 µM ATP. The presence of ATP triggered a shift in the inflection temperature for the wild-type protein, but not for the mutant protein. All experiments were performed in six replicates.
Microscale Thermophoresis
Microscale Thermophoresis (MST) analysis was performed on a NanoTemper Monolith NT.115 instrument using premium coated capillaries at 25°C. GST-GHR1 and GST-PRK5 intracellular domains were fluorescently labeled using the Monolith protein labeling kit RED-MALEIMIDE (Cysteine-reactive) according to the manufacturer’s instructions. After labeling, the proteins were eluted into MST buffer (75 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2), which was also used as assay buffer for the MST experiments. Thermophoresis was performed with excitation power 60% using 66 nM GST-GHR1ID or 40 nM GST-PRK5ID and CPK3 concentrations of 0.1–5000 nM. All experiments were repeated four times.
Generation of Constructs for Localization and BiFC Analysis in Nicotiana benthamiana
For subcellular localization studies, the coding sequences of GHR1 and GHR1W799* were amplified from Col-0 cDNA, and GHR1G108D and GHR1D293N were amplified from ghr1-7 and ghr1-8 cDNA, respectively, and cloned into the GatewayTM pDONRTM/Zeo vector (Thermo Scientific) according to the manufacturer’s instructions. The coding sequences were subsequently recombined into pEarleyGate101 (ABRC stock no. CD3-683) to generate fusions with C-terminal YFP-HA.
The BIFC experiments were performed using the pDOE-05 vector (ABRC stock No. CD3-1905) as described before (Gookin and Assmann, 2014). Cloning into MCS3 of pDOE-05 (35S:X:CmVen210) leads to the addition of at least three amino acids (MGS; when the SanDI enzyme is used for vector linearization) at the protein N terminus, potentially disrupting the signal peptides. To avoid extra N-terminal amino acids upstream of the MCS3 SanDI recognition site, the ATG codon was mutagenized into an ATT codon (Ile). The G to T mutation was introduced by PCR with plasmid-specific reverse complement primers (Supplemental Data Set 3) and Q5 High-Fidelity DNA Polymerase (New England BioLabs) according to the manufacturer’s instructions.
The coding sequences of GHR1, GHR1W799*, PRK5, SLAC1, and CPK3 were amplified from Col-0 cDNA with Phusion High-Fidelity DNA Polymerase (New England BioLabs) according to the manufacturer’s instructions with gene- and MCS-specific primers (Supplemental Data Set 3). To generate parent clones (pDOE5 35S:SLAC1/CPK3:NmVen210 – 35S:X:CmVen210), mutated pDOE-05 vector was linearized with NcoI and BcuI (Thermo Scientific) in MCS1 and recombined with the coding sequences using an In-Fusion HD Cloning Kit (Clontech Laboratories). Parent clones were cleaved with AatII and Eco105I (Thermo Scientific) in MCS3 and recombined with the respective coding sequences.
Expression of Fusion Proteins in Nicotiana benthamiana
Transient expression of fluorescent fusion proteins in Nicotiana benthamiana leaf tissue was performed as described (Wu et al., 2004) with minor modifications. Agrobacterium strain GV3101 containing the binary plasmid was grown in LB medium with the appropriate antibiotics at 28°C for 48 h. The cultures were diluted 10-fold into fresh LB and grown for another 24 h. The cells were centrifuged, washed once in infiltration medium (10 mM MES pH 5.6, 10 mM MgCl2 and 200 μM acetosyringone), and resuspended to a final OD600 of 1.0 (PRK5 constructs) or 0.1 (all other constructs). The cultures were mixed 1:1 (v/v) with P19 strain (Voinnet et al., 2003), which had been prepared the same way, and incubated at room temperature for 1 h before infiltration. Five-week-old N. benthamiana plants were inoculated with a 1 mL needleless syringe. Leaf discs were viewed under a confocal microscope at 2 d post-inoculation.
Confocal Microscopy
Imaging of plant samples was performed using a Leica TCS SP5 II HCS A inverted confocal laser-scanning microscope. YFP was excited with a 514 nm argon laser with the emission detection window set at 529-589 nm. GFP was excited with a 488 nm argon laser with the emission detection window set to 500-515 nm. HCX PL APO 63x/1.2 W Corr/0.17 CS (water) objective was used for imaging Arabidopsis guard cells, while HC PL APO 20x/0.7CS (air) objective was used for imaging Nicotiana benthamiana epidermal cells.
For BiFC imaging, the argon laser line excitation wavelengths 458 nm and 514 nm were used with emission window wavelengths 462-493 nm and 529-589 nm set for mTurquoise2 and mVenus, respectively. Sequential scanning alternating between turquoise and yellow channels was performed to avoid crosstalk between the fluorophores. Image acquisition parameters were kept identical within an experiment. The remainder of the leaf sample was collected and used for protein extraction and immunoblot analysis.
Immunoblot Analysis of Plant Tissue
Frozen leaf samples were ground by shaking using an Ivoclar Vivadent Silamat S6 mixer, followed by extraction in 2% SDS, 50 mM Tris-HCl, pH 7.5, and protease inhibitors (Sigma P9599, 1:100) at 37°C for 20 min. Total protein (70 µg) was loaded onto a 10% SDS polyacrylamide gel, and after electrophoresis, the gel was electroblotted onto a polyvinylidene difluoride (PVDF) membrane. For Nicotiana benthamiana studies, immunological reactions were performed with CmVen (1:10000; Agrisera AS11 1775, Lot 1606) or NmVen (1:2000; Agrisera AS11 1776, Lot 1605) polyclonal antisera before detection with a horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (1:10000; Agrisera AS09 602, Lot 1808). For Arabidopsis studies, immunological reactions were performed with Anti-GFP mouse IgG monoclonal antibody (1:1000; Roche Diagnostics Gmbh 11814460001, Lot 11,751,700) before detection with a Goat Anti-Mouse IgG (H+L) -HRP Conjugate secondary antibody (1:10000; Bio-Rad no. 170-6516, Lot 64,048,329).
Generation of Arabidopsis Transgenic Lines and Their Analysis
The coding sequences of GHR1, GHR1K798W, and GHR1KD were cloned into the pART7 vector in frame with the GFP sequence. Subsequently, the 35S expression cassettes with the GHR1, GHR1K798W, and GHR1KD sequences were re-cloned into the agrobacterial pMLBart vector. The final constructs were introduced into Agrobacterium tumefaciens strain GV3101. Agrotransformation of the ghr1-3 mutant plants was performed using the floral dip method (Bent, 2006). Transgenic plants were selected by spraying with Basta (glufosinate ammonium) as well as by detection of the GFP signal in guard cells. T2 plants were germinated in pots for the gas exchange experiments, sprayed with Basta at the age of five days, and studied at the age of four weeks using a custom-made gas exchange device (Kollist et al., 2007). The fresh weight loss of detached leaves after 2 h of drying was determined for the same plants. Leaves were cut, weighed, and left in open Petri dishes on a laboratory table. The leaves were weighed after 2 h, and the relative fresh weight loss was calculated.
Cloning of GHR1 and GHR1W799* into pEarleyGate101 was described above. The constructs were transformed into Col-0 plants by Agrotransformation, and transgenic lines were selected for Basta-resistance on selection plates (1/2× Murashige and Skoog Medium, 0.7% agar, 7.5 μg/ml Basta, 125 μg/ml Cefotaxime, pH 5.6) and subsequently transferred to soil. Fusion protein expression was confirmed by immunoblot analysis of nine transgenic T1 individuals for each construct. Two representative independent lines for each construct were selected for further work, and fusion protein expression and subcellular localization were confirmed in later generations.
Expression Analysis by qPCR
The expression of GHR1 was measured by RT-qPCR using three biological repeats. Rosettes of five three-week-old plants were pooled for each sample, and the tissue was ground in liquid nitrogen. Total RNA was extracted using a Genejet plant RNA isolation kit and treated with DNaseI (Thermo Fisher Scientific). Five micrograms of RNA was reverse-transcribed with Maxima H Minus Reverse Transcriptase (RT) and Ribolock RNase inhibitor (Thermo Fisher Scientific) in a 20 μL volume. The reactions were diluted to the final volume of 100 μL, 3 μL of which was used as template for PCR using HOT FIREPol EvaGreen qPCR Mix Plus (no ROX; Solis Biodyne). The PCR was performed on the ABI 7900HT Fast RT PCR System (Applied Biosystems) with the following cycle conditions: 95°C 10 min, 40 cycles with 95°C 15 s, 60°C 30 s, 72°C 30 s, and ending with melting curve analysis. Data normalization was performed in qBase 2.0 (Biogazelle) using YELLOW-LEAF-SPECIFIC GENE8 (YLS8), TAP42 INTERACTING PROTEIN OF 41 KDA (TIP41), and PROTEIN PHOSPHATASE 2A SUBUNIT A3 (PP2AA3) as reference genes. A plasmid DNA dilution series was used to determine primer amplification efficiencies in qBase. Primer sequences are listed in Supplemental Data Set 3. The data were log10-transformed and statistically analyzed to evaluate significant differences between the mutant and the wild type by independent-samples t test using IBM SPSS Statistics 24.
Modeling
All protein structure templates were taken from the Protein Data Bank (RCSB PDB; Bluhm et al., 2011), and modeling was performed using MODELER software (Webb and Sali, 2014). Resulting structures were analyzed using the BIOVIA (Accelrys) Discovery Studio molecular modeling environment (www.accelrys.com). Color figures were produced with MOLSCRIPT (Kraulis, 1991) and Raster3D (Merritt and Bacon, 1997). For modeling of the GHR1 kinase domain, residues 776 to 1053 were used (GenBank: AGT59499.1). The region includes the “PKc_like” protein kinase catalytic domain (Conserved Domain Database [CDD], accession no. cl21453; Marchler-Bauer et al., 2015). A search of the RCSB PDB with the sequence of the GHR1 kinase domain produced two major groups of available structures: (1) the group consisting of 3UIM, 3ULZ, and 3TL8, which were structures of the plant receptor-like kinase BAK1 in different settings, and (2) the group consisting of 4OA2, 4OH4, 4Q5J, 4OA6, 4OA9, 4OAB, and 4OAC, which were structures of the BRI1 kinase domain. The 4OA9 structure contained not only AMP, but also two Mn2+ cations, and thus, was chosen as a template for modeling of the kinase domain of GHR1. Alignment between the GHR1 and BRI1 kinase domains was performed manually using (1) the SALIGN multiple protein sequence/structure alignment program (Braberg et al., 2012), which is a general alignment module of the modeling program MODELER; and (2) the MULTALIN alignment program (Corpet, 1988). The resulting alignment contained 30% sequence identity and 48% sequence similarity.
Cloning and cRNA Generation
The cDNAs of SLAC1, OST1, and GHR1 were cloned into oocyte (BiFC) expression vectors (based on pGEM vectors) using an advanced uracil-excision-based cloning technique as described (Nour-Eldin et al., 2006). Site-directed mutations were introduced using a modified USER fusion method as described by (Dadacz-Narloch et al., 2011). For functional analysis, cRNA was prepared with an mMESSAGE mMACHINE T7 transcription kit (Ambion). Oocyte preparation and cRNA injection were performed as described (Becker et al., 1996). For the oocyte BiFC and electrophysiological experiments, 12.5 ng of SLAC1, OST1, or GHR1 (or mutants thereof) cRNA was injected.
BiFC Experiments in Xenopus Oocytes
To document the oocyte BiFC results, photographs were taken with a confocal laser-scanning microscope (Leica DM6000 CS, Leica Microsystems CMS GmbH) equipped with a Leica HCX IRAPO L25×/0.95W objective for oocyte images. For interaction studies in Xenopus oocytes, SLAC1 was fused to the C-terminal half of YFP and coexpressed with the respective interaction partner fused to the N-terminal half of YFP.
DEVC
For the double-electrode voltage clamp (DEVC) studies, oocytes were perfused with standard solutions containing 10 mM Tris/MES pH 5.6, 1 mM CaGluconate2, 1 mM MgGluconate2, 1 mM LaCl3 and either 100 or 30 mM NaNO3. To balance the ionic strength, nitrate variations were compensated for using sodium gluconate. Starting from a holding potential (VH) of 0 mV, single voltage pulses were applied in 20 mV decrements from +60 to -200 mV. The relative open probability PO was determined based on current responses to a constant voltage pulse to -120 mV, subsequent to the different 20 s test pulses. These currents were normalized to the saturation value of the calculated Boltzmann distribution. Instantaneous currents (IT) were extracted immediately after the voltage jump from the holding potential of 0 mV to 50-ms test pulses ranging from +70 to −150 mV.
Detection of GHR1 expression in oocytes by Immunoblot analysis
Isolation of GHR1 wild type or mutant proteins from oocytes was performed by homogenizing five oocytes in 200 μL buffer A (10 mM HEPES pH 7.9, 83 mM NaCl, 10 mM MgCl2, 5 mM PMSF, 1x protease inhibitor (Roche), 2 mM EDTA pH 8) by pipetting up and down. After 2x centrifugation for 10 min (4°C, 1000g), the supernatant was centrifuged for 20 min (4°C, 10000g) and the pellet was resuspended in water. To achieve the correct molecular weight of GHR1, putative N-linked oligosaccharides were removed by PNGase F digestion (New England Biolabs) and mixed with 2x loading buffer. Total proteins were loaded onto a 10% SDS polyacrylamide gel, and after electrophoresis, the gels were electroblotted onto a polyvinylidene difluoride (PVDF) membrane. Immunological reactions were performed with Anti-NT-YFP (Agrisera AS11 1776, Lot 1605) polyclonal antisera (diluted 1:2000) before detection with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody in a 1:2000 dilution (Cell Signaling Technology #7074S, Lot 20, Ref 02/2011). Molecular weight was defined via comparison with a prestained protein ladder (Thermo Scientific, PageRulerTM Plus Prestained Protein Ladder, #26619).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: At4g20940 (RCD7/GHR1), At4g33950 (OST1), At1g12480 (SLAC1), At4g23180 (CRK10), At1g50610 (PRK5), and At4g23650 (CPK3). The rcd7 genome resequencing data have been deposited at NCBI under SRA accession number PRJNA495717.
Supplemental Data
Supplemental Figure 1. Phenotypes of the rcd7 mutant, O3-sensitivity screen of T-DNA mutants for rcd7 candidate genes, and allelism tests
Supplemental Figure 2. GHR1 kinase activity assays
Supplemental Figure 3. Sequence and predicted structure of the kinase domain of GHR1 and GHR1 expression in Saccharomyces cerevisiae
Supplemental Figure 4. Characterization of stomatal phenotypes and subcellular localization of fusion proteins in transgenic lines expressing 35S-GHR1-GFP and 35S-GHR1K798W-GFP
Supplemental Figure 5. Characterization of ghr1-17 and transgenic lines expressing GHR1 variants
Supplemental Figure 6. In vitro binding of GHR1ID with CPK3 analyzed by microscale thermophoresis
Supplemental Figure 7. Subcellular localization of fusion proteins
Supplemental Figure 8. Immunoblot analysis of OST1:YN, GHR1 WT:YN, GHR1G108D:YN and GHR1D293N:YN protein levels in oocytes
Supplemental Table 1. Inheritance, mapping process and candidate genes for RCD7
Supplemental Table 2. Allele nomenclature for the rcd7/ghr1 mutants used
Supplemental Data Set 1. ANOVA Tables
Supplemental Data Set 2. MaxQuant Phospho(STY)Sites table of in vitro phosphorylation sites of GST-GHR1 by 6xHis-HT1 identified by mass-spectrometry
Supplemental Data Set 3. Primers used in this study
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
Leena Grönholm, Marjukka Uuskallio, and Susanna Tarkiainen are acknowledged for excellent technical assistance. Anna Huusari is acknowledged for generation and analysis of Arabidopsis lines expressing GHR1-YFP, GHR1W799*-YFP, and YFP. NanoTemper Technologies is acknowledged for the opportunity to use Tycho NT.6 and the support with data acquisition and analysis provided by Jakub Nowak. Thanks to Michael Wrzaczek for helpful discussions relating to kinases, Mikael Brosché for comments on the manuscript, and Teemu Teeri for Nicotiana benthamiana plants. Anne Dievart provided the HMM profile for LRRs, and Lauri Vaahtera ran the HMM analysis. Kirsi Lipponen and Eeva-Marja Turkki are acknowledged for their technical assistance on NGS sequencing. This work was supported by the University of Helsinki (K.O., J.K.); Academy of Finland Fellowships (Decisions no. 251397, 256073 and 283254; K.O.); the Academy of Finland Centre of Excellence programs (2006-11 and 2014-19, J.K.) and Research Grant (Decision 250336; JK); Academy of Finland post-doctoral fellowships (Decision 294580, C.W.; 266793, T.V.; and 140187, A.G.), Ella and Georg Ehrnrooth foundation (M.S.), Finnish Cultural Foundation (M.S., T.V.), work in the lab of HK was supported by Estonian Ministry of Science and Education (IUT2-21, HK; PUT331, DY; MJD323, TV) and European Regional Fund (Center of Excellence in Molecular Cell Engineering). R.H. and D.G. were supported by the D.F.G. within the TR/SFB166 “ReceptorLight” project B08. Elina Savolainen created the artwork associated with this article.
AUTHOR CONTRIBUTIONS
All authors wrote their respective methods sections, edited, commented and approved the manuscript. D.G., R.H., T.M., M.S., K.O., C.W., K.D., K.L., K.T., L.P., P.A., H.K., J.K., H.H., D.Y., and K.D. designed research; M.S., K.O., and J.K. wrote the paper; T.M., M.S., T.V., A.G., K.O., H.H., C.W., D.Y., K.D., K.L., K.T., J.P.V., and T.P. acquired data; and T.M., J.S., H.K., J.K., K.O., M.S., H.H., D.Y., K.D., K.L., K.T., and J.P.V. analyzed data.
Footnotes
Articles can be viewed without a subscription.
References
- Bauer H., Ache P., Lautner S., Fromm J., Hartung W., Al-Rasheid K.A., Sonnewald S., Sonnewald U., Kneitz S., Lachmann N., Mendel R.R., Bittner F., et al. (2013). The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr. Biol. 23: 53–57. [DOI] [PubMed] [Google Scholar]
- Becker D., Dreyer I., Hoth S., Reid J.D., Busch H., Lehnen M., Palme K., Hedrich R. (1996). Changes in voltage activation, Cs+ sensitivity, and ion permeability in H5 mutants of the plant K+ channel KAT1. Proc. Natl. Acad. Sci. USA 93: 8123–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A. (2006). Arabidopsis thaliana floral dip transformation method. Methods Mol. Biol. 343: 87–103. [DOI] [PubMed] [Google Scholar]
- Blaum B.S., Mazzotta S., Nöldeke E.R., Halter T., Madlung J., Kemmerling B., Stehle T. (2014). Structure of the pseudokinase domain of BIR2, a regulator of BAK1-mediated immune signaling in Arabidopsis. J. Struct. Biol. 186: 112–121. [DOI] [PubMed] [Google Scholar]
- Bluhm W.F., Beran B., Bi C., Dimitropoulos D., Prlic A., Quinn G.B., Rose P.W., Shah C., Young J., Yukich B., Berman H.M., Bourne P.E. (2011). Quality assurance for the query and distribution systems of the RCSB Protein Data Bank. Database (Oxford) 2011: bar003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojar D., Martinez J., Santiago J., Rybin V., Bayliss R., Hothorn M. (2014). Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 78: 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfield J.K., Smith Kf., Staden R. (1995). A new DNA sequence assembly program. Nucleic Acids Res. 23: 4992–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgström E., Lundin S., Lundeberg J. (2011). Large scale library generation for high throughput sequencing. PLoS One 6: e19119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdais G., Burdiak P., Gauthier A., Nitsch L., Salojärvi J., Rayapuram C., Idänheimo N., Hunter K., Kimura S., Merilo E., Vaattovaara A., Oracz K., et al. ; CRK Consortium (2015). Large-scale phenomics identifies primary and fine-tuning roles for CRKs in responses related to oxidative stress. PLoS Genet. 11: e1005373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braberg H., Webb B.M., Tjioe E., Pieper U., Sali A., Madhusudhan M.S. (2012). SALIGN: a web server for alignment of multiple protein sequences and structures. Bioinformatics 28: 2072–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt B., Brodsky D.E., Xue S., Negi J., Iba K., Kangasjärvi J., Ghassemian M., Stephan A.B., Hu H., Schroeder J.I. (2012). Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. USA 109: 10593–10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt B., Munemasa S., Wang C., Nguyen D., Yong T.M., Yang P.G., Poretsky E., Belknap T.F., Waadt R., Aleman F., Schroeder J.I. (2015). Calcium specificity signaling mechanisms in abscisic acid signal transduction in Arabidopsis guard cells. eLife 4: e03599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castells E., Casacuberta J.M. (2007). Signalling through kinase-defective domains: the prevalence of atypical receptor-like kinases in plants. J. Exp. Bot. 58: 3503–3511. [DOI] [PubMed] [Google Scholar]
- Chater C., Peng K., Movahedi M., Dunn J.A., Walker H.J., Liang Y.K., McLachlan D.H., Casson S., Isner J.C., Wilson I., Neill S.J., Hedrich R., et al. (2015). Elevated CO2-induced responses in stomata require ABA and ABA signaling. Curr. Biol. 25: 2709–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin P.L., Saracco S.A., Norris S.R., Last R.L. (2000). Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics 154: 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16: 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto D., Zipfel C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16: 537–552. [DOI] [PubMed] [Google Scholar]
- Cubero-Font P., Maierhofer T., Jaslan J., Rosales M.A., Espartero J., Díaz-Rueda P., Müller H.M., Hürter A.L., Al-Rasheid K.A., Marten I., Hedrich R., Colmenero-Flores J.M., et al. (2016). Silent S-type anion channel subunit SLAH1 gates SLAH3 open for chloride root-to-shoot translocation. Curr. Biol. 26: 2213–2220. [DOI] [PubMed] [Google Scholar]
- Dadacz-Narloch B., Beyhl D., Larisch C., López-Sanjurjo E.J., Reski R., Kuchitsu K., Müller T.D., Becker D., Schönknecht G., Hedrich R. (2011). A novel calcium binding site in the slow vacuolar cation channel TPC1 senses luminal calcium levels. Plant Cell 23: 2696–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir F., Horntrich C., Blachutzik J.O., Scherzer S., Reinders Y., Kierszniowska S., Schulze W.X., Harms G.S., Hedrich R., Geiger D., Kreuzer I. (2013). Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proc. Natl. Acad. Sci. USA 110: 8296–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I., Voss U., Jürgens G., Beeckman T. (2009). Receptor-like kinases shape the plant. Nat. Cell Biol. 11: 1166–1173. [DOI] [PubMed] [Google Scholar]
- Devireddy AR, Zandalinas SI, Gomez-Cadenas A, Blumwald E, Mittler R (2018). Coordinating the overall stomatal response of plants: Rapid leaf-to-leaf communication during light stress. Sci Signal 11: eaam9514. [DOI] [PubMed]
- Eyers P.A., Murphy J.M. (2013). Dawn of the dead: protein pseudokinases signal new adventures in cell biology. Biochem. Soc. Trans. 41: 969–974. [DOI] [PubMed] [Google Scholar]
- Geiger D., Scherzer S., Mumm P., Stange A., Marten I., Bauer H., Ache P., Matschi S., Liese A., Al-Rasheid K.A., Romeis T., Hedrich R. (2009). Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. USA 106: 21425–21430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D., Scherzer S., Mumm P., Marten I., Ache P., Matschi S., Liese A., Wellmann C., Al-Rasheid K.A., Grill E., Romeis T., Hedrich R. (2010). Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. USA 107: 8023–8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish L.A., Clark S.E. (2011). The RLK/Pelle family of kinases. Plant J. 66: 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gookin T.E., Assmann S.M. (2014). Significant reduction of BiFC non-specific assembly facilitates in planta assessment of heterotrimeric G-protein interactors. Plant J. 80: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X., He K., Yang H., Yuan T., Lin H., Clouse S.D., Li J. (2010). Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genomics 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greeff C., Roux M., Mundy J., Petersen M. (2012). Receptor-like kinase complexes in plant innate immunity. Front. Plant Sci. 3: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzel Deger A., Scherzer S., Nuhkat M., Kedzierska J., Kollist H., Brosché M., Unyayar S., Boudsocq M., Hedrich R., Roelfsema M.R. (2015). Guard cell SLAC1-type anion channels mediate flagellin-induced stomatal closure. New Phytol. 208: 162–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter T., Imkampe J., Blaum B.S., Stehle T., Kemmerling B. (2014a). BIR2 affects complex formation of BAK1 with ligand binding receptors in plant defense. Plant Signal. Behav. 9: e28944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter T., Imkampe J., Mazzotta S., Wierzba M., Postel S., Bücherl C., Kiefer C., Stahl M., Chinchilla D., Wang X., Nürnberger T., Zipfel C., et al. (2014b). The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 24: 134–143. [DOI] [PubMed] [Google Scholar]
- Hammarén H.M., Ungureanu D., Grisouard J., Skoda R.C., Hubbard S.R., Silvennoinen O. (2015a). ATP binding to the pseudokinase domain of JAK2 is critical for pathogenic activation. Proc. Natl. Acad. Sci. USA 112: 4642–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarén H.M., Virtanen A.T., Silvennoinen O. (2015b). Nucleotide-binding mechanisms in pseudokinases. Biosci. Rep. 36: e00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks S.K. (2003). Genomic analysis of the eukaryotic protein kinase superfamily: a perspective. Genome Biol. 4: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks S.K., Quinn A.M., Hunter T. (1988). The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241: 42–52. [DOI] [PubMed] [Google Scholar]
- Hashimoto M., Negi J., Young J., Israelsson M., Schroeder J.I., Iba K. (2006). Arabidopsis HT1 kinase controls stomatal movements in response to CO2. Nat. Cell Biol. 8: 391–397. [DOI] [PubMed] [Google Scholar]
- Hedrich R. (2012). Ion channels in plants. Physiol. Rev. 92: 1777–1811. [DOI] [PubMed] [Google Scholar]
- Hedrich R., Geiger D. (2017). Biology of SLAC1-type anion channels - from nutrient uptake to stomatal closure. New Phytol. 216: 46–61. [DOI] [PubMed] [Google Scholar]
- Hohmann U., Lau K., Hothorn M. (2017). The structural basis of ligand perception and signal activation by receptor kinases. Annu. Rev. Plant Biol. 68: 109–137. [DOI] [PubMed] [Google Scholar]
- Hohmann U., Nicolet J., Moretti A., Hothorn L.A., Hothorn M. (2018a). The SERK3 elongated allele defines a role for BIR ectodomains in brassinosteroid signalling. Nat. Plants 4: 345–351. [DOI] [PubMed] [Google Scholar]
- Hohmann U., Santiago J., Nicolet J., Olsson V., Spiga F.M., Hothorn L.A., Butenko M.A., Hothorn M. (2018b). Mechanistic basis for the activation of plant membrane receptor kinases by SERK-family coreceptors. Proc. Natl. Acad. Sci. USA 115: 3488–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hõrak H., Sierla M., Tõldsepp K., Wang C., Wang Y.S., Nuhkat M., Valk E., Pechter P., Merilo E., Salojärvi J., Overmyer K., Loog M., et al. (2016). A dominant mutation in the HT1 kinase uncovers roles of MAP kinases and GHR1 in CO2-induced stomatal closure. Plant Cell 28: 2493–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua D., Wang C., He J., Liao H., Duan Y., Zhu Z., Guo Y., Chen Z., Gong Z. (2012). A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis. Plant Cell 24: 2546–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idänheimo N., Gauthier A., Salojärvi J., Siligato R., Brosché M., Kollist H., Mähönen A.P., Kangasjärvi J., Wrzaczek M. (2014). The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against apoplastic oxidative stress. Biochem. Biophys. Res. Commun. 445: 457–462. [DOI] [PubMed] [Google Scholar]
- Jaspers P., Brosché M., Overmyer K., Kangasjärvi J. (2010). The transcription factor interacting protein RCD1 contains a novel conserved domain. Plant Signal. Behav. 5: 78–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.M., Xuan Y., Xu M., Wang R.S., Ho C.H., Lalonde S., You C.H., Sardi M.I., Parsa S.A., Smith-Valle E., Su T., Frazer K.A., et al. (2014). Border control--a membrane-linked interactome of Arabidopsis. Science 344: 711–716. [DOI] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J.D.G., Shirasu K., Menke F., Jones A., Zipfel C. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54: 43–55. [DOI] [PubMed] [Google Scholar]
- Kangasjärvi J., Jaspers P., Kollist H. (2005). Signalling and cell death in ozone-exposed plants. Plant Cell Environ. 28: 1021–1036. [Google Scholar]
- Kim T.W., Wang Z.Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61: 681–704. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Böhmer M., Hu H., Nishimura N., Schroeder J.I. (2010). Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61: 561–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Schumaker K.S., Zhu J.K. (2006). EMS mutagenesis of Arabidopsis. Methods Mol. Biol. 323: 101–103. [DOI] [PubMed] [Google Scholar]
- Koch E., Slusarenko A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollist H., Nuhkat M., Roelfsema M.R. (2014). Closing gaps: linking elements that control stomatal movement. New Phytol. 203: 44–62. [DOI] [PubMed] [Google Scholar]
- Kollist T., Moldau H., Rasulov B., Oja V., Rämma H., Hüve K., Jaspers P., Kangasjärvi J., Kollist H. (2007). A novel device detects a rapid ozone-induced transient stomatal closure in intact Arabidopsis and its absence in abi2 mutant. Physiol. Plant. 129: 796–803. [Google Scholar]
- Kornev A.P., Haste N.M., Taylor S.S., Eyck L.F. (2006). Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. USA 103: 17783–17788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis P.J. (1991). MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 24: 946–950. [Google Scholar]
- Kumar D., Kumar R., Baek D., Hyun T.K., Chung W.S., Yun D.J., Kim J.Y. (2017). Arabidopsis thaliana RECEPTOR DEAD KINASE1 functions as a positive regulator in plant responses to ABA. Mol. Plant 10: 223–243. [DOI] [PubMed] [Google Scholar]
- Kwak S.H., Woo S., Lee M.M., Schiefelbein J. (2014). Distinct signaling mechanisms in multiple developmental pathways by the SCRAMBLED receptor of Arabidopsis. Plant Physiol. 166: 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeberg L.K., Scott J.D. (2015). Signalling scaffolds and local organization of cellular behaviour. Nat. Rev. Mol. Cell Biol. 16: 232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C., Lan W., Buchanan B.B., Luan S. (2009). A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. USA 106: 21419–21424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ruan J., Durbin R. (2008). Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18: 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., Cai G., Gao L., Zhang X., Wang Y., Chen S., Zhou J.M. (2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338. [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068. [DOI] [PubMed] [Google Scholar]
- Maierhofer T., Diekmann M., Offenborn J.N., Lind C., Bauer H., Hashimoto K., S Al-Rasheid K.A., Luan S., Kudla J., Geiger D., Hedrich R. (2014). Site- and kinase-specific phosphorylation-mediated activation of SLAC1, a guard cell anion channel stimulated by abscisic acid. Sci. Signal. 7: ra86. [DOI] [PubMed] [Google Scholar]
- Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S. (2002). The protein kinase complement of the human genome. Science 298: 1912–1934. [DOI] [PubMed] [Google Scholar]
- Mansfield T.A. (1973). The role of stomata in determining the response of plants to air pollutants. Curr Advan Plant Sci 2: 11–20. [Google Scholar]
- Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I., Lanczycki C.J., Lu F., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43: D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam S.A., Brodribb T.J. (2016). Linking turgor with ABA biosynthesis: implications for stomatal responses to vapor pressure deficit across land plants. Plant Physiol. 171: 2008–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980. [DOI] [PubMed] [Google Scholar]
- Merilo E., Laanemets K., Hu H., Xue S., Jakobson L., Tulva I., Gonzalez-Guzman M., Rodriguez P.L., Schroeder J.I., Broschè M., Kollist H. (2013). PYR/RCAR receptors contribute to ozone-, reduced air humidity-, darkness-, and CO2-induced stomatal regulation. Plant Physiol. 162: 1652–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merilo E., Jalakas P., Kollist H., Brosché M. (2015). The role of ABA recycling and transporter proteins in rapid stomatal responses to reduced air humidity, elevated CO2, and exogenous ABA. Mol. Plant 8: 657–659. [DOI] [PubMed] [Google Scholar]
- Merilo E., Yarmolinsky D., Jalakas P., Parik H., Tulva I., Rasulov B., Kilk K., Kollist H. (2018). Stomatal VPD response: there is more to the story than ABA. Plant Physiol. 176: 851–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt E.A., Bacon D.J. (1997). Raster3D: photorealistic molecular graphics. Methods Enzymol. 277: 505–524. [DOI] [PubMed] [Google Scholar]
- Milne I., Stephen G., Bayer M., Cock P.J., Pritchard L., Cardle L., Shaw P.D., Marshall D. (2013). Using Tablet for visual exploration of second-generation sequencing data. Brief. Bioinform. 14: 193–202. [DOI] [PubMed] [Google Scholar]
- Mori I.C., Murata Y., Yang Y., Munemasa S., Wang Y.F., Andreoli S., Tiriac H., Alonso J.M., Harper J.F., Ecker J.R., Kwak J.M., Schroeder J.I. (2006). CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol. 4: e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J.M., Zhang Q., Young S.N., Reese M.L., Bailey F.P., Eyers P.A., Ungureanu D., Hammaren H., Silvennoinen O., Varghese L.N., Chen K., Tripaydonis A., et al. (2014). A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem. J. 457: 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli A.C., Merlot S., Vavasseur A., Fenzi F., Giraudat J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi J., Matsuda O., Nagasawa T., Oba Y., Takahashi H., Kawai-Yamada M., Uchimiya H., Hashimoto M., Iba K. (2008). CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452: 483–486. [DOI] [PubMed] [Google Scholar]
- Nour-Eldin H.H., Hansen B.G., Nørholm M.H., Jensen J.K., Halkier B.A. (2006). Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 34: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer D.G., Herman P.L., Sivakumaran S., Esch J., Marks M.D. (1991). A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67: 483–493. [DOI] [PubMed] [Google Scholar]
- Overmyer K., Tuominen H., Kettunen R., Betz C., Langebartels C., Sandermann H. Jr., Kangasjärvi J. (2000). Ozone-sensitive arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 12: 1849–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer K., Kollist H., Tuominen H., Betz C., Langebartels C., Wingsle G., Kangasjärvi S., Brader G., Mullineaux P., Kangasjärvi J. (2008). Complex phenotypic profiles leading to ozone sensitivity in Arabidopsis thaliana mutants. Plant Cell Environ. 31: 1237–1249. [DOI] [PubMed] [Google Scholar]
- Park S.Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T.F., Alfred S.E., Bonetta D., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelfsema M.R., Hedrich R., Geiger D. (2012). Anion channels: master switches of stress responses. Trends Plant Sci. 17: 221–229. [DOI] [PubMed] [Google Scholar]
- Scherzer S., Maierhofer T., Al-Rasheid K.A., Geiger D., Hedrich R. (2012). Multiple calcium-dependent kinases modulate ABA-activated guard cell anion channels. Mol. Plant 5: 1409–1412. [DOI] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B. (2003). Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132: 530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierla M., Rahikainen M., Salojärvi J., Kangasjärvi J., Kangasjärvi S. (2013). Apoplastic and chloroplastic redox signaling networks in plant stress responses. Antioxid. Redox Signal. 18: 2220–2239. [DOI] [PubMed] [Google Scholar]
- Sierla M., Waszczak C., Vahisalu T., Kangasjärvi J. (2016). Reactive oxygen species in the regulation of stomatal movements. Plant Physiol. 171: 1569–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich M., Ma Q., Weidtkamp-Peters S., Stahl Y., Felekyan S., Bleckmann A., Seidel C.A.M., Simon R. (2015). Real-time dynamics of peptide ligand-dependent receptor complex formation in planta. Sci. Signal. 8: ra76. [DOI] [PubMed] [Google Scholar]
- Somssich M., Bleckmann A., Simon R. (2016). Shared and distinct functions of the pseudokinase CORYNE (CRN) in shoot and root stem cell maintenance of Arabidopsis. J. Exp. Bot. 67: 4901–4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Miao Y., Song C.P. (2014). Behind the scenes: the roles of reactive oxygen species in guard cells. New Phytol. 201: 1121–1140. [DOI] [PubMed] [Google Scholar]
- Soon F.F., Ng L.M., Zhou X.E., West G.M., Kovach A., Tan M.H.E., Suino-Powell K.M., He Y., Xu Y., Chalmers M.J., Brunzelle J.S., Zhang H., et al. (2012). Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335: 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J.M., Walker J.C. (1995). Plant protein kinase families and signal transduction. Plant Physiol. 108: 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Miller G., Morales J., Shulaev V., Torres M.A., Mittler R. (2011). Respiratory burst oxidases: the engines of ROS signaling. Curr. Opin. Plant Biol. 14: 691–699. [DOI] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., Ishihama Y., Hirayama T., Shinozaki K. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaddepalli P., Fulton L., Batoux M., Yadav R.K., Schneitz K. (2011). Structure-function analysis of STRUBBELIG, an Arabidopsis atypical receptor-like kinase involved in tissue morphogenesis. PLoS One 6: e19730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T., Kollist H., Wang Y.F., Nishimura N., Chan W.Y., Valerio G., Lamminmäki A., Brosché M., Moldau H., Desikan R., Schroeder J.I., Kangasjärvi J. (2008). SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452: 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T., Puzõrjova I., Brosché M., Valk E., Lepiku M., Moldau H., Pechter P., Wang Y.S., Lindgren O., Salojärvi J., Loog M., Kangasjärvi J., et al. (2010). Ozone-triggered rapid stomatal response involves the production of reactive oxygen species, and is controlled by SLAC1 and OST1. Plant J. 62: 442–453. [DOI] [PubMed] [Google Scholar]
- Vainonen J.P., Jaspers P., Wrzaczek M., Lamminmäki A., Reddy R.A., Vaahtera L., Brosché M., Kangasjärvi J. (2012). RCD1-DREB2A interaction in leaf senescence and stress responses in Arabidopsis thaliana. Biochem. J. 442: 573–581. [DOI] [PubMed] [Google Scholar]
- Vlad F., Rubio S., Rodrigues A., Sirichandra C., Belin C., Robert N., Leung J., Rodriguez P.L., Laurière C., Merlot S. (2009). Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956. [DOI] [PubMed] [Google Scholar]
- Wang X., Goshe M.B., Soderblom E.J., Phinney B.S., Kuchar J.A., Li J., Asami T., Yoshida S., Huber S.C., Clouse S.D. (2005). Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kota U., He K., Blackburn K., Li J., Goshe M.B., Huber S.C., Clouse S.D. (2008). Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 15: 220–235. [DOI] [PubMed] [Google Scholar]
- Wang Y., Hills A., Vialet-Chabrand S., Papanatsiou M., Griffiths H., Rogers S., Lawson T., Lew V.L., Blatt M.R. (2017). Unexpected connections between humidity and ion transport discovered using a model to bridge guard cell-to-leaf scales. Plant Cell 29: 2921–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszczak C., Carmody M., Kangasjärvi J. (2018). Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 69: 209–236. [DOI] [PubMed] [Google Scholar]
- Webb B, Sali A (2014). Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics 47: 5.6.1–5.6.32. [DOI] [PubMed]
- Wrzaczek M., Vainonen J.P., Stael S., Tsiatsiani L., Help-Rinta-Rahko H., Gauthier A., Kaufholdt D., Bollhöner B., Lamminmäki A., Staes A., Gevaert K., Tuominen H., et al. (2015). GRIM REAPER peptide binds to receptor kinase PRK5 to trigger cell death in Arabidopsis. EMBO J. 34: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A.J., Andriotis V.M., Durrant M.C., Rathjen J.P. (2004). A patch of surface-exposed residues mediates negative regulation of immune signaling by tomato Pto kinase. Plant Cell 16: 2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Wang Y., Williamson L., Holroyd G.H., Tagliavia C., Murchie E., Theobald J., Knight M.R., Davies W.J., Leyser H.M., Hetherington A.M. (2006). The identification of genes involved in the stomatal response to reduced atmospheric relative humidity. Curr. Biol. 16: 882–887. [DOI] [PubMed] [Google Scholar]
- Xue S., Hu H., Ries A., Merilo E., Kollist H., Schroeder J.I. (2011). Central functions of bicarbonate in S-type anion channel activation and OST1 protein kinase in CO2 signal transduction in guard cell. EMBO J. 30: 1645–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Negi J., Wang C., Isogai Y., Schroeder J.I., Iba K. (2016). The transmembrane region of guard cell SLAC1 channels perceives CO2 signals via an ABA-independent pathway in Arabidopsis. Plant Cell 28: 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Costa A., Leonhardt N., Siegel R.S., Schroeder J.I. (2008). Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R., Hobo T., Ichimura K., Mizoguchi T., Takahashi F., Aronso J., Ecker J.R., Shinozaki K. (2002). ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 43: 1473–1483. [DOI] [PubMed] [Google Scholar]
- Zeqiraj E., van Aalten D.M. (2010). Pseudokinases-remnants of evolution or key allosteric regulators? Curr. Opin. Struct. Biol. 20: 772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T.Y., Li F.C., Fan C.M., Li X., Zhang F.F., He J.M. (2017). Role and interrelationship of MEK1-MPK6 cascade, hydrogen peroxide and nitric oxide in darkness-induced stomatal closure. Plant Sci. 262: 190–199. [DOI] [PubMed] [Google Scholar]
- Zhang X., Facette M., Humphries J.A., Shen Z., Park Y., Sutimantanapi D., Sylvester A.W., Briggs S.P., Smith L.G. (2012). Identification of PAN2 by quantitative proteomics as a leucine-rich repeat-receptor-like kinase acting upstream of PAN1 to polarize cell division in maize. Plant Cell 24: 4577–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]