Arabidopsis RRP7 encodes a ribosome biogenesis factor involved in 45S pre-rRNA processing and interacts synergistically with NUC1 and HDA6, which encode epigenetic regulators of 45S rDNA transcription.
Abstract
Ribosome biogenesis is fundamental to growth and development in eukaryotes and is linked to human diseases and cancer. Arabidopsis thaliana MORPHOLOGY OF ARGONAUTE1-52 SUPPRESSED 2 (MAS2) participates in splicing and 45S ribosomal DNA (rDNA) expression. In a screen for MAS2 interactors, we identified RIBOSOMAL RNA PROCESSING 7 (RRP7), an ortholog of yeast rRNA processing protein 7 (Rrp7), which is required for 18S ribosomal RNA (rRNA) maturation. Arabidopsis rrp7 mutants exhibit a pleiotropic phenotype including slow growth, altered shoot phyllotaxy, aberrant venation in lateral organs, partial infertility, and abscisic acid hypersensitivity in seedlings. In Arabidopsis, RRP7 localizes mainly to the nucleolus, the site of the 45S rDNA transcription that produces a 45S pre-rRNA primary transcript, precursor of the 25S, 18S and 5.8S rRNAs. Lack of RRP7 function perturbs 18S rRNA maturation, causes nucleolar hypertrophy, and results in an increased 25S/18S rRNA ratio. Arabidopsis contains hundreds of 45S rDNA genes whose expression is epigenetically regulated, and deregulated, in rrp7 mutants. Double mutant analysis revealed synergistic interactions between RRP7 alleles and alleles of MAS2, NUCLEOLIN1 (NUC1), and HISTONE DEACETYLASE 6 (HDA6), which encode epigenetic regulators of 45S rDNA transcription. Our results reveal the evolutionarily conserved but divergent roles of RRP7 as a ribosome biogenesis factor.
INTRODUCTION
The ribosome is a molecular machine that originated 3 to 4 billion years ago. It translates mRNAs (mRNAs) into proteins in a process universal to all life forms (Petrov et al., 2014). In eukaryotic cells, cytoplasmic, mitochondrial, and chloroplast ribosomes are composed of a large subunit (LSU) and a small subunit (SSU), each consisting of 1 to 3 rRNAs (rRNAs) and tens of ribosomal proteins (RPs) (Wilson and Doudna Cate, 2012). For example, the eukaryotic cytoplasmic (80S) ribosome is composed of ∼80 RPs and four rRNAs, which include one 28S/25S rRNA (28S in metazoans, and 25S in plants and yeast), together with 18S, 5.8S, and 5S rRNAs.
The 28S/25S, 18S, and 5.8S rRNAs are the final products of the transcription and complex processing of a single polycistronic precursor, the 47S/45S/35S pre-rRNA (47S in metazoans, 45S in plants, and 35S in yeast; Henras et al., 2015; Lafontaine, 2015). The 47S/45S/35S rDNA genes are transcribed by RNA polymerase I (RNA Pol I) in the nucleolus, resulting in a single 47S/45S/35S pre-rRNA, which undergoes chemical modifications, endonucleolytic cleavages, and exonucleolytic trimmings to render mature 28S/25S, 18S, and 5.8S rRNAs (Supplemental Figures 1 and 2).
Biogenesis of the 80S ribosome begins with the transcription of the 47S/45S/35S rDNA genes and processing of their primary transcripts to render mature 28S/25S, 18S, and 5.8S rRNAs. The SSU and LSU are assembled in the nucleus and exported to the cytoplasm in almost completely mature form. The 40S subunit contains the 18S rRNA and ∼33 RPs, whereas the 60S subunit contains the 28S/25S, 5.8S, and 5S rRNAs, and ∼47 RPs (Weis et al., 2015). The final eukaryotic 80S ribosome is assembled in the cytoplasm from a 40S SSU and a 60S LSU.
Work on the yeast (Saccharomyces cerevisiae) 80S ribosome has elucidated its composition, architecture, action, and the interactions of its components (Woolford and Baserga, 2013; Chaker-Margot et al., 2017). Much of the assembly and maturation of the yeast 40S ribosomal subunit is performed by a large ribonucleoprotein complex termed the SSU processome or 90S preribosomal particle (Phipps et al., 2011). The SSU processome is cotranscriptionally assembled during transcription of the 35S rDNA by the stepwise recruitment of ribosome biogenesis factors (RBFs). Many of these RBFs assemble as subcomplexes prior to their incorporation into the SSU processome. Some subcomplexes are small nucleolar ribonucleoprotein particles (snoRNPs) that include a snoRNA that is partially complementary to different regions of the 35S pre-rRNA. The snoRNAs guide site-specific cleavages, ribose 2′-O-methylation (box C/D family snoRNAs), and uridine-to-pseudouridine conversions (box H/ACA snoRNAs) (Kiss et al., 2002).
Examination of yeast snoRNPs has identified key factors in rRNA processing. Yeast U3 snoRNA is a conserved member of the box C/D family that plays a central role in SSU processome assembly and function (Hughes and Ares, 1991; Beltrame and Tollervey, 1992, 1995). RBFs identified based on their coprecipitation with U3 snoRNA are referred to as U-three proteins (Utps; Phipps et al., 2011). Together with U3 snoRNP, one subcomplex that joins the nascent SSU processome is UtpC, including the essential Ribosomal RNA processing protein 7 (Rrp7), which is required for 18S rRNA maturation (Baudin-Baillieu et al., 1997; Krogan et al., 2004; Phipps et al., 2011), U3 snoRNA-associated protein 22 (Utp22), and the four subunits of Casein kinase 2 (CK2) (Krogan et al., 2004; Phipps et al., 2011). Yeast UtpC components also bind to the transcription factor Interact with Fork Head 1 (Ifh1) to form the CURI (CK2, Utp22, Rrp7, and Ifh1) complex, which does not belong to the SSU processome (Rudra et al., 2007).
The CURI complex is thought to play a key role in coordinating the two parallel pathways required for ribosome biogenesis in yeast: (1) the transcription of rDNA and the processing of pre-rRNAs to produce mature rRNAs, and (2) the transcription of genes encoding RPs (Rudra et al., 2007). When 35S rDNA transcription is active, most Rrp7 and Utp22 molecules are engaged in processing 35S pre-rRNAs and are not available to form CURI complexes, and this in turn allows Ifh1 to bind Repressor/activator site binding protein 1 (Rap1) and the Fork head-like 1 (Fhl1) transcription factor in the promoters of RP-encoding genes, thus activating their transcription. Conversely, when 35S rDNA transcription is less active, lfh1 contributes to the formation of CURI complexes, it does not bind Rap1 and Fhl1, and transcription of RP-encoding genes is not induced. Furthermore, the loss of Rrp7 function reduces 40S ribosomal subunit production and increases the expression of RP-encoding genes (Rudra et al., 2007).
ARGONAUTE1 (AGO1; Baumberger and Baulcombe, 2005) is the main Arabidopsis thaliana (Arabidopsis) ribonuclease involved in posttranscriptional gene silencing pathways mediated by small RNAs, including microRNAs (miRNAs). We previously performed second-site mutagenesis of the ago1-52 hypomorphic and viable mutant and isolated 22 extragenic suppressors. We named the corresponding genes MORPHOLOGY OF ARGONAUTE1-52 SUPPRESSED (MAS; Micol-Ponce et al., 2014). One of these genes, AT4G02720 (MAS2), encodes a perinucleolar protein that negatively regulates 45S rDNA expression (Sánchez-García et al., 2015).
In a screen based on the yeast two-hybrid (Y2H) assay, we identified 14 interactors of MAS2, including two putative RBFs, encoded by the AT2G40430 and AT5G38720 genes. AT2G40430 encodes the putative ortholog of yeast Nucleolar protein 53 (Nop53) and human (Homo sapiens) Glioma Tumor-Suppressor Candidate Region Gene 2 (GLTSCR2), which was named SMALL ORGAN 4 (SMO4) by Zhang et al. (2015). AT5G38720 and its plant orthologs have not been studied; it is annotated to encode a rRNA processing 7 protein at TAIR10, Araport11 and NCBI. Here, we studied the action and interactions of AT5G38720, which we have named RRP7. We show that Arabidopsis RRP7 is important for 18S rRNA maturation and that it is functionally related to MAS2, as well as NUCLEOLIN1 (NUC1) and HISTONE DEACETYLASE 6 (HDA6), which are epigenetic regulators of 45S rDNA transcription.
RESULTS
RRP7 Orthologs Share an RRP7-like Domain Occupying Their C-terminal Half
RRP7 is annotated at The Arabidopsis Information Resource 10 (TAIR10) and Araport11 (Cheng et al., 2017) databases as encoding a 306 amino-acid ortholog of yeast Rrp7. Both yeast Rrp7 and its human RRP7A ortholog are essential proteins required for 18S rRNA maturation (Baudin-Baillieu et al., 1997; Krogan et al., 2004; Phipps et al., 2011; Tafforeau et al., 2013). The identities and similarities among the full-length sequences of these three proteins are low: 20.9% and 35.4% (Arabidopsis RRP7 versus human RRP7A), 14.1% and 25.6% (Arabidopsis RRP7 versus yeast Rrp7) and 16.4% and 32.7% (human RRP7A versus yeast Rrp7), respectively (Supplemental Figure 3).
Yeast Rrp7 has both an N-terminal domain (NTD; amino acids 1–156), through which it dimerizes with Utp22, and a C-terminal domain (CTD; amino acids 190–297) that has RNA binding capability and is required for its association with the central domain of the 18S rRNA coding sequence of the 35S pre-rRNA. Deletion of either NTD or CTD is lethal (Lin et al., 2013). Alignment of yeast Rrp7 with its human and Arabidopsis orthologs indicates that only the region containing the CTD is conserved to some extent (Supplemental Figure 3). According to HomoloGene (www.ncbi.nlm.nih.gov/HomoloGene), RRP7 proteins share an RRP7-like conserved domain occupying their C-terminal half, which includes the CTD defined by Lin et al. (2013). The RRP7-like domain of Arabidopsis RRP7 (135 aa) shares an identity and similarity of 31.6% and 52.5%, respectively, with its human ortholog (130 aa) and 23.6% and 38.9%, respectively, with that of yeast (128 aa). The amino acid sequence identity and similarity between the RRP7-like domain of human RRP7A and that of yeast Rrp7 are 21.9% and 43.1%, respectively.
For multiple alignment of plant RRP7s, we selected one representative species from different angiosperm lineages as classified in Myburg et al. (2014). We used one gene encoding an RRP7 factor in Arabidopsis (AT5G38720, this work), Oryza sativa (Os02g0728700), Vitis vinifera (VIT_14s0083g00590) and Nicotiana sylvestris (LOC104216441), two paralogs in Populus trichocarpa (POPTR_004G106900v3 and POPTR_0017s14290g) and Phoenix dactylifera (LOC103703294 and LOC103707756), and three in Glycine max (GLYMA_07G204700, GLYMA_13G171200 and GLYMA_19G087700). The rice RRP7 ortholog is annotated as splicing regulatory glutamine/lysine-rich protein 1, the Nicotiana sylvestris ortholog as DEAD-box ATP-dependent RNA helicase 42 or General transcription factor IIF subunit 1, and those of Phoenix dactylifera as RRP7A (LOC103703294) and RRP7B (LOC103707756). In the case of multiple RRP7 orthologs, we selected the longest isoform of the most similar protein to Arabidopsis RRP7, as shown by BLASTP searches, which were XP_002305975 of Populus trichocarpa (encoded by POPTR_004G106900v3), XP_008784326 (RRP7A) of Phoenix dactylifera (encoded by LOC103703294), and XP_003529364 (encoded by GLYMA_07G204700) of Glycine max (Supplemental Figure 4).
As expected, the level of conservation among plant RRP7 orthologs was higher than that shared with RRP7 orthologs from other kingdoms, considering both their full-length sequences and the RRP7-like domain only (Supplemental Figure 4 and Supplemental Tables 1 and 2). For the full-length plant RRP7 orthologs, the lowest identity and similarity were 29.2% and 37.6% (for Glycine max versus Oryza sativa) and the highest were 48.2% and 58.2% (for Populus trichocarpa versus Vitis vinifera), respectively. For the RRP7-like domains of plant RRP7 orthologs, the lowest identity and similarity were 63.0% and 78.5% (for Arabidopsis thaliana versus Populus trichocarpa) and the highest were 81.3% and 92.5% (for Phoenix dactylifera versus Vitis vinifera), respectively.
RRP7 Plays Several Roles in Arabidopsis Development
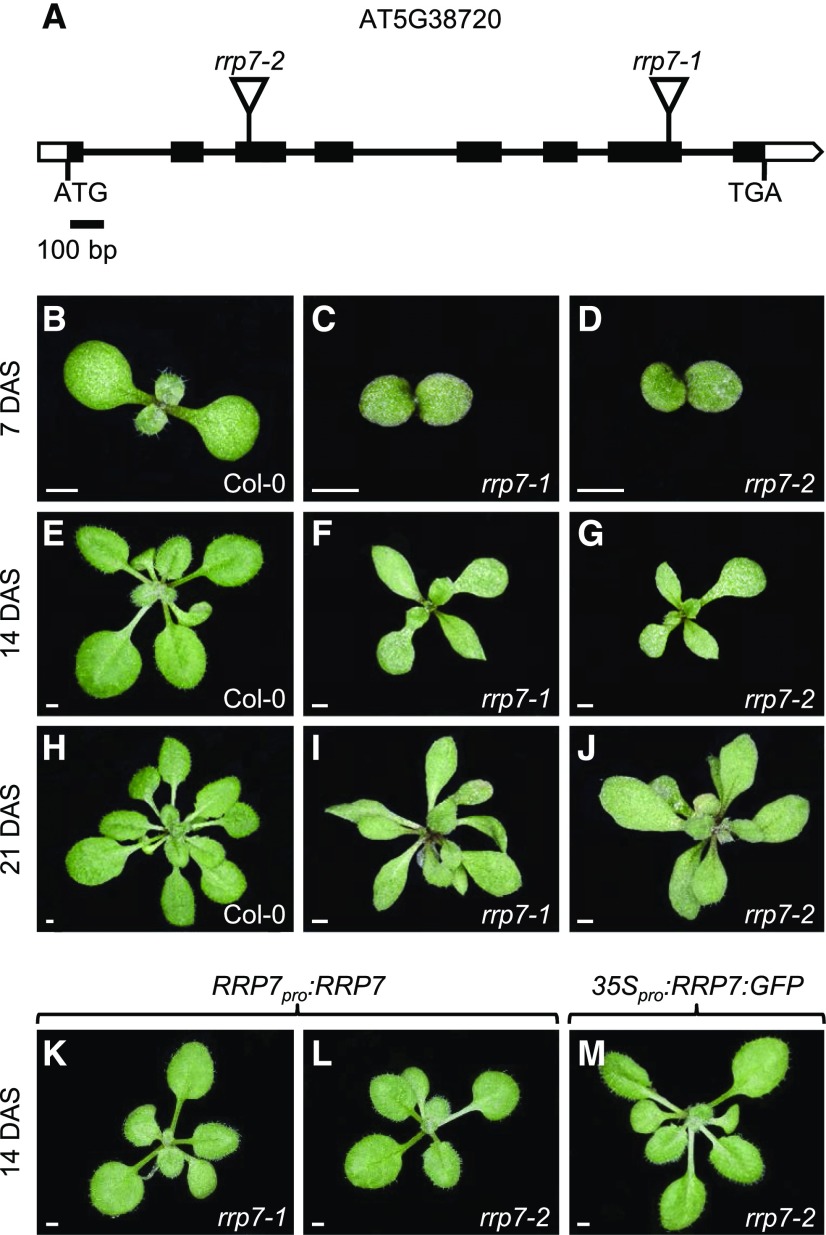
We obtained two publicly available lines carrying T-DNA insertions disrupting AT5G38720, SAIL_628_F08 and WISCDSLOX461-464C16, which we named rrp7-1 and rrp7-2, respectively (Figure 1A). Plants homozygous for these mutations were viable and indistinguishable, and they exhibited slow growth and pointed leaves (Figures 1B to 1J). This leaf phenotype is characteristic of mutations in genes encoding RPs (Van Lijsebettens et al., 1994; Horiguchi et al., 2011; Weis et al., 2015) and is similar, although a bit more severe, to the phenotype caused by null alleles of NUCLEOLIN1 (NUC1). NUC1 is a nucleolar protein involved in transcriptional regulation of 45S rDNA and 45S pre-rRNA processing (Petricka and Nelson, 2007; Pontvianne et al., 2007).
Figure 1.
Structure of RRP7 and the Molecular Nature, Rosette Phenotype, and Transgene-Mediated Complementation of its Mutant Alleles.
(A) Schematic representation of RRP7, including the positions and molecular nature of the rrp7 mutations. Black boxes represent exons and open boxes represent the 5′- and 3′-UTRs. Lines between boxes represent introns, and triangles represent T-DNA insertions.
(B) to (J) Rosettes of Col-0, rrp7-1, and rrp7-2 plants as indicated.
(K) to (M) Rosettes of rrp7-1 RRP7pro:RRP7 (K), rrp7-2 RRP7pro:RRP7 (L), and rrp7-2 35Spro:RRP7:GFP (M) plants.
Photographs were taken 7 (B) to (D), 14 (E) to (G) and (K) to (M), and 21 (H) to (J) days after stratification (DAS). Scale bars: 1 mm.
To confirm that the phenotype of rrp7 mutants was caused by the absence of AT5G38720 activity, we constructed RRP7pro:RRP7 and 35Spro:RRP7 transgenes and transferred them into rrp7 plants, and this fully complemented the mutant phenotypes (Figures 1K to 1M). The 35Spro:RRP7 and 35Spro:RRP7:GFP transgenes had no visible phenotypic effect when transferred to the wild-type Col-0 background, although both caused RRP7 overexpression (Supplemental Figure 5).
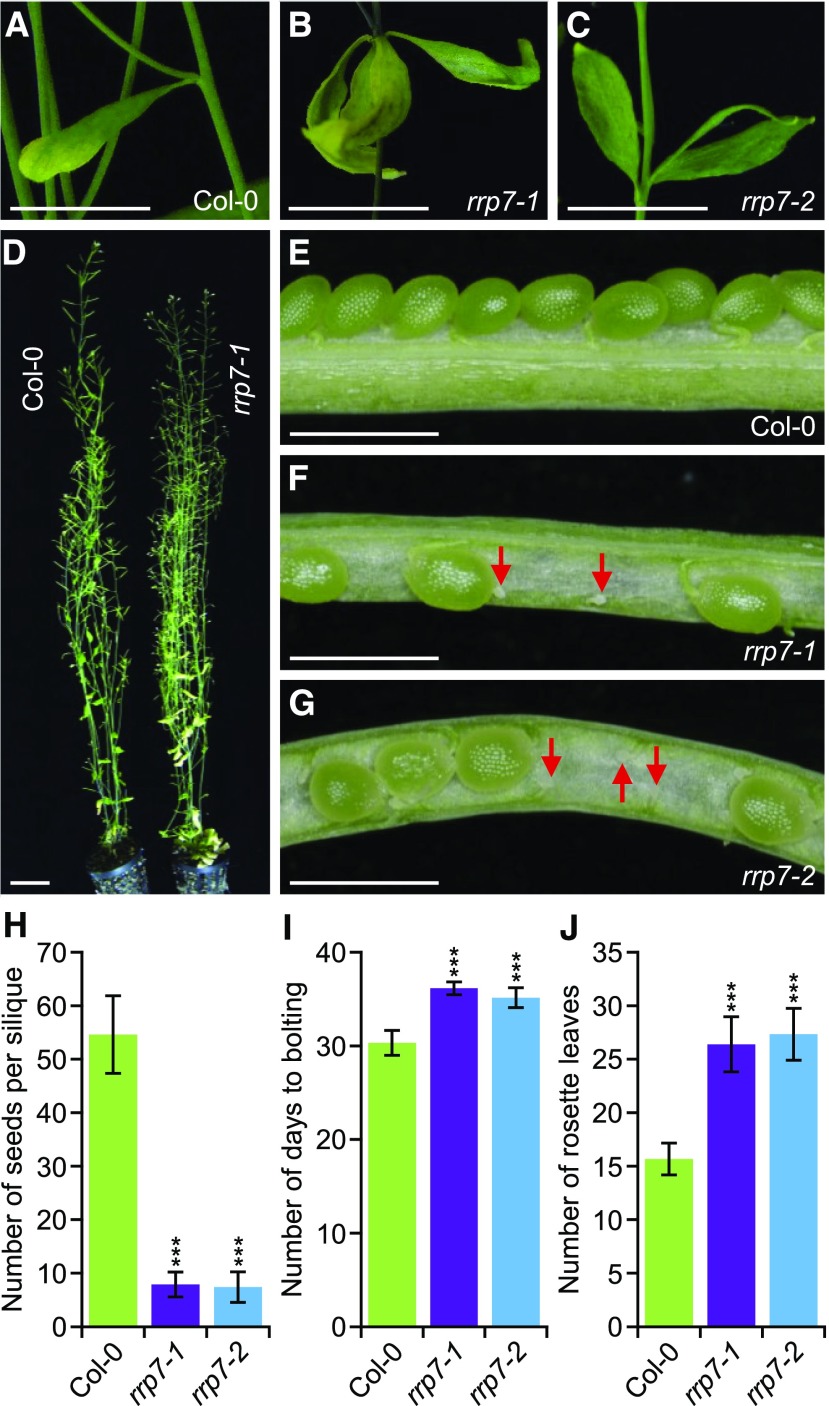
All rrp7-1 and rrp7-2 plants exhibited two or three cauline leaves emerging from each axil (Figures 2B and 2C), whereas wild-type Arabidopsis plants produce only one per axil (Figure 2A) (Müller-Xing et al., 2015). Despite their delayed growth, the rrp7 mutants reached a final stature similar to that of wild type (Figure 2D). Siliques of these plants contained many unfertilized ovules and only produced ∼10% viable seeds compared with Col-0 siliques (Figures 2E to 2H). Flowering occurred earlier in Col-0 than in rrp7, which also developed an increased number of vegetative leaves before bolting (Figures 2I and 2J), suggesting that the late flowering of the mutants is not simply a consequence of slow growth.
Figure 2.
Pleiotropic Morphological Phenotype of rrp7 Plants.
(A) to (C) Presence of a single cauline leaf in the axils of Col-0 stems (A), and three cauline leaves in the axils of rrp7 mutant stems (B) and (C).
(D) Adult Col-0 and rrp7-1 plants.
(E) to (G) Dissected siliques from Col-0 (E), rrp7-1 (F), and rrp7-2 (G) plants. Red arrows indicate unfertilized ovules.
(H) Fertility of rrp7 plants, expressed as the number of seeds per silique. Seeds from ten siliques collected from five plants per genotype were counted.
(I) Flowering time (number of days from stratification to bolting) in the rrp7 mutants.
(J) Number of rosette leaves that developed before bolting in rrp7 plants.
The assays shown in (I) and (J) were performed with at least 30 plants of each genotype. Photographs were taken at 60 DAS (A) to (G). Error bars in (H) to (J) indicate standard deviations. Asterisks indicate values significantly different from the corresponding wild type in a Student′s t test (***p < 0.001). Scale bars = 1 cm (A) to (C); 3 cm (D); and 1 mm (E) to (G).
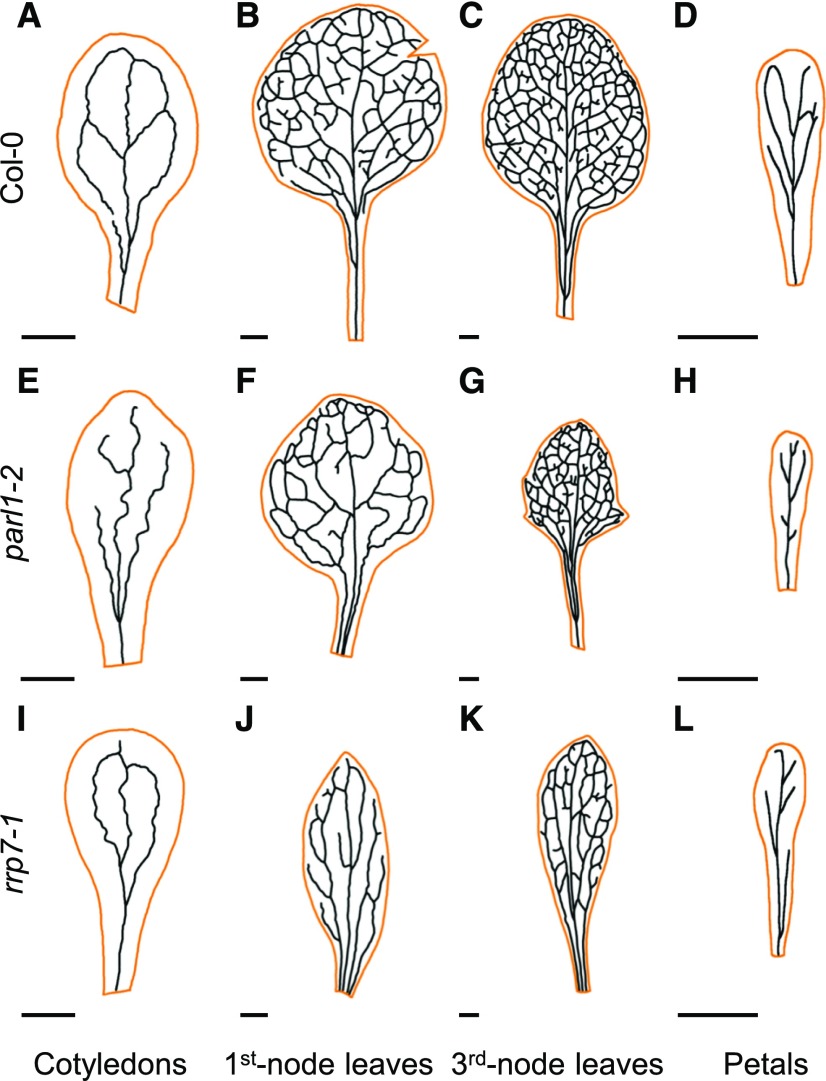
Arabidopsis NUC1 is also referred to as PARALLEL1 (PARL1) because null parl1 mutants exhibit parallel veins in their cotyledons, vegetative leaves, sepals, and petals, a pattern never observed in wild-type plants (Petricka and Nelson, 2007). We found that rrp7-1 cotyledons, first- and third-node leaves, and petals also exhibited aberrant venation patterns (Figure 3). The cotyledons of rrp7-1 have two areoles (closed loops) instead of four (as in Col-0), whereas parl1-2 cotyledons lack areoles. First- and third-node rrp7-1 leaves, collected 21 d after stratification (DAS), exhibited a reduced number of venation branching points. The basal region of the lamina of rrp7-1 leaves has parallel veins (Figures 3J and 3K). In Col-0, petal veins form some areoles (Figure 3D), which are absent from rrp7-1 and parl1-2 (Figures 3H and 3L). Morphometric analysis of leaf venation parameters commonly used to describe vascular pattern complexity (Supplemental Table 3) confirmed that vein patterning in leaves is more defective in rrp7-1 than in parl1-2.
Figure 3.
Aberrant Venation Patterns in parl1-2 and rrp7-1 Cotyledons, Vegetative Leaves, and Petals.
(A) to (L) Diagrams were drawn from micrographs taken from cotyledons (A), (E) and (I), first-node leaves (B), (F) and (J), third-node leaves (C), (G) and (K), and petals (D), (H) and (L). Cotyledons and leaves were collected at 21 DAS and petals were collected at 63 DAS from Col-0 (A) to (D), parl1-2 (E) to (H), and rrp7-1 (I) to (L) plants. Cotyledon, leaf and petal margins are shown in orange. Scale bars = 1 mm.
Loss of Function of RRP7 Causes Abscisic Acid Hypersensitivity During Seedling Establishment
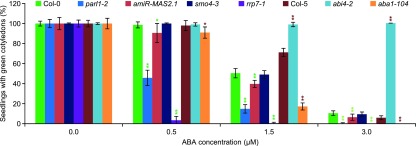
The role of abscisic acid (ABA) in the inhibition of seed germination under adverse conditions is well known (Lopez-Molina et al., 2001). Mutations in genes involved in several aspects of mRNA metabolism, such as pre-mRNA maturation and mRNA degradation, export, or translation cause hypersensitivity to ABA (Kuhn et al., 2008). To determine whether rrp7-1 plants exhibit ABA hypersensitivity, we sowed seeds of this mutant on ABA-supplemented medium. The genotypes of other seeds that we studied in this way were: parl1-2; smo4-3, a null allele of the SMO4 gene, which encodes a RBF that we found to interact with MAS2 in Y2H assays (Sánchez-García et al., 2015); and amiR-MAS2.1, a transgenic line producing an artificial microRNA partially inhibiting expression of MAS2 (Sánchez-García et al., 2015). As controls, we used abscisic acid deficient1-104 (aba1-104) and abscisic acid insensitive4-2 (abi4-2) seeds, which are hypersensitive and insensitive to ABA, respectively (Quesada et al., 2000).
As expected, the germination rates of these seeds did not differ—all seeds germinated like their respective wild types (data not shown). However, the seedling establishment rates clearly differed among lines (Figure 4). Whereas almost all Col-0 seedlings exhibited expanded cotyledons on 0.5 μM ABA, only 3.4 ± 3.9% of rrp7-1 seedlings did so, and no expanded cotyledons were detected at higher ABA concentrations. The values for parl1-2 were 45.8 ± 7.7% at 0.5 μM, 14.8 ± 4.2% at 1.5 μM (50.4 ± 4.7% of Col-0), and 0.2 ± 0.7% at 3 μM ABA (10.4 ± 2.4% of Col-0). The amiR-MAS2.1 plants were more sensitive to ABA than Col-0 at all concentrations tested, but to a lesser extent than parl1-2 and rrp7-1. No significant differences were detected between smo4-3 and Col-0.
Figure 4.
Effect of Exogenous ABA on rrp7-1, smo4-3, parl1-2, and amiR-MAS2.1 Plants.
Percentage of seedlings grown in medium supplemented with different concentrations of ABA that displayed green, fully expanded cotyledons when scored at 10 DAS. The abi4-2 and aba1-104 mutants were used as controls. The experiment was repeated three times, each with 156 seeds of each genotype, sown onto three different plates. Error bars indicate sd. Asterisks indicate values significantly different from the corresponding wild type in a Student′s t test (*P < 0.05, and **P < 0.01). The genetic background of abi4-2 and aba1-104 is Col-5, and that of all other mutants is Col-0.
RRP7 Expression Is Widespread, and Its Promoter Shares Regulatory Elements with SMO4 and MAS2
According to the eFP Browser and the TraVA databases (see Methods), RRP7 is expressed in all organs and at all developmental stages, with maximum expression at the beginning of seed germination. We constructed a transgene that had the GUS reporter gene driven by the RRP7 promoter (RRP7pro:GUS). We detected GUS staining in all tissues of RRP7pro:GUS plants, whose flowers and siliques showed no GUS activity (Supplemental Figure 6), perhaps because the GUS assay is much less sensitive than microarray and RNA-seq analyses. Another possibility is that the 1,026-bp genomic region upstream of the translation start codon of RRP7 does not contain all of regulatory elements. It has been shown that the majority of the regulatory elements (86%) in Arabidopsis are located proximal to the transcription start site (TSS; +1 bp position), from −1,000 bp to +200 bp (Yu et al., 2016).
We also analyzed the RRP7, SMO4 and MAS2 promoters using the PLACE and Athena databases (see “Methods” and Supplemental Figure 7). We found a TELOBOXATEEF1AA1 motif (AAACCCTAA) at the –20 to –12 bp position in the RRP7 promoter, and two identical GGCCCATTA motifs in the SMO4 promoter at –52 to –44 and –82 to –74 bp from its TSS. These motifs correspond to the UP1ATMSD (GGCCCAWWW, where W is A or T) element, which also appears twice in the MAS2 promoter, once (GGCCCAATA) at –65 to –57 and once (inverted) at –82 to –77 bp (TATTGGGCC). Enrichment of the UP1ATMSD motif has been found in the promoters of genes upregulated after main stem decapitation in Arabidopsis (Tatematsu et al., 2005). The TELOBOXATEEF1AA1 is a longer version of the UP2ATMSD (AAACCCTA) and Telo-box (Short interstitial telomere) motifs (AAACCCWA; where W is A or T). The latter sequence is identical to the repeat units of plant telomeres and has been preferentially found in 5′ flanking regions of Arabidopsis and rice genes encoding components of the ribosome biogenesis machinery, including RPs, snoRNAs, and RBFs (Tremousaygue et al., 1999; Gaspin et al., 2010). The UP1ATMSD and UP2ATMSD motifs are enriched among genes encoding RPs (Ma et al., 2012). These results reinforce the hypothesis of a functional relationship among RRP7, SMO4, and MAS2 and their activity in ribosome biogenesis.
RRP7 Is Mainly a Nucleolar Protein
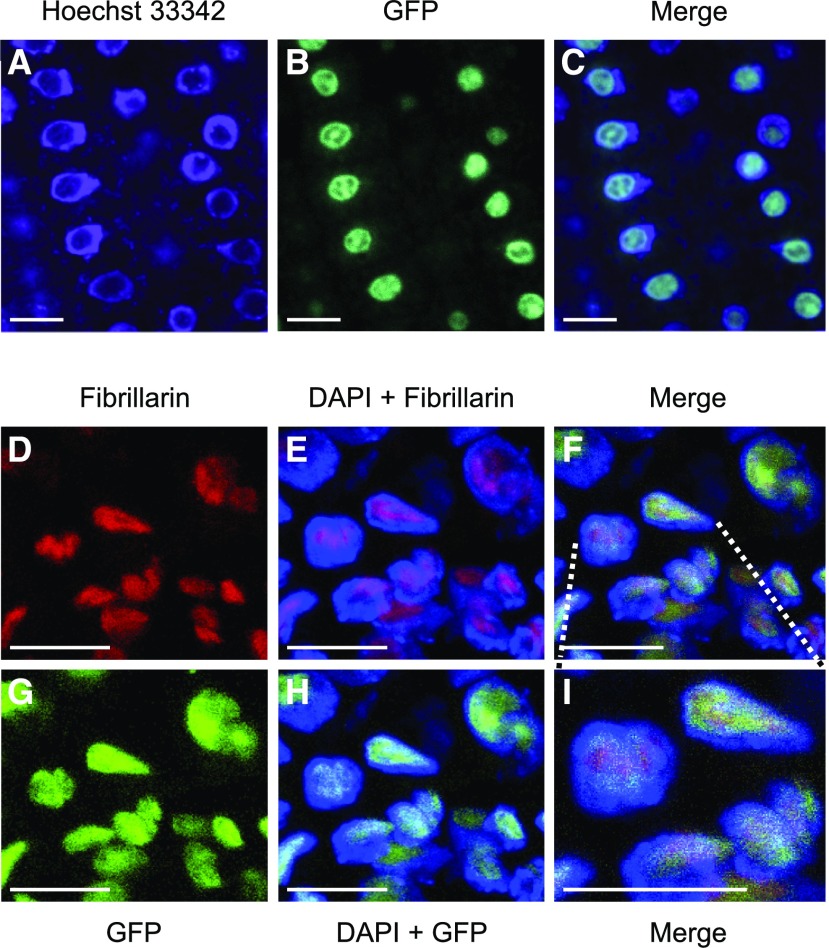
To visualize the subcellular localization of RRP7, we generated translational fusions of RRP7 and the gene encoding the green fluorescent protein (GFP), under the control of the RRP7 endogenous promoter (RRP7pro:RRP7:GFP) or the 35S promoter (35Spro:RRP7:GFP). The RRP7:GFP fusions were functional, as shown by full complementation of the mutant phenotype of rrp7 plants (Figure 1M). To visualize GFP fluorescence, we used roots of Col-0 plants stained with Hoechst 33,342. This dye binds strongly to double-stranded DNA and counterstains the nucleus, but not the nucleolus, which contains mainly RNA. As expected, identical GFP-fluorescence signals were detected in the nucleoli using both the RRP7pro:RRP7:GFP (Figure 5A to 5C) and 35Spro:RRP7:GFP transgenes. The GFP signal was broader than the nucleolus and slightly overlapped with the DAPI-stained region.
Figure 5.
Subcellular Localization of RRP7.
(A) to (C) Confocal laser-scanning micrographs of roots from plants homozygous for the RRP7pro:RRP7:GFP transgene. Fluorescence signals correspond to Hoechst 33342 (A), GFP (B), and the merged image (C).
(D) to (I) Immunolocalization of fibrillarin in plants homozygous for the RRP7pro:RRP7:GFP transgene. Fluorescence signals show: fibrillarin detection in red (D) and (E); DAPI staining in blue (E) and (H); GFP fluorescence in green (G) and (H); and the corresponding merged images (F) and (I). Scale bars = 10 µm.
To determine whether RRP7 is exclusively nucleolar, we performed a fluorescent immunoassay using an antibody against fibrillarin, a conserved nucleolar methyltransferase that directs 2′-O-ribose methylation of pre-rRNAs, a commonly used nucleolar marker. RRP7 and fibrillarin partially colocalized, because the GFP signal occupied a larger portion of the nucleolus than fibrillarin. In addition, a small portion of the GFP signal clearly colocalized with DAPI around of the nucleolus (Figures 5D to 5I).
18S rRNA Maturation Is Impaired in the rrp7 Mutants
In yeast, rrp7 null mutations cause lethality, and rrp7 conditional alleles reduce production of the mature 18S rRNA and 40S ribosomal subunits. In these mutants, depletion of 18S rRNA appears to be mainly due to the inhibition of cleavage at the A2 site in 32S pre-rRNA, which leads to a loss of 27SA2 and 20S pre-rRNAs (Supplemental Figure 1) (Baudin-Baillieu et al., 1997). In addition, yeast Rrp7 coimmunoprecipitates with 90S pre-ribosomes containing 35S or 23S pre-rRNAs (Lin et al., 2013).
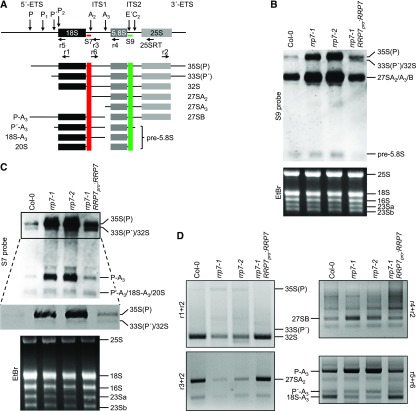
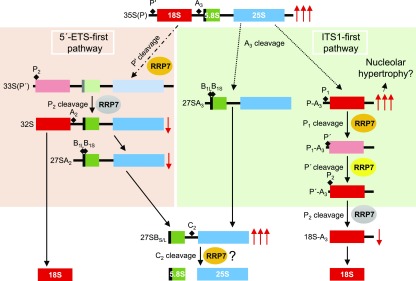
To determine whether RRP7 plays a role in Arabidopsis 45S pre-rRNA processing, we performed gel blot analyses of RNA extracted from rrp7 plants, using the S7 and S9 probes (Lange et al., 2011), which bind the ITS1 and ITS2 regions of 45S pre-rRNA processing intermediates, respectively (Figure 6A and Supplemental Figure 2). Using the S9 probe, we found that the rrp7-1 and rrp7-2 mutants overaccumulated the 35S(P), 33S(P′) and/or 32S pre-rRNAs, and had much higher levels of 27SA2, 27SA3, and/or 27SB pre-rRNAs than did wild-type plants or the complemented rrp7-1 RRP7pro:RRP7 plants (Figures 1K and 6B). Hybridization with the S7 probe indicated that the accumulated early precursor was 35S(P). In addition, we found a strong accumulation of P-A3 pre-rRNA—an intermediate of the ITS1-first pathway for 18S rRNA maturation—in rrp7-1 and rrp7-2 but not in Col-0 or rrp7-1 RRP7pro:RRP7 plants (Figure 6C and Supplemental Figure 2).
Figure 6.
Early Steps in 45S pre-rRNA Processing and 18S rRNA Maturation in the rrp7 Mutants.
(A) Diagram illustrating the pre-rRNAs that can be detected in an RNA gel blot using the S7 and S9 probes. Modified from Hang et al. (2014). 5′-ETS and 3′-ETS, external transcribed spacers. ITS1 and ITS2, internal transcribed spacers. Red and green vertical stripes mark the regions of the pre-rRNAs to which the S7 and S9 probes hybridize, respectively.
(B) and (C) RNA gel blots. Total RNA was separated in formaldehyde-agarose gels, transferred to a nylon membrane, and hybridized with the S9 (B) and S7 (C) probes. RNA was extracted from Col-0, rrp7-1, rrp7-2, and rrp7-1 RRP7pro:RRP7 plants. EtBr: photographs of ethidium bromide-stained gels taken before blotting as loading controls. The 16S, 23Sa and 23Sb bands correspond to chloroplast rRNAs. A magnified view of the selected region is shown in (C), corresponding with a very short exposure time, which allows distinction between the 35S(P), and 33(P′)/32S pre-rRNAs.
(D) Ethidium bromide-stained agarose gels (negative images) visualizing circular RT-PCR products. RNA was circularized and reverse transcribed using the 25SRT primer, and the cDNA obtained was PCR amplified with the r1+r2, r3+r2 and r4+r2 primer pairs. The cDNA PCR amplified with the r5+r6 primer pair was obtained from RNA circularized and reverse transcribed using the r1 primer.
An additional RNA gel blot analysis was performed using the p42-p43 probe, which hybridizes to the A2-A3 segment of ITS1 and allows detection of 27SA2 and 27SA3, but not 27SB pre-rRNAs (Hang et al., 2014). We did not detect any 27SA pre-rRNA species with the p42-p43 probe (Supplemental Figure 8); this observation suggests that the 27SB pre-rRNA is the major contributor to the overaccumulation of 27S pre-rRNAs visualized by the S9 probe in rrp7 mutants (Figures 6A and 6B). These results suggest that RRP7 participates in the P′ site cleavage at the 5′-ETS-first pathway, in the P1 cleavages at the ITS1-first pathway, and in the C2 cleavage, after the convergence of both pathways.
We performed several circular RT-PCR amplifications to unequivocally identify the products overaccumulated in rrp7 plants, discriminating among 35S(P), 33S(P′), and 32S pre-rRNAs; among 27SA2, 27SA3, and 27SB pre-rRNAs; and among P-A3 and the other 18S rRNA precursors. We circularized total RNA, and the circular molecules obtained were reverse transcribed with primers hybridizing into the mature 18S or 25S rRNA sequences (Supplemental Table 4). The cDNAs obtained from circular molecules containing the 18S and 25S rRNA sequences were PCR amplified with different primer sets (Supplemental Table 4).
The r1+r2 primer pair should allow the amplification of 35S(P), 33S(P′), and 32S pre-rRNAs (Figure 6A). We found reduced 32S and increased 35S(P) pre-rRNA levels in rrp7 plants, compared with Col-0 and rrp7-1 RRP7pro:RRP7 plants (Figure 6D). This result confirms that the band observed by RNA gel blotting using the S7 and S9 probes (Figure 6B and 6C) corresponds to the 35S(P) intermediate. The r3+r2 vs. r4+r2 primer pairs allowed us to discriminate among the 27SA2, 27SA3 (only with r4+r2), and 27SB pre-rRNAs (Figure 6A). With the r3+r2 primer pair, we found much less 27SA2 pre-rRNA in rrp7 mutants than in Col-0 or rrp7-1 RRP7pro:RRP7. On the other hand, the r4+r2 primer pair also revealed increased amounts of the 27SB intermediate in rrp7 plants (Figure 6D). We could not amplify the 27SA3 intermediate, probably because it is rapidly processed to 27SB. These results show that RRP7 participates in the earlier steps of 45S pre-rRNA processing, in which cleavage at the P′, P1 and P2 sites of 5′-EST occurs (Supplemental Figure 2).
To confirm the nature of the product that we found to be overaccumulated by RNA gel blots using the S7 probe (Figures 6A and 6C), we amplified the cDNA obtained from reverse transcription of circularized RNAs containing the 18S rRNA sequence using the r5+r6 primer pair. These primers should amplify the P-A3, P′-A3, and 18S-A3 pre-rRNAs, which are intermediates of the ITS1-first pathway of 18S rRNA maturation (Figure 6A and Supplemental Figure 2). We concluded that the overaccumulated intermediate detected by the S7 probe is the P-A3 pre-rRNA (Figure 6D), supporting the requirement of RRP7 for cleavage at the P1 site in the ITS1-first pathway (Supplemental Figure 2). We also found reduced the 18S-A3 pre-rRNA in the rrp7 mutants. These results also confirm that RRP7 participates in the P2 cleavage at the ITS1-first pathway.
The patterns of accumulation of intermediate species of the rRNA biogenesis pathway found in rrp7 plants were opposite to those found in Arabidopsis prmt3 plants, which carry a mutant allele of the Arabidopsis PROTEIN ARGININE METHYLTRANSFERASE 3 (PRMT3) gene. Loss of PRMT3 function causes over-accumulation of the 18S-A3 and 27SB intermediates, suggesting that PRMT3 promotes the major (ITS1-first) pathway and inhibits the minor (5′-EST-first) pathway for 45S pre-rRNA processing (Supplemental Figure 2; Hang et al., 2014).
In rrp7 Mutants, the 25S/18S Ratio Is Increased and 18S rRNA Species Accumulate in the Nucleolus
We examined the rRNA profiles of the rrp7 mutants using an Agilent 2100 Bioanalyzer, as previously used in a functional analysis of putative RBFs in human HeLa cells (Tafforeau et al., 2013). The 25S/18S mature rRNA ratios were 1.97 ± 0.06 in the rrp7 mutants and 1.33 ± 0.15 in the wild type (Supplemental Figure 9). Taken together, these results indicate that RRP7 is required for the synthesis and/or stabilization of mature 18S rRNA species. In addition, the rRNA ratios in rrp7 are reminiscent of the reduced 18S rRNA levels observed in yeast rrp7 conditional mutants by Baudin-Baillieu et al. (1997).
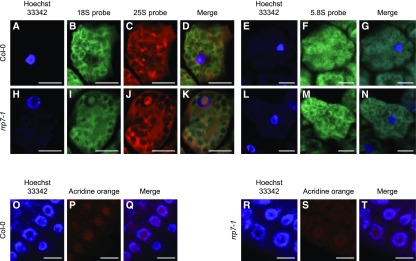
Because the above results suggest that 45S pre-rRNA processing is defective in the rrp7 mutants, we performed RNA fluorescence in situ hybridization (RNA-FISH), using probes that detect the mature 25S, 18S, or 5.8S rRNAs, as well as their respective precursors. No alteration in the subnuclear localization of 5.8S rRNA was detected (Figures 7E to 7G, 7L to 7N), but nucleolar hypertrophy and accumulation of one or more molecules containing the 18S rRNA sequence, probably the P-A3 precursor, were observed in the rrp7-1 mutant (Figures 7B and 7I). In addition, we observed a strong 25S rRNA signal within rrp7 nucleoli, which seems similar in intensity to that within the wild-type nucleoli, but with a larger area, due to nucleolar hypertrophy (Figures 7C to 7D, 7J to 7K)
Figure 7.
Subcellular Localization of 25S, 18S, and 5.8S rRNA Species in rrp7-1 Plants.
(A) to (T) Fluorescence signals correspond to Hoechst 33342 (A), (H), (O) and (R), an 18S probe (B) and (I), a 25S probe (C) and (J), a 5.8S probe (F) and (M), acridine orange (P) and (S), and their corresponding merged images (D), (K), (G), (N), (Q) and (T). Scale bars = 10 µm.
To quantify the size of the nucleolus, we stained cells with DAPI and acridine orange, which emits green or red fluorescence depending on whether it is bound to DNA or RNA, respectively. The ratio of the size of the nucleolus to the size of the whole cell was significantly higher (n = 200, p < 0.001) in rrp7-1 plants (0.28 ± 0.07; Figures 7R to 7T) than in Col-0 (0.21 ± 0.06; Figures 7O to 7Q). These results confirm that 18S rRNA maturation is impaired in rrp7-1 plants and suggest that accumulation of 18S rRNA precursors, in particular the P-A3 pre-rRNA, contributes to nucleolar hypertrophy, as has been proposed for other Arabidopsis mutants defective in rRNA maturation (Lahmy et al., 2004; Abbasi et al., 2010; Chen et al., 2016). We cannot exclude, however, that the other alterations of 45S pre-rRNA processing shown by rrp7 mutants contribute to nucleolar hypertrophy. In addition, it is likely that assembly and export of the pre-40S particles containing the precursors of the ITS1-first pathway are impaired in the rrp7 mutants.
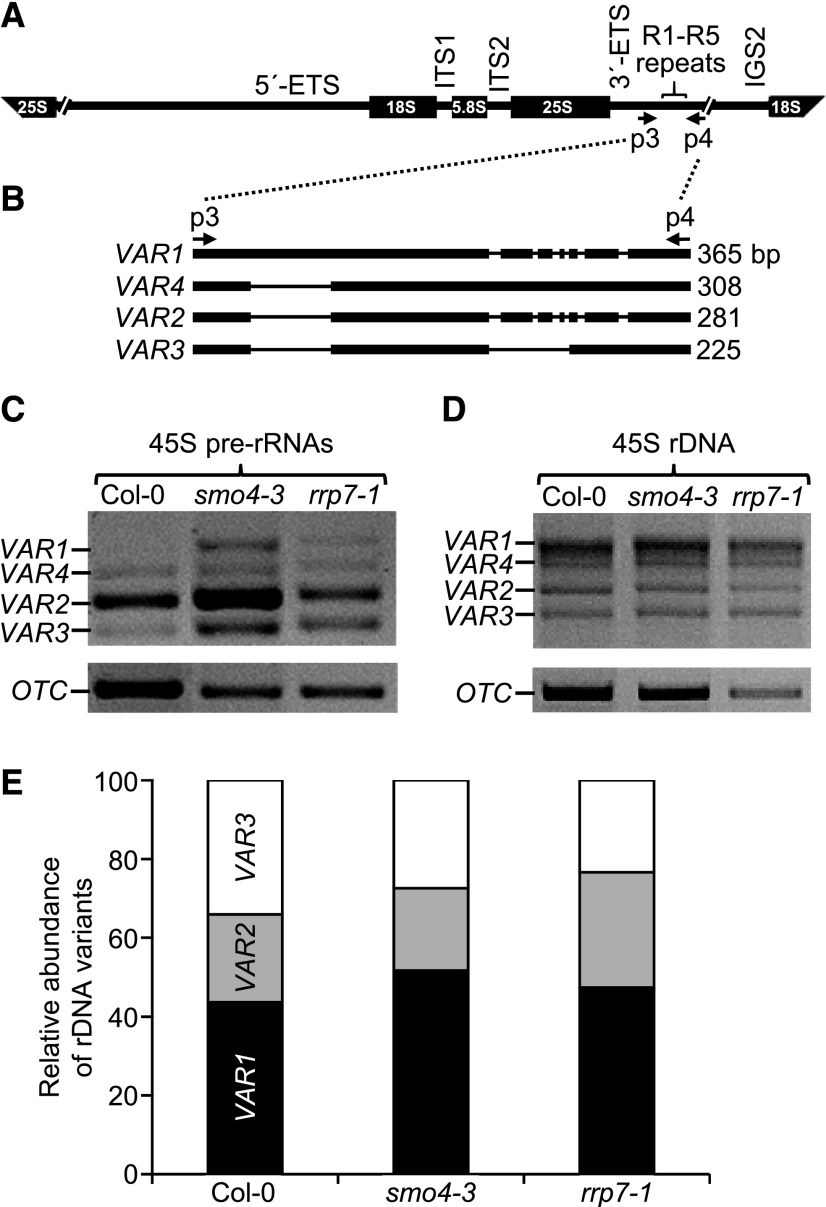
Expression of the 45S rDNA Variants Is Altered in rrp7 and smo4 Mutants
Four different variants (VARs) exist among the hundreds of copies of the 45S rDNA gene in Arabidopsis. These VARs differ in their 3′-ETS sequences and their temporal and spatial expression patterns, as observed in wild-type accessions (Figure 8A and 8B; Pontvianne et al., 2010). The number of genomic copies of each variant does not appear to be related to its expression levels. Indeed, VAR1 is the most abundant 45S rDNA variant in Col-0, but it is only expressed during seed germination, and this accession contains few copies of VAR2, but they are highly expressed. The temporal expression pattern of VAR1 is modified by mutant alleles of NUC1 (Pontvianne et al., 2010).
Figure 8.
45S rDNA VAR Expression in smo4-3 and rrp7-1.
(A) and (B) Schematic representation of the 45S pre-rRNA (A) and its 3′-ETS polymorphic region (B). 5′-ETS and 3′-ETS, external transcribed spacers. ITS1 and ITS2, internal transcribed spacers. The p3 and p4 primers were used for PCR amplifications (Supplemental Table 4).
(C) to (E) PCR analysis of the relative abundance of 45S rDNA variants (VAR1-VAR4) in reverse-transcribed RNA (C), and genomic DNA (D) and (E), from Col-0, smo4-3, and rrp7-1 plants as indicated. Relative amounts of each 45S rDNA variant (E) were determined using the Agilent DNA 1000 kit on an Agilent 2100 Bioanalyzer. VAR4 was not detected. The ORNITHINE TRANSCARBAMYLASE (OTC) housekeeping gene (Quesada et al., 1999) was used as an internal control in (C).
The similar phenotypes of parl1-2 and rrp7 mutants prompted us to analyze the abundance of each VAR and its expression level in the smo4-3 and rrp7-1 mutants. Using the p3+p4 primer pair (Figure 8A and Supplemental Table 4), we amplified by PCR the 45S rDNA 3′-ETS and measured its corresponding transcript levels by RT-PCR (Pontvianne et al., 2010) using RNA extracted from plants collected 14 DAS. There was no significant difference in the genomic content of each VAR among the mutants and wild type (Figure 8D and 8E). We did not detect VAR1 expression in Col-0, and VAR2 was much more highly expressed than VAR3 and VAR4, which were expressed at similar levels (Figure 8C; Pontvianne et al., 2010). However, heterochronic expression of VAR1 was detected in smo4-3 and rrp7-1, and increased expression of VAR2 and VAR3 was detected in smo4-3 at the same ratio as that of Col-0. VAR3 expression levels were higher in rrp7-1 than in Col-0. These results suggest that as already known for NUC1, SMO4 and RRP7 also participate in the regulation of 45S rDNA expression and are required for the negative regulation of VAR1. Changes in the relative abundance of 45S rRNA variants are usually interpreted as changes in their transcription (Kojima et al., 2007; Pontvianne et al., 2010; Durut et al., 2014). Our results, however, could also indicate differential turnover of the variants.
RRP7 Interacts Synergistically with MAS2, NUC1, and HDA6
Double mutant analysis is a classical genetic approach to the understanding of functional relationships between genes (Pérez-Pérez et al., 2009). To genetically confirm the physical interaction of RRP7 with MAS2 that we detected in our previous Y2H-based screen (Sánchez-García et al., 2015), we obtained double mutant combinations of rrp7-1 with mas2-1, a viable MAS2 allele that is a dominant informational suppressor of ago1-52 and lacks phenotypic effects per se in a wild-type genetic background (Sánchez-García et al., 2015). The rrp7-1 mas2-1 double mutant displayed a synergistic phenotype, with very much dwarfed rosettes and narrow, pointed cotyledons and leaves (Figure 9A and 9F, and Supplemental Figure 10). Despite their strong morphological aberrations, rrp7-1 mas2-1 double mutant plants completed their life cycles and produced some seeds.
Figure 9.
Genetic Interactions of rrp7-1 with mas2-1, smo4-3, parl1-2, nuc2-2, and hda6-7.
(A) to (J) Phenotypes of rosettes of single (A) to (E) and double (F) to (J) mutants. Images show rosettes of mas2-1 (A), smo4-3 (B), parl1-2 (C), nuc2-2 (D), and hda6-7 (E) plants (top row) along with rrp7-1 mas2-1 (F), rrp7-1 smo4-3 (G), rrp7-1 parl1-2 (H), rrp7-1 nuc2-2 (I), and rrp7-1 hda6-7 (J) plants (bottom row). Photographs were taken at 21 DAS. Scale bars = 1 mm.
The physical interaction of SMO4 and RRP7 with MAS2 in Y2H assays (Sánchez-García et al., 2015), the regulatory elements shared by their promoters, and the synergistic phenotypes of the rrp7-1 mas2-1 plants prompted us to obtain the rrp7-1 smo4-3 double mutant, which was morphologically indistinguishable from the rrp7-1 single mutant (Figure 9B, 9G and Supplemental Figure 10).
The abovementioned temporal expression pattern of VAR1 is also modified by mutant alleles of NUC2 (Pontvianne et al., 2007), a partially redundant paralog of NUC1, and the HISTONE DEACETYLASE6 (HDA6; Aufsatz et al., 2002); NUC1, NUC2 and HDA6 are epigenetic regulators of 45S rDNA transcription (Petricka and Nelson, 2007; Pontvianne et al., 2010; Layat et al., 2012; Durut et al., 2014). To determine whether any of these genes interacts with RRP7, we obtained double mutant combinations of rrp7-1 with parl1-2, nuc2-2, or hda6-7. The rrp7-1 parl1-2 plants exhibited very short roots and small rosettes with narrow leaves that accumulated anthocyanins (Figure 9C, 9H and Supplemental Figure 10). Rosettes of rrp7-1 nuc2-2 plants were morphologically similar to those of rrp7-1 but larger with pale leaves (Figure 9D, 9I and Supplemental Figure 10). Although hda6-7 plants were barely distinguishable from wild type (Figure 9E and Supplemental Figure 10), rrp7-1 hda6-7 plants displayed the most extreme phenotype among the double mutants obtained, with very much dwarfed rosettes (Figure 9J and Supplemental Figure 10). Unexpectedly, flowering was earlier in rrp7-1 hda6-7 than in the rrp7-1 or hda6-7 single mutants, both of which are late-flowering. Only some plants produced some seeds, most of which (83.33%; n = 20) failed to germinate. These synergistic phenotypes reveal a functional relationship of RRP7 with NUC1 and HDA6.
DISCUSSION
In Silico Analyses Support the Evolutionary Conservation of RRP7 as an RBF
Essential cellular functions, such as the translation of mRNA into protein, are evolutionarily conserved. The 80S ribosome is the molecular machine that performs protein synthesis in the cytoplasm in Eukarya; its mature structure is very similar in all organisms studied, although its biogenesis in fungi, plants, and metazoans exhibits both evolutionary conservation and diversification (Tafforeau et al., 2013; Woolford and Baserga, 2013).
A number of RBFs have been extensively studied in yeast and some have also been studied in human and/or mouse cells; comparative analyses have provided examples of both conservation and divergence. However, only a few RBFs have been experimentally studied in plants (Weis et al., 2015). For example, both yeast Rrp7 and human RRP7A share an NTD and a CTD, but only the CTD is conserved (to some extent) in Arabidopsis RRP7. These findings suggest that the ability of Rrp7 to bind pre-rRNAs within preribosomal particles is conserved in Arabidopsis RRP7 and human RRP7A, but dimerization with an Utp22 ortholog is conserved only in RRP7A. Indeed, mammalian Utp22 orthologs exist (Utama et al., 2002), including mouse nucleolar RNA-associated protein (Nrap) and human nucleolar protein 6 (NOL6), whose interaction with RRP7A was demonstrated by affinity-purification mass spectrometry. However, a role for NTD in such dimerization is yet to be demonstrated (Huttlin et al., 2015).
According to HomoloGene, AT1G63810 encodes the Arabidopsis ortholog of Utp22. This gene has not been studied and is annotated as encoding a nucleolar protein in Araport11 and as Nrap-like in the ARAMEMNON database (Schwacke et al., 2003). The protein shares 31.3% identity with yeast Utp22 and 33.8% with NOL6 and Nrap. As Arabidopsis RRP7 lacks an NTD, it likely does not interact with the AT1G63810 gene product, unless they dimerize in a way substantially different from that of RRP7A with NOL6 (and Rrp7 with Utp22). Both, RRP7 and Utp22 have recently been found in the Arabidopsis nucleolar proteome (Montacié et al., 2017).
We identified the RRP7 and SMO4 RBFs in a Y2H-based screen for interactors of MAS2, the Arabidopsis ortholog of human NKAP (Sánchez-García et al., 2015). MAS2, SMO4, and RRP7 appear to be expressed in all tissues and developmental stages, particularly in actively dividing cells (Sánchez-García et al., 2015; Zhang et al., 2015; this work). Cis regulatory elements usually map within the −1,000 to +200 bp (including introns) regions of Arabidopsis gene promoters relative to the TSS, with a peak at −50 bp (Yu et al., 2016). Indeed, we identified known regulatory elements within the −85 to −1 bp upstream regions of the MAS2, SMO4, and RRP7 promoters. These elements are shared by many genes related to the translational machinery, providing additional support for the assignment of MAS2, SMO4, and RRP7 to the functional class of proteins related to the translational apparatus.
Loss of RRP7 Function Causes Pleiotropic Developmental Effects, but Not Lethality
Most yeast, human, and Arabidopsis genes encoding RBFs are single-copy genes. Their null alleles, however, are lethal in yeast and human or mouse cells, but not in Arabidopsis, and this is the case for RRP7 (this work). A plausible explanation for the viability and relatively mild phenotypes of the rrp7 mutants is that yeast either does not use alternative pre-rRNA processing pathways or uses these pathways to a lesser extent than does Arabidopsis. In another example, the Arabidopsis mtr4 null alleles are viable and have mild phenotypes, whereas their mammalian and yeast equivalents are lethal. Accumulation of pre-rRNAs would occur as a consequence of rrp7 or mtr4 mutations, but the levels of mature rRNAs would be sufficient for viability owing to the alternative pre-rRNA processing pathway(s).
Some other Arabidopsis genes encoding RBFs mentioned in this work (NUC1 and XRN2) have a redundant paralog, which explains their mild mutant phenotypes. The severe phenotypes of the nuc1 nuc2 and xrn2 xrn3 double mutants confirm this explanation (Zakrzewska-Placzek et al., 2010; Durut et al., 2014). By contrast, null alleles of the genes encoding other Arabidopsis RBFs, such as orthologs of yeast Rrp5, Periodic tryptophan protein 2 (Pwp2), Nin one binding protein 1 (Nob1; Supplemental Figure 1), Essential nuclear protein 1 (Enp1), and Nucleolar complex protein 4 (Noc4) cause lethality in Arabidopsis, as do null alleles of their orthologs in yeast (Missbach et al., 2013).
Although RRP7 is not essential, it appears to play important roles in physiology and development as shown by the pleiotropic phenotype of the rrp7 mutants, which exhibit a pointed-leaf phenotype and defective leaf-vein patterning (two well-known traits of ribosomal mutants; Van Lijsebettens et al., 1994; Petricka and Nelson, 2007; Horiguchi et al., 2011; Weis et al., 2015), delayed growth, altered phyllotaxy, late flowering, partial infertility, and hypersensitivity to exogenous ABA at the seedling establishment stage. A deficiency in mature ribosome levels might explain these phenotypes, although it is not clear why a general reduction in mRNA translation causes specific defects in processes as diverse and specific as phyllotaxy regulation, flowering time control, embryogenesis, lateral organ venation patterning (or provascular cell differentiation), and ABA perception.
The absence of RRP7 or NUC1 or the depletion of MAS2 (but not the absence of SMO4) caused ABA hypersensitivity at the seedling establishment stage. The strongest effect was observed in rrp7 plants and the weakest was observed in amiR-MAS2.1 plants. These results suggest that RRP7, NUC1, and MAS2 contribute (to different extents) to the negative regulation of ABA responses. RECEPTOR FOR ACTIVATED C KINASE 1 (RACK1) and eIF6 are also required for normal ribosome production and negatively regulate ABA responses in Arabidopsis; rack1 mutants are hypersensitive to ABA (Guo and Chen, 2008; Guo et al., 2009). In addition to its known role as a seed germination inhibitor under adverse conditions, ABA appears to play a role in the regulation of ribosome biogenesis. During germination, chromatin is decondensed at nucleolar organizer regions, histone acetylation at the 45S rDNA promoters increases, and transcription of these genes is induced, but the opposite process occurs in plants grown in the presence of ABA (Zhang et al., 2012). ABA also induces the expression of DEAD-BOX ATP-DEPENDENT RNA HELICASE 57 (RH57), whose product is involved in 45S pre-rRNA processing; the rh57-1 null mutant is hypersensitive to ABA (Hsu et al., 2014). In addition, Arabidopsis NUC1 is likely a substrate of SUCROSE NONFERMENTING 1 (SNF1)-RELATED PROTEIN KINASE 2 (SNRK2) in response to ABA (Umezawa et al., 2013; Wang et al., 2013). The auxin gene expression1-5 (axe1-5) allele of the HDA6 gene also causes hypersensitivity to ABA (Chen et al., 2010). Notably, we found a synergistic phenotype in the rrp7-1 hda6-7 double mutant, indicating a genetic interaction between RRP7 and HDA6.
RRP7 Is a Mainly Nucleolar Protein that Is Required for 18S rRNA Maturation
An analysis of the nuclear proteome of Arabidopsis identified 1602 proteins in the nucleolar fraction and 2544 in the nuclear fraction, 1429 of which overlapped; RRP7 was detected in both fractions (Palm et al., 2016). Consistent with these findings, we found that RRP7 mainly localized to the nucleolus but also was found at its periphery, which might correspond to the perinucleolar compartment in human cells. This dynamic structure is enriched in RNA binding proteins, including RNA polymerase III, nucleolin, and factors involved in the regulation of alternative splicing (Pollock and Huang, 2010). The only overlapping subnuclear localization of the RRP7 and MAS2 interacting proteins is the periphery of the nucleolus, which could indicate a role for RRP7 in splicing, as already proposed for MAS2 (Sánchez-García et al., 2015).
The information about RRP7 factors is very scarce, with most information coming from Saccharomyces cerevisiae Rrp7. The function of yeast Rrp7 has been deduced based on the effects of its deletion, conditional depletion, and site-directed mutagenesis on growth and molecular phenotypes, including the accumulation of pre-rRNAs in rrp7 mutants (Lin et al., 2013). The yeast Rrp7 protein is part of the SSU processome, which synthesizes the 18S rRNA via cotranscriptional cleavage at the A0, A1, and A2 sites of the 35S pre-rRNA, generating 27SA2 and 20S pre-rRNAs (Supplemental Figure 1). By contrast, the 23S pre-rRNA, which includes most of the 5′-ETS, results from endonucleolytic cleavage at the A3 site of the 35S pre-rRNA prior to cleavage at the A0, A1, and A2 sites via an alternative pathway (Supplemental Figure 1; Granneman and Baserga, 2004; Chaker-Margot et al., 2017). Both the 23S and 21S pre-rRNAs accumulate in yeast rrp7 conditional mutants (Baudin-Baillieu et al., 1997). The 21S pre-rRNA results from cleavage at the A1 site (corresponding to the Arabidopsis P2 site; Supplemental Figures 1 and 2) of 22S pre-rRNA, which in turn is produced by cleavage of 23S pre-rRNA at A0 site (which could be equivalent to the Arabidopsis P′ site; Supplemental Figures 1 and 2). Accumulation of the 23S and 21S pre-rRNAs reveals that cleavage of A0 and A1 is delayed in rrp7 mutants. Therefore, Rrp7 is required primarily for cleavage at A2 site (A2 in Arabidopsis), but also to a lesser extent at A1 (P2) and A0 (P′) sites.
We demonstrated that Arabidopsis RRP7 is involved in 18S rRNA maturation, as is yeast Rrp7. In the Arabidopsis rrp7 mutants studied here, some 18S rRNA precursors overaccumulate, whereas others are depleted, compared with wild type. Overaccumulation or depletion of a pre-rRNA indicates that it is the substrate or the product, respectively, of a partially or completely blocked processing step that requires RRP7. We found increased levels of 35S(P) and reduced amounts of the 32S and 27SA2 pre-rRNAs, but the 27SA3 and P-A3 pre-rRNAs were not depleted. These observations clearly indicate that RRP7 is required for P′ site cleavage at the 5′-ETS-first pathway, but not for A3 site cleavage at the ITS1-first pathway. The participation of RRP7 in the cleavage of the P′ site of 35S(P) pre-rRNA suggests the possibility of a similar role at the P′ site of P1-A3 pre-rRNA. The levels of the substrate (P1-A3) and product (P′-A3) of this step of the ITS1-first pathway, however, were indistinguishable from those of the wild type (Supplemental Figure 2 and Figure 10).
Figure 10.
Proposed Roles for RRP7 in 45S pre-rRNA Processing Based on the Molecular Phenotype of the rrp7 Mutants.
Red arrows indicate over-accumulated (↑↑↑) or depleted (↓) pre-rRNAs, as detected by RNA gel blots and circular RT-PCR assays using the rrp7 mutants. In our model, Arabidopsis RRP7 is assumed to participate in the steps of 45S pre-rRNA processing in which the substrate is overaccumulated in the rrp7 mutants (P′, P1 and C2 cleavages; orange ovals). The question mark in C2 cleavage (bottom middle) indicates that overaccumulation of 27SB pre-rRNA can alternatively be explained as an indirect effect of impaired 18S rRNA production. RRP7 might also participate in the steps in which the product is depleted in the rrp7 mutants (P2 cleavages; gray ovals). Because RRP7 participates in the P′ cleavage at the 5′-ETS-first pathway, it might also cleave the same site at the ITS1-first pathway (yellow oval). Short-lived pre-rRNAs are shown in faint colors. Other details are as shown in Supplemental Figure 2.
Depletion of 32S and 27SA2 pre-rRNAs could be a consequence of defective cleavage at the P′ site of the 35S(P) precursor; nevertheless, it may also be caused by altered cleavage at the P2 site of 33S(P′) pre-rRNA. The latter hypothesis is supported by the reduction of the 18S-A3 pre-rRNA, which is likely to be due to defective cleavage at the P2 site of P′-A3 pre-rRNA. However, no increase in P′-A3 pre-rRNA was observed. We also detected substantial accumulation of the P-A3 intermediate, indicating defective P1 site cleavage at the ITS1-first pathway. Furthermore, overaccumulation of 27SB pre-rRNAs also suggests the participation of RRP7 in the cleavage at the C2 site, which would involve RRP7 not only in 18S rRNA maturation but also in the maturation of the 25S and 5.8S rRNAs (Supplemental Figure 2 and Figure 10). However, alteration in the mature 25S and 5.8S rRNA levels is not supported by the increased 25S/18S rRNA ratio that we have found (Supplemental Figure 9). An alternative explanation to the implication of RRP7 in C2 site cleavage is that the primary effect of its depletion is to impair 18S rRNA production in the rrp7 mutants. Thus, the increased 27SB pre-rRNA level would be an indirect effect of RRP7 depletion.
Compared with yeast Rrp7, less is known about mammalian Rrp7a, a single-copy gene that has also been named Gastric cancer antigen Zg14, since it was identified in a search for human immunogenic proteins for gastric cancer (Linē et al., 2002). Rrp7a was annotated in large-scale cDNA sequencing projects (Kawai et al., 2001; Okazaki et al., 2002). According to HomoloGene, the human RRP7A protein shares 83.9% identity and 91.8% similarity with its mouse Rrp7a ortholog. Mouse Rrp7a is expressed in preimplantation embryos and is required for the morula-to-blastocyst transition, as shown by the lethality caused by its knockdown by microinjection of double-stranded RNA in zygotes (Maserati et al., 2014). Surprisingly, Rrp7a is found throughout the cytoplasm but not in the nucleus of eighT cell blastomeres, prompting the authors to suggest that this protein is a structural component of the ribosome. More recently, however, human RRP7A has been identified as a member of the nucleolar proteome of HeLa cells, and its depletion by small interfering RNAs caused a reduction in the levels of 21S and 18S-E pre-rRNAs, both of which are precursors of the 18S rRNA (Tafforeau et al., 2013).
Our results indicate high functional conservation between yeast and Arabidopsis RRP7 proteins because in both cases their loss cause overaccumulation of 18S rRNA precursors of the ITS1-first pathway. However, due the differences of this pathway in yeast and Arabidopsis, it is not easy to identify the equivalent cleavage sites. Arabidopsis RRP7 seems to participate mainly in the cleavage of the P′ and P1 sites, both of which could correspond to A0 in yeast. However, yeast Rrp7 and Arabidopsis RRP7 also could be functionally divergent in their functions. Yeast Rrp7 is specifically involved in 18S rRNA maturation and has not been found to participate in 5.8S or 25S rRNA maturation, but Arabidopsis RRP7 may participate in their maturation. The inability of Arabidopsis rrp7 mutants to use both pathways to produce mature 18S rRNA would reduce the concentration of the 18S rRNA and consequently, the number of 40S pre-ribosomal particles able to assemble into mature 80S ribosomes. In this model, rrp7 cells would have an excess of 60S subunits compared with 40S subunits, and this would explain their increased 25S/18S rRNA ratio, which in turn leads to a deficiency in mature ribosomes, thereby causing delayed growth (Supplemental Figure 9 and Figure 1B to 1G).
In addition, the nucleolus is enlarged in rrp7 cells, likely due to the accumulation of 18S rRNA species, particularity the P-A3 intermediate, which we detected by RNA-FISH. Enlarged nucleoli have been observed in other mutants affected in genes encoding RBFs, such as arabidopsis pumilio23-1 (pum23-1) and RNA helicase10 (rh10-1; Matsumura et al., 2016) In apum23-1 plants, 18S and 5.8S rRNA precursors accumulate, but their mature rRNAs do not, indicating that, like RRP7, APUM23 is involved in 35S pre-rRNA processing and/or the degradation of pre-rRNA processing byproducts (Abbasi et al., 2010).
As mentioned in the Introduction, the loss of function of yeast Rrp7 reduces 18S rRNA production, which in turn induces the expression of RP-encoding genes and causes a deficiency in the formation of 40S ribosomal subunits and leads to subsequent lethality (Rudra et al., 2007). Our in silico analysis revealed putative Arabidopsis orthologs of the Utp22 and CK2 members of the CURI complex, but not of Ifh1, suggesting that Arabidopsis lacks a CURI complex. However, we cannot exclude the possibility that RRP7 forms part of another complex that coordinates the pathways for the biogenesis of rRNA and RP ribosome components.
Null mutations of yeast Rrp7 are lethal. By contrast, the lack of function of Arabidopsis RRP7 and human RRP7A impairs 18S rRNA biogenesis but the mutants are viable. This observation cannot be explained by functional redundancy because both are single-copy genes. More studies will be required to understand this difference and the absence of anomalies in ribosome biogenesis in Arabidopsis rrp7 mutants. Our results might shed light on the activity of the mammalian orthologs of Arabidopsis RRP7, given the scarcity of information available.
RRP7 Is Functionally Related to MAS2, HDA6, and NUC1
Several of our in silico and experimental results suggest that RRP7 and MAS2 are functionally related. In addition, the synergistic phenotypes of rrp7-1 parl1-2 and rrp7-1 mas2-1 are very similar, indicating that RRP7 genetically interacts with NUC1 and MAS2. The merely additive phenotype of rrp7-1 nuc2-2 plants was expected, given that NUC2 expression is almost undetectable in wild-type plants and that this gene is thought to play a marginal role in ribosome biogenesis compared with NUC1 (Durut et al., 2014). The extremely severe phenotype of rrp7-1 hda6-7 suggests a relationship of RRP7 with HDA6 closer than with NUC1 or MAS2.
MAS2 participates in the control of 45S rDNA expression, as well as splicing (Sánchez-García et al., 2015). NUC1 is involved in the regulation of 45S rDNA expression and 45S pre-rRNA processing (Petricka and Nelson, 2007; Pontvianne et al., 2007). HDA6 participates in several epigenetic processes, including 45S rDNA, transgene, and transposon silencing (Aufsatz et al., 2002; Probst et al., 2004; Earley et al., 2010), but it has not been associated with 45S pre-rRNA processing. The link between MAS2, NUC1, and HDA6 is their participation in the regulation of 45S rDNA transcription, a process that might involve RRP7. This genetic evidence reinforces the notion that RRP7 is involved in 45S rDNA transcriptional regulation. Indeed, we detected heterochronic activation of the VAR1 45S rDNA variant in the rrp7 mutants. We cannot rule out the possibility that this is an indirect effect caused by impaired 45S pre-rRNA processing if 45S rDNA transcription is subjected to negative feedback self-regulation. On the other hand, deregulation of 45S rDNA transcription does not appear to contribute to the phenotype of rrp7, because in hda6 and smo4 plants, VAR1 is also heterochronically misexpressed and their developmental phenotypes are very mild (Pontvianne et al., 2010; this work). In conclusion, our findings demonstrate that RRP7 acts as a plant RBF, functionally related to MAS2, NUC1 and HDA6, and they increase our understanding of the conservation and diversification of RRP7 function among eukaryotes.
METHODS
Plant Material and Growth Conditions
The Arabidopsis thaliana (L) Heynh. Columbia-0 (Col-0) wild-type accession was initially obtained from the Nottingham Arabidopsis Stock Center (NASC; Nottingham, UK) and propagated at our laboratory for further analysis. Seeds of the rrp7-1 (SAIL_628_F08), rrp7-2 (WISCDSLOX461-464C16), parl1-2 (SALK_002764; Petricka and Nelson, 2007), mtr4-2 (GK_048G02; Lange et al., 2011), hda6-7 (also named rts1-1; Aufsatz et al., 2002) and nuc2-2 (GABI_178D01; Durut et al., 2014) mutants were also provided by NASC. The mas2-1 allele was isolated in our laboratory (Sánchez-García et al., 2015). All T-DNA insertional mutants were in the Col-0 background. Plant culture, seed sterilization, and sowing were performed as previously described (Ponce et al., 1998; Berná et al., 1999).
Culture medium was supplemented, when required, with hygromycin (15 µg·mL−1). ABA sensitivity assays were conducted with three plates per genotype, each sown with 52 mutant and 52 wild-type seeds, and the assays were repeated three times. For these ABA assays, the medium was supplemented with 0, 0.5, 1.5, and 3 µM ABA, and non-germinated seeds, seeds exhibiting aborted germination, and seedlings with expanded, green cotyledons were scored 10 DAS. The scores of each line were obtained after calculating the percentages of germinated seedlings on supplemented medium versus the same line on non-supplemented medium.
Gene Nomenclature and Genotyping
RRP7 can be found under accession number AT5G38720 at TAIR (http://arabidopsis.org) and Araport (http://araport.org). NUC1 and NUC2 have been also termed PARL1 and PARALLEL1-LIKE 1 (PARLL1), respectively (Petricka and Nelson, 2007). HDA6 is also annotated as AXE1 (AUXIN GENE EXPRESSION 1; Murfett et al., 2001), RNA-MEDIATED TRANSCRIPTIONAL SILENCING 1 (RTS1; Aufsatz et al., 2002), and RPD3-LIKE HISTONE DEACETYLASE 3B (RPD3B).
The presence of T-DNA insertions in the genes under study was verified by PCR using the primers shown in Supplemental Table 4. Discrimination between the wild-type MAS2 and mutant mas2-1 alleles was performed as described in Sánchez-García et al. (2015).
RNA Isolation, RT-PCR, RNA Gel Blot, and Circular RT-PCR Assay
Each biological replicate of RNA was isolated with TRI RNA Isolation Reagent (Sigma-Aldrich). RNA samples were either treated twice with DNase using 2 U of TURBO DNase (TURBO DNA-free Kit, Thermo Fisher Scientific) per microgram of RNA or not treated, depending of their use for RT-PCR or RNA gel blots, respectively.
RT-PCR experiments were performed using three biological replicates, each replicate consisting in a pool of three plants. Each plant of a pool was selected from a different plate. PCR amplifications of 45S rDNA or RNA variants for the determination of its relative abundance in genome and expression were performed with the p3+p4 primer pair (Supplemental Table 4) using genomic DNA or cDNA as templates, respectively, from plants collected 15 DAS. Genomic DNA was isolated using the DNeasy Plant Mini Kit (Qiagen), and RNA and cDNA were prepared as previously described. RT-PCR products were visualized by electrophoresis in an agarose gel, whereas PCR products from genomic DNA were purified with the Agencourt AMPure XP system (Beckman Coulter), processed in a 2100 Bioanalyzer using the Agilent DNA 1000 kit and PCR products, and the relative abundance of each variant was determined with the 2100 Expert Software (Agilent Technologies).
RNA gel blot analyses were performed with digoxigenin (DIG)- labeled probes: the S7 and S9 probes are 5′-DIG labeled oligonucleotides (Eurofins Genomics), and the p42-p43 probe was obtained by PCR using 11-DIG-dUTP (Roche) and the p42+p43 primer pair. Primers for the synthesis of both probes (Supplemental Table 4) were described in Lange et al. (2011). RNA gel blot analysis of 3 µg of total RNA samples was performed in 1.2% (w/v) agarose/formaldehyde or 6% polyacrylamide/8 M urea gels. The RNA was visualized and transferred via cross-linking to an Amersham Hybond N+ nylon membrane (Thermo Fisher Scientific). Electrophoresis, hybridization, and detection were performed as previously described (Jover-Gil et al., 2014). In brief, the membrane was prehybridized for 2 h at 65°C and hybridized overnight at 65°C with 149 ng·mL−1 of probe. No blocking agent was added to the hybridization solution, as recommended by the manufacturer. The membrane was incubated with 0.05 U·mL−1 of Fab fragments from an anti-digoxin antibody from sheep, conjugated with alkaline phosphatase (α-DIG-AP, Fab fragments; Roche), washed twice, equilibrated in detection buffer, and incubated with 25 mM CDP-Star (Roche) diluted 1:200 in detection buffer for 5 min in the dark. Visualization of RNA bands was performed by developing Lumi-film chemiluminescent films (Roche) that were exposed to the membrane for 20 min or overnight.
Circular RT-PCR was performed as described in Hang et al. (2014), using 5 µg of total RNA. The 25SRT and r1 primers (Supplemental Table 4) were used for reverse transcription of circularized 25S and 18S rRNA precursors, respectively, using Maxima reverse transcriptase (Thermo Fisher Scientific) following the manufacturer′s instructions. PCR amplifications were performed with the r1+r2, r3+r2, r4+r2, and r5+r6 primer pairs (Supplemental Table 4).
Construction of Transgenic Lines
Transgenic plants were obtained as described in Sánchez-García et al. (2015). The transgenes were obtained using the pGEM-T Easy221 entry vector and the pMDC32, pMD164, pMDC83, and pMDC107 destination vectors (Curtis and Grossniklaus, 2003). To construct the 35Spro:RRP7 and 35Spro:RRP7:GFP overexpression transgenes, the full-length coding sequence (with stop codons removed to obtain the GFP translational fusions) of RRP7 was PCR amplified from Col-0. The 1,026-bp genomic region upstream of the translation start codon of RRP7 was PCR amplified and used as the promoter to drive the RRP7pro:GUS, and RRP7pro:RRP7:GFP constructs. All constructs were verified by sequencing before being transferred into plants. Primers used to obtain these constructs and for Sanger sequencing are described in Supplemental Table 4. Sanger sequencing was performed with ABI PRISM BigDye Terminator Cycle Sequencing kits on an ABI PRISM 3130xl Genetic Analyzer (Applied Biosystems [now Thermo Fisher Scientific], Waltham, MA, USA).
Morphometry, Histology, Histochemical Assays, and RNA-FISH
A Nikon D-Eclipse C1 confocal microscope was used for microscopy. Micrographs were digitally processed using EZ-C1 operation software (Nikon). For venation pattern morphometry, the phenoVein software (http://www.plant-image-analysis.org/) was used with samples of cotyledons, leaves, and petals, which were collected, cleared, and mounted on slides, and the corresponding micrographs taken and converted into diagrams as previously described (Candela et al., 1999; Robles et al., 2010; Jover-Gil et al., 2012).
GUS assays were performed as described by Robles et al. (2010): GUS activity was analyzed in plants homozygous for the GUS transgene, and photographs were taken from three plants from each of three independent lines per genotype.
Visualization of RRP7 subcellular localization was performed in roots from RRP7pro:RRP7:GFP or 35Spro:RRP7:GFP transgenic plants collected at 10 DAS. For immunolocalization with fibrillarin, RRP7pro:RRP7:GFP seedlings were collected at 10 DAS and squashed on slides in a mix of 1x PBS and 4% paraformaldehyde. Immunodetection of fibrillarin was performed using a 1:250 dilution of the mouse monoclonal anti-fibrillarin antibody [38F3] (Abcam), followed by detection with a 1:1,000 dilution of TRITC (tetramethylrhodamine-5-isothiocyanate)-conjugated anti-mouse IgG secondary antibody (Sigma-Aldrich). Nuclei were stained as previously described (Díaz-Tielas et al., 2012) with Hoechst 33342 and the Vectashield mounting medium (Vector Laboratories).
RNA-FISH was performed as described in Parry et al. (2006), with some modifications. Approximately 100 cells from first- and second-node leaves from 10 plants collected at 12 DAS per genotype were analyzed. The leaves were fixed in glass vials at 600 mbar for 25 min. Probes were obtained using the labeled oligonucleotides shown in Supplemental Table 4 and were used in a 0.5 µg·mL−1 hybridization solution. Samples were mounted on slides with a drop of Vectashield antifade mounting medium (Vector Laboratories, Burlingame, California, USA) containing 10 µg·mL−1 of Hoechst 33,342.
Acridine orange staining was performed as described by Hirano et al. (2011). Nucleoli outlines were drawn on a Wacom DTF-720 Pen Display with the Adobe Photoshop CS3 software, and outline areas were measured using the NIS Elements AR 3.1 image analysis package. The ratio between the nucleolus and whole cell areas was obtained from ∼200 cells from 10 plants per genotype.
Bioinformatic and Statistical Analyses
BLASTP searches were performed against the sequences for all organisms at the National Center for Biotechnology Information BLASTP server (NCBI; https://blast.ncbi.nlm.nih.gov/Blast.cgi; Altschul et al., 1997) using the NCBI non-redundant database with default settings. Alignments were obtained with ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalo/; Larkin et al., 2007) and shaded with BOXSHADE3.21 (http://www.ch.embnet.org/software/BOX_form.html). Pair-wise identity and similarity percentages were calculated from the alignments using the Ident and Sim feature of the Sequence Manipulation Suite (http://www.bioinformatics.org/sms2/ident_sim.html; Stothard, 2000).
For promoter analysis, the Scan tool of the PLACE (Plant cis-acting regulatory DNA elements; http://www.dna.affrc.go.jp/PLACE; Higo et al., 1998, 1999) and Athena (http://bioinformatics1.smb.wsu.edu/cgi-bin/Athena/cgi/home.pl; O’Connor et al., 2005) databases were used. The original PLACE website is no longer available, but its data set of regulatory motifs can be accessed at http://togodb.biosciencedbc.jp/togodb/view/place_main#en.
Spatiotemporal gene expression patterns were obtained from the TraVA (Transcriptome Variation Analysis) database (http://travadb.org/; Klepikova et al., 2016), which is based on RNA-seq data. For TraVA output visualization, the Raw Norm option was chosen for read counts number type, and default values were chosen for all other options. To compare the mutant, transgenic, and wild-type lines, the Mann-Whitney U-test (n ≤ 10) or Student′s t test (n > 10) was used.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: ABA1 (AT5G67030), ABI4 (AT2G40220), HDA6 (AT5G63110), NUC1 (AT1G48920), NUC2 (AT3G18610), MAS2 (AT4G02720), OTC (AT1G75330), RRP7 (AT5G38720), SMO4 (AT2G40430). Germplasm included seeds of rrp7-1 (SAIL_628_F08), rrp7-2 (WISCDSLOX461-464C16), parl1-2 (SALK_002764; Petricka and Nelson, 2007), mtr4-2 (GK_048G02; Lange et al., 2011), hda6-7 (also named rts1-1; Aufsatz et al., 2002) and nuc2-2 (GABI_178D01; Durut et al., 2014) that were provided by NASC.
Supplemental Data
Supplemental Figure 1. Overview of 35S pre-rRNA Processing in Yeast.
Supplemental Figure 2. Overview of 45S pre-rRNA Processing in Arabidopsis.
Supplemental Figure 3. Sequence Conservation among Human, Yeast, and Arabidopsis RRP7 Putative Orthologs.
Supplemental Figure 4. Sequence Conservation among Plant RRP7 Putative Orthologs.
Supplemental Figure 5. Morphological Phenotype and RRP7 mRNA Levels of 35Spro:RRP7:GFP and 35Spro:RRP7 Plants.
Supplemental Figure 6. Spatial Expression Pattern of RRP7.
Supplemental Figure 7. Motifs Related to the Translational Apparatus Found in the Promoters of MAS2, SMO4, and RRP7.
Supplemental Figure 8.Gel Blot Analysis of RNA Isolated from the rrp7 Mutants Using the p42-p43 Probe.
Supplemental Figure 9. Agilent 2100 Bioanalyzer Electropherogram Profiles of Total RNA Samples Extracted from Col-0, rrp7-1, and rrp7-1 RRP7pro:RRP7 Plants.
Supplemental Figure 1.0. Genetic Interactions of rrp7-1 with mas2-1, smo4-3, parl1-2, nuc2-2, and hda6-7.
Supplemental Table 1. Identity and Similarity Among Full-Length Representative Plant RRP7 Putative Orthologs.
Supplemental Table 2. Identity and Similarity among the RRP7-like Domains of Representative Plant RRP7 Putative Orthologs.
Supplemental Table 3. Morphometry of Leaf Venation in the rrp7-1 and parl1-2 Mutants.
Supplemental Table 4. Oligonucleotides Used in this Work.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank J.M. Serrano-García, J. Castelló-Bañuls, M.J. Ñíguez-Gómez and S.B. Ingham for their excellent technical assistance, and F.J. Medina for his help and advice with fibrillarin immunolocalization. We also thank B. Scheres for the pGEM-T Easy221 vector and J.L. Micol for useful discussions, comments on the manuscript, and the use of his facilities. This research was supported by grants from the Ministerio de Economía, Industria y Competitividad of Spain (BIO2014-56889-R and BIO2017-83629-R) and the Generalitat Valenciana (PROMETEOII/2014/006) to M.R.P., by a UPVD fellowship to C.M. and an ANR-17-CE12-0026-01 grant to J.S.V.
AUTHOR CONTRIBUTIONS
M.R.P. conceived and supervised the study, obtained funding and provided resources. M.R.P., R.M.-P., and R.S.-M. designed the methodology. All authors performed the research. M.R.P. and R.M.-P. wrote the original draft. All authors reviewed and edited the manuscript.
References
- Abbasi N., Kim H.B., Park N.I., Kim H.S., Kim Y.K., Park Y.I., Choi S.B. (2010). APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 64: 960–976. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufsatz W., Mette M.F., van der Winden J., Matzke M., Matzke A.J. (2002). HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 21: 6832–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin-Baillieu A., Tollervey D., Cullin C., Lacroute F. (1997). Functional analysis of Rrp7p, an essential yeast protein involved in pre-rRNA processing and ribosome assembly. Mol. Cell. Biol. 17: 5023–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102: 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M., Tollervey D. (1992). Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 11: 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M., Tollervey D. (1995). Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 14: 4350–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berná G., Robles P., Micol J.L. (1999). A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 152: 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela H., Martínez-Laborda A., Micol J.L. (1999). Venation pattern formation in Arabidopsis thaliana vegetative leaves. Dev. Biol. 205: 205–216. [DOI] [PubMed] [Google Scholar]
- Chaker-Margot M., Barandun J., Hunziker M., Klinge S. (2017). Architecture of the yeast small subunit processome. Science 355: aal1880. [DOI] [PubMed] [Google Scholar]
- Chen L.T., Luo M., Wang Y.Y., Wu K. (2010). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61: 3345–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.C., Wang H.J., Jauh G.Y. (2016). Dual Role of a SAS10/C1D Family Protein in Ribosomal RNA Gene Expression and Processing Is Essential for Reproduction in Arabidopsis thaliana. PLoS Genet. 12: e1006408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.Y., Krishnakumar V., Chan A.P., Thibaud-Nissen F., Schobel S., Town C.D. (2017). Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89: 789–804. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Tielas C., Graña E., Sotelo T., Reigosa M.J., Sánchez-Moreiras A.M. (2012). The natural compound trans-chalcone induces programmed cell death in Arabidopsis thaliana roots. Plant Cell Environ. 35: 1500–1517. [DOI] [PubMed] [Google Scholar]
- Durut N., Abou-Ellail M., Pontvianne F., Das S., Kojima H., Ukai S., de Bures A., Comella P., Nidelet S., Rialle S., Merret R., Echeverria M., et al. (2014). A duplicated NUCLEOLIN gene with antagonistic activity is required for chromatin organization of silent 45S rDNA in Arabidopsis. Plant Cell 26: 1330–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K., Pontvianne F., Wierzbicki A., Blevins T., Tucker S., Costa-Nunes P., Pontes O., Pikaard C. (2010). Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev. 24: 1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspin C., Rami J.F., Lescure B. (2010). Distribution of short interstitial telomere motifs in two plant genomes: putative origin and function. BMC Plant Biol. 10: 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S., Baserga S.J. (2004). Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 296: 43–50. [DOI] [PubMed] [Google Scholar]
- Guo J., Chen J.G. (2008). RACK1 genes regulate plant development with unequal genetic redundancy in Arabidopsis. BMC Plant Biol. 8: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Wang J., Xi L., Huang W.D., Liang J., Chen J.G. (2009). RACK1 is a negative regulator of ABA responses in Arabidopsis. J. Exp. Bot. 60: 3819–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang R., Liu C., Ahmad A., Zhang Y., Lu F., Cao X. (2014). Arabidopsis protein arginine methyltransferase 3 is required for ribosome biogenesis by affecting precursor ribosomal RNA processing. Proc. Natl. Acad. Sci. USA 111: 16190–16195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A.K., Plisson-Chastang C., O’Donohue M.F., Chakraborty A., Gleizes P.E. (2015). An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 6: 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Higo H. (1998). PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 26: 358–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Matsuzawa T., Takegawa K., Sato M.H. (2011). Loss-of-function and gain-of-function mutations in FAB1A/B impair endomembrane homeostasis, conferring pleiotropic developmental abnormalities in Arabidopsis. Plant Physiol. 155: 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G., Mollá-Morales A., Pérez-Pérez J.M., Kojima K., Robles P., Ponce M.R., Micol J.L., Tsukaya H. (2011). Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J. 65: 724–736. [DOI] [PubMed] [Google Scholar]
- Hsu Y.F., Chen Y.C., Hsiao Y.C., Wang B.J., Lin S.Y., Cheng W.H., Jauh G.Y., Harada J.J., Wang C.S. (2014). AtRH57, a DEAD-box RNA helicase, is involved in feedback inhibition of glucose-mediated abscisic acid accumulation during seedling development and additively affects pre-ribosomal RNA processing with high glucose. Plant J. 77: 119–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J.M., Ares M. Jr (1991). Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 10: 4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin E.L., Ting L., Bruckner R.J., Gebreab F., Gygi M.P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., Dong R., Guarani V., et al. (2015). The BioPlex Network: A systematic exploration of the human interactome. Cell 162: 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jover-Gil S., Candela H., Robles P., Aguilera V., Barrero J.M., Micol J.L., Ponce M.R. (2012). The microRNA pathway genes AGO1, HEN1 and HYL1 participate in leaf proximal-distal, venation and stomatal patterning in Arabidopsis. Plant Cell Physiol. 53: 1322–1333. [DOI] [PubMed] [Google Scholar]
- Jover-Gil S., Paz-Ares J., Micol J.L., Ponce M.R. (2014). Multi-gene silencing in Arabidopsis: a collection of artificial microRNAs targeting groups of paralogs encoding transcription factors. Plant J. 80: 149–160. [DOI] [PubMed] [Google Scholar]
- Kawai J., Shinagawa A., Shibata K., Yoshino M., Itoh M., Ishii Y., Arakawa T., Hara A., Fukunishi Y., Konno H., Adachi J., Fukuda S., et al. ; RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium (2001). Functional annotation of a full-length mouse cDNA collection. Nature 409: 685–690. [DOI] [PubMed] [Google Scholar]
- Kiss A.M., Jády B.E., Darzacq X., Verheggen C., Bertrand E., Kiss T. (2002). A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res. 30: 4643–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepikova A.V., Kasianov A.S., Gerasimov E.S., Logacheva M.D., Penin A.A. (2016). A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 88: 1058–1070. [DOI] [PubMed] [Google Scholar]
- Kojima H., Suzuki T., Kato T., Enomoto K., Sato S., Kato T., Tabata S., Sáez-Vasquez J., Echeverría M., Nakagawa T., Ishiguro S., Nakamura K. (2007). Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 49: 1053–1063. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Peng W.T., Cagney G., Robinson M.D., Haw R., Zhong G., Guo X., Zhang X., Canadien V., Richards D.P., Beattie B.K., Lalev A., et al. (2004). High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13: 225–239. [DOI] [PubMed] [Google Scholar]
- Kuhn J.M., Hugouvieux V., Schroeder J.I. (2008). mRNA cap binding proteins: effects on abscisic acid signal transduction, mRNA processing, and microarray analyses. Curr. Top. Microbiol. Immunol. 326: 139–150. [DOI] [PubMed] [Google Scholar]
- Lafontaine D.L. (2015). Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 22: 11–19. [DOI] [PubMed] [Google Scholar]
- Lahmy S., Guilleminot J., Cheng C.M., Bechtold N., Albert S., Pelletier G., Delseny M., Devic M. (2004). DOMINO1, a member of a small plant-specific gene family, encodes a protein essential for nuclear and nucleolar functions. Plant J. 39: 809–820. [DOI] [PubMed] [Google Scholar]
- Lange H., Sement F.M., Gagliardi D. (2011). MTR4, a putative RNA helicase and exosome co-factor, is required for proper rRNA biogenesis and development in Arabidopsis thaliana. Plant J. 68: 51–63. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Layat E., Sáez-Vásquez J., Tourmente S. (2012). Regulation of Pol I-transcribed 45S rDNA and Pol III-transcribed 5S rDNA in Arabidopsis. Plant Cell Physiol. 53: 267–276. [DOI] [PubMed] [Google Scholar]
- Lin J., Lu J., Feng Y., Sun M., Ye K. (2013). An RNA-binding complex involved in ribosome biogenesis contains a protein with homology to tRNA CCA-adding enzyme. PLoS Biol. 11: e1001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linē A., Stengrēvics A., Slucka Z., Li G., Jankevics E., Rees R.C. (2002). Serological identification and expression analysis of gastric cancer-associated genes. Br. J. Cancer 86: 1824–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98: 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Bachan S., Porto M., Bohnert H.J., Snyder M., Dinesh-Kumar S.P. (2012). Discovery of stress responsive DNA regulatory motifs in Arabidopsis. PLoS One 7: e43198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maserati M., Dai X., Walentuk M., Mager J. (2014). Identification of four genes required for mammalian blastocyst formation. Zygote 22: 331–339. [DOI] [PubMed] [Google Scholar]
- Matsumura Y., Ohbayashi I., Takahashi H., Kojima S., Ishibashi N., Keta S., Nakagawa A., Hayashi R., Saéz-Vásquez J., Echeverria M., Sugiyama M., Nakamura K., et al. (2016). A genetic link between epigenetic repressor AS1-AS2 and a putative small subunit processome in leaf polarity establishment of Arabidopsis. Biol. Open 5: 942–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micol-Ponce R., Aguilera V., Ponce M.R. (2014). A genetic screen for suppressors of a hypomorphic allele of Arabidopsis ARGONAUTE1. Sci. Rep. 4: 5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missbach S., Weis B.L., Martin R., Simm S., Bohnsack M.T., Schleiff E. (2013). 40S ribosome biogenesis co-factors are essential for gametophyte and embryo development. PLoS One 8: e54084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montacié C., Durut N., Opsomer A., Palm D., Comella P., Picart C., Carpentier M.C., Pontvianne F., Carapito C., Schleiff E., Sáez-Vásquez J. (2017). Nucleolar Proteome Analysis and Proteasomal Activity Assays Reveal a Link between Nucleolus and 26S Proteasome in A. thaliana. Front. Plant Sci. 8: 1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Xing R., Schubert D., Goodrich J. (2015). Non-inductive conditions expose the cryptic bract of flower phytomeres in Arabidopsis thaliana. Plant Signal. Behav. 10: e1010868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfett J., Wang X.J., Hagen G., Guilfoyle T.J. (2001). Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell 13: 1047–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myburg A.A., Grattapaglia D., Tuskan G.A., Hellsten U., Hayes R.D., Grimwood J., Jenkins J., Lindquist E., Tice H., Bauer D., Goodstein D.M., Dubchak I., et al. (2014). The genome of Eucalyptus grandis. Nature 510: 356–362. [DOI] [PubMed] [Google Scholar]
- O’Connor T.R., Dyreson C., Wyrick J.J. (2005). Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413. [DOI] [PubMed] [Google Scholar]
- Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Yamanaka I., Kiyosawa H., et al. ; FANTOM Consortium; RIKEN Genome Exploration Research Group Phase I & II Team (2002). Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563–573. [DOI] [PubMed] [Google Scholar]
- Palm D., Simm S., Darm K., Weis B.L., Ruprecht M., Schleiff E., Scharf C. (2016). Proteome distribution between nucleoplasm and nucleolus and its relation to ribosome biogenesis in Arabidopsis thaliana. RNA Biol. 13: 441–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G., Ward S., Cernac A., Dharmasiri S., Estelle M. (2006). The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18: 1590–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez J.M., Candela H., Micol J.L. (2009). Understanding synergy in genetic interactions. Trends Genet. 25: 368–376. [DOI] [PubMed] [Google Scholar]
- Petricka J.J., Nelson T.M. (2007). Arabidopsis nucleolin affects plant development and patterning. Plant Physiol. 144: 173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A.S., Bernier C.R., Hsiao C., Norris A.M., Kovacs N.A., Waterbury C.C., Stepanov V.G., Harvey S.C., Fox G.E., Wartell R.M., Hud N.V., Williams L.D. (2014). Evolution of the ribosome at atomic resolution. Proc. Natl. Acad. Sci. USA 111: 10251–10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps K.R., Charette J., Baserga S.J. (2011). The small subunit processome in ribosome biogenesis—progress and prospects. Wiley Interdiscip. Rev. RNA 2: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock C., Huang S. (2010). The perinucleolar compartment. Cold Spring Harb. Perspect. Biol. 2: a000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce M.R., Quesada V., Micol J.L. (1998). Rapid discrimination of sequences flanking and within T-DNA insertions in the Arabidopsis genome. Plant J. 14: 497–501. [DOI] [PubMed] [Google Scholar]
- Pontvianne F., Matía I., Douet J., Tourmente S., Medina F.J., Echeverria M., Sáez-Vásquez J. (2007). Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol. Biol. Cell 18: 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., Abou-Ellail M., Douet J., Comella P., Matia I., Chandrasekhara C., Debures A., Blevins T., Cooke R., Medina F.J., Tourmente S., Pikaard C.S., et al. (2010). Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet. 6: e1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst A.V., Fagard M., Proux F., Mourrain P., Boutet S., Earley K., Lawrence R.J., Pikaard C.S., Murfett J., Furner I., Vaucheret H., Mittelsten Scheid O. (2004). Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16: 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V., Ponce M.R., Micol J.L. (1999). OTC and AUL1, two convergent and overlapping genes in the nuclear genome of Arabidopsis thaliana. FEBS Lett. 461: 101–106. [DOI] [PubMed] [Google Scholar]
- Quesada V., Ponce M.R., Micol J.L. (2000). Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154: 421–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles P., Fleury D., Candela H., Cnops G., Alonso-Peral M.M., Anami S., Falcone A., Caldana C., Willmitzer L., Ponce M.R., Van Lijsebettens M., Micol J.L. (2010). The RON1/FRY1/SAL1 gene is required for leaf morphogenesis and venation patterning in Arabidopsis. Plant Physiol. 152: 1357–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D., Mallick J., Zhao Y., Warner J.R. (2007). Potential interface between ribosomal protein production and pre-rRNA processing. Mol. Cell. Biol. 27: 4815–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-García A.B., Aguilera V., Micol-Ponce R., Jover-Gil S., Ponce M.R. (2015). Arabidopsis MAS2, an essential gene that encodes a homolog of animal NF-kappa B activating protein, is involved in 45S ribosomal DNA silencing. Plant Cell 27: 1999–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W.B., Flügge U.I., Kunze R. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P. (2000). The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28: 1102–, 1104.. [DOI] [PubMed] [Google Scholar]
- Tafforeau L., Zorbas C., Langhendries J.L., Mullineux S.T., Stamatopoulou V., Mullier R., Wacheul L., Lafontaine D.L. (2013). The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol. Cell 51: 539–551. [DOI] [PubMed] [Google Scholar]
- Tatematsu K., Ward S., Leyser O., Kamiya Y., Nambara E. (2005). Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiol. 138: 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremousaygue D., Manevski A., Bardet C., Lescure N., Lescure B. (1999). Plant interstitial telomere motifs participate in the control of gene expression in root meristems. Plant J. 20: 553–561. [DOI] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Takahashi F., Anderson J.C., Ishihama Y., Peck S.C., Shinozaki K. (2013). Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6: rs8. [DOI] [PubMed] [Google Scholar]
- Utama B., Kennedy D., Ru K., Mattick J.S. (2002). Isolation and characterization of a new nucleolar protein, Nrap, that is conserved from yeast to humans. Genes Cells 7: 115–132. [DOI] [PubMed] [Google Scholar]
- Van Lijsebettens M., Vanderhaeghen R., De Block M., Bauw G., Villarroel R., Van Montagu M. (1994). An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J. 13: 3378–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Xue L., Batelli G., Lee S., Hou Y.J., Van Oosten M.J., Zhang H., Tao W.A., Zhu J.K. (2013). Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 110: 11205–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis B.L., Kovacevic J., Missbach S., Schleiff E. (2015). Plant-specific features of ribosome biogenesis. Trends Plant Sci. 20: 729–740. [DOI] [PubMed] [Google Scholar]
- Wilson D.N., Doudna Cate J.H. (2012). The structure and function of the eukaryotic ribosome. Cold Spring Harb. Perspect. Biol. 4: a011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford J.L. Jr., Baserga S.J. (2013). Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.P., Lin J.J., Li W.H. (2016). Positional distribution of transcription factor binding sites in Arabidopsis thaliana. Sci. Rep. 6: 25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska-Placzek M., Souret F.F., Sobczyk G.J., Green P.J., Kufel J. (2010). Arabidopsis thaliana XRN2 is required for primary cleavage in the pre-ribosomal RNA. Nucleic Acids Res. 38: 4487–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Hu Y., Yan S., Li H., He S., Huang M., Li L. (2012). ABA-mediated inhibition of seed germination is associated with ribosomal DNA chromatin condensation, decreased transcription, and ribosomal RNA gene hypoacetylation. Plant Mol. Biol. 79: 285–293. [DOI] [PubMed] [Google Scholar]
- Zhang X.R., Qin Z., Zhang X., Hu Y. (2015). Arabidopsis SMALL ORGAN 4, a homolog of yeast NOP53, regulates cell proliferation rate during organ growth. J. Integr. Plant Biol. 57: 810–818. [DOI] [PubMed] [Google Scholar]