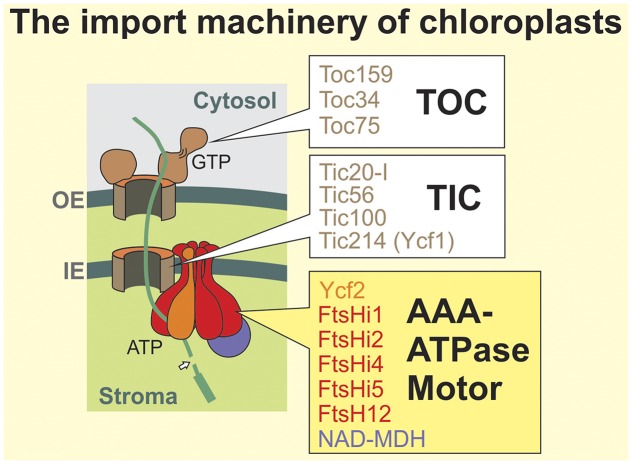
Most chloroplast proteins are encoded by the nuclear genome and synthesized on cytosolic ribosomes. N-terminal transit peptides serve as targeting sequences to direct precursors of chloroplast proteins to receptors on the chloroplast surface. These receptors are part of the translocase of the outer envelope, the TOC complex, and transport precursor proteins in a GTP-dependent reaction into the translocation channel that is formed by the β-barrel protein Toc75.
The translocation reaction across the inner envelope is much less well understood. Masato Nakai and colleagues developed an elegant biochemical approach to screen for the components of the inner envelope translocase: They arrested a translocation intermediate in the chloroplast import machinery that consisted of a ferredoxin precursor fused to a tobacco etch virus (TEV) cleavage sequence and a Protein A domain (Kikuchi et al., 2013). Upon native purification of this precursor with mild detergents and cleavage with TEV protease, they were able to recover and determine the components of the TOC and TIC complexes. In addition to the well-characterized subunits of the TOC complex, this approach identified four components as critical core constituents of the inner envelope translocase, the TIC complex (see figure, white boxes). Surprisingly, in addition to Tic20, which is presumed to form the protein translocation pore, a plastid-encoded large protein was identified, called Ycf1 or Tic214 (Kikuchi et al., 2013).
The Import Machinery of Chloroplasts. TIC/TOC components (white boxes) identified by Kikuchi et al. (2013). AAA-ATPase motor subunits (yellow box) identified by Kikuchi et al. (2018). OE, outer envelope; IE inner envelope.
Employing the same strategy, Kikuchi et al. (2018) have identified a motor that is associated with the TIC complex to drive precursor import into the stroma. This motor comprises a 2-MD complex of seven subunits (see figure, yellow box): six distinct ATPases that compose a heterohexameric AAA-ATPase and NAD-malate dehydrogenase (NAD-MDH). The six ATPases (Ycf2, which, like Ycf1, is plastid encoded, FtsHi1, FtsHi2, FtsHi4, FtsHi5, and FtsH12) are structurally and phylogenetically related to the bacterial inner membrane protease FtsH. Except for FtsH12, these proteins have lost their proteolytic activity (therefore, they are called FtsHi for FtsH-inactive) and even for the essential FtsH12 subunit, the proteolytic activity proved to be dispensable. The NAD-MDH subunit presumably plays a structural role in the motor because its catalytic activity is dispensable and it cannot be functionally replaced by unrelated malate dehydrogenases, even if they exhibit the same enzymatic activity (Schreier et al., 2018).
Previously, protein import into the stroma was thought to be driven by chaperones, such as Hsp70 and Hsp93 (ClpC), that prevent “backsliding” of import intermediates and drive their translocation by a Brownian ratchet mechanism. Such chaperone-driven import processes are well characterized for the translocation of proteins into the endoplasmic reticulum and the matrix of mitochondria (Matlack et al., 1999). Mechanistically, an AAA-ATPase motor would be very different from these Brownian motors: AAA-ATPases of the FtsH type as we know them from the bacterial and mitochondrial inner membrane are processive and power protein movements by hydrolyzing ATP in order to pull on their substrates. Thus, the mechanism by which these hexameric machines drive protein translocation is reminiscent of how helicases promote DNA movements. Hence, such a force-generating motor would be well suited to unfold cytosolic domains of translocation intermediates. A recent study proposed that chloroplasts, unlike mitochondria, can import even folded protein domains (Ganesan et al., 2018). A strong import motor might be necessary for such a process. However, this presumably would come at a high price: A processive import motor should be much slower than a Brownian ratchet-driven motor (at least for unfolded substrates) and would consume more ATP.
Import of the mitochondrial protein Pcp1 requires the mitochondrial m-AAA protease, which is a homolog of the newly discovered chloroplast AAA-ATPase motor (Tatsuta et al., 2007). Thus, a role of FtsH-like AAA-ATPases in protein translocation might not be restricted to chloroplasts. It is conceivable that these strong pullers are engaged only when import intermediates are stalled in the translocation machinery—exactly the situation that was used by Kikuchi et al. to screen for the chloroplast motor. Nevertheless, the observation that the 2-MD AAA-ATPase forms a stable supercomplex with the TIC translocon at the inner envelope membrane even without the stalled import intermediates suggests that this motor is a central device of the chloroplast import machinery to promote translocation of hundreds of precursor proteins.
The discovery of this novel 2-MD AAA-ATPase motor on the TIC translocon promises to revive debates of the past about whether power-generating pulling motors are necessary to drive protein translocation across cellular membranes. It will be exciting now to elucidate the mechanisms by which chloroplasts import their proteins in more detail.
Footnotes
Articles can be viewed without a subscription.
References
- Ganesan I., Shi L.-X., Labs M., Theg S.M. (2018). Evaluating the functional pore size of chloroplast TOC and TIC protein translocons: import of folded proteins. Plant Cell 30: 2161–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S., Bédard J., Hirano M., Hirabayashi Y., Oishi M., Imai M., Takase M., Ide T., Nakai M. (2013). Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339: 571–574. [DOI] [PubMed] [Google Scholar]
- Kikuchi S., Asakura Y., Imai M., Nakahira Y., Kotani Y., Hashiguchi Y., Nakai Y., Takafuji K., Bedard J., Hirabayashi-Ishioka Y., et al. (2018). A Ycf2-FtsHi heteromeric AAA-ATPase complex is required for chloroplast protein import. Plant Cell 30:2677–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack K.E., Misselwitz B., Plath K., Rapoport T.A. (1999). BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell 97: 553–564. [DOI] [PubMed] [Google Scholar]
- Schreier T.B., Cléry A., Schläfli M., Galbier F., Stadler M., Demarsy E., Albertini D., Maier B.A., Kessler F., Hörtensteiner S., Zeeman S.C., Kötting O. (2018). Plastidial NAD-dependent malate dehydrogenase: a moonlighting protein involved in early chloroplast development through its interaction with an FtsH12-FtsHi protease complex. Plant Cell 30: 1745–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T., Augustin S., Nolden M., Friedrichs B., Langer T. (2007). m-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria. EMBO J. 26: 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]