The transcription factor HB52 acts as an important node in the crosstalk between ethylene and auxin signaling by modulating the expression of auxin transport genes downstream of ethylene signaling.
Abstract
The gaseous hormone ethylene participates in many physiological processes in plants. Ethylene-inhibited root elongation involves PIN-FORMED2 (PIN2)-mediated basipetal auxin transport, but the molecular mechanisms underlying the regulation of PIN2 function by ethylene (and therefore auxin distribution) are poorly understood. Here, we report that the plant-specific and ethylene-responsive HD-Zip gene HB52 is involved in ethylene-mediated inhibition of primary root elongation in Arabidopsis thaliana. Biochemical and genetic analyses demonstrated that HB52 is ethylene responsive and acts downstream of ETHYLENE-INSENSITIVE3 (EIN3). HB52 knockdown mutants displayed an ethylene-insensitive phenotype during primary root elongation, while its overexpression resulted in short roots, as observed in ethylene-treated plants. In addition, root auxin distribution and gravitropism were impaired in HB52 knockdown and overexpression lines. Consistent with these findings, in vitro and in vivo binding experiments showed that HB52 regulates the expression of auxin transport-related genes, including PIN2, WAVY ROOT GROWTH1 (WAG1), and WAG2 by physically binding to their promoter regions. These findings suggest that HB52 functions in the ethylene-mediated inhibition of root elongation by modulating the expression of auxin transport components downstream of EIN3, revealing a mechanism in which HB52 acts as an important node in the crosstalk between ethylene and auxin signaling during plant growth and development.
INTRODUCTION
Ethylene, a gaseous phytohormone, triggers plant maturation, the senescence of plant organs, and responses to environmental cues, including leaf shedding and defense responses. Ethylene is also involved in various plant growth and developmental processes, including root growth (Alonso et al., 1999; Grbić and Bleecker, 2003; Chaves and Mello-Farias, 2006; Růzicka et al., 2007). For example, exogenous treatment with ethylene (C2H4) or its biosynthetic precursor 1-aminocyclopropane-1-carboxylic acid (ACC) induces inhibited elongation and increased widening of primary roots, as well as the ectopic formation of root hairs (Masucci and Schiefelbein, 1996; Smalle and Van Der Straeten, 1997; Le et al., 2001). These ethylene-induced responses promote soil penetration and greater anchorage of plants to the ground.
In the past decade, great progress in understanding ethylene signaling has been made using genetic approaches in Arabidopsis thaliana (Merchante et al., 2013) and rice (Oryza sativa) (Yang et al., 2015a, 2015b; Yin et al., 2015; Chen et al., 2018; Ma et al., 2018). In the absence of ethylene, ethylene receptors (ETR/ERS/EIN) and other accessory proteins recruit the Raf-like kinase CTR1 to phosphorylate the C-terminal end of ETHYLENE-INSENSITIVE2 (EIN2), which inhibits the C-terminal translocation of EIN2 into the nucleus and therefore stabilizes the downstream transcription factors EIN3/EIL1 (EIN3-LIKE1). In the presence of ethylene, the ethylene molecule binds to the receptors, leading to the inactivation of CTR1, allowing the unphosphorylated C-terminal end of EIN2 to be cleaved. EIN2 subsequently moves into the nucleus to stabilize EIN3/EIL1, ultimately leading to the release of the downstream transcriptional cascade (Gao et al., 2003; Ju et al., 2012; Qiao et al., 2012; Wen et al., 2012). Other EIN2-mediated ethylene signaling mechanisms were recently reported. EIN2 acts as a translational repressor of EBF1 (EIN3 BINDING F BOX PROTEIN1) and EBF2 mRNA (Li et al., 2015; Merchante et al., 2015) and mediates the direct regulation of histone acetylation during the ethylene response (Zhang et al., 2016, 2017, 2018).
Intriguingly, mutants of auxin biosynthesis, signaling, and transport display an aberrant response to ethylene treatment, indicating the existence of crosstalk between auxin and ethylene signaling. For example, mutations in auxin biosynthetic genes, such as ASA1, ASB1, and TAA1, lead to ethylene-insensitive root phenotypes (Stepanova et al., 2005, 2008). YUCCA (YUC) genes also play key roles in ethylene-mediated root responses (Won et al., 2011; Qin et al., 2017). Mutants of AXR2/IAA7 and AXR3/IAA17, encoding transcriptional regulators involved in auxin signaling, exhibit ethylene-insensitive root growth (Růzicka et al., 2007; Swarup et al., 2007). The auxin efflux transporter PIN-FORMED2 (PIN2) and the influx transporter AUX1 are also involved in ethylene-mediated root responses (Růzicka et al., 2007). AGCVIII kinase family members PINOID (PID) and PID homolog WAVY ROOT GROWTH1 (WAG1) and WAG2 and D6 PROTEIN KINASEs (D6PKs) regulate PIN polarity and PIN-mediated auxin transport activity, respectively, via phosphorylation (Michniewicz et al., 2007; Barbosa et al., 2014). Thus, the PID and D6PK subfamilies are important for auxin transport.
Plants possess numerous transcription factors to functionally regulate different growth and developmental processes. Plant-specific HD-Zip transcription factors display a singular combination of a homeodomain with a leucine zipper, which functions as a dimerization motif. This transcription factor family consists of 47 members in Arabidopsis, which are classified into four subfamilies (Ariel et al., 2007). ATHB1 participates in the determination of leaf cell fate, while ATHB13 and ATHB23 are involved in cotyledon and leaf development (Aoyama et al., 1995; Nakamura et al., 2006). HAT2 overexpression lines display a typical auxin-overproduced phenotype, indicating a role for HAT2 in auxin-mediated development (Delarue et al., 1998; Sawa et al., 2002). PHV, PHB, and REV have similar functions in embryogenesis and leaf polarity determination (Prigge and Clark, 2006). ATHB10, ATML1, and PDF2 play important roles in cell fate establishment by regulating cell layer-specific gene expression (Abe et al., 2003). In addition, EDT1/HDG11 confers drought resistance; plants overexpressing this gene exhibit reduced stomatal density and an improved root system (Yu et al., 2008, 2013, 2016a). However, the functional roles and modes of action of other members of the HD-Zip family are largely unknown.
In this study, we used genetic and biochemical approaches to dissect the functional role of the HD-Zip family member HOMEOBOX PROTEIN52 (HB52), which we previously identified as an ethylene-response factor during a study of the role of Arabidopsis ETHYLENE RESPONSE FACTOR1 (ERF1) (Mao et al., 2016) in the ethylene-mediated inhibition of root elongation. Our results demonstrate that HB52 acts downstream of EIN3 in ethylene signaling to regulate auxin transport by transcriptionally modulating PIN2, WAG1, and WAG2, suggesting that HB52 acts as a node in the crosstalk between ethylene and auxin signaling in Arabidopsis.
RESULTS
Expression Pattern and Subcellular Localization of HB52
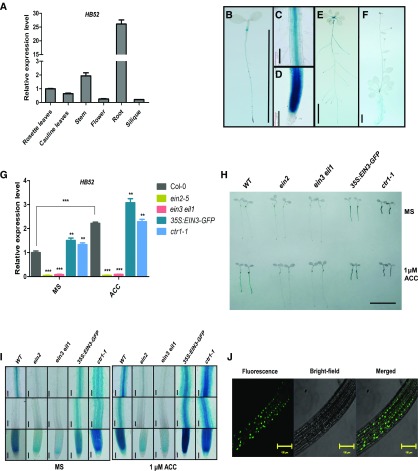
To determine the role of HB52 in plant growth and development, we examined the transcriptional expression patterns of HB52 in different tissues and organs of 4-week-old Arabidopsis plants by qRT-PCR. Relatively high expression levels of HB52 were detected in roots, stems, and rosette leaves (Figure 1A). Consistently, histochemical analysis of transgenic lines harboring the GUS gene driven by the HB52 promoter (HB52pro:GUS) showed that the HB52 promoter was primarily expressed in the root tips and hypocotyl bases of 4-d-old seedlings (Figures 1B to 1D). In 10-d-old seedlings, GUS signals were primarily observed in the roots and petioles of rosette leaves (Figure 1E). In mature plants, GUS signals were found only in the roots (Figure 1F).
Figure 1.
Expression Pattern and Subcellular Localization of HB52.
(A) HB52 transcript levels in different tissues, as determined using quantitative RT-PCR analysis. The values are mean ± sd (n = 3 experiments).
(B) to (F) GUS staining of HB52pro:GUS transgenic plants. GUS activity was observed in 4-d-old seedlings (B), the roots of 4-d-old seedlings ([C] and [D]), 10-d-old seedlings (E), and 4-week-old plants (F). The plants were incubated in GUS staining solution for 2 h before photographs were taken. Bar = 1 cm in (B), (E), and (F) and 100 μm in (C) and (D). At least 10 independent lines were used for GUS staining, and representative images are shown.
(G) Transcript levels of HB52 in the wild type (Col-0) and ethylene signaling mutants. The seeds were germinated on MS medium for 4 d and transferred to MS liquid medium with or without 1 μM ACC for 24 h. RNA was isolated from at least 20 individual seedlings for each mutant, and quantitative RT-PCR analysis was performed to detect HB52 expression levels. Values are the mean ± sd (n = 3 experiments, **P < 0.01, ***P < 0.001). Statistically significant differences were calculated based on Student’s t tests.
(H) and (I) GUS staining of HB52pro:GUS transgenic seedlings in the ethylene signaling mutant background. HB52pro:GUS was introduced into ethylene signaling mutants by genetic crossing. Seeds were germinated on MS medium for 4 d and transferred to MS liquid medium with or without or 1 μM ACC for 24 h. Seedlings were incubated in the GUS staining solution for 0.5 h before the photographs were taken (H). The roots of the stained seedlings were observed under a microscope (I). Bar = 1 cm in (H) and 100 μm in (I). At least 20 individual seedlings per mutant were used for GUS staining, and the images of two representative seedlings are shown.
(J) Subcellular localization of HB52 protein. 35S:HB52-GFP transgenic seeds were germinated on MS medium for 4 d, and fluorescence was observed under a confocal laser-scanning microscope (bar = 100 μm). At least 10 independent transgenic lines were observed for GFP signals, and a representative image is shown.
To address whether HB52 functions in ethylene signaling, we examined the transcript levels of HB52 in the wild-type (Col-0) and ethylene signaling mutant seedlings treated with mock and the ethylene precursor ACC via qRT-PCR. Upon exposure to ACC, HB52 expression levels increased in wild-type seedlings but not in the ethylene signaling mutants ein2-5 and ein3-1 eil1 relative to the corresponding mock control (Figure 1G). However, in the ethylene signaling-enhanced mutants ctr1-1 and 35S:EIN3-GFP, HB52 expression levels were significantly elevated regardless of ACC treatment (Figure 1G). To confirm these results, we introduced HB52pro:GUS into the ein2-5, ein3-1 eil1, 35S:EIN3-GFP, and ctr1-1 backgrounds by genetic crossing. As expected, GUS signals were much weaker in the ein2-5 and ein3-1 eil1 mutants than in the wild type (Figures 1H and 1I). By contrast, GUS signals were stronger in the 35S:EIN3-GFP and ctr1-1 backgrounds compared with the wild type regardless of ACC treatment (Figures 1H and 1I). These results indicate that HB52 is dependent on EIN3 and EIL1.
Next, we generated 35S:HB52-GFP transgenic lines to examine the subcellular localization of HB52. Live-cell imaging revealed strong fluorescent signals for HB52-GFP in the nucleus (Figure 1J), which is consistent with the notion that HD-Zip family members function as nuclear transcription factors.
HB52 Regulates Primary Root Elongation in Response to Ethylene
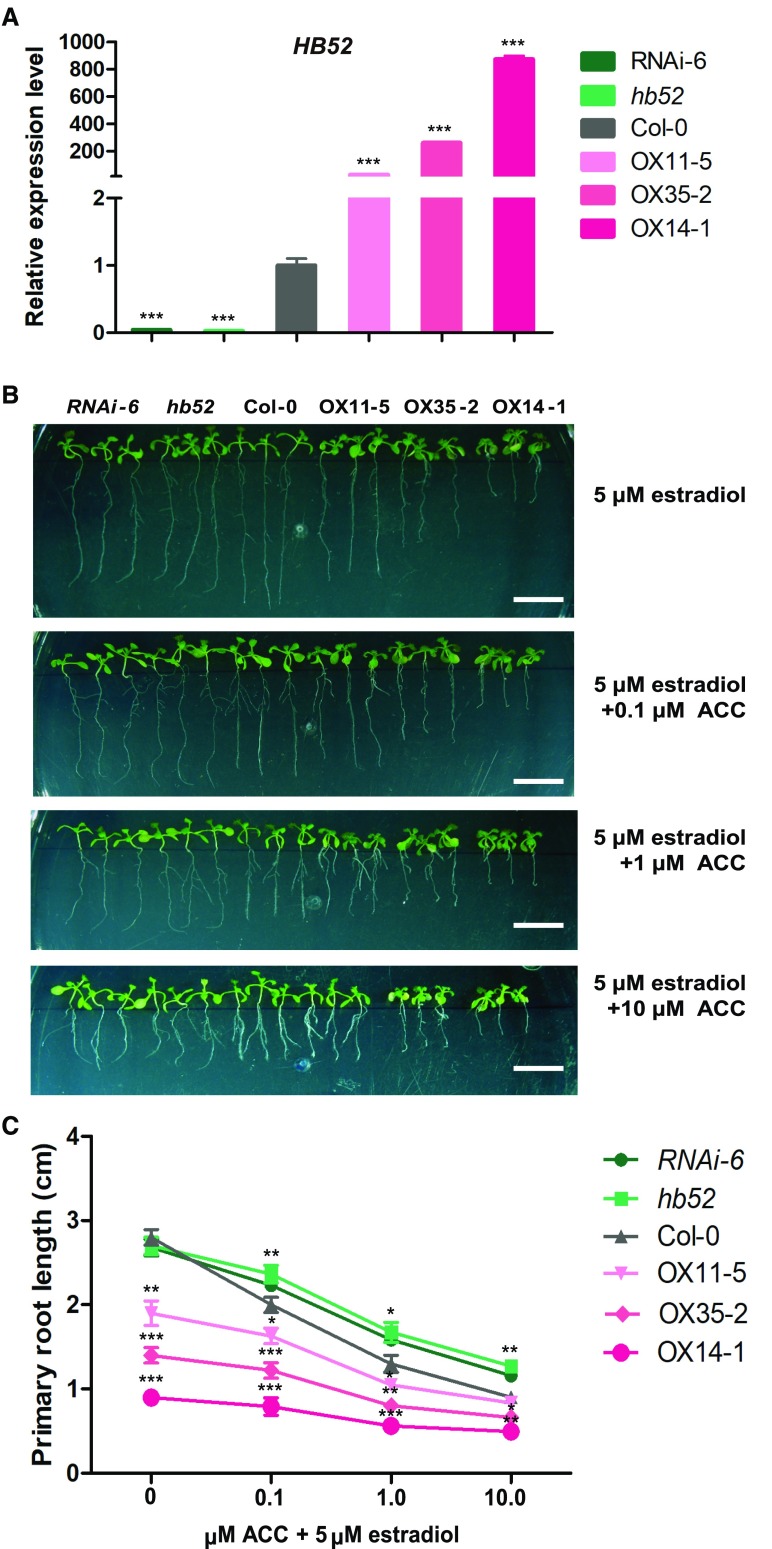
To explore whether HB52 is involved in ethylene-mediated inhibition of root elongation, we obtained the CS909234 mutant, which harbors a T-DNA insertion in the HB52 promoter, from the ABRC (Supplemental Figure 1) and generated the estradiol-inducible RNAi line RNAi-6. For clarity, the CS909234 mutant was renamed hb52. HB52 transcript levels were significantly reduced in hb52 and RNAi-6 compared with the wild type (Figure 2A). To obtain overexpression lines, we initially generated HB52 overexpression lines under the control of the constitutive CaMV 35S promoter, which displayed aberrant flower development and therefore an infertile phenotype. Thus, we generated HB52 overexpression lines driven by an estradiol-inducible promoter instead. The transcript levels of HB52 in three independent overexpression lines, OX11-5, OX35-2, and OX14-1, were 30-, 250-, and 870-fold higher than the wild type, respectively, following estradiol (5 μM) induction (Figure 2A). When germinated on MS medium with estradiol, the relative rates of primary root elongation of these three overexpression lines were reduced to 87, 66, and 43% of the wild type after induction, respectively (Supplemental Figure 2B, left panel). However, the overexpression line OX14-1 exhibited yellow cotyledons, likely due to too high an expression level of HB52 (Supplemental Figure 2A).
Figure 2.
Primary Root Elongation of HB52 Knockdown Mutants and Overexpression Lines in Response to Ethylene.
(A) HB52 transcript levels in knockdown mutants and inducible overexpression lines. The seeds were germinated on MS medium for 3 d, and the seedlings were transferred to liquid MS medium with 5 μM estradiol for 24 h to induce gene expression. The roots were then detached, and RNA was isolated from at least 20 seedlings per line for quantitative RT-PCR analysis. Values are the mean ± sd (n = 3 experiments, ***P < 0.001). Statistically significant differences were calculated based on Student’s t tests.
(B) and (C) Root elongation of knockdown mutants and inducible overexpression lines. The seeds were germinated on MS medium for 3 d, and the seedlings were transferred to MS medium with 5 μM estradiol to induce gene expression for 3 d. Afterwards, the seedlings were transferred to MS medium with 5 μM estradiol supplemented with 0.1, 1, or 10 μM ACC for 4 d. At least 30 seedlings per line were observed, photographs of three representative seedlings are shown (B), and the primary root lengths were measured (C). Values are the mean ± sd (n = 30 seedlings, *P < 0.05, **P < 0.01, and ***P < 0.001). Statistically significant differences were calculated based on Student’s t tests.
To examine the responses of HB52 mutants to ethylene while avoiding cotyledon yellowing, we germinated the seeds on MS medium for 3 d and transferred the seedlings to MS medium supplemented with estradiol (5 μM) and allowed them to grow for an additional 3 d. We then transferred the seedlings to MS medium supplemented with estradiol (5 μM) plus different concentrations of ACC for 4 d and measured primary root length (Figures 2B and 2C). Under mock treatment (0 μM ACC), the rate of primary root elongation of the knockdown mutants (RNAi-6 and hb52) was comparable to that of the wild-type control, while the overexpression lines displayed a considerable reduction in primary root elongation (Figure 2B, top panel), which is consistent with previous results (Supplemental Figure 2B). Root elongation in the two HB52 knockdown lines and three overexpression lines was less sensitive to ACC treatment than the wild type (Figures 3B and 3C). Among the three overexpression lines, root elongation in OX14-1 was the most resistant to ACC treatment (Figure 3C). These results indicate that HB52 plays an important role in the ethylene-mediated inhibition of primary root elongation.
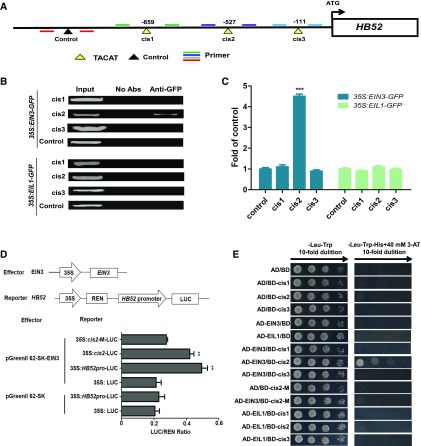
Figure 3.
Binding Assays of EIN3 and EIL1 with the HB52 Promoter.
(A) Schematic representation of the HB52 promoter showing putative EBSs upstream of the ATG codon. EBSs are indicated by yellow triangles, while the black triangle indicates a control with no EBS in this region. PCR-amplified fragments are indicated by different pairs of colored primers used for ChIP-PCR and quantitative ChIP-PCR.
(B) and (C) ChIP-PCR assays. Four-day-old 35S:EIN3-GFP and 35S:EIL1-GFP transgenic seedlings (>50) were treated with 1 μM ACC for 24 h for the ChIP assays. Approximately 200-bp HB52 promoter fragments containing the EBS were enriched by anti-GFP antibody in the ChIP-PCR analysis (B). A region of the HB52 promoter lacking an EBS was used as a control. The results of the ChIP-PCR were confirmed using quantitative ChIP-PCR (C). Values are the mean ± sd (n = 3 experiments, ***P < 0.001). Statistically significant differences were calculated based on Student’s t tests.
(D) Transient transactivation assay. The coding sequence of EIN3 was cloned into the pGreenII 62-SK vector to generate the effector. The HB52 promoter, the EIN3 binding site cis2, and the mutated site cis-M were cloned into the pGreenII0800-LUC vector to generate the reporters. Both the effector and reporter were transfected into Arabidopsis mesophyll protoplasts. Firefly LUC and REN activities were detected using a dual-luciferase reporter assay. Relative REN activity was used as an internal control, and the LUC/REN ratios were calculated. Values are the mean ± sd (n = 3 experiments, **P < 0.01). Statistically significant differences were calculated based on Student’s t tests.
(E) Yeast-one-hybrid assay. pGADT7/EIN3 (AD-EIN3) and pGADT7/EIL1 (AD-EIL1) constructs were cotransformed with pHIS2/HB52 (BD-cis) separately into yeast strain Y187. AD/BD, AD/BD-cis1, AD/BD-cis2, AD/BD-cis3, AD-EIN3/BD, and AD-EIL1/BD were used as negative controls. AD/BD-cis2-M and AD-EIN3/BD-cis2-M represent the mutated cis2 element.
In addition, we observed abnormal root meristems, reduced meristem cell number, and short elongation zone cell length in the overexpression lines (Supplemental Figures 2A and 3). The abnormal root meristems could be rescued by exogenous treatment with the polar auxin transport inhibitor NPA (N-1-naphthylphthalamic acid) (Supplemental Figure 2B, right panel). The root gravitropic response was also altered in the knockdown mutants and overexpression lines (Supplemental Figure 4).
We also performed a triple response assay. In the dark, the knockdown mutants (RNAi-6 and hb52) showed similar hypocotyl lengths and apical hooks to those of the wild type in the presence or absence of ACC (Supplemental Figures 5A and 5B). However, the overexpression lines (OX11-5, OX35-2, and OX14-1) displayed a significant reduction in hypocotyl length and open cotyledons compared with the wild type (Supplemental Figures 5A and 5B) under mock treatment (0 μM ACC) and less sensitive hypocotyl elongation responses under ACC treatment (Supplemental Figures 5A and 5B). The primary root lengths of lines grown in the dark (Supplemental Figures 5A and 5C) showed similar tendencies to those grown in the light (Figures 2B and 2C).
HB52 Is a Direct Target of EIN3
The above results indicate that the regulation of HB52 transcript level by ethylene depends on EIN3 and EIL1 (Figure 1). However, whether HB52 is a direct target of EIN3 and EIL1 remained to be elucidated. Three putative EIN3 binding sites (EBSs; TACAT and TTCAAA) have been identified in the promoter region of HB52 (Konishi and Yanagisawa, 2008; Zhong et al., 2009; An et al., 2012; Li et al., 2013) (Figure 3A). Thus, we performed chromatin immunoprecipitation (ChIP) assays using 35S:EIN3-GFP and 35S:EIL1-GFP transgenic plants. Marked enrichment of the region containing the cis2 site (TACAT) was detected in the 35S:EIN3-GFP transgenic plants using ChIP-PCR assays (Figures 3B and 3C), indicating that EIN3 but not EIL1 binds to this region in vivo. Consistent with this notion, a transient transactivation assay showed that EIN3 was able to activate the expression of the HB52 promoter- and cis2-driven luciferase (LUC) reporter in Arabidopsis mesophyll protoplasts but not the mutated cis2-m (Figure 3D). In addition, we conducted yeast-one-hybrid assays to determine whether EIN3 and EIL1 could directly bind to the EBS in the HB52 promoter. As expected, EIN3 indeed binds to the cis2 site in the HB52 promoter, while EIL1 did not (Figure 3E). Taken together, these results suggest that HB52 is a direct target of EIN3.
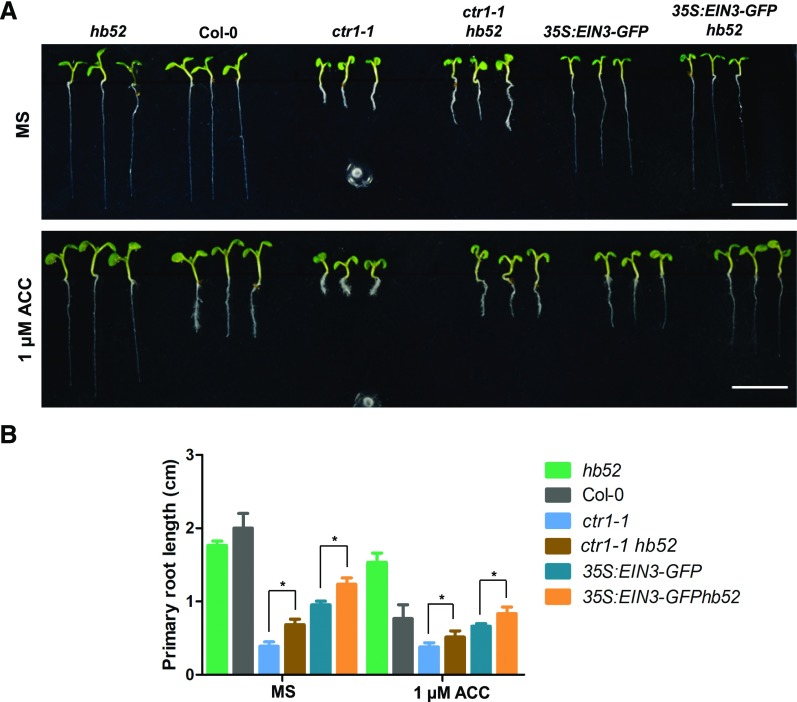
To confirm this genetically, we crossed the hb52 mutant with 35S:EIN3-GFP transgenic lines and the ctr1-1 mutant. The double mutant ctr1-1 hb52 had the same point mutation as the single mutant ctr1-1 at the CTR1 locus, and 35S:EIN3-GFP hb52 had the same expression level of EIN3 as 35S:EIN3-GFP (Supplemental Figures 6A and 6B). HB52 transcript levels remained low in ctr1-1 hb52 and 35S:EIN3-GFP hb52 as in hb52 (Supplemental Figure 6C). If HB52 is a direct target of EIN3, the increased EIN3 levels in ctr1-1 hb52 should have a less inhibitory effect on primary root elongation because the target HB52 is impaired in this double mutant. As expected, the primary roots of both 35S:EIN3-GFP hb52 and ctr1-1 hb52 were longer than those of 35S:EIN3-GFP and ctr1-1, regardless of ACC treatment (Figure 4).
Figure 4.
HB52 Genetically Acts Downstream of EIN3.
(A) Root elongation phenotype. Seeds of the indicated lines were germinated on MS medium with and without 1 μM ACC for 5 d before the photographs were taken. Three representative seedlings are shown for each line. Bar = 1 cm.
(B) Primary root length. Seeds of the indicated lines as in (A) were germinated on MS medium with and without 1 μM ACC for 5 d before the primary root length was measured. Values are the mean ± sd (n = 30 seedlings, *P < 0.05). Statistically significant differences were calculated based on Student’s t tests.
HB52 Directly Regulates PIN2, WAG1, and WAG2
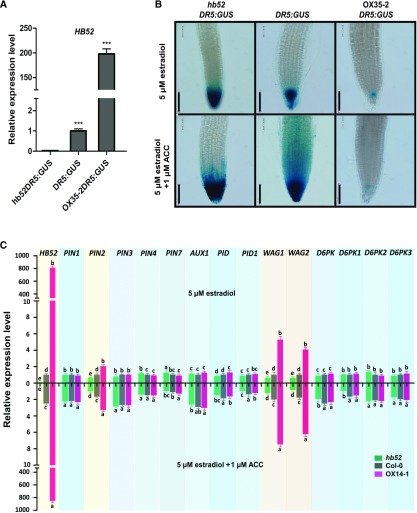
Although the evidence presented so far suggests that HB52 plays an important role in ethylene-mediated inhibition of root elongation, how HB52 functions in this process is yet to be resolved. To address this question, we crossed the hb52 mutants and OX35-2 transgenic lines with DR5-GUS reporter lines to examine whether the downregulation or overexpression of HB52 alters auxin distribution or accumulation in the root tip. We confirmed the identities of both lines by detecting the transcript levels of HB52 (Figure 5A). In the mock control (without ACC), GUS signals were stronger in the hb52 mutants and weaker in the OX35-2 lines than that in the wild type (Figure 5B). When treated with ACC, GUS signals displayed a smeared pattern in the wild-type elongation zone but not in the hb52 mutants or OX35-2 lines (Figure 5B). By contrast, the GUS signals were reduced in the OX35-2 lines compared with the mock control (Figure 5B). These results indicate that the partial loss and gain of function of HB52 affect auxin distribution in the root tip.
Figure 5.
HB52 Affects Auxin Transport by Regulating Auxin Transport-Related Genes.
(A) HB52 transcript levels in HB52 mutants harboring the DR5:GUS reporter. Seeds of the indicated lines were germinated on MS medium for 4 d and transferred to MS liquid medium with 5 μM estradiol for 24 h. The roots were detached from more 50 seedlings, and RNA was isolated for quantitative RT-PCR analysis. Values are the mean ± sd (n = 3 experiments, ***P < 0.001). Statistically significant differences were calculated based on Student’s t tests.
(B) GUS staining of DR5:GUS marker lines in various HB52 backgrounds. Seeds of the indicated lines were germinated on MS medium with 5 μM estradiol for 4 d, and the seedlings were transferred to liquid MS medium with 5 μM estradiol supplemented with and without 1 μM ACC for 24 h before staining. The seedlings (>20) were incubated in the GUS staining solution for 2 h before the photographs were taken. A representative seedling of each line is shown. Bar = 100 μm.
(C) Transcript levels of auxin transport-related genes in mutants with various HB52 backgrounds with or without ACC treatment. Seeds of the indicated lines were germinated on MS medium for 4 d and transferred to MS liquid medium with 5 μM estradiol or 5 μM estradiol + 1 μM ACC for 24 h. The roots from at least 30 seedlings for each line were detached, and RNA was isolated for quantitative RT-PCR analysis. The relative expression level of the ACC-treated samples was normalized to the wild-type control of the corresponding untreated samples. Values are the mean ± sd (n = 3 experiments). Different lowercase letters indicate significant difference based on Student’s t tests (P < 0.05).
The observation that auxin distribution and gravitropic growth are impaired in the roots of the HB52 knockdown and overexpression lines (Figure 5; Supplemental Figure 4) prompted us to explore how HB52 is involved in regulating auxin distribution. We first examined the transcript levels of the auxin transporters PINs and AUX1 and various PIN phosphorylation kinase genes, including PID and its homologs WAG1 and WAG2, as well as D6PKs (D6PK, D6PK1, D6PK2, and D6PK3), in the hb52 mutant and OX35-2 line in response to ACC treatment. We also measured HB52 levels to ensure its proper expression in the mutant and overexpression line. As shown in Figure 5C, the transcript levels of PIN2, WAG1, and WAG2 but not the other genes examined were significantly reduced in the hb52 mutant and elevated in OX35-2 relative to the wild type under control conditions (5 μM estradiol). All genes except PIN7 positively responded to ACC treatment (5 μM estradiol + 1 μM ACC) in the wild type and overexpression line, with significantly higher transcript levels than in the untreated wild-type control. Only PIN2, WAG1, and WAG2 showed both significantly reduced transcript levels in hb52 and elevated transcript levels in the overexpression line. These results suggest that HB52 directly or indirectly regulates PIN2, WAG1, and WAG2 at the transcriptional level.
To address whether HB52 directly regulates PIN2, WAG1, and WAG2 transcription, we analyzed potential HB52 binding sites in the promoter regions of these three genes and performed binding experiments using ChIP-PCR and transient transactivation assays combined with yeast one-hybrid assays and electrophoretic mobility shift assays (EMSAs). As shown in Figures 6A to 8A, we found multiple candidate binding sites. In addition, in vitro and in vivo binding experiments showed that HB52 physically binds to at least one homeodomain binding site in the promoter regions of these three genes (Figures 6 to 8).
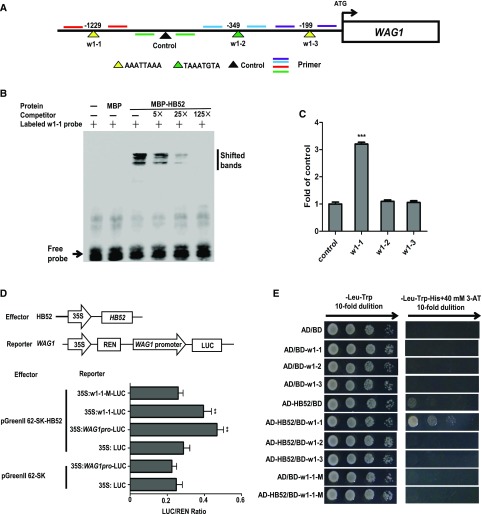
Figure 6.
Binding Assays of HB52 Protein with the WAG1 Promoter.
(A) Schematic representation of the WAG1 promoter showing putative HB52 binding sites upstream of the ATG codon. HB52 binding sites are indicated by yellow and green triangles, while the black triangle indicates a control with no HB52 binding sites in this region. Numbers above the black lines represent the precise HB52 binding sites. PCR-amplified fragments are indicated by different pairs of colored primers, which were used for quantitative RT-PCR.
(B) EMSA of in vitro binding. Biotin-labeled probe (w1-1 region) was incubated with HB52-MBP protein. As indicated, HB52-dependent mobility shifts were detected and competed with by the unlabeled probe in a dose-dependent manner. Similar results were obtained from 3 repeat experiments. The multiple shifted bands were due to the formation of multimers of the probe.
(C) ChIP-PCR assay. Four-day-old 35S:HB52-GFP transgenic seedlings were treated with 1 μM ACC for the ChIP-PCR assay. A region of the WAG1 promoter that does not contain HB52 binding sites was used as a control. Values are the mean ± sd (n = 3 experiments, ***P < 0.001). Statistically significant differences were calculated based on Student’s t tests.
(D) Transient transactivation assay. The coding sequence of HB52 was cloned into the pGreenII 62-SK vector to generate the effector. The WAG1 promoter, HB52 binding site w1-1, and the mutated binding site w1-1-M were cloned into the pGreenII0800-LUC vector to generate the reporters. Both the effector and reporter were transfected into Arabidopsis mesophyll protoplasts. Firefly LUC and REN activities were detected using a dual-luciferase reporter assay system. The relative REN activity was used as an internal control, and the LUC/REN ratios were calculated. Values are the mean ± sd (n = 3 experiments, **P < 0.01). Statistically significant differences were calculated based on Student’s t tests.
(E) Yeast-one-hybrid assay. pGADT7/HB52 (AD-HB52) was cotransformed with pHIS2/WAG1 (BD-w1) into yeast strain Y187. AD/BD, AD/BD-w1-1, AD/BD-w1-2, AD/BD-w1-3, and AD-HB52/BD were used as negative controls. AD/BD-w1-1-M and AD-HB52/BD-w1-1-M represent the mutated w1-1 site.
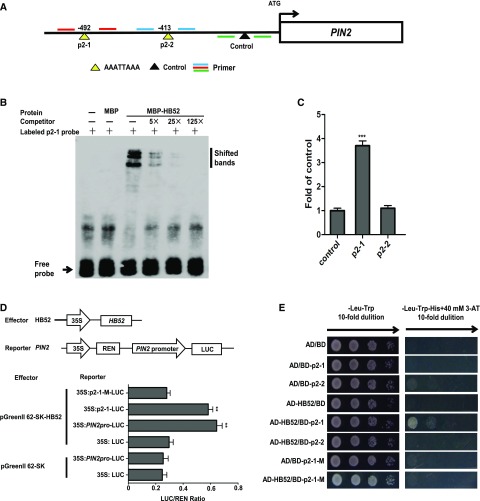
Figure 8.
Binding Assays of HB52 Protein with the PIN2 Promoter.
(A) Schematic representation of the PIN2 promoter showing putative HB52 binding sites upstream of the transcription start site. HB52 binding sites are indicated by yellow triangles, while the black triangle indicates a control that has no HB52 binding sites in this region. Numbers above the black lines represent the precise HB52 binding sites. PCR-amplified fragments are indicated by different pairs of colored primers, which were used for quantitative ChIP-PCR.
(B) EMSA of in vitro binding. Biotin-labeled probe (p2-1 region) was incubated with HB52-MBP protein. As indicated, HB52-dependent mobility shifts were detected and competed with by the unlabeled probe in a dose-dependent manner. Similar results were obtained from three repeat experiments. The multiple shifted bands were due to the formation of multimers of the probe.
(C) ChIP-PCR assay. Four-day-old 35S:HB52-GFP transgenic seedlings were treated with 1 μM ACC for the ChIP-PCR assay. A region of PIN2 that does not contain HB52 binding sites was used as a control. Values are the mean ± sd (n = 3 experiments, ***P < 0.001). Statistically significant differences were calculated based on Student’s t tests.
(D) Transient transactivation assay. The coding sequence of HB52 was cloned into the pGreenII 62-SK vector to generate the effector. The PIN2 promoter, the HB52 binding site p2-1, and the mutated binding site p2-1-M were cloned into the pGreenII0800-LUC vector to generate the reporters. Both the effector and reporter were transfected into Arabidopsis mesophyll protoplasts. Firefly LUC and REN activities were detected using a dual-luciferase reporter assay. Relative REN activity was used as an internal control, and the LUC/REN ratios were calculated. Values are the mean ± sd (n = 3 experiments, **P < 0.01). Statistically significant differences were calculated based on Student’s t tests.
(E) Yeast one-hybrid assay. pGADT7/HB52 (AD-HB52) was cotransformed with pHIS2/PIN2 (BD-p2) into yeast strain Y187. AD/BD, AD/BD-p2-1, AD/BD-p2-2, and AD-HB52/BD were used as negative controls. AD/BD-p2-1-M and AD-HB52/BD-p2-1-M represent the mutated p2-1 site.
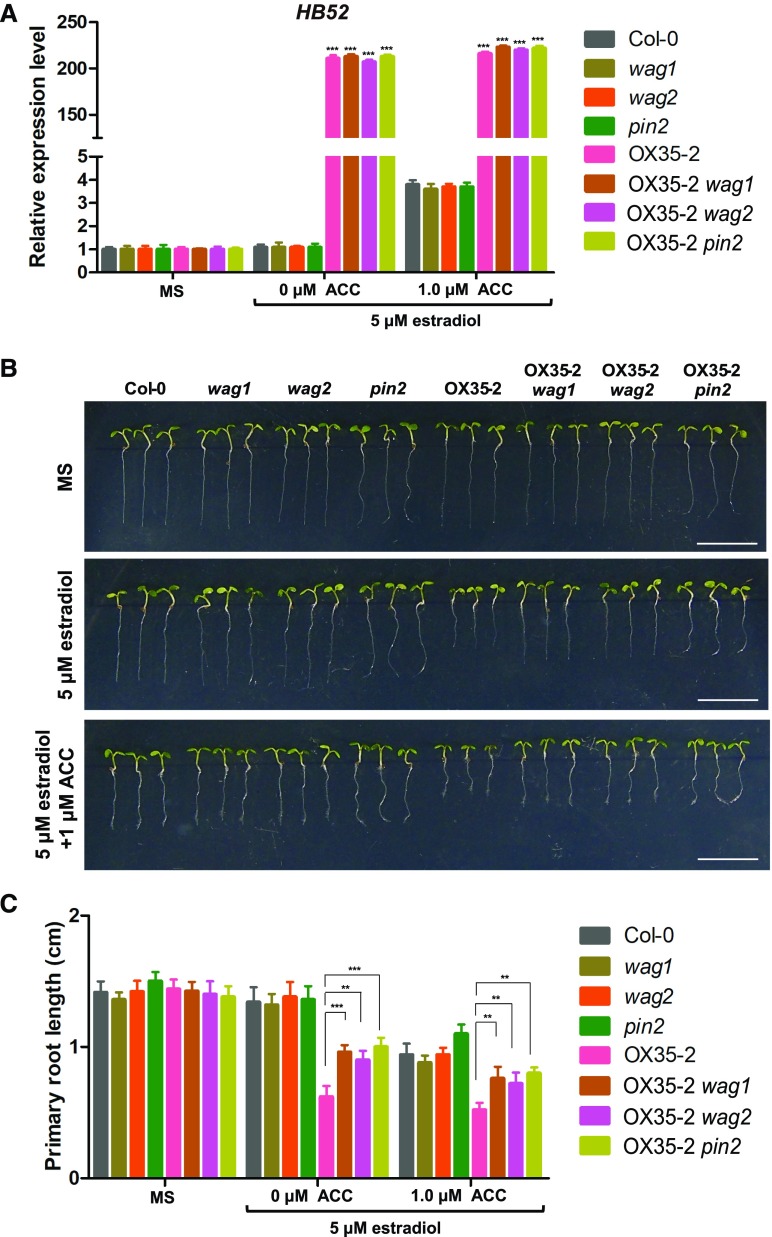
If HB52 indeed binds to these genes, one would expect that the loss of PIN2, WAG1, or WAG2 would alleviate the inhibitory effect of HB52 overexpression on root elongation. To test this hypothesis, we crossed knockout mutants of PIN2, WAG1, and WAG2 with OX35-2 line to construct the double mutants OX35-2 wag1, OX35-2 wag2, and OX35-2 pin2, respectively. HB52 transcript levels in these double mutants were similar to those in OX35-2 (Figure 9A). As expected, the primary roots of the double mutants were longer than those of OX35-2 regardless of ACC treatment (middle and bottom panels of Figures 9B and 9C). Taken together, these results suggest that HB52 functions in the ethylene-mediated inhibition of root elongation by transcriptionally modulating WAG1, WAG2, and PIN2.
Figure 9.
PIN2, WAG1, and WAG2 Genetically Act Downstream of HB52.
(A) HB52 transcript levels in various HB52 mutants. Seeds of the indicated lines were germinated on MS medium for 4 d and transferred to MS liquid medium with 5 μM estradiol plus or minus 1 μM ACC for 24 h. RNA was isolated from more than 30 seedlings for quantitative RT-PCR analysis. Values are the mean ± sd (n = 3 experiments, ***P < 0.001). Statistically significant differences were calculated based on Student’s t tests.
(B) and (C) Root elongation. Seeds (>30) of the indicated lines were separately germinated on MS medium, MS + 5 μM estradiol, and MS + 5 μM estradiol + 1 μM ACC for 5 d before photographs were taken. Three representative seedlings are shown for each line (B) (bar = 1 cm). The primary root length was measured (C). Values are the mean ± sd (n = 30 seedlings, **P < 0.01 and ***P < 0.001). Statistically significant differences were calculated based on Student’s t tests.
DISCUSSION
The synergistic effects of auxin and ethylene on root elongation have been extensively studied. Ethylene enhances auxin biosynthesis, auxin transport, and signaling, thereby inhibiting root elongation (Pickett et al., 1990; Alonso et al., 2003; Stepanova et al., 2005, 2008; Růzicka et al., 2007; Swarup et al., 2007; Mao et al., 2016). Exactly how ethylene interacts synergistically with auxin in the regulation of root elongation remains to be elucidated. The HD-Zip transcription factor family is unique to plants and is divided into four subfamilies (I–IV) primarily based on protein structure and function. HB52 is a member of the HD-Zip I subfamily. Some members of this subfamily are involved in responses to environmental cues, including abiotic stress, abscisic acid, and light (Ariel et al., 2007). In this study, we demonstrated that HB52 plays a functional role in the ethylene and auxin-induced regulation of root elongation.
HB52 Acts Downstream of EIN3 in Ethylene Signaling
Our study revealed that HB52 is transcriptionally regulated by ethylene in an ethylene signaling-dependent manner (Figure 1). In addition, in vitro and in vivo binding experiments (Figure 3) indicated that EIN3 effectively binds to the HB52 promoter region. Our finding that HB52 acts downstream of EIN3 in ethylene signaling is consistent with the public data, including the e-FP browser, which shows that HB52 is upregulated by ethylene, as well as previous data for EIN3 obtained by ChIP-seq (Chang et al., 2013).
To further explore the role of HB52 in ethylene-mediated root elongation, we generated partial loss-of-function and gain-of-function mutants of HB52. These partial loss and gain-of-function mutants displayed resistance to the ethylene precursor ACC during root elongation relative to the wild type (Figure 2). In addition, our genetic analysis showed that primary root elongation was enhanced in the 35S:EIN3-GFP hb52 and ctr1-1 hb52 mutants compared with 35S:EIN3-GFP and ctr1-1 seedlings, respectively, in both the presence and absence of ACC treatment (Figure 4). Thus, our findings demonstrate a role for HB52 in ethylene-mediated root elongation. We noticed that hb52 only partially suppressed the ctr1-1 and 35S:EIN3 phenotypes. Considering that the ethylene signaling pathway regulates many downstream genes besides HB52, it is not surprising that the phenotypes of these plants were only partially suppressed in the hb52 background.
Mode-of-Action of HB52 in the Regulation of Root Elongation via Ethylene and Auxin
PIN2 mediates root gravitropic growth (Chen et al., 1998; Jones, 1998; Luschnig et al., 1998; Müller et al., 1998). The loss of PIN2 function inhibits basipetal auxin transport in the root, therefore leading to auxin accumulation in the root tip (Abas et al., 2006), suggesting that PIN2 mediates root gravitropic growth by regulating basipetal auxin transport in the root. However, the mechanism underlying the PIN2-mediated regulation of auxin transport is not yet fully understood. Our study showed that the downregulation of HB52 caused auxin accumulation in the root tip, while the gain-of-function of HB52 reduced auxin accumulation in the root tip (Figure 5). Upon ACC treatment, wild-type plants showed more auxin accumulation in the root tip and elongation zone compared with the mock control, as indicated by the expression of the DR5-GUS marker. By contrast, the GUS signal was slightly increased in the root tip but not in the elongation zone of the hb52 mutant, while it was significantly reduced in the HB52 overexpression line, likely due to the enhanced transport of auxin out of the root tip (Figure 5). These results suggest that HB52 is likely involved in basipetal auxin transport. These findings are consistent with the ethylene-insensitive root elongation of HB52 partial loss-of-function and gain-of-function mutants (Figure 2). Notably, we also cannot rule out the possibility that the aberrant development of the meristematic zone in gain-of-function mutant roots affects auxin distribution and accumulation in the root tip and therefore leads to the insensitivity of mutant root elongation to ethylene (Figure 5B; Supplemental Figure 2A). The increased auxin accumulation in the root tip of the wild type upon ACC treatment might be partially due to increased auxin biosynthesis (Figure 5B), which is known to be enhanced by the presence of the ethylene-responsive ERF1 gene (Mao et al., 2016) or by OsEIL1-induced activation of YUC8M in rice (Qin et al., 2017).
In addition, the root phenotype of the HB52 overexpression lines is very similar to those of the PID, WAG1, and WAG2 overexpression lines, which also display reduced DR5:GUS expression, the loss of gravitropism, and abnormal root meristems (Christensen et al., 2000; Benjamins et al., 2001; Santner and Watson, 2006; Dhonukshe et al., 2010; Li et al., 2011). Abnormal root meristems were observed in the overexpression lines and could be rescued by the polar auxin inhibitor NPA (Supplemental Figure 2B, right panel), which is consistent with the finding that this type of abnormal root meristem can be rescued by NPA treatment (Benjamins et al., 2001; Dhonukshe et al., 2010),
Exogenous ACC treatment upregulates the transcriptional expression of PIN2, and the loss-of-function pin2/eir1 mutants are insensitive to ethylene during root elongation (Růzicka et al., 2007), suggesting that PIN2 is involved in the ethylene-mediated inhibition of root elongation. Since PIN2 is not a direct target of EIN3 (Benjamins et al., 2001; Chang et al., 2013), the link between ethylene and PIN2 remains unclear. PID, WAG1, and WAG2 belong to the plant-specific AGCVIII family of kinases and function redundantly in regulating the establishment of PIN polarity during root gravitropic growth. The most distal cells of the pid wag1 wag2 root epidermis displays basal localization of PIN2 compared with its apical localization in the wild type, while the overexpression of these three genes leads to the apical localization of PIN1 in the root stele, PIN2 in the cortex, and PIN4 in the root meristem (Dhonukshe et al., 2010). Our transcript profiling revealed that the partial loss of HB52 reduced the expression of PIN2, WAG1, and WAG2, whereas the gain of function of HB52 enhanced their expression (Figure 5C). In addition, our in vitro and in vivo binding experiments (Figures 6 to 8) showed that HB52 could physically bind to the promoter regions of PIN2, WAG1, and WAG2. Thus, our findings demonstrate that PIN2, WAG1, and WAG2 are direct targets of HB52 during the ethylene-mediated inhibition of root elongation. We noticed that in each of the three promoters, only one out of the two/three putative HD binding sites was bound by HB52 in planta (Figures 6C, 7C, and 8C) and in yeast (Figures 6E, 7E, and 8E), which indicates that the other predicted binding sites are not functional or are less likely to be bound by other homeodomain proteins. In the double mutants, pin2, wag1, and wag2 only partially reversed the HB52ox phenotype (Figure 9), which is likely due to the functional redundancy among PID, WAG1, and WAG2. Alternatively, HB52 might regulate other genes related to the auxin signal pathway in addition to PIN2, WAG1, and WAG2. Thus pin2, wag1, and wag2 could only partially reverse the phenotype of the HB52 overexpression lines.
Figure 7.
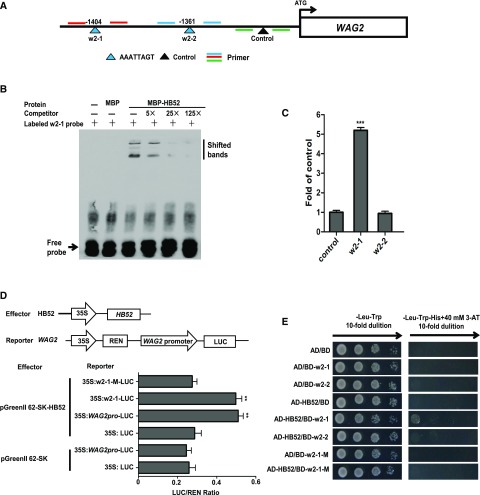
Binding Assays of HB52 Protein with the WAG2 Promoter.
(A) Schematic representation of the WAG2 promoter showing putative HB52 binding sites upstream of the ATG codon. HB52 binding sites are indicated by yellow triangles, while the black triangle indicates a control that has no HB52 binding sites in this region. Numbers above the black lines represent the precise HB52 binding sites. PCR-amplified fragments are indicated by different pairs of colored primers, which were used for quantitative RT-PCR.
(B) EMSA of in vitro binding. Biotin-labeled probe (w2-1 region) was incubated with the HB52-MBP protein. As indicated, HB52-dependent mobility shifts were detected and competed with by the unlabeled probe in a dose-dependent manner. Similar results were obtained from three repeat experiments. The multiple shifted bands were due to the formation of multimers of the probe.
(C) ChIP-PCR assay. Four-day old 35S:HB52-GFP transgenic seedlings were treated with 1 μM ACC for the ChIP-PCR assay. A region of the WAG2 promoter that does not contain HB52 binding sites was used as a control. Values are the mean ± sd (n = 3 experiments, ***P < 0.001). Statistically significant differences were calculated based on Student’s t tests.
(D) Transient transactivation assay. The coding sequence of HB52 was cloned into the pGreenII 62-SK vector to generate the effector. The WAG2 promoter, HB52 bind site w2-1, and the mutated binding site w2-1-M were cloned into the pGreenII0800-LUC vector to generate the reporters. Both the effector and reporter were transfected into Arabidopsis mesophyll protoplasts. Firefly LUC and REN activities were detected using a dual-luciferase reporter assay system. Relative REN activity was used as an internal control, and the LUC/REN ratios were calculated. Values are the mean ± sd (n = 3 experiments, **P < 0.01). Statistically significant differences were calculated based on Student’s t tests.
(E) Yeast one-hybrid assay. pGADT7/HB52 (AD-HB52) was cotransformed with pHIS2/WAG2 (BD-w2) into yeast strain Y187. AD/BD, AD/BD-w2-1, AD/BD-w2-2, and AD-HB52/BD were used as the negative controls. AD/BD-w2-1-M and AD-HB52/BD-w2-1-M represent the mutated w2-1 site.
Taken together, our results support a model in which HB52 acts downstream of ethylene signaling to affect root elongation. According to this model, ethylene stabilizes EIN3 and therefore upregulates HB52. Subsequently, HB52 enhances the transcriptional expression of PIN2, WAG1, and WAG2. WAG1 and WAG2 enhance PIN2 activity. Consequently, more auxin in the root tip is transported to the elongation zone, thereby leading to inhibited root elongation (Figure 10). Thus, our findings reveal a mechanism in which HB52 acts as an important node between ethylene and auxin signaling during plant growth and development. Further analysis of the role of HB52 in auxin signaling will provide a more thorough understanding of the roles of ethylene and auxin in the regulation of root elongation. In addition to auxin transport, auxin biosynthesis might be equally important in this process. The biosynthesis and transport of auxin must be well coordinated in order to inhibit root elongation in response to ethylene.
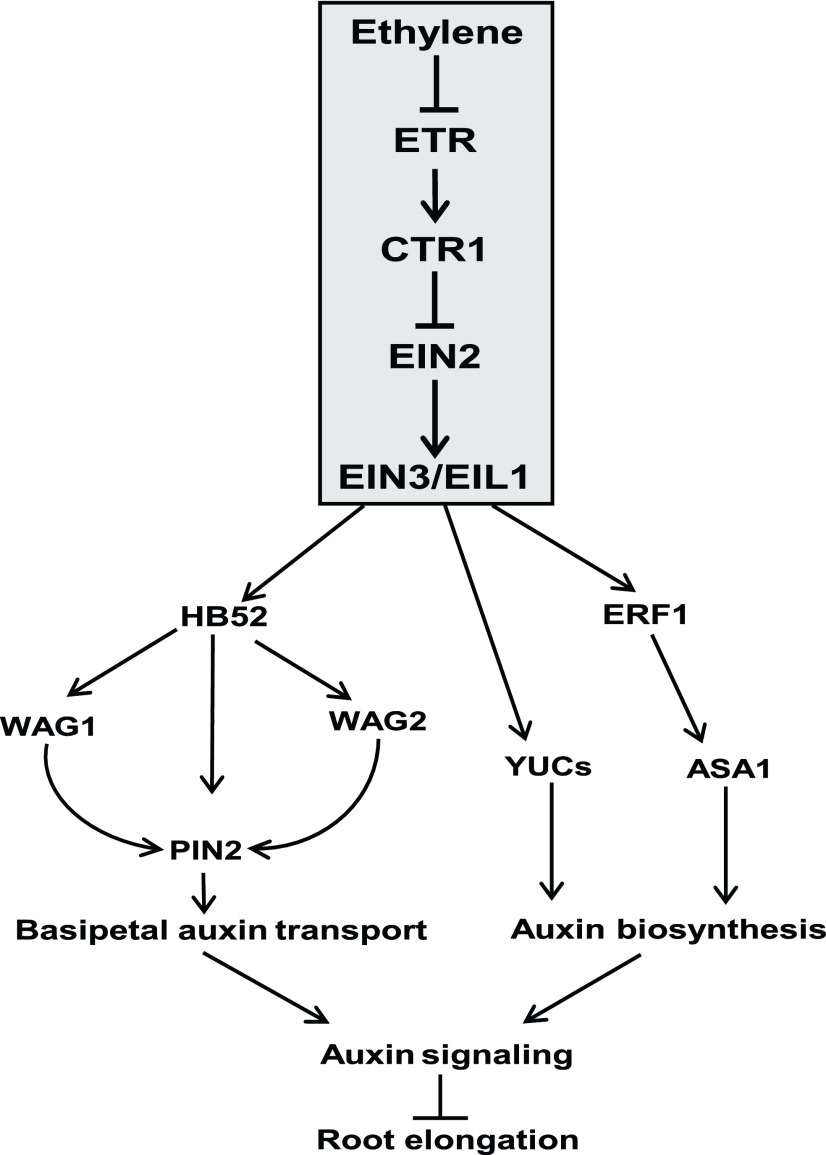
Figure 10.
Model for the Role of HB52 in Mediating the Crosstalk between Ethylene Signaling and Auxin Transport during Primary Root Elongation.
HB52 is a downstream target of the ethylene signaling pathway. When ethylene is produced, it stabilizes EIN3 and therefore upregulates HB52 expression. Subsequently, HB52 binds to the promoters of PIN2, WAG1, and WAG2 and increases their expression. WAG1 and WAG2 phosphorylate PIN2, and phosphorylated PIN2 gathers at the apical side of the cell. Thus, more auxin is transported to the elongation zone, and primary root elongation is inhibited. In addition to auxin transport, ethylene signaling also modulates auxin biosynthesis, for instance, by upregulating auxin biosynthesis-related genes ASA1 and YUCs via EIN3/EIL1 or ethylene-responsive ERF1. Auxin biosynthesis might be equally important in this process. Both processes must be well coordinated in order to inhibit root elongation in response to ethylene.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used as the wild type. A homozygous HB52 knockdown mutant, CS909234 (hb52), was ordered from the ABRC. The OX11-5, 35-2, 14-1, 18-4, RNAi-6, HB52pro:GUS, 35S:HB52-GFP, and 35S:EIN3-GFP transgenic plants were obtained by Agrobacterium tumefaciens (C58C1)-mediated transformation using the Arabidopsis floral-dip method (Clough and Bent, 1998). For OX11-5, 35-2, 14-1, and 18-4, the HB52 coding sequence was amplified using the primers pER8-HB52-P1 and pER8-HB52-P2 (Supplemental Data Set 1) and cloned into pER8 (Zuo et al., 2000) between the XhoI and SpeI sites. For RNAi-6, ∼200 bp of the HB52 coding sequence was amplified using RNAi-P1 and RNAi-P2, followed by RNAi-P3 and RNAi-P4. Both segments were cloned into pUC18RNAi vector (constructed by the Zhong Zhao laboratory, School of Life Sciences, USTC, Hefei, China) and shuttled into pER8. For HB52pro:GUS, the HB52 promoter was amplified using GUS-HB52-P1 and GUS-HB52-P2, cloned into pDONR207, and shuttled into pCB308R via the Gateway cloning system (Invitrogen). For 35S:HB52-GFP, the HB52 coding sequence without a stop codon was amplified using GFP-HB52-P1 and GFP-HB52-P2, cloned into pDONR207, and shuttled into pGWB5 via the Gateway cloning system.
Several plant materials were previously described: ein2-5 (Alonso et al., 1999), ein3-1 eil1-1 (Alonso et al., 2003), ctr1-1 (Kieber et al., 1993), 35S:EIN3-GFP, pin2 (CS8058), wag1 (Salk_002056), wag2 (Salk_070240). HB52pro:GUS ein2-5, HB52pro:GUS ein3-1 eil1, HB52pro:GUS 35S:EIN3-GFP, and HB52pro:GUS ctr1-1 were constructed by crossing HB52pro:GUS with ein2-5, ein3-1 eil1, 35S:EIN3-GFP, and ctr1-1 separately. ctr1-1 hb52 and 35S:EIN3-GFP hb52 double mutants were constructed by separately crossing hb52 with ctr1-1 and 35S:EIN3-GFP.
Arabidopsis seeds were surface sterilized in 10% bleach for 15 min and washed six times with distilled water. The seeds were vernalized at 4°C for 3 d and vertically germinated on MS medium (Murashige and Skoog) supplemented with and without 0.1, 1, or 10 μM ACC or 0.1 μM NPA. If transferred to soil, all plants were grown under long-day conditions with a 16-h-light/8-h-dark cycle and light intensity of 150 μmol m-2s−1 using a 30-W fluorescent lamp at 22 to 24°C.
Measurement of Root Gravitropic Responses
Seeds were germinated on MS medium plates with 5 μM estradiol for 4 d vertically under long-day conditions under a 16-h-light/8-h-dark cycle and light intensity of 150 μmol m−2 s−1 using a 30-W fluorescent lamp at 22 to 24°C. Each root was assigned to one of twelve 30° sectors according to its angle relative to a perpendicular line, and the number of roots was represented by the length of each line segment (Supplemental Figure 4) as previously described (Pan et al., 2009).
Histochemical GUS Staining and Fluorescence Observation
Histochemical GUS staining of transgenic plants was performed as previously described (Mao et al., 2016; Yu et al., 2016b). Images were captured using an Olympus IX81 microscope and a HiROX MX5040RZ digital optical microscope (Questar China Limited).
Fluorescence from the GFP transgenic plants was imaged using a ZEISS710 confocal laser scanning microscope: 543 nm for excitation and 620 nm for emission. For fluorescence observation of propidium iodide (PI)-stained plants, seedlings were incubated in 10 mg/mL PI for 3 min and washed twice with water. The stained seedlings were imaged using a Zeiss 710 confocal laser scanning microscope: 488 nm for excitation and 510 nm for emission.
RT-PCR and quantitative RT-PCR analysis
Total RNA was isolated using the TRIzol reagent (Invitrogen) and reverse transcribed using a TransScript RT kit (Invitrogen). cDNA was used for RT-PCR and quantitative RT-PCR. For the RT-PCR analysis, the PCR products were amplified and examined on a 2% agarose gel. Quantitative RT-PCR was performed with a StepOne real-time PCR system using a 2×SYBR Premix Ex Taq II (TaKaRa), gene-specific primers described in Supplemental Data Set 1, cDNA, and deionized water with two steps cycling method (stage 1, 95°C for 30 s; stage 2, 95°C for 5 s; and 60°C for 30 s, 40 cycles). The expression level was normalized to that of UBIQUITIN5 (UBQ5). Three replicates were used for each biological sample, and three technical replicates were used for each experiment.
Yeast One-Hybrid Assay
The yeast-one-hybrid assay was conducted as described previously (Mao et al., 2016). The coding sequences of the proteins were cloned into pAD-GAL4-2.1 (AD vector), and the putative protein binding sites were cloned into pHIS2 (BD vector).
Starch Granule Staining
Starch granule staining was performed as described previously (Sabatini et al., 1999). Images were captured using a HiROX MX5040RZ digital optical microscope (Questar China Limited).
ChIP Assay
The ChIP assay was conducted as described previously (Cai et al., 2014). An anti-GFP antibody (Abmart; M20004L, lot 253971) was added to the sonication products based on 1:500 dilution concentration for an overnight incubation at 4°C.
EMSA
Probes and competitors were commercially synthesized (Sangon Biotech). The probes were synthesized with biotin labels at their 5′ end. The coding sequence of HB52 was amplified with the specific primers described in Supplemental Data Set 1 and cloned into pMAL-C2 between the SalI and PstI sites via T4 DNA ligase system (TaKaRa), and the HB52-MBP fusion protein was expressed in the Rosetta 2 strain of Escherichia coli and affinity purified. The EMSA was performed using a LightShift EMSA Optimization and Control Kit (20148×) according to the manufacturer’s instructions (Thermo Fisher Scientific; lot QL225867).
Transient Transactivation Assay
The coding sequences of EIN3 and HB52 were cloned into pGreenII 62-SK vectors to generate effectors (Hellens et al., 2005). The promoters of HB52, WAG1, WAG2, and PIN2 were cloned into pGreenII0800-LUC vectors to generate reporters (Hellens et al., 2005). The effector and reporter constructs were transfected into Arabidopsis mesophyll protoplasts as described (Yoo et al., 2007). Firefly LUC and REN activities were detected using the dual-luciferase reporter assay system (Promega). The relative REN activity was used as an internal control, and the LUC/REN ratios were calculated.
Triple Response Assay
Triple response assays were conducted as described (Guzmán and Ecker, 1990). Briefly, the seeds were surface sterilized and germinated on MS medium (pH 5.8, solidified with 0.5% agar) with 0, 0.1, 1, and 10 μM ACC (Sigma-Aldrich) in the dark for 3 d at 4°C. The plates were transferred to the light for 6 h and subsequently returned to the dark for 4 d at 21°C before measuring the root and hypocotyl length.
Statistical Analysis
Statistical analyses were conducted using Student’s t tests. Values are the mean ± sd and P < 0.05 was considered statistically significant (*P < 0.05, **P < 0.01, and ***P < 0.001).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: HB52 (At5g53980), UBQ5 (At3g62250), EIN3 (AT3g20770), EIN2 (AT5g03280), CTR1 (AT5g03730), EIL1 (AT2g27050), PIN2 (AT5g57090), WAG1 (AT1g53700), WAG2 (AT3g14370), PIN1 (AT1g73590), PIN3 (AT1g70940), PIN4 (AT2g01420), PIN7 (AT1g23080), AUX1 (AT2g38120), PID (AT2g34650), PID2 (AT2g26700), D6PK (AT5g55910), D6PKL1 (AT4g26610), D6PKL2 (AT5g47750), and D6PKL3 (AT3g27580).
Supplemental Data
Supplemental Figure 1. Identification of the T-DNA insertions in CS909234 (hb52).
Supplemental Figure 2. Phenotypes of HB52 overexpression lines.
Supplemental Figure 3. Phenotypes of the meristematic and elongation zones.
Supplemental Figure 4. Histogram of root gravitropic responses in HB52 knockdown mutants and overexpression lines.
Supplemental Figure 5. Triple response assay.
Supplemental Figure 6. Analysis of ctr1-1 hb52 and 35S:EIN3-GFP hb52.
Supplemental Data Set 1. Primers used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This study was supported by grants from Ministry of Science and Technology of China (2016YFD0100701) and National Natural Science Foundation of China (31572183). We thank Yunde Zhao and Jianwei Pan for their critical review and editing of the manuscript and the ABRC for providing the mutant seeds.
AUTHOR CONTRIBUTIONS
C.-B.X. and Z.-Q.M. designed the experiments. Z.-B.M., P.-X.Z., J.-L.M., L.-H.Y., Y.Y., H.T., and Z.-B.L. performed the experiments and data analyses. Z.-Q.M. wrote the manuscript. C.-B.X supervised the project and revised the manuscript.
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wiśniewska J., Moulinier-Anzola J.C., Sieberer T., Friml J., Luschnig C., Luschnig C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8: 249–256. [DOI] [PubMed] [Google Scholar]
- Abe M., Katsumata H., Komeda Y., Takahashi T. (2003). Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130: 635–643. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., Hirayama T., Roman G., Nourizadeh S., Ecker J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., Stepanova A.N., Solano R., Wisman E., Ferrari S., Ausubel F.M., Ecker J.R. (2003). Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 2992–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F., Zhang X., Zhu Z., Ji Y., He W., Jiang Z., Li M., Guo H. (2012). Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 22: 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T., Dong C.H., Wu Y., Carabelli M., Sessa G., Ruberti I., Morelli G., Chua N.H. (1995). Ectopic expression of the Arabidopsis transcriptional activator Athb-1 alters leaf cell fate in tobacco. Plant Cell 7: 1773–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F.D., Manavella P.A., Dezar C.A., Chan R.L. (2007). The true story of the HD-Zip family. Trends Plant Sci. 12: 419–426. [DOI] [PubMed] [Google Scholar]
- Barbosa I.C.R., Zourelidou M., Willige B.C., Weller B., Schwechheimer C. (2014). D6 PROTEIN KINASE activates auxin transport-dependent growth and PIN-FORMED phosphorylation at the plasma membrane. Dev. Cell 29: 674–685. [DOI] [PubMed] [Google Scholar]
- Benjamins R., Quint A., Weijers D., Hooykaas P., Offringa R. (2001). The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128: 4057–4067. [DOI] [PubMed] [Google Scholar]
- Cai X.T., Xu P., Zhao P.X., Liu R., Yu L.H., Xiang C.B. (2014). Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 5: 5833. [DOI] [PubMed] [Google Scholar]
- Chang K.N., et al. (2013). Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2: e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves A.L.S., Mello-Farias P.C. (2006). Ethylene and fruit ripening: from illumination gas to the control of gene expression, more than a century of discoveries. Genet. Mol. Biol. 29: 508–515. [Google Scholar]
- Chen H., Ma B., Zhou Y., He S.J., Tang S.Y., Lu X., Xie Q., Chen S.Y., Zhang J.S. (2018). E3 ubiquitin ligase SOR1 regulates ethylene response in rice root by modulating stability of Aux/IAA protein. Proc. Natl. Acad. Sci. USA 115: 4513–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Hilson P., Sedbrook J., Rosen E., Caspar T., Masson P.H. (1998). The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. USA 95: 15112–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S.K., Dagenais N., Chory J., Weigel D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100: 469–478. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Delarue M., Prinsen E., Onckelen H.V., Caboche M., Bellini C. (1998). Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 14: 603–611. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Huang F., Galvan-Ampudia C.S., Mähönen A.P., Kleine-Vehn J., Xu J., Quint A., Prasad K., Friml J., Scheres B., Offringa R. (2010). Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137: 3245–3255. [DOI] [PubMed] [Google Scholar]
- Gao Z., Chen Y.F., Randlett M.D., Zhao X.C., Findell J.L., Kieber J.J., Schaller G.E. (2003). Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J. Biol. Chem. 278: 34725–34732. [DOI] [PubMed] [Google Scholar]
- Grbić V., Bleecker A.B. (2003). Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 8: 595–602. [Google Scholar]
- Guzmán P., Ecker J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.M. (1998). Auxin transport: down and out and up again. Science 282: 2201–2203. [DOI] [PubMed] [Google Scholar]
- Ju C., et al. (2012). CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 19486–19491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441. [DOI] [PubMed] [Google Scholar]
- Konishi M., Yanagisawa S. (2008). Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J. 55: 821–831. [DOI] [PubMed] [Google Scholar]
- Le J., Vandenbussche F., Van Der Straeten D., Verbelen J.P. (2001). In the early response of Arabidopsis roots to ethylene, cell elongation is up- and down-regulated and uncoupled from differentiation. Plant Physiol. 125: 519–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Ma M., Feng Y., Li H., Wang Y., Ma Y., Li M., An F., Guo H. (2015). EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 163: 670–683. [DOI] [PubMed] [Google Scholar]
- Li Y., Dai X., Cheng Y., Zhao Y. (2011). NPY genes play an essential role in root gravitropic responses in Arabidopsis. Mol. Plant 4: 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Peng J., Wen X., Guo H. (2013). Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25: 3311–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C., Gaxiola R.A., Grisafi P., Fink G.R. (1998). EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12: 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., Zhou Y., Chen H., He S.J., Huang Y.H., Zhao H., Lu X., Zhang W.K., Pang J.H., Chen S.Y., Zhang J.S. (2018). Membrane protein MHZ3 stabilizes OsEIN2 in rice by interacting with its Nramp-like domain. Proc. Natl. Acad. Sci. USA 115: 2520–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J.L., Miao Z.Q., Wang Z., Yu L.H., Cai X.T., Xiang C.B. (2016). Arabidopsis ERF1 mediates cross-talk between ethylene and auxin biosynthesis during primary root elongation by regulating ASA1 expression. PLoS Genet. 12: e1005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci J.D., Schiefelbein J.W. (1996). Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8: 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C., Alonso J.M., Stepanova A.N. (2013). Ethylene signaling: simple ligand, complex regulation. Curr. Opin. Plant Biol. 16: 554–560. [DOI] [PubMed] [Google Scholar]
- Merchante C., Brumos J., Yun J., Hu Q., Spencer K.R., Enríquez P., Binder B.M., Heber S., Stepanova A.N., Alonso J.M. (2015). Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 163: 684–697. [DOI] [PubMed] [Google Scholar]
- Michniewicz M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056. [DOI] [PubMed] [Google Scholar]
- Müller A., Guan C., Gälweiler L., Tänzler P., Huijser P., Marchant A., Parry G., Bennett M., Wisman E., Palme K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17: 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Katsumata H., Abe M., Yabe N., Komeda Y., Yamamoto K.T., Takahashi T. (2006). Characterization of the class IV homeodomain-Leucine Zipper gene family in Arabidopsis. Plant Physiol. 141: 1363–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Fujioka S., Peng J., Chen J., Li G., Chen R. (2009). The E3 ubiquitin ligase SCFTIR1/AFB and membrane sterols play key roles in auxin regulation of endocytosis, recycling, and plasma membrane accumulation of the auxin efflux transporter PIN2 in Arabidopsis thaliana. Plant Cell 21: 568–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett F.B., Wilson A.K., Estelle M. (1990). The aux1 mutation of Arabidopsis confers both auxin and ethylene resistance. Plant Physiol. 94: 1462–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge M.J., Clark S.E. (2006). Evolution of the class III HD-Zip gene family in land plants. Evol. Dev. 8: 350–361. [DOI] [PubMed] [Google Scholar]
- Qiao H., Shen Z., Huang S.S., Schmitz R.J., Urich M.A., Briggs S.P., Ecker J.R. (2012). Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Zhang Z.J., Wang J., Chen X.B., Wei P.C., Huang R.F. (2017). The activation of OsEIL1 on YUC8M transcription and auxin biosynthesis is required for ethylene-inhibited root elongation in rice early seedling development. PLoS Genet. 13: e1006955.https://doi.org/10.1371/journal.pgen.1006955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růzicka K., Ljung K., Vanneste S., Podhorská R., Beeckman T., Friml J., Benková E. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472. [DOI] [PubMed] [Google Scholar]
- Santner A.A., Watson J.C. (2006). The WAG1 and WAG2 protein kinases negatively regulate root waving in Arabidopsis. Plant J. 45: 752–764. [DOI] [PubMed] [Google Scholar]
- Sawa S., Ohgishi M., Goda H., Higuchi K., Shimada Y., Yoshida S., Koshiba T. (2002). The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J. 32: 1011–1022. [DOI] [PubMed] [Google Scholar]
- Smalle J., Van Der Straeten D. (1997). Ethylene and vegetative development. Physiol. Plant. 100: 593–605. [Google Scholar]
- Stepanova A.N., Hoyt J.M., Hamilton A.A., Alonso J.M. (2005). A Link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17: 2230–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., Dolezal K., Schlereth A., Jürgens G., Alonso J.M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191. [DOI] [PubMed] [Google Scholar]
- Swarup R., Perry P., Hagenbeek D., Van Der Straeten D., Beemster G.T., Sandberg G., Bhalerao R., Ljung K., Bennett M.J. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19: 2186–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Zhang C., Ji Y., Zhao Q., He W., An F., Jiang L., Guo H. (2012). Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 22: 1613–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., Kasahara H., Kamiya Y., Chory J., Zhao Y. (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 18518–18523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Lu X., Ma B., Chen S.Y., Zhang J.S. (2015a). Ethylene signaling in rice and Arabidopsis: conserved and diverged aspects. Mol. Plant 8: 495–505. [DOI] [PubMed] [Google Scholar]
- Yang C., Ma B., He S.J., Xiong Q., Duan K.X., Yin C.C., Chen H., Lu X., Chen S.Y., Zhang J.S. (2015b). MAOHUZI6/ETHYLENE INSENSITIVE3-LIKE1 and ETHYLENE INSENSITIVE3-LIKE2 regulate ethylene response of roots and coleoptiles and negatively affect salt tolerance in rice. Plant Physiol. 169: 148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C.C., et al. (2015). Ethylene responses in rice roots and coleoptiles are differentially regulated by a carotenoid isomerase-mediated abscisic acid pathway. Plant Cell 27: 1061–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yu H., Chen X., Hong Y.Y., Wang Y., Xu P., Ke S.D., Liu H.Y., Zhu J.K., Oliver D.J., Xiang C.B. (2008). Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20: 1134–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Chen X., Wang Z., Wang S., Wang Y., Zhu Q., Li S., Xiang C. (2013). Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 162: 1378–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L.H., Wu S.J., Peng Y.S., Liu R.N., Chen X., Zhao P., Xu P., Zhu J.B., Jiao G.L., Pei Y., Xiang C.B. (2016a). Arabidopsis EDT1/HDG11 improves drought and salt tolerance in cotton and poplar and increases cotton yield in the field. Plant Biotechnol. J. 14: 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Zhang Y., Wang J., Yan X., Wang C., Xu J., Pan J. (2016b). Clathrin-mediated auxin efflux and maxima regulate hypocotyl hook formation and light-stimulated hook opening in Arabidopsis. Mol. Plant 9: 101–112. [DOI] [PubMed] [Google Scholar]
- Zhang F., Qi B., Wang L., Zhao B., Rode S., Riggan N.D., Ecker J.R., Qiao H. (2016). EIN2-dependent regulation of acetylation of histone H3K14 and non-canonical histone H3K23 in ethylene signalling. Nat. Commun. 7: 13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Wang L., Qi B., Zhao B., Ko E.E., Riggan N.D., Chin K., Qiao H. (2017). EIN2 mediates direct regulation of histone acetylation in the ethylene response. Proc. Natl. Acad. Sci. USA 114: 10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Wang L., Ko E.E., Shao K., Qiao H. (2018). Histone deacetylases SRT1 and SRT2 interact with ENAP1 to mediate ethylene-induced transcriptional repression. Plant Cell 30: 153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Zhao M., Shi T., Shi H., An F., Zhao Q., Guo H. (2009). EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 106: 21431–21436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]