Abstract
Peroxisome proliferator-activated receptor-γ (PPARγ) is a master regulator of adipogenesis, and alterations in its function are associated with various pathological processes related to metabolic syndrome. Recently, we found that the chicken PPARγ gene is regulated by three alternative promoters (P1, P2 and P3), producing five different transcript isoforms and two protein isoforms. In this study, the P1 promoter structure was characterized. Bioinformatics identified six putative nuclear respiratory factor 1 (NRF1) binding sites in the P1 promoter, and a reporter assay showed that NRF1 inhibited the activity of the P1 promoter. Of the six putative NRF1 binding sites, individual mutations of three of them abolished the inhibitory effect of NRF1 on P1 promoter activity. Furthermore, a ChIP assay indicated that NRF1 directly bound to the P1 promoter, and real-time quantitative RT-PCR analysis showed that NRF1 mRNA expression was negatively correlated with PPARγ1 expression (Pearson’s r = -0.148, p = 0.033). Further study showed that NRF1 overexpression inhibited the differentiation of the immortalized chicken preadipocyte cell line (ICP1), which was accompanied by reduced PPARγ1 mRNA expression. Taken together, our findings indicated that NRF1 directly negatively regulates the P1 promoter of the chicken PPARγ gene and inhibits adipogenesis.
Keywords: nuclear respiratory factor 1, chicken, PPARγ, transcription, adipogenesis
Introduction
Adipogenesis plays central roles in energy homeostasis and is significantly associated with obesity. Adipogenesis is tightly controlled by a complex network of transcriptional regulators (Rosen et al., 2002; Garinshkolnik et al., 2014). PPARγ is a master regulator of adipogenesis and regulates the transcription of various genes involved in adipocyte differentiation and lipid accumulation (de la Rosa Rodriguez and Kersten, 2017; Kim et al., 2017; Park et al., 2017). Ectopic expression of PPARγ is sufficient to induce adipocyte differentiation in fibroblasts, and no factor has been reported to promote adipogenesis in the absence of PPARγ (Lee and Ge, 2014).
In mammals, the PPARγ gene is regulated by multiple alternative promoters. Because of alternative promoter usage and alternative splicing, the PPARγ gene produces various transcript isoforms, encoding two different protein isoforms: PPARγ1 and PPARγ2 (Zhu et al., 1995; Lee and Ge, 2014; Chandra et al., 2017). The human PPARγ gene has four alternative promoters designated the PPARγ1, PPARγ2, PPARγ3 and PPARγ4 promoters while the mouse PPARγ gene has two alternative promoters designated the PPARγ1 and PPARγ2 promoters (Zhu et al., 1995; Al-Shali et al., 2004; Lee and Ge, 2014; Chandra et al., 2017). To date, a number of transcription factors and coregulators have been identified that regulate the alternative promoters of the PPARγ gene in humans and mice. For example, E2F transcription factor 1 (E2F1), early B-cell factor 1 (EBF1), and sterol regulatory element-binding protein-1 (SREBP1) directly bind to mouse PPARγ1 promoter and enhance the expression of the PPARγ1 transcript (Lee and Ge, 2014). Nuclear factor E2-related factor 2 (NRF2), Krüppel-like factors 5 (KLF5), KLF9 and KLF15 directly bind to mouse PPARγ2 promoter and enhance the expression of the PPARγ2 transcript, while KLF2 directly binds to mouse PPARγ2 promoter and decreases PPARγ2 transcript expression (Pi et al., 2010; Lee and Ge, 2014). Forkhead box class O1 (FOXO1) can directly bind to and reduce human PPARγ2 promoter activity, and in addition, it can indirectly inhibit human PPARγ1 promoter activity (Armoni et al., 2006).
Recently, we demonstrated for the first time that the chicken PPARγ gene is regulated by three alternative promoters designated P1, P2 and P3, producing five different transcript isoforms (PPARγs 1-5) due to alternative splicing and promoter usage (Duan et al., 2015). PPARγ1 initiates from the P1 promoter, PPARγs 2-4 from the P2 promoter and PPARγ5 from the P3 promoter. Among these five different transcript isoforms, PPARγ1 is highly expressed in various chicken tissues, including adipose tissue, liver, kidney, spleen and duodenum (Duan et al., 2015). The transcriptional regulation of the P1 promoter is still unknown. In the present study, we characterized the P1 promoter organization and demonstrated that nuclear respiratory factor 1 (NRF1) negatively regulates the P1 promoter of the PPARγ gene and inhibits chicken adipogenesis.
Materials and Methods
Ethics Statement
The research project was approved by the Institutional Biosafety Committee of Northeast Agricultural University (Harbin, P. R. China). Plasmid construction and transfection were performed in accordance with the guidelines for the Regulation on Safety Administration of Agricultural Genetically Modified Organisms (RSAGMO) established by the People’s Republic of China (Revised version 2017).
Tissues
The abdominal fat tissue samples used in this study were from a previous study by our group (Zhang et al., 2017). These samples were obtained from generation 19 of Northeast Agricultural University broiler lines divergently selected for abdominal fat content (NEAUHLF) and composed of a total of 70 abdominal fat tissues of male birds (five birds per line and per time point) from 1 to 7 weeks of age. These samples were stored in liquid nitrogen until total RNA extraction.
Plasmid Construction
The chicken PPARγ P1 promoter and its subsequent 5′-truncation mutants were generated by PCR from chicken genomic DNA using the designated forward primers and a common reverse primer (Cloning P1+108 promoter R) as shown in Table 1 and then subcloned into the promoterless luciferase expression vector pGL3-Basic (Promega, United States). Site-directed mutagenesis was performed by DNA synthesis (GENEWIZ, China). The site-mutated P1 promoters were cloned into a pGL3-Basic vector. For the NRF1 overexpression vector, the full-length coding region of chicken NRF1 was amplified by RT-PCR from chicken abdominal fat tissue total RNA and cloned into an empty pCMV-HA vector (Clontech, United States). All primers are shown in Table 1, and all final constructs were confirmed by DNA sequencing.
Table 1.
PCR primers used in this study.
| Primer name | Sequence |
|---|---|
| Cloning P1-1891 promoter F | F:5′-ATTTCTCTATCGATAGGTACCTCCTGCCTCAATCTGCTAAAATA-3′ |
| Cloning P1-950 promoter F | F:5′-ATTTCTCTATCGATAGGTACCGTAATTTGATGTATTCAACTCTAATCTATAA-3′ |
| Cloning P1-450 promoter F | F:5′-ATTTCTCTATCGATAGGTACCGCGACTACACGCAGCGCA-3′ |
| Cloning P1-327 promoter F | F:5′-ATTTCTCTATCGATAGGTACCCCAGATCCACTCCAGGGC-3′ |
| Cloning P1-231 promoter F | F:5′-ATTTCTCTATCGATAGGTACCGAACAGATTGGGTCTAAAGGG-3′ |
| Cloning P1+1 promoter F | F:5′-ATTTCTCTATCGATAGGTACCGGCGGTGCCCCGGCGGGG-3′ |
| Cloning P1+108 promoter R | R:5′-CAGTACCGGAATGCCAAGCTTTGGCGCTGTCAAGTCTCA-3′ |
| Cloning full-length NRF1 CDS | F: 5′-TGGCCATGGAGGCCCGAATTCGGATGGAAGAACACGGCGTG-3′ |
| R: 5′-GATCCCCGCGGCCGCGGTACCTCACTGTTCCAAAGTTACCA-3′ | |
| qRT-PCR NRF1 | F: 5′-ACCCATCCATCCGTAAGAG-3′ |
| R: 5′-CTTGCGTACCACATTCTCC-3′ | |
| qRT-PCR PPARγ1 | F: 5′-GGAGTTTATCCCACCAGAAG-3′ |
| R: 5′-AATCAACAGTGGTAAATGGC-3′ | |
| qRT-PCR NONO | F: 5′-AGAAGCAGCAGCAAGAAC-3′ |
| R: 5′-TCCTCCATCCTCCTCAGT-3′ | |
| qRT-PCR FABP4 | F: 5′-ATGTGCGACCAGTTTGT-3′ |
| R: 5′-TCACCATTGATGCTGATAG-3′ | |
| qRT-PCR G0S2 | F: 5′-GACGGCAAGGATGGAAAAGAT-3′ |
| R: 5′-GTCGTAGTGGTTCTGCTCGTTGTA-3′ | |
| qRT-PCR GPDH | F: 5′-ACCTCCCATCCCATACCGA-3′ |
| R: 5′-CCACTCCACGCTGCCAACA-3′ | |
| qRT-PCR AdipoQ | F: 5′-GCAACAACAACGGGGTCT-3′ |
| R: 5′-AGGGGAATTTTCTGGTACATAG-3′ | |
| ChIP-qPCR P1 promoter | F: 5′-GAGCCCCGACCCGCGCAGCGCCCAC-3′ |
| R: 5′-ATAAACTCCCCGGGCCGGCCCATCC-3′ | |
| ChIP-qPCR Fluc | F: 5′-AAAACGGATTACCAGGGATTTCAGT-3′ |
| R: 5′-AGCGACACCTTTAGGCAGACCAGT-3′ |
Cell Culture
DF1 cells, chicken stromal vascular fraction (SVF) cells and an immortalized chicken preadipocyte (ICP1) cell line (Wang et al., 2017) were maintained in Dulbecco’s-modified Eagle’s medium (DMEM) with high glucose (Gibco, United States) or DMEM: nutrient mixture F12 (DMEM/F12) (Gibco) supplemented with 10% fetal bovine serum (FBS) (BI, Germany) plus 100 units/ml penicillin and 100 units/ml streptomycin in a humidified incubator at 37°C and 5% CO2.
Transfection and Luciferase Assays
After reaching 70–80% confluence, the DF1, ICP1 and SVF cells were washed with PBS, and transient transfections were performed using Lipofectamine 2000 reagent (Invitrogen, United States). A reporter luciferase assay was performed according to the manufacturer’s instructions with a Dual-Luciferase Reporter Assay System (Promega, United States) after 48 h of transfection. Firefly luciferase (Fluc) activity was normalized to Renilla luciferase (Rluc) activity.
Western Blot Analysis
Cultured cells were washed three times with cold PBS and lysed in 6-well plates by using 100 μl RIPA Buffer (Beyotime Biotechnology, China) supplemented with 1 μl phenylmethanesulfonyl fluoride (Beyotime Biotechnology, China). Cell lysates were subjected to SDS-PAGE followed by Western blot analysis using an HA-specific antibody (Abmart, China) and a β-actin antibody (Beyotime Biotechnology, China). The blots were visualized using an ECL Plus detection kit (Beyotime Biotechnology, China).
Chromatin Immunoprecipitation (ChIP) Assay
Chromatin immunoprecipitation was performed using a ChIP assay kit (Cell Signaling Technology, United States) as previously described (Funato et al., 2003; Ouyang et al., 2011; Deng et al., 2012). DF1 cells were cotransfected with pGL3P1-1891/+108 and pCMV-HA-NRF1 or an empty pCMV-HA vector for 48 h; 1 × 107 transfected cells were then fixed with 1% formaldehyde at room temperature for 10 min and quenched in 125 mM glycine at room temperature for 5 min. The Chromatin was digested with 0.5 μl micrococcal nuclease into 100–900 bp DNA/protein fragments, and immunoprecipitated using with 5 μg of HA-specific antibody (Abcam, United States) or 5 μg of mouse IgG (Beyotime, China). The purified DNA fragments were analyzed by qPCR. The qPCR was performed on the 7500 Real-Time PCR System (Applied Biosystems, United States) with SYBR Green PCR Master Mix (Roche Molecular Systems, United States). A specific pair of primers was designed to detect the coding region of the firefly luciferase gene, which was used as a negative control. The primers used in the ChIP-qPCR assay are shown in Table 1. Non-immunoprecipitated DNA (2%) was used as an input control. ChIP qPCR data were normalized to input chromatin and then presented as fold enrichment over the negative control using the ΔΔCt equation (Tatler et al., 2016).
ICP1 Cell Differentiation
Immortalized chicken preadipocyte cell lines were cultured to 50% confluence, and then, the cells were induced to differentiate by 160 μM sodium oleate (Sigma, United States). Subsequently, the medium was removed every 24 h and replaced with fresh medium containing DMEM/F12 containing 10% FBS together with 160 μM sodium oleate. Cells were harvested for RT-PCR every 24 h. Differentiation of ICP1 cells was continued for a total of 96 h.
Oil Red O Staining
Oil red O staining was performed as described previously (Wang et al., 2017). Briefly, the differentiated ICP1 cells were washed twice with PBS and fixed by 4% paraformaldehyde for 30 min at 4°C, washed with PBS twice and distilled water twice, and stained with 0.5% Oil Red O staining solution in isopropanol and water (3:2) for 15 min at room temperature, followed by washing with PBS twice. Alternatively, for a quantitative assay, the stained cells were destained with isopropanol, and the optical density was determined at a 510 nm wavelength with a spectrophotometer (Ultrospec 1000, United Kingdom).
RNA Isolation and Quantitative Real-Time RT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen, United States) following the manufacturer’s instructions, and cDNA was generated with a Transcriptor First Strand cDNA Synthesis kit (Roche Molecular Systems, United States), followed by analysis using a 7500 Real-Time PCR System (Applied Biosystems, United States) with SYBR Green PCR Master Mix (Roche Molecular Systems, United States). The primers used are shown in Table 1. Relative mRNA levels were normalized to the NONO gene (non-POU domain containing) using the 2-ΔΔCt method.
Statistical Analysis
Data are expressed as the mean ± SE of the mean. Student’s t-test was performed using Graph Pad Prism 5. Pearson’s r was used to determine the degree of correlation between PPARγ1 and NRF1 mRNA expression levels as described (Sun et al., 2014). Statistical significance was indicated by ∗p < 0.05, ∗∗p < 0.01.
Results
Characterization of the Chicken PPARγ P1 Promoter
To understand the transcriptional regulation of the chicken PPARγ P1 promoter, an ∼2-kb genomic DNA fragment spanning 1,891 bp upstream and 108 bp downstream of the transcription start site of PPARγ1 was amplified by PCR and cloned into luciferase reporter vector pGL3-Basic. The resultant construct was named pGL3P1-1891/+108. A reporter assay showed that the luciferase activities of pGL3P1-1891/+108 were 110-, 66- and 75-fold higher than those of empty pGL3-Basic vector in a chicken embryo fibroblast (DF1) cell line, chicken SVF cells and an ICP1 cell line, respectively (Figure 1A), confirming that we had cloned the active P1 promoter. The P1 promoter was active in these three different cell lines, consistent with our previous finding that PPARγ1 was widely expressed in various chicken tissues (Duan et al., 2015).
FIGURE 1.
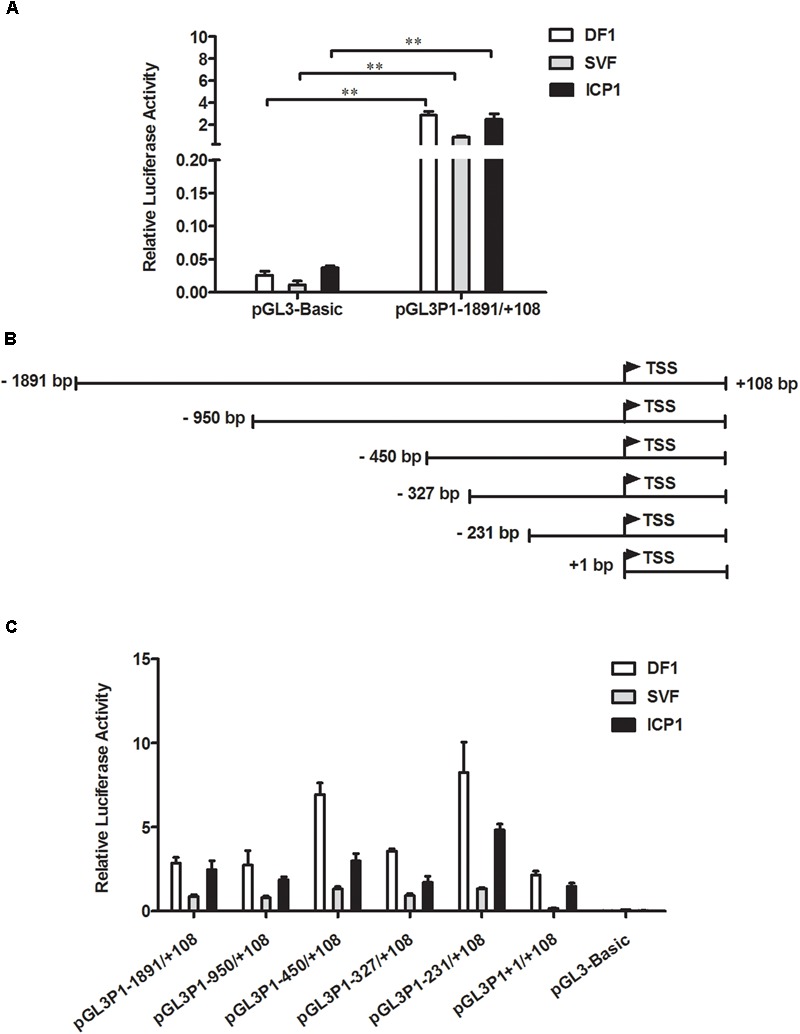
Characterization of the chicken PPARγ gene P1 promoter. (A) Luciferase activity of the P1 promoter reporter construct pGL3P1-1891/+108 in DF1, SVF, and ICP1 cells. Cells were transfected with pGL3P1-1891/+108 along with the pRL-TK Renilla luciferase vector by using Lipofectamine 2000 reagent, and luciferase activity was determined at 48 h after transfection. (B) Schematic diagram of the reporter constructs of the P1 promoter and its 5′ truncation mutants. The transcription start site (TSS) of chicken PPARγ1 is represented by the bent arrow. The positions are numbered relative to the TSS. (C) Truncation analysis of the P1 promoter in DF1, SVF, and ICP1 cells. The indicated P1 promoter constructs and pRL-TK vector were cotransfected into DF1, SVF, and ICP1 cells. Luciferase activity was detected 48 h after cotransfection. The pRL-TK vector was used for normalization of transfection efficiency. All data represent the mean ± SE. Statistical significance was determined by Student’s t-test. ∗∗p < 0.01.
To delineate the sequences required for the P1 promoter, we generated a total of five 5’-deletion fragments of the P1 promoter by PCR and constructed their respective reporter constructs (pGL3P1-950/+108, pGL3P1-450/+108, pGL3P1-327/+108, pGL3P1-231/+108, and pGL3P1+1/+108) (Figure 1B). All five of these P1 truncation mutants had higher luciferase activities than did the empty pGL3-Basic vector (p < 0.05) (Figure 1C) and displayed a similar promoter activity pattern in DF1, SVF and ICP1 cells (Figure 1C). The promoter activity of pGL3P1-1891/+108 was essentially similar to that of pGL3P1-950/+108. Progressive 5′-truncation from -950 to -450 (pGL3P1-450/+108) or from -327 to -231 (pGL3P1-231/+108) significantly increased the promoter activity compared with that of pGL3P1-950/+108 or pGL3P1-327/+108 (p < 0.05) (Figure 1C), indicating the presence of strong repressive elements in the P1 promoter regions from -950 to -450 and from -327 to -231 (Figure 1C). However, compared with pGL3P1-231/+108, further 5’-truncation of 231 bp (pGL3P1+1/+108) resulted in a significant decrease in the promoter activity (p < 0.05) (Figure 1C), indicating the presence of positive regulatory elements in the P1 promoter region from -231 to +1. Among all P1 promoter reporter constructs, pGL3P1-231/+108 exhibited the highest luciferase activity, whereas pGL3P1+1/+108 had the minimum luciferase activity that still retained basal promoter activity compared with the empty pGL3-Basic vector (p > 0.05) (Figure 1C), suggesting that the +1/+108 region is the core P1 promoter region.
To understand the transcriptional regulation of the P1 promoter, using JASPAR (Roy et al., 2017), we predicted the transcription factor binding sites in the -231/+108 region, which had the highest promoter activity (Figure 1C). The result showed that there were a number of putative binding sites for various transcription factors, including GATA binding protein 3 (GATA3), activating enhancer binding protein 2 (AP2), transcription factor Sp1 (Sp1) and NRF1. Of these transcription factors, NRF1 interested us. NRF1 has been known as a key transcription factor for mitochondrial biogenesis and functions (Scarpulla, 2002). Moreover, several independent lines of evidence indicated that NRF1 is implicated in lipid droplet accumulation, cell proliferation and apoptosis of the mouse preadipocyte cell line 3T3-L1 and the pathogenesis of type 2 diabetes (Cho et al., 2005; Liu et al., 2008; Tienen et al., 2010). This evidence inspired us to determine whether NRF1 regulates transcription of the P1 promoter and is involved in the regulation of adipogenesis. A detailed bioinformatics analysis showed that a total of six putative NRF1 binding sites were present in the P1 promoter region from -1891 to +108; five of them were clustered in the region -231/+108, and the other one was present at the position -285/-275 (Figure 2A). In this report, these six putative NRF1 binding sites were designated N1 (-285 to -275), N2 (-32 to -22), N3 (+1 to +11), N4 (+7 to +17), N5 (+18 to +28), and N6 (+42 to +52), relative to the transcription start site of PPARγ1 (Figure 2A), and the mutated binding sites were referred to as M1, M2, M3, M4, M5, and M6 (Figure 2A).
FIGURE 2.
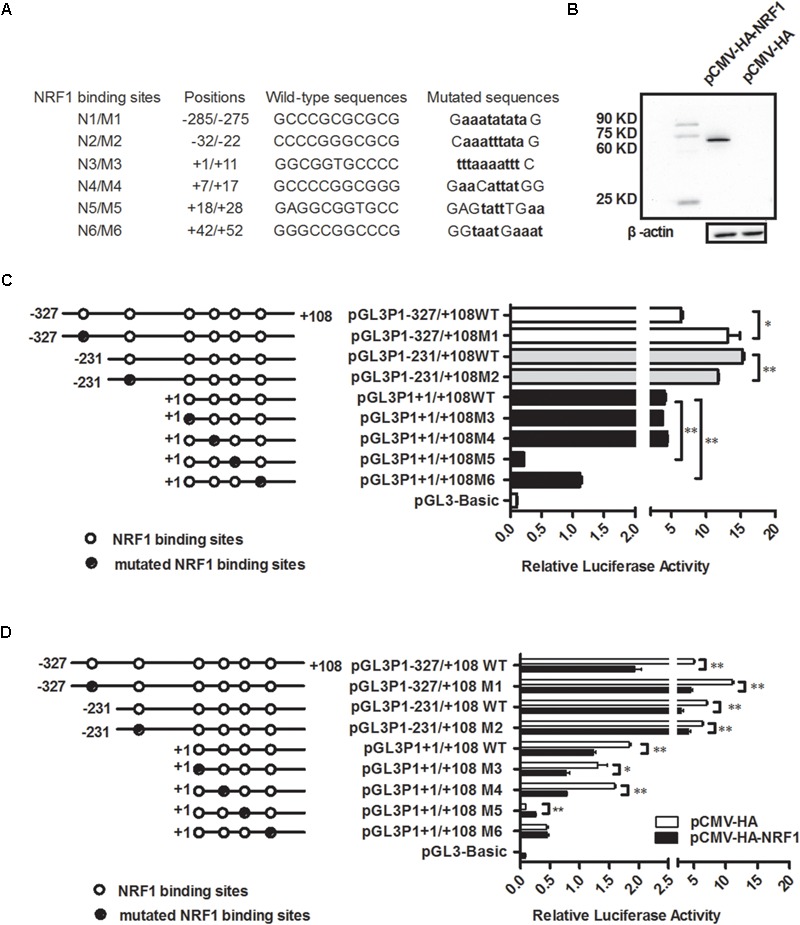
NRF1 represses P1 promoter activity. (A) The nucleotide sequences of the six putative NRF1 binding sites and their respective mutations in the P1 promoter. The P1 promoter mutants were created by direct DNA synthesis and subsequent cloning, and the positions of the six putative NRF1 binding sites in the P1 promoter region were numbered relative to the transcription start site (TSS). NRF1 binding sites are indicated by capital letters, and their mutated nucleotides are indicated by bold lowercase letters. (B) Western blot identification of chicken NRF1 expression vector (pCMV-HA-NRF1). The pCMV-HA-NRF1 or empty pCMV-HA vector was transfected into DF1 cells. The cell lysates were harvested 48 h after transfection and immunoblotted with an HA-specific antibody. (C) The effects of mutation of the six individual putative NRF1 binding sites on basal P1 promoter activity. The indicated P1 promoter constructs (pGL3P1-327/+108, pGL3P1-231/+108, and pGL3P1+1/+108) or indicated P1 promoter mutant constructs (pGL3P1-327/+108M1, pGL3P1-231/+108M2, pGL3P1+1/+108M3, pGL3P1+1/+108M4, pGL3P1+1/+108M5, and pGL3P1+1/+108M6), along with pRL-TK, were cotransfected into DF1 cells, and luciferase activity was determined 48 h after transfection. The open circles indicate the wild-type NRF1 binding sites, and the filled circles indicate the mutated NRF1 binding sites. All data represent mean ± SE. Statistical significance was determined by Student’s t-test comparing mutated versus wild-type NRF1 binding site. ∗p < 0.05, ∗∗p < 0.01. (D) Effects of mutation of the six individual NRF1 binding sites on NRF1-mediated inhibition of the P1 promoter. DF1 cells were cotransfected with the indicated reporter constructs along with pCMV-HA-NRF1 or empty pCMV-HA vector and pRL-TK. Luciferase activity was determined 48 h after cotransfection. All data represent the mean ± SE. Statistical significance was determined by Student’s t-test comparing the cotransfection of the designated reporter constructs and pCMV-HA-NRF1 versus the cotransfection of the designated reporter constructs and empty pCMV-HA vector. ∗p < 0.05, ∗∗p < 0.01.
NRF1 Inhibits the P1 Promoter Activity
To test whether NRF1 regulates the P1 promoter, we initially constructed and confirmed an NRF1 expression vector, pCMV-HA-NRF1 (Figure 2B). Then, DF1 cells were transiently cotransfected with the P1 reporter constructs (pGL3P1-327/+108, pGL3P1-231/+108, or pGL3P1+1/+108) and pCMV-HA-NRF1 or pCMV-HA vector plus pRL-TK Renilla luciferase vector. The P1 reporter construct pGL3P1-327/+108 contained all six putative NRF1 binding sites, pGL3P1-231/+108 contained five of six putative NRF1 binding sites (except for N1), and pGL3P1+1/+108 contained four of these NRF1 binding sites (except for N1 and N2). A reporter assay showed that transfection with pCMV-HA-NRF1 significantly inhibited the luciferase reporter activity of these three reporters (pGL3P1-327/+108, pGL3P1-231/+108, and pGL3P1+1/+108) (p < 0.01) (Figure 2D), suggesting that NRF1 negatively regulates the P1 promoter.
To define which NRF1 binding sites are required for the NRF1-mediated inhibition of the P1 promoter, we mutated individual NRF1 binding sites by site-directed mutagenesis using DNA synthesis and generated their reporter constructs based on the aforementioned promoter constructs (Figures 2C,D). In brief, the mutated N1 and N2 binding site promoter reporter constructs were generated based on pGL3P1-327/+108 and pGL3P1-231/+108, respectively, and their mutant reporter constructs were designated pGL3P1-327/+108M1 and pGL3P1-231/+108M2. The mutated N3, N4, N5, and N6 binding site promoter constructs were generated based on pGL3P1+1/+108, and the resultant mutant reporter constructs were designated pGL3P1+1/+108M3, pGL3P1+1/+108M4, pGL3P1+1/+108M5, and pGL3P1+1/+108M6, respectively. First, we tested the effect of these individual NRF1 binding site mutations on the basal activity of the P1 promoter. A reporter gene assay showed that compared with their respective wild-type reporters, N1 mutation caused a 104.68% increase in basal promoter activity (p < 0.05) (Figure 2C), but N2 mutation led to a 23.41% reduction in P1 promoter activity (p < 0.01) (Figure 2C). Both N3 and N4 mutations had no effect on basal promoter activity (p > 0.05) (Figure 2C). In contrast, N5 and N6 mutations caused a 94.70 and 72% reduction, respectively, in basal promoter activity (p < 0.01) (Figure 2C). These results suggest that these NRF1 binding sites have different regulatory roles in basal P1 promoter activity.
Then, we investigated which NRF1 binding sites are required for the NRF1-mediated inhibition of the P1 promoter. Our mutation analysis showed that N2 mutation markedly reduced the inhibitory effect of NRF1 on the promoter activity of pGL3P1-231/+108 by 38.57% (p < 0.05) (Figure 2D). Both N5 and N6 mutations almost entirely abolished the inhibitory effect of NRF1 on the promoter activity of pGL3P1+1/+108 (Figure 2D). However, N1 mutation had no obvious effect on the inhibitory effect of NRF1 on the promoter activity of pGL3P1-327/+108 (p > 0.05) (Figure 2D). In contrast, N3 and N4 mutations enhanced the inhibitory effect of NRF1 on the promoter activity by 26 and 55.27%, respectively (Figure 2D). Collectively, these results suggested that the binding sites N2, N5 and N6, but not N1, N3 and N4, are required for NRF1-mediated inhibition of the P1 promoter.
NRF1 Binds to the P1 Promoter
To investigate whether NRF1 directly binds to and regulates the P1 promoter, we employed a chromatin immunoprecipitation (ChIP) assay. The P1 reporter construct (pGL3P1-1891/+108) and pCMV-HA-NRF1 vector were cotransfected into DF1 cells, and ChIP was performed with an HA-specific antibody or mouse IgG (negative control). Two additional negative controls (A and B) were prepared by the cotransfection of DF1 cells with empty pCMV-HA vector and pGL3P1-1891/+108 reporter construct and immunoprecipitation with either mouse IgG (A) or HA-specific antibody (B). Enriched DNA was analyzed using quantitative PCR with a specific pair of primers (ChIP-qPCR P1 promoter, Table 1), which was designed to amplify the -60/+52 region of the P1 promoter. The ChIP-qPCR results showed that the P1 promoter fragment was significantly enriched (9-, 9- and 12-fold) in the DNA immunoprecipitated by the HA-specific antibody compared with the negative controls (mouse IgG, A and B) (p < 0.01) (Figure 3), but not enriched in any one of the three negative controls (mouse IgG, A and B; enrichment folds: 1:1:0.7) (p > 0.05) (Figure 3). The coding region of the firefly luciferase gene (Fluc) was used as a negative control in this study. As expected, the coding region of Fluc was not enriched in the DNA immunoprecipitated by HA-specific antibody compared with that by the negative controls (mouse IgG, A and B) in the ChIP-qPCR analysis using a specific pair of primers to amplify the coding region of Fluc (p > 0.05) (Figure 3). Taken together, these findings suggest that NRF1 can directly bind to the P1 promoter.
FIGURE 3.
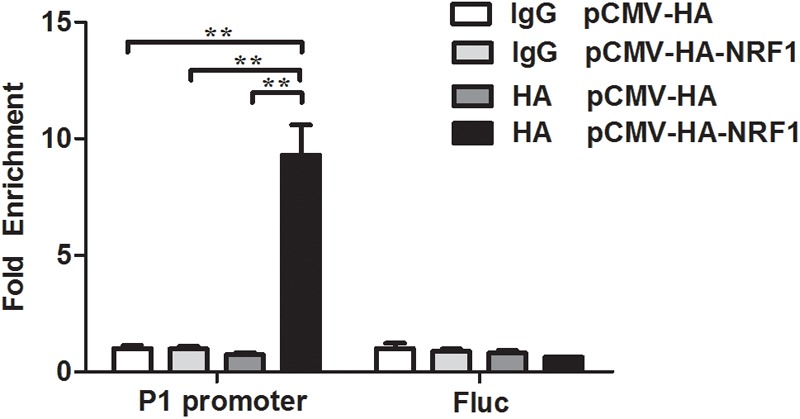
NRF1 directly binds to the P1 promoter. DF1 cells were cotransfected with the P1 promoter construct (pGL3P1-1891/+108) and either pCMV-HA-NRF1 or empty pCMV-HA vector. At 48 h after transfection, ChIP was performed with an HA-specific antibody or mouse IgG. Immunoprecipitated DNA was quantified by qPCR using two specific pairs of primers. One pair of primers for the P1 promoter region from –60 to +52 was used to determine the enrichment of NRF1 binding to P1 promoter, and the other pair of primers for the coding region of the firefly luciferase gene (Fluc) was used to determine the enrichment of NRF1 binding to the Fluc gene coding region and was used as a negative control. Non-immunoprecipitated DNA (2%) was used as an input control. Data were presented as fold enrichment over the negative control, which was prepared by cotransfection of DF1 cells with empty pCMV-HA vector and pGL3P1-1891/+108 and immunoprecipitated with mouse IgG (set to 1). All data represent the mean ± SE. Statistical significance was determined by Student’s t-test. ∗∗p < 0.01.
Expression Patterns of NRF1 and PPARγ1 During Chicken Adipose Development
To investigate whether NRF1 regulates the P1 promoter in vivo, we analyzed the expression patterns of chicken NRF1 and PPARγ1 in abdominal fat tissues from Northeast Agricultural University broiler lines divergently selected for abdominal fat content (NEAUHLF) from 1 to 7 weeks of age using real-time RT-PCR. The results showed that PPARγ1 mRNA expression was remarkably higher in fat than in lean chicken lines from 2 to 7 weeks of age (p < 0.05) (Figure 4A), which was consistent with our previous study (Duan et al., 2015). Conversely, NRF1 mRNA expression was lower in fat than in lean lines from 1 to 7 weeks of age; particularly, NRF1 mRNA expression was significantly lower in fat than in lean chicken lines at 3 and 6 weeks of age (p < 0.05) (Figure 4A). Correlation analysis showed that NRF1 and PPARγ1 mRNA expression levels were significantly negatively correlated (Pearson’s r = -0.148, p = 0.033) from 1 to 7 weeks of age in the lean and fat lines of NEAUHLF. These expression data support our finding that NRF1 negatively regulates the P1 promoter (Figure 4A).
FIGURE 4.
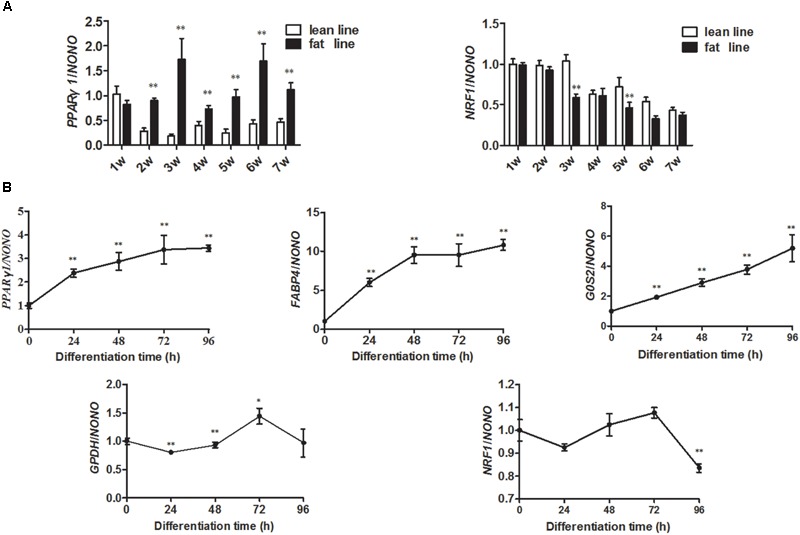
Expression levels of PPARγ1 and NRF1 genes during chicken abdominal adipose tissue development and ICP1 cell differentiation. (A) Real-time RT-PCR analysis of the expression levels of PPARγ1 and NRF1 in the abdominal adipose tissues of NEAUHLF (each line, n = 5) from 1 to 7 weeks of age. Statistical significance was determined by Student’s t-test comparing lean to fat lines. ∗p < 0.05, ∗∗p < 0.01. (B) Real-time RT-PCR analysis of the expression levels of NRF1, PPARγ1 and adipogenic differentiation markers (FABP4, G0S2, GPDH, and AdipoQ) during differentiation of ICP1 cells. At 50% confluence, ICP1 cells were induced to differentiate by adding 160 μM sodium oleate with medium changes every 24 until 96 h of culture. The cells were harvested at 0, 24, 48, 72, and 96 h of differentiation. Chicken NONO was used as the internal control. The expression levels of PPARγ1, FABP4, G0S2, GPDH, and NRF1 at the indicated time points are expressed relative to the expression of the respective genes in ICP1 cells at 0 h of differentiation. All data represent the mean ± SE. Statistical significance was determined by Student’s t-test comparing expression at the indicated time points versus 0 h of ICP1 cell differentiation. ∗p < 0.05, ∗∗p < 0.01.
NRF1 Overexpression Inhibits Chicken Adipocyte Differentiation
PPARγ is a master regulator of adipogenesis. To test whether NRF1 is also involved in the regulation of adipogenesis, we first assayed the expression of NRF1 and PPARγ1 during differentiation of ICP1 cells induced by sodium oleate. Differentiation was evident as indicated by increasing expression levels of FABP4, G0S2, GPDH, and AdipoQ from 0 to 96 h after induction of differentiation (Figure 4B). The NRF1 mRNA level remained relatively constant up to 72 h after induction of differentiation (p > 0.05) but significantly decreased at 96 h to a level lower than that in ICP1 cells at 0 h of differentiation (p < 0.01) (Figure 4B). By contrast, PPARγ1 mRNA expression was continuously upregulated throughout the 96 h time course of differentiation (Figure 4B). Then, we examined the effect of NRF1 overexpression on chicken preadipocyte differentiation by transient transfection of pCMV-HA-NRF1 into ICP1 cells. After 24 h of transfection, the cells were induced to differentiate by sodium oleate. Western blot confirmed that the NRF1 protein was overexpressed at 24, 48, and 72 h of differentiation of ICP1 cells transfected with pCMV-HA-NRF1 compared to the cells transfected with empty pCMV-HA vector (Figure 5A).
FIGURE 5.
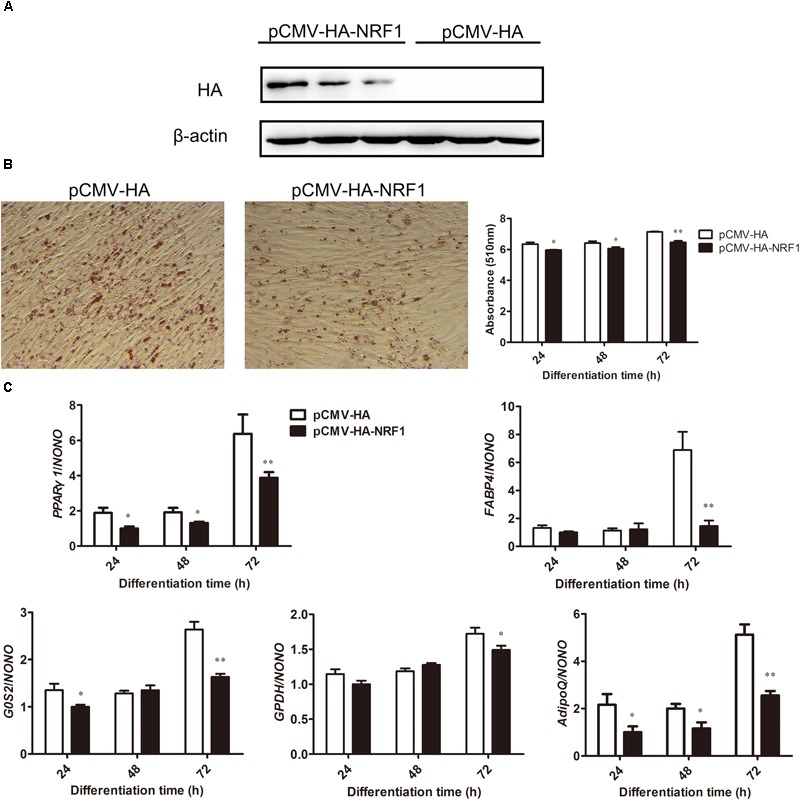
NRF1 overexpression inhibits chicken preadipocyte differentiation. (A) Western blot analysis of NRF1 overexpression during chicken preadipocyte differentiation. ICP1 cells were transfected with either pCMV-HA-NRF1 or empty pCMV-HA vector as a control. At 24 h after transfection, the cells were induced to differentiate by adding 160 μM sodium oleate for 24, 48, and 72 h, and the cell lysates were harvested and immunoblotted with an HA-specific antibody. (B) The effects of NRF1 overexpression on lipid droplet accumulation during ICP1 cell differentiation. Oil Red O staining of the ICP1 cells transfected with pCMV-HA or pCMV-HA-NRF1 was performed at 72 h of differentiation (left two panels) and its quantification at 24, 48, and 72 h (right panel). Statistical significance was determined by Student’s t-test comparing empty pCMV-HA vector versus pCMV-HA-NRF1 transfection. ∗p < 0.05, ∗∗p < 0.01. (C) Real-time RT-PCR analysis of expression levels of PPARγ1 and adipogenic differentiation markers (FABP4, G0S2, GPDH, and AdipoQ) during the differentiation of ICP1 cells transfected with either empty pCMV-HA vector or pCMV-HA-NRF1 at the indicated times points. All data represent the mean ± SE. Statistical significance was determined by Student’s t-test comparing empty pCMV-HA vector versus pCMV-HA-NRF1 transfection. ∗p < 0.05, ∗∗p < 0.01.
Oil red O staining and its quantification showed that compared with the empty vector-transfected cells, NRF1 overexpression significantly decreased lipid droplet accumulation at 24, 48, and 72 h of differentiation (p < 0.05) (Figure 5B). Consistent with the Oil red O staining results, the expression levels of adipogenic markers FABP4, G0S2, GPDH, and AdipoQ were significantly reduced during differentiation when NRF1 was overexpressed (p < 0.05) (Figure 5C). These results suggest that NRF1 inhibits chicken preadipocyte differentiation. In addition, gene expression analysis showed that compared with empty vector-transfected cells, NRF1 overexpression decreased the PPARγ1 mRNA level at 24, 48, and 72 h of differentiation (p < 0.05), which is consistent with our finding that NRF1 negatively regulates the P1 promoter (Figure 2D).
Discussion
In this study, we demonstrated for the first time that NRF1 directly negatively regulates the P1 promoter of the PPARγ gene and inhibits chicken adipogenesis. A luciferase reporter assay demonstrated that NRF1 inhibits P1 promoter activity (Figure 2D). The site-directed mutagenesis analysis revealed that a mutation of any one of the three putative NRF1 binding sites (N2, N5, and N6) abolished NRF1-mediated inhibition of P1 promoter activity (Figure 2D). The finding that NRF1 directly bound to the P1 promoter (Figure 3) suggests that NRF1 directly regulates transcription of the P1 promoter. Negative regulation of the P1 promoter by NRF1 was supported by our finding that PPARγ1 and NRF1 mRNA expression levels were negatively correlated in the fat and lean chicken lines of NEAUHLF (Figure 4A) and that NRF1 overexpression significantly decreased PPARγ1 mRNA expression during ICP1 cell differentiation (Figure 5C). Therefore, our findings indicate that NRF1 can specifically bind to the P1 promoter, resulting in decreased expression of cPPARγ1.
Nuclear respiratory factor 1 is an important transcription factor that regulates mitochondrial biogenesis, functioning as an activator of nuclear-encoded mitochondrial genes such as cytochrome, mitochondrial transcription factor A (TFAM) and transcription factor B proteins (TFBs) (Scarpulla, 2012; Qi and Ding, 2012; Seo et al., 2015). Recent studies demonstrated that in addition to its roles in mitochondria, NRF1 also targets several genes such as the integrin-associated protein gene (IAP), insulin-degrading enzyme gene (IDE), and proton-coupled folate transporter gene (PCFT) and is involved in the control of cell growth (Cam et al., 2004), neurite outgrowth (Chang and Huang, 2004), diabetes mellitus (DM), Alzheimer’s disease (AD) (Zhang et al., 2012), and folate transport (Gonen and Assaraf, 2010). Our present study demonstrated that NRF1 inhibits adipogenesis, providing new evidence that NRF1 can perform multiple functions.
Mitochondria play an essential role in the differentiation and maturation of adipocytes (Wilsonfritch et al., 2003; Koh et al., 2007; De Pauw et al., 2009; Kusminski and Scherer, 2012; Boudina and Graham, 2015; Liu et al., 2015). NRF1 plays important roles in regulating mitochondrial biogenesis and function (Koh et al., 2007; De Pauw et al., 2009; Kusminski and Scherer, 2012). PPARγ is a master regulator of adipogenesis, and both PPARγ1 and PPARγ2 are capable of inducing adipogenesis (Wilsonfritch et al., 2004). Like NRF1, PPARγ1 also regulates mitochondrial gene expression, biogenesis and function (Boudina and Graham, 2015; Li et al., 2016). In the present study, our results demonstrated that NRF1 directly negatively regulates the P1 promoter of the PPARγ gene and inhibits chicken adipogenesis (Figures 2–5). Taking into consideration all these data, we presume that NRF1 may regulate chicken adipogenesis via directly controlling the transcription of PPARγ1 and indirectly controlling mitochondrial biogenesis and function.
In the present study, we demonstrated that NRF1 negatively regulates the P1 promoter. Considering that a transcription factor generally has multiple target genes, we cannot exclude the possibility that NRF1 inhibits chicken preadipocyte differentiation in part by directly and/or indirectly regulating the expression of other adipogenesis regulators, including the remaining PPARγ transcript isoforms. For a better understanding of the molecular mechanisms of NRF1 in adipogenesis, it is worth performing ChIP-seq to identify other potential targets of NRF1 that may contribute to the inhibition of chicken preadipocyte differentiation by NRF1.
In the present study, our results showed that NRF1 overexpression inhibited ICP1 cell differentiation, as demonstrated by Oil red O staining and mRNA expression of adipogenic genes (FABP4, G0S2, GDPH, and AdipoQ). Similar to our results, Tienen et al. also found that NRF1 overexpression led to less lipid accumulation in differentiated 3T3-L1 cells; however, in contrast to our results, they found that neither increased expression of adipogenesis inhibitors, such as Delta-like kinase (DLK) and GATA binding protein 2 (GATA2), nor decreased expression of key adipogenic markers, such as PPARγ and C/EBPα (Tienen et al., 2010), was observed when NRF1 was overexpressed during the differentiation of 3T3-L1 preadipocytes, suggesting that NRF1 overexpression seems to have no effect on 3T3-L1 preadipocyte differentiation. This discrepancy may be due to species specificity and different experimental conditions (e.g., different adipogenesis inducers and different levels of NRF1 overexpression).
Excessive abdominal fat deposition within the carcass is a challenge for the broiler chicken industry (Resnyk et al., 2013, 2017), decreasing feed efficiency and reducing yield and the nutritional and commercial value of carcass parts (Resnyk et al., 2013, 2017). Our findings provide new insight into the mechanisms underlying chicken adipogenesis and excessive fat deposition. This improved understanding may be exploited in the future to control excessive fat deposition in chickens.
In conclusion, we characterized chicken PPARγ P1 promoter and demonstrated that NRF1 negatively regulates the PPARγ P1 promoter and inhibits chicken adipogenesis.
Author Contributions
TC performed the experiments, analyzed the data, and wrote the manuscript. TX, JH, FM, YJ, XY, YC, and HL critically revised the manuscript. NW designed the experiments, provided funding support, and critically revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the members of the poultry breeding group at Northeast Agricultural University for helping with managing the birds and collecting the data.
Abbreviations
- AdipoQ
adiponectin
- FABP4
fatty acid-binding protein 4
- Fluc
firefly luciferase
- G0S2
G0/G1 switch 2
- GPDH
glycerol-3-phosphate dehydrogenase 2
- NRF1
nuclear respiratory factor 1
- PPARγ
peroxisome proliferator-activated receptor-γ
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (Nos. 31572392 and 31372299) and the China Agriculture Research System (No. CARS-41).
References
- Al-Shali K., Cao H., Knoers N., Hermus A. R., Tack C. J., Hegele R. A. (2004). A single-base mutation in the peroxisome proliferator-activated receptor gamma4 promoter associated with altered in vitro expression and partial lipodystrophy. J. Clin. Endocrinol. Metab. 89 5655–5660. 10.1210/jc.2004-0280 [DOI] [PubMed] [Google Scholar]
- Armoni M., Harel C., Karni S., Chen H., Baryoseph F., Ver M. R., et al. (2006). FOXO1 represses peroxisome proliferator-activated receptor-γ1 and -γ2 gene promoters in primary adipocytes. J. Biol. Chem. 281 19881–19891. [DOI] [PubMed] [Google Scholar]
- Boudina S., Graham T. E. (2017). Mitochondrial function/dysfunction in white adipose tissue. Exp. Physiol. 99 1168–1117. 10.1074/jbc.M600320200 [DOI] [PubMed] [Google Scholar]
- Cam H., Balciunaite E., Blais A., Spektor A., Scarpulla R. C., Young R., et al. (2004). A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell 16 399–411. 10.1113/expphysiol.2014.081414 [DOI] [PubMed] [Google Scholar]
- Chandra M., Miriyala S., Panchatcharam M. (2017). PPARγ and its role in cardiovascular diseases. PPAR Res. 2017:6404638. 10.1155/2017/6404638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W. T., Huang A. M. (2004). Alpha-Pal/NRF-1 regulates the promoter of the human integrin-associated protein/CD47 gene. J. Biol. Chem. 279 14542–14550. 10.1016/j.molcel.2004.09.037 [DOI] [PubMed] [Google Scholar]
- Cho Y. M., Shin H. D., Park B. L., Kim J. H., Park K. S., Kim S. Y., et al. (2005). Association between polymorphisms in the nuclear respiratory factor 1 gene and type 2 diabetes mellitus in the Korean population. Diabetologia 48 2033–2038. 10.1074/jbc.M309825200 [DOI] [PubMed] [Google Scholar]
- de la Rosa Rodriguez M. A., Kersten S. (2017). Regulation of lipid droplet-associated proteins by peroxisome proliferator-activated receptors. Biochim. Biophys. Acta 1862 1212–1220. 10.1007/s00125-005-1855-7 [DOI] [PubMed] [Google Scholar]
- De Pauw A., Tejerina S., Raes M., Keijer J., Arnould T. (2009). Mitochondrial (dys)function in adipocyte (de)differentiation and systemic metabolic alterations. Am. J. Pathol. 175 927–939. 10.2353/ajpath.2009.081155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Zhang W., O-Sullivan I., Williams J. B., Dong Q., Park E. A., et al. (2012). FoxO1 inhibits sterol regulatory element-binding protein-1c (SREBP-1c) gene expression via transcription factors Sp1 and SREBP-1c. J. Biol. Chem. 287 20132–20143. 10.1074/jbc.M112.347211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K., Sun Y., Zhang X., Zhang T., Zhang W., Zhang J., et al. (2017). Identification and characterization of transcript variants of chicken peroxisome proliferator-activated receptor gamma. Poult. Sci. 94 2516–2527. 10.3382/ps/pev229 [DOI] [PubMed] [Google Scholar]
- Funato N., Ohyama K., Kuroda T., Nakamura M. (2003). Basic helix-loop-helix transcription factor epicardin/capsulin/Pod-1 suppresses differentiation by negative regulation of transcription. J. Biol. Chem. 278 7486–7493. 10.3382/ps/pev229 [DOI] [PubMed] [Google Scholar]
- Garinshkolnik T., Rudich A., Hotamisligil G. S., Rubinstein M. (2014). FABP4 attenuates PPARγ and adipogenesis and is inversely correlated with PPARγ in adipose tissues. Diabetes Metab. Res. Rev. 63 900–911. 10.2337/db13-0436 [DOI] [PubMed] [Google Scholar]
- Gonen N., Assaraf Y. G. (2010). The obligatory intestinal folate transporter PCFT (SLC46A1) is regulated by nuclear respiratory factor 1. J. Biol. Chem. 285:33602–33613. 10.1074/jbc.M110.135640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. R., Lee G. Y., Yu H., Maeng H. J., Oh T. J., Kim K. M., et al. (2017). Suppression of Nrf2 attenuates adipogenesis and decreases FGF21 expression through PPAR gamma in 3T3-L1 cells. Biochem. Biophys. Res. Commun. 497 1149–1153. 10.1016/j.bbrc.2017.01.107 [DOI] [PubMed] [Google Scholar]
- Koh E. H., Park J. Y., Park H. S., Jeon M. J., Ryu J. W., Kim M., et al. (2007). Essential role of mitochondrial function in adiponectin synthesis in adipocytes. Diabetes Metab. Res. Rev. 56 2973–2981. 10.1016/j.bbrc.2017.01.107 [DOI] [PubMed] [Google Scholar]
- Kusminski C. M., Scherer P. E. (2012). Mitochondrial dysfunction in white adipose tissue. Trends Endocrinol. Metab. 23 435–443. 10.2337/db07-0510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. E., Ge K. (2014). Transcriptional and epigenetic regulation of PPARγ expression during adipogenesis. Cell Biosci. 4:29. 10.1016/j.tem.2012.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Zhang F., Zhang X., Xue C., Namwanje M., Fan L., et al. (2016). Distinct functions of PPAR (isoforms in regulating adipocyte plasticity. Biochem. Biophys. Res. Commun. 481 132–138. 10.1016/j.bbrc.2016.10.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Ji H., Li C., Chen L. Q., Du Z. Y. (2017). Morphology, mitochondrial development and adipogenic-related genes expression during adipocytes differentiation in grass carp (Ctenopharyngodon idellus). Sci. Bull. 60 1241–1251. 10.1016/j.bbrc.2016.10.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Niu N., Zhu X., Du T., Wang X., Chen D., et al. (2008). Genetic variation and association analyses of the nuclear respiratory factor 1 (nRF1) gene in Chinese patients with type 2 diabetes. Diabetes Metab. Res. Rev. 57 777–782. 10.1007/s11434-015-0833-9 [DOI] [PubMed] [Google Scholar]
- Ouyang X., Zhang R., Yang J., Li Q., Qin L., Zhu C., et al. (2011). Transcription factor IRF8 directs a silencing programme for TH17 cell differentiation. Nat. Commun. 2:314. 10.1038/ncomms1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. K., Wang L., Giampietro A., Lai B., Lee J. E., Ge K. (2017). Distinct roles of transcription factors KLF4, KROX20, and peroxisome proliferator-activated receptor γ in adipogenesis. Mol. Cell. Biol. 37 e554–e516. 10.1128/MCB.00554-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi J., Leung L., Xue P., Wang W., Hou Y., Liu D., et al. (2010). Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. J. Biol. Chem. 285 9292–9300. 10.1074/jbc.M109.093955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Ding S. (2012). Transcriptional regulation by nuclear corepressors and PGC-1α: implications for mitochondrial quality control and insulin sensitivity. PPAR Res. 2012:348245. 10.1155/2012/348245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnyk C. W., Carré W., Wang X., Porter T. E., Simon J., Bihanduval E. L., et al. (2013). Transcriptional analysis of abdominal fat in genetically fat and lean chickens reveals adipokines, lipogenic genes and a link between hemostasis and leanness. BMC Genomics 14:557. 10.1186/1471-2164-14-557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnyk C. W., Carré W., Wang X., Porter T. E., Simon J., Bihan-Duval E. L., et al. (2017). Transcriptional analysis of abdominal fat in chickens divergently selected on bodyweight at two ages reveals novel mechanisms controlling adiposity: validating visceral adipose tissue as a dynamic endocrine and metabolic organ. BMC Genomics 18:626. 10.1186/s12864-017-4035-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen E. D., Hsu C. H., Wang X., Sakai S., Freeman M. W., Gonzalez F. J., et al. (2002). C/EBPα induces adipogenesis through PPARγ: a unified pathway. Genes Dev. 16 22–26. 10.1101/gad.948702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D., Farabaugh K. T., Wu J., Charrier A., Smas C., Hatzoglou M., et al. (2017). Coordinated transcriptional control of adipocyte triglyceride lipase (Atgl) by transcription factors Sp1 and PPARγ during adipocyte differentiation. J. Biol. Chem. 292 14827–14835. 10.1074/jbc.M117.783043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R. C. (2002). Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim. Biophys. Acta. 1576 1–14. 10.1016/S0167-4781(02)00343-3 [DOI] [PubMed] [Google Scholar]
- Scarpulla R. C. (2012). Nucleus-encoded regulators of mitochondrial function: integration of respiratory chain expression, nutrient sensing and metabolic stress. Biochim. Biophys. Acta 1819 1088–1097. 10.1016/j.bbagrm.2011.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S., Lee M. S., Chang E., Shin Y., Oh S., Kim I. H., et al. (2017). Rutin increases muscle mitochondrial biogenesis with AMPK activation in high-fat diet-induced obese rats. Nutrients 7 8152–8169. 10.3390/nu7095385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. N., Gao Y., Qiao S. P., Wang S. Z., Duan K., Wang Y. X., et al. (2014). Epigenetic DNA methylation in the promoters of peroxisome proliferator-activated receptor in chicken lines divergently selected for fatness. J. Anim. Sci. 92 48–53. 10.2527/jas.2013-6962 [DOI] [PubMed] [Google Scholar]
- Tatler A. L., Habgood A., Porte J., John A. E., Stavrou A., Hodge E., et al. (2016). Reduced ets domain-containing protein elk1 promotes pulmonary fibrosis via increased integrin αvβ6 expression. J. Biol. Chem. 18 9540–9553. 10.1074/jbc.M115.692368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tienen F. H. V., Lindsey P. J., Kallen C. J. V. D., Smeets H. J. (2010). Prolonged Nrf1 overexpression triggers adipocyte inflammation and insulin resistance. J. Cell. Biochem. 111 1575–1585. 10.1002/jcb.22889 [DOI] [PubMed] [Google Scholar]
- Wang W., Zhang T., Wu C., Wang S., Wang Y., Li H., et al. (2017). Immortalization of chicken preadipocytes by retroviral transduction of chicken TERT and TR. PLoS One 12:e0177348. 10.1371/journal.pone.0177348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsonfritch L., Burkart A., Bell G., Mendelson K., Leszyk J., Nicoloro S., et al. (2003). Mitochondrial biogenesis and remodeling during Adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol. Cell. Biol. 23 1085–1094. 10.1128/MCB.23.3.1085-1094.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsonfritch L., Nicoloro S., Chouinard M., Lazar M. A., Chui P. C., Leszyk J., et al. (2004). Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J. Clin. Investig. 114 1281–1289. 10.1172/JCI21752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Cheng B. H., Yang L. L., Wang Z. P., Zhang H. L., Xu S. S., et al. (2017). Identification of a potential functional single nucleotide polymorphism for fatness and growth traits in the 3′-untranslated region of the PCSK1 gene in chickens. J. Anim. Sci. 95 4776–4786. 10.2527/jas2017.1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ding Q., Wang Z. (2012). Nuclear respiratory factor 1 mediates the transcription initiation of insulin-degrading enzyme in a TATA box-binding protein-independent manner. PLoS One 7:e42035. 10.1371/journal.pone.0042035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Qi C., Korenberg J. R., Chen X. N., Noya D., Rao M. S., et al. (1995). Structural organization of mouse peroxisome proliferator-activated receptor gamma (mPPAR gamma) gene: alternative promoter use and different splicing yield two mPPAR gamma isoforms. Proc. Natl. Acad. Sci. U.S.A. 92 7921–7925. 10.1073/pnas.92.17.7921 [DOI] [PMC free article] [PubMed] [Google Scholar]


