We demonstrate that Plasmodium vivax parasites in Cambodia are fully susceptible to chloroquine and that relapse of infection due to dormant parasites, not antimalarial drug resistance, constitutes the greatest challenge to P. vivax malaria elimination.
Keywords: Malaria, Plasmodium vivax, chloroquine, drug resistance, relapse, recrudescence, reinfection, next generation sequencing, Cambodia
Abstract
Background
Plasmodium vivax resistance to chloroquine (CQ) has been reported worldwide, although the World Health Organization clinical drug efficacy studies protocol does not permit classification of patient outcomes.
Methods
We enrolled 40 patients with P. vivax malaria in northeastern Cambodia, where >17% treatment failures were previously reported. Patients were treated with CQ (30 mg/kg) and followed for 2 months, with frequent clinical examination and capillary blood sample collection for microscopy, molecular parasite detection and genotyping, and drug concentration measurements. Reinfections were prevented by relocating patients to a transmission-free area.
Results
P. vivax parasites were eliminated in all patients by day 3. Genomic analyses revealed that all clones in polyclonal infections were cleared at the same rate, indicating their equal susceptibility to CQ. CQ blood concentrations were below the therapeutic level in all recurrent infections (24 of 40 patients), which were efficiently cleared by a second course of CQ treatment. Genotyping (128 SNPs barcode) and sequences of entire parasite genome (Whole-Genome Sequencing, Illumina) indicated that two thirds (6 of 8) of the recurrent parasites resulted from heterologous relapses whose 50% are from by sibling/recombinant clones.
Conclusions
No evidence of CQ resistance was observed. Our data suggest that P. vivax antimalarial drug resistance is likely overestimated and that the current guidelines for clinical drug studies of P. vivax malaria need to be revised.
Plasmodium vivax is the most widespread human malaria parasite, with approximately 2.5 billion people at risk of infection and >15 million clinical cases annually [1, 2]. Although traditionally considered as causing benign malaria, P. vivax is responsible for significant morbidity and, sometimes, severe cases of clinical malaria caused by anemia [3–8]. Chloroquine (CQ) has been used for decades as first-line treatment in uncomplicated P. vivax malaria. CQ treatment failures in P. vivax malaria were first detected in Papua, Indonesia, in 1989 [9, 10], >30 years after the first reports of CQ resistance in Plasmodium falciparum malaria and have since been described in many other areas of endemicity, including Cambodia [11]. In Cambodia, clinical drug efficacy studies reported high rates of CQ treatment failure (at day 28 follow-up) in 2009 [12] and, artemisinin combination therapy (ACT) is now recommended as first-line treatment for uncomplicated P. vivax malaria.
Currently, rigorous clinical assessment of drug efficacy in P. vivax malaria is complicated by our inability to reliably classify patient outcomes and, specifically, to distinguish recrudescence (ie, P. vivax malaria due to real treatment failure) from reinfection (ie, due to new infectious mosquito bites) or relapse (ie, due to reactivation of dormant parasites in the liver) in patients experiencing recurrence [11]. In particular, we lack host biomarkers or parasite signatures to confirm the reason for parasite recurrence (recrudescence, relapse, or reinfection). In addition, it is complicated to assess P. vivax in vitro susceptibility because (1) long-term cultures of P. vivax isolates are not routinely feasible and (2) ex vivo drug susceptibility assays are difficult to implement and are difficult to interpret because P. vivax infections are typically asynchronous and the parasite’s susceptibility depends on its stage [13, 14]. In the current 2009 World Health Organization guidelines [2], P. vivax CQ resistance is defined by the presence of parasites at a blood concentration of >100 ng/mL, regardless of whether recurrent parasitemia represents recrudescence, relapse, or reinfection [3, 15]. However, the CQ blood concentration is rarely determined in antimalarial drug efficacy studies.
Here, we describe a clinical drug efficacy study designed to control, as best as possible, for reinfection and relapse and to rigorously assess CQ clinical efficacy. We enrolled 40 patients with P. vivax malaria in northeastern Cambodia, where high treatment failure rates were recorded in 2009 [12], and relocated half to a malaria-transmission-free area for 2 months, to exclude reinfection as a cause of recurrence. We did not treat enrolled patients with primaquine (an 8-aminoquinoline) regimen (high or low daily doses over 14 days or single weekly doses for 8 weeks), the only widely available hypnozoitocidal antimalarial drug, since this drug is not currently recommended by the National Malaria Control Program in Cambodia in the absence of available G6PD testing. We conducted frequent molecular tests for detection of P. vivax after treatment and during follow-up, to provide early and accurate detection of recurrences. We also measured the CQ concentration in whole-blood specimens collected on day 7 and the day of recurrence (if any). We complemented these clinical data with extensive genetic characterization of the parasites, using whole-genome sequencing and high-throughput genotyping, to (1) determine, in polyclonal infections, whether all clones were cleared at the same rate and (2) identify whether the recurring parasites were genetically identical to those present in the same patient at enrollment. Overall, our study describes an analytical framework to rigorously evaluate antimalarial drug efficacy in P. vivax malaria. In contrast to previous studies [12], we detected no evidence of CQ resistance in northeast Cambodia.
METHODS
Study Site
We conducted a prospective, open-label drug efficacy study in Ratanakiri Province, an area of low-intensity malaria transmission in northeast Cambodia, in 2014. Local transmission intensity and epidemiological characteristics has been described previously [16–19]. Patients from villages located within 50 km of BanLung were enrolled. The fieldwork was supported by a mobile laboratory installed in BanLung for the duration of the project [20].
Patients
Patients with a current fever or a history of fever in the last 48 hours and who were seeking antimalarial treatment after diagnosis of non–P. falciparum infection by a malaria rapid diagnostic test (CareStart Malaria HRP2/pLDH Pf/PAN Combo, Access Bio) were offered the opportunity to participate in the study. Pregnant or lactating woman and patients with signs of severe malaria, known other illness, inability to provide informed written consent, or aged <15 years old were excluded. Only patients with real-time polymerase chain reaction (PCR)–confirmed P. vivax monoinfection were eligible for the study. The study was approved by the National Ethic Committee at the National Institute of Public Health (Phnom Penh, Cambodia) and registered at ClinicalTrials.gov (identifier NCT02118090).
Study Design and Procedures
Participants were offered relocation to a malaria-free area for the entire 2-month study duration. Participants were treated with a 3-day supervised regimen of CQ (30 mg/kg Nivaquine; Sanofi, Gentilly, France). At enrollment, medical history was recorded and physical, clinical, and biological examinations were performed. A 4-mL blood specimen was collected from all patients at day 0 and in case of recurrence. Follow-up consisted of clinical examinations, axillary temperature measurements, and capillary blood specimen collection (volume, 500 µL). Nonrelocated patients were followed up on days 1–3, 5, and 7 and then weekly until day 60. Relocated patients were followed every 8 hours until 2 consecutive negative results of real-time PCR for detection of P. vivax and then every 48 hours until day 60. If any patient presented malaria symptoms during the monitoring period, a full regimen of CQ was readministered after confirmation of P. vivax monoinfection by real-time PCR, and the patient was followed for a minimum of 14 days. On completion of the study, all patients testing positive for P. vivax infection (symptomatic and asymptomatic) were treated with a standard 3-day regimen of dihydroartemisinin-piperaquine (Duo-Cotecxin; Zhejiang Holley Nanhu Pharmaceutical, Jiaxing City, China).
Microscopy
The parasitemia level was determined on Giemsa-stained thick films as the number of parasites per 200 white blood cells (assuming a white blood cell count of 8000 cells/µL). For relocated patients, regression models of the log-transformed parasite counts were fitted to estimate the parasite clearance rate, using the online Parasite Clearance Estimator Tool [21].
DNA Extraction and Real-Time PCR
Parasite DNA was extracted from a 50-µL whole-blood specimen, using the QIAamp DNA Blood Mini Kit (Qiagen, Courtaboeuf, France). Molecular detection and identification of Plasmodium parasites were performed by real-time PCR, using primers targeting the gene encoding cytochrome b [20]. The limit of the detection is estimated to be 1 parasite/µL of blood [20, 22].
Drug Concentration Measurement in Blood
Drug concentrations (CQ and N-desethyl chloroquine [DCQ], an active metabolite of CQ) were determined on day 7 by liquid chromatography–tandem mass spectrometry [23]. Fifty-microliter blood specimens were stored at −80°C within 2 hours after collection at all follow-up time points, and complete pharmacokinetic profiles were generated for patients with recurrences.
Genotyping and Whole-Genome Sequencing
Parasites from all blood samples were genotyped at 128 single-nucleotide polymorphisms (SNPs), using a custom sequencing-based assay [24], and allele frequency was determined for each variable position covered by >100 reads.
After leukocyte depletion was performed using cellulose columns [25], DNA from samples with high parasitemia levels were used to sequence the parasites’ entire genomes [26], and the relative proportion of each allele for all nucleotide positions covered by at least 50 reads in a given sample were analyzed (Supplementary Materials). All sequence data are freely available in the National Center for Biotechnology Information Sequence Read Archive (BioProject SUB3268581).
Statistical Analysis
Data were analyzed with MedCalc, version 12 (Mariakerke, Belgium). Mann-Whitney or Kruskal-Wallis tests were conducted for nonparametric comparisons. For proportions (expressed with percentages and 95% confidence intervals [CIs]), we used χ2 or Fisher exact tests. Statistical analyses for genomic investigations are detailed in Appendix 1. P values of <.05 were considered statistically significant.
RESULTS
Forty patients (20 relocated and 20 nonrelocated) completed the 2-month follow-up period (Table 1). A flow chart of the clinical study is presented in the Supplementary Figure 1.
Table 1.
Baseline Characteristics of Plasmodium vivax–Infected Patients Enrolled in the Clinical Drug Efficacy Study, Ratanakiri, Cambodia, 2014
| Characteristic | Relocated Patients (n = 20) | Nonrelocated Patients (n = 20) |
|---|---|---|
| Sex, patients, no. | ||
| Male | 11 | 16 |
| Female | 9 | 4 |
| Age, y | 27.5 (15–52) | 27.1 (16–44) |
| Weight, kg | 52.6 (42–64) | 54.8 (40–70) |
| Initial asexual parasitemia level, parasites/μLa,c | 11090 (400–30000) | 5559 (100–19000) |
| Gametocytemia density, gametocytes/µLb,c | 1354 (0–5500) | 244 (0–1600) |
| Gametocytes detected, patients, no. (%) | 17 (85) | 16 (80) |
Data are mean (range), unless otherwise indicated. The mean initial proportion of ring-stage parasites among all 40 patients was 51.5% (range, 0%–100%), and >95% of parasites in 8 patients (20%) were ring stage.
a P< .05. In both groups, the majority of patients (80% and 85%, respectively) had gametocytes detected by microscopy at inclusion.
b P < .05.
cThe initial asexual parasitemia levels were correlated with gametocyte densities (r = 0.603; P < .0001).
All P. vivax Parasites Were Cleared Rapidly After Standard CQ Treatment
P. vivax parasites from all patients were cleared by day 3 (microscopy) or day 6 (real-time PCR; Table 2 and Supplementary Table 1). Parasite clearance half-lives ranged from 2 to 7 hours, except for 1 patient (BL17) with a slower parasite clearance (11 hours; Supplementary Figure 2). No significant associations were observed between parasite clearance half-lives and patient age (P = .85) or initial parasitemia level (P = .41; Table 1). Despite P. vivax infections being typically asynchronous and some P. vivax stages more susceptible than others to CQ (eg, ring stages are more susceptible than trophozoite stages [13]), we did not observe significant associations between the proportion of parasite stages at enrollment and (1) the presence of detectable parasites 24 or 48 hours after CQ treatment (P = .14 and P = .44, respectively) or (2) parasite clearance half-lives (P = .78).
Table 2.
Proportion of Patients with Plasmodium vivax Parasites Detected by Microscopy or Real-Time Polymerase Chain Reaction (PCR) in the First 5 Days Following a 3-Day Supervised Regimen of Chloroquine (30 mg/kg), Ratanakiri, Cambodia, 2014
| Day of Positivity, Test | Relocated Patients, % (No.) (n = 20) | Nonrelocated Patients, % (No.) (n = 20) | P a |
|---|---|---|---|
| Day 1 | |||
| Microscopy | 85 (17) | 85 (17) | 1.0 |
| Real-time PCR | 100 (20) | 100 (20) | 1.0 |
| Day 2 | |||
| Microscopy | 10 (2) | 25 (5) | .41 |
| Real-time PCR | 80 (16) | 95 (19) | .34 |
| Day 3 | |||
| Microscopy | 0 | 0 | 1.0 |
| Real-time PCR | 70 (14) | 55 (11) | .51 |
| Day 5 | |||
| Microscopy | 0 | 0 | 1.0 |
| Real-time PCR | 20 (4) | 35 (7) | .48 |
aBy the Fisher exact test.
All Clones Were Cleared at Similar Rates by CQ in Polyclonal Infections
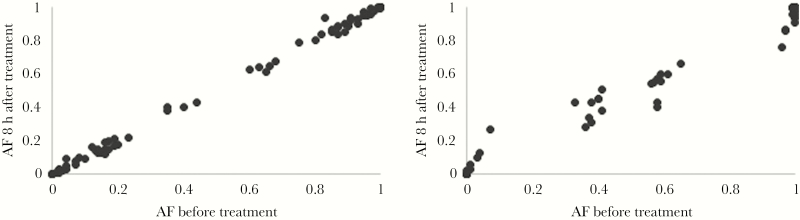
In polyclonal infections, if some clones are less susceptible to CQ than others, these clones should increase in proportion during the CQ treatment course [27]. Such analyses are particularly appealing since they control for interindividual differences that could affect parasite clearance (eg, drug catabolism or acquired immunity). We genotyped, for all relocated patients, P. vivax parasites present before treatment (ie, during the initial infection) and every 8 hours after treatment. The genotyping success decreased after treatment and was correlated with the parasite density (Supplemental Figure 3), indicating that DNA from dead parasites did not persist long in blood. Half of the initial infections for relocated patients (10 of 20) were polyclonal and further analyzed. Allele frequencies at variable positions were highly correlated between consecutive time points (eg, R2 > 0.98 for all individuals between 0 and 8 hours after treatment), indicating similar susceptibility of all clones to CQ (Figure 1 and Supplementary Table 2). Of the 10 polyclonal infections successfully genotyped, 9 did not have any SNP for which the allele frequency significantly changed in the 8 hours following CQ treatment. Only 1 infection (in patient BL12) showed variations in allele frequency greater than expected on the basis of stochastic noise (at 3 SNPs), but the change in allele frequency was <20%, suggesting that the relative proportion of the clones in these infections was only moderately affected by CQ treatment. When later time points were considered, the pattern remained identical, although the variability increased because of the lower parasitemia level (Supplementary Table 2).
Figure 1.
Relative proportion of clones in polyclonal infections upon a 3-day supervised regimen of chloroquine (30 mg/kg), Ratanakiri, Cambodia, 2014. Allele frequencies (AFs) for 2 patients with complex infections (patient BL12 [A] and patient BL13 [B]) at each single-nucleotide polymorphism (black dots) before (x-axis) and 8 hours after (y-axis) treatment. See also Supplementary Table 2.
P. vivax Recurrences Were Frequent After CQ Treatment
In both groups (ie, relocated and nonrelocated patients), P. falciparum was not detected by microscopy or real-time PCR in any recurrent infections (24 of 40). The proportion of patients with recurrence, the clinical presentation (ie, symptomatic vs asymptomatic), and the time to recurrence were similar between relocated and nonrelocated patients, suggesting that reinfections contributed little to the recurrences in nonrelocated patients (Figure 2 and Table 3), consistent with the low malaria transmission profile in this region [19]. Four patients had submicroscopic P. vivax parasites only detected by real-time PCR. Among the 20 microscopy-positive recurrences (involving 11 relocated and 9 nonrelocated patients), 10 patients (5 in each group) developed malaria symptoms and were retreated with CQ. The time to recurrence varied greatly among patients from day 25 to day 61 according to real-time PCR findings and from day 33 to day 59 according to microscopy findings (Table 3). Neither the parasitemia level at enrollment nor the parasite clearance half-lives of the initial infection were associated with recurrence (P = .25 and P = .62, respectively) or the time to recurrence (P = .07 and P = .19, respectively). Patient BL17, who displayed a slightly delayed parasite clearance, remained free of parasites until day 50.
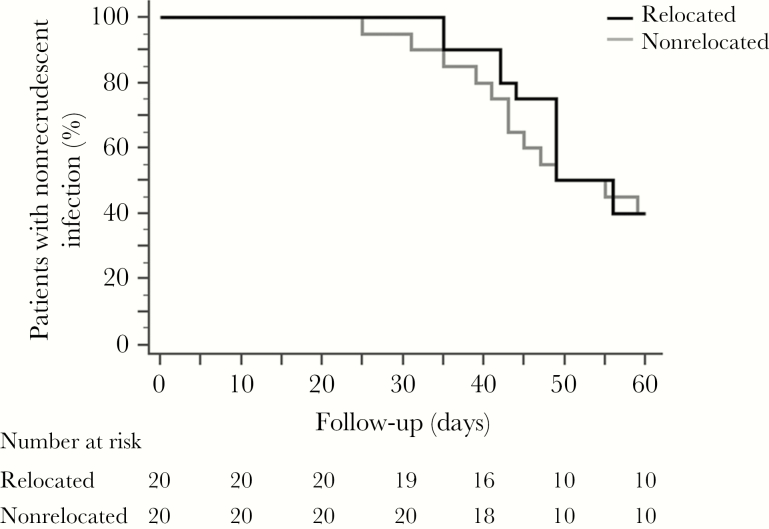
Figure 2.
Cumulative proportion of patients with nonrecrudescent infection detected by real-time polymerase chain reaction, Ratanakiri, Cambodia, 2014. Patients were or were not relocated to a malaria-free area and were treated with a 3-day supervised regimen of chloroquine (30 mg/kg). The ratio of the hazards for recrudescence for nonrelocated versus relocated patients was 0.86 (95% confidence interval [CI], .38–1.92; P = .7 by the log-rank test); The median time to recurrence (in days) was similar between nonrelocated patients (49.0; 95% CI, 49.0–56.0) and relocated patients (49.0; 95% CI, 43.0–59.0).
Table 3.
Cumulative Number of Recurrences of Plasmodium vivax Infection Detected by Microscopy or Real-Time Polymerase Chain Reaction (PCR) Following a 3-Day Supervised Regimen of Chloroquine (30 mg/kg) Over the 2-Month Monitoring Period, Ratanakiri, Cambodia, 2014.
| Variable | Relocated Patients (n = 20) | Nonrelocated Patients (n = 20) |
|---|---|---|
| Day 28 positivity | ||
| By microscopy | 0 | 0 |
| By real-time PCR | 1 (5) | 1 (5) |
| Patients with recurrence | ||
| Symptomatic (real-time PCR+/ microscopy+) | 5 (25) | 5 (25) |
| Asymptomatic (real-time PCR+/ microscopy+) | 6 (30) | 4 (20) |
| Submicroscopic (real-time PCR+) | 1 (5) | 3 (15) |
| Day of recurrence detection | ||
| By real-time PCR | 42.8 (25–59) | 45.7 (28–56) |
| By microscopy | 47 (33–61) | 48.9 (42–56) |
Data are number (%) of patients or mean value (range).
Abbreviation: +, positive.
Recurrences Occurred When the CQ Blood Level Was Below the Therapeutic Blood Concentration
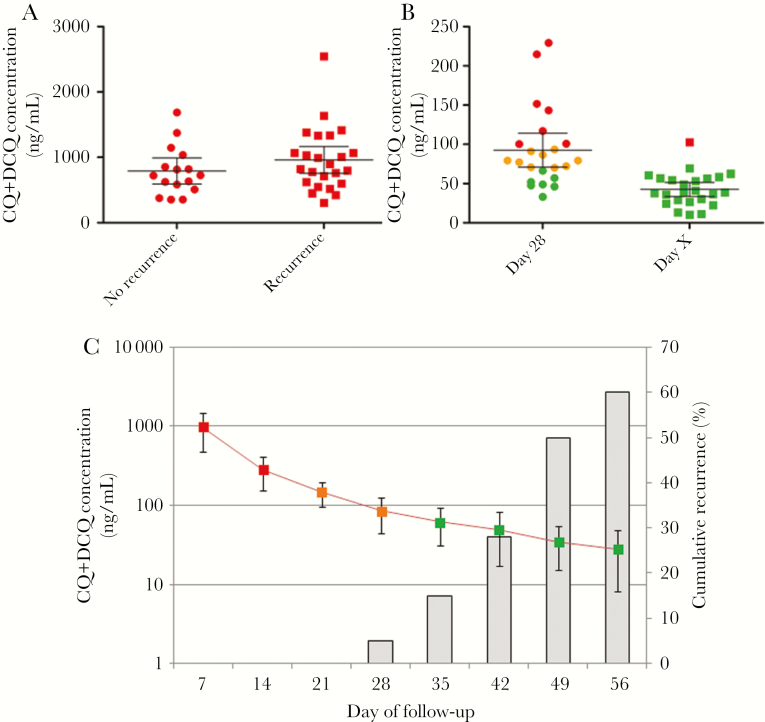
The blood CQ concentration at day 7 was not significantly different between patients with and those without recurrences (P = .27; Figure 3A). For patients with recurrences (Figure 3C), the mean drug concentration in blood on the day of recurrence (based on real-time PCR positivity) was 42.6 ng/mL (range, 10.5–102.7 ng/mL). All concentrations were below the threshold used to define CQ resistance (100 ng/mL) [2, 15], except in 1 patient, who had a slightly higher drug concentration (V23; 102.7 ng/mL on day 42). Two patients with early recurrences (BL08 and V16; on days 25 and 28, respectively) displayed concentrations of 56.0 and 57.0 ng/mL, respectively, at the time of the recurrences. Interestingly, while in clinical drug efficacy studies, resistance is defined by the presence of parasites in the presence of CQ blood concentrations of >100 ng/mL, the mean CQ+DCQ concentration in our study population at day 28 was 92.3 ng/mL (16 of 23 patients had a concentration of <100 ng/mL; Figure 3B).
Figure 3.
Whole-blood drug concentrations in Plasmodium vivax–infected patients receiving a 3-day supervised regimen of chloroquine (CQ; 30 mg/kg), Ratanakiri, Cambodia, 2014. Data are expressed as the sum of the concentrations of CQ and N-desethyl chloroquine (DCQ). A, Concentrations on day 7 for patients with and those without recurrences during the monitoring period. B, CQ blood concentrations on day 28 and the day of recurrence (detected by real-time polymerase chain reaction [PCR]) in patients experiencing recurrences during the 2-month monitoring period. C, Mean pharmacokinetic profile (±SD) over time (line) and cumulative proportion of parasite recurrences detected by real-time PCR (histograms). In panels A and B, CQ blood concentrations above the threshold for resistance (ie, 100 ng/mL) are in red, concentrations from 70 to 100 ng/mL are in orange, and concentrations <70 ng/mL are in green.
Recurrent Parasites Were Fully Susceptible to Retreatment With CQ
Ten patients developed symptomatic recurrence and were retreated with a second regimen of CQ (30 mg/kg). They all cleared parasites within 48 hours, similar to their initial episode or to observations for patients with no recurrences (Supplementary Table 3). The parasite clearance half-lives were similar to those following the initial treatment (P = .99), suggesting that the recurring parasites were equally susceptible to CQ. In 2 polyclonal recurrences, genotyping after the second treatment showed that all clones had similar clearance rates (Supplementary Table 2), confirming their equal susceptibility to CQ.
Many Recurring P. vivax Parasites Were Not Present in the Initial Infections
We genotyped parasites for 12 pairs of initial and recurrent infections and tested whether some recurring clones were absent from the initial infections. In 6 cases, we observed clear evidence of new alleles in the recurrence. Since only approximately 100 SNPs were investigated by genotyping, closely related clones might appear identical. We therefore also generated high-coverage genome sequence data (>100 reads over 20 million bases, or >80% of the P. vivax genome) for 8 pairs of initial and recurrent infections and confirmed the presence of numerous new alleles, indicating new clones, in each recurring infection (Table 4 and Supplementary Materials).
Table 4.
Genetic Differences Investigated by Genotyping and Whole-Genome Sequencing Between Parasites Collected From the Day of Initial Infection Identification (D0) and the Day of Recurrent Infection Identification (Dx), Ratanakiri, Cambodia, 2014
| Relocation Status, Patient ID | Dx | Genotyping | Whole-Genome Sequencing | Potential Origin of Recurrent Parasitesc | ||||
|---|---|---|---|---|---|---|---|---|
| Coverage, No. of Reads, D0 | Coverage, No. of Reads, Dx | New Allelesa | Coverage, No. of Reads, D0 | Coverage, No. of Reads, Dx | New Allelesb | |||
| Relocated | ||||||||
| BL02 | 41 | 2184 | 1575 | 0/73 | 299 | 122 | 5437/21277399 | Heterologous relapse (from related clones) |
| BL03 | 60 | 2921 | 1425 | 0/87 | 313 | … | … | Homologous relapse |
| BL07 | 59 | 2472 | 403 | 5/51 | 271 | 7 | … | Heterologous relapse |
| BL08 | 36 | 2713 | 1572 | 1/96 | … | 171 | … | Homologous relapse |
| BL10 | 56 | 1546 | 1956 | 22/86 | 205 | 177 | 35400/21105486 | Heterologous relapse |
| BL11 | 48 | 3438 | 1797 | 1/96 | 431 | 146 | 4442/20705930 | Heterologous relapse (from related clones) |
| BL12 | 47 | 2292 | 1334 | 18/101 | 153 | 377 | 19930/21279386 | Heterologous relapse |
| BL18 | 58 | 2155 | 1145 | 6/82 | 374 | 163 | 3614/21299372 | Heterologous relapse (from related clones) |
| Nonrelocated | ||||||||
| V04 | 53 | 1544 | 2645 | 14/102 | 131 | 182 | 20627/20313219 | Heterologous relapse (or reinfection) |
| V10 | 53 | 1958 | … | … | 357 | 181 | 37455/20167617 | Heterologous relapse (or reinfection) |
| V16 | 49 | 1330 | 2078 | 0/91 | 262 | … | … | Homologous relapse |
| V17 | 58 | 1650 | 1462 | 20/81 | 247 | 175 | 19743/21381922 | Heterologous relapse (or reinfection) |
| V27 | 62 | 1820 | 1208 | 0/64 | 217 | … | … | Homologous relapse |
Data are average sequence coverage (Cov) and number of new alleles detected in the recurrent infection obtained for each patient by genotyping and whole-genome sequencing, respectively.
Abbreviations: ID, identifier; SNP, single-nucleotide polymorphism.
aGenotyping of 128 SNPs was attempted; the number of SNPs successfully genotyped in both initial and recurrent infections is indicated as the denominator. The number of new alleles detected is further influenced by the polyclonality of the infections [26].
bThe number of nucleotides analyzed is indicated as the denominator. The number of new alleles detected is further influenced by the polyclonality of the infections [26].
cHeterologous relapses can be caused by relapse of clones unrelated to those present in the initial infections (leading to >15000 new alleles) or by sibling/recombinant clones (leading to only 3000–6000 new alleles detected by whole-genome sequencing and, typically, to only a handful of differences detected by genotyping).
Polymorphisms in Candidate Genes Suspected to Be Associated With CQ Resistance
To examine reported associations between P. vivax resistance to CQ and polymorphisms in candidate genes, we determined the proportion of these resistance-associated mutations in our set of CQ-susceptible parasites. Mutations at 3 loci, including those in the genes encoding multidrug resistance protein 1 (M908L, T958M, Y976F, and F1076L), multidrug resistance–associated protein 1 (T234M), and chloroquine resistance transporter (in an intron at positions 331151T→C, 332453T→C, and 332874A→C) were observed in >70% of isolates with CQ-susceptible parasites, indicating that these polymorphisms are not causally associated with CQ resistance (Supplementary Table 4).
DISCUSSION
Drug resistance, one of the greatest challenges of malaria elimination, has been reported in P. falciparum for most antimalarial drugs [28]. In P. vivax, clinical assessment of drug efficacy is complicated by the difficulty to disentangle recrudescence of resistant parasites from reinfections or relapses [11]. Using a combination of modern fieldwork and genomic analyses, we showed that standard CQ treatment was effective, resulting in full clearance of blood-stage parasites from and cure of patients with P. vivax malaria in northeastern Cambodia (in contrast to previous reports [12]). In polyclonal infections, all clones were removed at the same rate, indicating their equal susceptibility to CQ. However, we observed a high proportion of recurrence in the weeks following apparently successful treatment. Reinfections were prevented for half of patients who had been relocated to a transmission-free area. A similar proportion of recurrence in nonrelocated patients suggested that reinfections contributed little to recurrences, consistent with the low malaria transmission rate in Cambodia [19]. Importantly, all recurrences occurred once the CQ blood concentration was <100 ng/mL and were effectively cleared with a second CQ regimen. Sequences of entire parasite genome (Whole-Genome Sequencing, Illumina) indicated that two thirds (6/8) of the recurrent parasites resulted from heterologous relapses whose 50% are from by sibling/recombinant clones (Table 4).
Two reasons could explain the differences of CQ treatment failure rates observed between our study and a previous report [12], although the small number of patients enrolled in 2014 (8 of 46 in 2009 vs 0 of 40 in 2014; P = .02). First, less fit CQ-resistant parasites may have been eliminated from P. vivax populations between 2009 and 2014, as CQ was withdrawn in 2012 and replaced by dihydroartemisinin-piperaquine, as it has been observed in Malawi with the reemergence of CQ-susceptible P. falciparum malaria after cessation of CQ use [29, 30]. Alternatively, previous clinical assessments may have been confounded by relapses, leading to an overestimation of the treatment failure rate. Because the CQ blood concentration was not previously measured, we speculate that most recurrences were caused by relapses of CQ-susceptible parasites by day 28. In our study, on day 28, two patients were PCR positive for parasites, and 70% of patients had CQ levels below the therapeutic threshold. Moreover, while delayed parasite clearance has been suggested to be associated with reduced CQ susceptibility [11], we observed a similar proportions of patients with parasites in the first 3 days following CQ treatment in our study and in the 2009 study.
As demonstrated in this study, whole-genome sequencing and high-throughput genotyping in clinical trials aiming to assess the efficacy of antimalarial drugs are useful complementary tools to detect the presence of clones in recurrent isolates, absent from initial infections, as shown previously [31]. Data from relocated patients (for whom reinfections were prevented) showed that, in all successfully sequenced infections, the recurrent parasites resulted from relapses, excluding firmly the likelihood of CQ treatment failures. However, we are aware that these tools remain limited because they cannot differentiate relapse resulting from homologous parasite populations from recrudescence resulting from incomplete clearance of the primary asexual parasitemia.
Our study illustrates how knowledge from P. falciparum influences our thinking about other malaria parasite species. Owing to the inherent difficulties in studying P. vivax, most of our understanding about P. vivax malaria is derived from P. falciparum clinical studies. However, specific biological features of P. falciparum and P. vivax likely drive different adaptive responses to similar drug pressures. In P. falciparum malaria, an effective antimalarial treatment will eliminate all parasites, imposing tremendous selective pressure and eventually leading to the selection of resistant parasites. In P. vivax malaria, a lower selective drug pressure is likely acting on the parasites is mainly due to (1) the early production of gametocytes before the onset of the clinical symptoms (increasing the likelihood of transmission) and (2) the inactivity of antimalarial treatment on dormant hypnozoites, leading to new clinical episodes caused by relapsing parasites once the drug has been cleared. In this regard, it is fascinating to recall that CQ resistance was first reported in P. vivax malaria 30 years after P. falciparum resistance, despite their coendemicity and their similar drug exposure. We provide here convincing data showing that the combination of relocation of patients (excluding those with reinfection) to a malaria-free area, assessment of CQ blood concentration, and genome sequencing/high-throughput genotyping are needed to help decipher clinical outcomes of P. vivax infection and control the usual overestimation of CQ resistance in areas where relapses are frequent. Although our study needs to be replicated and validated in larger cohorts and other settings, such as high-transmission areas, our findings suggest that the assumption of CQ resistance in Cambodia may have been premature and that CQ might remain an effective therapeutic option in uncomplicated P. vivax malaria in several areas. We propose that guidelines for clinical drug studies in P. vivax malaria will be improved by using such an approach.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We thank all patients and healthcare workers involved in this study, as well as the staff of the National Center for Parasitology, Entomology, and Malaria Control in Cambodia and the Malaria Molecular Epidemiology Unit at the Institut Pasteur in Cambodia. All authors declare no conflicts of interest.
Disclaimer. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (award R01 AI103228 to D. S.); the Case Western Reserve University (grant T32 A107024 to L. R. F.); and the Institut Pasteur (grant PTR 2014–490 to D. M.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Gething PW, Elyazar IRF, Moyes CL, et al. . A long neglected world malaria map: Plasmodium vivax endemicity in 2010. PLoS Negl Trop Dis 2012; 6:e1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization. Methods for surveillance of antimalarial drug efficacy. In: Data WLC-i-P, ed, 2009. [Google Scholar]
- 3. Baird JK. Evidence and implications of mortality associated with acute Plasmodium vivax malaria. Clin Microbiol Rev 2013; 26:36–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barber BE, William T, Grigg MJ, et al. . Parasite biomass-related inflammation, endothelial activation, microvascular dysfunction and disease severity in vivax malaria. PLoS Pathog 2015; 11:e1004558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Douglas NM, Pontororing GJ, Lampah DA, et al. . Mortality attributable to Plasmodium vivax malaria: a clinical audit from Papua, Indonesia. BMC Med 2014; 12:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Naing C, Whittaker MA, Nyunt Wai V, Mak JW. Is Plasmodium vivax malaria a severe malaria?: a systematic review and meta-analysis. PLoS Negl Trop Dis 2014; 8:e3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rahimi B, Thakkinstian A, White N, Sirivichayakul C, Dondorp A, Chokejindachai W. Severe vivax malaria: a systematic review and meta-analysis of clinical studies since 1900. Malaria J 2014; 13:481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Siqueira AM, Lacerda MV, Magalhães BM, et al. . Characterization of Plasmodium vivax-associated admissions to reference hospitals in Brazil and India. BMC Med 2015; 13:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rieckmann KH, Davis DR, Hutton DC. Plasmodium vivax resistance to chloroquine?Lancet 1989; 2:1183–4. [DOI] [PubMed] [Google Scholar]
- 10. Schwartz IK, Lackritz EM, Patchen LC. Chloroquine-resistant Plasmodium vivax from Indonesia. N Engl J Med 1991; 324:927. [DOI] [PubMed] [Google Scholar]
- 11. Price RN, von Seidlein L, Valecha N, Nosten F, Baird JK, White NJ. Global extent of chloroquine-resistant Plasmodium vivax: a systematic review and meta-analysis. Lancet Infect Dis 2014; 14:982–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leang R, Barrette A, Bouth DM, et al. . Efficacy of dihydroartemisinin-piperaquine for treatment of uncomplicated Plasmodium falciparum and Plasmodium vivax in Cambodia, 2008 to 2010. Antimicrob Agents Chemother 2013; 57:818–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kerlin DH, Boyce K, Marfurt J, et al. . An analytical method for assessing stage-specific drug activity in Plasmodium vivax malaria: implications for ex vivo drug susceptibility testing. PLoS Negl Trop Dis 2012; 6:e1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Russell B, Chalfein F, Prasetyorini B, et al. . Determinants of in vitro drug susceptibility testing of Plasmodium vivax. Antimicrob Agents Chemother 2008; 52:1040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baird JK, Leksana B, Masbar S, et al. . Diagnosis of resistance to chloroquine by Plasmodium vivax: timing of recurrence and whole blood chloroquine levels. Am J Trop Med Hyg 1997; 56:621–6. [DOI] [PubMed] [Google Scholar]
- 16. Durnez L, Pareyn M, Mean V, et al. . Identification and characterization of areas of high and low risk for asymptomatic malaria infections at sub-village level in Ratanakiri, Cambodia. Malar J 2018; 17:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gryseels C, Uk S, Erhart A, et al. . Injections, cocktails and diviners: therapeutic flexibility in the context of malaria elimination and drug resistance in Northeast Cambodia. PLoS One 2013; 8:e80343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sluydts V, Durnez L, Heng S, et al. . Efficacy of topical mosquito repellent (picaridin) plus long-lasting insecticidal nets versus long-lasting insecticidal nets alone for control of malaria: a cluster randomised controlled trial. Lancet Infect Dis 2016; 16:1169–77. [DOI] [PubMed] [Google Scholar]
- 19. Sluydts V, Heng S, Coosemans M, et al. . Spatial clustering and risk factors of malaria infections in Ratanakiri Province, Cambodia. Malar J 2014; 13:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Canier L, Khim N, Kim S, et al. . An innovative tool for moving malaria PCR detection of parasite reservoir into the field. Malar J 2013; 12:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Flegg JA, Guerin PJ, White NJ, Stepniewska K. Standardizing the measurement of parasite clearance in falciparum malaria: the parasite clearance estimator. Malar J 2011; 10:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Canier L, Khim N, Kim S, et al. . Malaria PCR detection in Cambodian low-transmission settings: dried blood spots versus venous blood samples. Am J Trop Med Hyg 2015; 92:573–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang LZ, Ong RY, Chin TM, et al. . Method development and validation for rapid quantification of hydroxychloroquine in human blood using liquid chromatography-tandem mass spectrometry. J Pharm Biomed Anal 2012; 61:86–92. [DOI] [PubMed] [Google Scholar]
- 24. Friedrich LR, Popovici J, Kim S, et al. . Complexity of infection and genetic diversity in Cambodian Plasmodium vivax. PLoS Negl Trop Dis 2016; 10:e0004526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Venkatesan M, Amaratunga C, Campino S, et al. . Using CF11 cellulose columns to inexpensively and effectively remove human DNA from Plasmodium falciparum-infected whole blood samples. Malar J 2012; 11:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Popovici J, Friedrich LR, Kim S, et al. . Genomic analyses reveal the common occurrence and complexity of Plasmodium vivax relapses in Cambodia. MBio 2018; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mideo N, Bailey JA, Hathaway NJ, et al. . A deep sequencing tool for partitioning clearance rates following antimalarial treatment in polyclonal infections. Evol Med Public Health 2016; 2016:21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Menard D, Dondorp A. Antimalarial drug resistance: a threat to malaria elimination. Cold Spring Harb Perspect Med 2017; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kublin JG, Cortese JF, Njunju EM, et al. . Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis 2003; 187:1870–5. [DOI] [PubMed] [Google Scholar]
- 30. Laufer MK, Thesing PC, Eddington ND, et al. . Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med 2006; 355:1959–66. [DOI] [PubMed] [Google Scholar]
- 31. Lin JT, Hathaway NJ, Saunders DL, et al. . Using amplicon deep sequencing to detect genetic signatures of Plasmodium vivax relapse. J Infect Dis 2015; 212:999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.