Abstract
Background
MicroRNA-562 (miR-562) has been found to possess anti-cancer function in certain tumors. However, the function of miR-562 in glioblastoma (GBM) is still not fully understood.
Purpose
The aim at present study is to analyze the function of miR-562 and its possible target in GBM cells.
Patients and methods
In the present study, a total of 80 GBM samples and 16 adjacent noncancerous tissues were used to examine the expression of miR-562 and c-MET. In order to gain a deep insight into the molecular network of miR-562 and c-MET in GBM, the miR-562 mimic and inhibitor were transfected into two GBM cell lines (U251 and U87), respectively. Meanwhile, lentiviral vector was used to mediate overexpression of c-MET. Cell proliferation was examined via Cell Counting Kit-8 (CCK-8) assays. Meanwhile, cell apoptosis was analyzed by Annexin V-FTTC/PI staining assay.
Results
Our results indicated that the level of miR-562 was downregulated in GBM tissues and the expression of c-MET was upregulated in tumors. Cell proliferation analysis indicated that miR-562 was an anti-proliferation effector in GBM cells. Moreover, cell apoptosis analysis suggested the pro-apoptosis function of miR-562 in GBM cells.
Conclusion
Our results demonstrated that miR-562 negatively regulated the c-MET/AKT signal pathway. In addition, caspase-3 might also serve as another target for miR-562 in GBM cells. This research not only obtained a deep understanding of miR-562 but also provided evidence in terms of developing new prognostic biomarker for GBM.
Keywords: glioblastoma, miR-562, c-MET, AKT, p-AKT, caspase-3
Introduction
Glioblastomas (GBMs) are one of the malignant brain tumors all over the world that are most commonly diagnosed in adults.1 More than half of GBM sufferers died within 1 year because of its worst prognosis. Much attention has been paid to determining the effective therapy method for GBM. However, the survival rate of GBM patients is still very low.2–4 Therefore, gaining a deep insight into the GBM molecule mechanisms is urgently needed.
MicroRNAs (miRs) are a group of small noncoding RNAs, which serve as a valuable post-transcription mediator through binding on the 3′UTRs of the targeting miRs.5,6 Growing evidence has indicated that miRs play an essential regulatory role in tumorigenesis and cellular processes of GBM.7–9 Reports have demonstrated that miR-562 is downregulated in Wilms tumor, which is capable of inducing tumorigenesis.10 However, it is rather less known about the function of miR-562 in GBM.
c-MET is the receptor for hepatocyte growth or scatter factor, which is mapped on chromosome 7 at band 7q21-q31.11 The c-MET receptor plays a critical role in many human cancers. Previous report has indicated that knockdown of the expression of c-MET leads to the inhibition of cell growth and the induction of apoptosis in GBM cells.12 Moreover, c-MET has been suggested as a direct target of miR-449b-5p and miR-410 in the pathogenesis of GBM.13,14 Therefore, there is a close relationship between c-MET and miRs in GBM. However, whether there is a correlation between c-MET and miR-562 in GBM is still not clear.
Cleaved caspase-3 belongs to the cysteine protease family, which is the active form of caspase-3.15 Previous report has indicated that cleaved caspase-3 shows the potential value as a biomarker for tumor prognosis.16 Moreover, cleaved caspase-3 contributes to the apoptosis process in tumor.17 In addition, cleaved caspase-3 has the function to inhibit chemical-induced hepatocarcinogenesis.18 However, it is still less known about the molecular network of cleaved caspase-3 in GBM cells.
The AKT pathway plays an important role in various biological functions, which is usually abnormal in human cancer.19 The phosphorylation of AKT (p-AKT) is essential for its biological function. Previous report has indicated that the p-AKT correlates with the activity of AKT in human non-small-cell lung cancer.20 Moreover, the PI3K/AKT pathway has participated in the cell cycle and apoptosis of lung cancer cells.21 Co-activation of AKT and c-MET has triggered rapid hepatocellular cancer in mice.22 Therefore, it is valuable to examine the function of AKT in GBM cells.
In this study, we systemically analyzed the function of miR-562 and its possible target gene in GBM cells. Our research not only gained a deep insight into the molecular network of miR-562 but also provided evidence in terms of developing a new prognostic biomarker for GBM.
Materials and methods
Tissue specimen and cell culture
GBM samples (n=80) and adjacent noncancerous tissues (n=16) were obtained from Huzhou Central Hospital, Huzhou, Zhejiang, P.R. China. Samples were snap-frozen in liquid nitrogen, stored at −80°C for further analysis. All patients were informed and gave written consent. This study was approved by the independent ethics committee of Huzhou Central Hospital, Huzhou, Zhejiang and was in accordance with the Declaration of Helsinki. The median expression value of miR-562 was used to define the high expression group (above the median) and the low expression group (below the median).
The GBM cell lines (U87 and U251) used in this study were obtained from cell bank of Shanghai Biology Institute (Shanghai, P.R. China). A 10% FBS (Thermo Fisher Scientific) mixed into all culture media, 2 mM l-glutamine, and 1% penicillin/streptomycin (Solarbio, Beijing, P.R. China). Cells were grown in DMEM (Trueline, Kaukauna, WI, USA) and were maintained under a 5% CO2 atmosphere, at 37°C.
RNA isolation and real-time PCR
Total RNA from GBM samples was extracted by TRIzol Reagent (Invitrogen, Waltham, MA, USA). Then, RNA was reverse transcribed into cDNA by cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The conditions of real-time PCR were as follows: 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds, 60°C for 45 seconds and normalized to glyceraldehyde 3-phosphate dehydrogenase. The gene relative expression was calculated by the 2−ΔΔCt method. All data represent the average of three replicates. The primers used in this study were listed in Supplementary material S1.
Western blot
Whole protein lysates were extracted from indicated cells (U87 and U251) by RIPA lysis buffer (JRDUN, Shanghai, P.R. China) with EDTA-free Protease inhibitor Cocktail (Roche, Heidelberg, Germany). The concentration was estimated by an enhanced BCA protein assay kit (Thermo Fisher Scientific). Equal amounts of total protein (25 µg) were fractionated on 10% SDS-PAGE and transferred to a nitrocellulose membrane (Millipore, Billerica, MA, USA) overnight. Then, after being blocked with 5% nonfat dry milk for 1 hour at room temperature, the membranes were probed at 4°C overnight with the primary antibodies followed by secondary antibody anti-mouse IgG (1:1,000; Beyotime, Shanghai, P.R. China) for 1 hour at 37°C. An enhanced chemiluminescence system (Tanon, Shanghai, P.R. China) was used for detection protein expression value. All the detail information of primary antibodies was provided in Table S1. The caspase-3 inhibitor Z-VAD was purchased from Selleck, Houston, TX, USA.
Cell transfection
The miR-562 mimics, inhibitor, and negative control (NC) were purchased from GenePharma (Shanghai, P.R. China). Cells were grown on 80% confluence prior to transfection. Transfection of the miR-562 mimics or inhibitor into glioma U251 and U87 cells was performed by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. After 48 hours posttransfection, transfection efficacy was measured by qRT-PCR or Western blot. The sequence information was provided in Table S2.
Lentiviral-mediated overexpression of c-MET
Lentiviral plasmid (pLVX-puro) containing the full length of human c-MET (BC130420.1) cDNA sequence and the empty plasmid acted as its overexpressing negative control (oeNC). All of them were transiently transfected into cells by Lipofectamine 2000 (Invitrogen) according to the instruction of the manufacturer. Experiments were performed 48 hours after being transfected.
Cell proliferation assay
Cell Counting Kit-8 (CCK-8) assay kits (SAB, College Park, MD, USA) was used to examine cell proliferation according to the protocol of the manufacturer. Briefly, cells transfected as indicated were seeded in 96-well plates and cultured for 0, 24, 48, and 72 hours, CCK-8 solution (1:10) was mixed to each well and incubated for 1 hour. Microplate reader (Pulangxin, Beijing, P.R. China) was used to measure the ODs at wave length 450 nm. Triplicates were performed at each time point.
Cell apoptosis assay
In brief, cells (U87 and U251) were collected and stained by Annexin V-fluorescein isothiocyanate’ apoptosis detection kit (Beyotime) according to the instructions of the manufacturer at 48 hours after viral infection. Then, flow cytometer (BD, San Diego, CA, USA) were used to determine cells. All data represented the average of three replicates.
Dual-luciferase reporter assays
The c-MET 3′-UTR, containing the NC, wild type (WT), and mutated target sequences of miR-562, was cloned into Dual-Luciferase Expression Vector pGL3-Promoter (Promega, Madison, WI, USA) and all vectors were transfected to U87 cells. Then, cells were harvested after 48 hours of transfection and then detected using the Dual-GLO Luciferase Assay Kit (Promega).
Statistical analysis
GraphPad Prism software Version 7.0 (La Jolla CA, USA) was used for statistical analyses. Data were displayed as mean ± SD for at least three samples. Statistical significance was determined by one-way ANOVA for multiple comparisons. A P-value <0.05 indicates statistical significance.
Results
miR-562 negatively correlated with c-MET in GBM tissues
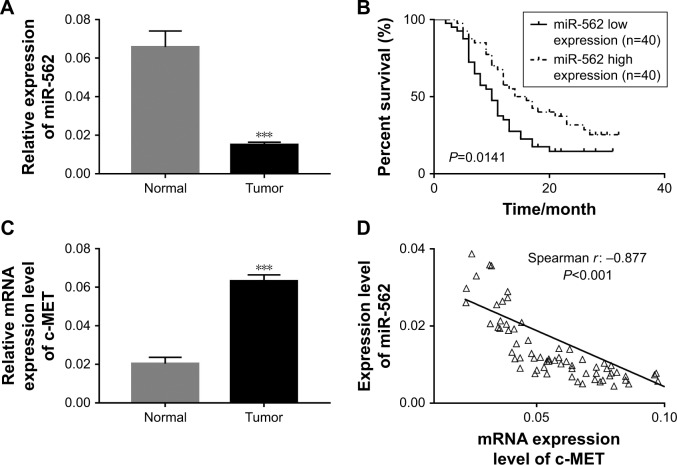
The expression level of miR-562 and c-MET was examined by qRT-PCR in 80 GBM tissues and 16 normal brain samples, respectively. As shown in Figure 1A, the expression level of miR-562 was significantly downregulated in GBM tissues compared with that in normal tissues. Kaplan–Meier survival analysis (Figure 1B) indicated that the survival time of patients with higher expression of miR-562 was significantly longer than that of patients with lower miR-562 expression (P<0.05). On the contrary, the level of c-MET was significantly higher in GBM than that of normal tissues (Figure 1C). Moreover, Spearman correlation analysis indicated a negative correlation between miR-562 and c-MET (Figure 1D).
Figure 1.
The negative correlation between miR-562 and c-MET in GBM tissues.
Notes: (A) The miR expression level of miR-562 in GBM tissues (n=80) and para-cancerous tissues (n=16). ***P<0.001 vs normal. (B) The overall survival time of 80 patients with GBM. (C) The mRNA expression level of c-MET in GBM tissues (n=80) and para-cancerous tissues (n=16). ***P<0.001 vs normal. (D) Correlation between the expression of miR-562 and c-MET in GBM samples.
Abbreviations: GBM, glioblastoma; miR, microRNA.
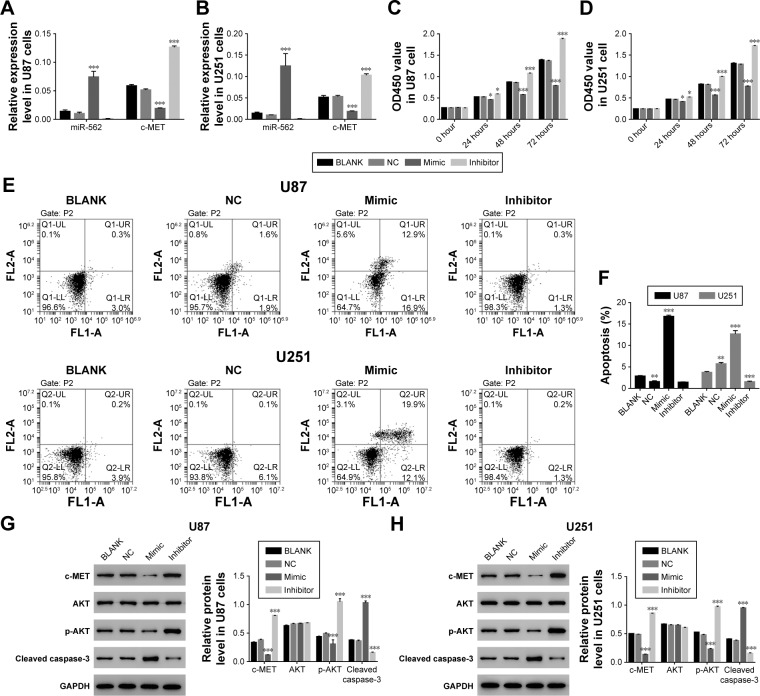
miR-562 suppressed cell proliferation and induced cell apoptosis in GBM cells
In order to investigate the effect of miR-562 on cell proliferation and cell apoptosis, we utilized gain-of-function and loss-of-function assay by transiently transfecting the miR-410-3p mimic and inhibitor, as well as their NC into U87 and U251 cells, respectively. The untreated cells served as blank control (BLANK).
As shown in Figure 2A and B, overexpression of miR-562 strongly inhibited the expression of c-MET. On the other side, the expression of c-MET was significantly upregulated in miR-562 inhibitor transfected cells. Then, cell proliferation was examined via CCK-8 assays among different transfected cells as indicated. From the results, the cell proliferation rate was deeply suppressed in miR-562 mimic transfected cells compared with that of NC cells. Meanwhile, the miR-562 inhibitor transfected cells showed a significantly upregulated in cell proliferation. Therefore, miR-562 inhibited the proliferation of GBM cells (Figure 2C and D). Next, cell apoptosis was analyzed by Annexin V-FTTC/PI staining assay among different transfected cells as indicated, the cell apoptosis ratio of miR-562 mimic transfected cells was significantly increased compared with that of NC cells in U87 and U251 cell lines, which reflected the pro-apoptosis role of miR-562 on GBM cells (Figure 2E and F).
Figure 2.
The effect of miR-562 mimic and inhibitor in GBM cell.
Notes: (A, B) Stands for the expression level of miR-562 and c-MET in different transfected cells of GBM, respectively. ***P<0.001 vs NC. (C, D) Stands for cell proliferation was detected at 12, 24, 48, and 72 hours in different transfected cells of GBM, respectively. *P<0.05 vs NC, ***P<0.001 vs NC. (E, F) Stands for the cell apoptosis profile of GBM cells transfected with NC, miR-562 mimic and inhibitor, the untreated cells acted as BLANK. **P<0.01 vs NC, ***P<0.001 vs NC. (G, H) The protein level of c-MET, AKT, p-AKT, and caspase-3 in different transfected cells of GBM as indicated, respectively, ***P<0.001 vs NC.
Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GBM, glioblastoma; NC, negative control; p-AKT, phosphorylation of AKT.
In addition, Western blot was performed to determine the protein level of c-MET, AKT, p-AKT, and cleaved caspase-3 in the cells as indicated. As shown in Figure 2G and H, the protein level of c-MET was significantly decreased in miR-562 mimic transfected cells and showed a negative correlation with miR-562. Moreover, our results suggested that the p-AKT was deeply suppressed by miR-562. In addition to that the protein level of cleaved caspase-3 presented a positive correlation with the expression of miR-562 in GBM cells.
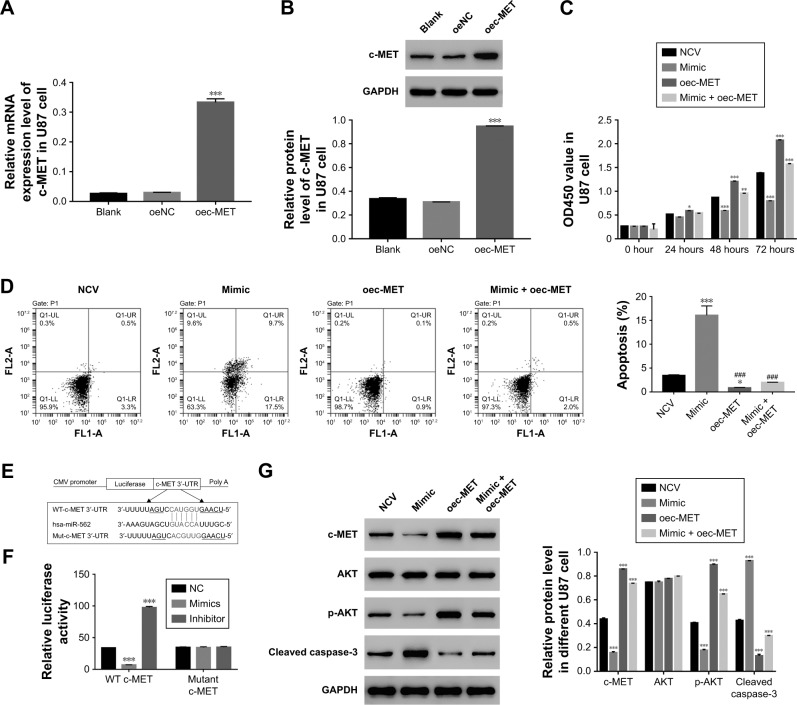
c-MET neutralized the effect of miR-562 on GBM cells
To further explore the connection between c-MET and miR-562, U87 cells was transfected with a plasmid over-expressing c-MET (oec-MET), mock plasmid (oeNC), and mock plasmid with miR-562 NC (NCV). As shown in Figure 3A and B, the mRNA and protein level of c-MET was significantly upregulated in oec-MET transfected cells compared with oeNC transfected cells. Moreover, the cell proliferation rate was significantly upregulated in oec-MET transfected cells. However, this effect was deeply neutralized by miR-562 mimic (Figure 3C). As shown in Figure 3D, the cell apoptosis rate was downregulated in oec-MET transfected cells compared with that of NCV and miR-562 mimic transfected cells. Interestingly, the cell apoptosis rate showed no significant difference between oec-MET and miR-562 mimic + oec-MET transfected cells. However, the cell apoptosis rate of the double-transfected (miR-562 mimic + oec-MET) cells was significantly decreased compared with miR-562 mimic, which indicated that the c-MET suppressed the effect of miR-562 on cell apoptosis. Taken together, these results suggested that c-MET neutralized the effect of miR-562 on GBM cells.
Figure 3.
c-MET neutralized the effect of miR-562 in GBM cells.
Notes: (A, B) Stands for the mRNA expression and protein level of c-MET by oec-MET in GBM cells, respectively, ***P<0.001 vs oeNC. (C) Cell proliferation was detected 12, 24, 48, and 72 hours in U87 cells transfected with NCV, miR-562 mimic, oec-MET, and miR-562 mimic + oec-MET, respectively, **P<0.01 vs NCV, ***P<0.001 vs NCV. (D) The cell apoptosis profile of transfected cells as indicated above. *P<0.05 vs NCV, ###P<0.001 vs mimic and ***P<0.001 vs NCV. (E) Schematic of the luciferase construct with the c-MET 3′-UTR containing a miR-562 binding sequence. (F) Dual-luciferase assays of miR-562 and c-MET 3′-UTR. WT or mutant 3′-UTR of c-MET was cloned into luciferase reporter vectors. U87 cells were transfected with miR-562 mimics or inhibitor in addition to the luciferase reporter vector and incubated for 48 hours. ***P<0.001 vs NC. (G) The protein level of c-MET, AKT, p-AKT, and caspase-3 in different transfected cells of GBM as indicated, respectively, ***P<0.001 vs NCV.
Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GBM, glioblastoma; p-AKT, phosphorylation of AKT; WT, wild type.
In addition, some highly conserved regions in 3′-UTR of c-MET were investigated using miRDB database (http://www.mirdb.org), and predicted the possible binding sites for miR-562. To further determine the interaction between c-MET and miR-562, luciferase reporters that contained the WT or mutant 3′-UTR of c-MET was generated (Figure 3E). As shown in Figure 3F, the results showed that the luciferase activity of the WT c-MET 3′-UTR reporter vector was markedly decreased by miR-562 mimic and significantly enhanced by miR-562 inhibitor. However, the luciferase activity of the mutant 3′-UTR of c-MET reporter vector showed no significant difference among different transfected cells. Taken together, these results indicated that miR-562 regulated c-MET expression by directly binding on its 3′-UTR. In addition, Western blot was used to examine the protein level of c-MET, AKT, p-AKT, and cleaved caspase-3 in different transfected cells as indicated. Our results suggested that miR-562 inhibited the expression of c-MET and p-AKT in GBM cells. Meanwhile, the level of cleaved caspase-3 was positively correlated with the expression of miR-562 in GBM cells (Figure 3G).
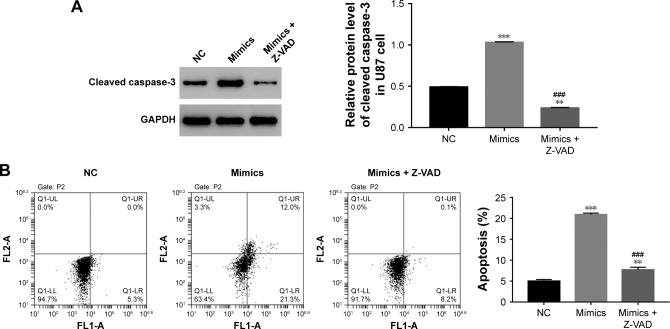
The effect of miR-562 was suppressed by a specific caspase-3 inhibitor Z-VAD on GBM cells
In order to further examine the correlation between miR-562 and caspase-3, a specific caspase-3 inhibitor Z-VAD was used to culture the miR-562 mimic transfected cells. As shown in Figure 4A, the protein level of cleaved caspase-3 was deeply downregulated in Z-VAD cultured cells compared with that of miR-562 mimic transfected cells. Moreover, the cell apoptosis rate also showed significantly decreased in Z-VAD-treated cells (Figure 4B). These results indicated that miR-562 might regulate the apoptosis of GBM cells through activating caspase-3.
Figure 4.
The effect of miR-562 mimic was suppressed by Z-VAD in GBM cells.
Notes: (A) The protein level of caspase-3 in U87 cells transfected with miR-562 mimic and miR-562 mimic treat by a specific caspase-3 inhibitor Z-VAD, respectively, ***P<0.001 vs NC. **P<0.01 vs NC and ###P<0.001 vs mimics. (B) Cell apoptosis profile of transfected cells as indicated above, ***P<0.001 vs NC.
Abbreviations: GBM, glioblastoma; miR, microRNA; NC, negative control.
Discussion
As in the case of common malignant brain tumors, the survival rate of GBM is usually limited to 2 years. Although the traditional therapy (tumor resection and radiotherapy) can help to fight against the development of GBM, the prognosis is usually disappointing. Therefore, gaining a deep insight into the molecular mechanism of GBM would contribute to develop novel therapy approach for its treatment.
In the present study, we systemically analyzed the function of the miR-562 on GBM cells. In order to obtain a reliable result, the miR-562 mimic and inhibitor were used for overexpression and suppression of miR-562. Results from these two experimental phases were in agreement. Hence, our results were more understandable.
From the CCK-8 assay, the cell proliferation rate was deeply suppressed in miR-562 mimic transfected cells, which was upregulated in miR-562 inhibitor transfected cells. Therefore, these results demonstrated the anti-proliferation function of miR-562. Apoptosis is the basic essential process for eliminating damaged or unnecessary cells. Dysregulation of the apoptotic pathway has been confirmed as a key event in cancer development and progression.23 In this study, results from cell apoptosis assay indicated a pro-apoptosis role of miR-562 in GBM cells. Taken together, these results indicated the potential value of miR-562 as a promising prognostic biomarker for GBM.
The c-MET signaling pathway is essential for cell proliferation and apoptosis. Overexpression of c-MET often leads to the tumor growth. Therefore, inhibiting the expression of c-MET is believed as a promising target for cancer treatment. Growing evidence has indicated that miRs are a novel and additional system to regulate c-MET expression in tumors.24 Previous reports have demonstrated that miR-139-5P and miR-206 inhibit proliferation and causes apoptosis through downregulating c-Met in lung cancer cells.25,26 Moreover, it has been reported that miR-34a has suppressed the expression of c-MET through binging on its 3′-UTR.27 In addition, miR-23b can recognize four sites in the c-MET 3′-UTR, which decreases the migration and proliferation of hepatocellular carcinoma cells.28 In the present study, our results also reflected that miR-562 negatively regulated the c-MET through binding on its 3′-UTR. Therefore, all these miRs binding sites on the 3′-UTR of c-MET contributed to develop novel approach for certain cancers treatment.
It has been reported that HGF/c-MET/AKT pathway regulates the cell apoptosis process in tumors.29 Previous report has demonstrated that the main function of miR-206 is depended on suppressing the c-MET/AKT signaling pathway.30 In this study, the p-AKT was downregulated by miR-562. Therefore, we suspected that miR-562 might regulate the proliferation and apoptosis of GBM cells via mediating the c-MET/AKT signaling pathway as well. In addition to that the caspase-3 might serve as another target by miR-562 in the apoptosis of GBM cells. Hence, miR-562 might regulate apoptosis through inhibiting the c-MET/AKT pathway and activating the downstream effectors caspase-3.
Conclusion
In this study, we examined the effect of miR-562 mimic and inhibitor on GBM cells. Our results not only gained a better understanding of the miR-562 function but also provided evidence to indicate its potential value as a prognostic biomarker for GBM.
Supplementary materials
Supplementary material S1
Primer sequence information
1.1 hsa-miR-562 MIMAT0003226
RT-Primer: 5′ GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCAAAT 3′
PCR Primer:
Primer F 5′ CGCGCGAAAGTAGCTGTACC 3′
Primer R 5′ AGTGCAGGGTCCGAGGTATT 3′
1.2 Homo sapiens MET proto-oncogene, receptor tyrosine kinase (MET), transcript variant 2, mRNA
NCBI Reference Sequence: NM_000245.3
Primer F: 5′ TTGGGACATCAGAGGGTCG 3′
Primer R: 5′ AACAAAGGGTGGGGCAGAG 3′
Pos: 1364–1608
Amplified product: Size: 245 bps
1.3 Homo sapiens RNA, U6 small nuclear 1 (RNU6-1), small nuclear RNA NR_004394.1
Primer F: 5′ CTCGCTTCGGCAGCACA 3′
Primer R: 5′ AACGCTTCACGAATTTGCGT 3′
Pos: 4–97
Amplified product: Size: 94 bps
Table S1.
The primary antibodies information
| Antibody name | Source | Dilution factor |
|---|---|---|
|
| ||
| c-MET | Abcam, UK | 1:500 |
| Caspase-3 | Abcam, UK | 1:500 |
| AKT | CST, USA | 1:1,000 |
| p-AKT | CST, USA | 1:2,000 |
| GAPDH | CST, USA | 1:2,000 |
Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase; p-AKT, phosphorylation of AKT.
Table S2.
The sequence information of miR-562 mimic and inhibitor
| Name | Sequence |
|---|---|
|
| |
| hsa-miR-562 | 5′-AAAGUAGCUGUACCAUUUGC-3′ |
| hsa-miR-miR-562 mimic | 5′-AAAGUAGCUGUACCAUUUGC-3′ |
| hsa-miR-miR-562 inhibitor | 5′-GCAAAUGGUACAGCUACUUU-3′ |
| microRNA NC | 5′-CAGUACUUUUGUGUAGUACAA-3′ |
Abbreviation: NC, negative control.
Acknowledgments
This research was supported by the Science Technology Department of Zhejiang province (No 2016C3220) and the National Natural Science Foundation of China (No 81703752).
Footnotes
Author contributions
Xiaohu Nie and Yue Zhou are co first authors. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Sturm D, Bender S, Jones DT, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer. 2014;14(2):92–107. doi: 10.1038/nrc3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerber NK, Goenka A, Turcan S, et al. Transcriptional diversity of long-term glioblastoma survivors. Neuro Oncol. 2014;16(9):1186–1195. doi: 10.1093/neuonc/nou043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun G, Shi L, Yan S, et al. miR-15b targets cyclin D1 to regulate proliferation and apoptosis in glioma cells. Biomed Res Int. 2014;2014:1–9. doi: 10.1155/2014/687826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xia B, Yang S, Liu T, Lou G. miR-211 suppresses epithelial ovarian cancer proliferation and cell-cycle progression by targeting Cyclin D1 and CDK6. Mol Cancer. 2015;14:57. doi: 10.1186/s12943-015-0322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chai C, Song LJ, Han SY, Li XQ, Li M. MicroRNA-21 promotes glioma cell proliferation and inhibits senescence and apoptosis by targeting SPRY1 via the PTEN/PI3K/AKT signaling pathway. CNS Neurosci Ther. 2018;24(5):369–380. doi: 10.1111/cns.12785. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Zhang LX, Jin W, Zheng J, et al. MicroRNA-375 regulates proliferation and apoptosis of glioma cancer cells by inhibiting CTGF-EGFR signaling pathway. Bratisl Lek Listy. 2018;119(1):17–21. doi: 10.4149/BLL_2018_004. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Zhang J, Qiu W, et al. MicroRNA-1231 exerts a tumor suppressor role through regulating the EGFR/PI3K/AKT axis in glioma. J Neurooncol. 2018;139(3):547–562. doi: 10.1007/s11060-018-2903-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drake KM, Ruteshouser EC, Natrajan R, et al. Loss of heterozygosity at 2q37 in sporadic Wilms’ tumor: putative role for miR-562. Clin Cancer Res. 2009;15(19):5985–5992. doi: 10.1158/1078-0432.CCR-09-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li MY, Yang P, Liu YW, et al. Low c-Met expression levels are prognostic for and predict the benefits of temozolomide chemotherapy in malignant gliomas. Sci Rep. 2016;6(1):21141. doi: 10.1038/srep21141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu SH, Feng DF, Zhang H, et al. c-Met-targeted RNA interference inhibits growth and metastasis of glioma U251 cells in vitro. J Neurooncol. 2009;93(2):183–189. doi: 10.1007/s11060-008-9772-5. [DOI] [PubMed] [Google Scholar]
- 13.Zhen L, Yun-Hui L, Hong-Yu D, Jun M, Yi-Long Y. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumour Biol. 2016;37(1):673–683. doi: 10.1007/s13277-015-3843-y. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Zhang J, Feng Y, et al. miR-410 regulates MET to influence the proliferation and invasion of glioma. Int J Biochem Cell Biol. 2012;44(11):1711–1717. doi: 10.1016/j.biocel.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 15.Motta CM, Frezza V, Simoniello P. Caspase 3 in molluscan tissues: localization and possible function. J Exp Zool A Ecol Genet Physiol. 2013;319(10):548–559. doi: 10.1002/jez.1817. [DOI] [PubMed] [Google Scholar]
- 16.Hu Q, Peng J, Liu W, et al. Elevated cleaved caspase-3 is associated with shortened overall survival in several cancer types. Int J Clin Exp Pathol. 2014;7(8):5057–5070. [PMC free article] [PubMed] [Google Scholar]
- 17.Carrillo García C, Riedt T, Li J, Dotten M, Brossart P, Janzen V. Simultaneous deletion of p21Cip1/Waf1 and caspase-3 accelerates proliferation and partially rescues the differentiation defects of caspase-3 deficient hematopoietic stem cells. PLoS One. 2014;9(10):e109266. doi: 10.1371/journal.pone.0109266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shang N, Bank T, Ding X, et al. Caspase-3 suppresses diethylnitrosamine-induced hepatocyte death, compensatory proliferation and hepatocarcinogenesis through inhibiting p38 activation. Cell Death Dis. 2018;9(5):558. doi: 10.1038/s41419-018-0617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshizawa A, Fukuoka J, Shimizu S, et al. Overexpression of phospho-eIF4E is associated with survival through AKT pathway in non-small cell lung cancer. Clin Cancer Res. 2010;16(1):240–248. doi: 10.1158/1078-0432.CCR-09-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vincent EE, Elder DJ, Thomas EC, et al. Akt phosphorylation on Thr308 but not on Ser473 correlates with Akt protein kinase activity in human non-small cell lung cancer. Br J Cancer. 2011;104(11):1755–1761. doi: 10.1038/bjc.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X, Zhang Y, Qu D, Jiang T, Li S. Osthole induces G2/M arrest and apoptosis in lung cancer A549 cells by modulating PI3K/Akt pathway. J Exp Clin Cancer Res. 2011;30:33. doi: 10.1186/1756-9966-30-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu J, Che L, Li L, et al. Co-activation of AKT and c-Met triggers rapid hepatocellular carcinoma development via the mTORC1/FASN pathway in mice. Sci Rep. 2016;6:20484. doi: 10.1038/srep20484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porebska I, Wyrodek E, Kosacka M, Adamiak J, Jankowska R, Harłozińska-Szmyrka A. Apoptotic markers p53, Bcl-2 and Bax in primary lung cancer. In Vivo. 2006;20(5):599–604. [PubMed] [Google Scholar]
- 24.Giglio S, Vecchione A. c-Met and miRs in cancer. Biomedicines. 2015;3(1):32–44. doi: 10.3390/biomedicines3010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun C, Sang M, Li S, et al. Hsa-miR-139-5p inhibits proliferation and causes apoptosis associated with down-regulation of c-Met. Oncotarget. 2015;6(37):39756–92. doi: 10.18632/oncotarget.5476. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Sun C, Liu Z, Li S, et al. Down-regulation of c-Met and Bcl2 by microRNA-206, activates apoptosis, and inhibits tumor cell proliferation, migration and colony formation. Oncotarget. 2015;6(28):25533–74. doi: 10.18632/oncotarget.4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan D, Zhou X, Chen X, et al. MicroRNA-34a inhibits uveal melanoma cell proliferation and migration through downregulation of c-Met. Invest Ophthalmol Vis Sci. 2009;50(4):1559–1565. doi: 10.1167/iovs.08-2681. [DOI] [PubMed] [Google Scholar]
- 28.Salvi A, Sabelli C, Moncini S, et al. MicroRNA-23b mediates urokinase and c-met downmodulation and a decreased migration of human hepatocellular carcinoma cells. FEBS J. 2009;276(11):2966–2982. doi: 10.1111/j.1742-4658.2009.07014.x. [DOI] [PubMed] [Google Scholar]
- 29.Yao Y, Dou C, Lu Z, Zheng X, Liu Q. MACC1 suppresses cell apoptosis in hepatocellular carcinoma by targeting the HGF/c-MET/AKT pathway. Cell Physiol Biochem. 2015;35(3):983–996. doi: 10.1159/000369754. [DOI] [PubMed] [Google Scholar]
- 30.Dai C, Xie Y, Zhuang X, Yuan Z. miR-206 inhibits epithelial ovarian cancer cells growth and invasion via blocking c-Met/AKT/mTOR signaling pathway. Biomed Pharmacother. 2018;104:763–770. doi: 10.1016/j.biopha.2018.05.077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
The primary antibodies information
| Antibody name | Source | Dilution factor |
|---|---|---|
|
| ||
| c-MET | Abcam, UK | 1:500 |
| Caspase-3 | Abcam, UK | 1:500 |
| AKT | CST, USA | 1:1,000 |
| p-AKT | CST, USA | 1:2,000 |
| GAPDH | CST, USA | 1:2,000 |
Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase; p-AKT, phosphorylation of AKT.
Table S2.
The sequence information of miR-562 mimic and inhibitor
| Name | Sequence |
|---|---|
|
| |
| hsa-miR-562 | 5′-AAAGUAGCUGUACCAUUUGC-3′ |
| hsa-miR-miR-562 mimic | 5′-AAAGUAGCUGUACCAUUUGC-3′ |
| hsa-miR-miR-562 inhibitor | 5′-GCAAAUGGUACAGCUACUUU-3′ |
| microRNA NC | 5′-CAGUACUUUUGUGUAGUACAA-3′ |
Abbreviation: NC, negative control.