Abstract
Background
Long non-coding RNAs (lncRNAs) play important roles in human cancers. However, the functional roles of lncRNAs in non-small-cell lung cancer (NSCLC) and the underlying mechanisms are not well understood.
Methods
We examined the expression of lncRNA DANCR in NSCLC by qRT-PCR and explored its biological roles in NSCLC progression by cell and molecular biology studies.
Results
DANCR expression level was increased in human NSCLC. The knockdown of DANCR inhibited NSCLC cell proliferation by inducing cell apoptosis and cell cycle arrest. In addition, DANCR knockdown suppressed NSCLC cell migration and invasion via inhibition of epithelial–mesenchymal transition (EMT). On the contrary, DANCR overexpression had the opposite effects. DANCR knockdown inhibited EZH-2-mediated epigenetic silencing of p21 promoter and increased p21 expression. Moreover, DANCR knockdown inhibited NSCLC cell proliferation, migration, and invasion in a p21-dependent manner.
Conclusion
DANCR plays oncogenic roles in NSCLC and may provide a novel biomarker for NSCLC diagnosis and prognosis.
Keywords: DANCR, NSCLC, progression, biomarker
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide. Approximately 70% of patients with lung cancer present with locally advanced or metastatic disease at the time of diagnosis.1 Non-small-cell lung cancer (NSCLC) accounts for 80% of diagnosed lung cancer. Despite recent advances in disease diagnosis and treatment, the long-term prognosis of NSCLC patients remains poor.2 The elucidation of molecular mechanisms underlying NSCLC progression will provide new strategies for NSCLC diagnosis and therapy.
The important roles of long non-coding RNAs (lncRNAs) in health and diseases have been revealed in the past decade. LncRNAs have emerged as important regulators of gene expression. LncRNAs have critical roles in various biological processes including cell proliferation, apoptosis, migration, and invasion. Increasing evidence suggests that many lncRNAs are aberrantly expressed in human cancers including NSCLC.3,4 Previous studies have shown that lncRNAs could function as either tumor suppressors or oncogenes, contributing to the development and progression of NSCLC.5–7 Therefore, further study of the roles of lncRNAs and their mechanisms of action may provide novel diagnostic and prognostic biomarkers for NSCLC.
DANCR has previously been shown to be abnormally expressed in several human cancers. For instance, DANCR is upregulated in gastric cancer,8 colon cancer,9 esophageal cancer,10 osteosarcoma,11 and glioma.12 High expression of DANCR is associated with disease progression and poor clinical outcome in cancer patients, indicating that DANCR may act as an oncogene. DANCR has been shown to promote cancer cell proliferation, migration, and invasion by acting as a competitive endogenous RNA.13,14 However, whether DANCR is involved in NSCLC growth and metastasis has not been well characterized.
In this study, we reported that DANCR was highly expressed in human NSCLC, and DANCR knockdown inhibited NSCLC cell proliferation, migration, and invasion. DANCR knockdown induced cell cycle arrest and cell apoptosis in NSCLC cells. Furthermore, DANCR knockdown reversed EMT and inhibited NSCLC cell migration and invasion. DANCR exerted promoting roles in NSCLC cell proliferation, migration, and invasion by epigenetically silencing p21 expression. These findings provide a basis for a better understanding of the roles of lncRNAs in NSCLC progression and a new biomarker for NSCLC diagnosis and therapy.
Materials and methods
Clinical specimens
A total of 40 paired cancer and adjacent non-cancerous tissues (5 cm away from the tumor edge) were obtained from Nantong Tumor Hospital between May 2016 and April 2017. Written informed consent was obtained from all the patients and this study was approved by the Institutional Ethics Committee of Nantong Tumor Hospital. The study was conducted in accordance with the Declaration of Helsinki. All of the tissues were frozen in liquid nitrogen and then stored at −80°C for further use. The patients included in this study had not received any preoperative therapies.
Cell culture
Human NSCLC cell lines (A549, H1299, and H358) and human lung epithelial cell line (BEAS-2B) were purchased from the Institutes for Biological Sciences at the Chinese Academy of Sciences (Shanghai, China) and cultured in high-glucose DMEM, supplemented with 10% FBS (Gibco®; Thermo Fisher Scientific, Waltham, MA, USA). All the cells were cultured in a humidified incubator with 5% CO2 at 37°C.
Gene transfection
Cells were seeded in 6-well plates at a density of 2×105/well and cultured at 37°C in an incubator overnight. The overexpressing plasmid and silencing shRNAs (Hanbio, Shanghai, China) were transfected into the cells by using LipoFiter transfection reagent (Hanbio) in a serum-free medium. Cells were changed to complete the medium at 6 hours after transfection and cultured for another 30 hours. The target sequences of shRNAs were provided in Table 1.
Table 1.
The target sequences of shRNAs
| Sequence | |
|---|---|
|
| |
| sh-DANCR | Sense: GATCCGGAGCTAGAGCAGTGACAATGTTCAAGAGACATTGTCACTGCTCTAGCTCCTTTTTTC |
| Antisense: AATTGAAAAAAGGAGCTAGAGCAGTGACAATGTCTCTTGAACATTGTCACTGCTCTAGCTCCG | |
| sh-Ctrl | Sense: GATCCGGAGCTCATGGGTCCTTTGTATCGGTACCGATACAAAGGACCCATGAGCTTTTTTG |
| Antisense: AATTCAAAAAAGCTCATGGGTCCTTTGTATCGGTACCGATACAAAGGACCCATGAGCTCCG | |
Cell apoptosis assay
Cell apoptosis was determined by using the Annexin V-Alexa Fluor 647/propidium iodide (PI) apoptosis detection kit (Fcmacs, Jiangsu, China). The transfected cells were digested with collagenase, collected, and stained with Annexin V-Alexa Fluor 647 and PI for 15 minutes at room temperature. The cell apoptosis rate was analyzed by flow cytometry.
Cell cycle analysis
Cell cycle analysis was conducted with a cell cycle detection kit (Fcmacs). The transfected cells were collected and fixed in 95% ethanol overnight. Afterwards, the cells were stained with 50 µg/mL PI for 30 minutes in the dark. The cell cycle distribution was analyzed on a flow cytometer (FACS Cali-bur, BD, Franklin Lakes, NJ, USA) using CellQuest software.
Cell counting and cell colony formation assays
For cell counting assay, the transfected cells were seeded in 24-well plates (1×104/well) and counted for 6 consecutive days. For cell colony formation assay, the transfected cells were seeded in 6-well plates (1×103/well) and cultured for 10 consecutive days. The medium was changed every 3 days. At the end of experiments, the cells were fixed with 4% para-formaldehyde and stained with crystal violet. The number of colonies was calculated under a microscope.
Transwell migration assay
The transfected cells were collected and seeded into the upper chamber (8 µm) at a density of 1×105 cells/well (Corning Incorporated, Corning, NY, USA). The lower chamber was filled with 500 µL culture medium supplemented with 10% FBS. Twelve hours later, the cells on the upper surface of the membrane were removed with a cotton swab. Then, the lower cells were fixed with formaldehyde and stained with crystal violet for 30 minutes. The number of migrated cells was counted under a microscope.
Matrigel invasion assay
The diluted basement Matrigel (BD, Franklin Lakes, NJ, USA) was added into each chamber and left to polymerize at 37°C for 30 minutes. The transfected cells were seeded into the upper chamber at a density of 2×105 cells/well. The lower chamber was filled with 500 µL culture medium supplemented with 10% FBS. The cells were allowed to invade to the lower membrane for 24 hours. Subsequently, the cells on the upper surface of the membrane were removed with a cotton swab. The lower cells were then fixed with formaldehyde and stained with crystal violet for 30 minutes. The number of migrated cells was counted under a microscope.
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific) and reverse transcribed into cDNA using miScript reverse transcription kit (Bio-Rad Laboratories Inc., Hercules, CA, USA). The relative expression of target genes was detected on a Bio-Rad CFX96 qRT-PCR system with the SYBR Green method (β-actin served as an internal control). The sequences of the primers are listed in Table 2.
Table 2.
Primers used for quantitative RT-PCR and chromatin immunoprecipitation (ChIP)
| Gene | Sequence (5′–3′) | Size (bp) | Temperature (°C) |
|---|---|---|---|
|
| |||
| E-cadherin | F: 5′-CGCATTGCCACATACACTCT-3′ | 252 | 55 |
| R: 5′-TTGGCTGAGGATGGTGTAAG-3′ | |||
| N-cadherin | F: 5′-AGTCAACTGCAACCGTGTCT-3′ | 337 | 55 |
| R: 5′-AGCGTTCCTGTTCCACTCAT-3′ | |||
| Vimentin | F: 5′-GAGCTGCAGGAGCTGAATG-3′ | 344 | 55 |
| R: 5′-AGGTCAAGACGTGCCAGAG-3′ | |||
| Slug | F: 5′-CCTGGTTGCTTCAAGGACAC-3′ | 395 | 55 |
| R: 5′-TCCATGCTCTTGCAGCTCTC-3′ | |||
| Twist | F: 5′-ACGAGCTGGACTCCAAGATG-3′ | 484 | 55 |
| R: 5′-GGCACGACCTCTTGAGAATG-3′ | |||
| Snail | F: 5′-GCGAGCTGCAGGACTCTAAT-3′ | 310 | 55 |
| R: 5′-GCCTCCAAGGAAGAGACTGA-3′ | |||
| Bcl-2 | F: 5′-GGATCCAGGATAACGGAGGC-3′ | 150 | 55 |
| R: 5′-CCAGATAGGCACCCAGGGT-3′ | |||
| Cyclin D1 | F: 5′-CCGAGAAGCTGTGCATCTAC-3′ | 221 | 55 |
| R: 5′-CTTCACATCTGTGGCACAGAG-3′ | |||
| p21 | F: 5′-AAGTCAGTTCCTTGTGGAGCC-3′ | 246 | 55 |
| R: 5′-GGTTCTGACGGACATCCCCA-3′ | |||
| p21 (ChIP) | F: 5′-CTGCCTCTGCTCAATAATGTTCT-3′ | 85 | 55 |
| R: 5′-GGAATTCACCTTCACACAGGC-3′ | |||
Western blot analysis
The cells were washed twice with PBS and lysed with RIPA buffer containing 1% protease inhibitors. Equal amounts of proteins were separated on 12% SDS-polyacrylamide gels and transferred onto polyvinylidene fluoride membranes, followed by blocking with 5% non-fat milk for 1 hour. The membranes were incubated with primary antibodies overnight at 4°C. The following primary antibodies were used: anti-E-cadherin (4695S; Cell Signaling Technology, Beverly, MA, USA), anti-N-cadherin (4370S; Cell Signaling Technology), anti-Vimentin (5741S; Cell Signaling Technology), anti-Slug (9585S; Cell Signaling Technology), anti-Snail (3879S; Cell Signaling Technology), anti-Twist (46702S; Cell Signaling Technology), anti-Bax (5023S; Cell Signaling Technology), anti-Bcl2 (H-108; Santa Cruz Biotechnology Inc., Dallas, TX, USA), anti-Cyclin D1 (H-108; Santa Cruz Biotechnology Inc.), and anti-GAPDH (MB001; Bioworld Technology, St Louis Park, MN, USA). After incubation with the secondary antibodies (Bioworld Technology) at 37°C for 1 hour, the bands were visualized with an enhanced chemiluminescence detection system.
RNA immunoprecipitation (RIP)
RIP assay was performed by using a Magna RIP kit (EMD Millipore, Billerica, MA, USA) according to the instructions of the manufacturer. Whole cell lysate was incubated with RIP buffer containing magnetic beads which had been conjugated with human anti-EZH2 antibody or normal mouse IgG as negative control. The immunoprecipitated RNAs were extracted, purified, and analyzed by using qRT-PCR to detect the binding of target RNAs.
Chromatin immunoprecipitation
The chromatin immunoprecipitation assay was performed in A549 cells by using a commercial kit (EMD Millipore). After cross-linking with 1% formaldehyde at 37°C for 10 minutes, the cells were harvested in sodium dodecyl sulfate lysis buffer and the DNA was shredded to fragments of 200 bp by sonication. The pre-cleared chromatin was incubated with the antibodies against EZH2, H3K27me3 or non-specific IgG overnight. Protein G-agarose beads were added and incubated at 4°C for 1 hour. After reversing the cross-links, the DNA was isolated and used for PCR. The specific primers for PCR detection of p21 gene were shown in Table 2.
Statistical analysis
All data are presented as means ± SD. Differences between groups were evaluated using the Student’s t-test, chi-squared test, or Mann–Whitney analysis. P<0.05 was considered as statistically significant.
Results
DANCR is upregulated in human NSCLC
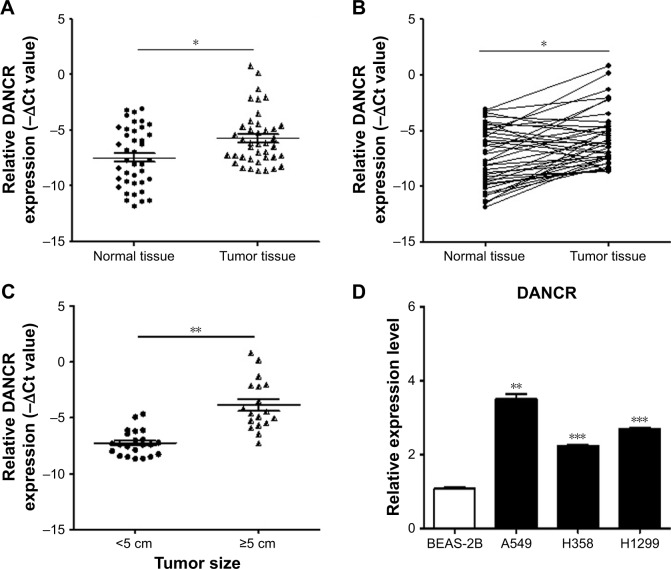
We first examined the expression of DANCR in normal and tumor tissues from NSCLC patients by using qRT-PCR. The results showed that the expression levels of DANCR in the tumor tissues of NSCLC patients were significantly higher than those in normal tissues (Figure 1A and B). The association between DANCR expression levels and clinicopathological features of NSCNLC patients was shown in Table 3. DANCR expression was significantly higher in NSCLC patients with large tumors (≥5 cm) than in those with small tumors (<5 cm) (Figure 1C). Moreover, DANCR expression levels were detected in the normal lung epithelial cell line (BEAS-2B) and NSCLC cell lines (A549, H358, and H1299). The expression levels of DANCR in A549, H358, and H1299 cells were significantly higher than that in BEAS-2B cells (Figure 1D). Taken together, these findings suggest that DANCR is upregulated in NSCLC.
Figure 1.
DANCR is upregulated in human NSCLC tumor tissues and cell lines.
Notes: (A, B) Analysis of DANCR expression in paired primary tumor and non-tumor tissues (n=40) from NSCLC patients by qRT-PCR. (C) DANCR expression levels in the tumor tissues of NSCLC patients with different tumor sizes. (D) The expression profiles of DANCR in A549, H358, H1299, and BEAS-2B cells were determined by qRT-PCR. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: NSCLC, non-small-cell lung cancer; qRT-PCR, quantitative RT-PCR.
Table 3.
The association between DANCR expression levels (−ΔCt) in tumor tissues and the clinicopathological features of non-small-cell lung cancer patients
| Features | Number | DANCR
|
Fisher’s exact test (P-value) | |
|---|---|---|---|---|
| High | Low | |||
|
| ||||
| Gender | 0.43 | |||
| Male | 26 | 19 | 7 | |
| Female | 14 | 9 | 5 | |
| Age (years) | 0.65 | |||
| <60 | 10 | 7 | 3 | |
| ≥60 | 30 | 21 | 9 | |
| Tumor size (cm) | ,0.01 | |||
| <5 | 22 | 13 | 9 | |
| ≥5 | 18 | 15 | 3 | |
| Lymphatic metastasis | 0.42 | |||
| N0 | 19 | 13 | 6 | |
| N1-3 | 21 | 15 | 6 | |
| Tumor infiltration | 0.53 | |||
| Absent | 23 | 16 | 7 | |
| Present | 17 | 12 | 5 | |
| TNM stage | 0.38 | |||
| I/II | 27 | 19 | 8 | |
| III/IV | 13 | 9 | 4 | |
DANCR knockdown inhibits the proliferation of NSCLC cells by inducing apoptosis and cell cycle arrest
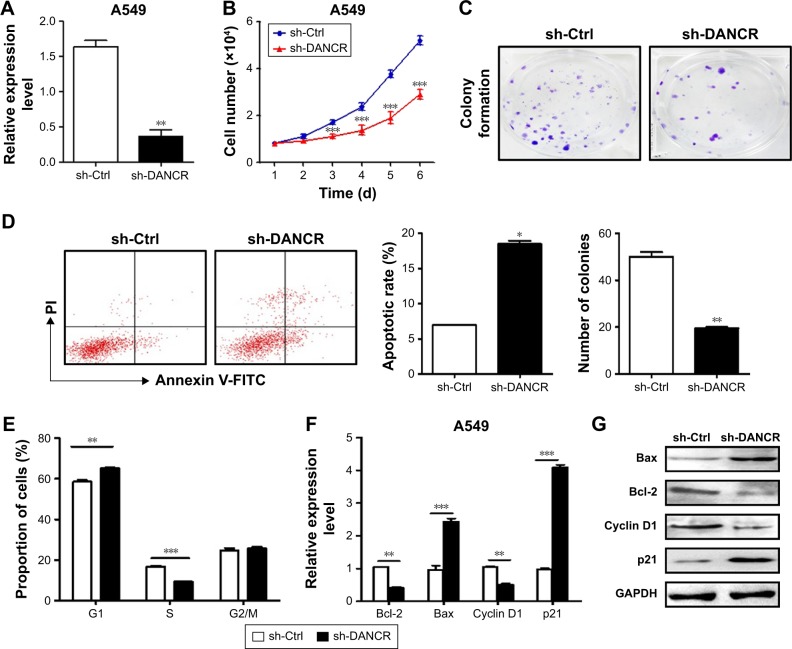
To investigate the roles of DANCR in NSCLC, we knocked down DANCR expression in NSCLC cells by using shRNA. The efficacy of gene silencing was validated (Figure 2A). We then determined the proliferation abilities of A549 cells by using cell counting and colony formation assays. The knockdown of DANCR significantly inhibited the proliferation rate of A549 cells (Figure 2B). The results of colony formation assay showed that A549 cells with DANCR knockdown formed significantly less colonies than control cells (P<0.01, Figure 2C).
Figure 2.
DANCR knockdown inhibits the proliferation of NSCLC cells.
Notes: (A) qRT-PCT analyses of DANCR expression in Ctrl and DANCR shRNA-transfected A549 cells. (B) Cell counting assay for the growth of A549 cells in Ctrl and DANCR knockdown groups. (C) Colony formation assays for A549 cells in Ctrl and DANCR knockdown groups. (D) Flow cytometric analyses of cell apoptosis in Ctrl and DANCR shRNA-transfected A549 cells. (E) Cell cycle distribution of Ctrl and DANCR shRNA-transfected A549 cells was determined by flow cytometry. (F) qRT-PCR of cell cycle- and apoptosis-related genes in Ctrl and DANCR shRNA-transfected A549 cells. (G) Western blot assays for the expression of cell cycle- and apoptosis-related proteins in Ctrl and DANCR shRNA-transfected A549 cells. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: Ctrl, control; NSCLC, non-small-cell lung cancer; PI, propidium iodide; qRT-PCR, quantitative RT-PCR.
We determined the effect of DANCR knockdown on cell apoptosis in A549 cells by flow cytometry. As shown in Figure 2D, DANCR knockdown in A549 cells signifi-cantly increased the percentage of apoptotic cells. We then evaluated the effect of DANCR knockdown on cell cycle distribution. The results of cell cycle analysis revealed that DANCR knockdown in A549 cells significantly increased the proportion of cells in G1 phase (P<0.01), while it decreased that in S phase (P<0.01) (Figure 2E). We determined the expression of apoptosis and cell cycle-related genes and proteins by using qRT-PCR and Western blot. As shown in Figure 2F and G, DANCR knockdown decreased the expression of cyclin D1 and Bcl-2 while it increased that of Bax and p21 in A549 cells. Taken together, these results indicate that DANCR knockdown could inhibit NSCLC cell proliferation by inducing cell cycle arrest and apoptosis.
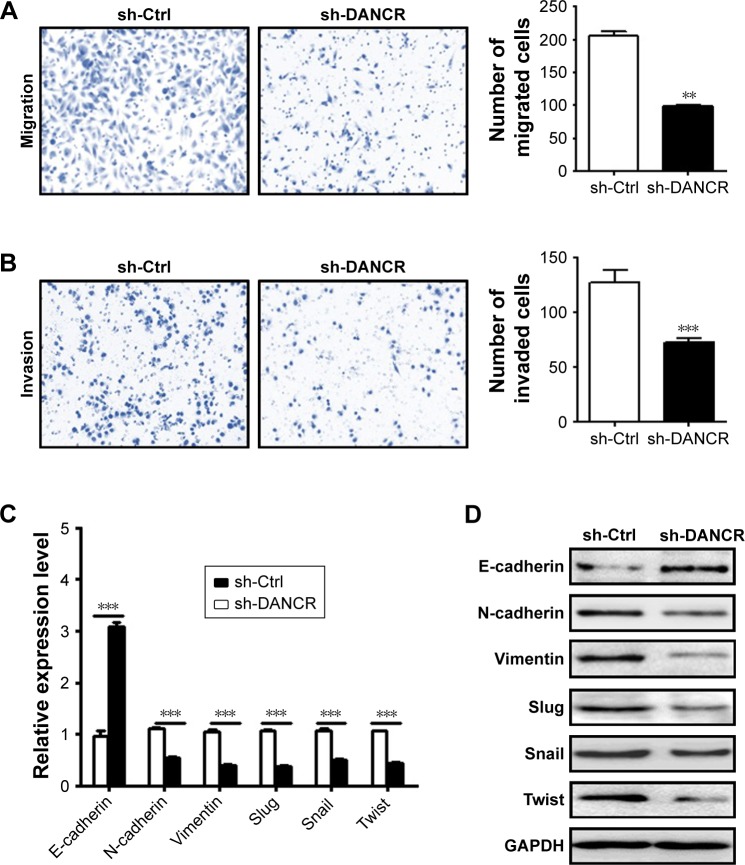
DANCR knockdown suppresses the migration and invasion of NSCLC cells
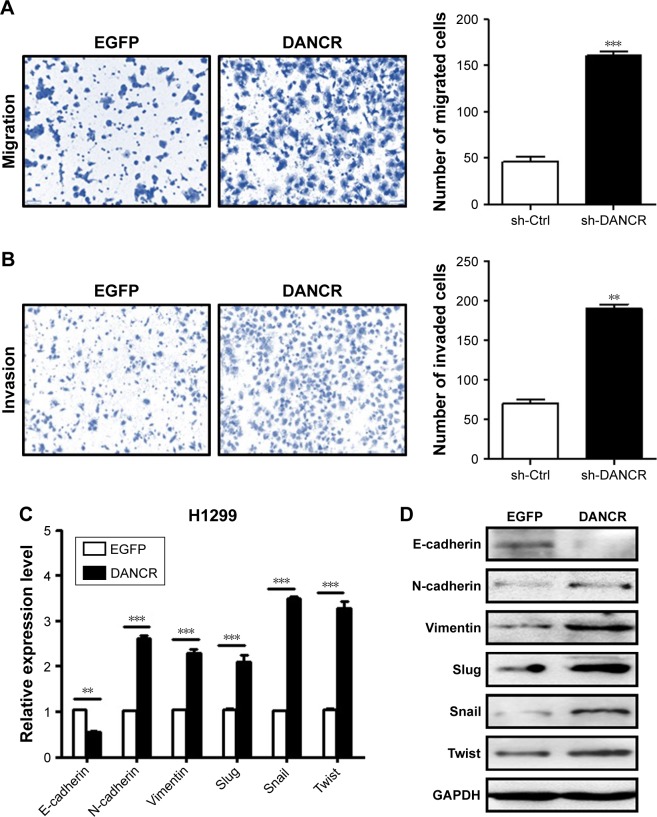
We next determined the effects of DANCR knockdown on the metastatic potential of NSCLC cells in vitro. Our transwell migration assay results showed that DANCR knockdown group had less migrated cells than that of control group (Figure 3A). In addition, the results of Matrigel invasion assays showed that DANCR knockdown also inhibited the invasion abilities of A549 cells (Figure 3B). We detected the expression of EMT-related factors in A549 cells with or without DANCR knockdown. We found that DANCR knockdown upregulated the expression of E-cadherin gene, while it downregulated that of N-cadherin, vimentin, slug, snail, and twist genes in A549 cells (Figure 3C). Western blot results also confirmed the same changes (Figure 3D). These data indicate that DANCR knockdown inhibits the metastatic potential of NSCLC cells through the suppression of EMT.
Figure 3.
DANCR knockdown inhibits the migration and invasion of NSCLC cells in vitro.
Notes: (A) Transwell migration assays were performed to investigate the migratory abilities of A549 cells with DANCR knockdown. (B) Matrigel invasion assays were performed to establish the invasive abilities of DANCR shRNA-transfected A549 cells. (C) qRT-PCR and (D) Western blot analyses of EMT-specific markers in A549 cells with DANCR knockdown. **P<0.01, ***P<0.001.
Abbreviations: Ctrl, control; EMT, epithelial–mesenchymal transition; NSCLC, non-small-cell lung cancer; qRT-PCR, quantitative RT-PCR.
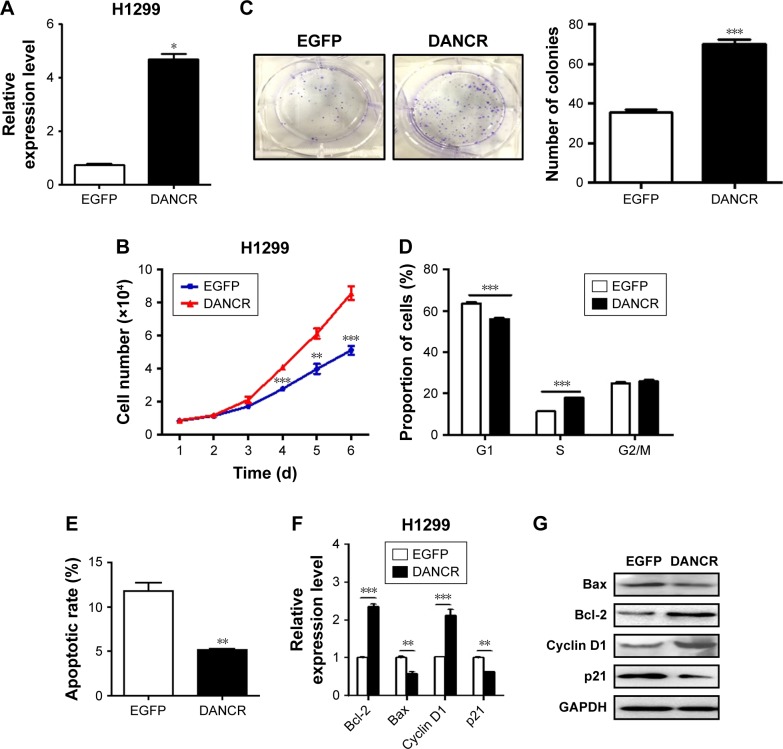
DANCR overexpression promotes the proliferation, migration, and invasion of NSCLC cells
We then determined the biological roles of DANCR overexpression in NSCLC progression. The results of cell counting and colony formation assays showed that DANCR-overexpressing H1299 cells grew faster and formed more colonies than control cells (Figure 4A–C). DANCR over-expression increased the percentage of cells at S phase, while it decreased that of cells at G1 phase (Figure 4D). The rate of apoptotic cells decreased when DANCR was overexpressed in H1299 cells (Figure 4E). DANCR overexpression also led to the downregulation of Bax and p21 genes but the upregulation of Bcl-2 and cyclin D1 genes and proteins in H1299 cells (Figure 4F and G). In addition, DANCR overexpression promoted the migration and invasion abilities of H1299 cells (Figure 5A and B). DANCR overexpression decreased the expression of E-cadherin gene while it increased that of N-cadherin, vimentin, slug, snail, and twist genes in H1299 cells (Figure 5C and D). Taken together, these findings suggest that DANCR overexpression promotes the proliferation, migration, and invasion of NSCLC cells.
Figure 4.
DANCR overexpression promotes the proliferation of NSCLC cells.
Notes: (A) qRT-PCT analyses of DANCR expression in Ctrl and DANCR-overexpressing plasmid-transfected H1299 cells. (B) Cell counting assay for the growth of H1299 cells in Ctrl and DANCR overexpression groups. (C) Colony formation assays for H1299 cells in Ctrl and DANCR overexpression groups. (D) The cell cycles of H1299 cells with DANCR overexpression were analyzed by flow cytometry. (E) Cell apoptosis in H1299 cells transfected with DANCR was determined by flow cytometry. (F) qRT-PCR analyses of Bcl-2, Bax, Cyclin D1, and p21 gene expression in DANCR-overexpressing H1299 cells. (G) Western blot analyses for Bcl-2, Bax, Cyclin D1, and p21 proteins in DANCR-overexpressing H1299 cells. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: NSCLC, non-small-cell lung cancer; qRT-PCR, quantitative RT-PCR.
Figure 5.
DANCR overexpression promotes the metastatic potential of NSCLC cells in vitro.
Notes: (A) Transwell migration assays were performed to investigate the migratory abilities of H1299 cells with DANCR overexpression. (B) Matrigel invasion assays for the invasive abilities of DANCR-overexpressing A549 cells. (C) qRT-PCR and (D) Western blot analyses of EMT-specific markers in H1299 cells with DANCR overexpression. **P<0.01, ***P<0.001.
Abbreviations: Ctrl, control; EMT, epithelial–mesenchymal transition; NSCLC, non-small-cell lung cancer; qRT-PCR, quantitative RT-PCR.
DANCR promotes NSCLC cell proliferation, migration, and invasion by epigenetically silencing p21 expression
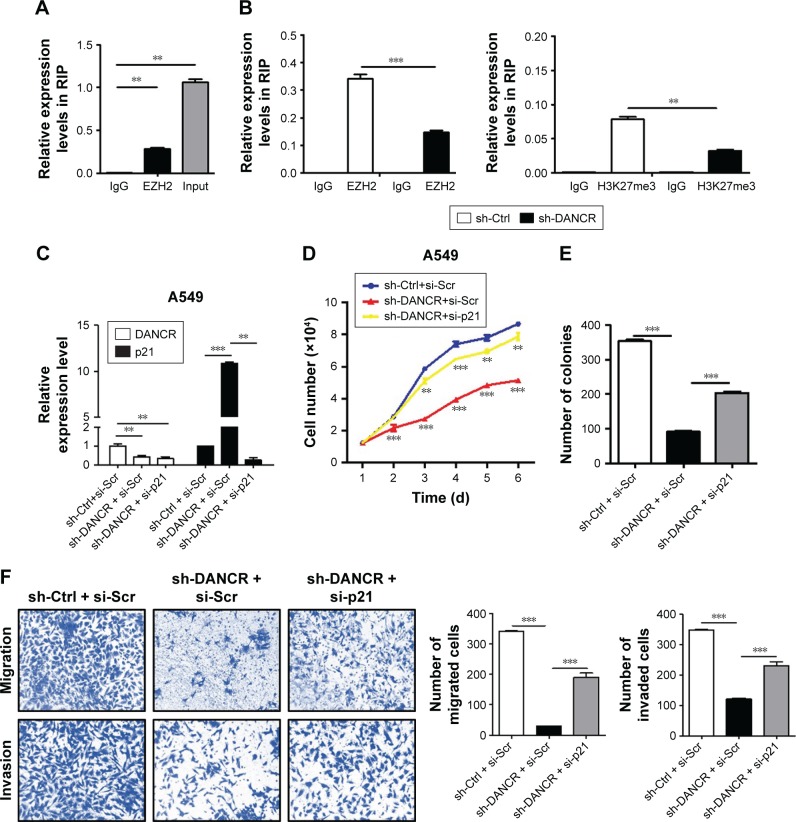
We performed RIP assay to determine the binding of DANCR to EZH2 and found that DANCR was immunoprecipitated by EZH2 antibody (Figure 6A). We further compared the relative level of EZH2 binding to p21 gene and the associated H3K27me3 level of p21 gene in control and DANCR knockdown groups. As shown in Figure 6B, DANCR knockdown decreased the amount of EZH2 binding to p21 gene and the associated H3K27me3 level of p21 gene. To demonstrate the importance of p21 regulation by DANCR in NSCLC progression, we then co-transfected p21 siRNA with DANCR shRNA into H1299 cells. As expected, DANCR knockdown increased p21 expression while p21 siRNA co-transfection reversed this effect (Figure 6C). The results of cell counting and colony formation assays showed that A549 cells with both DANCR knockdown and p21 knockdown grew faster and formed more colonies than cells with DANCR knockdown alone (Figure 6D and E). In addition, the inhibition of cell migration and invasion in A549 cells by DANCR knockdown was also reversed by p21 siRNA co-transfection (Figure 6F). These results indicate that DANCR epigenetically silences p21 expression to promote the proliferation, migration, and invasion of NSCLC cells.
Figure 6.
DANCR epigenetically silences p21 expression to promote NSCLC cell proliferation, migration, and invasion.
Notes: (A) RIP assays for the binding of DANCR and EZH2. (B) ChIP assays for the binding of EZH2 to p21 gene and the levels of H3K27me3 in p21 gene in Ctrl and DANCR knockdown groups. (C) qRT-PCR analyses of DANCR and p21 expression in A549 cells from Ctrl, DANCR knockdown alone, and p21 siRNA co-transfection groups. (D) Cell counting and (E) colony formation assays for A549 cells in Ctrl, DANCR knockdown alone, and p21 siRNA co-transfection groups. (F) Transwell migration assay and Matrigel invasion assay for A549 cells in Ctrl, DANCR knockdown alone, and p21 siRNA co-transfection groups. **P<0.01, ***P<0.001.
Abbreviations: ChIP, chromatin immunoprecipitation; Ctrl, control; NSCLC, non-small-cell lung cancer; RIP, RNA immunoprecipitation; qRT-PCR, quantitative RT-PCR.
Discussion
In the past decade, various lncRNAs have been identified to play critical roles in the pathogenesis of cancer.15–17 Several lncRNAs have been suggested as independent risk factors of prognosis in NSCLC patients. In this study, we showed that DANCR was frequently upregulated in NSCLC patients. We also found that the elevated DANCR expression in NSCLC was positively associated with tumor size. We further investigated the function of DANCR in NSCLC and reported that DANCR knockdown inhibited, while DANCR overexpression promoted, the proliferation, migration, and invasion of NSCLC cells in vitro, indicating that DANCR plays oncogenic roles in NSCLC.
Our data suggest that the upregulation of DANCR may contribute to the advanced progression of NSCLC. Similar findings have been reported in other cancers. Yuan et al demonstrated that DANCR is overexpressed in stem-like hepatocellular carcinoma (HCC) cells. DANCR increases the stemness features of HCC cells to promote tumorigenesis and metastasis through the upregulation of CTNNB1 expression.18 In esophageal cancer, DANCR promotes cell proliferation, migration, invasion, and resistance to apoptosis.10 Pan et al demonstrated that DANCR expression is elevated in the serum of gastric cancer patients compared to healthy controls. DANCR knockdown inhibits gastric cancer cell proliferation, migration, and invasion in vitro and suppresses gastric cancer growth in vivo.19 Moreover, the elevated expression of DANCR is associated with advanced tumor progression and poor prognosis in colorectal cancer, and the silencing of DANCR promotes apoptosis and inhibits tumor growth.20 These findings suggest that DANCR is a potential diagnostic biomarker and therapeutic target in various cancers.
Previous studies have shown that lncRNAs are critical regulators of p21 expression. Nie et al demonstrated that ANRIL promotes NSCLC cell proliferation and inhibits apoptosis by silencing KLF2 and p21 expression.21 Zhang et al suggested that TUG1 associates with PRC2 to epigenetically repress the expression of CDKIs including p21, thus contributing to the regulation of gastric cancer cell proliferation.22 Qi et al demonstrated that AGAP2-AS1 interacts with LSD1 and EZH2 to suppress p21 and E-cadherin transcription, promoting gastric cancer cell proliferation, migration, and invasion in vitro and tumor growth in vivo.23 Lv et al demonstrated that SBF2-AS1 is upregulated in NSCLC and promotes the proliferation of NSCLC cells. SBF2-AS1 interacts with SUZ12 to increase trimethylation of H3K27 at the promoter region of p21.24 SNHG20 promotes NSCLC cell proliferation and migration by epigenetically silencing p21 expression.25 Recently, Lu et al demonstrated that DANCR could inhibit the expression of p21 and that the inhibitory effects of DANCR loss on cell proliferation could be partially rescued by p21 silencing.26 DANCR has been shown to promote invasion of prostate cancer cells through epigenetically silencing the expression of TIMP2/3.27 In this study, we showed that DANCR knockdown decreased, while DANCR overexpression increased, p21 expression in NSCLC cells. We identified p21 as a novel target gene for DANCR-regulated and EZH2-mediated epigenetic silencing.
We further showed that p21 knockdown could reverse the inhibitory effects of DANCR knockdown on NSCLC cell proliferation, migration, and invasion. Altogether, these data suggest that DANCR facilitates EZH2-mediated trimethylation at the H3K27 site of p21 promoter, consequently inhibiting p21 expression and promoting the growth and metastasis of NSCLC cells in vitro.
DANCR has also been suggested to function as an miRNA sponge. Jiang et al showed that DANCR upregulates the expression of receptor tyrosine kinase AXL by competitively binding to miR-33a-5p and activates PI3K/Akt signaling pathway, promoting osteosarcoma cell proliferation, migration, and invasion in vitro as well as xenograft tumor growth and lung metastasis in vivo.11 In addition, DANCR promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma.28 Moreover, DANCR contributes to NSCLC progression by sponging miR-496,29 miR-758-3p,30 and miR-216a.31 Whether DANCR could regulate p21 expression by sequestering miRNAs warrants further investigation in future studies.
In conclusion, we demonstrated in this study that DANCR promotes NSCLC cell proliferation, migration, and invasion, partly at least, via EZH2-mediated silencing of p21. The present study adds new evidence for the role of DANCR in NSCLC progression and provides a potential marker for NSCLC diagnosis and therapy.
Acknowledgments
The present study was supported by the Guidance Project of Zhenjiang Municipal Science and Technology Bureau (FZ2017038), the Guidance Project of Nantong Municipal Science and Technology Bureau (HS13922), and the Foundation of Clinical Science and Technology of Wuxi (MS201609).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Chheang S, Brown K. Lung cancer staging: clinical and radiologic perspectives. Semin Intervent Radiol. 2013;30(2):099–113. doi: 10.1055/s-0033-1342950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang Q, Ni Z, Cheng Z, Xu J, Yu H, Yin P. Three circulating long non-coding RNAs act as biomarkers for predicting NSCLC. Cell Physiol Biochem. 2015;37(3):1002–1009. doi: 10.1159/000430226. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Wang R, Zhang K, Chen LB. Long non-coding RNAs in non-small cell lung cancer as biomarkers and therapeutic targets. J Cell Mol Med. 2014;18(12):2425–2436. doi: 10.1111/jcmm.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z, Li JL, Lin S, et al. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J Clin Invest. 2016;126(6):2267–2279. doi: 10.1172/JCI85250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang EB, Yin DD, Sun M, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:e1243. doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li K, Sun D, Gou Q, et al. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018;420:80–90. doi: 10.1016/j.canlet.2018.01.060. [DOI] [PubMed] [Google Scholar]
- 8.Hao YP, Qiu JH, Zhang DB, Yu CG, Cg Y. Long non-coding RNA DANCR, a prognostic indicator, promotes cell growth and tumorigenicity in gastric cancer. Tumour Biol. 2017;39(6):101042831769979. doi: 10.1177/1010428317699798. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Zhang M, Liang L, Li J, Chen YX. Overexpression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2015;8(9):11480–11484. [PMC free article] [PubMed] [Google Scholar]
- 10.Shi H, Shi J, Zhang Y, et al. Long non-coding RNA DANCR promotes cell proliferation, migration, invasion and resistance to apoptosis in esophageal cancer. J Thorac Dis. 2018;10(5):2573–2582. doi: 10.21037/jtd.2018.04.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang N, Wang X, Xie X, et al. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46–55. doi: 10.1016/j.canlet.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Ma Y, Zhou G, Li M, et al. Long noncoding RNA DANCR mediates cisplatin resistance in glioma cells via activating AXL/PI3K/Akt/NF-κB signaling pathway. Neurochem Int. 2018;118:233–241. doi: 10.1016/j.neuint.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Sun X, Chen S, et al. Long noncoding RNA DANCR regulates miR-1305-Smad 4 axis to promote chondrogenic differentiation of human synovium-derived mesenchymal stem cells. Biosci Rep. 2017;37(4):BSR20170347. doi: 10.1042/BSR20170347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Lu Z, Wang N, et al. Long noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 sponging. Exp Mol Med. 2018;50(5):57. doi: 10.1038/s12276-018-0082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han L, Zhang EB, Yin DD, et al. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis. 2015;6:e1665. doi: 10.1038/cddis.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nie W, Ge HJ, Yang XQ, et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371(1):99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 17.Yu T, Zhao Y, Hu Z, et al. MetaLnc9 facilitates lung cancer metastasis via a PGK1-activated AKT/mTOR pathway. Cancer Res. 2017;77(21):5782–5794. doi: 10.1158/0008-5472.CAN-17-0671. [DOI] [PubMed] [Google Scholar]
- 18.Yuan SX, Wang J, Yang F, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63(2):499–511. doi: 10.1002/hep.27893. [DOI] [PubMed] [Google Scholar]
- 19.Pan L, Liang W, Gu J, et al. Long noncoding RNA DANCR is activated by SALL4 and promotes the proliferation and invasion of gastric cancer cells. Oncotarget. 2018;9(2):1915–1930. doi: 10.18632/oncotarget.23019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang XJ, Zhao JJ, Chen WJ, Zhang GG, Wang W, Tao HC. Silencing long non-coding RNA, differentiation antagonizing non-protein coding RNA promotes apoptosis and inhibits tumor growth in colon cancer. Oncol Lett. 2018;16(3):2865–2872. doi: 10.3892/ol.2018.9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nie FQ, Sun M, Yang JS, et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14(1):268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 22.Zhang E, He X, Yin D, et al. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016;7:e2109. doi: 10.1038/cddis.2015.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi F, Liu X, Wu H, et al. Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer. J Hematol Oncol. 2017;10(1):48. doi: 10.1186/s13045-017-0420-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lv J, Qiu M, Xia W, et al. High expression of long non-coding RNA SBF2-AS1 promotes proliferation in non-small cell lung cancer. J Exp Clin Cancer Res. 2016;35:75. doi: 10.1186/s13046-016-0352-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, Chen X, Chen P, et al. Long non-coding RNA SNHG20 promotes non-small cell lung cancer cell proliferation and migration by epigenetically silencing of P21 expression. Cell Death Dis. 2017;8(10):e3092. doi: 10.1038/cddis.2017.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Y, Hu Z, Mangala LS, et al. MYC targeted long noncoding RNA DANCR promotes Cancer in part by reducing p21 levels. Cancer Res. 2018;78(1):64–74. doi: 10.1158/0008-5472.CAN-17-0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia J, Li F, Tang XS, et al. Long noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3. Oncotarget. 2016;7(25):37868–37881. doi: 10.18632/oncotarget.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Zeng X, Wang N, et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17(1):89. doi: 10.1186/s12943-018-0837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu QC, Rui ZH, Guo ZL, Xie W, Shan S, Ren T. LncRNA-DANCR contributes to lung adenocarcinoma progression by sponging miR-496 to modulate mTOR expression. J Cell Mol Med. 2018;22(3):1527–1537. doi: 10.1111/jcmm.13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhen Q, Gao LN, Wang RF, et al. LncRNA DANCR promotes lung cancer by sequestering miR-216a. Cancer Control. 2018;25(1):107327481876984. doi: 10.1177/1073274818769849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Jiang M. The long non-coding RNA-DANCR exerts oncogenic functions in non-small cell lung cancer via miR-758-3p. Biomed Pharmacother. 2018;103:94–100. doi: 10.1016/j.biopha.2018.03.053. [DOI] [PubMed] [Google Scholar]