Abstract
Purpose
Pseudomonas aeruginosa is the leading cause of contact lens–associated bacterial keratitis. Secreted bacterial proteases have a key role in keratitis, including the P. aeruginosa small protease (PASP), a proven corneal virulence factor. We investigated the mechanism of PASP and its importance to corneal toxicity.
Methods
PASP, a serine protease, was tested for activity on various substrates. The catalytic triad of PASP was sought by bioinformatic analysis and site-directed mutagenesis. All mutant constructs were expressed in a P. aeruginosa PASP-deficient strain; the resulting proteins were purified using ion-exchange, gel filtration, or affinity chromatography; and the proteolytic activity was assessed by gelatin zymography and a fluorometric assay. The purified PASP proteins with single amino acid changes were injected into rabbit corneas to determine their pathological effects.
Results
PASP substrates were cleaved at arginine or lysine residues. Alanine substitution of PASP residues Asp-29, His-34, or Ser-47 eliminated protease activity, whereas PASP with substitution for Ser-59 (control) retained activity. Computer modeling and Western blot analysis indicated that formation of a catalytic triad required dimer formation, and zymography demonstrated the protease activity of the homodimer, but not the monomer. PASP with the Ser-47 mutation, but not with the control mutation, lacked corneal toxicity, indicating the importance of protease activity.
Conclusions
PASP is a secreted serine protease that can cleave proteins at arginine or lysine residues and PASP activity requires dimer or larger aggregates to create a functional active site. Most importantly, proteolytic PASP molecules demonstrated highly significant toxicity for the rabbit cornea.
Keywords: catalytic triad, active site, protease, P. aeruginosa, keratitis
Pseudomonas aeruginosa is an opportunistic human pathogen that also is pathologic for animals and plants.1 In humans, this gram-negative bacterium can cause infection at any compromised tissue or body site and it can lead to severe morbidity in immunocompromised patients.2 P. aeruginosa is a common nosocomial pathogen, accounting for 10% of all hospital-acquired infections.3 It also is the leading cause of contact lens–associated bacterial keratitis, causing a sight-threatening disease of the human eye.4,5 Prompt antibiotic therapy is crucial to limit corneal damage and subsequent scarring to preserve corneal transparency. However, P. aeruginosa has the propensity to develop antibiotic resistance.6 Furthermore, tissue damage caused by P. aeruginosa can continue after antibiotics kill the bacteria because this bacterium secretes numerous virulence factors, including proteases that continue to damage the tissue.7–14
Among the well-studied Pseudomonas-secreted proteases are four metalloproteases whose role in keratitis is described as nonessential.15–19 P. aeruginosa also secretes four serine proteases, including protease IV and P. aeruginosa small protease (PASP), that have been demonstrated as important corneal virulence factors.9,12,20–23
PASP has an important role in the pathogenesis of keratitis and is required to obtain full virulence.12–14 The PASP-deficient mutant of P. aeruginosa produces significantly reduced virulence in the rabbit or mouse cornea compared to the parental or rescued strain with restored PASP production.14 PASP also can degrade corneal structural proteins, such as types I and IV collagen, contributing to corneal ulcers.13 Host defense molecules, such as complement C3 and antimicrobial peptide LL-37, are cleaved by PASP, as were a range of proteins in the rabbit tear film.14 Tosyl-L-lysine chloromethylketone (TLCK), a serine protease inhibitor, can inhibit PASP activity in a dose-dependent manner.13 PASP has little sequence homology with other known proteases, but there is a sequence similarity between PASP and Escherichia coli YceI protein.12 E. coli YceI protein is a periplasmic protein whose crystal structure has been determined.24
Serine proteases are the best-known class of proteases (E.C. 3.4.21), comprising approximately one-third of all known proteases.25 Serine proteases use an Asp-His-Ser triad in the active site.26 The triad of amino acids can be found scattered in the primary sequence of a serine protease, but are in close proximity in the tertiary structure.27
In our study, the catalytic residues comprising the active site of PASP have been identified and the substrate specificity and kinetics of its catalytic activity also have been studied. The findings demonstrated that a single amino acid change in the catalytic triad can eliminate protease activity and corneal toxicity. This study also showed that PASP undergoes dimerization to achieve its enzymatic activity.
Methods
Bacterial Strains, Plasmids, and Growth Conditions
P. aeruginosa strain PA103-29 was kindly provided by Dennis E. Ohman (Medical College of Virginia Campus of Virginia Commonwealth University, Richmond, VA, USA). E. coli JM109 competent cells were purchased from Promega (Madison, WI, USA). E. coli strains harboring plasmids were grown and maintained on Luria-Bertani (LB; Difco; BD Biosciences, Sparks, MD, USA) agar plates or in LB broth with appropriate antibiotics. Pseudomonas cultures were grown at 37°C with agitation (180–200 rpm) in dialyzed tryptic soy broth (TSB; Difco) with addition of 60 mM monosodium glutamate, 1 mM MgSO4, and 1% glycerol as described by Marquart et al.12 Bacteria were removed by centrifugation at 5000g for 30 minutes. Culture supernatants were filtered through a 0.22-μm filter and concentrated using an ultrafiltration cell with a 10-kDa molecular weight cutoff filter (Amicon; Millipore, Billerica, MA, USA).
Site-Directed Mutagenesis
To generate targeted mutations in the pasP gene, a PCR-mediated method of extending overlapping gene segments was used. A pair of master flanking primers of the pasP gene and two internal primers with overlapping sequences and mutations of interest were used in this method (Table 1). The first round PCR was to amplify two gene segments with overlapping sequences using the pair of one flanking and one internal primer. PCR products were analyzed by agarose gel electrophoresis and reaction mixtures were diluted 1/10 and combined to use as template DNA for the second round PCR. In the second round PCR (overlap extension), the two flanking primers and 1 μL of the template DNA were used in a 25-μL reaction. The PCR reaction initially underwent 10 cycles with the annealing temperature based on the calculated melting temperature (Tm) of the overlapping sequence (higher than the Tm of the flanking primers), followed by 20 cycles with the annealing temperature based on the Tm of the flanking primers. The PCR products of the mutated pasP gene were cloned into plasmid pUCP20.14 To make the constructs for the recombinant PASP protein with a C-terminal histidine tag, the wild-type (WT) or mutated pasP gene was PCR-amplified using a reverse primer with the codons for six histidines incorporated. All constructs were confirmed by sequencing in both directions (SeqWright, Inc., Houston, TX, USA).
Table 1.
Primers Used for Cloning of PASP and its Site-Directed Mutants
Purification of the WT and Mutant PASP Proteins
Since E. coli and P. putida failed to secrete the active PASP protein (unpublished findings), we chose P. aeruginosa PA103-29 PASP(-), a mutant created by allelic replacement of the pasP gene, as the expression host.14 P. aeruginosa PA103-29 PASP(-) was chemically transformed with a construct of the WT or mutated pasP gene on the plasmid pUCP20 using the method described by Traidej et al.21 Plasmid-containing PA103-29 PASP(-) was grown at 37°C in 4 liters of dialyzed TSB containing 100 μg/mL carbenicillin for 40 hours. Culture supernatants were collected by centrifugation and concentrated. The concentrates then were dialyzed overnight against 50 mM Tris-HCl buffer (pH 8.0) and applied to a pre-packed Q sepharose column (HiTrap Q XL; GE Healthcare Life Sciences, Piscataway, NJ, USA). Fractions were eluted with a pH gradient generated by addition of 50 mM Tris-HCl buffer (pH 6.0). Further purification was achieved by gel filtration chromatography using a sephacryl S-200 column. Recombinant PASP proteins containing histidine tags were purified from the supernatants of Pseudomonas cultures by affinity chromatography (Talon resin; BD-Clontech).
Thin Layer Chromatography
Purified P. aeruginosa elastase B (LasB) and alkaline protease (AP) were purchased from Elastin Products Company, Inc. (Owensville, MO, USA) and United States Biochemical (Cleveland, OH, USA), respectively. Elastase A (LasA) was a kind gift from Dennis E. Ohman. Native protease IV was purified as described previously.12 Poly-L-arginine (1 mg/mL; Sigma-Aldrich Corp., St. Louis, MO, USA), and poly-L-lysine (4 mg/mL; Sigma-Aldrich Corp.) were mixed 1:1 (vol/vol) with either 10 μg rPASP or 0.1 μg protease IV, AP, LasA, or LasB at 37°C for 8 hours. Arginine (4 mg/mL; Sigma-Aldrich Corp.) and lysine (4 mg/mL; Sigma-Aldrich Corp.) were incubated with equal volumes of dH2O. An aliquot (7 μL) of each sample was spotted onto a silica gel-coated plate (PE SIL G; Whatman Ltd., Kent, England). The plate was developed in a solvent consisting of propanol (20 mL), acetic acid (20 mL), and dH2O (10 mL). The plate then was visualized by spraying 0.2% ninhydrin (Sigma-Aldrich Corp.) solution made in n-butanol.
SDS-PAGE, Western Blotting, and Mass Spectrometry
Polyacrylamide gels were prepared according to the method described by Sambrook et al.28 Western blot analysis was performed as described previously.13 For mass spectrometry, protein bands on SDS-PAGE were excised and analyzed at the University of Texas Medical Branch Biomolecular Resource Facility (Galveston, TX, USA), using the method of matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF).
Zymography
Protein samples mixed with a nonreducing loading buffer were electrophoresed through a 10% SDS-polyacrylamide gel containing 0.1% gelatin at 4°C and 20 mA (constant current setting). Subsequently, the gel was washed twice for 15 minutes each in a 2.5% Triton X-100 solution to remove SDS. The gel then was incubated in a reaction buffer consisting of 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 10 mM CaCl2, and 1 μM ZnCl2 at 37°C overnight. After incubation, the gel was fixed and stained with Coomassie blue.
Colorimetric and Fluorometric Assays of Protease Activity
A library of 29 chromogenic substrates (Sigma-Aldrich Corp.) was screened for their susceptibility to PASP by incubating the substrate with active rPASP (10 μg in 10 μL) at 37°C overnight. Positive reactions form a yellow color and can be measured spectrophotometrically at 410 nm. Dye-quenched (DQ)-gelatin, fluorescein conjugate (Molecular Probes, Eugene, OR, USA) was chosen as the substrate for PASP because it is highly sensitive and has been previously developed into a fluorometric assay.23 DQ-gelatin was dissolved in water at 1 mg/mL as a stock solution. To calculate the molar concentration of the substrate, a molecular weight of 100,000 g/mol for gelatin was used.29 All dilutions of the substrate and enzyme were made in a reaction buffer (50 mM Tris-HCl pH 7.6, 150 mM NaCl, and 5 mM CaCl2) and the final reaction volume was 100 μL. For end-point assays, the WT or mutant recombinant PASP protein (250 ng each) was incubated with 0.1 μM DQ-gelatin in triplicate in a 96-well microtiter plate at 37°C for 24 hours. Fluorescence was measured using a multimode microplate reader (FLUOstar Omega; BMG Labtech, Ortenberg, Germany) with excitation at 485 nm and emission at 520 nm. Background values derived from the enzyme-free (negative) controls were subtracted from the fluorescence measurements.
Kinetic analysis was performed on the WT enzyme and the S59A mutant because the fluorescence signal for the mutant D29A, H34A, or S47A was undetectable. Each enzyme variant (250 ng) was tested against a range of DQ-gelatin concentrations (0.025–0.4 μM) in triplicate. Fluorescence was monitored every 10 minutes for 2 hours at 37°C. The changes in relative fluorescence unit per minute (ΔRFU/min) versus the substrate concentrations (μM) were plotted and fitted by nonlinear regression to the Michaelis-Menten equation using GraphPad Prism 6 software (MathWorks, Natick, MA, USA). Km and Vmax were calculated by the software.
Injection of Mutated PASP Protein into Rabbit Corneas and Histologic Analysis
New Zealand white rabbits were used and maintained according to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Animal protocols were approved and monitored by the Animal Use and Care Committee of the University of Mississippi Medical Center. Recombinant PASP derived from cloned pasP gene with a site-directed mutation at Ser-47 or Ser-59 was purified and intrastromally injected into rabbit corneas (n = 4 per group). Slit-lamp examination (SLE) was performed after 24 hours and eyes were photographed.13 Corneas were subsequently harvested, fixed, and processed by Excalibur Pathology, Inc. (Norman, OK, USA). The tissue sections were stained with hematoxylin and eosin. Each slide was examined microscopically and photographed under low and high magnifications.13
Bioinformatics
The protein sequence and crystal structure of E. coli YceI were obtained from the RCSB Protein Data Base (PDB # 1y0g; available in the public domain at http://www.rcsb.org/pdb/explore/explore.do?structureId=1y0g). Pairwise sequence alignment between PASP and YceI was performed using the ClustalW2 program (available in the public domain at http://www.ebi.ac.uk/Tools/msa/clustalw2/). Modeling of PASP was performed using the SWISS-MODEL Protein Modeling Server (available in the public domain at http:/www.expasy.org/swissmod/SWISS-MODEL.html) with YceI as the template. Catalytic site prediction was performed by searching the Catalytic Site Atlas (available in the public domain at http://www.ebi.ac.uk/thornton-srv/databases/CSA/) using YceI as a query structure.
Statistical Analysis
The methods used have been described previously.13 Mean and SEM were calculated with commercial statistical analysis software (SAS, Inc., Cary, NC). For comparison of SLE scores, nonparametric 1-way ANOVA (Kruskal-Wallis test) was performed. P ≤ 0.05 was considered significant.
Results
Substrate Specificity of PASP
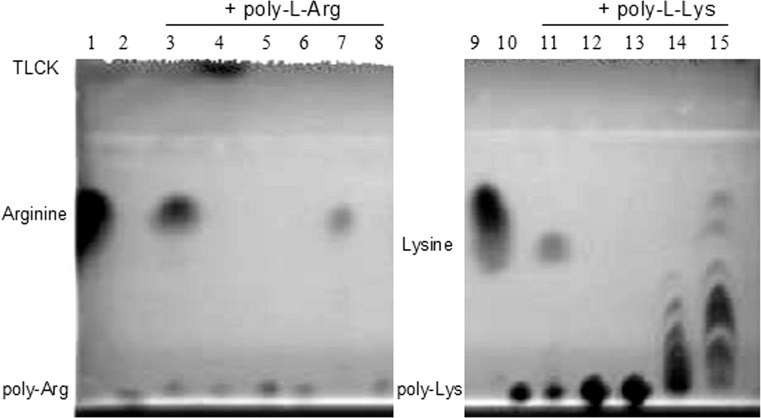
To determine the substrate specificity of PASP, 29 chromogenic substrates were screened for PASP susceptibility by incubating each substrate with active rPASP (10 μg) at 37°C overnight (Table 2). The release of the yellow chromophore pNA (para-nitroaniline) was defined as positive. The four positive substrates, L-lysine p-nitroanilide dihydrobromide, L-arginine p-nitroanilide dihydrochloride, Gly-Arg p-nitroanilide dihydrochloride, and Tosyl-Gly-Pro-Lys p-nitroanilide, have either lysine or arginine at the cleavage (P1) position. Additionally, poly-L-arginine and poly-L-lysine peptides were tested with rPASP and other Pseudomonas proteases by thin layer chromatography (Fig. 1). The cleavage of poly-L-arginine or poly-L-lysine by PASP was indicated by appearance of arginine or lysine monomers or intermediate products (lanes 3 and 11). The cleavage of poly-L-arginine by PASP was inhibited by TLCK (lane 4). Alkaline protease also was found to cleave both polypeptides (lanes 7 and 14) and protease IV cleaved poly-L-lysine (lane 15) as reported previously.20
Table 2.
Chromogenic Substrates Tested for Susceptibility to PASP

Figure 1.
Degradation of poly-L-arginine and poly-L-lysine by Pseudomonas proteases. Poly-L-arginine (lanes 3–8) or poly-L-lysine (lanes 11–15) was incubated with rPASP (10 μg/10 μL; lanes 3 and 11), rPASP + 20 mM TLCK (lane 4), 0.1 μg LasA (lanes 5 and 12), LasB (lanes 6 and 13), AP (lanes 7 and 14), or protease IV (lanes 8 and 15) at 37°C for 8 hours. Lane 1, single arginine alone; lane 2, poly-L-arginine alone; lane 9, single lysine alone; and lane 10, poly-L-lysine alone. An aliquot of the reaction was spotted on a thin layer chromatography plate and developed in a propanol: acetic acid: dH2O (2:2:1) solvent. The plate was visualized by ninhydrin staining.
Prediction of the Catalytic Triad of PASP as a Serine Protease
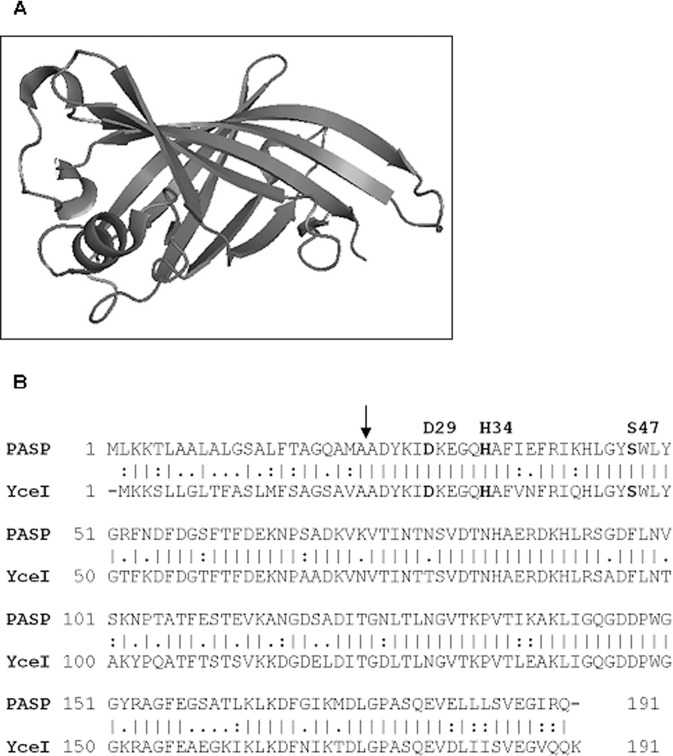
Because PASP activity was inhibited by TLCK,13 PASP was postulated to be a serine protease with the classic Asp-His-Ser catalytic triad. The three-dimensional (3-D) structure of E. coli YceI protein (Fig. 2A; PDB # 1y0g), a PASP homolog, is known and was used as a query structure. A catalytic site prediction was performed using the Catalytic Site Atlas, which documents catalytic residues in enzymes of known 3-D structures. The catalytic residues of the porcine trypsin molecule (in complex with soybean trypsin inhibitor; PDB # 1avw) matched to YceI. Using the structure and triad information from YceI and porcine trypsin, respectively, a catalytic triad for PASP was postulated (Fig. 2B). Based on the postulated catalytic residues of PASP, alanine was substituted via site-directed mutagenesis into the PASP sequence at amino acids Asp-29, His-34, and Ser-47, as well as a control residue Ser-59.
Figure 2.
The prediction of the PASP catalytic triad. (A) The structure of E. coli YceI protein (PDB ID: 1y0g), pictured as a ribbon diagram showing YceI as an eight-stranded antiparallel β-barrel. (B) Alignment of the amino acid sequences of PASP and YceI. The proposed triad residues are listed above the PASP sequence. The vertical lines (|) represent identical amino acids whereas the colons (:) and the periods (.) represent strongly and weakly conserved amino acids, respectively. The arrow indicates where the cleavage of the type II secretory signal peptide of PASP occurs.
Effect of Mutations at the Proposed Catalytic Triad on PASP Enzymatic Activity
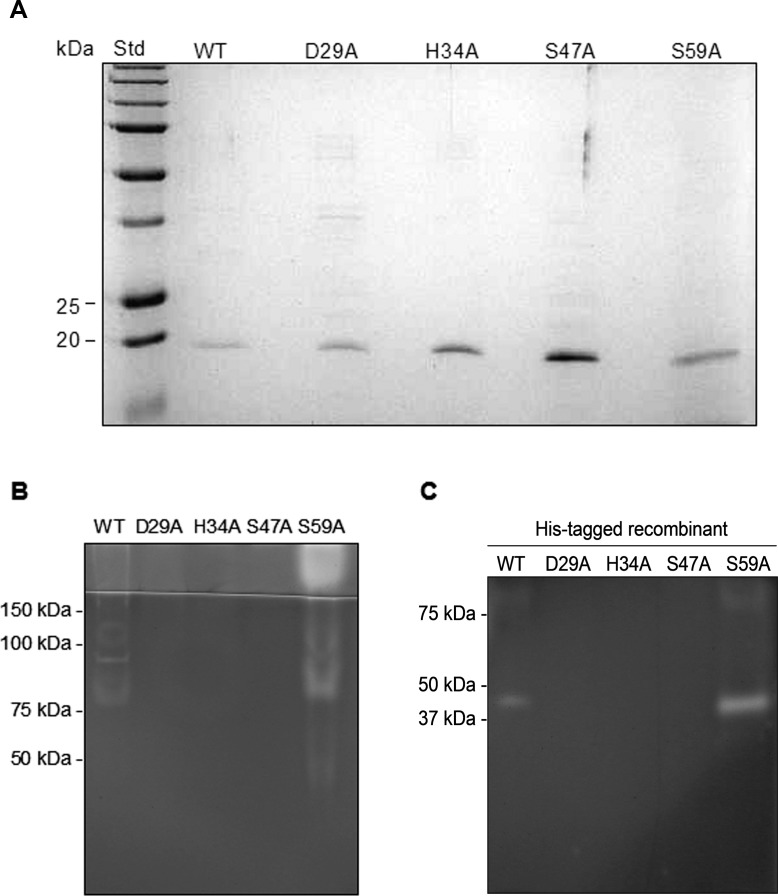
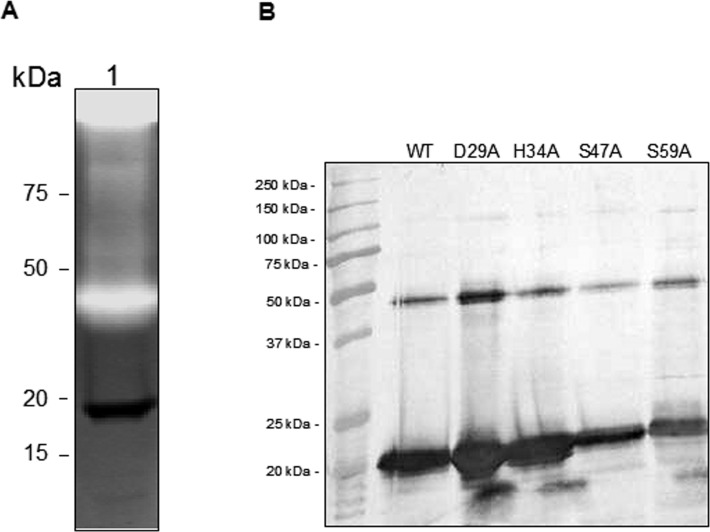
For purified native PASP and its alanine substituted mutants, each had a band of approximately 19 kDa on reduced SDS-PAGE (Fig. 3A). The identity of the purified proteins as PASP was confirmed by Western blot analysis and mass spectrometry (data not shown). When subjected to gelatin zymography, only the WT protein and the mutant S59A demonstrated protease activity as evidenced by the protease bands at approximately 80 kDa and in the stacking gel area (Fig. 3B). The protease seen at 80 kDa and in the stacking gel could be due to aggregates known to form under zymography conditions.12 An alanine substitution of PASP at Asp-29, His-34, or Ser-47, which comprises the postulated catalytic triad, abolished the enzymatic activity (Fig. 3B). Site-directed mutations also were prepared in the recombinant PASP proteins containing a C-terminal 6-His tag. On the zymogram, the recombinant PASP molecule with a mutation at Asp-29, His-34, or Ser-47 lacked protease activity, but the rPASP protein with the S59A mutation or the WT protein retained protease activity (Fig. 3C).
Figure 3.
Effects of mutations of the proposed catalytic triad on enzyme activity. (A) SDS-PAGE analysis of the purified native PASP proteins. The WT and mutant PASP proteins were purified from the culture supernatants by ion-exchange and gel filtration chromatography. An aliquot of each purified protein (approximately 100 ng) was electrophoresed through a 12% SDS-polyacrylamide gel. All five variants of PASP contained a 19-kDa band by Coomassie staining under reducing conditions. (B) Gelatin zymography of the proteins. The same amount of such protein was subjected to gelatin zymography. The PASP proteins with mutations at the proposed catalytic triad (Asp-29, His-34, and Ser-47) showed no gelatinolytic activity on the zymogram. The WT and the S59A mutant were active with activities mainly located at approximately 80 kDa and in the stacking gel area. WT, wild-type PASP; D29A, H34A, S47A, and S59A, PASP with the alanine substitution at Asp-29; His-34, Ser-47, or Ser-59, respectively. (C) Gelatin zymography of the His-tagged recombinant proteins. The WT and the S59A showed activity at approximately 40 kDa whereas D29A, H34A, and S47A demonstrated to be enzymatically inactive.
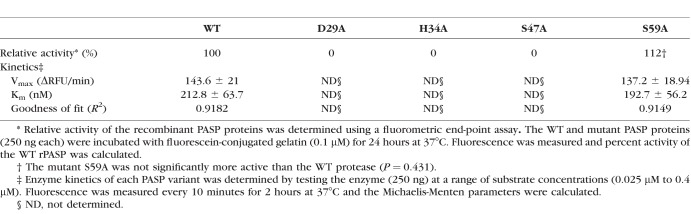
Quantitative analysis of the effects of the site-directed mutations on enzyme activity was performed using the substrate DQ-gelatin and the recombinant PASP proteins. Relative activities of the recombinant proteases were determined by testing each protease variant at a single substrate concentration. The results (Table 3) showed that the mutants D29A, H34A, and S47A were inactive, whereas the WT enzyme and the mutant S59A were equivalent in relative activity. Furthermore, kinetic analysis was performed on the WT and S59A enzyme (Table 2). The Km for the WT and S59A enzyme was 0.21 and 0.19 μM, respectively.
Table 3.
Gelatinolytic Activity of Recombinant PASP and its Site-Directed Mutants
Formation of the Active Site of PASP by Dimerization
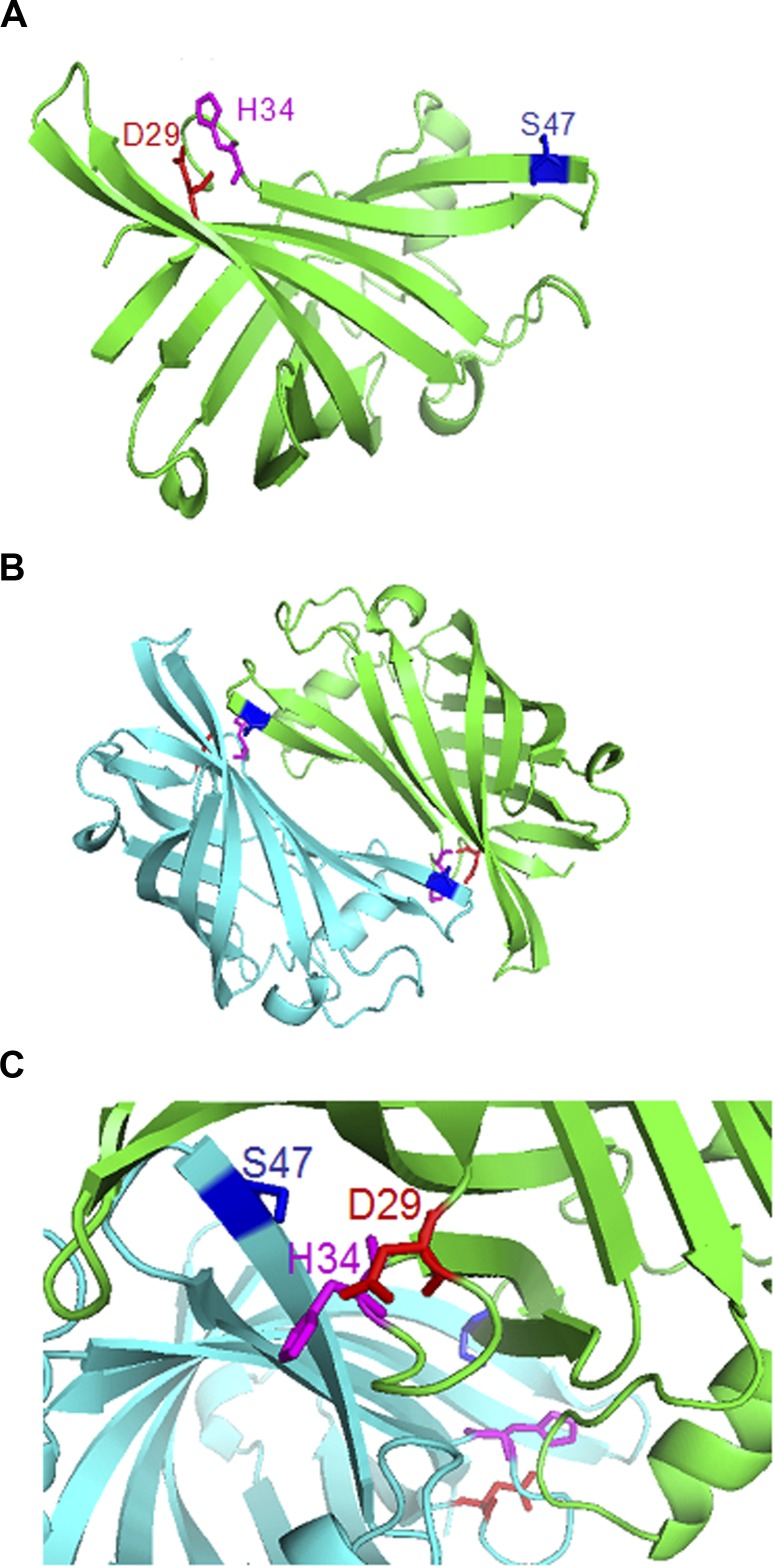
A computer model of PASP structure was generated using YceI as a structural template (Fig. 4A). The modeled PASP structure has an eight-stranded antiparallel β-barrel fold and is predicted to form a homodimer as its biological assembly (Fig. 4B). On the model of the monomeric PASP molecule (Fig. 4A), residue Ser-47 of the catalytic triad is not located near residues Asp-29 and His-34 preventing the formation of a catalytic triad. In the model of the dimeric PASP, residue Ser-47 appears to converge with residues Asp-29 and His-34 from the adjoining PASP molecule forming a catalytic triad (Figs. 4B, 4C).
Figure 4.
Computer modeling of PASP structure. (A) The predicted structure of PASP. The residues of the catalytic triad are indicated on the model. (B) The predicted biological assembly (functional form) of PASP. PASP forms a homodimer with one monomer in green and the other in cyan. The residues of the catalytic triad are shown in color with their side chains as sticks. Magenta, His-34; red, Asp-29; and blue, Ser-47. (C) The proposed active site of PASP is enlarged. The residues His-34 and Asp-29 from one PASP monomer (green) are positioned in close proximity to the residue Ser-47 from another PASP monomer (cyan).
When the WT rPASP protein was analyzed by zymography (Fig. 5A, lane 1), an inactive band of approximately 20 kDa was observed, which corresponds to the PASP monomer. This inactive PASP monomer band is distinct from the higher molecular weight band with proteolytic activity seen at <50 kDa, a size consistent with that expected for the PASP dimer. To detect the presence of PASP dimers or multimers, the purified recombinant PASP variants were analyzed by Western blotting. On the Western blot (Fig. 5B), two major bands were present in each of the purified PASP proteins, which relate to the monomeric and dimeric forms of PASP proteins, respectively. The alanine substitutions of these proteins do not change or disturb the dimer formation.
Figure 5.
Dimerization of PASP. (A) Presence of the inactive PASP monomer. On the zymogram (lane 1), the 20-kDa band of rPASP protein was inactive, whereas the majority of the protease activity was located at approximately 40 kDa, corresponding to the size of the dimeric rPASP protein. (B) Detection of the PASP monomers and dimers by Western blot analysis. Two major bands, instead of a single band, were present for each of the purified recombinant PASP proteins. Protein samples were subjected to SDS-PAGE under nonreducing conditions and subsequently Western blot analysis was performed using anti-rPASP antibody.
Corneal Toxicity of Mutated PASP
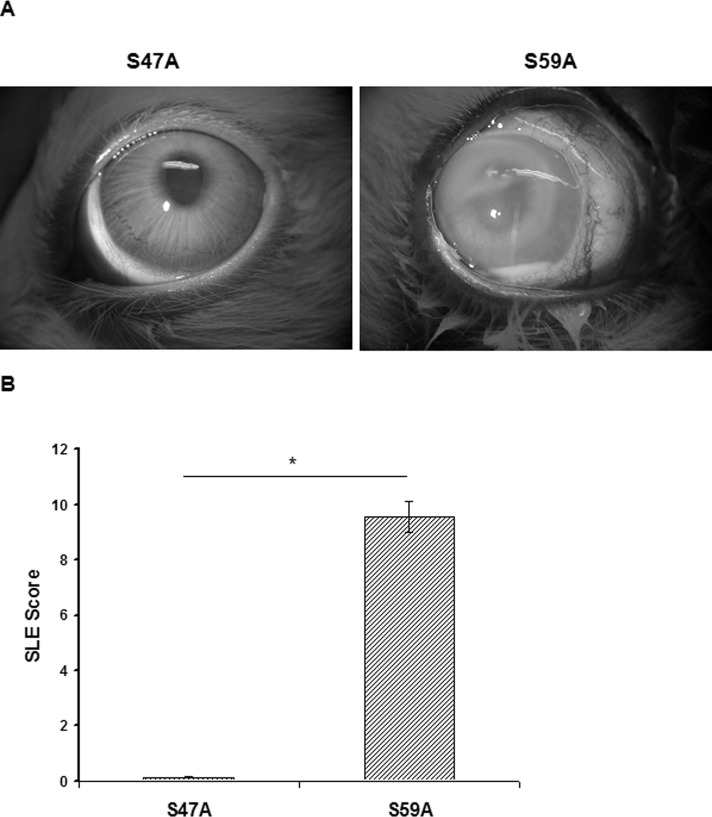
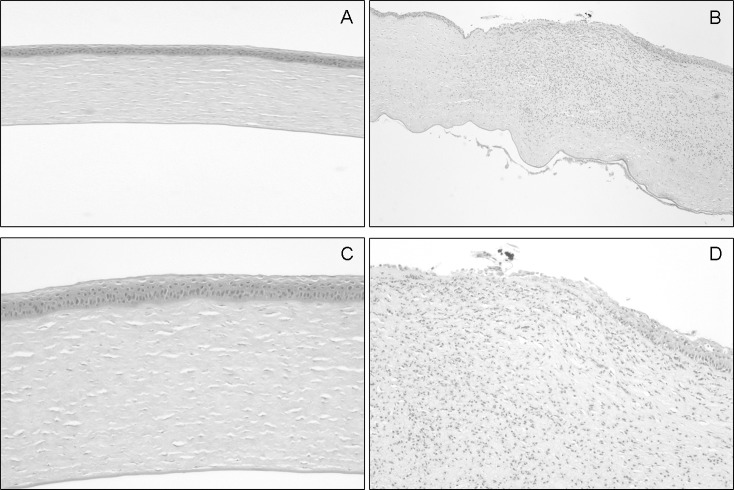
The injection of recombinant PASP with an alanine substitution in Ser-47 (S47A; 8 μg), which lacked protease activity, caused minimal pathologic changes in the rabbit eye (Fig. 6A). In contrast, injection of the active rPASP mutant (S59A; 8 μg) demonstrated severe conjunctival swelling, iritis, and corneal infiltrate at 24 hours after injection. The eyes injected with the inactive S47A mutant had a significantly lower SLE score (0.13 ± 0.08) than eyes injected with the active S59A mutant (9.56 ± 0.56; P = 0.019; Fig. 6B). Histologic analysis of corneas injected with the active S59A mutant demonstrated substantial pathology, including edema, epithelial erosion, and neutrophil infiltrate (Figs. 7B, 7D), whereas corneas injected with the inactive S47A mutant appeared to be normal (Figs. 7A, 7C). These findings demonstrated that protease activity is required for the toxicity of PASP in the rabbit cornea.
Figure 6.
Effects of mutated PASP injection in rabbit eyes. (A) Rabbit eyes at 24 hours after intrastromal injection of the inactive S47A mutant or the active S59A mutant (8 μg in 10 μL). (B) Mean SLE scores of rabbit eyes at 24 hours after injection. The eyes injected with the active S59A protein had a significantly higher SLE score than the eyes injected with the inactive S47A protein. *P = 0.019.
Figure 7.
Histologic analysis of rabbit corneas injected with mutated PASP. Corneas were harvested at 24 hours after injection and corneal sections were stained with hematoxylin and eosin, microscopically examined, and photographed under low power (×40; [A, B]) and high power (×100, [C, D]). The cornea injected with S47A (A, C) showed normal structure whereas the cornea injected with S59A demonstrated epithelial erosion (detachment), severe neutrophil infiltration, and corneal edema (B, D).
Discussion
We analyzed the substrate specificity and catalytic residues of PASP, a corneal virulence factor and protease that shows no sequence similarity to any other known protease. A structure-based prediction of three amino acids, Asp-29, His-34, and Ser-47, that comprise the active site of PASP was confirmed by site-directed mutagenesis studies. An alanine substitution at any of these three amino acids resulted in a loss of enzymatic activity and the mutation in Ser-47 completely eliminated PASP corneal toxicity. Our analysis of PASP indicated that dimerization is required to form an active site and the PASP monomer lacks enzymatic activity. Most importantly, the molecule with an active site mutation lacks protease activity and toxicity for the cornea.
Pseudomonas virulence for the cornea has been proven to be mediated by two proteases, protease IV and PASP.23 Inactivation of the gene for either of these proteases significantly reduces the corneal virulence, but does not reduce the growth of the bacteria in the corneal stroma.14,22 Protease IV is able to degrade key host defense molecules, but has little activity on collagen.11,13 PASP does have considerable collagen degrading activity and fully active preparations of PASP can cause epithelial and stromal erosions of the cornea.13 Virulence of P. aeruginosa appears to require protease IV activity on the host defense and PASP activity on a spectrum of host proteins, including collagens.
The association of protease IV and PASP with virulence also has been found in studies of Pseudomonas lung infections.30–32 Protease IV has been shown to degrade the surfactant proteins of the lung and cytokine IL-22, and to augment the virulence of pneumococcus in the lung.33–35 Studies of Pseudomonas strains isolated from the lungs of cystic fibrosis patients have shown that these isolates produce more PASP and more protease IV than prototropic strains.30–32,36 Antibodies to PASP and protease IV have been detected in the sera of cystic fibrosis patients who had a Pseudomonas infection demonstrating the production of these proteases during infection.37 Biofilms are found commonly in the lungs of infected cystic fibrosis patients and can form on contact lenses contaminated with Pseudomonas and also on abiotic surfaces in the eye.38 Biofilms have been shown to have increased production of protease IV and PASP.39
In the modeled PASP monomer structure, the residue Ser-47 of the triad is not located in proximity to the other two residues of the catalytic triad (His-34 and Asp-29). So, the lack of proximity of the triad residues in this structure is not consistent with forming an active site of a serine protease, as evidenced by the lack of protease activity of the PASP monomer. However, PASP is predicted to form a homodimer, and in the dimerized structure of PASP, the residue Ser-47 of one PASP molecule is located close to the residues His-34 and Asp-29 of the other PASP molecule. The postulated nonproteolytic monomer and the proteolytic dimer of PASP were detected by zymography (Fig. 5A). The mutations of the catalytic residues did not prevent the formation of PASP dimers (Fig. 5B), indicating that residues other than the catalytic triad are involved in dimerization. While the activity of the native PASP protein was detected mainly at approximately 80 kDa on zymograms, the activity of the recombinant PASP protein was at approximately 50 kDa. This suggested that the formation of a dimer is required for PASP activity and a possibility exists of PASP forming a higher-order multimer (tetramer) with proteolytic activity.
Dimerization is common among a wide variety of serine proteases. For example, factor XI is a homodimeric zymogen of a serine protease that is involved in the blood clotting cascade.40,41 Another example of dimeric proteases is a family of homologous serine proteases from all nine known human herpesviruses (HHV).42 Inhibitors targeting the active site or the dimeric interface of the serine proteases of HHV are under development.43,44 In the case of PASP, an inhibitor that disrupts the dimerization process could prevent protease formation and subsequently reduce the tissue damage during Pseudomonas keratitis.
In summary, PASP is a distinct and unique serine protease from P. aeruginosa with the classical Asp-His-Ser catalytic triad. This novel protease joins the wide-spread family of YceI-like proteins whose functions are poorly understood. The function of PASP has been demonstrated to be a protease and virulence factor in two animal models of Pseudomonas keratitis.
Acknowledgments
Supported by funds from National Institutes of Health (NIH; Bethesda, MD, USA) Grant EY12961.
Disclosure: A. Tang, None; A.R. Caballero, None; M.E. Marquart, None; M.A. Bierdeman, None; R.J. O'Callaghan, None
References
- 1.He J, Baldini RL, Déziel E, et al. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc Natl Acad Sci U S A. 2004;101:2530–2535. doi: 10.1073/pnas.0304622101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Driscoll JA, Brody SL, Kollef MH. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs. 2007;67:351–368. doi: 10.2165/00003495-200767030-00003. [DOI] [PubMed] [Google Scholar]
- 3.NNIS System. NNIS. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2003, issued August 2003. Am J Infect Control. 2003;31:481–498. doi: 10.1016/j.ajic.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Green M, Apel A, Stapleton F. Risk factors and causative organisms in microbial keratitis. Cornea. 2008;27:22–27. doi: 10.1097/ICO.0b013e318156caf2. [DOI] [PubMed] [Google Scholar]
- 5.Cheng KH, Leung SL, Hoekman HW, et al. Incidence of contact-lens-associated microbial keratitis and its related morbidity. Lancet Lond Engl. 1999;354:181–185. doi: 10.1016/S0140-6736(98)09385-4. [DOI] [PubMed] [Google Scholar]
- 6.Alexandrakis G, Alfonso EC, Miller D. Shifting trends in bacterial keratitis in south Florida and emerging resistance to fluoroquinolones. Ophthalmology. 2000;107:1497–1502. doi: 10.1016/s0161-6420(00)00179-2. [DOI] [PubMed] [Google Scholar]
- 7.Fleiszig SMJ, Evans DJ. The pathogenesis of bacterial keratitis: studies with Pseudomonas aeruginosa. Clin Exp Optom. 2002;85:271–278. doi: 10.1111/j.1444-0938.2002.tb03082.x. [DOI] [PubMed] [Google Scholar]
- 8.Twining SS, Kirschner SE, Mahnke LA, Frank DW. Effect of Pseudomonas aeruginosa elastase, alkaline protease, and exotoxin A on corneal proteinases and proteins. Invest Ophthalmol Vis Sci. 1993;34:2699–2712. [PubMed] [Google Scholar]
- 9.O'Callaghan RJ, Engel LS, Hobden JA, Callegan MC, Green LC, Hill JM. Pseudomonas keratitis. The role of an uncharacterized exoprotein, protease IV, in corneal virulence. Invest Ophthalmol Vis Sci. 1996;37:534–543. [PubMed] [Google Scholar]
- 10.O'Callaghan RJ. Role of exoproteins in bacterial keratitis: the Fourth Annual Thygeson Lecture, presented at the Ocular Microbiology and Immunology Group Meeting, November 7, 1998. Cornea. 1999;18:532–537. doi: 10.1097/00003226-199909000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Engel LS, Hill JM, Caballero AR, Green LC, O'Callaghan RJ, Protease IV. a unique extracellular protease and virulence factor from Pseudomonas aeruginosa. J Biol Chem. 1998;273:16792–16797. doi: 10.1074/jbc.273.27.16792. [DOI] [PubMed] [Google Scholar]
- 12.Marquart ME, Caballero AR, Chomnawang M, Thibodeaux BA, Twining SS, O'Callaghan RJ. Identification of a novel secreted protease from Pseudomonas aeruginosa that causes corneal erosions. Invest Ophthalmol Vis Sci. 2005;46:3761–3768. doi: 10.1167/iovs.04-1483. [DOI] [PubMed] [Google Scholar]
- 13.Tang A, Marquart ME, Fratkin JD, et al. Properties of PASP: a Pseudomonas protease capable of mediating corneal erosions. Invest Ophthalmol Vis Sci. 2009;50:3794–3801. doi: 10.1167/iovs.08-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang A, Caballero AR, Marquart ME, O'Callaghan RJ. Pseudomonas aeruginosa small protease (PASP), a keratitis virulence factor. Invest Ophthalmol Vis Sci. 2013;54:2821–2828. doi: 10.1167/iovs.13-11788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alcorn JF, Wright JR. Degradation of pulmonary surfactant protein D by Pseudomonas aeruginosa elastase abrogates innate immune function. J Biol Chem. 2004;279:30871–30879. doi: 10.1074/jbc.M400796200. [DOI] [PubMed] [Google Scholar]
- 16.Henke MO, John G, Rheineck C, Chillappagari S, Naehrlich L, Rubin BK. Serine proteases degrade airway mucins in cystic fibrosis. Infect Immun. 2011;79:3438–3444. doi: 10.1128/IAI.01252-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Berre R, Nguyen S, Nowak E, et al. Relative contribution of three main virulence factors in Pseudomonas aeruginosa pneumonia. Crit Care Med. 2011;39:2113–2120. doi: 10.1097/CCM.0b013e31821e899f. [DOI] [PubMed] [Google Scholar]
- 18.Hobden JA. Pseudomonas aeruginosa proteases and corneal virulence. DNA Cell Biol. 2002;21:391–396. doi: 10.1089/10445490260099674. [DOI] [PubMed] [Google Scholar]
- 19.Thibodeaux BA, Caballero AR, Marquart ME, Tommassen J, O'Callaghan RJ. Corneal virulence of Pseudomonas aeruginosa elastase B and alkaline protease produced by Pseudomonas putida. Curr Eye Res. 2007;32:373–386. doi: 10.1080/02713680701244181. [DOI] [PubMed] [Google Scholar]
- 20.Engel LS, Hill JM, Moreau JM, Green LC, Hobden JA, O'Callaghan RJ. Pseudomonas aeruginosa protease IV produces corneal damage and contributes to bacterial virulence. Invest Ophthalmol Vis Sci. 1998;39:662–665. [PubMed] [Google Scholar]
- 21.Traidej M, Caballero AR, Marquart ME, Thibodeaux BA, O'Callaghan RJ. Molecular analysis of Pseudomonas aeruginosa protease IV expressed in Pseudomonas putida. Invest Ophthalmol Vis Sci. 2003;44:190–196. doi: 10.1167/iovs.02-0458. [DOI] [PubMed] [Google Scholar]
- 22.Caballero A, Thibodeaux B, Marquart M, Traidej M, O'Callaghan R. Pseudomonas keratitis: protease IV gene conservation, distribution, and production relative to virulence and other Pseudomonas proteases. Invest Ophthalmol Vis Sci. 2004;45:522–530. doi: 10.1167/iovs.03-1050. [DOI] [PubMed] [Google Scholar]
- 23.Marquart ME, O'Callaghan RJ. Infectious keratitis: secreted bacterial proteins that mediate corneal damage. J Ophthalmol. 2013;2013:369094. doi: 10.1155/2013/369094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stancik LM, Stancik DM, Schmidt B, Barnhart DM, Yoncheva YN, Slonczewski JL. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J Bacteriol. 2002;184:4246–4258. doi: 10.1128/JB.184.15.4246-4258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ekici OD, Paetzel M, Dalbey RE. Unconventional serine proteases: variations on the catalytic Ser/His/Asp triad configuration. Protein Sci Publ Protein Soc. 2008;17:2023–2037. doi: 10.1110/ps.035436.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 27.Markland FS, Smith EL, Subtilisin BPN., VII. Isolation of cyanogen bromide peptides and the complete amino acid sequence. J Biol Chem. 1967;242:5198–5211. [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory;; 1989. [Google Scholar]
- 29.Vandooren J, Geurts N, Martens E, et al. Gelatin degradation assay reveals MMP-9 inhibitors and function of O-glycosylated domain. World J Biol Chem. 2011;2:14–24. doi: 10.4331/wjbc.v2.i1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hare NJ, Solis N, Harmer C, et al. Proteomic profiling of Pseudomonas aeruginosa AES-1R, PAO1 and PA14 reveals potential virulence determinants associated with a transmissible cystic fibrosis-associated strain. BMC Microbiol. 2012;12:16. doi: 10.1186/1471-2180-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hare NJ, Soe CZ, Rose B, et al. Proteomics of Pseudomonas aeruginosa Australian epidemic strain 1 (AES-1) cultured under conditions mimicking the cystic fibrosis lung reveals increased iron acquisition via the siderophore pyochelin. J Proteome Res. 2012;11:776–795. doi: 10.1021/pr200659h. [DOI] [PubMed] [Google Scholar]
- 32.Scott NE, Hare NJ, White MY, Manos J, Cordwell SJ. Secretome of transmissible Pseudomonas aeruginosa AES-1R grown in a cystic fibrosis lung-like environment. J Proteome Res. 2013;12:5357–5369. doi: 10.1021/pr4007365. [DOI] [PubMed] [Google Scholar]
- 33.Malloy JL, Veldhuizen RAW, Thibodeaux BA, O'Callaghan RJ, Wright JR. Pseudomonas aeruginosa protease IV degrades surfactant proteins and inhibits surfactant host defense and biophysical functions. Am J Physiol Lung Cell Mol Physiol. 2005;288:L409–L418. doi: 10.1152/ajplung.00322.2004. [DOI] [PubMed] [Google Scholar]
- 34.Guillon A, Brea D, Morello E, et al. Pseudomonas aeruginosa proteolytically alters the interleukin 22-dependent lung mucosal defense. Virulence. 2017;8:810–820. doi: 10.1080/21505594.2016.1253658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradshaw JL, Caballero AR, Bierdeman MA, et al. Pseudomonas aeruginosa protease IV exacerbates pneumococcal pneumonia and systemic disease. mSphere. 2018;3 doi: 10.1128/mSphere.00212-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fothergill JL, Walshaw MJ, Winstanley C. Transmissible strains of Pseudomonas aeruginosa in cystic fibrosis lung infections. Eur Respir J. 2012;40:227–238. doi: 10.1183/09031936.00204411. [DOI] [PubMed] [Google Scholar]
- 37.Upritchard HG, Cordwell SJ, Lamont IL. Immunoproteomics to examine cystic fibrosis host interactions with extracellular Pseudomonas aeruginosa proteins. Infect Immun. 2008;76:4624–4632. doi: 10.1128/IAI.01707-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zegans ME, Becker HI, Budzik J, O'Toole G. The role of bacterial biofilms in ocular infections. DNA Cell Biol. 2002;21:415–420. doi: 10.1089/10445490260099700. [DOI] [PubMed] [Google Scholar]
- 39.Passmore IJ, Nishikawa K, Lilley KS, Bowden SD, Chung JCS, Welch M. Mep72, a metzincin protease that is preferentially secreted by biofilms of Pseudomonas aeruginosa. J Bacteriol. 2015;197:762–773. doi: 10.1128/JB.02404-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley J, McEwan PA, Gailani D. Structure and function of factor XI. Blood. 2010;115:2569–2577. doi: 10.1182/blood-2009-09-199182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geng Y, Verhamme IM, Smith SB, et al. The dimeric structure of factor XI and zymogen activation. Blood. 2013;121:3962–3969. doi: 10.1182/blood-2012-12-473629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimba N, Nomura AM, Marnett AB, Craik CS. Herpesvirus protease inhibition by dimer disruption. J Virol. 2004;78:6657–6665. doi: 10.1128/JVI.78.12.6657-6665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waxman L, Darke PL. The herpesvirus proteases as targets for antiviral chemotherapy. Antivir Chem Chemother. 2000;11:1–22. doi: 10.1177/095632020001100101. [DOI] [PubMed] [Google Scholar]
- 44.Lee GM, Shahian T, Baharuddin A, Gable JE, Craik CS. Enzyme inhibition by allosteric capture of an inactive conformation. J Mol Biol. 2011;411:999–1016. doi: 10.1016/j.jmb.2011.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]