Abstract
Nucleolus is viewed as a plurifunctional center in the cell, tightly linked to ribosome biosynthesis. As a non-membranous structure, how the size of nucleolus is determined is a long outstanding question, and the possibility of “direct size scaling to the nucleus” was raised by genetic studies in fission yeast. Here, we used the model organism Caenorhabditis elegans to test this hypothesis in multi-cellular organisms. We depleted ani-2, ima-3, or C27D9.1 by RNAi feeding, which altered embryo sizes to different extents in ncl-1 mutant worms. DIC imaging provided evidence that in size-altering embryo nucleolar size decreases in small cells and increases in large cells. Furthermore, analyses of nucleolar size in four blastomeres (ABa, ABp, EMS, and P2) within the same embryo of ncl-1 mutants consistently demonstrated the correspondence between cell and nucleolar sizes – the small cells (EMS and P2) have smaller nucleoli in comparison to the large cells (ABa).
Keywords: Nucleolar size, Embryonic size, ani-2, ima-3, C27D9.1
Size scaling, from organisms to organelles, is an interesting and important issue in biology. Genetics is known to play a role in maintaining the sizes of organisms and organelles. However, under different physiological conditions, the same organelles adapt various sizes in correspondence to their functional activities [1]. Inside the nucleus, nucleolus is a condensed structure intimately correlated with ribosome biosynthesis, aging, and cell stress sensing [2]. Enlarged nucleolus size and/or increased numbers of nucleolus are often used as a biomarker of cancer cells [2], [3]. Using fission yeast, Neumann and Nurse [4] demonstrated a proportional relationship between nuclear (N) and cellular (C) size, indicating that the large cells have a large nucleus. A positive correlation in size between nucleus and nucleolus was likewise observed [4]. Therefore, the ratios between nucleus to cell (N/C) and between nucleolus to cytoplasm (No/C) are near a constant. Skewing of this ratio may be pathogenic, as enlarged nucleoli likely reflect an increased tempo of ribosome biogenesis and protein synthesis, contributing to situations such as the premature aging syndrome Hutchinson-Gilford progeria [5] and many tumors [3].
Caenorhabditis elegans is a good model for studying cell biology, neurobiology and behavior because it is easily cultivated and has a short lifespan and a large repertoire of genetic variants. It is particularly suitable for studying nucleolar size regulation because this model organism has a limited number of cells, which are transparent and easily visualized and photographed [6]. Furthermore, embryos at the 2-cell stage have two asymmetric blastomeres, in which the anterior AB cell is larger than the posterior P cells. The AB cell descents (ABa and ABp cell) are always larger than the P cell descents (EMS and P2 cells) in the 4-cell embryos [Fig. 1A]. However, the nucleolus of four cells is invisible in wild-type worm (N2) embryos, in contrast to an instantly recognizable structure in ncl-1 mutant embryos, an alteration that is advantageous to deciphering the size ratio between nucleolus to cell (No/C). Lastly, the RNAi feeding method to knock down particular genes, such as ani-2, ima-3 and C27D9.1 genes, provides a very convenient means to obtain various sizes of embryos in C. elegans [7]. In this capacity, loss-of-function mutant of either ani-2 or ima-3 is known to exhibit a smaller embryonic size: ANI-2 is one of the three C. elegans anillins that has been implicated in nuclear sequestration during interphase and microtubule-driven cell membrane separation during mitosis [8], [9], while IMA-3 is one of the three importin α nuclear transport factors required for normal embryonic, larval, and germline development [10]. By contrast, C27D9.1, which is a negative regulator of embryonic size, is an ortholog of the human fucosyltransferase and plays a role in body morphogenesis, embryo development and reproduction via yet unknown mechanism [11], [12], [13].
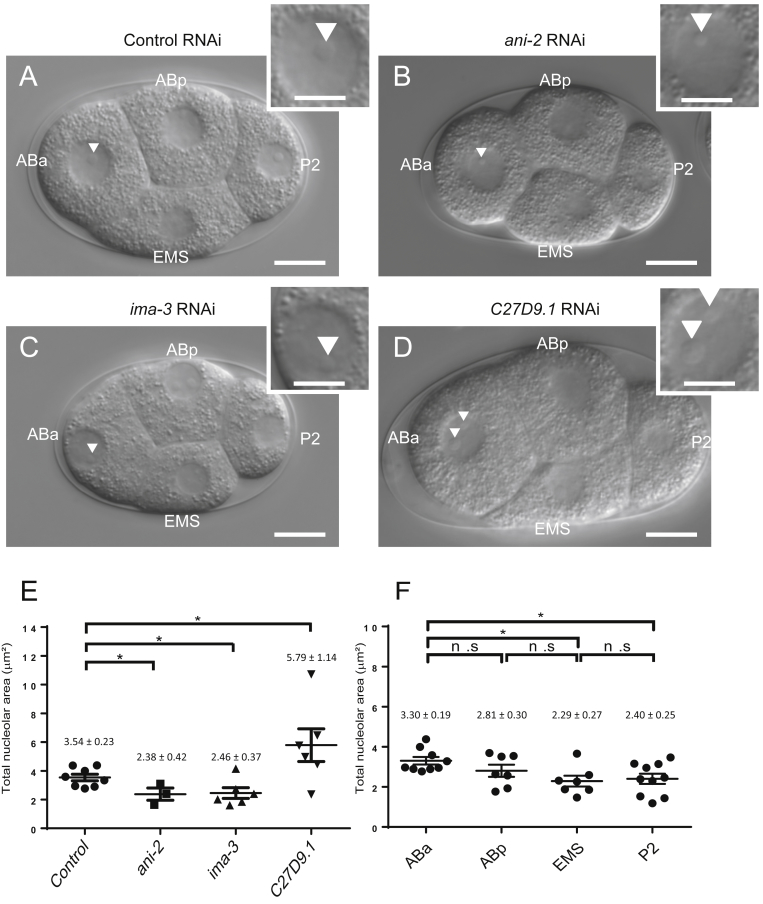
Fig. 1.
DIC images and nucleolus size comparison of four-cell embryos from ncl-1 worms treated with various RNAi knockdown. Mutant worms (ncl-1) at the L4 stage were treated with L4440 (control) (A), ani-2 (B), ima-3 (C) and C27D9.1 (D) RNAi. Embryos were separated by needle dissection and four-cell stage embryos were transferred onto 5% agar pads for observation. The nucleolus of four-cell stage embryos was then recorded by DIC image of the Zeiss Axio Imager 2 microscopy. Magnified images of ABa cell are shown in the upper right. Arrowheads denote the location of nucleolus. The scale bar is 10 μm in the original DIC image and 5 μm in the magnified image. (E) & (F) Total nucleolar areas within a nucleus were measured and summed by the outline tool of the AxioVision software, subsequently represented by a scatter plot using GraphPad Prism. (E) Distribution of total nucleolar areas of the ABa cells from various embryos as indicated. (F) Comparison of the nucleolar sizes in distinct cells within a four-cell embryo of the ncl-1 mutant. *p < 0.05; n.s., non-significant.
In addition to using DIC microscopy to obtain images of the nucleolus, worm researchers frequently exploit a fusion protein (FIB-1::GFP) – nucleolar protein fibrillarin (FIB-1) fused with green fluorescence protein (GFP) – as a reporter to study nucleolus size control [7], [14]. In most cases, the intensity of FIB-1::GFP closely correlates the sizes of nucleoli [14], [15]; however, presence of FIB-1::GFP in early embryos of wild-type worms does not correspond to the size of nucleolus because no nucleolus is detectable at this stage under a DIC microscope. In this study, we used ncl-1 mutants that exhibit prominent nucleolus structure at the four-cell stage, performed RNAi of various size related genes, and used DIC microscopy to record four-cell stage embryos. As expected, the embryos derived from ncl-1 worms fed with E. coli expressing double-stranded RNA of ani-2 and ima-3 genes became smaller, in contrast to the enlarged embryos from C27D9.1-knockdown worms. Consistently, we found that the nucleolus size of the ABa cell undergoes 32.7% and 30.5% reduction in the smaller embryos of ani-2- and ima-3-knockdown worms, respectively, but becomes 63.6% larger in the C27D9.1-depleted, enlarged embryos [Fig. 1A–E]. Additional analyses of nucleolar size in each blastomere cell (ABa, ABp, EMS, and P2) within the ncl-1 mutant embryo revealed smaller nucleoli in the two smaller cells (EMS and P2) while that the two larger cells (ABa and ABp) exhibited significantly bigger nucleoli, with about 22.7%–44.1% increase in size [Fig. 1F].
Our current finding is contradictory to a previous report [7], in which the authors reported an inverse correlation between nucleolar size change and cell size in C. elegans embryos. The unexpected discrepancies in findings by two similar studies are explicable by the following scenarios: Firstly, given that the nucleoli are indiscernible in blastomeres of N2 worm background by DIC microscopy (also see Fig. S1 of the Current Biology report) [7], the green dots in the fluorescence images shown in Figures 1 and 2 of the report [7] presumably are not bona fide nucleoli but rather aggregates of FIB-1::GFP at two loci of rDNA chromosomes. Because two bright spots, instead of one, are frequently seen in their images [7], the FIB-1::GFP-positive dots are likely the pre-nucleolar structures that reportedly encompass rDNA sequences, RNA polymerase I, and endogenous fibrillarin. In contrast, mature nucleoli in ncl-1 mutant embryonic cells that materialize at the 2-cell stage are constantly in singular as observed by DIC microscopy [Fig. 1A]. Secondly, the Weber and Brangwynne study [7] did not exclude the possibility that the variable intensities of FIB-1::GFP observed in the different RNAi embryos could actually be attributed to the loss of ANI-2, IMA-3 or C27D9.1. In other words, it remains formally possible that the abundance of the ectopic fusion protein is directly under the control of these genes, rendering it an ineffective structural reporter for the nucleolus. Finally, the inverse relationship between intensity of FIB-1::GFP and embryonic cell size observed by Weber and Brangwynne could also be explained by a protein density effect: When the fixed amount of FIB-1::GFP synthesized in oocytes is distributed equally to the subsequent daughter cells, the signals may appear dimmer in cells with larger volume, whereas a denser distribution in the smaller cells yields a brighter signal. While the issue of nucleolus size control remains unsettled, our observations and another report by the Brangwynne's group [1] provide strong support to the model of direct size scaling of nucleolus in early embryos as well as in growing intestine cells of C. elegans.
Conflicts of interest
There is no conflict of interest.
Acknowledgements
We would like to express our sincere thanks to Professors Throu Pederson and Simon Sliver for their critical review and comments on the manuscript. We also like to thank CGC (Caenorhbdatitis Genetics Center) for providing strains of worms. The nucleolus research works were supported by grants from the Minister of Education (EMRPD1A043, EMRPD1B0081, and EMRPD1C0051), the Minister of Science and Technology (MOST103-231-B-182-003 to SJL; MOST104-2320-B-182-029-MY3 and MOST105-2314-B-182-061-MY4 to BC-MT) and the Chang Gung Memorial Hospital (BMRP742 to SJL; CMRPD1F0442, CMRPD1H0021 and BMRP960 to BC-MT). This work was financially supported by the Research Center for Emerging Viral Infections from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan and the Ministry of Science and Technology (MOST), Taiwan (MOST 107-3017-F-182-001).
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bj.2018.07.003.
Contributor Information
Bertrand Chin-Ming Tan, Email: btan@mail.cgu.edu.tw.
Szecheng J. Lo, Email: losj@mail.cgu.edu.tw.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Uppaluri S., Weber S.C., Brangwynne C.P. Hierarchical size scaling during multicellular growth and development. Cell Rep. 2016;17:345–352. doi: 10.1016/j.celrep.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 2.Pederson T., Powell K. Thoru Pederson: spotting novel roles for the nucleolus. J Cell Biol. 2015;208:384–385. doi: 10.1083/jcb.2084pi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hein N., Hannan K.M., George A.J., Sanij E., Hannan R.D. The nucleolus: an emerging target for cancer therapy. Trends Mol Med. 2013;19:643–654. doi: 10.1016/j.molmed.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Neumann F.R., Nurse P. Nuclear size control in fission yeast. J Cell Biol. 2007;179:593–600. doi: 10.1083/jcb.200708054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchwalter A., Hetzer M.W. Nucleolar expansion and elevated protein translation in premature aging. Nat Commun. 2017;8:328. doi: 10.1038/s41467-017-00322-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee L.W., Lee C.C., Huang C.R., Lo S.J. The nucleolus of Caenorhabditis elegans. J Biomed Biotechnol. 2012;2012:601274. doi: 10.1155/2012/601274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber S.C., Brangwynne C.P. Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Curr Biol. 2015;25:641–646. doi: 10.1016/j.cub.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen A., Akhshi T.K., Lavoie B.D., Wilde A. Importin beta2 mediates the spatio-temporal regulation of anillin through a noncanonical nuclear localization signal. J Biol Chem. 2015;290:13500–13509. doi: 10.1074/jbc.M115.649160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maddox A.S., Habermann B., Desai A., Oegema K. Distinct roles for two C. elegans anillins in the gonad and early embryo. Development. 2005;132:2837–2848. doi: 10.1242/dev.01828. [DOI] [PubMed] [Google Scholar]
- 10.Geles K.G., Adam S.A. Germline and developmental roles of the nuclear transport factor importin alpha3 in C. elegans. Development. 2001;128:1817–1830. doi: 10.1242/dev.128.10.1817. [DOI] [PubMed] [Google Scholar]
- 11.Piano F., Schetter A.J., Morton D.G., Gunsalus K.C., Reinke V., Kim S.K. Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr Biol. 2002;12:1959–1964. doi: 10.1016/s0960-9822(02)01301-5. [DOI] [PubMed] [Google Scholar]
- 12.Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 13.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Barrell D., Bateman A. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 2003;31:315–318. doi: 10.1093/nar/gkg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi Y.H., Ma T.H., Lee L.W., Chiou P.T., Chen P.H., Lee C.M. A genetic cascade of let-7-ncl-1-fib-1 modulates nucleolar size and rRNA pool in Caenorhabditis elegans. PLoS Genet. 2015;11:e1005580. doi: 10.1371/journal.pgen.1005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin R.M., Tunnemann G., Leonhardt H., Cardoso M.C. Nucleolar marker for living cells. Histochem Cell Biol. 2007;127:243–251. doi: 10.1007/s00418-006-0256-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.