Abstract
Background
Enhancer RNAs (eRNAs) are a group of lncRNAs transcribed from enhancers, whose regulatory effects on gene expression are an emerging area of interest. However, the role of eRNAs in regulating trophoblast cells and unexplained recurrent pregnancy loss (URPL) remains elusive.
Methods
We profiled eRNAs in villi from URPL patients and matched controls by RNA-seq. Functions of URPL-related eRNAs were further investigated in vitro.
Results
We identified lnc-SLC4A1-1, which was transcribed from an active enhancer marked with H3K27ac and H3K4me1 and so-called eRNA, highly expressed in URPL patients. Gain-of-function experiments indicated that lnc-SLC4A1-1 facilitated trophoblast cell migration and apoptosis. Mechanistically, as an eRNA, lnc-SLC4A1-1 was retained in the nuclei and recruited transcription factor NF-κB to bind to CXCL8, resulting in increased H3K27ac in the CXCL8 promoter and subsequent elevation of CXCL8 expression. Activation of CXCL8 exacerbated inflammatory reactions in trophoblast cells by inducing TNF-α and IL-1β, which could be blocked by an antagonist of lnc-SLC4A1-1.
Interpretation
These findings indicate that an eRNA, lnc-SLC4A1-1, alters trophoblast function via activation of immune responses and by regulating the NF-κB/CXCL8 axis. Our study provides new insights in understanding lncRNA/eRNA function in pathological pregnancy, potentially informing on therapeutic strategies for URPL.
Fund
National Natural Science Foundation of China, Natural Science Foundation of Jiangsu Province, National Key Research and Development Program, the Priority Academic Program for the Development of Jiangsu Higher Education Institutions.
Keywords: Recurrent pregnancy loss, Enhancer RNA, Inflammation
Research in context.
Evidence before this study
Although increasing risk factors have been identified to be related to recurrent pregnancy loss (RPL), the reasons of approximately half of these cases still remain unexplained. We searched PubMed with the terms “recurrent pregnancy loss” and “enhancer RNA” for original research articles up to March 1, 2017. There were no language restrictions. No documents match our search terms.
Added value of this study
Our results indicated that lnc-SLA4A1-1, which was characterized as an enhancer RNA (eRNA), was upregulated with increased H3k27ac modification in unexplained recurrent pregnancy loss (URPL) patients. This upregulation altered trophoblast cells migrations and apoptosis. Mechanistically, we demonstrated that eRNA lnc-SLA4A1 interacted with the transcriptional factor NF-κB to promote the expression of CXCL8 and activated the inflammation response, which might affect the trophoblast cell functions and eventually lead to URPL.
Implications of all the available evidence
eRNA lnc-SLC4A1-1 recruits NF-κB and binds to the prompter of CXCL8, which leading to upregulation of CXCL8 and may cause immune response and eventually lead to URPL. Our work provides new insights in understanding the etiology of URPL. Characterization and analysis of eRNAs provided possibilities of identification of new biomarkers of URPL for early diagnosis and targeted treatment, especially eRNAs in the serum which is easy to access. Current study was limited by sample type and small size and future studies with larger sample sizes are needed to verify our findings.
Alt-text: Unlabelled Box
1. Introduction
Recurrent pregnancy loss (RPL) is viewed as a distinct disorder of pregnancy loss and it is estimated that 5% of women experience two clinical miscarriages and approximately 1% experience three or more losses [[1], [2], [3]]. The etiology of RPL is complicated, and known causes include anatomic factors (uterine malformations), genetic factors (chromosome inversion, deletion, duplication, etc.), and immune and endocrine factors (luteal insufficiency, hypothyroidism, etc.). Although increasing risk factors have been identified to be related to RPL, the reasons for approximately half of these cases still remain unexplained (unexplained recurrent pregnancy loss, URPL) [[4], [5], [6]].
Long noncoding RNAs (lncRNAs) are a class of noncoding RNAs that are >200 bp in length and are transcribed from exons, introns and gene deserts [7]. Recent studies have demonstrated that lncRNA functions in a cell type-specific or tissue-specific manner, indicating that lncRNA may be the driver of cell specific response [8,9]. Consequently, lncRNA is involved in a variety of biological processes, including genomic imprinting, cell proliferation and differentiation, and cellular developmental processes [[10], [11], [12]]. Several studies have shown that lncRNA-related endocrine and immune pathways may be the major causes of URPL, but the underlying mechanisms remain incompletely understood [13,14].
A small portion of lncRNA is transcribed from the enhancer regions, is nucleus-retained, and has gene regulatory function; thus, it is part of the so-called enhancer-derived lncRNAs, or eRNAs [[15], [16], [17]]. eRNAs can recruit transcription factors, transcriptional co-activators, and chromatin remodelers in cis or in trans to regulate gene transcription and biological process [18,19]. Unlike common lncRNAs, eRNAs exhibit similar transcription rates but produce fewer stable transcripts [20]. To date, the regulatory roles of eRNAs in URPL, especially in trophoblast cells, have not been explored.
During normal pregnancy, trophoblast cells can maintain maternal immune tolerance through generating ligands to active receptors in specific immune cells [21]. Those activated lymphocytes take up residence at the maternal-fetal interface and build an appropriate uterine microenvironment to promote fetal growth [22]. In the early stages of pregnancy, trophoblast cells invade the decidualized endometrium and remodel uterine spiral arteries to increase maternal blood flow to the placenta villi, which is essential for the exchange of waste and nutrients between maternal and fetal blood [23]. The failure of transitions from progenitors to different trophoblast cells or compromised trophoblast function will cause significant adverse pregnancy outcomes, including URPL [24]. Previous studies had identified several non-coding RNAs that could regulate trophoblast cell functions, including lncRNAs H19, HOXA11-AS and RPAIN [[25], [26], [27]]. However, the molecular mechanisms by which the eRNAs regulate trophoblast cell functions and URPL remain elusive.
In this study, we profiled lncRNAs in villi from URPL patients and matched normal controls by RNA-seq. Our results indicated that lnc-SLA4A1-1, which was characterized as an eRNA, was upregulated with increased H3K27ac modification in URPL patients. This upregulation altered trophoblast cell migration and apoptosis. Mechanistically, we demonstrated that eRNA lnc-SLA4A-1 interacted with NF-κB to promote the expression of CXCL8 and activate the inflammatory response, which might affect trophoblast cell functions and eventually lead to URPL. Our work provides new insights in understanding the etiology of URPL.
2. Materials and methods
2.1. Participating cohorts
This study was approved by the Institutional Ethics Committee of Nanjing Medical University. The study cohort included women with confirmed URPL with 2 or more consecutive pregnancy losses before 20 weeks of undetermined etiology. We excluded patients with abnormal karyotype, infection, endocrine disorders, thyroid dysfunction or abnormal uterine anatomy of URPL. The control group consisted of randomly selected women who underwent legal termination of an apparently normal early pregnancy at the same hospital during the same period, without medical reasons, history of pregnancy loss or any other pregnancy complication. A questionnaire was used to collect clinical and characteristic information, such as personal information, lifestyle factors and medical history. All women enrolled in the study provided signed informed consent. Clinical characteristics of URPL patients and controls are shown in Supplementary Material, Table S1. As expected, there was no significant differences between two groups with respect to baseline characteristic factors, including age, body mass index (BMI), gestational age and childbirth frequency. Villi samples were isolated from products of conception (POC) under a dissecting microscope at the time of dilation and curettage and stored at −80 °C immediately after being washed thoroughly. All activities involved in this study were done under full compliance with government policies and the Helsinki Declaration. All experiment protocols were approved by the Institutional Review Board (IRB) of Nanjing Medical University (NMU) prior to the study (IRB No. NMU (2016)132).
2.2. RNA sequencing and analysis
Total RNA was extracted using RNeasy Kits (Qiagen, Duesseldorf, Germany) and treated with DNase I (Life Technologies, Gaithersburg, USA) according to standard protocols. Tissue RNA-seq (3 URPL patients and 3 controls) was done in Genesky using TruSeq Stranded Total RNA kits (Genesky, Shanghai, China). Cell RNA-seq (3 replicates each group) was done in Ribobio (Ribobio, Guangzhou, China). Briefly, intact RNA was fragmented, end repaired, adapter ligated and PCR amplified following the Illumina protocol. Libraries were sequenced by Illumins Hiseq 2000 (Illumina, San Diego, USA). The sequenced reads were aligned to the human reference genome (H19) using TopHat v1.4.1. Differential gene expression (DEG) analysis was performed with Cuffdiff and DEseq (tissue RNA-seq), or DEseq and DEGseq (cell RNA-seq). LncRNAs/genes with fold change >2 or <−2 and FDR P value <0.05 were selected as DEGs. The data reported in this study have been uploaded in the Gene Ex- pression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo) (Tissue RNA-seq: GSE No. 121950, cell RNA-seq: GSE No. 121951).
2.3. Quantitative PCR (qPCR)
Total RNA was extracted from villi or cells with Trizol (Invitrogen, Carlsbad, USA); nuclear and cytoplasmic RNA was prepared using a PARIS™ Kit (Invitrogen Carlsbad, USA). We used 20 URPL patients and 20 controls in the cohort for the validation. First strand cDNA was synthesized (Vazyme, Nanjing, China) and quantitative PCR was performed. All reactions were performed in triplicate with SYBR Green master mix (Vazyme, Nanjing, China) under the following conditions: 5 min at 95 °C for initial denaturation, followed by 40 cycles of segments of 95 °C for 30 s and 60 °C for 30 s in the ABI Prism 7900HT/FAST (Applied Biosystems, Foster City, USA). The expression levels of GAPDH were used to normalize the expression levels of the tested genes. All the primer sequences used were listed in Supplementary Material, Table S2.
2.4. In vitro experiments
The HTR-8/SVneo cell line was cultured in 1640 medium with 10% FBS (Gibco, Carlsbad, USA). The JEG-3 cell line was cultured in MEM medium (Gibco, Carlsbad, USA) with 10% FBS (Gibco, Carlsbad, USA), 100 U/ml penicillin (Gibco, Carlsbad, USA), and 100 μg/ml streptomycin (Gibco, Carlsbad, USA) at 37 °C under 5% CO2. Cells were co-transfected with pcDNA-lncRNAs or pcDNA-control using Lipofectamine 2000 (Invitrogen, Carlsbad, USA) for overexpression of lncRNAs. The relative expression of transfected lncRNAs after 24 h was detected by qRT-PCR.
After 24 h transfection, cells were harvested for cell proliferation, apoptosis, cell cycle and invasion analysis. For cell proliferation analysis, approximately 3000 cells were seeded in each well of a 96-well plate. After transfection, cells were treated with 10 μl Cell Counting Kit-8 solution in each well and cultured for 0.5–1 h before measuring absorbance at 450 nm. For invasion analysis, 500 μl medium containing 10% fetal bovine serum was added per well in a 24-well plate. Transwell chamber inserts were put into the wells with 100 μl cell suspension mixed with serum free medium. The cells were immobilized with Polyoxymethylene and photographed after 24 h. For cell apoptosis analysis, cell suspensions were prepared and analyzed immediately by a FACS Calibur Flow Cytometer (BD Medical Technology, Lake Franklin, USA) 24 h after transfection. For cell cycle analysis, single cell suspensions were immobilized with 70% ethanol, stained with PI and analyzed using a FACS Calibur Flow Cytometer (BD Medical Technology, Lake Franklin, USA) 24 h after transfection.
2.5. Western blot analysis
Western blots were performed using antibodies against P65 (1:1000, RRID: AB_10859369) (Cell Signaling Technology, Beverly, USA), CXCL8 (1:10, RRID: AB_444617) (Abcam, Cambridge, UK) and GAPDH (1:1000, RRID: AB_2715590) (Beyotime Biotechnology, Shanghai, China) following standard protocols. Briefly, 80 μg protein was applied to SDS-PAGE and transferred to PVDF membrane. Non-specific binding of antibody was blocked by 5% skim milk for 1 h. After incubation with primary antibody overnight, the membrane was washed and incubated with HRP-conjugated secondary antibody for 1 h. GAPDH was used as an internal control. The blots were visualized using the chemiluminescent detection method (Pierce, Thermo Scientific, Waltham, USA). Three repeats were set up for each treatment. The intensity of the western blot bands was quantified by ImageJ.
2.6. ChIP-qPCR
The ChIP methodology was described previously [28]. Briefly, cells were fixed with 1% formaldehyde for 10 min at room temperature. Cross-linking was terminated by addition of 0.125 M glycine. Cells were lysed and sonicated using a Bioruptor (Diagenode, Denville, USA) to generate short fragments for immunoprecipitation. The chromatin was immunoprecipitated by incubating overnight with 1 μg of H3K4me1 (RRID: AB_310614) (Millipore, Bedford, USA) and H3K27ac (RRID: AB_2118291) (Abcam, Cambridge, UK) antibody. After incubating with either protein A or G (EMD Millipore, Bedford, USA) beads for 2 h, the samples were washed with low salt buffers, then high salt buffers, then LiCl buffers and finally TE buffer. The protein–DNA complexes were reverse crosslinked at 65 °C overnight with Proteinase K. Three repeats were set up for each treatment. All the primer sequences used for ChIP-qPCR are listed in Supplementary Material, Table S3.
2.7. RIP-qPCR
Cells were fixed with formaldehyde at a final concentration of 1%. Cells were lysed with 500 μl lysis buffer (50 mM Tris, 150 mM NaCl, 0.1% NP-40, 1 mM EDTA, and RNAse inhibitor) and incubated overnight at 4 °C with 50 μl of Protein G Dynabeads that were prewashed and premixed with 5 μl antibodies P50/P105 (RRID: AB_ 330564), P65 (RRID: AB_2118291), AP-1 (RRID: AB_ 253112) and c-Jun (RRID: AB_ 2,130,165) (Cell Signaling Technology, Beverly, USA) or IgG control (Millipore, Bedford, USA). After washing three times, RNA was extracted by Trizol and reverse transcribed for qPCR analysis. Three repeats were set up for each treatment. All the primer sequences used for RIP-qPCR are listed in Supplementary Material, Table S4.
2.8. Elisa
TNF-α, IL-1β and CXCL8 in culture supernatants were analyzed by enzyme-linked immunosorbent assay (ELISA), following the Elabscience ™ ELISA kit instructions (Elabscience, Wuhan, China). Three repeats were set up for each treatment. Absorbance values were measured using a microplate reader and a two-tailed Student's t-test was used for statistical analysis.
2.9. Statistical analysis
All statistical analyses were performed using Graphpad Prism 5.0 (GraphPad Software, San Diego, USA). The Kolmogorov-Smirnov test was used to test the assumption of normal distribution. Student's t-test was used to show differences in continuous variables such as age and body mass index (BMI).
For data that do not conform to the normal distribution, we used the Mann-Whitney U test. Multiple comparisons were subjected to one-way analysis of variance (one-way ANOVA) followed by ad-hoc testing. The significance was set at p < 0.05. All data were expressed as mean ± SEM.
3. Results
3.1. Dysregulation of lncRNAs in URPL
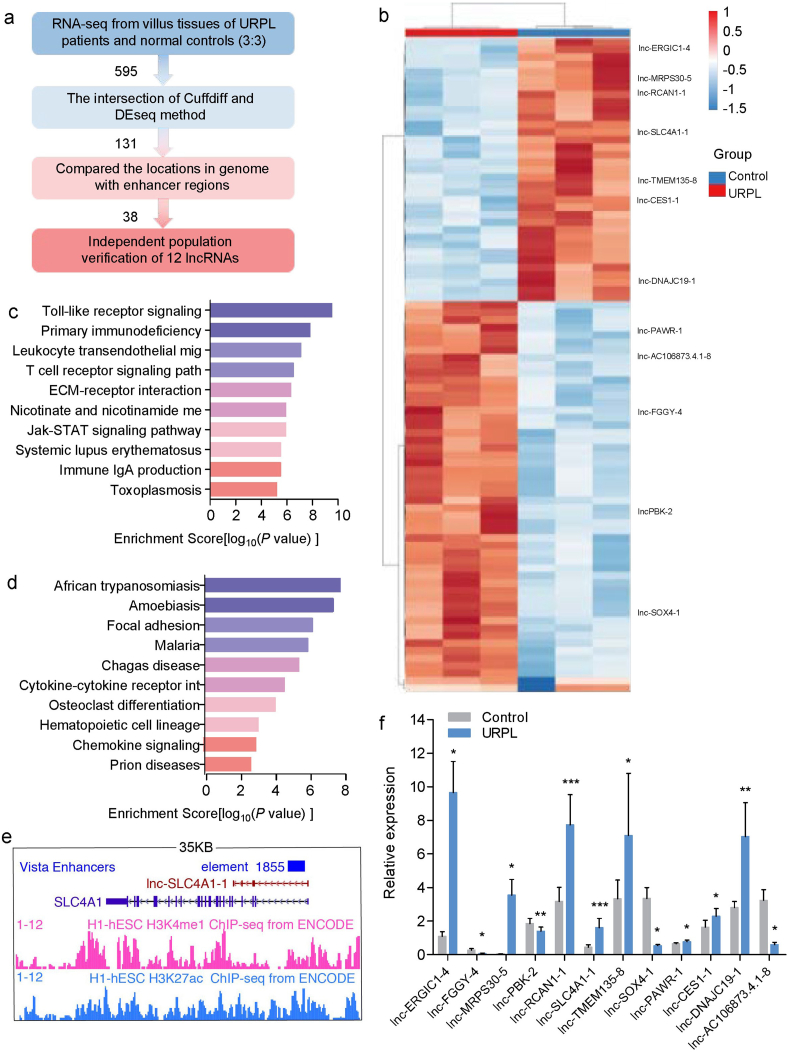
To identify the lncRNAs that are potentially involved in URPL, we profiled the transcriptome by RNA-seq and analyzed the data following the pipeline illustrated in Fig. 1a. Comparing URPL with matched controls, 476 and 250 differentially expressed lncRNAs (DELs) were identified in villus tissue using Cuffdiff and DESeq with 2-fold change and an FDR p value <0.05, respectively. One hundred and thirty-one lncRNAs were selected as confident DELs from the intersection of Cuffdiff and DEseq results (Fig. 1b). KEGG pathway analysis was used to predict the enriched biological pathways of URPL-related DELs. The top 10 up-regulated and top 10 down-regulated pathways are shown in Fig. 1c,d. Most of the pathways were associated with immune function, including Toll-like receptor signaling pathway, T cell receptor signaling pathways and Jak-STAT signaling pathways.
Fig. 1.
Dysregulation of lncRNAs in URPL. (a) The screening process pipeline used in our study. (b) Heatmap of differentially expressed lncRNAs between URPL and controls by RNA-seq (n = 3 per group). (c) Top 10 upregulated KEGG pathways. (d) Top 10 downregulated KEGG pathways. (e) Example of eRNA in UCSC genome browser. (f) Validation of differentially expressed lncRNAs in URPL and controls (n = 20 per group). (Two-tailed paired Student's t-test.) *P < 0.05, **P < 0.01, ***P < 0.001. Data are presented as mean ± SEM. Three repeats were set up for each treatment.
LncRNAs transcribed from enhancer regions are called enhancer RNAs (eRNAs), and have been shown to recruit transcription factors and co-activators to promote gene activation. To identify potential eRNAs among URPL-related DELs, we compared their locations with annotated enhancer regions in VISTA Enhancer Browser (https://enhancer.lbl.gov/) (Fig. 1e). There are approximately 55,000 potential enhancers in the human genome and among the 131 URPL-related DELs (Supplementary Material, Table S5), 38 DELs were found to overlap with enhancer regions (32 up-regulated and 6 down-regulated). Most of these potential eRNAs have not been studied, and their functions are still largely unknown. We then selected 12 potential eRNAs with the largest fold change for validation in an independent cohort as described in methods. The results were highly consistent except for lnc-PAWR-1 (t = 2.761, p = 0.028) (Fig. 1f). Seven eRNAs were upregulated, including lnc-ERGIC1-4(t = 4.531, p = 0.011), lnc-MRPS30-5(t = 3.717, p = 0.021), lnc-RCAN1-1(t = 6.107, p = 6.117*10−5), lnc-SLC4A1-1(t = 6.078, p = 3.549*10−4), lnc-TMEM135-8(t = 2.092, p = 0.045), lnc-CES1-1(t = 2.874, p = 0.011) and lnc-DNAJC19-1(t = 2.978, p = 0.007). Four eRNAs, including lnc-FGGY-4(t = −3.920, p = 0.017), lncPBK-2(t = −3.183, p = 0.003), lnc-SOX4-1(t = −4.431, p = 0.013) and lnc-AC106873.4.1-8(t = −4.053, p = 0.015), were down-regulated (Fig. 1f).
3.2. URPL-related eRNAs regulated trophoblast cell functions
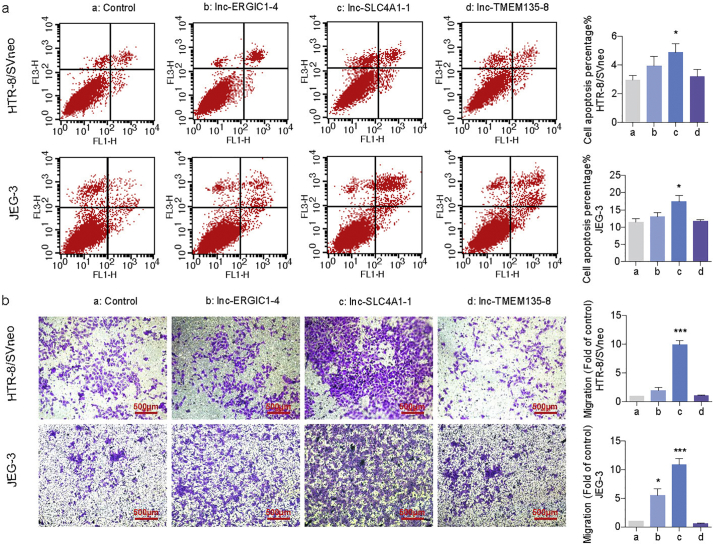
We used the JEG-3 (derived from a choriocarcinoma) and HTR-8/SVneo (derived from normal first trimester placenta) cell lines to study the eRNA's function [29]. In an attempt to determine whether URPL-related eRNAs have effects on trophoblast cell function, we overexpressed selected URP-related eRNAs (lnc-ERGIC1-4, lnc-SLC4A1-1 and lnc-TMEM135-8), which showed large differences (fold change >5) in both screening and validation stages in HTR-8/SVneo and JEG-3. After transfection, the expression levels of lnc-ERGIC1-4, lnc-SLC4A1-1 and lnc-TMEM135-8 were significantly higher than that in controls (Supplementary Material, Fig. S1a). Cell proliferation was significantly decreased in both HTR-8/SVneo (t = 11.420, p = 2.422*10−5) and JEG-3 (t = 2.579, p = 0.032) cells after overexpression of lnc-SLC4A1-1, while the other lncRNAs had no effects (Fig. S2a, b). Correspondingly, cell apoptosis was significantly increased in HTR-8/SVneo (t = 2.282, p = 0.045) and JEG-3 (t = 2.883, p = 0.045) cells with high expression of lnc-SLC4A1-1, revealing that growth inhibition was accompanied by increased apoptosis (Fig. 2a). We also found a significantly increased number of cells migrating through transwell pores after lnc-SLC4A1-1 (t = 11.520, p = 7.019*10−6) and lnc-ERGIC1-4 (t = 11.930, p = 7.361*10−6) overexpression in both HTR-8/SVneo and JEG-3 cells (Fig. 2b). All three lncRNAs had no effects on cell cycle (Supplementary Material, Fig. S2c, d). Collectively, our results suggested that URPL-related eRNAs inhibited cell proliferation, but promoted apoptosis and migration.
Fig. 2.
URPL related eRNAs regulated trophoblast cell function. (a) Cell apoptosis analysis in HTR-8/SVneo and JEG-3 cells. (b) Transwell assay in HTR-8/SVneo and JEG-3 cells. (Two-tailed paired Student's t-test.) *P < 0.05, **P < 0.01, ***P < 0.001. Data are presented as mean ± SEM. Three repeats were set up for each treatment. a: Control, b: lnc-ERGIC1-4, c: lnc-SLC4A1-1, d: lnc-TMEM135-8.
Since lnc-ERGIC1-4 and lnc-TMEM135-8 had mild effects on cell function, subsequent functional study was focused on lnc-SLC4A1-1. Lnc-SLC4A1-1(ENST00000498270) (http://www.lncipedia.org/) locates on Chr.17 with a full length of 460 bp and containing four exons in total. To ascertain the eRNA potency of lnc-SLC4A1-1, we performed ChIP-qPCR at the lnc-SLC4A1-1 transcription locus and detected a robust enrichment of H3K27ac (t = −4.051, p = 0.015) (Supplementary Material, Fig. S3a). In addition, subcellular localization analysis showed that lnc-SLC4A1-1 localized in the nucleus rather than in the cytoplasm (t = 2.941, p = 0.042), which indicated that lnc-SLC4A1-1 might be an eRNA and have gene regulatory effect within the nucleus (Supplementary Material, Fig. S3b).
3.3. Lnc-SLC4A1-1 enhanced CXCL8 transcription in trophoblast cells
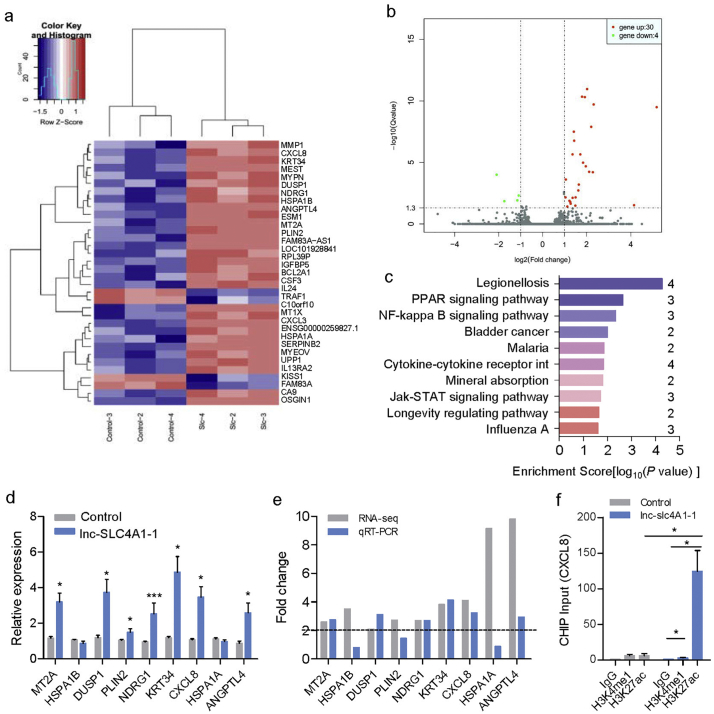
Lnc-SLC4A1-1 was transcribed from an enhancer near the SLC4A1 gene. Thus, we first tested whether lnc-SLC4A1-1 played a cis regulatory role in regulating SLC4A1. However, after overexpression of lnc-SLC4A1-1, no cis-regulatory effect of lnc-SLC4A1-1 on SLC4A1 was found (t = 0.716, p = 0.514) (Supplementary Material, Fig. S3c). To further evaluate the function of lnc-SLC4A1-1 in trophoblast cells, we performed RNA-seq in HTR-8/SVneo cells overexpressing lnc-SLC4A1-1. Using a fold change >2 and FDR P value <0.05 as a cut off, we identified 30 genes that were upregulated and 4 genes that were downregulated upon overexpression of lnc-SLC4A1-1 (Fig. 3a, b). Among those 34 genes, MT2A, ANGPTL4, HSPA1B, DUSP1, PLIN2, NDRG1, KRT34, CXCL8 and HSPA1A were identified by both DEseq and DEGseq. A cytokine-cytokine receptor interaction pathway, JAK − STAT signaling pathway and NF-kappa B signaling pathway were enriched in the KEGG analysis. These findings suggested that lnc-SLC4A1-1 regulated the expression of cytokine and inflammation-related genes (Fig. 3c).
Fig. 3.
Lnc-SLC4A1-1 enhanced CXCL8 transcription in trophoblast cells. (a) Heat map of differentially expressed genes after lnc-SLC4A1-1 overexpression. (b) Volcano plot of differentially expressed genes after lnc-SLC4A1-1 overexpression. (c) KEGG pathway analysis and number of differentially expressed genes in enriched pathway was shown on the right side. (d, e) Validation of differentially expressed genes by qRT-PCR. (f) ChIP-qPCR analysis of H3K4me1, H3K27ac enrichment near CXCL8 in HTR-8/SVneo cells. The IgG was used as negative control. (Two-tailed paired Student's t-test.) *P < 0.05, **P < 0.01, ***P < 0.001. Results were presented as mean ± SEM. Three repeats were set up for each treatment.
In addition, we used qRT-PCR to validate the differentially expressed genes identified in RNA-seq. Compared with the control, we found that lnc-SLC4A1-1 significantly promoted the expression levels of MT2A(t = −4.016, p = 0.016), ANGPTL4(t = −3.031, p = 0.039), DUSP1(t = −3.389, p = 0.028), PLIN2(t = −2.799, p = 0.010), NDRG1(t = −5.559, p = 1.592*10−4), KRT34(t = −4.153, p = 0.014) and CXCL8(t = −4.074, p = 0.015) in HTR-8/SVneo cells (Fig. 3d). Among these genes, MT2A, ANGPTL4, DUSP1, NDRG1, KRT34 and CXCL8 had >2-fold changes in both the RNA-seq and validation stages (Fig. 3e). Enhancers can regulate gene expression through transcription factor binding and histone modifications. H3K4me1 is considered a marker of poised enhancers and H3K27ac is a mark of active enhancers and promoters. We carried out ChIP-qPCR to determine whether there was a change of H3K27ac or H3K4me1enrichment near differentially expressed genes (Fig. 3f and Supplementary Material, Fig. S4). We found relatively high enrichment of H3K27ac near the promoter region of CXCL8 rather than near the other genes (t = −3.101, p = 0.036) (Fig. 3f). These results suggested that lnc-SLC4A1-1 might recruit transcription factors for active transcription of CXCL8.
3.4. Lnc-SLC4A1-1 interacted with NF-κB to promote CXCL8 expression
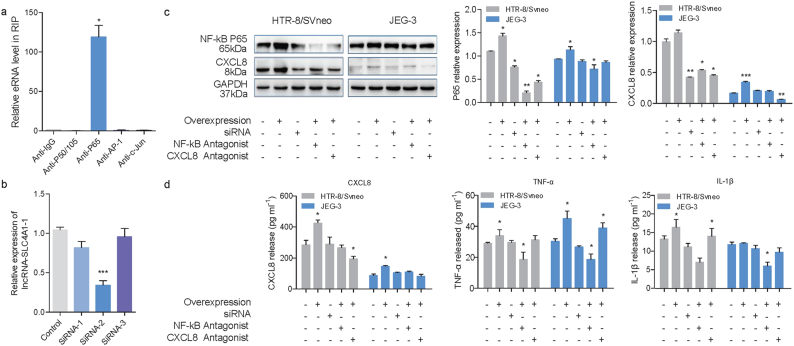
Given the enrichment of H3K27ac near CXCL8, we hypothesized that transcription factors might be recruited to CXCL8 by lnc-SLC4A1-1. To test this hypothesis, we performed RNA immunoprecipitation followed by reverse transcription and quantitative PCR (RIP-qPCR) with antibodies against P50/P105, P65, AP-1 and c-Jun, factors reported to regulate CXCL8 expression [30] (Fig. 4a). Compared with other transcription factors, P65 was highly enriched near CXCL8 (t = 8.088, p = 0.015) (Fig. 4a). This demonstrated that lnc-SLC4A1-1 might specifically interact with P65.
Fig. 4.
Lnc-SLC4A1-1 interacted with NF-κB to promote CXCL8 expression. (a) RNA immunoprecipitation (RIP) followed by qRT-PCR with antibodies anti P50/P105, P65, AP-1 and c-Jun to identify transcription factors that are associated with lnc-SLC4A1-1. (b) Relative expression of lnc-SLC4A1-1 after transfection of siRNA. (c) Western blot of NF-κB and CXCL8 in HTR-8/SVneo and JEG-3 cells under different condition. (d) The levels of CXCL8, TNF-α and IL1β were determined by ELISA in HTR-8/SVneo and JEG-3 cells under different condition. (Two-tailed paired Student's t-test and Mann-Whitney U test.) *P < 0.05, **P < 0.01, ***P < 0.001. Data were shown as mean ± SEM. Three repeats were set up for each treatment.
To determine whether lnc-SLC4A1-1 interacts with P65 to promote CXCL8 expression, we designed a group of siRNA antisense inhibitors against lnc-SLC4A1-1; siRNA-2 worked well (t = 10.820, p = 4.238*10−4) (Fig. 4b). We observed that knockdown of lnc-SLC4A1–1 attenuated CXCL8 expression (Fig. 4c, d) (Z = −2.913, p = 0.002). In addition, P65 was also decreased after knockdown of lnc-SLC4A1–1(Z = −2.923, p = 0.002). Moreover, blocking the NF-κB pathway with antagonists also downregulated CXCL8, even with overexpressed lnc-SLC4A1–1, in both HTR-8/SVneo and JEG-3 cells (Fig. 4c). These findings indicated that the activation of CXCL8 by lnc-SLC4A1-1 depended on P65.
3.5. Lnc-SLC4A1-1/CXCL8 mediated immune responses in trophoblast cells
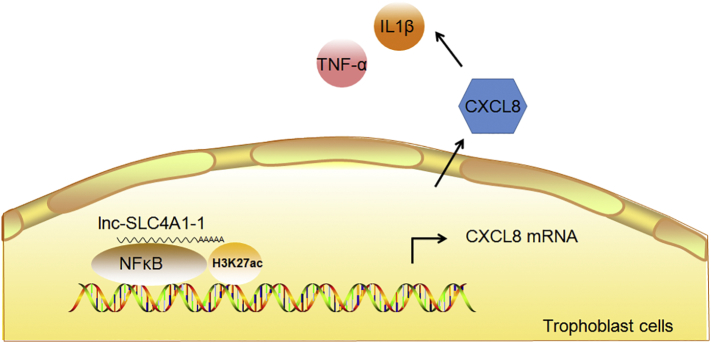
Immune responses have been reported to be associated with URPL. To determine whether lnc-SLC4A1-1/CXCL8 had roles in the inflammatory response, we also measured levels of proinflammatory factors including TNF-α and IL1β by ELISA in cells overexpressing lnc-SLC4A1-1. As expected, significantly higher levels of TNF-α (Z = −2.353, p = 0.024) and IL1β (Z = −2.598, p = 0.010) were induced after overexpression of lnc-SLC4A1-1 (Fig. 4d). This response indicated the activation of inflammation in HTR-8/SVneo and JEG-3 cells after overexpression of lnc-SLC4A1-1. In addition, when lnc-SLC4A1-1 was knocked down by siRNA or when lnc-SLC4A1-1 was overexpressed but we simulataneously added inhibitors of the NF-κB pathway, the induction of CXCL8, TNF-α and IL1β was attenuated (Fig. 4d). Overall, our data indicated that lnc-SLC4A1-1 recruited NF-κB to upregulate CXCL8 and induced the release of TNF-α and IL1β following activation of the inflammation response in trophoblast cells, which might be involved in the URPL (Fig. 5).
Fig. 5.
Proposed working model. Lnc-SLC4A1-1 recruits NF-κB to upregulate CXCL8 and induces the release of TNF-α and IL1β following activation of the inflammation response in trophoblast cells, which might be involved in the URPL.
4. Discussion
URPL is a common pathological problem in pregnancy and the etiology is complicated. In this study, we sought to identify URPL-associated lncRNAs as potential biomarkers for diagnosis and therapy. For the first time, we showed lnc-SLC4A1-1, which is an eRNA, to be dysregulated in the villi of URPL-derived trophoblast cells. Additionally, we explored the potential mechanisms by which lnc-SLC4A1-1 binding to NF-κB enhanced the expression level of CXCL8, leading to the release of inflammatory factors TNF-α and IL-1β, which might contribute to URPL.
Recently, numerous studies suggested that certain lncRNAs, which were transcribed from active enhancer regions and are referred to as eRNAs, could interact with transcription factors to regulate gene expression [[31], [32], [33], [34]]. It is estimated that >55,000 possible enhancers exist in the human genome. This number is greater than the number of protein-encoding genes, indicating the complexity of the roles of enhancers in gene regulation [35]. Enhancer regions usually harbor different histone markers, for example, poised enhancers enriched with H3K4me1, while H3K27ac and H3K4mel are enriched in active enhancers [36,37]. eRNAs transcribed from active enhancers generally exert regulatory effects in two modes: one in which eRNAs recruit transcription machinery to local promoters (cis); the other in which eRNA-induced enhancer–promoter looping activates gene transcription (trans) [38]. eRNAs were reported to interact with CBP, which is a transcription co-activator at enhancers and modifies local histone acetylation, to regulate the target genes [39]. Additionally, in breast cancer cells, estrogen stimulation could active the ERa and enhance the transcription of eRNAs. Those estrogen dependent eRNAs exerted important roles in forming enhancer-promoter looping to active target genes [31,34].
In this study, we found lnc-SLC4A1-1 had a trans regulatory effect on its target gene. Lnc-SLC4A1-1 was transcribed from the region containing H3K27ac and H3K4me1 and was retained in the nuclei. Instead of activating SLC4A1 nearby, lnc-SLC4A1-1 upregulated the expression level of CXCL8 in trans, which induced downstream release of cytokines. Although limited DEGs were observed after lnc-SLC4A1-1 was overexpressed, we noted an alteration of trophoblast cell apoptosis and migration. Rescue experiments revealed that the regulatory role of lnc-SLC4A1-1 was mediated, at least in part, via recruiting transcription factor NF-κB.
CXCL8, also known as interleukin-8 (IL-8), plays an important role in inflammation-related diseases including chronic obstructive pulmonary disease, asthma and atherosclerosis [40,41]. CXCL8 regulates inflammatory responses in lung, stomach, pancreas, liver and some cancerous tissues [40,42]. Blocking CXCL8 and CXCL8 receptor with antagonists could reduce the inflammatory response and improve the symptoms of inflammation-related diseases [43]. In our study, CXCL8 also induced inflammatory factors, such as TNF-α and IL-1β, which further exacerbate the inflammatory response. Using CXCL8 antagonists can attenuate the TNF-α and IL-1β levels even when lnc-SLC4A1-1 was overexpressed in trophoblast cells. Inflammation and inflammatory cytokines during pregnancy were associated with trophoblast function [44]. Elevated concentrations of inflammatory cytokines in the trophoblast are associated with preterm delivery (PTD), labor induction, and premature rupture of the membrane [[45], [46], [47]]. For instance, Sherief et al. reported relationships between inflammatory cytokines and PTD, showing significantly increased levels of IL-2, IFNγ, and IL-12 in trophoblast cells from PTD compared with controls [45]. The release of inflammatory cytokines can activate inflammatory pathways, resulting in increased apoptosis and decreased proliferation of trophoblast cells [48,49]. Researchers found that microRNA-145-5p was stimulated by the inflammatory cytokine TNF-α, and overexpression of microR-145-5p significantly impaired the migration and invasion of HTR-8/SVneo cells.
eRNA lnc-SLC4A-1 was found to recruit NF-κB, indicating it participated in the activation of the NF-κB signaling pathway. The NF-κB signaling pathway has been shown to regulate the expression of downstream inflammatory cytokines [[50], [51], [52]]. The activation of NF-κB was also observed in some pathological pregnancy [53]: increase of NF-κB level in the placenta was related to the downregulation of anti-apoptotic factor Bcl-2 and the increase of caspase-3, suggesting that the apoptosis of trophoblast cells depends on the NF-κB pathway [54]; another study reported that p53's role in the regulation of trophoblast proliferation and apoptosis was mediated by NF-κB [55].
Although we validated lnc-SLC4A-1's function in vitro, there were still some limitations in our study. First of all, the sample size used for sequencing was relatively small and we didn't perform functional enhancer screening for all candidate eRNAs. Secondly, we didn't have the spatiotemporal expression data of lnc-SLC4A-1, thus it was difficult to determine the exact role of lnc-SLC4A-1 during URPL. Thirdly, we didn't use the in vivo model and further study is warranted to fully characterize the causative relationship between lnc-SLC4A-1 and URPL.
In summary, our findings suggest that eRNA lnc-SLC4A1-1 recruits NF-κB and binds to the prompter of CXCL8, which leads to upregulation of CXCL8. Increased CXCL8 exacerbates inflammatory reaction by inducing TNF-α and IL-1β and affects trophoblast function, which may impact the immune response and eventually lead to URPL.
Acknowledgments
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81671461, 81402706 and 81630085), Natural Science Foundation of Jiangsu Province of China (BK20181366), National Key Research and Development Program (2016YFC1000207) and the Priority Academic Program for the Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine). Funders had no roles in study design, data collection, data analysis, interpretation, and writing of the report.
Declaration of interests
All the authors have no conflict of interests.
Authors' contributions
C.L. and X.W. directed the study, obtained financial support and were responsible for study design. Z.H. performed overall project management along with G.D. and X.H. and drafted the initial manuscript. Y.Q. performed statistical analysis. L.H. and Y.Z. were responsible for sample collection and processing. G.D. were responsible for population screening and verification. X.H. and M.Y. were responsible for functional analysis in HTR-8/SVneo and JEG-3 cells. X.H., B.X. and Y.X. conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Institutional Ethics Committee of Nanjing Medical University. All activities involved in this study were done under full compliance with government policies and the Helsinki Declaration. All experiments protocol was approved by the Institutional Review Board of Nanjing Medical University prior to the study (IRB No. NMU(2016)132).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.11.015.
Appendix A. Supplementary data
Supplementary material
References
- 1.Nelen W.L.D.M., Blom H.J., Steegers E.A.P., den Heijer M., Thomas C.M.G., Eskes T.K.A.B. Homocysteine and folate levels as risk factors for recurrent early pregnancy loss. Obstet Gynecol. 2000;95(4):519–524. doi: 10.1016/s0029-7844(99)00610-9. [DOI] [PubMed] [Google Scholar]
- 2.Van den Boogaard E., Kaandorp S., Franssen M., Mol B., Leschot N., Wouters C. Consecutive or non-consecutive recurrent miscarriage: is there any difference in carrier status? Hum Reprod. 2010;25(6):1411–1414. doi: 10.1093/humrep/deq089. [DOI] [PubMed] [Google Scholar]
- 3.Rai R., Regan L. Recurrent miscarriage. Lancet. 2006;368(9535):601–611. doi: 10.1016/S0140-6736(06)69204-0. [DOI] [PubMed] [Google Scholar]
- 4.Ford H.B., Schust D.J. Recurrent pregnancy loss: etiology, diagnosis, and therapy. Rev Obstet Gynecol. 2009;2(2):76–83. [PMC free article] [PubMed] [Google Scholar]
- 5.de La Rochebrochard E., Thonneau P. Paternal age and maternal age are risk factors for miscarriage; results of a multicentre European study. Hum Reprod. 2002;17(6):1649–1656. doi: 10.1093/humrep/17.6.1649. [DOI] [PubMed] [Google Scholar]
- 6.Andersen A.-M.N., Andersen P.K., Olsen J., Grønbæk M., Strandberg-Larsen K. Moderate alcohol intake during pregnancy and risk of fetal death. Int J Epidemiol. 2012;41(2):405–413. doi: 10.1093/ije/dyr189. [DOI] [PubMed] [Google Scholar]
- 7.Anastasiadou E., Jacob L.S., Slack F.J. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crawford G.E., Holt I.E., Whittle J., Webb B.D., Tai D., Davis S. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16(1):123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Werner M.S., Sullivan M.A., Shah R.N., Nadadur R.D., Grzybowski A.T., Galat V. Chromatin-enriched lncRNAs can act as cell-type specific activators of proximal gene transcription. Nat Struct Mol Biol. 2017;24(7):596. doi: 10.1038/nsmb.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng Y., Huang H. High maternal estradiol programming dyslipidemia in offspring via altered long non-coding rnas in fetal livers. Fertil Steril. 2017;108(3) (e324-e5) [Google Scholar]
- 12.Kallen A.N., Zhou X.-B., Xu J., Qiao C., Ma J., Yan L. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52(1):101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L., Tang H., Xiong Y., Tang L. Differential expression profile of long noncoding RNAs in human chorionic villi of early recurrent miscarriage. Clin Chim Acta. 2017;464:17–23. doi: 10.1016/j.cca.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Wang H., Cao Q., Ge J., Liu C., Ma Y., Meng Y. Lnc RNA-regulated infection and inflammation pathways associated with pregnancy loss: genome wide differential expression of lnc rna s in early spontaneous abortion. Am J Reprod Immunol. 2014;72(4):359–375. doi: 10.1111/aji.12275. [DOI] [PubMed] [Google Scholar]
- 15.Natoli G., Andrau J.-C. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet. 2012;46(1):1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 16.Kim T.-K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Santa F., Barozzi I., Mietton F., Ghisletti S., Polletti S., Tusi B.K. A large fraction of extragenic RNA Pol II transcription sites overlap enhancers. PLoS Biol. 2010;8(5) doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaikkonen Minna U., Spann Nathanael J., Heinz S., Romanoski Casey E., Allison Karmel A., Stender Joshua D. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51(3):310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hah N., Danko Charles G., Core L., Waterfall Joshua J., Siepel A., Lis John T. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145(4):622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W., Lam M.T.Y., Notani D. Enhancer RNAs. Cell Cycle. 2014;13(20):3151–3152. doi: 10.4161/15384101.2014.962860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mor G., Romero R., Aldo P.B., Abrahams V.M. Is the trophoblast an immune regulator? Role Toll-Like Recept During Pregnancy. 2005;25(5):375–388. doi: 10.1615/critrevimmunol.v25.i5.30. [DOI] [PubMed] [Google Scholar]
- 22.Fu B., Zhou Y., Ni X., Tong X., Xu X., Dong Z. Natural killer cells promote fetal development through the secretion of growth-promoting factors. Immunity. 2017;47(6) doi: 10.1016/j.immuni.2017.11.018. (1100–13.e6) [DOI] [PubMed] [Google Scholar]
- 23.Okae H., Toh H., Sato T., Hiura H., Takahashi S., Shirane K. Derivation of human trophoblast stem cells. Cell Stem Cell. 2018;22(1):50–63. doi: 10.1016/j.stem.2017.11.004. [e6] [DOI] [PubMed] [Google Scholar]
- 24.Brosens I., Pijnenborg R., Vercruysse L., Romero R. The “great obstetrical syndromes” are associated with disorders of deep placentation. Am J Obstet Gynecol. 2011;204(3):193–201. doi: 10.1016/j.ajog.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song X., Rui C., Meng L., Zhang R., Shen R., Ding H. Long non-coding RNA RPAIN regulates the invasion and apoptosis of trophoblast cell lines via complement protein C1q. Oncotarget. 2017;8(5):7637. doi: 10.18632/oncotarget.13826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun L., Xu Y., Wu D., Liu J., Ma Z., Hui B. Down-regulated Long Noncoding RNA HOXA11-AS affects trophoblast cell proliferation and migration by regulating RND3 and HOXA7 expression in preeclampsia. bioRxiv. 2018:285643. doi: 10.1016/j.omtn.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Y., Sun N., Sheng F., Ji Y., Ding H., Zhang Q. Long noncoding RNA H19 inhibits the growth and invasion of trophoblasts by inactivating Wnt/beta-catenin signaling via downregulation of DDX3X. Int J Clin Exp Pathol. 2017;10(6):6560–6567. [Google Scholar]
- 28.Qin Y., Roberts J.D., Grimm S.A., Lih F.B., Deterding L.J., Li R. An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome Biol. 2018;19(1):7. doi: 10.1186/s13059-018-1389-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abou-Kheir W., Barrak J., Hadadeh O., Daoud G. HTR-8/SVneo cell line contains a mixed population of cells. Placenta. 2017;50:1–7. doi: 10.1016/j.placenta.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Cotton J., Platnich J., Muruve D., Jijon H., Buret A., Beck P. Interleukin-8 in gastrointestinal inflammation and malignancy: induction and clinical consequences. Int J Interferon Cytokine Mediat Res. 2016;8:13–34. [Google Scholar]
- 31.Li W., Notani D., Ma Q., Tanasa B., Nunez E., Chen A.Y. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y., Su Z., Song X., Liang B., Zeng F., Chang X. Enhancer RNA-driven looping enhances the transcription of the long noncoding RNA DHRS4-AS1, a controller of the DHRS4 gene cluster. Sci Rep. 2016;6:20961. doi: 10.1038/srep20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Léveillé N., Melo C.A., Rooijers K., Díaz-Lagares A., Melo S.A., Korkmaz G. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat Commun. 2015;6:6520. doi: 10.1038/ncomms7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai P.-F., Dell'Orso S., Rodriguez J., Vivanco K.O., Ko K.-D., Jiang K. A muscle-specific enhancer RNA mediates cohesin recruitment and regulates transcription in trans. Mol Cell. 2018;71(1) doi: 10.1016/j.molcel.2018.06.008. (129–41.e8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2010;470:279. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H., Du G., Song X., Li L. Non-coding transcripts from enhancers: new insights into enhancer activity and gene expression regulation. Genomics Proteomics Bioinformatics. 2017;15(3):201–207. doi: 10.1016/j.gpb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bose D.A., Donahue G., Reinberg D., Shiekhattar R., Bonasio R., Berger S.L. RNA binding to CBP stimulates histone acetylation and transcription. Cell. 2017;168(1–2) doi: 10.1016/j.cell.2016.12.020. (135–49.e22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russo R.C., Garcia C.C., Teixeira M.M., Amaral F.A. The CXCL8/IL-8 chemokine family and its receptors in inflammatory diseases. Expert Rev Clin Immunol. 2014;10(5):593–619. doi: 10.1586/1744666X.2014.894886. [DOI] [PubMed] [Google Scholar]
- 41.Graham G., Locati M., Mantovani A., Rot A., Thelen M. The biochemistry and biology of the atypical chemokine receptors. Immunol Lett. 2012;145(1–2):30–38. doi: 10.1016/j.imlet.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 42.Horuk R. The Duffy antigen receptor for chemokines DARC/ACKR1. Front Immunol. 2015;6:279. doi: 10.3389/fimmu.2015.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ha H., Debnath B., Neamati N. Role of the CXCL8-CXCR1/2 axis in cancer and inflammatory diseases. Theranostics. 2017;7(6):1543–1588. doi: 10.7150/thno.15625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brien M.-E., Duval C., Palacios J., Boufaied I., Hudon-Thibeault A.-A., Nadeau-Vallée M. Uric acid crystals induce placental inflammation and alter trophoblast function via an IL-1–dependent pathway: implications for fetal growth restriction. J Immunol. 2017;198(1):443–451. doi: 10.4049/jimmunol.1601179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El-Shazly S., Makhseed M.A., Azizieh F., Raghupathy R. Increased expression of pro-inflammatory cytokines in placentas of women undergoing spontaneous preterm delivery or premature rupture of membranes. Am J Reprod Immunol. 2004;52(1):45–52. doi: 10.1111/j.1600-0897.2004.00181.x. [DOI] [PubMed] [Google Scholar]
- 46.Romero R., Mazor M., Brandt F., Sepulveda W., Avila C., Cotton D.B. Interleukin-1α and Interleukin-1 β in preterm and term human parturition. Am J Reprod Immunol. 1992;27(3–4):117–123. doi: 10.1111/j.1600-0897.1992.tb00737.x. [DOI] [PubMed] [Google Scholar]
- 47.Koh Y.Q., Chan H.-W., Nitert M.D., Vaswani K., Mitchell M.D., Rice G.E. Differential response to lipopolysaccharide by JEG-3 and BeWo human choriocarcinoma cell lines. Eur J Obstet Gynecol Reprod Biol. 2014;175:129–133. doi: 10.1016/j.ejogrb.2013.12.032. [DOI] [PubMed] [Google Scholar]
- 48.Banerjee P., Malik A., Malhotra S.S., Gupta S.K. Role of STAT signaling and autocrine action of chemokines during H2O 2 induced HTR-8/SVneo trophoblastic cells invasion. J Cell Physiol. 2018:1–18. doi: 10.1002/jcp.26934. [DOI] [PubMed] [Google Scholar]
- 49.Chen Y., Xue F., Han C., Yang H., Han L., Li K. Ferulic acid ameliorated placental inflammation and apoptosis in rat with preeclampsia. Clin Exp Hypertens. 2018:1–7. doi: 10.1080/10641963.2018.1516773. [DOI] [PubMed] [Google Scholar]
- 50.Baldwin A.S., Jr. The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14(1):649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 51.Pahl H.L. Activators and target genes of Rel/NF-κB transcription factors. Oncogene. 1999;18(49):6853. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Q., Lenardo M.J., Baltimore D. 30 years of NF-κB: a blossoming of relevance to human pathobiology. Cell. 2017;168(1–2):37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakowicz A. The role of NFκB in the three stages of pregnancy – implantation, maintenance, and labour: a review article. BJOG. 2018;0(0) doi: 10.1111/1471-0528.15172. [DOI] [PubMed] [Google Scholar]
- 54.Aban M., Cinel L., Arslan M., Dilek U., Kaplanoglu M., Arpaci R. Expression of nuclear factor-kappa b and placental apoptosis in pregnancies complicated with intrauterine growth restriction and preeclampsia: an immunohistochemical study. Tohoku J Exp Med. 2004;204(3):195–202. doi: 10.1620/tjem.204.195. [DOI] [PubMed] [Google Scholar]
- 55.Ryan K.M., Ernst M.K., Rice N.R., Vousden K.H. Role of NF-κB in p53-mediated programmed cell death. Nature. 2000;404:892. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material