Abstract
Background
Amyloid precursor protein (APP), best known for its association with Alzheimer disease, has recently been implicated in breast cancer progression. However, the precise mechanism involved remains unclear. Here, we investigated the role of APP proteolytic cleavage in breast cancer functions.
Methods
The presence of APP proteolytic cleavage products was examined in breast cancer cell lines. The functional roles of APP in breast cancer were studied in vitro and tumor xenograft model using siRNA. The effects of full length APP and the α-secretase cleaved ectodomain fragment, soluble APPα (sAPPα) were further investigated for their overexpression in breast cancers. The α-secretase involved was identified. The α-secretase expression together with APP was examined in clinical breast cancers.
Results
We showed that APP underwent proteolytic cleavage in breast cancer cells to generate sAPPα. The sAPPα and full length protein mediated breast cancer migration and proliferation, but in different functional extent. This proteolytic cleavage was mediated by ADAM10. Downregulation of APP and ADAM10 brought about similar functional effects. Overexpression of sAPPα reversed the effects of ADAM10 downregulation. Interestingly, in patients with non-luminal breast cancers, APP and ADAM10 expression correlated with each other and their co-expression was associated with the worst outcome.
Conclusions
These results demonstrated the contributory role of APP cleavage on its oncogenic roles in breast cancer. ADAM10 was the key α-secretase. APP and ADAM10 co-expression was associated with worse survival in non-luminal breast cancers. Targeting of APP or its processing by ADAM10 might be a promising treatment option in these cancers.
Keywords: Amyloid precursor protein, ADAM10, Breast cancer, Proliferation, Migration, Patients' survival
Research in context section.
Evidence before this study
Amyloid precursor protein (APP), best known for its association with Alzheimer disease, has recently suggested to play oncogenic roles in breast cancer. However, the precise mechanism involved remains unclear.
Added value of this study
Here, we provided evidence on its proteolytic cleavage in breast cancer functions, particularly in non-luminal breast cancers. APP was demonstrated to undergo proteolytic cleavage by ADAM10 to promote proliferation and migration in breast cancer cell lines. Overexpression of soluble APPα fragment could rescue the effects of ADAM10 inhibition. Importantly, their co-expression was particularly associated with adverse outcome in non-luminal breast cancers (included both HER2-overexprsesing and triple negative cancers).
Implications of all the available evidence
ADAM10 inhibition has been tested in a clinical trial for treatment of HER2 positive breast cancer. The current results may suggest a boarder application of ADAM10 inhibition also in triple negative cancers. Our observations suggested the potentials in targeting of APP or its processing by ADAM10 for the treatment in these cancers and further support on APP as a biomarker in clinical breast cancer.
Alt-text: Unlabelled Box
1. Introduction
Aberrant processing of amyloid precursor protein (APP) to release amyloid-β is a crucial event in the pathogenesis of Alzheimer's disease (AD). APP is a highly pleiotropic protein involved in a number of cellular functions, including cell survival, cellular adhesion, differentiation and migration [1]. All these processes are also essential for carcinogenesis. Apart from pathogenesis of AD, the oncogenic role(s) of APP have been suggested recently in breast cancers [[2], [3], [4]]. Indeed, APP knockdown (KD) in breast cancer cells caused growth inhibition in vitro and in vivo with the induction of p27 and caspase-3-mediated apoptosis [2]. Moreover, its downregulation also reduced breast cancer motility. In clinical samples, we and others have shown an unfavorable prognostic role of APP expression in patients with different subtypes of breast cancers [3,5]. However, it is yet to be defined how APP mediates these various functional effects in breast cancer.
APP can undergo sequential cleavage via two mutually exclusive pathways into biologically active fragments. In the amyloidogenic pathway which is associated with AD, APP is cleaved by β-secretase and γ-secretase to generate soluble N-terminal ectodomain APP-β (sAPPβ), pathogenic amyloid-β peptide and APP intracellular domain (AICD). In the non-amyloidogenic pathway, α-secretase (ADAM10 and/or ADAM17) cleaves APP within the amyloid-β sequence, following by γ-secretase cleavage to generate sAPPα, P3 fragment and AICD [1]. These APP cleavage products may contribute to carcinogenesis. In lung cancers, the C-terminal AICD fragment was indicated in regulation of cell cycle progression [6]. In other cancers, the N-terminal sAPP fragment can be detected in conditioned medium from cancer cell lines [[7], [8], [9], [10]]. The sAPP fragment was suggested to promote cancer cell proliferation [8,9,11]. In breast cancers, both α-secretases, ADAM10 and ADAM17, have been implicated in cancer progression and were found to be aberrantly expressed. It is possible that APP may undergo α-cleavage to mediate its oncogenic functions in breast cancers.
In the current study, we examined the expression of APP, ADAM10 and ADAM17 in different breast cancer cell lines and explored the role of APP processing in breast cancer pathogenesis. The relationship between APP and α-secretase expression was also examined in clinical breast cancers. We showed that expression of APP and the proteolytic fragments from α-cleavage could be detected in breast cancer cells. Inhibition of APP expression using RNA interference decreased breast cancer growth and migration in vitro and in vivo. In contrast, opposite effects were observed with sAPPα or full length APP overexpression. Knockdown of ADAM10, but not ADAM17, inhibited α-cleavage of APP. Overexpression of sAPPα can reverse the effect of ADAM10 knockdown in tumor cells. Importantly, there was a significant correlation of APP and ADAM10 expression in non-luminal breast cancer samples and co-expression of both proteins conferred unfavorable clinical outcome. This preclinical study and clinical data analysis supported the involvement of APP and its processing in the development of breast cancer.
2. Materials and methods
2.1. Cell lines and materials
MDA-MB231, MCF7, BT-549, MDA-MB468 and ZR-75-1 were grown in RPMI supplemented with 10% (vol/vol) fetal bovine serum (FBS). SK-BR-3 was grown in McCoy5A supplemented with 10% FBS. Cell lines were obtained from the American Tissue Culture Collection and maintained in a 37 °C CO2 humidified incubator. Cell line identity was confirmed by analysis of Short Tandem Repeat Loci. Cells were routinely tested for mycoplasma infection.
The primary antibodies used were as follows: anti-α-tubulin (DMIA; Sigma), anti-β-actin (C4, Santa Cruz); anti-APP N terminus (22C11; Millipore), anti-sAPPα (2B3, Immuno-Biological Laboratories), anti-sAPPβ (poly8134, Biolegend), anti-ADAM10 (polyclonal; Origene), anti-ADAM17 (H-300; Santa Cruz) and Ki67 (41,912; Ventana). Validation of sAPP antibodies was shown in Supplementary Fig. S1.
The mammalian expression constructs for APP (pCAX-APP751) and sAPPα (pCAX-APPs-751-alpha) were obtained from Addgene. On-Targetplus siRNA oligonucleotides against APP (SMARTpool L-003731-00-0020) and control non-target siRNA pool (D-001810-10-50) were purchased from Dharmacon. siRNA sequences for knockdown of ADAM10 and ADAM17 reported previously (ADAM10: AGACAUUAUGAAGGAUUAUdTdT and ADAM17: GAGAAGCUUGAUUCUUUGCdTdT) were synthesized by Dharmacon [12]. As a negative control, an unspecific scrambled siRNA duplex (5′-AGG UAG UGU AAU CGC CUU GTT-3′) was applied.
2.2. Patients and tissues
Consecutive excision cases from patients with invasive breast cancer were selected from 3 involved institutions over a period of 4 (2002–2005), 7 (2003–2009), and 4 (2003–2006) years. Information on patients' age, sex, tumor size, pT stage, pN stage and outcome data were obtained from medical reports. The original H&E slides for each case were retrieved and reviewed by two pathologists independently. Histologic diagnosis was made according to the WHO Classification of Tumors of the Breast (4th ed) [13]. The tumors were graded basing on modified Bloom and Richardson grading [14]. Any discrepancies were resolved at a multi-head microscope by discussion to reach a consensus. Breast cancer specific survival (BCSS) was defined as the time from surgery to death from breast cancer. Disease free survival (DFS) was defined as the duration from the time of surgery to initial diagnosis to the first detection of breast cancer specific relapse or death. Tissue microarray was constructed from representative tumor regions of paraffin embedded specimens for immunohistochemistry. The study was approved by Joint Chinese University of Hong Kong-New Territories East Cluster clinical research ethics committee.
2.3. Cell transfection
For knockdown of APP, ADAM10 or ADAM17, siRNA to the respective mRNA were transfected into cells using Lipofectamine RNAiMAX according to the manufacturer's protocols. Briefly, cells were seeded in 24-well plates at 1 × 104 cells/well and transfected with 6 pmol of siRNA. Cells were collected at 48–72 h post-transfection for further analysis. For over-expression, cells were seeded in 24 well plates at 1 × 105 cells/well and transfected with 500 ng of plasmid using Lipofectamine2000 (Invitrogen) according to the manufacturer's instruction. For co-transfection experiments, same protocol for over-expression was employed but with 250 ng of plasmid and 6 pmol of siRNA. The efficiency of knockdown/overexpression was determined using immunoblot analysis.
2.4. Immunoblotting
Cells were lysed in ice-cold RIPA buffer [50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate and 0.1%SDS] containing protease inhibitors (Biotool) and 1 mM Na3VO4. Protein quantitation was performed against BSA standard using RC DC Protein Assay (Bio-Rad). Samples containing equal amounts of protein were separated under reducing conditions using SDS-PAGE and the proteins transferred to PVDF membranes (Millipore, Watford, UK). Blots were blocked with 5% milk in TBST and primary antibody was incubated overnight. Bound antibodies were detected with peroxidase-conjugated secondary antibodies and Enhanced Chemiluminescence reagents (Amersham, Menlo Park, CA, USA).
2.5. Cell proliferation assay
The cell proliferation reagent CCK-8 (Biotool) was used to determine the rate of cell proliferation. Cells were seeded into a 96-well plate at a density of 1 × 103 cells per well. Cell viability was estimated at day 3–5. The absorbance in each well was measured at a wavelength of 450 nm and normalized using a wavelength of 690 nm, detected by the Victor 3 (Perkin Elmer).
2.6. Invasion and migration
Transwell migration and invasion assays were carried out using Boyen chamber system. Cells were cultured in serum-free medium overnight. For migration assays, 1 × 105 cells in 0.2 ml serum free medium were placed in the upper chamber with a non-coated polycarbonate membrane (24 well insert; 8 μm pore size, Corning, Cambridge, MA, USA) and 0.5 ml medium containing 10% FBS were added into the lower chamber. The cells were allowed to migrate for 8 h in the lower chamber. Migrated cells to the lower surface of the filter were stained with 0.5% crystal violet and counted under bright field microscope. For invasion assays, cells were placed in the upper chamber with a matrigel-coated membrane (8 μm pore size, BD BioCoat Matrigel) and allowed migrated for 22 h.
2.7. Xenograft mouse model
The control and APP knockdown MDA-MB231 (1 × 106) cells were prepared in the solution (1:1) of PBS and growth factor-reduced matrigel and followed by injection into mammary fat pads of female athymic nude mice (5-week-old nu/nu strain BALB/cAnN.Cg-Foxn1nu/CrlNarl). Tumor growth was monitored weekly by measuring perpendicular tumor diameters, length (L) and width (W), with a Vernier caliper. The tumor volume (V) was calculated as V = LW2/2. Mice were sacrificed at the end of the experiments by cervical dislocation and the tumors were preserved for further analysis. Ethical approval was obtained from the University Animal Experimentation Ethics Committee (AEEC), CUHK, and the animal license was approved by the Hong Kong Government, Department of Health.
2.8. Immunohistochemistry
4-μm sections from tissue microarrays (TMA) were used for IHC analysis of APP and ADAM10. Immunohistochemical (IHC) staining of APP (clone 22C11) and ADAM10 were performed on the TMA slides using Ultraview Universal DAB Detection Kit (Ventana, Tucson, AZ) after deparaffinization, rehydration, and antigen retrieval. They were counterstained with hematoxylin. Protein expression was assessed for staining intensity and the actual percentage of stained cells in tumor cells by two of the authors blinded to the clinical information and the staining results of other markers.
2.9. Statistical analysis
The findings were analyzed using the statistical software SPSS for Windows, Version 23. Chi-square analysis or Fisher exact test was used to test for the association of APP expression with categorical variables. Survival data were evaluated with Kaplan–Meier analysis with a log-rank test. The Student's t-test was used in the in vitro and in vivo experiments. One-way ANOVA followed by Turkey post-hoc comparisons were performed for comparing means of multiple groups. Statistical significance was established at p < 0.05.
3. Results
3.1. APP undergoes proteolytic cleavage in breast cancer cell lines
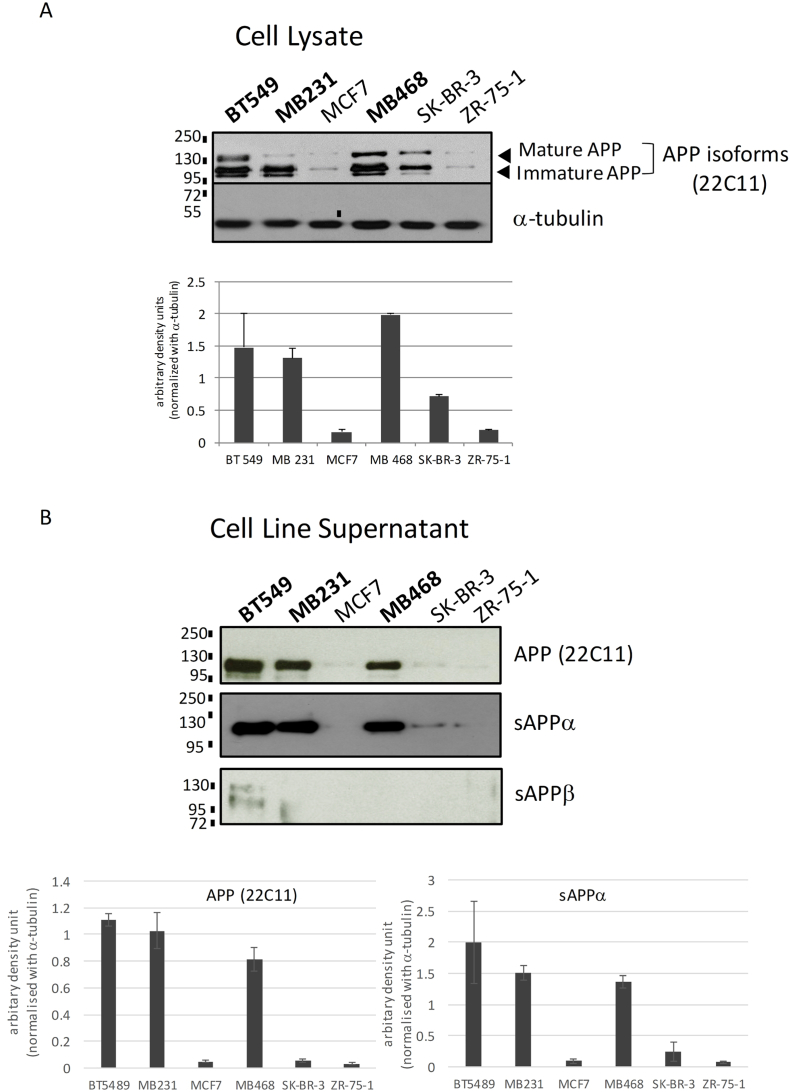
We examined APP protein levels in breast cancer cell lines, MCF7, BT-549, MDA-MB231, MDA-MB468, SK-BR-3 and ZR-75-1 using antibody recognizing the APP N-terminus (clone 22C11). The results showed that three triple negative breast cancer cell lines, BT-549, MDA-MB231 and MDA-MB468, showed the highest level of APP expression while SK-BR-3 (ER-HER2+) showed intermediate APP level and MCF7 and ZR-75-1 had the lowest APP expression, indicating that APP showed a differential expression in different breast cancer cell lines (Fig. 1A). Multiple bands observed corresponded to the mature and immature forms of APP with different degree of post-translational modifications [15]. In addition, soluble APP (sAPP) can be secreted by breast cancer cells into the culture supernatant. APP in the conditioned supernatant can be detected using antibodies against the N-terminal domain of APP (22C11) and sAPPα specific antibody, but not antibody specific for APPβ indicating that APP underwent α-cleavage to produce sAPPα in breast cancers (Fig. 1B).
Fig. 1.
Expression of APP and its proteolytic product in breast cancer cell lines. (A) Immunoblot analysis using an antibody against APP N-terminal (clone 22C11) showing differential expression of APP on breast cancer cell lines BT-549, MDA-MB231, MCF7, MDA-MB468, SK-BR-3 and ZR-75-1. α-tubulin was used as a control. Densitometric analysis of APP expression from triplicate experiments was shown at the lower panel. (mean ± SD; n = 3). (B) Conditioned medium from cell line culture was examined using antibodies specific for sAPPα and sAPPβ, in addition to clone 22C11. Secretion of sAPPα, but not sAPPβ into cell culture supernatant can be detected. All experiments were performed at least three times.
3.2. Knockdown of APP affected breast cancer functions
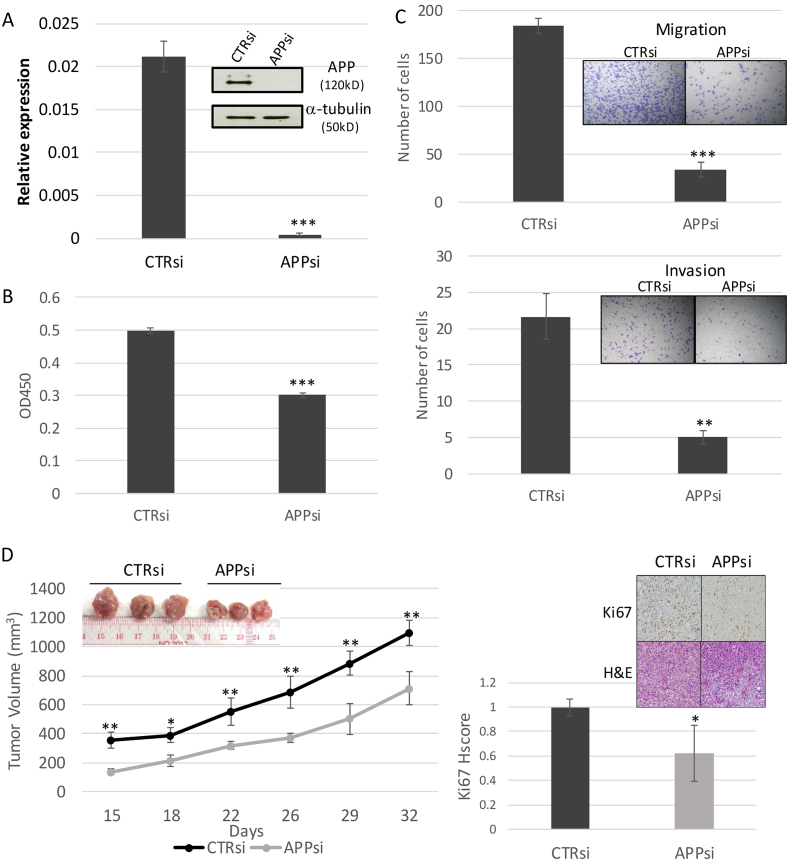
Functional roles of APP in breast cancer were further examined by the knockdown approach using APP specific siRNAs. Cells transfected with non-targeting siRNAs were used as controls. Both mRNA and protein expression of APP were markedly reduced in APP-KD BT-549 compared with controls after siRNA transfection (Fig. 2A). The effect of APP KD in cell proliferation was examined by CKK-8 assay. A significant reduction of cell growth in the KD cells was observed, compared to the control (Fig. 2B). Since APP expression has also been associated with metastasis in breast cancer patients [5], we examined the efficiency of APP KD cells in migration and invasion across Boyen chamber. APP KD significantly reduced both cell migration and invasive ability of breast cancer (Fig. 2C). Similar results were observed using MDA-MB231 cell lines (data not shown). To further confirm the in vitro findings, we examined the effect of APP KD in the tumor xenograft mouse model using MDA-MB231. Control or APP KD MDA-MB231 cells were injected into the mammary fat pad of nude mice. The mice were maintained for 4–6 weeks. Consistent with the in vitro findings, there was a significant difference in tumor volume between APP KD and control cells. Additionally, the tumor xenografts from APP KD cells showed a lower level of Ki67 expression (Fig. 2D). The data supported the multiple functional roles of APP in breast cancer progression.
Fig. 2.
APP Knockdown affects breast cancer functions. (A) Knockdown of APP by siRNA in breast cancer cell line (BT-549) resulted in marked reduction of APP expression in both mRNA and protein level. Knockdown of APP was verified in qRT-PCR using SYBR green and western blot of the cell lysate (insert) using anti-APP antibody clone 22C11 with β-actin as control were shown. (mean ± SD; n = 3). Statistical analysis was performed using student t-test. ***p < 0.001. All experiments were performed at least three times. (B) APP knockdown resulted in a reduction in cell proliferation. KD or control cells (1 × 103) were seeded in a 96 well plate and cell proliferation was measured at day 4 by CCK8 assay. (mean ± SD; n = 3). Statistical analysis was performed using student t-test. ***p < 0.001. All experiments were performed at least three times. (C) Inhibition of APP reduced both migration and invasion of breast cancer. Cells (1 × 105) were seeded with 0.2 ml serum free medium in the upper chamber of Boyen chamber after overnight serum starvation. 0.5 ml medium supplemented with 10% FBS was added to the lower chamber. Migration was evaluated for 22 h. For invasion, cells were placed in an upper chamber with matrigel coated membrane. (mean ± SD; n = 3). Statistical analysis was performed using student t-test. ***p < 0.001; **p < 0.01. All experiments were performed at least three times. (D) APP knockdown modulated breast cancer growth in vivo. APP KD MDA-MB231 or control cells (1 × 106) were injected into the mammary fat pad of nude mice and allowed to grow for 5 weeks. Tumor growth was measured twice weekly with a caliper (left panel). Ki67 level of the xenograft was analyzed by immunohistochemistry. Representative Ki67 and H&E stainings from each group were shown at the insert. (mean ± SD; n = 3). Statistical analysis was performed using student t-test. **p < 0.01; *p < 0.05. The in vivo data were reproduced in independent xenograft studies.
3.3. Overexpression of APP enhances breast cancer function
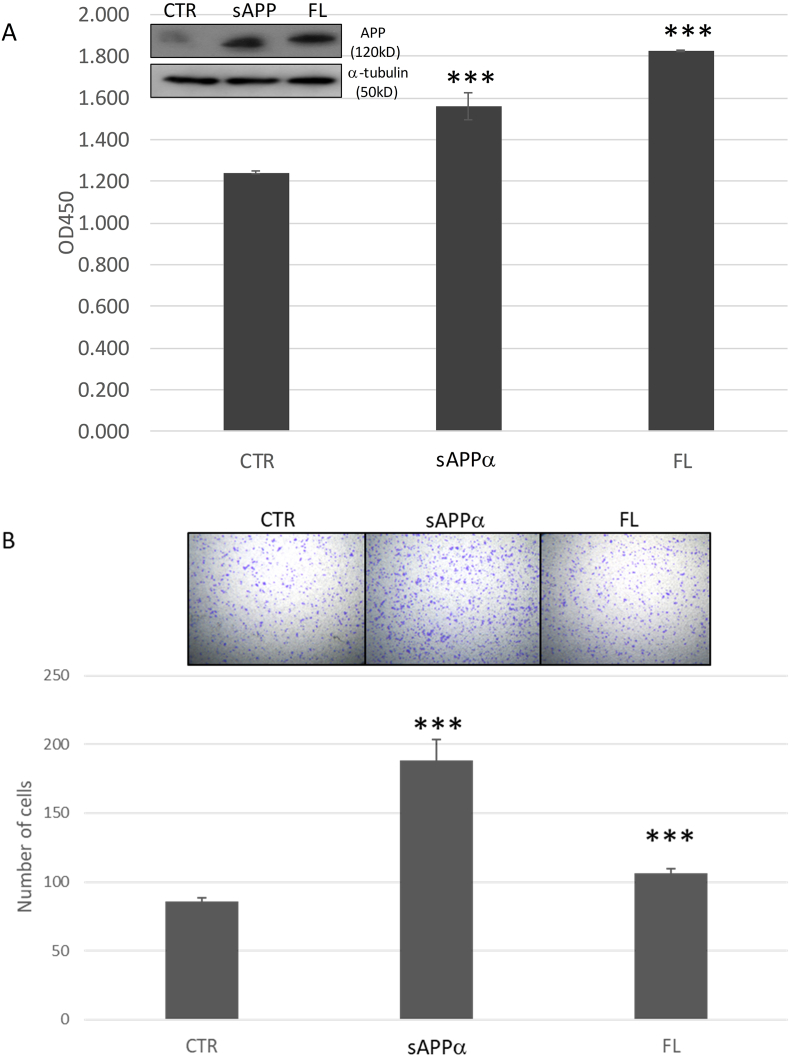
To examine whether full length or cleaved soluble protein is required for the functional effects of APP in breast cancer, MDA-MB231 was overexpressed with either full length APP or sAPPα and the effects on proliferation and migration were examined. Overexpression of full length protein significantly enhanced the proliferation of MDA-MB231, similarly for its migratory capacity. Interestingly, transfection with sAPPα construct can also significantly promote proliferation and migration in breast cancer compared to control (ANOVA p < 0.05; Supplementary Table S1) (Fig. 3). Of note, the extent of their effects appeared to be different in different breast cancer functions. Similar results were also obtained with MCF7 cells which expressed a low level of APP (Supplementary Fig. S2).
Fig. 3.
Overexpression of full length APP and sAPPα enhanced breast cancer proliferation and migration. (A) Expression of full length or sAPPα promoted proliferation in breast cancer cell line (MDA-MB231). MDA-MB231 was transfected with full length (FL), sAPP or control (CTR) construct and were seeded (1 × 103) in a 96 well plate. Western blotting analysis for APP expression after transfection was shown (insert). Cell proliferation was measured at day 4 by CCK8 assay. (mean ± SD; n = 3). Statistical analysis was performed using student t-test. ***p < 0.001. (B) APP overexpression enhanced migration of breast cancer. Cells transfected with different APP constructs (1 × 105) were seeded with 0.2 ml serum free medium in the upper chamber of Boyen chamber after overnight serum starvation. 0.5 ml medium supplemented with 10% FBS was added to the lower chamber. Migration was evaluated for 22 h. (mean ± SD; n = 3). Statistical analysis was performed using student t-test. ***p < 0.001. All experiments were performed at least three times.
3.4. ADAM10 but not ADAM17 is responsible for APP processing in breast cancer
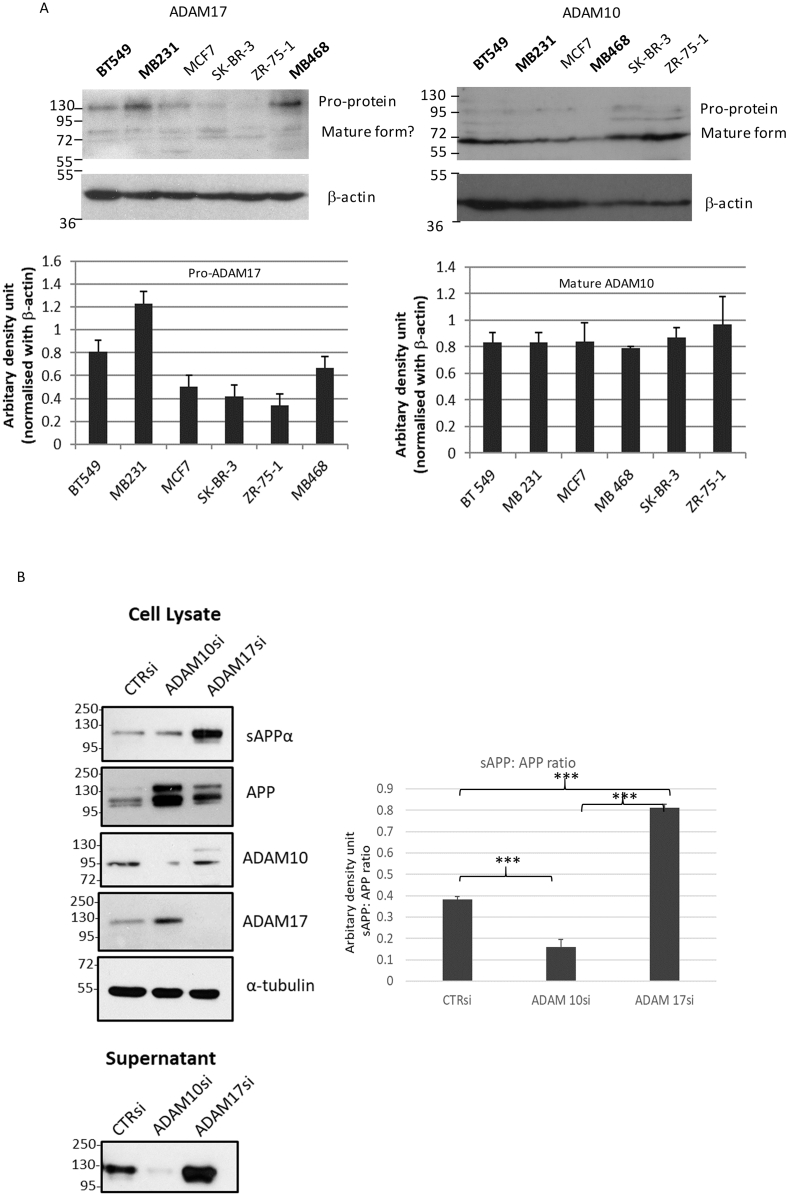
Both ADAM10 and ADAM17 were implicated in α-cleavage of APP. To investigate their role(s) in APP processing in breast cancers, we first examined their expression in breast cancer cell lines by immunoblotting. Both ADAM10 and ADAM17 can be detected in breast cancer cell lines. Using the antibody mainly detected the pro-protein, a differential expression of ADAM17 in different cell lines was observed, with the highest expression in MDA-MB231, followed by BT-549. ADAM10 showed high expression across different lines (Fig. 4A). The ADAM10 antibody supposedly recognizes both mature and pro-protein. It appears that the mature ADAM10 was predominantly observed. In line, a high level of ADAM10 expression could be detected also on the cell surface in different cell lines by flow cytometry. However, the surface expression was low for ADAM17 in all cell lines (Supplementary Fig. S3). In order to determine which ADAM is required for APP processing in breast cancers, siRNA specific for ADAM10 or ADAM17 was transfected into breast cancer cell lines and the release of sAPPα was determined. Effective downregulation of ADAM10 and ADAM17 expression was shown by immunoblotting. ADAM10 KD, but not ADAM17 KD reduced the release of sAPPα into culture supernatant (Fig. 4B). Similar results were obtained using ADAM10 (GI254023X) and ADAM17 (TAPI-2) inhibitors (Supplementary Fig. S4). Of note, a higher level of APP expression could be detected in cell lysates after ADAM10 KD, probably indicating an accumulation of full length APP. In addition, upon ADAM17 KD, an increased level of sAPP was detected in the supernatant, possibly due to the compensatory upregulation of ADAM10 upon ADAM17 KD [16]. The residual level of sAPPα detected upon ADAM10 siRNA transfection could be due to incomplete knockdown and possibly ADAM10 independent cleavage of APP by other proteases [17].
Fig. 4.
ADAM10 and ADAM17 in APP processing of breast cancer cell lines. (A) Immunoblot analysis using antibodies against ADAM10 and ADAM17 showed the expression of both proteins in breast cancer cell lines (BT-549, MDA-MB231, MCF7, MDA-MB468, SK-BR-3 and ZR-75-1). β-actin was used as a control. Densitometric analysis of protein expression on the predominant form detected (pro-ADAM17 and mature ADAM10) from replicate experiments were shown at the lower panel. (mean ± SD; n = 3). (B) Knockdown of ADAM 10 but not ADAM17 showed reduced sAPP level in conditioned medium from cell line culture. Breast cancer cell line (MDA-MB231) was transfected with ADAM10 or ADAM17 siRNA showed a marked reduction in protein and mRNA expression. A decreased level of sAPP in the conditioned supernatant was found with ADAM10 but not ADAM17 knockdown. Densitometric analysis of APP and sAPP was shown at the right panel. (mean ± SD; n = 3). Statistical analysis was performed using student t-test. ***p < 0.001. All experiments were performed at least three times.
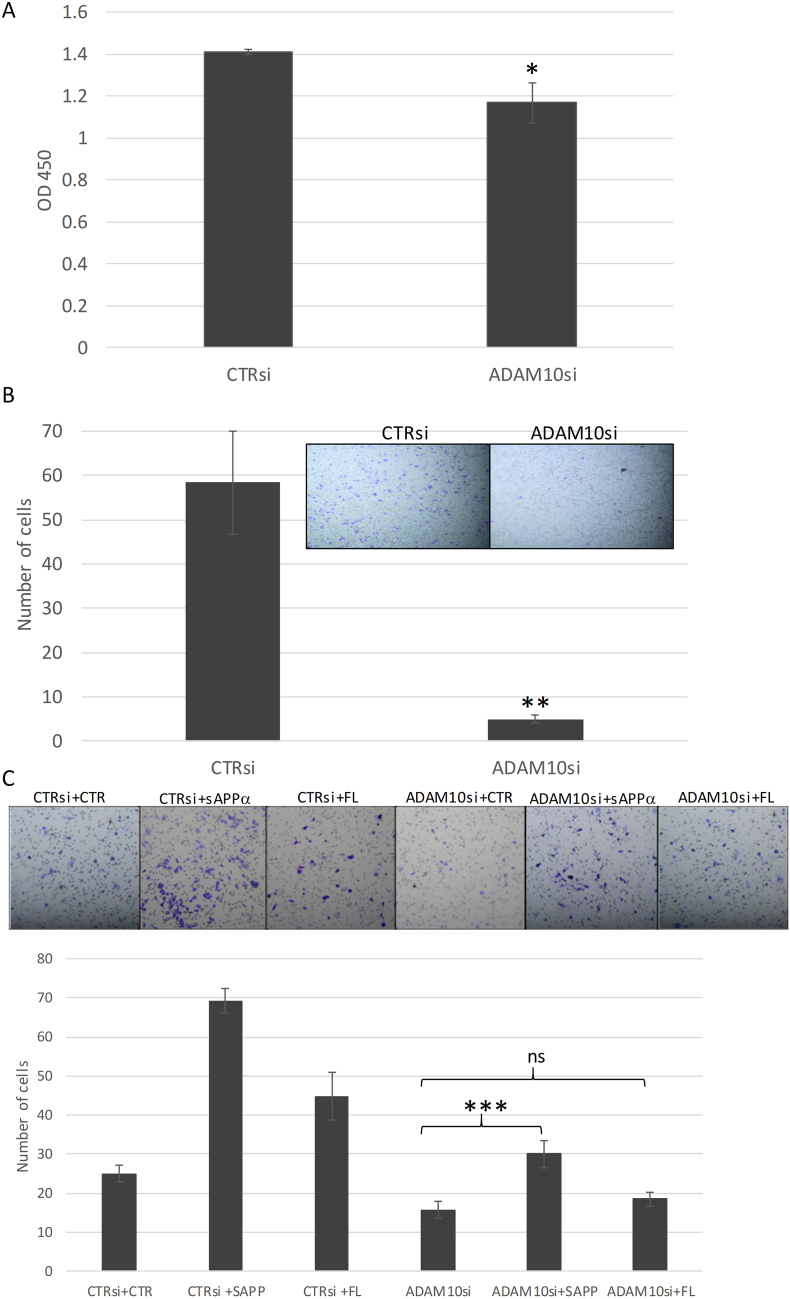
3.5. Functional effect of ADAM10 knockdown can be reversed by sAPPα expression
Having confirmed the effect of ADAM10 on sAPPα production, next, we examined whether ADAM10 KD showed similar functional effects as APP KD. MDA-MB231 was transfected with ADAM10 siRNA and the effects on proliferation and transwell migration were determined. Similar to APP KD, there was a reduction in proliferation and migratory capacities of ADAM10 KD cells as compared to the controls (Fig. 5A and B). To investigate whether the effects of ADAM10 KD were mediated by inhibition of sAPPα generation, ADAM10 siRNA was co-transfected with sAPPα construct into MDA-MB231 cells. In cells with co-transfection of control plasmid, a reduction in migration was found after ADAM10 KD compared to control siRNA. Interestingly, co-transfection of sAPPα can revert the inhibitory effect of ADAM10 KD (Fig. 5C). In contrast, the overexpression of the full length construct did not appear to have a similar effect (ANOVA p < 0.05; Post-hoc tests showed significant differences between group except CTRsi+CTR Vs ADAM10si + sAPP and ADAM10si + CTR Vs ADAM10si + FL, Supplementary Table S2). Rescue effects were also observed for sAPPα and full length APP in proliferation (Supplementary Fig. S5).
Fig. 5.
Effect of ADAM10 knockdown in breast cancer partly reversed by sAPPα expression. (A) ADAM10 knockdown reduced proliferation in breast cancer cell line (MDA-MB231). MDA-MB231 was transfected with ADAM10 siRNA and were seeded (1 × 103) in a 96 well plate. Cell proliferation was measured at day 4 by CCK8 assay. (mean ± SD; n = 3). Statistical analysis was performed using student t-test. *p < 0.05. (B) ADAM10 knockdown reduced migration of breast cancer. Cells transfected ADAM10 siRNA (1 × 105) were seeded with 0.2 ml serum free medium in the upper chamber of Boyen chamber after overnight serum starvation. 0.5 ml medium supplemented with 10% FBS was added to the lower chamber. Migration was evaluated for 22 h. (C) ADAM10 knockdown effect can be reversed by sAPP overexpression. MDA-MB231 cells co-transfected with ADAM10 siRNA and sAPPα/full length (FL) construct were seeded (1 × 105) with 0.2 ml serum free medium in the upper chamber of Boyen chamber after overnight serum starvation. 0.5 ml medium supplemented with 10% FBS was added to the lower chamber. Migration was evaluated for 22 h. (mean ± SD; n = 3). Statistical analysis was performed using student t-test. ***p < 0.001; **p < 0.01; *p < 0.05; ns-no statistical significance. All experiments were performed at least three times.
3.6. Relationship of APP and ADAM10 in clinical breast cancers
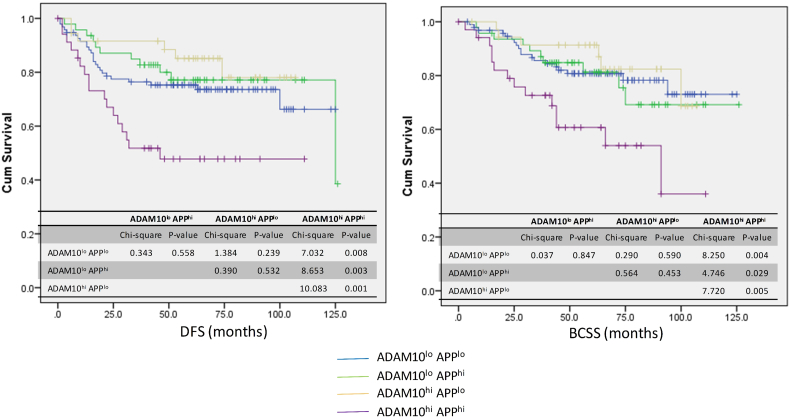
We have observed a high level and unfavorable prognostic role of APP expression in non-luminal breast cancers [4,5]. In order to investigate the involvement of APP processing on the prognostic role of APP, the expression of ADAM10 in a series of non-luminal breast cancers was analyzed by immunohistochemistry and correlated with APP expression and its prognosis. In total, 277 cases of non-luminal breast cancers were involved. High ADAM10 and APP expression were found in 37.8% (105/277) and 38.2% (96/251) of cancers respectively. No significant correlations were found between ADAM10 with grade, pT stage, pN stage, patients' age, tumor size and HER2 status. A significant positive correlation was found with APP expression (p = 0.004) and a marginal significance for Ki67 (p = 0.079) (Table 1). The clinico-pathological association of ADAM10 was similar to that observed for APP in our previous report [4]. Further, we have examined whether ADAM10 affected the prognostic value of APP in non-luminal breast cancers. Of note, cases co-expressing ADAM10 and APP (ADAM10hi APPhi) showed the worst breast cancer specific survival (chi-square = 11.519, p = 0.009) and disease free survival (chi-square = 14.252, p = 0.003) compared to the ADAM10lo APPlo cases (Fig. 6). In contrast, those cases with high expression of either APP or ADAM10 expression did not show any significant difference.
Table 1.
Clinico-pathological association of ADAM10 expression in non-luminal breast cancers.
| ADAM10 N(%) |
p-Value | ||||
|---|---|---|---|---|---|
| Low | High | Total | |||
| Grade | 1 | 5 | 3 | 8 | 0.426 |
| 2 | 26 | 21 | 47 | ||
| 3 | 141 | 81 | 222 | ||
| Total | 172 | 105 | 277 | ||
| pT | 1 | 52 | 35 | 87 | 0.944 |
| 2 | 103 | 54 | 157 | ||
| 3 | 11 | 11 | 22 | ||
| 4 | 6 | 3 | 9 | ||
| Total | 172 | 103 | 275 | ||
| pN | 0 | 85 | 47 | 132 | 0.986 |
| 1 | 42 | 34 | 76 | ||
| 2 | 23 | 14 | 37 | ||
| 3 | 19 | 9 | 28 | ||
| Total | 169 | 104 | 273 | ||
| Ki67 | Low | 111 | 56 | 167 | 0.079a |
| High | 61 | 48 | 109 | ||
| Total | 172 | 104 | 276 | ||
| HER2 | Negative | 98 | 69 | 167 | 0.150 |
| Positive | 74 | 36 | 110 | ||
| Total | 172 | 105 | 277 | ||
| APP | Negative | 110 | 45 | 155 | 0.004 |
| Positive | 51 | 45 | 96 | ||
| Total | 161 | 90 | 251 | ||
| Patients' age | Mean | 55.3 | 54.8 | 54.9 | 0.662 |
| SD | 13.3 | 12.3 | 12.9 | ||
| Range | 23–94 | 30–91 | |||
| Tumor size | Mean | 2.96 | 3.09 | 3.00 | 0.742 |
| SD | 1.43 | 1.64 | 1.51 | ||
| Range | 0.2–8.0 | 0.5–8.0 | |||
Indicated features showed significant association with APP in our previous study.
Fig. 6.
ADAM10 and APP co-expression associated with poor outcome in non-luminal breast cancers. Disease free survival and breast cancer specific survival of non-luminal breast cancer according to combined analysis of ADAM10 and APP expression by IHC using Kaplan-meier analysis. ADAM10hiAPPhi cases (purple line) showed the worst DFS and BCSS compared to ADAM10loAPPlo cases (blue line), ADAM10loAPPhi cases (green line) and ADAM10hiAPPlo cases (beige line).
4. Discussion
Overexpression of APP in breast cancers has been reported by us and others recently [[2], [3], [4], [5]]. APP has been demonstrated to mediate breast cancer cell proliferation and migration [2,3,5]. However, the precise mechanism of how APP is involved is not clear. It has been shown that the proteolytic products of APP processing have growth promoting qualities and regulate cell cycle in various cancers [6,8,9,11]. Previous studies in breast cancer demonstrated that APP is mechanistically linked to the AKT/FOXO and AKT/GSK3-β pathways [2]. Of note, sAPPα from APP processing has also reported to stimulate AKT/GSK3β pathway in neuronal cells and consequently resulted in its neuroprotective effect [18]. For that reason, it could be of interest to investigate the role of APP processing in breast cancer. Our data indicated that the pathological role of APP in breast cancer could at least partly attributable to its proteolytic product. Knockdown of APP expression in breast cancer cells reduced in vitro and in vivo cell growth and migration. The cleavage product of APP, namely sAPPα, could contribute significantly to these effects. Overexpression of not only full length APP but also sAPPα fragment could promote cell growth and migration in breast cancer cells. Among the ADAM implicated in APP processing, ADAM 10, but not ADAM 17, was found to be essential for APP cleavage in breast cancer. Inhibition of ADAM10 expression blocked the formation of sAPPα and suppressed cell growth as well as migration. Transfection of sAPPα to ADAM10 KD cells was able to reverse some effects of ADAM10 KD. ADAM10 in APP processing could have clinical relevance. We have previously found that APP was highly expressed in non-luminal breast cancers [5]. Interestingly, in this subset of breast cancers, there was a significant correlation between APP and ADAM10 expression. More importantly, we showed that cases co-expressing ADAM10 and APP were associated with the worst outcome.
In fact, sAPPα which comprises almost the entire extracellular region has been shown to have mitogenic action in fibroblasts, colon and pancreatic cancers [8,9,19]. It may interact with integrin β1 leading to an increase in neurite outgrowth via integrin signaling. Moreover, sAPPα modulates PI3K/AKT-GSK3β pathway in AD model [18]. Both AKT [20] and β-integrin [21] pathways play important role in cancer cell growth and migration. In line with that, in addition to its mitogenic effect, overexpression of sAPPα can also promote breast cancer cell migration, indicating the involvement of APP processing in controlling both proliferation and migratory capacities of cancer. Yet, the precise underlying mechanisms and physiological receptor(s) interacted with sAPPα on the cell remain unclear. Of interest, the extent of the effects for full length APP and sAPPα overexpression on migration and proliferation as well as the rescue experiments appears to be different. ADAM10 mediated APP cleavage to sAPPα appeared to have a greater relative impact on migration than proliferation; vice versa for full length APP transfection. The data indicated that sAPPα cannot account for all the functional effects of APP in breast cancers and different forms of APP could be involved in different functions. Additional cellular effects of APP could confer by other metabolites or the full length protein itself. APP α-cleavage also generated APP-carboxyl terminal domain which may subsequently cleaved by γ-secretase to release AICD and extracellular P3 peptide. AICD could translocate to the nucleus, analogous to Notch receptor signaling, engaging in the regulation of gene expression. In fact, APP was shown to regulate early cell cycle progression via its C-terminal domain in lung cancer [6]. Furthermore, APP could function analogously to a G protein coupled receptor, where membrane-bound full length APP or membrane-tethered AICD facilitate interactions and the recruitment of cytosolic adaptors/effectors to promote intracellular signaling [22]. The functional role(s) of other APP metabolites or as a full-length molecule warrants further investigations in breast cancer.
Our data also suggested that ADAM10, rather than ADAM17, is required for APP processing in breast cancer and the clinical relevance of a link between APP and ADAM10. Ectodomain cleavage by ADAMs has been recognized as a key mechanism for regulating receptor functions. It is increasingly recognized as attractive novel therapeutic targets. ADAM 10 and ADAM17 are structurally and functionally related and share similar substrate in vitro, including APP [23]. Despite that, the protease specificity was tightly controlled in cancer cells as shown here by the preferential cleavage of APP by ADAM10 in the breast cancer cellular environment. Several validated cellular substrates of ADAM10 have been reported [24]. Regarding ADAM10 substrate in breast cancer, ADAM10 had mainly been examined for its role in shedding HER2 receptor ectodomain, generating a constitutively active truncated form of the receptor [25]. The ADAM10 expression is upregulated after Trastuzumab treatment and mediates resistance of cancer cells [26]. In a similar way, APP processing by ADAM10 could contribute to the etiology of breast cancer malignancy. Recently, ADAM10 has been shown to promote in vitro migration/invasion of triple negative breast cancer (TNBC) cell lines but no substrate has been identified in that study [27]. In our previous analysis, APP was found to be highly expressed in all non-luminal cancers, including both HER2-OE and TNBC [5]. Moreover, using the TNBC cell lines BT-549 and MDA-MB231, we demonstrated that ADAM10 KD could inhibit sAPPα generation. Thus, APP processing could account for at least some effects of ADAM10 in TNBCs. More importantly, the co-expression of APP and ADAM10 in non-luminal cancers (both HER2-OE and TNBCs) was associated with worse patients' outcome, indicating the relevance of in vitro findings in clinical breast cancers. In term of therapy, ADAM10 inhibitor has been tested in phase I/II trial as a potential therapeutic target for HER2 positive breast cancer [28]. The current results may suggest a boarder application of ADAM10 inhibition also in TNBCs. In fact, inhibition of sAPPα generation by ADAM10 inhibitor has been shown to have a synergistic effect with a chemotherapeutic drug in inhibition of pancreatic cancer growth [10]. The combination treatment of sAPPα inhibition and chemotherapy may greatly reduce the dose of chemotherapy, thus could substantially reduce treatment-associated side effects in cancer patients. Interestingly, recent evidence suggested that mitogen activated protein kinase (MAPK) inhibitor could reduce shedding of APP in TNBC cell line (MDA-MB231) by promoting the binding of TIMP1 (tissue inhibitor of metalloproteinases 1) with ADAMs and subsequently inhibited their catalytic activities [29]. The data suggested sAPP processing and ADAM activity as a downstream event of MAPK signaling. In addition to MAPK, protein kinase C, phosphatidylinositol 3-kinase and cAMP signaling implicated in the regulation of the alpha cleavage process [30]. Inhibition of these pathways may also have a similar synergistic therapeutic effect with chemotherapeutic drugs.
5. Conclusions
In summary, the results of this study and our previous work support the role of APP, in particular, its proteolytic product in breast cancer progression. Both APP and ADAM10 knockdown led to a reduction of breast cancer cell migration and proliferation. Overexpression of sAPPα showed an opposite effect. Nonetheless, sAPPα cannot fully account for all the functional effects of APP in breast cancers. Different forms of APP could contribute differently to various cellular functions. In non-luminal cancers, we found a significant correlation between APP and ADAM10 expression and co-expression of the proteins correlated with the worst outcome in patients. Here, we provide further evidence for the use of APP as a biomarker for breast cancer, whereas the combined targeting of APP and ADAM10 might be a promising target for non-luminal breast cancer therapy. The precise molecular pathway for APP/its proteolytic products in mediating the progression of breast cancers remains unknown. Better understanding the mechanisms involved may aid in exploiting APP as a novel therapeutic target in breast cancer management.
Acknowledgments
Acknowledgements
This study was supported by the Health and Medical Research Fund of Hong Kong (02130746).
Funding sources
The funding source had no involvements in study design, data collection, data analysis, data interpretation, manuscript preparation and submission.
Declaration of interests
All authors declare no conflict of interests.
Author contributions
JT analyzed data and wrote the paper; ML, TC, and YS performed the experiments; SKC, SYC, YN and JL collected and arranged clinicopathological data of cases; KL contributed to analyzing data and revising the manuscript; GT conceived the idea for the paper, provided guidance and critically revised the paper. All authors read and approved the final version to be published.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.11.012.
Appendix A. Supplementary data
Supplementary material
References
- 1.Dawkins E., Small D.H. Insights into the physiological function of the beta-amyloid precursor protein: beyond Alzheimer's disease. J Neurochem. 2014;129(5):756–769. doi: 10.1111/jnc.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim S., Yoo B.K., Kim H.S., Gilmore H.L., Lee Y., Lee H.P. Amyloid-beta precursor protein promotes cell proliferation and motility of advanced breast cancer. BMC Cancer. 2014;14:928. doi: 10.1186/1471-2407-14-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takagi K., Ito S., Miyazaki T., Miki Y., Shibahara Y., Ishida T. Amyloid precursor protein in human breast cancer: an androgen-induced gene associated with cell proliferation. Cancer Sci. 2013;104(11):1532–1538. doi: 10.1111/cas.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsang J.Y., Lee M.A., Ni Y.B., Chan S.K., Cheung S.Y., Chan W.W. Amyloid precursor protein was associated with aggressive behavior in non-luminal breast cancers. Oncologist. 2018 Nov;23(11):1273–1281. doi: 10.1634/theoncologist.2018-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ni Y.B., Lee M.A., Tsang J.Y., Lau K.F., Tse G.M. Amyloid precursor protein as a potential marker of malignancy and prognosis of ER negative breast cancer. Mod Pathol. 2017;30(supp 2):62A. [Google Scholar]
- 6.Sobol A., Galluzzo P., Liang S., Rambo B., Skucha S., Weber M.J. Amyloid precursor protein (APP) affects global protein synthesis in dividing human cells. J Cell Physiol. 2015;230(5):1064–1074. doi: 10.1002/jcp.24835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botelho M.G., Wang X., Arndt-Jovin D.J., Becker D., Jovin T.M. Induction of terminal differentiation in melanoma cells on downregulation of beta-amyloid precursor protein. J Invest Dermatol. 2010;130(5):1400–1410. doi: 10.1038/jid.2009.296. [DOI] [PubMed] [Google Scholar]
- 8.Hansel D.E., Rahman A., Wehner S., Herzog V., Yeo C.J., Maitra A. Increased expression and processing of the Alzheimer amyloid precursor protein in pancreatic cancer may influence cellular proliferation. Cancer Res. 2003;63(21):7032–7037. [PubMed] [Google Scholar]
- 9.Meng J.Y., Kataoka H., Itoh H., Koono M. Amyloid beta protein precursor is involved in the growth of human colon carcinoma cell in vitro and in vivo. Int J Cancer. 2001;92(1):31–39. [PubMed] [Google Scholar]
- 10.Woods N.K., Padmanabhan J. Inhibition of amyloid precursor protein processing enhances gemcitabine-mediated cytotoxicity in pancreatic cancer cells. J Biol Chem. 2013;288(42):30114–30124. doi: 10.1074/jbc.M113.459255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takayama K., Tsutsumi S., Suzuki T., Horie-Inoue K., Ikeda K., Kaneshiro K. Amyloid precursor protein is a primary androgen target gene that promotes prostate cancer growth. Cancer Res. 2009;69(1):137–142. doi: 10.1158/0008-5472.CAN-08-3633. [DOI] [PubMed] [Google Scholar]
- 12.Murai T., Miyazaki Y., Nishinakamura H., Sugahara K.N., Miyauchi T., Sako Y. Engagement of CD44 promotes Rac activation and CD44 cleavage during tumor cell migration. J Biol Chem. 2004;279(6):4541–4550. doi: 10.1074/jbc.M307356200. [DOI] [PubMed] [Google Scholar]
- 13.Lakhani S.R., Ellis I.O., Schnitee S.J., Tan P.H., van de Vijver M.J., editors. World health organisation classification of tumors of the breast. 4th ed. IARC Press; Lyon: 2012. [Google Scholar]
- 14.Elston C.W., Ellis I.O. Pathological prognostic factors in breast cancer. I. the value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 15.Tomita S., Kirino Y., Suzuki T. Cleavage of Alzheimer's amyloid precursor protein (APP) by secretases occurs after O-glycosylation of APP in the protein secretory pathway. Identification of intracellular compartments in which APP cleavage occurs without using toxic agents that interfere with protein metabolism. J Biol Chem. 1998;273(11):6277–6284. doi: 10.1074/jbc.273.11.6277. [DOI] [PubMed] [Google Scholar]
- 16.Doberstein K., Pfeilschifter J., Gutwein P. The transcription factor PAX2 regulates ADAM10 expression in renal cell carcinoma. Carcinogenesis. 2011;32(11):1713–1723. doi: 10.1093/carcin/bgr195. [DOI] [PubMed] [Google Scholar]
- 17.Weskamp G., Cai H., Brodie T.A., Higashyama S., Manova K., Ludwig T. Mice lacking the metalloprotease-disintegrin MDC9 (ADAM9) have no evident major abnormalities during development or adult life. Mol Cell Biol. 2002;22(5):1537–1544. doi: 10.1128/mcb.22.5.1537-1544.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jimenez S., Torres M., Vizuete M., Sanchez-Varo R., Sanchez-Mejias E., Trujillo-Estrada L. Age-dependent accumulation of soluble amyloid beta (Abeta) oligomers reverses the neuroprotective effect of soluble amyloid precursor protein-alpha (sAPP(alpha)) by modulating phosphatidylinositol 3-kinase (PI3K)/Akt-GSK-3beta pathway in Alzheimer mouse model. J Biol Chem. 2011;286(21):18414–18425. doi: 10.1074/jbc.M110.209718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saitoh T., Sundsmo M., Roch J.M., Kimura N., Cole G., Schubert D. Secreted form of amyloid beta protein precursor is involved in the growth regulation of fibroblasts. Cell. 1989;58(4):615–622. doi: 10.1016/0092-8674(89)90096-2. [DOI] [PubMed] [Google Scholar]
- 20.Larue L., Bellacosa A. Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene. 2005;24(50):7443–7454. doi: 10.1038/sj.onc.1209091. [DOI] [PubMed] [Google Scholar]
- 21.Seguin L., Desgrosellier J.S., Weis S.M., Cheresh D.A. Integrins and cancer: regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 2015;25(4):234–240. doi: 10.1016/j.tcb.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deyts C., Thinakaran G., Parent A.T. APP receptor? To be or not to be. Trends Pharmacol Sci. 2016;37(5):390–411. doi: 10.1016/j.tips.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asai M., Hattori C., Szabo B., Sasagawa N., Maruyama K., Tanuma S. Putative function of ADAM9, ADAM10, and ADAM17 as APP alpha-secretase. Biochem Biophys Res Commun. 2003;301(1):231–235. doi: 10.1016/s0006-291x(02)02999-6. [DOI] [PubMed] [Google Scholar]
- 24.Pruessmeyer J., Ludwig A. The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin Cell Dev Biol. 2009;20(2):164–174. doi: 10.1016/j.semcdb.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Liu P.C., Liu X., Li Y., Covington M., Wynn R., Huber R. Identification of ADAM10 as a major source of HER2 ectodomain sheddase activity in HER2 overexpressing breast cancer cells. Cancer Biol Ther. 2006;5(6):657–664. doi: 10.4161/cbt.5.6.2708. [DOI] [PubMed] [Google Scholar]
- 26.Feldinger K., Generali D., Kramer-Marek G., Gijsen M., Ng T.B., Wong J.H. ADAM10 mediates trastuzumab resistance and is correlated with survival in HER2 positive breast cancer. Oncotarget. 2014;5(16):6633–6646. doi: 10.18632/oncotarget.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullooly M., McGowan P.M., Kennedy S.A., Madden S.F., Crown J., N O.D. ADAM10: a new player in breast cancer progression? Br J Cancer. 2015;113(6):945–951. doi: 10.1038/bjc.2015.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newton R.C., Bradley E.C., Levy R.S., Doval D., Bobdarde S., Sahoo T.P. Clinical benefit of INCB7839, a potent and selective ADAM inhibitor, in combination with trastuzumab in patients with metastatic HER2+ breast cancer. J Clin Oncol. 2010;28(15_supp):3025. [Google Scholar]
- 29.Miller M.A., Oudin M.J., Sullivan R.J., Wang S.J., Meyer A.S., Im H. Reduced proteolytic shedding of receptor tyrosine kinases is a post-translational mechanism of kinase inhibitor resistance. Cancer Discov. 2016;6(4):382–399. doi: 10.1158/2159-8290.CD-15-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Postina R. Activation of alpha-secretase cleavage. J Neurochem. 2012;120 Suppl 1:46–54. doi: 10.1111/j.1471-4159.2011.07459.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material