Abstract
Background
PARP-1 plays a critical role in DNA damage repair and contributes to progression of cancer. To explore the role of PARP-1 in acute myeloid leukemia (AML), we analyzed the expression of PARP-1 in AML and its relation to the clinical prognosis. Then, we investigated the efficacy and mechanism of PARP inhibitor BMN673 (Talazoparib) combined with NL101, a novel SAHA-bendamustine hybrid in vitro and in vivo.
Methods
The expression of PARP-1 in 339 cytogenetically normal AML (CN-AML) cases was evaluated using RT-PCR. According to the expression of PARP-1, the clinical characteristics and prognosis of the patients were grouped and compared. The combination effects of BMN673 and NL101 were studied in AML cells and B-NSG mice xenograft model of MV4-11.
Findings
We found patients in high PARP-1 expression group had higher levels of blast cells in bone marrow (P = .003) and white blood cells (WBC) in peripheral blood (P = .008), and were associated with a more frequent FLT3-ITD mutation (28.2% vs 17.3%, P = .031). The overall survival (OS) and event free survival (EFS) of the high expression group were significantly shorter than those in the low expression group (OS, P = .005 and EFS, P = .004). BMN673 combined with NL101 had a strong synergistic effect in treating AML. The combination significantly induced cell apoptosis and arrested cell cycle in G2/M phase. Mechanistically, BMN673 and NL101 combinatorial treatment promoted DNA damage. In vivo, the combination effectively delayed the development of AML and prolonged survival.
Interpretation
High PARP-1 expression predicts poor survival in CN-AML patients. The synergistic effects of PARP inhibitor BMN673 in combination with SAHA-bendamustine hybrid, NL101, provide a new therapeutic strategy against AML.
Fund
National Natural Science Foundation of China and Zhejiang Provincial Key Innovation Team.
Keywords: PARP-1, Acute myeloid leukemia, PARP inhibitor, SAHA-bendamustine hybrid
Research in context.
Evidence before this study
There is an urgent need for new therapeutic strategies to improve survival in patients with acute myeloid leukemia (AML). PARP inhibitors (PARPis) have shown significant benefits in a variety of malignancies patients and are considered as potential target drugs.
Added value of this study
We found that AML patients had elevated PARP-1 expression and patients with higher expression had poor prognosis. Therefore, we speculate that the use of PARPis in AML, especially relapse/refractory patients, may improve clinical outcomes. In this study, we found that PARPi BMN673 (Talazoparib) combined with novel SAHA-bendamustine NL101 had a synergistic inhibitory effect on AML cell lines and clinical patient specimens. Our experiments provide theoretical basis for the clinical treatment of PARPis.
Implications of all the available evidence
Our study suggested that PARP-1 is an independent prognostic biomarker for AML. In addition, our findings provide a powerful rationale for the clinical investigation of the BMN673 and NL101 combination therapy.
Alt-text: Unlabelled Box
1. Introduction
DNA repair pathways have been extensively studied in solid tumors [1]. Two important enzymes that facilitate DNA damage repair are poly (ADP-ribose) polymerase 1 (PARP-1) and 2 (PARP-2) [2]. PARP-1 is a cell cycle regulated protein. The transition of the cell cycle from G1 to S phase leads to transcription of PARP-1 [3]. PARP-1 is overexpressed in many cancers such as testicular and other germ cell tumors [4], neuroblastoma [5], malignant lymphoma [6], Ewing's sarcoma [7], breast cancer [8], and colon cancer [9]. PARP-1 also contributes to progression of endometrial cancer [10], BRCA-mutated ovarian cancer [11], and BRCA-mutated serous ovarian cancer [12].
When single-strand DNA breaks (SSBs) occur, PARP-1 binds to the damaged DNA sites and initiates the formation of a poly-ADP scaffold that recruits other members of the base excision repair (BER) pathway, such as XRCC1 [13]. Blocking PARP-1 inhibits BER, leading to the accumulation of SSBs and double-strand breaks (DSBs), which in turn activates homologous recombination (HR) repair [14,15]. The most critical proteins are BRCA1 [16,17] and BRCA2 [18] in HR, however, these two genes are often mutated in tumors leading to defects in HR [19,20]. Without effective HR repair, cells use non-conservative forms of DNA repair such as non-homologous end joining (NHEJ), which may generate large-scale genomic rearrangements leading to the lethality of tumor cells [21]. In 2005, Farmer H and Bryant HE found that BRCA-1/2 deficient tumors were sensitive to PARP inhibitors (PARPis) [22,23]. FDA has approved PARPis Olaparib (2014) [24] and Rucaparib (2016) [25] monotherapy in patients with BRCA-mutated advanced ovarian cancer. To date, there are a great deal of research on the use of PARP inhibitors in cancers with BRCA mutations in clinical trials [[26], [27], [28]].
Acute myeloid leukemia (AML) is a highly heterogeneous disease with poor clinical prognosis. DNA damage response (DDR) in hematological malignancies has been extensively studied but not fully understood [29,30]. It has been reported BRCA1 expression level was reduced in AML samples [31]. When AML was treated with DNA-damaging agents, the loss of BRCA1 function leads to the accumulation of genomic alterations, and even to synthetic lethality. A study by Esposito et al. demonstrated for the first time a potential utility of PARPi-induced lethality for leukemia driven by AML1-ETO and PML-RARa [32]. AML cells with low expression of key members of the DDR pathway such as Rad51, ATM, BRCA1, and BRCA2, displayed extremely sensitivity to PARPi Furthermore, they showed that combined PARPi with GSK3 inhibitor treatment was an effective therapeutic strategy for PARPi-resistant AML.
Currently, the studies combining PARPi with other inhibitors, particularly those that enhance DNA damage, have been successfully applied in both pre-clinical and clinical trials. Gojo et al. demonstrated that a combination therapy of veliparib, a PARPi, plus the DNA-alkylating agent temozolomide was efficacious against advanced AML using doses that were well-tolerated [33]. In another study, combining PARPis with DNA demethylating agents showed synergy in treating AML [34]. NL101, is a hybrid in which the side chain of bendamustine was replaced with the hydroxamic acid of HDACi vorinostat (SAHA) [35]. Both bendamustine [36,37] and SAHA [35,38]can activate DDR pathways as reported. NL101 [39] presented both the properties of HDAC inhibition and DNA damaging, prolong the survival of leukemia mice. Rasmussen RD et al. [40] performed a research that combined HDACi and PARPi could enhance the efficacy of targeting in glioblastoma. Therefore, we hypothesized that these two agents may have a strong synergistic effect through causing DNA damage in AML.
New treatment strategies are urgently needed to improve the survival of AML patients. PARPis have shown significant benefits in a variety of malignancies and are considered as a potential treatment for AML. In our study, we showed that high PARP-1 expression correlates with poor clinical outcome in AML. In particular, we explored the combination treatment of PARPi BMN673 with a novel SAHA-bendamustine hybrid NL101 in AML.
2. Materials & methods
2.1. Patients
Clinical data were collected from the medical records of AML patients at Zhejiang Institute of Hematology, China. From July 2010 to April 2016, 339 patients were included in this study with detailed diagnostic and treatment information. Cytogenetically normal acute myeloid leukemia (CN-AML) was defined as AML with the karyotype 46 XY [20] or 46 XX [20] in all 20 metaphase cells analyzed. Gene mutations of NPM1, FLT3-ITD, CEBPA, DNMT3a, IDH1 and IDH2 were analyzed by whole-gene sequencing. Patients with secondary AML or acute promyelocytic leukemia were excluded. Patient characteristics were summarized using descriptive statistics, which include frequency counts, median, and range. This study was approved by the Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University (Hangzhou, China). Informed consent was obtained from all patients according to institutional guidelines.
2.2. Antibodies and reagents
The antibodies GAPDH (#5174), Caspase3 (#9662), PARP (#9532), CyclinB1 (#4135), CDC2 (#9116), p-CDC2 (#9111), CDC25A (#3652), DAN damage kit (#9947) including p-ATM (Ser1981), p-CHK1 (ser345), p-CHK2 (Thr68), phospho-Histone H2AX (Ser139), and 488 Conjugate secondary antibody (#4412) were purchased from CST (Danvers, MA). CHK1 (10362–1-AP), CHK2 (13954–1-AP) and ATM (27156–1-AP) antibodies were purchase from ProteinTech (Rosemont, USA). Anti-Poly (ADP-Ribose) Polymer antibody (ab14459) and Anti-human CD45-FITC (ab134199) antibody was purchased from Abcam (Cambridge, MA). BMN673 was obtained from MedChemExpress (Monmouth Junction, NJ). NL101 was gifted by Hangzhou Minsheng Institute of Pharmaceutical Research (Hangzhou, China).
2.3. RNA expression by real time reverse transcription PCR (qRT-PCR)
Mononuclear cells (MNC) were separated from the bone marrow (BM) of AML patients at the time of initial diagnosis by Ficoll-Hypaque (TBD Science, Tianjin,China) density gradient centrifugation. RNA was extracted using the TRIzol reagent (Takara, Japan) and was reverse transcribed with PrimerSctipt RT agent Kit (Takara, Japan). Quantitative assessments of cDNA amplification for PARP-1, BRCA1 and GAPDH were performed in triplicate using SYBR-Green PCR Master Mix kit (Takara, Japan) on an IQ5 real time PCR instrument (Bio-Rad, Hercules, CA). The primers sequences were as follows: PARP-1 5′-TCT GAG CTT CGG TGG GAT GA-3′ (forward) and 5′-TTG GCA TAC TCT GCT GCA AAG-3′ (reverse); BRCA1 5′-GAA ACC GTG CCA AAA GAC TTC-3′ (forward) and 5′-CCA AGG TTA GAG AGT TGG ACA C-3′ (reverse); GAPDH 5′-ACC ACC CTG TTG CTG TAG CCA A-3′ (forward) and 5′-GTC TCC TCT GAC TTC AAC AGC G-3′ (reverse).
2.4. Cell lines and primary patient cells
The AML cell lines MV4-11 and MOLM-13 were kindly gifted by Professor Ravi Bhatia (City of Hope National Medical Center, Duarte, CA). HL-60 and Kasumi-1 were obtained from Cell Bank of Type Culture Collection of Chinese Academy of Sciense (Shanghai, China). MV4-11-luciferase was gifted by Professor Xu Rongzhen (The Second Affiliated Hospital of Zhejiang University). These cell lines were authenticated by DNA short-tandem repeat analysis by Shanghai Biowing Applied Biotechnology (Shanghai, China). MV4-11, MOLM-13 cell lines and primary AML cells were cultured in IMDM medium (Corning) supplemented with 10% fetal bovine serum (Gibco) at 37 °C in a humidified incubator containing 5% CO2. HL-60 and Kasumi-1 cell lines were cultured in RPMI1640 medium supplemented with 10% FBS. Primary AML cells were isolated by Ficoll-Hypaque density gradient centrifugation from the bone marrow after obtaining written informed consent.
2.5. Cell viability assay
Cells were seeded in 96-well plates at 1–2 × 104 (AML cell lines) or 1 × 105 (primary AML cells) per well. At the end of the drugs treatment, 20 μl MTS solution (Promega, Madison, WI) was added to each well and the cells were incubated for an additional 4 h at 37 °C. The plates were read at a wavelength of 490 nm. Experiments using AML cell lines were done in three independent replicates.
2.6. Flow cytometric analysis
To analyze cell cycle distribution, cells were treated with drugs for 24 h and fixed with 75% ethanol at 4 °C overnight. The next day, the cells were resuspended in buffer with 50 μg/ml propidium iodide (PI) and 100 μg/ml RNase A for 30 min at room temperature. For apoptosis assessment, cells were treated with drugs for 48 h and then co-stained with 10 μl Annexin V-Fluorescein Isothiocyanate (FITC) and 5 μl PI using an apoptosis detection kit (Mulisciences, Hangzhou, China). The engrafted MV4-11 cells were analyzed using anti-human FITC-labeled CD45 antibody (abcam, USA). The DNA content, apoptic cells and hCD45+ cells were analyzed by FACScan flow cytometer (Becton Dickinson, San Diego, CA).
2.7. Western blot analysis
Cells were lysed in RIPA buffer (Thermo Fisher Scientific, Waltham, MA) on ice for 30 min, and centrifuged at 12,000 ×g for 15 min at 4 °C to pellet cell debris. The protein concentration in the supernatant was determined using BCA reagent (BBI life science, Shanghai, China). Protein samples were separated by SDS-PAGE gel (Thermo Fisher Scientific, Waltham, MA) and transferred to PVDF membranes (Millipore, Burlington, MA). Membranes were blocked using Tris-buffered saline (TBS) containing 5% non-fat milk for 1 h and incubated with primary antibodies overnight at 4 °C. After washing with TBS buffer containing 1% Tween-20 three times, membranes were incubated with secondary antibodies (CST, Danvers, MA) for 1 h. The target proteins were visualized using an ECL kit (Thermo Fisher Scientific, Waltham, MA) and imaged using the ChemiDoc MP Imaging System (Bio-rad, Hercules, CA).
2.8. Immunofluorescence microscopy
Cells were cytospun onto a glass slide at 400 ×g for 5 min and then fixed for 30 min in blocking solution containing 5% BSA and 0.3% Triton X-100 at room temperature. Anti-human γ-H2AX (ser139) (CST, Danvers, MA) was diluted in PBS containing 1% BSA and incubated overnight at 4 °C. Slides were then washed three times with PBS and incubated in 488 Conjugate secondary antibody (CST, Danvers, MA) for 1-2 h at room temperature in the dark. After washed three times with PBS, nuclei were counterstained with DAPI (Sigma-Aldrich, St. Louis, MO). Cells were counted in at least three fields, with >100 cells per condition. The frequency of cells with >5 foci per cell was scored [41,42].
2.9. Mice models
For AML xenografts, female B-NSG mice (6–8 weeks old) (Biocytogen, China) were used. MV4-11-luc cells (1 × 106 cells) were injected into the tail vein of mice. Cell engraftment was assessed by intraperitoneal injection of luciferin (100 mg/kg) followed by imaging using IVIS Lumina LT system (PerkinElmer, CA, USA). Mice were randomly sorted into four groups before treatment. Mice were observed and weighed daily, and leukemic burden was assessed by bioluminescence imaging every 7 days. Mice were treated with either 0.3 mg/kg BMN673, 12 mg/kg NL101, in combination at indicated concentrations, or vehicle. NL101 was diluted in PBS to 12 mg/mL and stored at −20 °C. BMN673 was prepared in 0.01% carboxymethyl cellulose (CMC) and stored at −20 °C in the dark. BMN673 was administered orally, 5 days per week for 3 weeks. NL101 was administered by intravenous injection twice (day 11–12) post-transplant. Animals experiments were approved by the Ethics Committee for Laboratory Animals of the First Affiliated Hospital, College of Medicine, Zhejiang University (Hangzhou, China) and were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
2.10. Statistical analysis
Kaplan-Meier survival curves were compared using the log-rank test. The multivariate Cox proportional hazards model was used to identify independent outcome predictors after adjustment for confounders. Statistical analyses for biological assays were performed using the two-tailed unpaired t-test or multiple-group ANOVA. The combination index (CI) was calculated using CalcuSyn software (Biosoft, Cambridge, UK). For in vivo experiments, we used GraphPad Prism software (San Diego, CA) to calculate statistical significance using ANOVA or Mann-Whitney tests.
3. Results
3.1. PARP-1 expression positively correlates with clinical characteristics
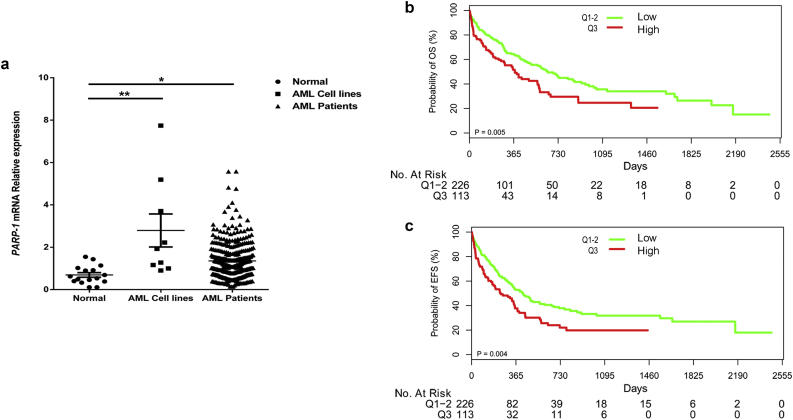
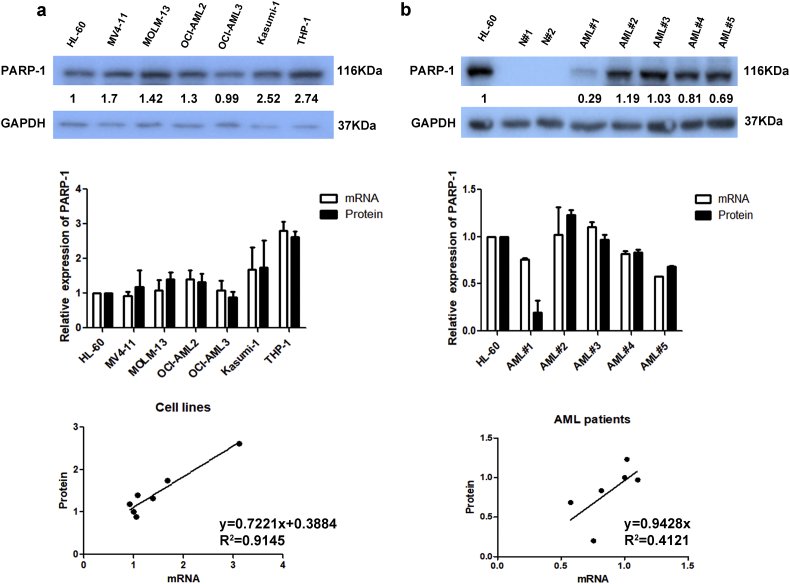
We compared PARP-1 expression in normal bone marrow (BM) cells, AML cell lines and cytogenetically normal AML (CN-AML) patient samples by real-time PCR. PARP-1 expression was significantly increased in CN-AML patients (P < .05, t-test) and AML cell lines (P < .01, t-test) compared to normal BM cells (Fig. 1a). PARP-1 protein levels had a strong correlation with mRNA levels (Fig. S1). The expression of PARP-1 in two normal control specimens (N#1 and N#2) was relatively low (Fig. S1b). The median PARP-1 expression value of AML patients was 1.16 (Range: 0.14–5.58). Patients were classified into high expression group (113, 33.3%) and low expression group (226, 66.7%) according to the mRNA expression level of PARP-1. Patients with high PARP-1 expression had more BM blasts (72.00 [Range: 51.50–85.00] vs 63.00 [Range: 37.00–78.38], P = .003, t-test), elevated peripheral blood WBC levels (17.00 × 109/L [Range: 3.60 × 109–87.20 × 109] vs 9.65 × 109/L [Range: 2.42 × 109–38.60 × 109], P = .008, t-test), and a higher incidence of FLT3-ITD mutation (28.2% vs 17.3%, P = .031, Pearson Chi-Square) than those with low expression (Table 1). There was no statistically significant correlation between PARP-1 expression and other variables such as gender, age, hemoglobin levels, platelet counts, FAB classifications, bone marrow transplantation condition, remission condition, treatment regimen, and gene mutations in NPM1, CEBPA, DNMT3a, IDH1, and IDH2.
Fig. 1.
PARP-1 is overexpressed and associated with poor clinical outcome in CN-AML patients. (a) qRT-PCR analysis of PARP-1 mRNA expression (mean ± SEM)in normal BM cells (n = 16), AML cell lines (n = 9), and CN-AML samples (n = 339) (*P < .05, **P < .01, t-test). Kaplan-Meier curves of overall survival (OS) (b) and event free survival (EFS) (c) in high and low PARP-1 expressing groups.
Table 1.
Characteristics of high and low PARP-1 expressing AML patients.
| Variables | Low expression | High expression | P value |
|---|---|---|---|
| Number | 226 | 113 | |
| Gender = 1 (%) | 133 (58.8) | 64 (56.6) | 0.727 |
| Age (median [IQR]), years | 54.50 [40.00, 64.00] | 57.00 [39.00, 64.00] | 0.801 |
| Blast (median [IQR]) % | 63.00 [37.00, 78.38] | 72.00 [51.50, 85.00] | 0.003 |
| WBC (median [IQR])a, ×10^9/L | 9.65 [2.42, 38.60] | 17.00 [3.60, 87.20] | 0.008 |
| Hb (median [IQR])b, g/L | 84.00 [66.25, 104.00] | 88.00 [70.00, 102.00] | 0.600 |
| PLT (median [IQR]d)c, ×10^9/L | 49.50 [26.00, 92.00] | 44.00 [25.00, 80.00] | 0.393 |
| FAB (%)e | 0.237 | ||
| 0 | 21 (9.3) | 13 (11.5) | |
| 1 | 15 (6.6) | 16 (14.2) | |
| 2 | 116 (51.3) | 50 (44.2) | |
| 4 | 14 (6.2) | 4 (3.5) | |
| 5 | 55 (24.3) | 27 (23.9) | |
| 6 | 5 (2.2) | 3 (2.7) | |
| FLT3-ITD = 1 (%) | 39 (17.3) | 31 (28.2) | 0.031 |
| NPM1 = 1 (%) | 52 (23.6) | 35 (33.7) | 0.061 |
| CEBPA = 1 (%)DM,f | 28 (13.3) | 16 (15.4) | 0.608 |
| DNMT3a = 1 (%) | 27 (13.2) | 17 (16.5) | 0.491 |
| IDH1 = 1 (%) | 40 (20.2) | 19 (17.8) | 0.651 |
| IDH2 = 1 (%) | 28 (14.6) | 15 (14.4) | 1 |
| Favorable = 1 (%)g | 58 (25.8) | 31 (27.9) | 0.773 |
| Treatment (%)h | 0.786 | ||
| DA | 57 (25.2) | 25 (22.1) | |
| HAA | 34 (15.0) | 16 (14.2) | |
| IA | 135 (59.7) | 72 (63.7) | |
| BMT = 1 (%)i | 17 (7.5) | 9 (8.0) | 1 |
| CR = 1 (%) | 131 (58.0) | 55 (48.7) | 0.135 |
| PARP-1 (median [IQR]) | 0.87 [0.55, 1.16] | 2.20 [1.79, 2.62] | <0.001 |
WBC: white blood cell.
HB: hemoglobin.
PLT: platelet counts.
IQR: interquartile.
FAB: French-American-British classification system.
DM: Double-allele mutation.
Favorable genotype represents NPM1 mutant and FLT3-ITD negative or double allele CEBPA mutations.
The protocols used for induction therapy in different groups including daunorubicin/Ara-C (DA)-based treatment group, idarubicin/Ara-C (IA)-based, and homoharringtonine/Ara-C/aclarubicin (HAA)-based treatment group.
BMT: bone marrow transplantation.
3.2. Overexpression of PARP-1 is associated with poor clinical outcome in CN-AML patients
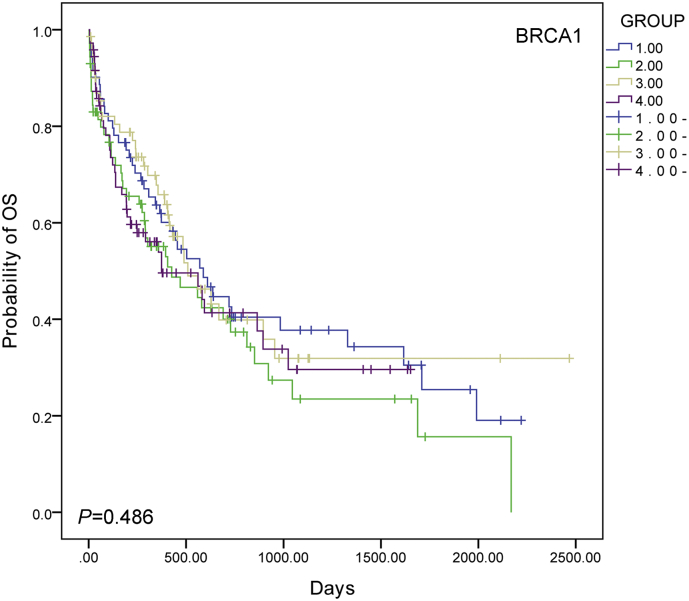
AML patients with high PARP-1 expression (n = 113) had a relatively short overall survival (OS) (P = .005, log-rank test) and event free survival (EFS) (P = .004, log-rank test) compared to patients in low expression group (n = 226) (Fig. 1b and c). To identify the potential confounders and interactions, we conducted interactive analyses. In the multivariable analysis for OS and EFS, high PARP-1 expression is associated with poor survival after adjusting for age, WBC, FLT3-ITD mutation, NPM1 and DMNT3a mutations, BMT, and treatment protocols regardless of OS [HR (95% CI), 1.949 (Range: 1.384–2.745); P < .001, cox proportional hazards regression model; Table 2] or EFS [HR (95% CI), 1.822 (Range: 1.323–2.509); P < .001, cox proportional hazards regression model; Table 2]. In addition, we examined the expression of BRCA1, another gene that plays an important role in DNA damage repair. However, there was no difference in OS among different BRCA1 expression groups (Fig. S2).
Table 2.
Multivariable analysis for OS and EFS in CN-AML patients.
| Variables | Overall survival |
Event free survival |
||
|---|---|---|---|---|
| HR (95%CI) | P | HR (95%CI) | P | |
| PARP-1expresion (High vs Low) | 1.949 (1.384,2.745) | <0.001 | 1.822 (1.323,2.509) | <0.001 |
| Age (>60) | 2.526 (1.774,3.596) | <0.001 | 2.29 (1.648,3.182) | <0.001 |
| WBC (>10) | 1.393 (0.988,1.963) | 0.058 | 1.218 (0.883,1.679) | 0.229 |
| FLT3-ITD | 2.592 (1.744,3.852) | <0.001 | 2.108 (1.448,3.07) | <0.001 |
| NPM1 | 0.701 (0.462,1.062) | 0.094 | 0.85 (0.578,1.251) | 0.41 |
| DNMT3a | 2.029 (1.307,3.149) | 0.002 | 1.827 (1.2,2.781) | 0.005 |
| BMT | 0.493 (0.233,1.043) | 0.064 | 0.794 (0.428,1.473) | 0.464 |
| HAA treatment | 0.518 (0.313,0.858) | 0.011 | 0.576 (0.356,0.931) | 0.024 |
| IA treatment | 0.499 (0.343,0.726) | <0.001 | 0.574 (0.4,0.824) | 0.003 |
3.3. PARPi BMN673 and a novel SAHA-bendamustine hybrid, NL101, synergistically inhibited growth of AML cells
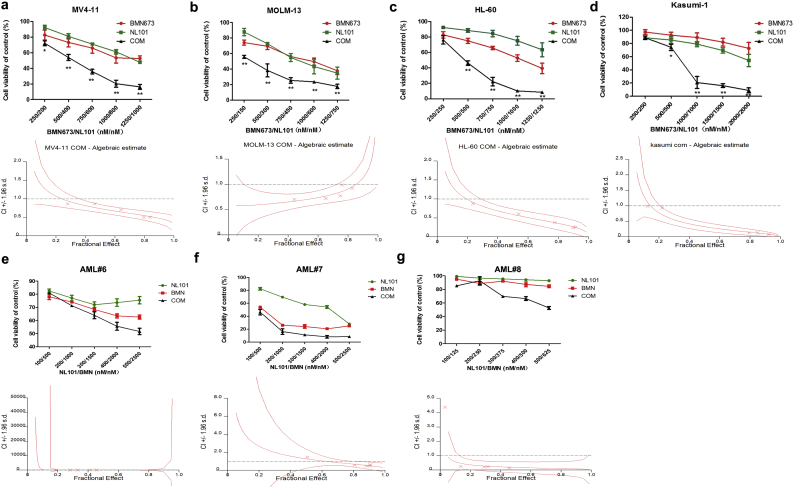
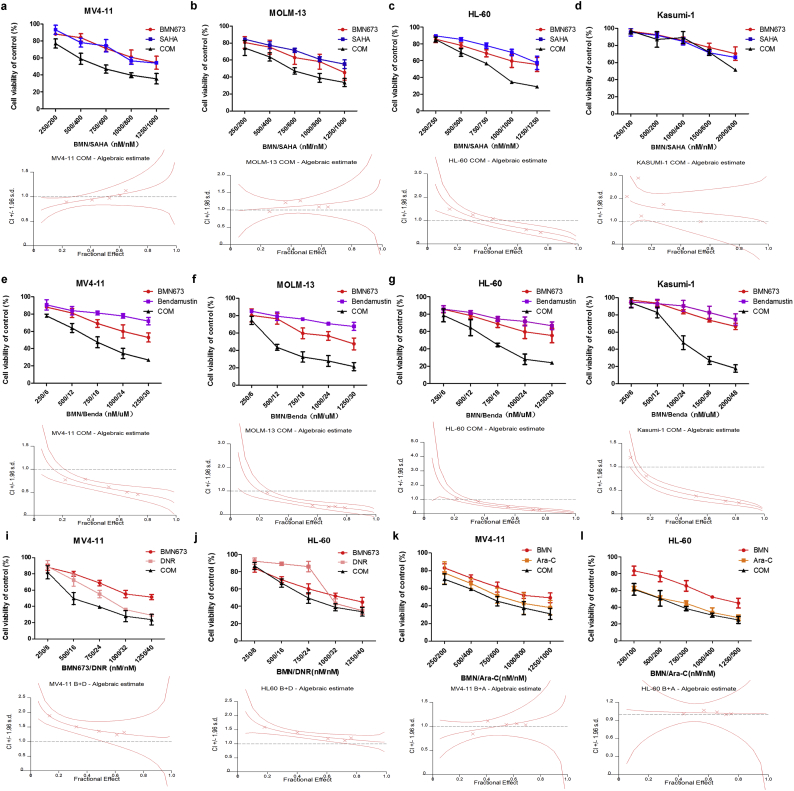
Given the poor prognosis of patients with high PARP-1 expression, we first tested if PARPi BMN673 in combination with HDACi SAHA or bendamustine could improve the outcome of AML patients. We first found minimal growth inhibition of AML cell lines by BMN673 in combination with SAHA after treatment for 48 h (Fig. S3a–d, up panel). We found that BMN673 combined with SAHA had a synergistic inhibitory effect only in HL-60 but not in MV4-11, MOLM-13 and Kasumi-1 (Fig. S3a–d, down panel, and Table S1). However, BMN673 combined with bendamustine displayed synergistic inhibition in all AML cell lines (Fig. S3e–h, Table S1). Moreover, we studied the combination of BMN673 with cytarabine (Ara-C) and daunorubicin (DNR) in MV4-11 and HL-60 cells (Fig. S3i–l), and found no significant synergistic effects. Then, AML cell lines were treated with BMN673, NL101, or in combination for 48 h. Compare to SAHA or bendamustine, NL101 at lower doses in combination with BMN673 was more efficacious at inhibiting the growth of all AML cell lines (Fig. 2a–d, up panel). The combination also effectively inhibited primary AML cells growth compared to single agent treatment (Fig. 2e–g, up panel). In addition, FLT3-ITD mutation in MV4-11, MOLM-13 cell lines and one of the patient samples did not affect cell sensitivity to the treatment. The characteristics of the patient samples are presented in Table S2. The dose-effect curves were determined by CalcuSyn analyses (Fig. 2a–g and Fig. S3a–l, down panel). The CI values were presented in Table S1. We demonstrated that BMN673 combined with NL101 had a strong synergistic effect (CI < 1.0) on AML cell lines and primary AML cells in vitro.
Fig. 2.
Cell viability after treatment with BMN673, NL101 or combination in AML cell lines (a-d, up panel) and primary AML cells (e-g, up panel) measured by MTS assay (*P < .05, **P < .01, One-way ANOVA,combination treatments versus single treatments). The combination index (CI) (a-g, down panel) was calculated using CalcuSyn software. The data are presented as mean ± SD from at least three independent experiments for cell lines.
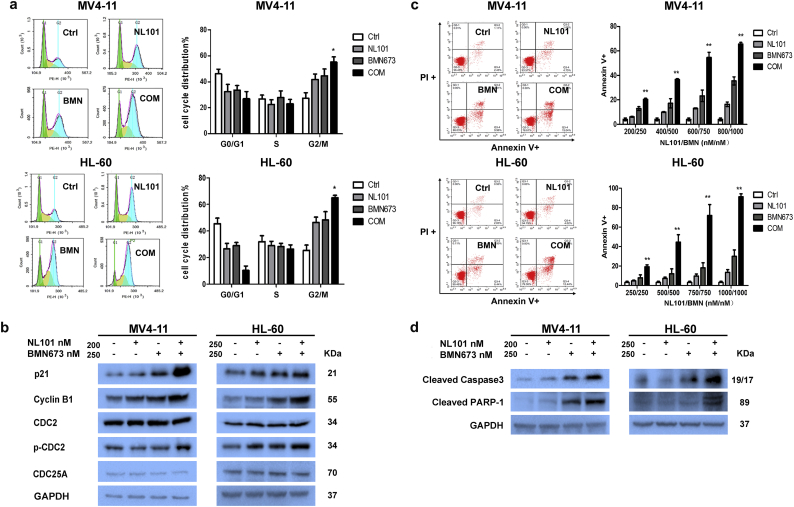
3.4. Combination treatment of BMN673 and NL101 induce G2/M cell cycle arrest and trigger apoptosis
To explore the mechanism of synergistic effect, we studied the effects of BMN673 and NL101 combination on cell cycle after exposure to drugs for 24 h. Significant G2/M accumulation and downregulation of the G0/G1 peak were observed after treatment with the drug combination in MV4-11and HL-60 cells compared to untreated cells and single agent treatment cells (P < .05,One-way ANOVA) (Fig. 3a). Western blot analysis showed an increase in cell cycle regulator P21cip/waf1, G2/M regulatory molecules, cyclin B1 and p-CDC2 (Tyr-15) in cells treated with the drug combination compared to single agent (Fig. 3b). Furthermore, the combination resulted in a significant increase of apoptosis as evident by Annexin V staining (P < .01, One-way ANOVA) (Fig. 3c) and an increase in the active form of Caspase-3 (Cleaved Caspase3) and inactive form of PARP-1 (Cleaved PARP-1) (Fig. 3d) compare to BMN673 or NL101.
Fig. 3.
Combination of BMN673 and NL101 significantly impeded cell cycle progression and induced apoptosis in MV4–11 and HL-60 cells. (a) After drug treatment for 24 h, cell cycle distribution was analyzed using flow cytometry(*P < .05, One-way ANOVA,combination treatments versus control and single treatments). (b) Western blot analysis of p21 and G2/M regulatory molecules, cyclin B1, CDC2, p-CDC2 (Tyr-15) and CDC25A. GAPDH served as a loading control. (c) Apoptosis induced by various treatments at 48 h (**P < .01, One-way ANOVA,combination treatments versus control and single treatments). (d) Western blot of cleaved Caspase-3 and cleaved PARP-1 in AML cells. GAPDH served as a loading control.
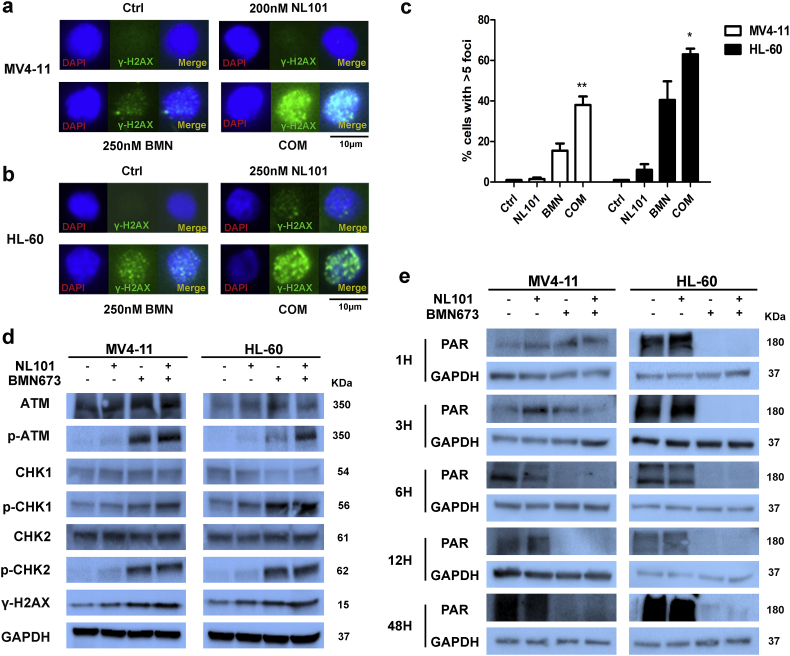
3.5. The combination of BMN673 and NL101 causes an increase in DNA-damage response and inhibition of poly (ADP)ribosylation
Inhibition of DNA damage repair is the major mechanism of PARPi. Considering that NL101 can cause DNA damage [39], we hypothesized that co-treatment of BMN673 and NL101 would yield higher levels of DNA damage. Indeed, we observed significantly higher levels of γ-H2AX foci in MV4-11 and HL-60 cells treated with both BMN673 and NL101, compared to single drug and control (Fig. 4a–c). Likewise, combination treatment increased levels of DNA damage markers p-ATM (Ser1981), p-CHK1 (Ser317), p-CHK2 (Thr68) and γ-H2AX (Fig. 4d). Moreover, we checked the inhibition of poly(ADPribosyl)ation (PAR) in AML cells at different time point. Both BMN673 and the combination treatment could significantly inhibit PAR activity at early and late stages (Fig. 4e).
Fig. 4.
Induction of cytotoxic DSBs by BMN673 and NL101. Representative images of γ-H2AX foci in control, NL101, BMN673 (BMN), and combination treated (COM) MV4-11 (a) and HL-60 (b) cells. (c) Quantification of γ-H2AX foci from immunofluorescent images shown in a and b (*P < .05, **P < .01, One-way ANOVA,combination treatments versus control and single treatments). (d) Western blot analysis of the DNA damage sensors ATM, p-ATM (Ser1981), CHK1, p-CHK1 (Ser317), CHK2 and p-CHK2 (Thr68) in MV4-11 and HL-60 cells. GAPDH served as a loading control.(e)Western blot analysis of PAR at different time points in MV4-11 and HL-60 cells. GAPDH served as a loading control.
3.6. The combination of BMN673 and NL101 improves survival of AML xenograft models
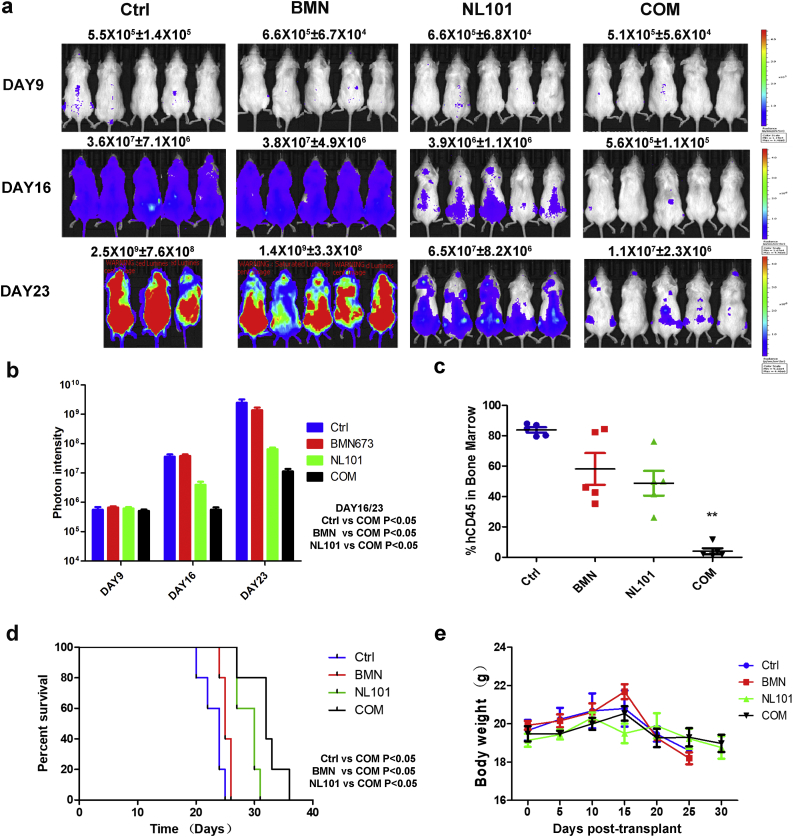
To clarify in vivo efficacy of the combination of BMN673 and NL101, we used an intravenous MV4-11-luc xenograft mouse model. Drugs were administered 9 days after injection of cells and bioluminescence imaging was performed on days 9, 16, and 23 post transplantation. Mice were observed daily and using hind limb paralysis as an endpoint [43]. Combination treatment of BMN673 (0.3 mg/kg) and NL101 (12mg/kg) showed the largest reduction in tumor burden on days 16 and 23 (Fig. 5a and b). Treatment using NL101 alone showed a modest reduction, but mice treated with BMN673 (0.3 mg/kg) alone had no significant reduction in tumor burden (Fig. 5a and b). hCD45-positive blast cells were significantly diminished in the bone marrow of mice treated with the drug combination (Fig. 5c) and tumor infiltration was reduced in the spleen (Fig. S4). All treatment groups prolonged survival, with the combination group providing the best survival (Fig. 5d). Drug doses were well tolerated, and there is no obivious effect on body weight throughout the duration of treatment in all groups (Fig. 5e). Therefore, our data demonstrated that the combination of NL101 and BMN673 could be a novel treatment regimen for AML.
Fig. 5.
Anti-leukemia effects of BMN 673 and NL101 in an AML xenograft model. (a) Images showing the leukemia burden of B-NSG mice transplanted with MV4-11-luc cells treated with drugs. (b) Quantification of images shown in a. Bars represent mean ± SEM. (c) The proportion of hCD45 positive blasts in the bone marrow of mice from different treatment groups (**P < .01, One-way ANOVA). Bars represent mean ± SEM. (d) Kaplan-Meier survival curves of the four different treatment groups. (e) Measurement of mouse body weight during the course of the study. Bars represent mean ± SEM.
4. Discussion
Chemotherapy is the standard treatment for AML despite several decades of clinical efforts to improve outcomes of this disease. However, long-term survival of AML patients remains poor for refractory/relapse cases [[44], [45], [46]]. We hope to explore new treatments that induce DNA damage and perturb cellular DDR in AML cells by understanding the role of PARP-1 in AML cells.
PARP-1 plays a pivotal role in DNA repair, particularly in response to DNA-damaging agents [47,48]. We found that PARP-1 was highly expressed in CN-AML patients and AML cell lines compared to normal BM donors. Furthermore, survival of CN-AML patients with higher PARP-1 expression was poor (Fig. 1). We hypothesized that high PARP-1 expression may be related to the insensitivity to chemotherapy, particularly DNA-damaging agents.
To date, two studies have shown lower BRCA1 expression in haematologic malignancies such as AML [31,49]. Our study found that the expression of BRCA1 in CN-AML patients had no significant correlation with prognosis (Fig.S2). This may indicate that PARP-1 plays a more critical role in the development of AML. In addition, patients with high PARP-1 expression had higher FLT3-ITD mutation rate (Table 1). However, there is a lack of more evidence for an interaction between FLT3-ITD mutation and PARP-1 expression in current literature, which requires further investigation.
PARPis have shown substantial efficacy in the treatment of breast and ovarian cancers with hereditary BRCA1/2 deficiency [50,51]. Defects in HR are not restricted to BRCA-associated tumors, other cancer types may be enriched for HR defects and are therefore sensitive to PARPis [52]. This has spurred research into the use of PARPis in tumors without BRCA1/2 mutations [32,53,54]. BMN673, a novel PARP1/2 inhibitor has a stronger inhibitory effect on PARP1/2 than other PARPis [55,56]. Since we found that PARP-1 is highly expressed in AML (Fig. 1a), we elected PARPis to treat AML patients, especially in refractory or relapse patients. However, PARPis alone are not efficacious in AML [34]. The cytotoxic effects of PARPis need to be enhanced by combination with other chemotherapeutics. Combined effects of PARPis and HDACis were found in prostate cancer [57], glioblastoma [40], and breast cancer [58] in previous reports, but in our current study we observed no synergistic effect of BMN673 combined with SAHA in AML (Fig. S2). Treatment regimens using other PARPis such as olaparib in combination with HDACis in AML remain to be explored.
In addition, we found a significant synergistic inhibition of AML cell growth using BMN673 and bendamustine. We hypothesized that NL101 may enhance the DNA damaging properties of BMN673 as previously reported [39]. NL101, also called ESO-101, is an alkylating HDAC inhibitor fusion molecule, displaying bi-functional activity against tumors. The agent showed strong preclinical activity in vitro and in vivo against multiple myeloma (MM) [59] alone or combined with proteasome inhibition [60] and AML [39]. In our study, we combined four monomeric drugs (SAHA, bendamustin, Ara-C, DNR) with BMN673 and found only bendamustine combined with BMN673 had a synergistic effect. However, the concentration of bendamustine is large and there is nearly no single agent activity in AML, while NL101 shows good synergy and lower concentration, so we believe that there is sufficient evidence to use the combination of NL101 and BMN673 as a new therapy for AML.
BMN673 combined with NL101 in AML cell lines inhibited cell survival, impaired cell cycle progression, and induced apoptosis (Fig. 2, Fig. 3). In agreement with the mechanisms of BMN673 and NL101, we observed an increase of DNA damage in AML cells (Fig. 4). In vivo experiments supported the vitro results and demonstrated that the effective dose had minimal side effects (Fig. 5). The cyclin inhibitor p21 is normally induced by p53 and other p53-independent pathways leading to arrest cell cycle. In our study, we found that the combination treatment of NL101 and BMN673 resulted in G2/M phase arrest and significant upregulation of p21 and G2/M regulatory molecules cyclin B1 and p-CDC2 (Tyr-15).
The findings from our present study indicate that PARP-1 expression negatively impacts the prognosis of AML patients. PARPi BMN673 combined with SAHA-Bendamustine Hybrid NL101 showed strong synergistic inhibitory effects on AML in vitro and in vivo. Our work paves the way for the potential use of BMN673 and NL101 in AML therapy.
The following are the supplementary data related to this article.
Supplementary Fig. 1.
PARP-1 mRNA and protein levels of AML cell lines (a) and AML patients (b).
Supplementary Fig. 2.
Kaplan-Meier curves of overall survival (OS) in different BRCA 1 expressing groups (Group 1-4: lower quarter-upper quarter, group1: n = 71, group 2: n = 71, group 3: n = 71 and group 4: n = 72).
Supplementary Fig. 3.
(a–d) The rate of cell viability induced by BMN673, SAHA and combination in MV4-11, MOLM-13, HL-60, Kasumi-1 cells. (e–h) The rate of cell viability induced by BMN673, Bendamustin and combination in MV4-11, MOLM-13, HL-60, Kasumi-1 cells. (i and j) The rate of cell viability induced by BMN673, DNR and combination in MV4-11, HL-60 cells. (k and I) The rate of cell viability induced by BMN673, Ara-C and combination in MV4-11, HL-60 cells.
Supplementary Fig. 4.
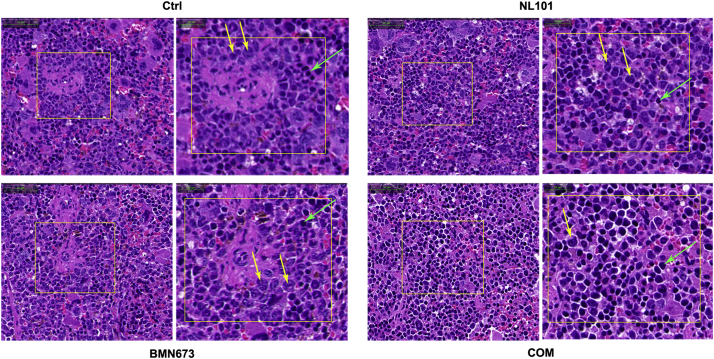
Hematoxylin and eosin stain of spleen sections from xenografted mice treated with vehicle (Ctrl), NL 101, BMN673, or combination (COM). Yellow arrows indicate leukemia cells and green arrows indicate mouse splenocytes.
CI values were calculated at the effective dose (ED) for 50%, 75% and 90% of the fraction affected.
Characteristics of primary AML patients.
Acknowledgments
Acknowledgements
We would like to thank Professor Ravi Bhatia for providing the MV4-11 and MOLM-13 cell lines, Professor Xu Rongzhen for providing us the MV4-11- luciferase cell line, and Hangzhou Minsheng Institute of Pharmaceutical Research for providing us the agent NL101.
Funding
This work was supported by the National Natural Science Foundation of China (Grant number: 81370643, 81470305, 81670124 and 81700137) and Zhejiang Provincial Key Innovation Team (Grant number: 2011R50015).
Authors' contributions
LX, LCY and JJR performed most experiments, analyzed the data, wrote the manuscript; MZX, WJH and HX participated in the data analysis; WJH, WYG, ZYL, XY, YX, YMX and HSJ provided animals, acquired and managed patients, provided facilities support; HJS, LFL and PJJ helped to correct the manuscript. JJ and YYP designed the overall study and supervised the experiments. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bartkova J., Horejsi Z., Sehested M. DNA damage response mediators MDC1 and 53BP1: constitutive activation and aberrant loss in breast and lung cancer, but not in testicular germ cell tumours. Oncogene. 2007;26(53):7414–7422. doi: 10.1038/sj.onc.1210553. [DOI] [PubMed] [Google Scholar]
- 2.Satoh M.S., Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356(6367):356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 3.Pietrzak J., Spickett C.M., Ploszaj T., Virag L., Robaszkiewicz A. PARP1 promoter links cell cycle progression with adaptation to oxidative environment. Redox Biol. 2018;18:1–5. doi: 10.1016/j.redox.2018.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mego M., Cierna Z., Svetlovska D. PARP expression in germ cell tumours. J Clin Pathol. 2013;66(7):607–612. doi: 10.1136/jclinpath-2012-201088. [DOI] [PubMed] [Google Scholar]
- 5.Newman E.A., Lu F., Bashllari D., Wang L., Opipari A.W., Castle V.P. Alternative NHEJ pathway components are therapeutic targets in high-risk neuroblastoma. Molecular Cancer Research : MCR. 2015;13(3):470–482. doi: 10.1158/1541-7786.MCR-14-0337. [DOI] [PubMed] [Google Scholar]
- 6.Tomoda T., Kurashige T., Moriki T., Yamamoto H., Fujimoto S., Taniguchi T. Enhanced expression of poly(ADP-ribose) synthetase gene in malignant lymphoma. Am J Hematol. 1991;37(4):223–227. doi: 10.1002/ajh.2830370402. [DOI] [PubMed] [Google Scholar]
- 7.Newman R.E., Soldatenkov V.A., Dritschilo A., Notario V. Poly(ADP-ribose) polymerase turnover alterations do not contribute to PARP overexpression in Ewing's sarcoma cells. Oncol Rep. 2002;9(3):529–532. [PubMed] [Google Scholar]
- 8.Rojo F., Garcia-Parra J., Zazo S. Nuclear PARP-1 protein overexpression is associated with poor overall survival in early breast cancer. Annals of oncology: official journal of the European Society for Medical Oncology. 2012;23(5):1156–1164. doi: 10.1093/annonc/mdr361. [DOI] [PubMed] [Google Scholar]
- 9.Dziaman T., Ludwiczak H., Ciesla J.M. PARP-1 expression is increased in colon adenoma and carcinoma and correlates with OGG1. PloS one. 2014;9(12) doi: 10.1371/journal.pone.0115558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi F.F., Li D., Yang Q. Hypomethylation of ETS transcription factor binding sites and upregulation of PARP1 expression in endometrial cancer. Biomed Res Int. 2013;2013:946268. doi: 10.1155/2013/946268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li D., Bi F.F., Cao J.M. Poly (ADP-ribose) polymerase 1 transcriptional regulation: a novel crosstalk between histone modification H3K9ac and ETS1 motif hypomethylation in BRCA1-mutated ovarian cancer. Oncotarget. 2014;5(1):291–297. doi: 10.18632/oncotarget.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bi F.F., Li D., Yang Q. Promoter hypomethylation, especially around the E26 transformation-specific motif, and increased expression of poly (ADP-ribose) polymerase 1 in BRCA-mutated serous ovarian cancer. BMC Cancer. 2013;13:90. doi: 10.1186/1471-2407-13-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishnakumar R., Kraus W.L. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39(1):8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murai J., Huang S.Y., Das B.B. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72(21):5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eustermann S., Wu W.F., Langelier M.F. Structural basis of detection and signaling of DNA single-strand breaks by human PARP-1. Mol Cell. 2015;60(5):742–754. doi: 10.1016/j.molcel.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isono M., Niimi A., Oike T. BRCA1 directs the repair pathway to homologous recombination by promoting 53BP1 dephosphorylation. Cell Rep. 2017;18(2):520–532. doi: 10.1016/j.celrep.2016.12.042. [DOI] [PubMed] [Google Scholar]
- 17.Kass E.M., Helgadottir H.R., Chen C.C. Double-strand break repair by homologous recombination in primary mouse somatic cells requires BRCA1 but not the ATM kinase. Proc Natl Acad Sci U S A. 2013;110(14):5564–5569. doi: 10.1073/pnas.1216824110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ying S., Myers K., Bottomley S., Helleday T., Bryant H.E. BRCA2-dependent homologous recombination is required for repair of Arsenite-induced replication lesions in mammalian cells. Nucleic Acids Res. 2009;37(15):5105–5113. doi: 10.1093/nar/gkp538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pothuri B. BRCA1- and BRCA2-related mutations: therapeutic implications in ovarian cancer. Annals of oncology : official journal of the European Society for Medical Oncology. 2013;24(Suppl. 8) doi: 10.1093/annonc/mdt307. viii22-viii7. [DOI] [PubMed] [Google Scholar]
- 20.Telli M.L., Hellyer J., Audeh W. Homologous recombination deficiency (HRD) status predicts response to standard neoadjuvant chemotherapy in patients with triple-negative or BRCA1/2 mutation-associated breast cancer. Breast Cancer Res Treat. 2017;168(3):625–630. doi: 10.1007/s10549-017-4624-7. [DOI] [PubMed] [Google Scholar]
- 21.Ceccaldi R., Rondinelli B., D'Andrea A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26(1):52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bryant H.E., Schultz N., Thomas H.D. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 23.Farmer H., McCabe N., Lord C.J. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 24.Kim G., Ison G., McKee A.E. FDA approval summary: olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(19):4257–4261. doi: 10.1158/1078-0432.CCR-15-0887. [DOI] [PubMed] [Google Scholar]
- 25.Balasubramaniam S., Beaver J.A., Horton S. FDA approval summary: Rucaparib for the treatment of patients with deleterious BRCA mutation-associated advanced ovarian cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2017;23(23):7165–7170. doi: 10.1158/1078-0432.CCR-17-1337. [DOI] [PubMed] [Google Scholar]
- 26.de Bono J., Ramanathan R.K., Mina L. Phase I, dose-escalation, two-part trial of the PARP inhibitor talazoparib in patients with advanced germline BRCA1/2 mutations and selected sporadic cancers. Cancer Discov. 2017;7(6):620–629. doi: 10.1158/2159-8290.CD-16-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matulonis U.A., Wulf G.M., Barry W.T. Phase I dose escalation study of the PI3kinase pathway inhibitor BKM120 and the oral poly (ADP ribose) polymerase (PARP) inhibitor olaparib for the treatment of high-grade serous ovarian and breast cancer. Annals of oncology : official journal of the European Society for Medical Oncology. 2017;28(3):512–518. doi: 10.1093/annonc/mdw672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kummar S., Chen A., Ji J. Phase I study of PARP inhibitor ABT-888 in combination with topotecan in adults with refractory solid tumors and lymphomas. Cancer Res. 2011;71(17):5626–5634. doi: 10.1158/0008-5472.CAN-11-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearsall E.A., Lincz L.F., Skelding K.A. The role of DNA repair pathways in AML chemosensitivity. Curr Drug Targets. 2018;19(10):1205–1219. doi: 10.2174/1389450119666180110093713. [DOI] [PubMed] [Google Scholar]
- 30.Rosen D.B., Leung L.Y., Louie B. Quantitative measurement of alterations in DNA damage repair (DDR) pathways using single cell network profiling (SCNP) J Transl Med. 2014;12:184. doi: 10.1186/1479-5876-12-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scardocci A., Guidi F., D'Alo F. Reduced BRCA1 expression due to promoter hypermethylation in therapy-related acute myeloid leukaemia. Br J Cancer. 2006;95(8):1108–1113. doi: 10.1038/sj.bjc.6603392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esposito M.T., Zhao L., Fung T.K. Synthetic lethal targeting of oncogenic transcription factors in acute leukemia by PARP inhibitors. Nat Med. 2015;21(12):1481–1490. doi: 10.1038/nm.3993. [DOI] [PubMed] [Google Scholar]
- 33.Gojo I., Beumer J.H., Pratz K.W. A phase 1 study of the PARP inhibitor veliparib in combination with temozolomide in acute myeloid leukemia. Clinical Cancer Research : an official journal of the American Association for Cancer Research. 2017;23(3):697–706. doi: 10.1158/1078-0432.CCR-16-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muvarak N.E., Chowdhury K., Xia L. Enhancing the cytotoxic effects of PARP inhibitors with DNA demethylating agents - a potential therapy for cancer. Cancer Cell. 2016;30(4):637–650. doi: 10.1016/j.ccell.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Bernardo G., Alessio N., Dell'Aversana C. Impact of histone deacetylase inhibitors SAHA and MS-275 on DNA repair pathways in human mesenchymal stem cells. J Cell Physiol. 2010;225(2):537–544. doi: 10.1002/jcp.22236. [DOI] [PubMed] [Google Scholar]
- 36.Leoni L.M., Bailey B., Reifert J. Bendamustine (Treanda) displays a distinct pattern of cytotoxicity and unique mechanistic features compared with other alkylating agents. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(1):309–317. doi: 10.1158/1078-0432.CCR-07-1061. [DOI] [PubMed] [Google Scholar]
- 37.Cai B., Lyu H., Huang J. Combination of bendamustine and entinostat synergistically inhibits proliferation of multiple myeloma cells via induction of apoptosis and DNA damage response. Cancer Lett. 2013;335(2):343–350. doi: 10.1016/j.canlet.2013.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conti C., Leo E., Eichler G.S. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res. 2010;70(11):4470–4480. doi: 10.1158/0008-5472.CAN-09-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu J., Qiu S., Ge Q. A novel SAHA-bendamustine hybrid induces apoptosis of leukemia cells. Oncotarget. 2015;6(24):20121–20131. doi: 10.18632/oncotarget.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasmussen R.D., Gajjar M.K., Jensen K.E., Hamerlik P. Enhanced efficacy of combined HDAC and PARP targeting in glioblastoma. Mol Oncol. 2016;10(5):751–763. doi: 10.1016/j.molonc.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tu W.Z., Li B., Huang B. gammaH2AX foci formation in the absence of DNA damage: mitotic H2AX phosphorylation is mediated by the DNA-PKcs/CHK2 pathway. FEBS Lett. 2013;587(21):3437–3443. doi: 10.1016/j.febslet.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 42.Daniel R., Ramcharan J., Rogakou E. Histone H2AX is phosphorylated at sites of retroviral DNA integration but is dispensable for postintegration repair. J Biol Chem. 2004;279(44):45810–45814. doi: 10.1074/jbc.M407886200. [DOI] [PubMed] [Google Scholar]
- 43.O'Farrell A.M., Abrams T.J., Yuen H.A. SU11248 is a novel FLT3 tyrosine kinase inhibitor with potent activity in vitro and in vivo. Blood. 2003;101(9):3597–3605. doi: 10.1182/blood-2002-07-2307. [DOI] [PubMed] [Google Scholar]
- 44.Fiedler W., Serve H., Dohner H. A phase 1 study of SU11248 in the treatment of patients with refractory or resistant acute myeloid leukemia (AML) or not amenable to conventional therapy for the disease. Blood. 2005;105(3):986–993. doi: 10.1182/blood-2004-05-1846. [DOI] [PubMed] [Google Scholar]
- 45.Dombret H., Seymour J.F., Butrym A. International phase 3 study of azacitidine vs conventional care regimens in older patients with newly diagnosed AML with >30% blasts. Blood. 2015;126(3):291–299. doi: 10.1182/blood-2015-01-621664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cooper B.W., Kindwall-Keller T.L., Craig M.D. A phase I study of midostaurin and azacitidine in relapsed and elderly AML patients. Clin Lymphoma Myeloma Leuk. 2015;15(7):428–432. doi: 10.1016/j.clml.2015.02.017. [e2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valdez B.C., Li Y., Murray D. The PARP inhibitor olaparib enhances the cytotoxicity of combined gemcitabine, busulfan and melphalan in lymphoma cells. Leuk Lymphoma. 2017;58(11):2705–2716. doi: 10.1080/10428194.2017.1306647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park H.J., Bae J.S., Kim K.M. The PARP inhibitor olaparib potentiates the effect of the DNA damaging agent doxorubicin in osteosarcoma. Journal of experimental & clinical cancer research : CR. 2018;37(1):107. doi: 10.1186/s13046-018-0772-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Podszywalow-Bartnicka P., Wolczyk M., Kusio-Kobialka M. Downregulation of BRCA1 protein in BCR-ABL1 leukemia cells depends on stress-triggered TIAR-mediated suppression of translation. Cell Cycle. 2014;13(23):3727–3741. doi: 10.4161/15384101.2014.965013. [DOI] [PubMed] [Google Scholar]
- 50.Wolf D.M., Yau C., Sanil A. DNA repair deficiency biomarkers and the 70-gene ultra-high risk signature as predictors of veliparib/carboplatin response in the I-SPY 2 breast cancer trial. NPJ Breast Cancer. 2017;3:31. doi: 10.1038/s41523-017-0025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaye S.B. Progress in the treatment of ovarian cancer-lessons from homologous recombination deficiency-the first 10 years. Annals of Oncology : official journal of the European Society for Medical Oncology. 2016;27(Suppl. 1):i1–i3. doi: 10.1093/annonc/mdw082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Javle M., Curtin N.J. The role of PARP in DNA repair and its therapeutic exploitation. Br J Cancer. 2011;105(8):1114–1122. doi: 10.1038/bjc.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao L., So C.W.E. PARPi potentiates with current conventional therapy in MLL leukemia. Cell Cycle. 2017;16(20):1861–1869. doi: 10.1080/15384101.2017.1288325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lok B.H., Gardner E.E., Schneeberger V.E. PARP inhibitor activity correlates with SLFN11 expression and demonstrates synergy with temozolomide in small cell lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2017;23(2):523–535. doi: 10.1158/1078-0432.CCR-16-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen Y., Rehman F.L., Feng Y. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(18):5003–5015. doi: 10.1158/1078-0432.CCR-13-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murai J., Huang S.Y., Renaud A. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther. 2014;13(2):433–443. doi: 10.1158/1535-7163.MCT-13-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chao O.S., Goodman O.B., Jr. Synergistic loss of prostate cancer cell viability by coinhibition of HDAC and PARP. Molecular cancer research : MCR. 2014;12(12):1755–1766. doi: 10.1158/1541-7786.MCR-14-0173. [DOI] [PubMed] [Google Scholar]
- 58.Baldan F., Mio C., Lavarone E. Epigenetic bivalent marking is permissive to the synergy of HDAC and PARP inhibitors on TXNIP expression in breast cancer cells. Oncol Rep. 2015;33(5):2199–2206. doi: 10.3892/or.2015.3873. [DOI] [PubMed] [Google Scholar]
- 59.Lopez-Iglesias A.A., Herrero A.B., Chesi M. Preclinical anti-myeloma activity of EDO-S101, a new bendamustine-derived molecule with added HDACi activity, through potent DNA damage induction and impairment of DNA repair. J Hematol Oncol. 2017;10(1):127. doi: 10.1186/s13045-017-0495-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Besse L., Kraus M., Besse A. The first-in-class alkylating HDAC inhibitor EDO-S101 is highly synergistic with proteasome inhibition against multiple myeloma through activation of multiple pathways. Blood cancer journal. 2017;7(7) doi: 10.1038/bcj.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CI values were calculated at the effective dose (ED) for 50%, 75% and 90% of the fraction affected.
Characteristics of primary AML patients.