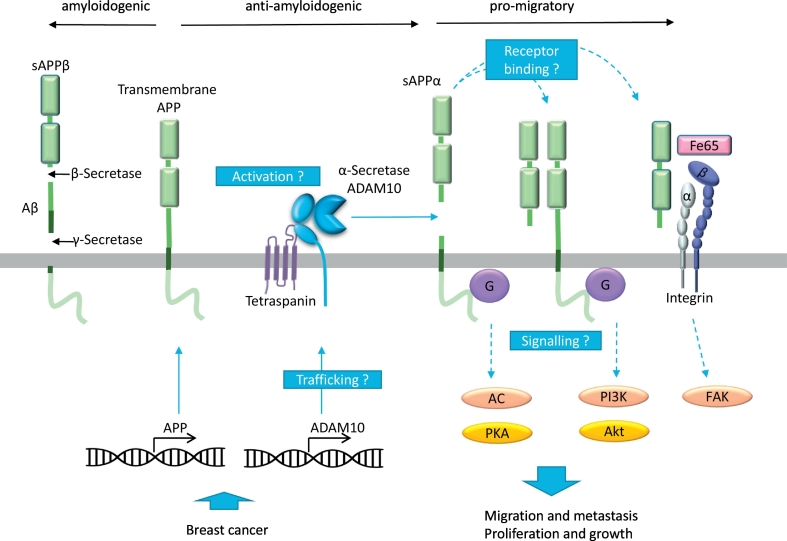
Cleavage of the amyloid precursor protein (APP) at the membrane-proximal extracellular position, termed the alpha secretase site, prevents the generation of amyloidogenic Aβ peptide (Fig. 1). Since the aggregation of Aβ peptides has been associated with the development of Alzheimer's disease the responsible α-secretase became an important subject of research. Almost 20 years ago this protease was identified as ADAM10 which stands for a disintegrin and a metalloproteinase [1]. Since then, researchers identified an increasing number of other ADAM10 substrates most prominently including Notch, EGF-like growth factors, some adhesion molecules, and ephrins and their receptors. Besides its function in development which largely depends on the cleavage of Notch, ADAM10 was found to play crucial roles in physiologic and pathophysiologic processes. Accumulating evidence suggests an important involvement of ADAM10 in migration of various cell types including leukocytes, various tissue cells, and cancer cells [2]. It seems that the underlying mechanisms can be very diverse owing to ADAM10's numerous substrates including ephrins, growth factors, adhesion molecules, transmembrane chemokines or respective receptors.
Fig. 1.
Proposed model for the interplay of APP and ADAM10 in breast cancer cell migration. APP and ADAM10 are upregulated in breast cancer tissue increasing the chance of APP cleavage. Soluble APPα formation is relevant for induction of breast cancer cell migration via yet undefined pathways. Further research (blue boxes) is warranted to clarify the trafficking and activation of ADAM10 leading to APP shedding e.g. the involvement of tetraspanins. Furthermore, APP receptor binding, formation of heterooligomers and signalling need to be investigated including integrin binding via FE65, activation of G-proteins and further downstream kinases (e.g. PKA, PI3K, Akt and FAK) involved in the regulation of cell motility and proliferation.
The study by Tsang et al. published in this issue of EBioMedicine indicates that there exists an additional mechanism by which ADAM10 can promote cell migration involving the cleavage of APP (Fig. 1). The authors previously reported that breast cancer tumor cells express elevated levels of APP. Their present study investigates APP shedding in different breast cancer cell lines and its role in breast cancer cell proliferation and migration [3]. APP and the proteolytic fragments from alpha-cleavage were detected in different breast cancer cell lines. Knockdown of APP expression using RNA interference decreased breast cancer growth and migration in vitro and in a murine tumor xenograft model, whereas overexpression of soluble APPα or full-length APP had the opposite effect. Knockdown of ADAM10 inhibited alpha-cleavage of APPα and reduced cell migration. The latter effect was reversed in part by overexpression of soluble APPα but not of transmembrane full-length APP. Expression of APP and ADAM10 was examined in clinical non-luminal breast cancer samples and a significant correlation of both APP and ADAM10 expression was found. Importantly, co-expression of both proteins indicated unfavorable clinical outcome.
These interesting and potentially very relevant results should prompt further investigations to study and compare the role of APP in different cancer cell types as well as in different in vivo models of cancer development and metastasis. It would also be important to know whether a similar activity accounts for the closely related ADAM10 substrate APLP2 which has been reported to modulate JNK-Dependent cell migration in Drosophila [4]. Importantly, the receptor and signalling mechanism needs to be elucidated in more detail. At the cell membrane APP has been described to associate with beta 1 integrins via the APP binding protein FE65 and formation of this complex correlates with enhanced cell movement [5]. Several studies indicated that the transmembrane APP can modulate G-protein coupled receptor signalling leading to activation of the kinases PKA and PI3K both of which could be required for cell migration [6]. Moreover, APP was reported to form multimers and by this drive JAK-mediated signalling towards metastasis and tumor growth. Interestingly, the present study indicates that the promigratory activity but not proliferation of breast cancer cells is mediated to a large extent by soluble APP rather than the intact molecule. It remains to be clarified whether other soluble APP fragments (e.g. generated from β-secretase) or the remaining transmembrane fragment can interfere with such a migration-promoting mechanism. One would further question what type of receptor for soluble APP could be responsible here. It is possible that oligomer formation of soluble and transmembrane APP is involved resulting in signalling via the cytoplasmic tail of transmembrane APP. By this mechanism, soluble APPα has been proposed to promote neuronal Akt activation and cell survival [7] while other responses such as modulation of BAG3 expression do not require transmembrane APP. Yet, for cell migration this still needs to be studied in detail. Analysis of the gene expression signature will potentially reveal further downstream transcriptional responses induced by soluble APP.
Another direction of research could address the regulation of APP shedding. The present study indicates upregulation of the protease ADAM10 in breast cancer cells. Besides this upregulation, trafficking and activity regulation may represent even more crucial steps. ADAM10 has been described to associate with tetraspanins of the TspanC8 family which are required for surface expression and also contribute to substrate selectivity of the protease [8]. Furthermore, ADAM10 is regulated by cell activation and calcium influx leading to exposure of negative charges at the membrane that are thought to promote a conformational switch removing the inhibitory prodomain of the protease from the active site [9]. Thus, it will be important not only to correlate the protease with its substrate but also determine the exact localization and activation status of the protease.
Solving these open questions will help to tailor more specific intervention therapies using APP as a potential therapeutic target. Inhibitors blocking APP may not only suppress migration and metastasis, but also cell survival of other cancer cell types expressing high levels of APP such as prostate cancer cells [10].
Conflicts of interest
The authors declare no conflicts of interest.
Contributor Information
Justyna Wozniak, Email: aludwig@ukaachen.de.
Andreas Ludwig, Email: aludwig@ukaachen.de.
References
- 1.Lammich S., Kojro E., Postina R., Gilbert S., Pfeiffer R., Jasionowski M. Constitutive and regulated alpha-secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci U S A. 1999 Mar 30;96(7):3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dreymueller D., Theodorou K., Donners M., Ludwig A. Fine tuning cell migration by a disintegrin and metalloproteinases. Mediators Inflamm. 2017;2017 doi: 10.1155/2017/9621724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsang J., Lee M., Chan T.H., Li J., Ni J.B., Shao Y. Proteolytic cleavage of amyloid precursor protein by ADAM10 mediates proliferation and migration in breast cancer. EBioMedicine. 2018 doi: 10.1016/j.ebiom.2018.11.012. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X., Guo X., Ma Y., Wu C., Li W., Xue L. APLP2 modulates JNK-dependent cell migration in drosophila. Biomed Res Int. 2018 Jul 29;2018 doi: 10.1155/2018/7469714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabo S.L., Ikin A.F., Buxbaum J.D., Greengard P. The Alzheimer amyloid precursor protein (APP) and FE65, an APP-binding protein, regulate cell movement. J Cell Biol. 2001 Jun 25;153(7):1403–1414. doi: 10.1083/jcb.153.7.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Copenhaver P.F., Kögel D. Role of APP interactions with heterotrimeric G proteins: physiological functions and pathological consequences. Front Mol Neurosci. 2017 Jan 31;10:3. doi: 10.3389/fnmol.2017.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milosch N., Tanriover G., Kundu A., Rami A., Francois J.C., Baumkotter F. Holo-APP and G-protein-mediated signaling are required for sAPPalpha-induced activation of the Akt survival pathway. Cell Death Dis. 2014;5:e1391. doi: 10.1038/cddis.2014.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matthews A.L., Koo C.Z., Szyroka J., Harrison N., Kanhere A., Tomlinson M.G. Regulation of leukocytes by TspanC8 tetraspanins and the "molecular scissor" ADAM10. Front Immunol. 2018 Jul 2;9:1451. doi: 10.3389/fimmu.2018.01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reiss K., Bhakdi S. The plasma membrane: penultimate regulator of ADAM sheddase function. Biochim Biophys Acta Mol Cell Res. 2017 Nov;1864(11 Pt B):2082–2087. doi: 10.1016/j.bbamcr.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Miyazaki T., Ikeda K., Horie-Inoue K., Inoue S. Amyloid precursor protein regulates migration and metalloproteinase gene expression in prostate cancer cells. Biochem Biophys Res Commun. 2014 Sep 26;452(3):828–833. doi: 10.1016/j.bbrc.2014.09.010. [DOI] [PubMed] [Google Scholar]