Abstract
Background
Hypoxia commonly occurs in solid tumors. The hypoxia in the center of solid tumors considerably decreases the chemosensitivity of tumor cells and induces epithelial–mesenchymal transition (EMT) as well as drug resistance of antitumor drugs.
Methods
Here, the effects of salidroside (Sal) combined with platinum drugs on human hepatocellular carcinoma were examined in vitro and in vivo. We investigated the antitumor effects of Sal by inhibiting the drug resistance and explained its mechanism in inhibiting tumor growth.
Findings
The results showed that Sal co-administration reverses the drug resistance of platinum drugs and suppressed metastasis induced by the hypoxic tumor microenvironment. Sal promoted the degradation of HIF-1α. In conclusion, Sal significantly increased the sensitivity to platinum drugs and inhibited hypoxia-induced EMT in hepatocellular carcinoma (HCC) through inhibiting HIF-1α signaling pathway.
Interpretation
Therefore, Sal may be an effective platinum drug sensitizer that can improve the chemotherapeutic efficacy in patients with HCC.
Keywords: Salidroside, HIF-1α, Hypoxic tumor microenvironment, Drug resistance
Abbreviations: Sal, salidroside; OXA, oxaliplatin; DDP, cisplatin; HCC, hepatocellular carcinomas; HIF-1α, hypoxia-inducible factor 1α; EMT, epithelial–mesenchymal transition; MDR, multi-drug resistance; MRP, multidrug resistance-associated protein; CI, combination index; PCNA, proliferating cell nuclear antigen; ECM, extracellular matrix
Research in context.
Evidence before this study
The hypoxia in the center of solid tumors considerably decreases the chemosensitivity of tumor cells and induces epithelial–mesenchymal transition (EMT) as well as drug resistance of antitumor drugs.
Added value of this study
Findings from this study indicate that Sal significantly increased the sensitivity to platinum drugs and inhibited hypoxia-induced EMT in hepatocellular carcinoma (HCC) through inhibiting HIF-1α signaling pathway.
Implications of all the available evidence
Sal may be an effective platinum drug sensitizer that can improve the chemotherapeutic efficacy in patients with HCC.
Alt-text: Unlabelled Box
1. Introduction
Since the approval of cancer therapy in the late 1970s, platinum-based drugs have been widely used, and they are often called the penicillin of cancer [1]. Cisplatin, the first platinum anticancer drug, has become the main chemotherapeutic agent in the treatment of various solid tumors [2,3]. Oxaliplatin (OXA), a third-generation platinum compound introduced in clinics, showed excellent anti-advanced HCC activity with tolerable toxicity. However, the overall effectiveness of platinum-based drug therapies is limited by their tumor cell resistance, which often develops and causes patients to become refractory to further treatment [4].
Platinum-based drugs can target highly proliferating cancer cells, but the strong quiescent cell fraction typically associated with hypoxia is nearly unaffected by various treatments [5,6]. Hypoxia is considered a common characteristic of tumors due to the imbalance between oxygen consumption in tumor tissues and oxygen supply to blood vessels [7]. Platinum is unevenly distributed in solid tumors, and its low dose can induce epithelial–mesenchymal transition (EMT) and metastasis. Hypoxia can also stimulate the aggressiveness of neoplasms and induce distant metastasis [8,9].
HIF-1 comprises α and β subunits, which are basic helix-loop-helix factors that are key gene regulatory factors involved in cell hypoxia response. The α subunit expression is significantly increased at hypoxia, but most cells remain at low level under normoxic condition [10]. HIF-1α regulates cell responses to hypoxia and tumor biological behavior by influencing apoptotic/proliferative activity, vasomotor function, energy metabolism, and angiogenesis [[11], [12], [13]]. This subunit also increases the expression of proteins that confer multi-drug resistance (MDR), including MDR1 and MRP [14,15]. HIF-1α can also transcriptionally regulate many EMT-related transcription factors, which all play indispensable roles in EMT induction. For example, HIF-1α induces EMT through the transcriptional regulation of E-cadherin, SNAIL, Zeb1, and Twist1. Therefore, HIF-1α is considered a target for tumor chemotherapy and metastasis [16].
Salidroside (Sal), which is isolated from Rhodiola rosea L and has long been used in preventing mountain sickness [17]. Sal has various pharmacological properties including neuroprotective, cardiovascular protective, and antiviral effects [[18], [19], [20], [21]]. Sal also concentration- and time-dependently inhibit the growth of different human cancer cell lines, and these cancer cells exhibit different sensitivities to Sal [22].
This study aims to explore the antitumor effects of Sal under hypoxic environment and explain the anti-tumor mechanism of Sal. We found that Sal enhanced the effects of OXA on HCC and reversed the drug resistance of OXA and EMT via HIF-1α signaling pathway.
2. Materials and methods
2.1. Materials
Salidroside was purchased from Meilun Biotechnology (Dalian, China) and was resuspended in phosphate buffer saline (in vitro) and normal saline (in vivo). 100 μM salidroside was used in all in vitro experiments except the cell viability assay. The antibodies to beta-actin (mAbcam 8226, 1/10000 dilution), PCNA (PC10, 1/1000 dilution), HIF-1 alpha (ab2185, 1/200 dilution for IHC and 1/1000 dilution for WB), HA tag (ab1424, 1/1000 dilution) were purchased from Abcam (Cambridge, MA, USA). The antibodies to Twist1 (AF4009, 1/1000 dilution), Zeb1 (DF7414, 1/1000 dilution), E-cadherin (AF0131, 1/2000 for WB, 1/100 for IHC and 1/500 for IF), Vimentin (BF0071, 1/1000 for WB and 1/500 for IHC) were purchased from Affinity (Cincinnati, USA).
2.2. Cell culture
Human liver cancer cell lines, namely, PLC/PRF/5, SMMC-7721, and HepG2, were purchased from KeyGen Biotech (Nanjing, China). All the cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (HyClone, USA) and 1% penicillin–streptomycin in a humidified atmosphere (37 °C, 5% CO2). To obtain a hypoxic condition, the cells were cultured in a CO2 incubator with 94% N2, 5% CO2, and 1% O2. Cells were put in hypoxic conditions for 24 h concurrently treated with salidroside (the time of wound-healing assay is 48 h).
2.3. Cell viability assay
The cells were resuspended in a complete medium and cultured in a 96-well plate with an initial density of 5 × 103 cells/well for 24 h. Various drugs in different concentrations were used to treat the cells after overnight incubation. After 48 h, 20 μL of MTT was added to each well, and the cells were cultured for 4 h. Finally, 150 μL of dimethyl sulfoxide was added. The absorbance at 590 nm and the 50% inhibitory concentration (IC50) value of each drug was measured (Multiskan™ FC, Thermo Scientific, Waltham, MA, USA). All samples were prepared in triplicate to ensure reproducibility. Data are presented as mean ± standard deviation.
2.4. Cell activation and intracellular staining
Human liver cancer cell lines, namely, PLC/PRF/5, SMMC-7721, and HepG2 (20,000 cells/well), were seeded in a 96-well plate. After overnight incubation, the cells were treated with different drugs (Sal: 100 μM, OXA: 5 μM). Prior to cell fixation, Live/Dead Fixable Dead Cell Stain Kit was used to stain all the cell lines. Flow cytometry was used to determine the amount of living and dead cells (easyCyte HT, Millipore, USA).
2.5. Wound-healing assay
Sal inhibitory effects on cell migration were examined by wound-healing assay. The cells were seeded into a 35 mm dish to form 100% confluent monolayers. The cells were wounded with 100 μL tips, washed with PBS, and incubated with different drugs. After drug treatment (Sal: 100 μM; OXA: 5 μM), the wound healing speed was measured at 0, 24, and 48 h. Wound images were obtained by using a Nikon microscope (200× magnification).
2.6. Transwell assays
Cell invasion ability was assessed through Transwell assays. Cells treated under different conditions (1 × 105 cells/mL) were suspended in 300 μL of 1640 medium without FBS and seeded into Matrigel-coated upper wells covered with an 8.0 μm pore size polyethylene terephthalate filter membrane. A total of 500 μL of RPMI 1640 medium containing 10% FBS was placed in the lower chamber. Cotton swabs were used to remove the cells on the upper surface of the filter after 24 h incubation. The cells that were fixed with 4% paraformaldehyde for 20 min migrated or invaded through the filter. Afterward, these cells were stained with 0.1% crystal violet for 10 min. Invading cells were imaged by an inverted microscope (Nikon, Japan) (at 100× magnification), and the cell number was manually counted.
2.7. Western blot analysis
The cells with a density of 3 × 105 cells/well were incubated overnight in a six-well plate. After incubation with different drugs for 24 h, 50 μL of cell lysis buffer that contained protease inhibitors was used to harvest and lyse the cells. SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was used to resolve the cells, and the resolved proteins were transferred into the PVDF membranes. These membranes were subsequently incubated in TBST (TBS contacting 0.1% Tween-20), which contained 5% skim milk powder, for 2 h and with primary antibodies against HIF-1α, PCNA, Twist1, Snail, Zeb1, E-cadherin, and GAPDH at 4 °C overnight.
2.8. Quantitative RT-PCR
5 × 106 Hela cells were prepared for the experiment. The total RNA was extract using RNAprep Pure Cell/Bacteria Kit (TIANGEN BIOTECH, Beijing, China) according the manufacturer's instructions. Reverse transcription was performed using the SuperScript RT-PCR kit (Invitrogen) for cDNA synthesis. Quantitative RT-PCR was performed as previously described [23]. The primers used in the current study included the following: HIF-1α forward 5’-CTC AAA GTC GGA CAG CCT CA-3′ and reverse 5’-CCC TGC AGT AGG TTT CTG CT-3′.
2.9. Immunoprecipitation
PLC/PRF/5 cells were treated with Sal (100 μM) or Sal (100 μM) combined with 5 μM of MG132. The cell lysates were incubated with anti-HIF-1α antibody after 24 h. Protein A-Sepharose beads were used to precipitate the immune complex. The precipitates were washed with IP buffer (25 mM HEPES pH 7.5, 150 mM NaCl, 0.5% NP-40, 25 mM NaF, 1 mM Na3VO4, 50 mM β-glycerolphosphate, 14 mM EGTA, 10 mM MgCl2, 1 mM PMSF, and PCL protease inhibitors), fractionated by SDS-PAGE, and immunoblotted with antiubiquitin antibody.]
2.10. Animal studies
BALB/c nude mice (18–20 g) were obtained from the Animal Center Academy of the Military Medical Science (Beijing, China). All animals in this experiment were bred under specific pathogen-free conditions. All animal procedures were approved on the basis of guidelines of the Animal Ethics Committee of the Tianjin International Joint Academy of Biotechnology and Medicine.
2.10.1. The effect of Sal on the PLC/PRF/5 xenograft model.
Human liver cancer cells PLC/PRF/5 were cultured in vitro to the log growth phase, centrifuged and washed with PBS, and resuspended in a 50% mixture of Matrigel (BD Biosciences) in PBS to a final cell count of 2 × 107 cells/mL. A volume of 0.2 mL of the cell suspension was injected in the right flank of each mouse. 14 days after tumor inoculation (the tumor volumn is about 100 mm3), the mice were categorized into the following groups (n = 5): (1) model group (saline by oral), (2) OXA group (5 mg/kg/2 day, intraperitoneal injection), (3) Sal group (Sal: 60 mg/kg/day, intragastric administration) and (4) Sal + OXA group (Sal: 60 mg/kg/day; OXA: 5 mg/kg/). Tumor volume and body weight were measured daily after tumor inoculation. Tumor volumes were calculated in accordance with the formula V = ab2/2 (a = length, b = width). After 3 weeks, all mice were euthanized. The xenografts were resected and measured. Another 20 mice were allocated randomly to 4 groups as described above (5 mice per group), in order to measure survival rates. The survival time of every mouse were recorded lasted for 60 days.
2.10.2. The effect of Sal on the PLC/PRF/5 orthotopic transplantation tumor model.
The model was established by using the intrahepatic tunnel implantation. When the PLC-PRF-5 xenograft tumor grew to 1.0 cm in diameter, the fresh tumor tissue was cut into 1.0 mm × 2.0 mm pieces and implanted into the tunnel under the capsule of the left lateral liver lobe by using a pair of ophthalmologic forceps. After 3 days, the mice were categorized into the following groups: (1) control group (saline by oral), (2) model group (saline by oral), (3) OXA group (5 mg/kg), and (4) Sal + OXA group (Sal: 60 mg/kg, OXA: 5 mg/kg). Mouse liver tissues were collected 2 weeks after modeling. About the survival analysis experiment, the methods of group dividing and model making were the same as described above, except that the drug administration time was 60 days.
2.11. Data statistics
All data are expressed as means ± standard deviation. Comparisons between groups were performed by one-way analysis of variance followed by Bonferroni post hoc test (SPSS software package version 17.0, SPSS Inc., Chicago, IL, USA). The level of significance was set at P < .05.
3. Results
3.1. Sal enhanced the effects of OXA on HCC
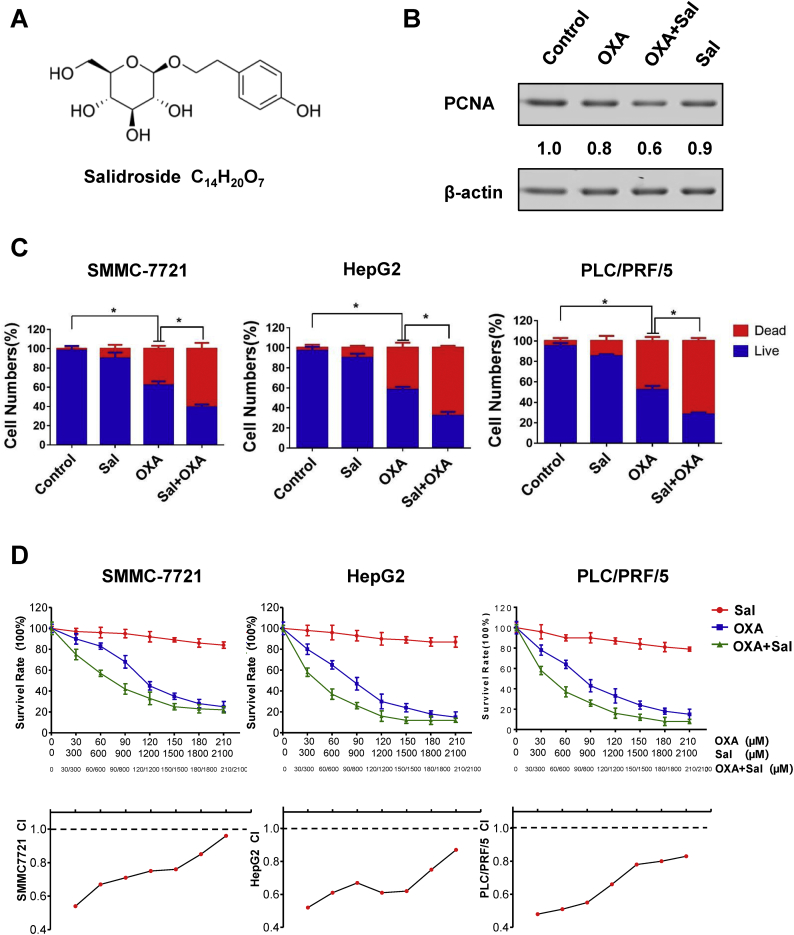
The molecular Sal formula is shown in Fig. 1A. We analyzed the expression of proliferating cell nuclear antigen (PCNA) using Western blot, a marker of cell proliferation. Fig. 1B shows that the cotreatment could reduce the expression of the PCNA. We also found that Sal and OXA can reduce the survival rate of HCC by Live/Dead assay. Results showed that the percentage of dead cells significantly increased in the co-treatment group compared with those in other groups (Fig. 1C). The effects of Sal, OXA, and Sal combined with OXA (co-treatment) on cell activity were determined by MTT assay (treated for 48 h). Fig. 1D shows that the combination of Sal with OXA dose-dependently inhibited the proliferation of SMMC-7721, HepG2, and PLC/PRF/5 cells. Sal can enhance the effects of OXA in inhibiting cell proliferation compared with that of OXA treatment alone. In addition, the combination index (CI) value in the combined treatment group was <1 at different doses, thereby indicating that Sal and OXA displayed synergistic effects. Fig. S1 indicated that Sal and OXA combination may have benefit clinical practices, with DRI values well above 1.
Fig. 1.
Sal enhanced the effects of OXA on HCC. (A) Molecular formula of salidroside (Sal). (B) Expression of the proliferating cell nuclear antigen (PCNA) in different groups. (C) Percentage of dead cells in different groups was determined by the Live/Dead Fixable Dead Cell Stain Kits. (D) Inhibitory effects of Sal, OXA, or Sal combined with OXA on PLC/PRF/5, HepG2, and SMMC-7721 cells for 48 h. Error bars represent the standard deviation of experiments performed in triplicate (*P < .05, **P < .01).
Fig. S1.
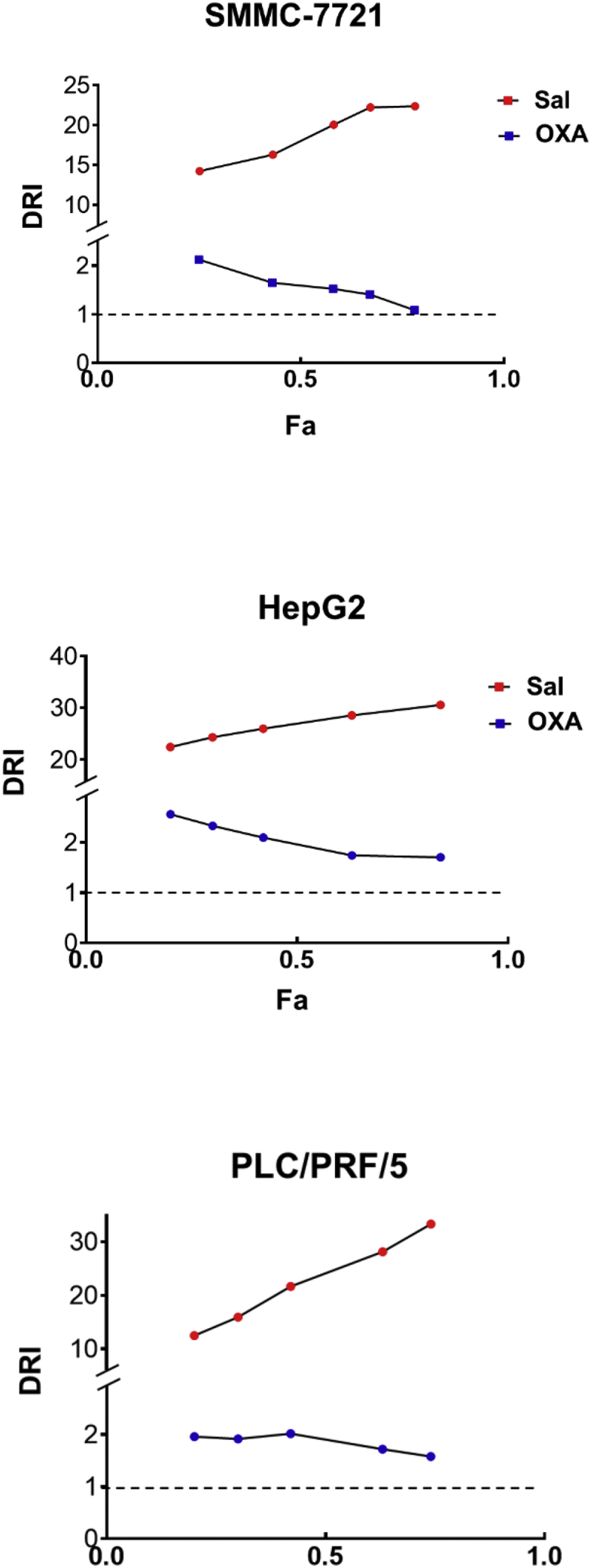
DRI of combination treatment.
3.2. Sal enhanced the antitumor effects of OXA in vivo
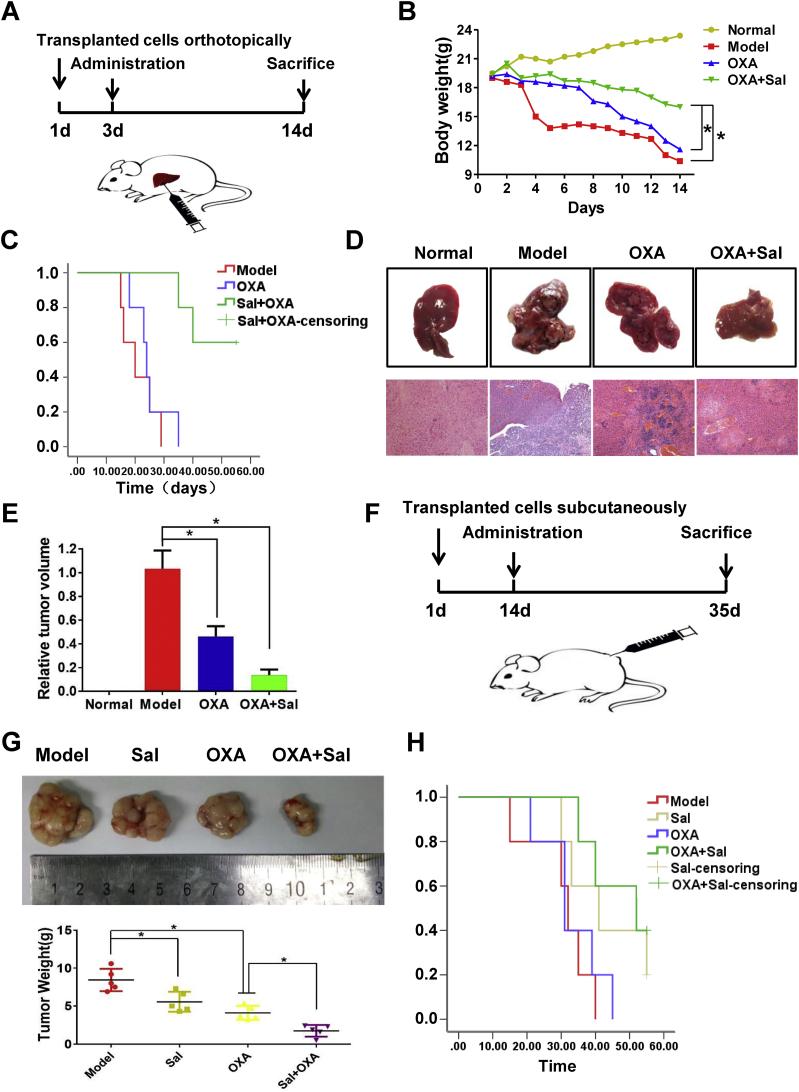
The co-treatment effects on PLC/PRF/5 orthotopically implanted tumors were also examined in nude mice (Fig. 2A). The body weights of the mice in co-treatment group increased compared with those in the OXA treatment group (Fig. 2B). The survival curve showed that the combination of Sal and OXA increased the survival rate of mice compared with that in OXA group (Fig. 2C). The tumor growth was inhibited in the OXA treatment group and the co-treatment group (Figs 2D and 2E), and the co-treatment group showed better effects. In addition, we established subcutaneously transplanted tumor model to detect the effects of Sal and OXA combination(Fig. 2F). Fig. 2G shows that the tumor in the combined administration group was significantly inhibited compared with that in administration group alone. The survival time was also significantly prolonged (Fig. 2H). These results demonstrated that Sal can improve the treatment effect of OXA and reverse the drug resistance of OXA.
Fig. 2.
Sal enhanced the antitumor effects of OXA in vivo. (A) Schematic diagram of the tumor model of orthotopic transplantation in mice. (B) Effects of different drugs on the body weights of orthotopically transplanted tumor mice. (C) Effects of different drugs on the survival of orthotopically transplanted tumor mice. (D–E) Effects of different drugs on tumor volume in orthotopically transplanted tumor mice. (F) Schematic diagram of a mouse model with subcutaneous transplantation tumor. (G) Effects of different drugs on tumor volume in subcutaneously transplanted tumor mice. (H) Effects of different drugs on survival in subcutaneously transplanted tumor mice. Error bars represent the standard deviation of experiments performed in triplicate (*P < .05, **P < .01).
3.3. Sal reversed the drug resistance and inhibited HIF-1-α signaling pathway
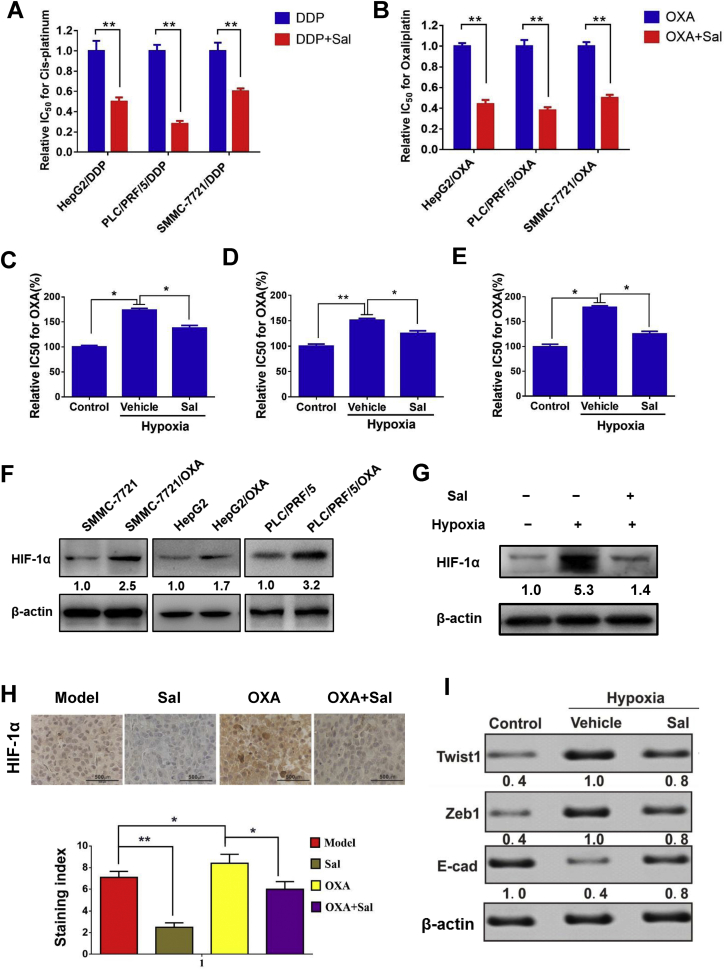
Next, the reversal effects of Sal on drug resistance were evaluated by using drug-resistant cell lines. As shown in Figs 3A and 3B, Sal can effectively reverse the drug resistance in DDP- and OXA-resistant cells (HepG2/DDP, PLC/PRF/5/DDP, SMMC-7721/DDP, HepG2/OXA, PLC/PRF/5/OXA and SMMC-7721/OXA). Cells can develop drug resistance under hypoxic conditions. Therefore, we established an anoxic cell model to detect the reversal effects of Sal on drug resistance. As shown in Figs 3C, 3D, and 3E, the IC50 value of OXA in hypoxia-treated group was evidently higher than that in the untreated group. Hypoxia treatment can reduce the drug sensitivity of cells. After Sal treatment, the proliferation ability of cells was significantly reduced, and the IC50 value of OXA was lower than that of hypoxia-treated group. Next, the expression of HIF-1α in HCC cells and OXA-resistant HCC cells were detected. The results showed that there was a higher expression of HIF-1α in OXA-resistant cells (Fig. 3F). Our results showed that the expression level of HIF-1α increased in hypoxia microenvironment, which could be decreased after Sal treatment (Fig. 3G). Similar results were observed in subcutaneous xenotransplanted tumor (Fig. 3H). The expression levels of Twist1 and Zeb1 were increased after hypoxia induction, whereas the E-cadherin expression was decreased. The Sal treatment could reverse the expression changes of these proteins induced by hypoxia in vitro (Fig. 3I). Furthermore, these proteins are all the downstream proteins of the HIF-1α pathway.
Fig. 3.
Sal reversed the drug resistance and inhibited HIF-1α signaling pathway. (A-B) Relative 50% inhibitory concentration (IC50) values of DDP/OXA-resistant cells for OXA or DDP in the treatment or co-treatment group. (C-E) IC50 values of PLC/PRF/5, HepG2, and SMMC-7721 cells treated with hypoxia alone and hypoxia combined with Sal. (F) Protein expression level of HIF-1α in cancer cells and OXA-resistant cancer cells. (G) Protein expression level of HIF-1α in PLC/PRF/5 cells treated with hypoxia and hypoxia combined with Sal. (H) Effect of different drugs on the expression levels of HIF-1α in tumor tissues of subcutaneously transplanted tumor mice. (I) Protein expression level of Twist1, Zeb1 and E-cadherin in PLC/PRF/5 cells treated hypoxia and hypoxia combined with Sal. Error bars represent the standard deviation of experiments performed in triplicate (*P < .05, **P < .01).
3.4. Sal inhibited migration, invasion, and EMT of HCC cells
To further study the motility potential and migration ability of HCC cells after treated with Sal and OXA, we performed wound-healing and transwell assays.
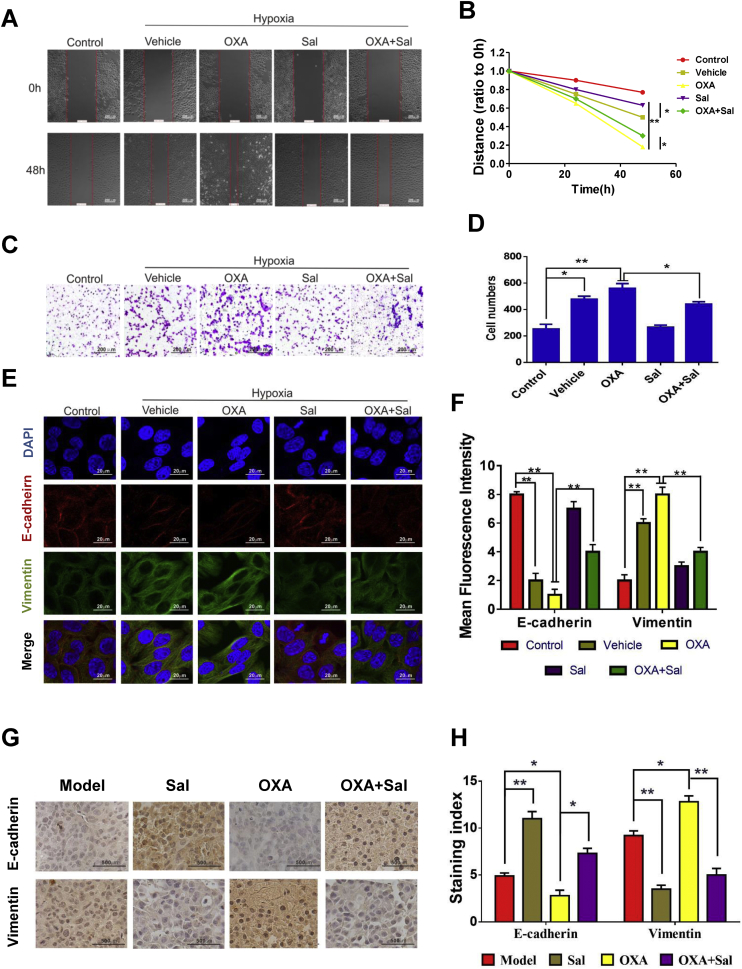
The motility potential and migration ability of cells in OXA-treated groups (hypoxia) were even more enhanced compared with the cells in only hypoxia-treated groups (Figs 4A-4D). In Sal-treated group and the co-treatment group (Sal+OXA), the migration ability of cells was markedly inhibited. E-cadherin and vimentin, the EMT biomarkers, were used to analyze the effects of co-treatment group on the EMT process. Immunofluorescence double staining results showed that the vimentin level was increased, whereas the E-cadherin level was decreased under hypoxic environment. In OXA group, the vimentin level increased, and the E-cadherin level decreased, which revealed that the EMT process was promoted by OXA. By contrast, the vimentin level significantly decreased, and the E-cadherin level significantly increased in the co-treatment group. These results indicated that the combination of Sal and OXA effectively inhibited the EMT (Figs 4E and 4F). Also immunohistochemistry results of the subcutaneous xenotransplanted tumors indicated that Sal treatement can effectively inhibit the decrease of E-cadherin and the increase of vimentin induced by OXA (Fig. 4G and H).
Fig. 4.
Sal inhibited migration and invasion and reversed the changes in EMT biomarkers. (A–B) Cell migration was measured after treatment with various drugs for 48 h. (C–D) Representative images of Transwell cell invasion assays were obtained at 200× magnification. (E–F) Double immunofluorescence staining for E-cadherin and vimentin after treated with Sal. (G-H) The expression levels of HIF-1α in tumor tissues of subcutaneously transplanted tumor mice. Error bars represent the standard deviation of experiments performed in triplicate (*P < .05, **P < .01).
3.5. Sal inhibits HIF-1α signaling pathway
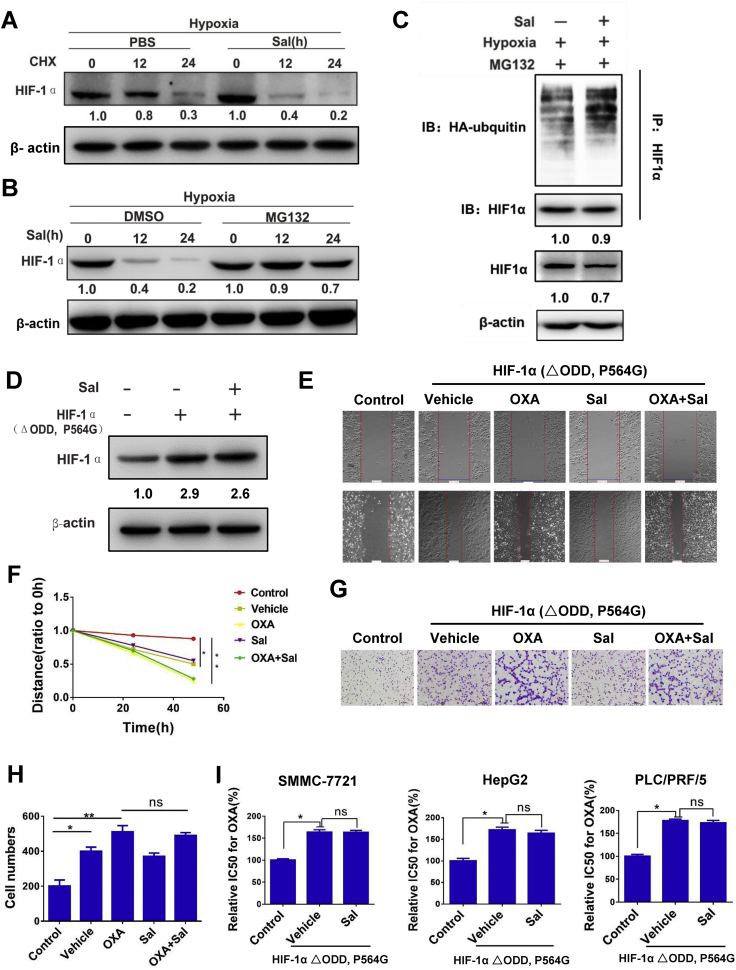
The Sal-induced reduction in HIF-1α was not affected by the protein synthesis inhibitor (CHX, 20 μg/mL) (Fig. 5A) but was prevented by MG132, a proteasome inhibitor (Fig. 5B). These results indicated that Sal decreased the level of HIF-1α through the ubiquitin-proteasome protein degradation pathway. To investigate whether Sal promotes HIF-1α ubiquitination, we performed IP experiments to pull down HIF-1α from the cell lysis and subsequently identified ubiquitinated HIF-1α by immunoblot analysis with anti-ubiquitin monoclonal antibody. As shown in Fig. 5C, these conjugated proteins significantly accumulated when the cells were incubated with the proteasome inhibitor in the Sal treatment group. A HIF-1α ΔODD, namely, P564G, was engineered from the human HIF-1α cDNA to prevent hydroxylation and to further define the role of Sal on HIF-1α signaling. As expected, Sal cannot reduce the level of overexpression of mutational HIF-1α (Fig. 5D) and inhibit the migration and invasion induced by the mutational HIF-1α (Figs 5E–H). The overexpression of mutational HIF-1α could induce the drug resistance of OXA. However, Sal could not reverse the drug resistance induced by mutational HIF-1α (Fig. 5I). These results showed that the HIF-1α degradation by ubiquitination pathway was promoted by Sal.
Fig. 5.
Sal inhibits HIF-1 α signaling pathway. (A) Protein expression level of HIF-1α after treatment with the protein synthesis inhibitor CHX in PLC/PRF/5 cells. (B) Protein expression level of HIF-1α after treatment with the proteasome inhibitor MG132 in PLC/PRF/5 cells. (C) Protein expression level after incubation with the proteasome inhibitor in PLC/PRF/5 cells. (D) Protein expression level of HIF-1α after overexpression of mutational HIF-1α in PLC/PRF/5 cells. (E-F) Cell migration was measured after overexpression of mutational HIF-1α in PLC/PRF/5 cells. (G-H) Representative images of Transwell cell invasion assays after overexpression of mutational HIF-1α in PLC/PRF/5 cells. (I) The relative IC50 values for OXA of PLC/PRF/5 after overexpression of mutational HIF-1α. Error bars represent the standard deviation of experiments performed in triplicate (*P < .05, **P < .01).
3.6. Sal affected multiple signaling pathways on HCC
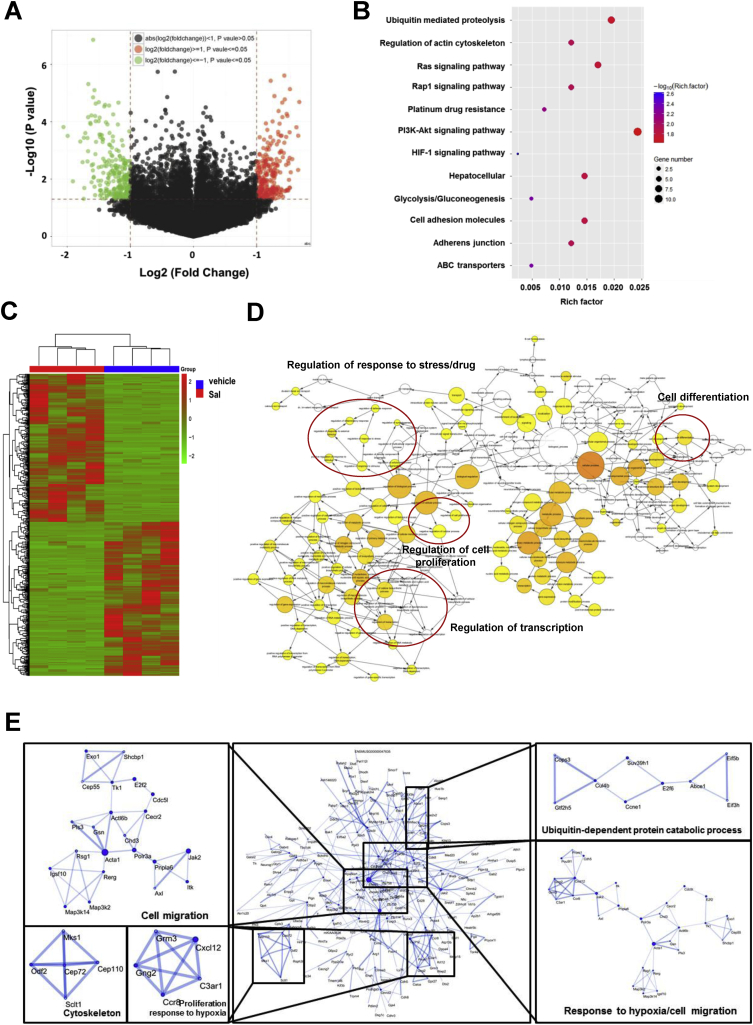
Multidimensional liquid chromatography-tandem mass spectrometry was performed to evaluate differentially expressed proteins after Sal treatment. As shown in Fig. 6A, the −log (P) of each gene was plotted against the log2 ratio of model group intensity to Sal treatment intensity. The downregulated genes are shown in green and upregulated genes are shown in red. The hierarchical clustering of the significant genes is shown in Fig. 6C. Gene ontology (GO) analysis revealed that differentially expressed genes were mainly enriched in the ubiquitination, platinum drug resistance and HIF-1α pathway (Fig. 6B). The molecular function, cellular component, and biological process were analyzed by BiNGO and visualized in Cytoscape (Fig. 6D). The types of interactions between the model and Sal treatment groups were examined by STRING database. Results showed that Sal influenced many proteins associated with cell migration, response to hypoxia and protein ubiquitination (Fig. 6E).
Fig. 6.
Sal affected multiple signaling pathways on HCC. (A) Volcano plot of all genes. (B) Gene ontology functional enrichment analyses. (C) Hierarchical clustering of the significant genes. (D) Function analysis using BiNGO and visualized in Cytoscape. (E) Protein–protein interaction networks and significant modules in the protein–protein interaction network.
3.7. HIF-1α plays an important role in prognosis of HCC.
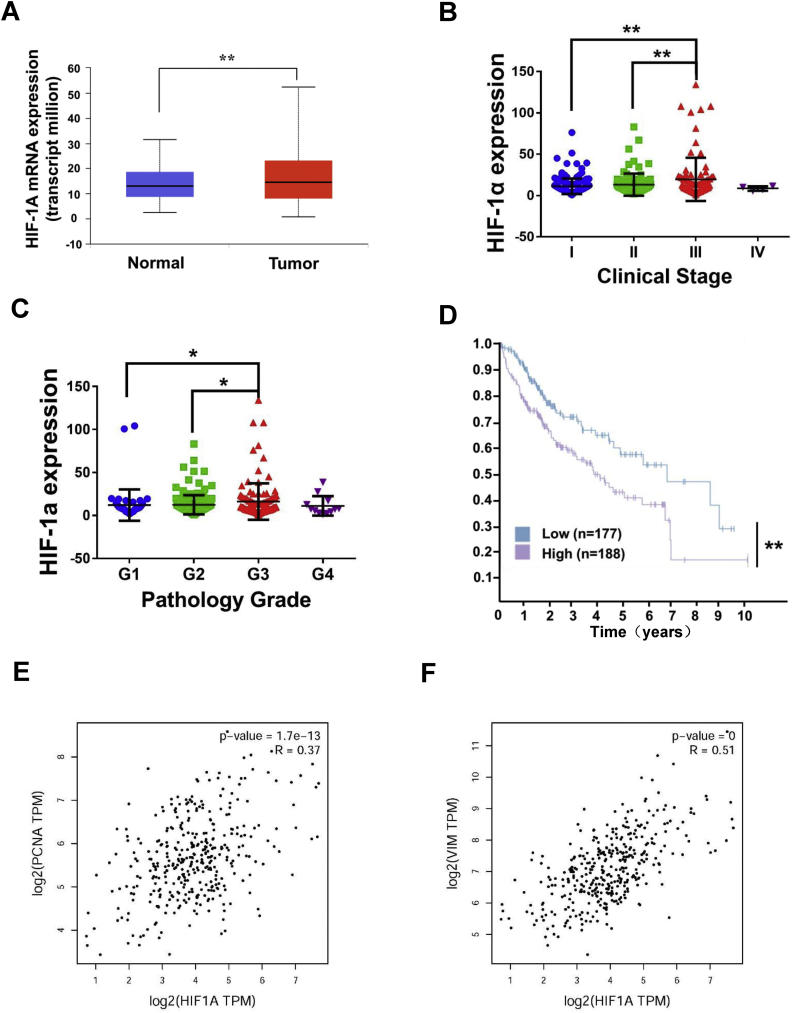
We analyzed the data of 346 patients in the TCGA database and obtained the clinical expression of HIF-1α. As shown in Fig. 7A, the HIF-1α expression in HCC tissues was significantly higher than that in normal liver tissues. We also analyzed the samples in TCGA database according to pathology grade and clinical stage. As demonstrated in Figs 7B and 7C, the expression level was positively correlated with pathology grade and clinical stage. Therefore, the HIF-1α expression increased with tumor malignancy. We analyzed the effects of HIF-1α expression on survival status. HIF-1α overexpression indicated poor prognosis (Fig. 7D). To further investigate the relationship between the HIF-1α expression and proliferation and metastasis of HCC, we analyzed the correlation between the expression level of HIF-1α expression and those of PCNA and Vimentin (Figs 7E–7F). Results showed that HIF-1α was closely related to these proteins.
Fig. 7.
HIF-1α plays an important role in prognosis of HCC. (A) HIF-1α expression in clinically normal liver and hepatocellular carcinoma tissues. (B) Correlation between HIF-1α expression and clinical stage. (C) Correlation between HIF-1α expression and pathology grade. (D) Effects of HIF-1α expression on survival status. (E) Correlation between the expression level of HIF-1α and PCNA. (F) Correlation between the expression level of HIF-1α and Vimentin.
4. Discussion
Although platinum-based chemotherapeutic drugs are the primary options for cancer treatment, many patients who receive such treatment are at risk of relapse [24,25]. Currently, numerous studies aim to improve the drug sensitivity of patients with HCC [26,27]. Nonetheless, the poor prognosis of HCC remains unresolved due to limited understanding of drug resistance mechanisms [28,29].
New therapeutic strategies that safely potentiate chemotherapeutic sensitivity provide a promising approach for effective HCC treatment. Our results showed that the Sal and OXA combination can increase the sensitivity of OXA in HCC and reverse the drug resistance of OXA and DDP in drug-resistant HCC cells.
Hypoxia commonly occurs in solid tumors because cells proliferate faster than angiogenesis. In hypoxic conditions, cell signaling and gene expression changes in cells promote chemotherapeutic drug tolerance, cell survival, and invasion [30]. Hypoxia in the center of solid tumors generally decreases the chemosensitivity of tumor cells and experimental hypoxia can induce EMT and promote the drug resistance of antitumor drugs in various cell lines [31]. EMT, an important stage in the progression of malignant tumors, can promote MDR. The residual cells after OXA treatment possess markedly increased metastasis ability in vitro and in vivo with the changes in EMT-related transcription factors and proteins [[32], [33], [34]]. The EMT induced by platinum anticancer drugs promotes chemotherapeutic resistance and metastasis [35]. Our results also showed that in all three different HCC cell lines, hypoxia induced drug resistance to OXA, and Sal reversed the hypoxia-induced drug resistance. Furthermore, in vivo results showed that the body weights in OXA treatment group were reduced compared with those in the control group, and those in the co-treatment group (Sal+OXA) were higher than those in the OXA treatment group. The survival time of the co-treatment group was extended compared with those of the other groups.
HIF-1α, as a transcription factor in hypoxia induction, is involved in the expression of various hypoxia-induced target genes, and it is a principal component of the hypoxic adaptation of the transcription factor. HIF-1α can promote hypoxic tumor cell survival and increase the invasive-related protein content. In particular, HIF-1α activation is related to the development of multidrug resistance and hypoxia-induced EMT in tumor cells [14]. Our results showed that the HIF-1α degradation by ubiquitination pathway was promoted by Sal, which was verified by using nondegradable HIF-1α. Multidimensional liquid chromatography–tandem mass spectrometry showed that Sal influenced a series of signaling pathways, in which most biological functions were related to HIF-1α. The clinical data also showed that HIF-1α plays an important role in prognosis of HCC.
Our results showed that Sal could inhibit HIF-1α induced by hypoxic conditions. However, some other preliminary studies showed Sal could increase the level of HIF-1α in some normal body cells [21,36,37]. According to these results, the Sal function differed between normal and malignant cells. Hypoxic condition is also a critical factor that must be evaluated.
In conclusion, Sal significantly increased the sensitivity of HCC cells to platinum-based drugs and inhibited hypoxia-induced EMT in HCC through inhibiting HIF-1 α signaling pathway. Therefore, Sal may be an effective platinum-based drug sensitizer that can improve the chemotherapeutic efficacy in patients with HCC.
Competing interests
The authors declare no competing interests.
Funding and acknowledgments
This study was founded by Foundation for National Natural Science Funds of China (Grant No. 81703581, 81572838, and 81402973), National Science and Technology Major Project (Grant No. 2018ZX09736005), Tianjin science and technology innovation system and the condition of platform construction plan (Grant No.14TXSYJC00572), Post doctoral innovative talent support program (Grant No. BX20180150). None of these funding sources had any role in writing the manuscript nor the decision to submit for publication. The authors attest they have not been paid towrite this article by a pharmaceutical company or other agency.
Author contributions
Cheng Yang, Tao Sun and Guang Yang conceived and designed the experiments. Yuan Qin, Huijuan Liu, Meng Li, Denghui Zhai and Jiahuan Yang performed all the experiments., Yuan Qin, Huijuan Liu, Yuanhao Tang, Lan Yang, Kailiang Qiao Wei-long Zhong, Qiang Zhang and Yan-rong Liu analyzed the data. Yuan Qin, Huijuan Liu wrote the manuscript. All authors read and approved the final manuscript.
The following are the supplementary data related to this article.
Contributor Information
Guang Yang, Email: sunshine@tju.edu.cn.
Tao Sun, Email: sunrockmia@hotmail.com.
Cheng Yang, Email: cheng.yang@nankai.edu.cn.
References
- 1.Schreiber-Brynzak E. Behavior of platinum(iv) complexes in models of tumor hypoxia: cytotoxicity, compound distribution and accumulation. Metallomics. 2016;8(4):422–433. doi: 10.1039/c5mt00312a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohmichi M. Mechanisms of platinum drug resistance. Trends Pharmacol Sci. 2005;26(3):113–116. doi: 10.1016/j.tips.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Wang J. Mapping sites of aspirin-induced acetylations in live cells by quantitative acid-cleavable activity-based protein profiling (QA-ABPP) Sci Rep. 2015;5:7896. doi: 10.1038/srep07896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson S.W., Ferry K.V., Hamilton T.C. Recent insights into platinum drug resistance in cancer. Drug Resist Updat. 1998;1(4):243–254. doi: 10.1016/s1368-7646(98)80005-8. [DOI] [PubMed] [Google Scholar]
- 5.Olive P.L. Analysis of DNA damage in individual cells. Methods Cell Biol. 2001;64:235–249. doi: 10.1016/s0091-679x(01)64016-0. [DOI] [PubMed] [Google Scholar]
- 6.Shannon A.M. Tumour hypoxia, chemotherapeutic resistance and hypoxia-related therapies. Cancer Treat Rev. 2003;29(4):297–307. doi: 10.1016/s0305-7372(03)00003-3. [DOI] [PubMed] [Google Scholar]
- 7.Vaupel P. Tumor microenvironmental physiology and its implications for radiation oncology. Semin Radiat Oncol. 2004;14(3):198–206. doi: 10.1016/j.semradonc.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Fischer K.R. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527(7579):472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erler J.T. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440(7088):1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 10.Lee J.W. Hypoxia-inducible factor (HIF-1)alpha: its protein stability and biological functions. Exp Mol Med. 2004;36(1):1–12. doi: 10.1038/emm.2004.1. [DOI] [PubMed] [Google Scholar]
- 11.Chun Y.S., Kim M.S., Park J.W. Oxygen-dependent and -independent regulation of HIF-1alpha. J Korean Med Sci. 2002;17(5):581–588. doi: 10.3346/jkms.2002.17.5.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang J. HIF-1alpha-mediated up-regulation of vascular endothelial growth factor, independent of basic fibroblast growth factor, is important in the switch to the angiogenic phenotype during early tumorigenesis. Cancer Res. 2001;61(15):5731–5735. [PubMed] [Google Scholar]
- 13.Raheja L.F. Hypoxic regulation of mesenchymal stem cell migration: the role of RhoA and HIF-1alpha. Cell Biol Int. 2011;35(10):981–989. doi: 10.1042/CBI20100733. [DOI] [PubMed] [Google Scholar]
- 14.Comerford K.M. Hypoxia-inducible factor-1-dependent regulation of the multidrug resistance (MDR1) gene. Cancer Res. 2002;62(12):3387–3394. [PubMed] [Google Scholar]
- 15.Xia S. Effects of hypoxia on expression of P-glycoprotein and multidrug resistance protein in human lung adenocarcinoma A549 cell line. Zhonghua Yi Xue Za Zhi. 2004;84(8):663–666. [PubMed] [Google Scholar]
- 16.Wu F. HIF1alpha genetic variants and protein expressions determine the response to platinum based chemotherapy and clinical outcome in patients with advanced NSCLC. Cell Physiol Biochem. 2013;32(6):1566–1576. doi: 10.1159/000356593. [DOI] [PubMed] [Google Scholar]
- 17.Zheng K.Y. The extract of Rhodiolae Crenulatae Radix et Rhizoma induces the accumulation of HIF-1alpha via blocking the degradation pathway in cultured kidney fibroblasts. Planta Med. 2011;77(9):894–899. doi: 10.1055/s-0030-1250627. [DOI] [PubMed] [Google Scholar]
- 18.Chen X. Salidroside attenuates glutamate-induced apoptotic cell death in primary cultured hippocampal neurons of rats. Brain Res. 2008;1238:189–198. doi: 10.1016/j.brainres.2008.07.051. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L. Protective effects of salidroside on hydrogen peroxide-induced apoptosis in SH-SY5Y human neuroblastoma cells. Eur J Pharmacol. 2007;564(1–3):18–25. doi: 10.1016/j.ejphar.2007.01.089. [DOI] [PubMed] [Google Scholar]
- 20.Wang H. The in vitro and in vivo antiviral effects of salidroside from Rhodiola rosea L. against coxsackievirus B3. Phytomedicine. 2009;16(2–3):146–155. doi: 10.1016/j.phymed.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J. Salidroside protects cardiomyocyte against hypoxia-induced death: a HIF-1alpha-activated and VEGF-mediated pathway. Eur J Pharmacol. 2009;607(1–3):6–14. doi: 10.1016/j.ejphar.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 22.Hu X. A preliminary study: the anti-proliferation effect of salidroside on different human cancer cell lines. Cell Biol Toxicol. 2010;26(6):499–507. doi: 10.1007/s10565-010-9159-1. [DOI] [PubMed] [Google Scholar]
- 23.Li J. Heparanase promotes radiation resistance of cervical cancer by upregulating hypoxia inducible factor 1. Am J Cancer Res. 2017;7(2):234–244. [PMC free article] [PubMed] [Google Scholar]
- 24.Cunningham D. Colorectal cancer. Lancet. 2010;375(9719):1030–1047. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 25.Li P. MALAT1 is Associated with Poor Response to Oxaliplatin-based Chemotherapy in Colorectal Cancer patients and Promotes Chemoresistance through EZH2. Mol Cancer Ther. 2017;16(4):739–751. doi: 10.1158/1535-7163.MCT-16-0591. [DOI] [PubMed] [Google Scholar]
- 26.Qin Y. Apigenin inhibits NF-kappaB and snail signaling, EMT and metastasis in human hepatocellular carcinoma. Oncotarget. 2016;7(27):41421–41431. doi: 10.18632/oncotarget.9404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J. Mechanistic Investigation of the specific Anticancer Property of Artemisinin and its Combination with Aminolevulinic Acid for Enhanced Anticolorectal Cancer activity. ACS Cent Sci. 2017;3(7):743–750. doi: 10.1021/acscentsci.7b00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong H. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36(25):3528–3540. doi: 10.1038/onc.2016.521. [DOI] [PubMed] [Google Scholar]
- 29.Gade T.P.F. Ischemia Induces Quiescence and Autophagy Dependence in Hepatocellular Carcinoma. Radiology. 2017;283(3):702–710. doi: 10.1148/radiol.2017160728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou J. Tumor hypoxia and cancer progression. Cancer Lett. 2006;237(1):10–21. doi: 10.1016/j.canlet.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 31.Teicher B.A. Hypoxia and drug resistance. Cancer Metastasis Rev. 1994;13(2):139–168. doi: 10.1007/BF00689633. [DOI] [PubMed] [Google Scholar]
- 32.Jiao L. Reactive oxygen species mediate oxaliplatin-induced epithelial-mesenchymal transition and invasive potential in colon cancer. Tumour Biol. 2016;37(6):8413–8423. doi: 10.1007/s13277-015-4736-9. [DOI] [PubMed] [Google Scholar]
- 33.Uchibori K. Establishment and characterization of two 5-fluorouracil-resistant hepatocellular carcinoma cell lines. Int J Oncol. 2012;40(4):1005–1010. doi: 10.3892/ijo.2011.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin Y. Dihydroartemisinin inhibits EMT induced by platinum-based drugs via Akt-Snail pathway. Oncotarget. 2017;8(61):103815–103827. doi: 10.18632/oncotarget.21793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma J.L. Epithelial-mesenchymal transition plays a critical role in drug resistance of hepatocellular carcinoma cells to oxaliplatin. Tumour Biol. 2016;37(5):6177–6184. doi: 10.1007/s13277-015-4458-z. [DOI] [PubMed] [Google Scholar]
- 36.Li L. Protective effect of salidroside against bone loss via hypoxia-inducible factor-1alpha pathway-induced angiogenesis. Sci Rep. 2016;6:32131. doi: 10.1038/srep32131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng K.Y. Salidroside stimulates the accumulation of HIF-1alpha protein resulted in the induction of EPO expression: a signaling via blocking the degradation pathway in kidney and liver cells. Eur J Pharmacol. 2012;679(1–3):34–39. doi: 10.1016/j.ejphar.2012.01.027. [DOI] [PubMed] [Google Scholar]