Abstract
Background
Reduced activity of proprotein convertase subtilisin/kexin type 9 (PCSK9) has been associated with decreased short-term death in patients with septic shock. Whether PCSK9 genotype influences long-term outcomes in sepsis survivors is unknown.
Methods
We evaluated the impact of PCSK9 loss-of-function (LOF) genotype on both 1-year mortality and infection-related readmission (IRR) after an index sepsis admission. The Derivation cohort included 342 patients who survived 28 days after a sepsis admission in a tertiary hospital (Vancouver/Canada, 2004–2014), while an independent Validation cohort included 1079 septic shock patients admitted at the same hospital (2000–2006). All patients were genotyped for three common missense PCSK9 LOF variants rs11591147, rs11583680, rs562556 and were classified in 3 groups: Wildtype, single PCSK9 LOF, and multiple PCSK9 LOF, according to the number of LOF alleles per patient. We also performed a meta-analysis using both cohorts to investigate the effects of PCSK9 genotype on 90-day survival.
Findings
In the Derivation cohort, patients carrying multiple PCSK9 LOF alleles showed lower risk for the composite outcome 1-year death or IRR (HR: 0.40, P = 0.006), accelerated reduction on neutrophil counts (P = 0.010), and decreased levels of PCSK9 (P = 0.037) compared with WT/single LOF groups. Our meta-analysis revealed that the presence of multiple LOF alleles was associated with lower 90-day mortality risk (OR = 0.69, P = 0.020).
Interpretation
The presence of multiple PCSK9 LOF alleles decreased the risk of 1-year death or IRR in sepsis survivors. Biological measures suggest this may be related to an enhanced resolution of the initial infection.
Funding
Canadian Institutes of Health Research (PJT-156056).
Keywords: PCSK9, Sepsis, Septic shock, Mortality, Readmission
Research in context.
Evidence before this study
Studies using animal models of sepsis have demonstrated that reduced activity of proprotein convertase subtilisin/kexin type 9 (PCSK9) decreases bacterial dissemination. PCSK9 loss-of-function (LOF) genotype has also been associated with decreased short-term death among patients with septic shock. In this study we investigated the impact of PCSK9 LOF genotype upon 1-year outcomes in sepsis survivors, specifically its effect upon 1-year death and the probability of suffering a recurrent infection-related readmission (IRR).
Added value of this study
Among patients who survived an episode of sepsis, those carrying multiple PCSK9 LOF alleles (two or more alleles) had significant protection against the composite of 1-year death or IRR compared with patients carrying no or one PCSK9 LOF allele. Importantly, this effect was driven entirely by reductions in IRRs. Improvements in the resolution of the initial infection may be involved in this protection.
Implications of all the available evidence
Unplanned hospital readmissions after sepsis due to infections are frequent and costly. PCSK9 genotype may be a novel prognostic biomarker for septic patients as it may identify those at increased risk of a serious IRR over the next 12 months. Further studies including randomized controlled trials with PCSK9 inhibitors for acute and long-term treatment of sepsis are still needed to define whether the observations in this study can be translated clinically.
Alt-text: Unlabelled Box
1. Introduction
Sepsis is a major cause of hospitalization, morbidity, and mortality worldwide [1,2]. Although the short-term mortality rate associated with sepsis is falling [3,4], a substantial number of sepsis survivors experience adverse long-term outcomes, such as hospital readmissions [5], increased late mortality [6], greater incidence of major cardiovascular events [7], and impaired quality of life [8]. Factors that contribute to adverse long-term outcomes are advanced age [9], comorbidity burden [6], organ dysfunction during sepsis [10], persistent inflammation [11] and chronic catabolism [12]. However, to our knowledge, no study has investigated the impact of specific genetic variants on long-term outcomes from sepsis.
Recently, proprotein convertase subtilisin kexin type 9 (PCSK9) loss-of-function (LOF) genotype has been found to be associated with decreased short-term mortality from septic shock and protection against bacterial dissemination in animal models [13]. PCSK9 inhibits the clearance of low-density lipoprotein cholesterol (LDL-C) from the blood by decreasing the density of LDL receptors (LDLR) on hepatic cells [14]. PCSK9 LOF leads to higher hepatic LDLR expression, increased clearance of LDL-C, and protection from coronary heart disease [15,16]. Pathogen lipids, such as endotoxins, are the trigger for the host inflammatory response in sepsis [17]. These pathogen lipids are incorporated into lipoprotein fractions (HDL, LDL, VLDL) and eventually cleared from the blood by the liver; a process mediated by hepatic LDLR [18]. In this way, clearance of pathogen lipids during sepsis is similar to clearance of cholesterol. Consequently, PCSK9 LOF variants are associated with increased clearance of pathogen lipids, decreased systemic inflammatory response, and decreased short-term mortality in sepsis [19].
One factor that may contribute to adverse long-term outcomes associated with sepsis is the increased number of new infection(s) in the weeks and months following the index admission [20]. There are potential beneficial effects of decreased PCSK9 function in decreasing 28-day mortality in patients with severe sepsis/septic shock [19]; however, the literature is still scant regarding the long-term effects of PCSK9 LOF genotype, including hospital readmissions due to an infectious reason.
The primary objectives of this study were to investigate the relationship between three common missense PCSK9 LOF variants: rs11591147 (R46L), rs11583680 (A53V), rs562556 (I474V) [21] and the risk of infection-related readmission (IRR) and/or all-cause mortality within one year after an episode of sepsis.
2. Methods
2.1. Ethics
This study was approved by the University of British Columbia Clinical Research Ethics Board (Research Ethics Board Number: H11-00505). Written informed consent was obtained from all patients, their next of kin, or another surrogate decision maker, as appropriate.
2.2. Study design
This was a retrospective observational study involving 1481 patients diagnosed with sepsis.
2.3. Patients – inclusion criteria
The Derivation cohort was composed of 402 patients with sepsis who were admitted to St. Paul's Hospital in Vancouver, Canada between July 2004 and June 2014. Patients presenting a clinically defined infection and at least 2 of the following: (i) Temperature >38°C or <36 °C; (ii) Heart rate >90 beats per minute; (iii) Respiratory rate >20 breaths per minute; (iv) White blood cell count >12,000 per mm3 or <4,000 per mm3, and had survived 28 days after admission were included (N = 342). The majority of these patients were recruited within the first 3 h in the Emergency Department, upon activation of the Institutional severe sepsis pathway. Validation Cohort was composed of 1079 patients with septic shock, including subjects from 2 distinct cohorts merged together: Cohort #1 was composed of 628 septic shock patients enrolled into VASST [22] who had DNA available. Approval, enrollment, and consent in the VASST Cohort have been described previously [22]. Briefly, inclusion criteria were age older than 16 years, presence of septic shock defined by the presence of two or more diagnostic criteria for the systemic inflammatory response syndrome, proven or suspected infection, and hypotension despite adequate fluid resuscitation; Cohort #2 was composed of 451 patients enrolled with septic shock at St Paul's Hospital in Vancouver Canada between January 2000 and December 2004. Sepsis was classified in accordance with the American College of Chest Physicians and the Society of Critical Care Medicine consensus [23]. Shock was defined as norepinephrine treatment and/or a mean arterial pressure of <70 mmHg within the first 5 days after ICU admission.
The three cohorts evaluated in this study were composed of a multiethnic population, in which ethnicity was determined phenotypically by the study coordinator or research assistant.
2.4. Exclusion criteria
Patients known to be on chronic use of statins were excluded from the analysis associations between PCSK9 genotype and plasma PCSK9, and post-sepsis LDL-C levels because statins upregulate PCSK9 [24]. Exclusion criteria for VASST (Validation, cohort #1) have been reported [22].
2.5. Measurements
Clinical and laboratory data were retrospectively obtained by reviewing the province of British Columbia electronic records from the index hospitalization for up to one year. For all cohorts, Acute Physiology and Chronic Health Evaluation (APACHE) II score [25] was calculated at the onset of sepsis.
2.6. Blood collection and PCSK9 genotyping
For PCSK9 genotyping, blood was collected from discarded clinical blood samples and spun at 1400 × g for 12 min to separate the plasma and cellular fractions. Total genomic DNA was extracted from the buffy coat fraction using QIAGEN DNeasy Blood & Tissue Kits (QIAGEN 69506). PCSK9 genotyping was performed in all samples for three common PCSK9 missense LOF variants (minor allele frequency (MAF) > 0.5%): R46L, A53V, and I474V, using pre-validated TaqMan SNP Genotyping Assays (ThermoScientific, Probe IDs rs11591147, rs11583680, rs562556 respectively, catalog number 4351379). Assays were run on a ViiA7 platform (software QuantStudio Real-Time PCR software V 1.3) using the system's single-nucleotide polymorphisms (SNP) genotyping software and protocol. All alleles were called using the software's clustering algorithm, with a 100% success rate. For quality control, 10% of samples were randomly repeated for each SNP to ensure complete reproducibility.
2.7. PCSK9 measurements, lipids and blood cell counts
Plasma PCSK9 was measured via ELISA (R&D Systems DPC900) using the manufacturer's recommended protocol. Absolute white blood cell and neutrophil counts were measured daily from admission to 14 days at the central hospital laboratory on Sysmex XN analyzer. To assess LDL-C measurements collected after the sepsis event, provincial health records of all patients were reviewed, and those patients with post-sepsis LDL-C measurements available were analyzed according to PCSK9 genotype.
2.8. PCSK9 genotyping and definitions
Patients were classified according to their PCSK9 genotyping into three groups: i. Wild-type group (WT): patients who did not carry any of the three LOF alleles; ii. Single LOF group: patients who carried exactly one LOF allele; iii. Multiple LOF group: patients who carried two or more LOF alleles, including either one homozygous LOF SNP or multiple different heterozygous LOF SNPs.
These definitions were based on the genotyping technique used by us and on the literature related to LDL-lowering effects (in humans) of the PCSK9 polymorphisms evaluated here [26]. As each SNP only partially diminishes the protein function, we considered two mutations in the same copy as roughly equivalent to one mutation each in two copies, in terms of total protein functional capacity.
2.9. Outcomes
The primary outcome was a composite outcome defined as death (all-cause mortality after 28 days) or infection-related readmission (IRR) up to 12 months thereafter. We also analyzed all-cause mortality after 28 days or IRR up to 12 months separately.
2.10. Statistical analysis
Results are expressed as interquartile range for continuous variables and absolute number (%) for categorical variables. Association among the PCSK9 genotype groups and demographic, physiological and laboratory parameters were tested using Mann-Whitney (or t-test when appropriate), and Chi-square (or Fisher's test when appropriate) tests for continuous variables and categorical variables, respectively [27].
In order to evaluate the association between PCSK9 genotype and the risk of 1-year all-cause mortality or IRR, we applied Kaplan-Meier estimates of time to event function for the composite outcome death or readmission within one year. The log-rank test was used for the comparison between curves. Adjusted Cox regression model (adjusted for age, gender, and APACHE II score) was performed to determine the independent relationship of the presence of PCSK9 genotype and the risk for 1-year death of IRR. Additional Kaplan-Meier estimates of time to event function analyses were performed considering each single outcome: 1-year mortality and 1-year IRR. To avoid double counting, patients who died were considered a mortality in the composite analysis and patients who survived 1 year were considered in the IRR analysis.
A meta-analysis for 90-day mortality according to PCSK9 genotype that included the Derivation and Validation cohorts was carried out for validation of our long-term findings. For this, we meta-analyzed using a two-step process and used the results of a first meta-analysis with the two original Validation cohorts for our final analysis. We assessed the derivation for 90-day mortality to make it comparable to the Validation cohorts. This outcome (90-day mortality) was used because that was the longest follow-up time for mortality available in all study cohorts. Data were pooled using the Mantel-Haenszel (M-H) method with a random effects model, and heterogeneity was evaluated using the χ2 and I2 statistics. All 402 patients from the Derivation cohort were included for this analysis.
To investigate relevant clinical factors associated with mortality and hospital readmission we compared white blood cell counts and absolute number of neutrophils according to PCSK9 genotype using repeated measures ANOVA in a subset of patients with laboratory data available during their first 14 days of sepsis. Specifically for this analysis, we used the Last-Observation-Carried-Forward method for patients with missing data [28].
Statistical analyses were performed using SPSS Statistics version 24.0 for Windows (IBM Corp., Armonk, NY, USA), Review Manager 5.3 was used for the meta-analysis, and aov statistical function in R (version 3.4.3, available http://www.R-project.org) was used for the repeated measures ANOVA. Statistical significance was set at α = 0.05 using two-sided P values.
3. Results
3.1. The risk of 1-year death or infection-related readmission (composite outcome) was decreased in patients carrying multiple PCSK9 LOF alleles
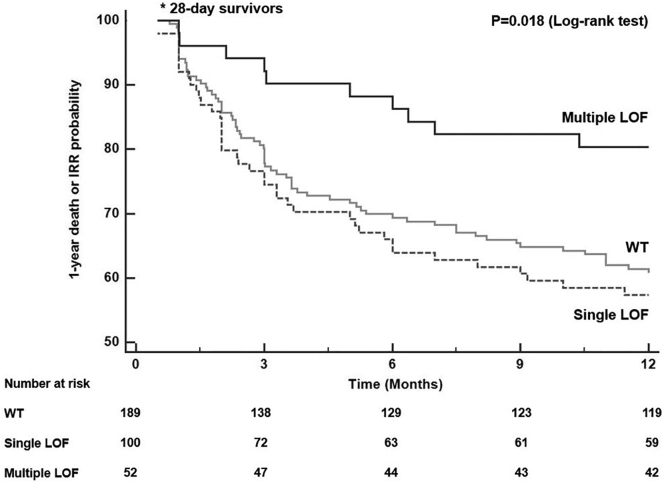
Thirty-seven percent (70/189) of WT patients, 41% (41/100) from single LOF group, and 19.2% (10/52) who had multiple LOF died or were readmitted within 1 year (P = 0.018, overall Log-rank test, Fig. 1). Overall, patients carrying multiple PCSK9 LOF alleles had the greatest protection against 1-year death or IRR compared with WT patients (P = 0.011, pairwise Log-rank test) and with single LOF patients (P = 0.005, pairwise Log-rank test). No differences were found between patients carrying single LOF and WT subjects (P = 0.511, pairwise Log-rank test).
Fig. 1.
Derivation (early sepsis) cohort (N = 342). Time to event curves for 1-year death or infection-related readmission according to PCSK9 genotype groups. Patients carrying multiple PCSK9 LOF alleles showed significantly decreased risk for this outcome in comparison to patients carrying no (wildtype) or one PCSK9 LOF allele (P = 0.018). One patient (WT group) was excluded from this analysis as the time to the composite outcome was not ascertained.
Based on these findings demonstrating substantial protection against death or readmission in patients carrying multiple LOF alleles, further analyses were done by comparing 2 PCSK9 genotype groups: WT/single LOF (indicated here as reference Ref. group), vs. multiple LOF.
3.2. Characteristics of the study subjects
Baseline demographic, physiological and clinical characteristics were similar according to the two PCSK9 genotype groups (Ref. vs. multiple LOF) (Table 1). Most patients (290/342, 84.7%) were classified as WT/single LOF, while 52 (15.2%) had multiple LOF. All SNPs were in Hardy-Weinberg equilibrium (HWE) (Table 2). Minor allele frequency (MAF) of each PCSK9 variant is described in Table 2. A53V (rs11583680), I474V (rs562556), and R46L (rs11591147) variants were present in 93 (27.1%), 94 (27.4%), and 7 (2.0%) patients, respectively. Tables S1 and S2 describe baseline characteristics and MAF/HWE, respectively, in the Validation cohort.
Table 1.
Derivation Cohort: Baseline characteristics according to PCSK9 genotype.
| Variable | All patients N = 342 |
Ref.a N = 290 |
Multiple LOF N = 52 |
P value |
|---|---|---|---|---|
| Age, Median (IQR) | 59 (44–69) | 59 (44–70) | 57 (43–65) | 0.515 |
| Gender (N, % male) | 226 (66.1) | 190 (65.5) | 36 (69.2) | 0.717 |
| Ethnicity (N, % Caucasians)b | 209 (61.1) | 173 (59.7) | 36 (69.2) | 0.250 |
| HR, Median (IQR)c | 108 (91–127) | 107 (91–126) | 115 (90–136) | 0.316 |
| MAP, Median (IQR)c | 58 (53–67) | 59 (53–67) | 58 (55–70) | 0.482 |
| Temperature, Median (IQR)c | 37.5 (36.5–38.4) | 37.5 (36.5–38.3) | 37.8 (36.4–39.0) | 0.568 |
| APACHE II score, Median (IQR) | 14 (7–19) | 14 (8–19) | 12 (7–21) | 0.989 |
| ICU (N, %) | 245 (71.6) | 207 (71.4) | 38 (73.1) | 0.934 |
| HGB g/L, Median (IQR)d | 112 (92–130) | 112 (93–130) | 106 (90–127) | 0.385 |
| WBC (x109), Median (IQR)d | 10.6 (6.3–15.0) | 10.7 (6.4–15.1) | 10.4 (5.9–13.3) | 0.520 |
| Platelets (x109), Median (IQR)d | 186 (131–271) | 190 (133–267) | 175 (110–284) | 0.542 |
| Creatinine (mmol/l), Median (IQR)d | 89 (65–154) | 89 (66–152) | 90 (55–158) | 0.651 |
| INR, Median (IQR)d | 1.2 (1.1–1.5) | 1.2 (1.1–1.5) | 1.2 (1.1–1.5) | 0.616 |
| Lactate (mmol/l), Median (IQR)d | 1.6 (1.1–2.6) | 1.6 (1.2–2.6) | 1.3 (1.0–2.7) | 0.424 |
| COPD (N, %) | 60 (17.5) | 48 (16.6) | 12 (23.1) | 0.347 |
| Chronic renal failure (N, %) | 15 (4.4) | 11 (4.0) | 4 (7.7) | 0.271 |
| Cirrhosis (N, %) | 10 (2.9) | 9 (3.1) | 1 (1.9) | 0.985 |
| CHF NYHA Class 3 or 4 (N, %) | 29 (8.5) | 25 (8.6) | 4 (7.6) | 0.982 |
| Hypertension (N, %) | 88 (25.7) | 78 (26.9) | 10 (19.2) | 0.321 |
| Diabetes mellitus (N, %) | 54 (15.8) | 46 (15.8) | 9 (15.4) | 0.913 |
| Statins use (N, %)e | 51 (27.9) | 42 (27.8) | 9 (28.1) | 0.972 |
Abbreviations: LOF: Loss-of-function; IQR: Interquartile Range; HR: Heart Rate; RR: Respiratory Rate; SpO2: arterial Oxygen Saturation; MAP: Mean Arterial Pressure; APACHE: Acute Physiology and Chronic Health Evaluation; ICU: Intensive Care Unit; AKI: Acute Kidney Injury; HGB: Hemoglobin; WBC: White Blood Cell; COPD: Chronic Obstructive Pulmonary Disease; CHF: Congestive Heart Failure; NYHA: New York Heart Association.
Ref. indicates WT/Single PCSK9 LOF allele group.
Determined by the study coordinator or research assistant.
Parameters measured at admission.
Measurements in the first 24 h of ED admission; e Available in 183 patients.
Table 2.
Derivation Cohort: PCSK9 genotype - allele frequency and Hardy-Weinberg equilibrium.
| PCSK9 SNPs | Major (minor)allele | Minor AlleleFrequency – N (%)a | HWE P-value |
|---|---|---|---|
| A53V (rs11583680) | G (A) | 100 (14.61%) | 0.99 |
| I474V (rs562556) | A (G) | 105 (15.35%) | 0.47 |
| R46L (rs11591147) | C (A) | 7 (1.02%) | 0.98 |
Abbreviations: PCSK9: Proprotein Convertase Subtilisin/kexin type 9; SNPs: single nucleotide polymorphisms; HWE: Hardy-Weinberg Equilibrium.
N refers to minor allele counts in relation to the total number of alleles in the Derivation Cohort (N = 684).
3.3. The presence of multiple PCSK9 LOF alleles was associated with significantly decreased risk of death at 1 year or infection-related readmission (composite outcome) after sepsis
Composite endpoint (death or IRR at 1 year): Considering 2 PCSK9 genotype groups (Ref. vs. multiple LOF), the adjusted HR for death or IRR within one year after sepsis in patients carrying two or more PCSK9 LOF alleles was 0.40 (95% C.I. 0.21–0.77, P = 0.006, Table 3).
Table 3.
Derivation Cohort: Adjusted Hazard Ratios (aHR)d for 1-year death or IRR (composite outcome), 1-year IRR, and 1-year mortality according to PCSK9 genotype (two or more alleles).
| Cohort | aHR (95% CI) | P value |
|---|---|---|
| Death or IRRa,b | 0.40 (0.21–0.77) | 0.006 |
| IRRa,c | 0.20 (0.05–0.86) | 0.031 |
| Mortalitya,b | 0.50 (0.24–1.04) | 0.064 |
Abbreviations: IRR: infection-related readmission.
Within one year.
Excluding patients who died within 28 days.
Excluding patients who died within one year.
Adjusted for age, sex and APACHE II score.
1-year mortality: In relation to Ref. group, the adjusted HR for this outcome in patients carrying multiple PCSK9 LOF alleles was 0.50 (15.3% vs. 24.8%, 95% CI = 0.24–1.04, P = 0.064, Table 3). No statistically significant difference between our study groups was found for this outcome.
Infection-related readmission: We found that, among patients who did not die within one year (N = 262), those who carried multiple PCSK9 LOF alleles had a significant risk reduction for 1-year IRR in comparison to patients who carried none (WT) or single PCSK9 LOF allele (Ref. group) (adjusted HR = 0.20, 95% C.I. 0.05–0.86, P = 0.031, Table 3). In the Ref. group, 17.8% of patients (39/218) were readmitted due to infection while only 4.5% (2/44) of patients were readmitted in the multiple LOF group (P = 0.023, Chi-square test).
3.4. The biological effects of multiple PCSK9 LOF alleles: PCSK9 plasma levels at sepsis admission and post-sepsis LDL-C levels
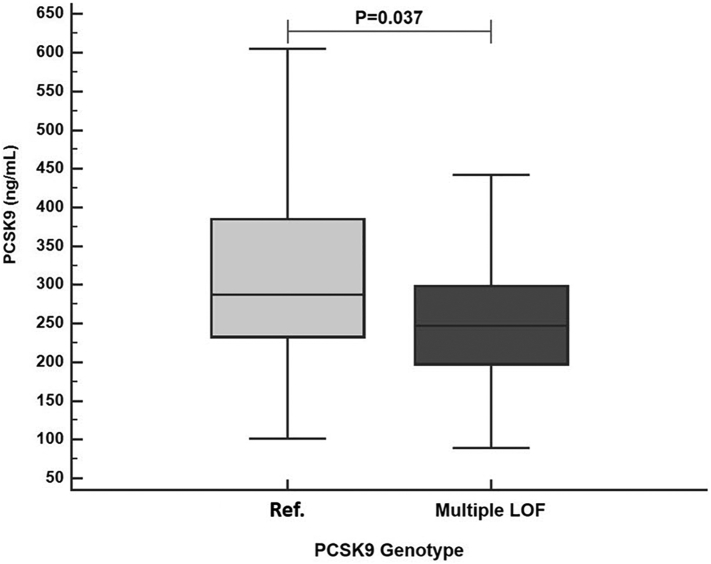
Plasma PCSK9 levels at sepsis admission were lower in patients carrying 2 or more PCSK9 LOF alleles than Ref. group patients. In the subset of patients who had PCSK9 levels measured at sepsis admission and who were not taking statins (N = 132) the presence of multiple LOF alleles was associated with lower plasma PCSK9 compared to Ref. group (247 ng/mL vs. 287 ng/mL, P = 0.037, Fig. 2).
Fig. 2.
Derivation (early sepsis) cohort (N = 132). PCSK9 levels at sepsis admission according to PCSK9 genotype (Ref. vs. multiple LOF group). Patients carrying multiple PCSK9 LOF alleles (N = 23) had lower PCSK9 levels than patients from the Ref. group (N = 109) (P = 0.037). Ref. indicates WT/Single PCSK9 LOF allele group.
Patients carrying multiple PCSK9 LOF alleles had a strong trend toward reduced LDL-C levels measured after sepsis discharge (19 ± 16 months) compared to Ref. patients. LDL-C levels were available in 55 patients: 41 in Ref. group and 14 in multiple LOF group. Post sepsis LDL-C levels were 94.8 ± 5.6 mg/dL in Ref. group vs. 77.1 ± 7.29 mg/dL in multiple LOF group (mean ± SEM, P = 0.09, data not shown).
3.5. The presence of multiple PCSK9 LOF alleles was associated with 90-day mortality
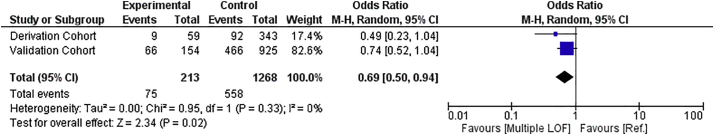
In order to validate our findings related to the association of multiple PCSK9 LOF alleles and long-term mortality, we did a meta-analysis using a two-step approach (describe in the Methods section) of the association of PCSK9 genotype with 90-day mortality. All patients from Derivation and Validation cohorts were included (N = 1481). Septic patients from the Validation cohort (N = 1079) were genotyped for PCSK9 LOF alleles, and 90-day mortality data was analyzed. We confirmed a significant association between the presence of multiple LOF alleles and significantly lower 90-day mortality (Odds Ratio 0.69; 95% CI: 0.50–0.94; p = 0.02, Fig. 3).
Fig. 3.
Meta-analysis of 90-day mortality of Derivation and Validation cohorts (N = 1481). The presence of multiple PCSK9 alleles had an overall protective effect for this outcome (OR = 0.69, 95% C.I. 0.50–0.94, P = 0.02). (M-H: Mantel-Haenszel). Ref. indicates WT/Single PCSK9 LOF allele group.
3.6. The effects of multiple PCSK9 LOF alleles upon white blood cell counts throughout sepsis admission
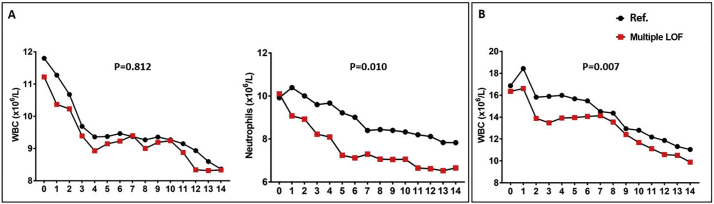
In the Derivation Cohort, white blood cell (WBC) and absolute neutrophil counts within the first 14 days of sepsis were available in 170 patients: Ref. group (N = 142), multiple LOF (N = 28). Patients carrying multiple PCSK9 LOF alleles demonstrated greatly neutrophil count reductions over time in comparison to Ref. patients (P = 0.01, Fig. 4A). No differences in WBC counts were found between these two groups (P = 0.812, Fig. 4A).
Fig. 4.
Blood counts during sepsis (from day 0 to day 14) according to PCSK9 genotype. X-axis represents time in day (0 to 14); Y-axis represents the respective blood cell counts for each panel; black circles represent mean values over time in Ref. patients; red squares represent mean values over time in multiple LOF patients. A) Derivation Cohort. Left panel: White blood cell (WBC) counts. Right panel: Neutrophil counts; B) Validation Cohort: WBC counts. Patients carrying multiple PCSK9 LOF alleles demonstrated greatly decreased absolute neutrophil counts in the Derivation Cohort (P = 0.01), and WBC counts in the Validation Cohort (P = 0.007) in comparison to Ref. patients. Ref. indicates WT/Single PCSK9 LOF allele group.
WBC counts from admission for septic shock to day 14 were available in 406 patients who survived 28 days in the Validation Cohort: Ref. group (N = 332), multiple LOF (N = 74). We found that patients carrying multiple PCSK9 LOF alleles demonstrated a more rapid and complete normalization of their presenting (elevated) WBC counts than Ref. patients (P = 0.007, Fig. 4B). These findings suggest that PCSK9 genotype might influence the host immune response to sepsis.
4. Discussion
The present study showed that reduced PCSK9 function according to PCSK9 genotype was associated with better long-term outcomes: composite of 1-year death or infection-related readmission in 28-day survivors of sepsis. The presence of multiple PCSK9 LOF alleles was associated with decreased hazard-ratio for the composite outcome by >50% and by 80% for 1-year infection-related readmission when compared with patients with carrying none or only a single PCSK9 LOF (Ref. group). Furthermore, the presence of multiple PCSK9 LOF alleles was associated with significantly lower plasma PCSK9 levels at sepsis admission than the Ref. group. We can then interpret that the “clinically relevant” PCSK9 LOF group was composed of patients who carry multiple PCSK9 LOF alleles. Prior studies of the role of PCSK9 genotype in infectious disease have only interrogated acute illness [13,19]. To our knowledge, this is the first study to find that common PCSK9 LOF SNPs are associated with improved long-term outcomes following an episode of sepsis.
The three PCSK9 LOF variants analyzed here are relatively common with reported minor allele frequency of >0.5% [21]. R46L (rs11591147), A53V (rs11583680), and I474V (rs562556) variants have been associated with decreased levels of LDL-cholesterol that ranges from modest to more pronounced reductions: around 10%, 15% and 20% for I474V, A53V, and R46L, respectively [15,16,29,30]. Based on their LDL-cholesterol-lowering effects, they have been described as LOF [26]. However, functional in vitro studies are available only for R46L [31,32].
Our study suggests that PCSK9 LOF genotype (defined here by the presence of multiple LOF alleles) is a novel protective factor for long-term outcomes in 28-day survivors of sepsis, such as infection-related readmission. Sepsis is associated with increased risk of all-cause late mortality rates from 1 to 10 years after index sepsis hospitalization [6,7,[33], [34], [35], [36]]. Additionally, hospital readmissions after sepsis are frequent [20,34,36] and expensive [37]. Recent studies have shown that a new event of sepsis was the most common reason for unplanned readmissions among sepsis survivors [20,34]. How the index infection influences readmission rates is still unclear. In the present study, the presence of multiple PCSK9 LOF alleles was independently associated with decreased HR for the composite outcome death or infection-related readmission within one year, and most important, this finding was particularly driven by a reduction in infection-related readmission rates.
We speculate that the presence of multiple PCSK9 LOF alleles plays an important role in the process of resolution of infection and/or bacterial clearance, and therefore decreases the risk of unplanned readmissions due to infection in 28-day sepsis survivors for the following reasons. Relapse or recrudescence of the initial infection in sepsis survivors is one of the most relevant factors associated with infection-related readmissions [39,39]. It has been suggested that patients who survive an episode of sepsis are more prone to re-infection [36], but the reasons for that are not fully elucidated. Dwived et al., using a cecal ligation and puncture model of sepsis, found that PCSK9 knockout (KO) mice had lower bacterial concentrations in the blood, lungs, and peritoneal cavity fluid than WT animals, suggesting that PCSK9 KO genotype is associated with improved bacterial suppression or clearance [13]. This effect may be directly relevant to our observation that infection-related readmission is decreased in patients carrying multiple PCSK9 LOF alleles. Even though this association was not evaluated in our study, the reduced neutrophil counts (Derivation cohort) and WBC counts (Validation cohort) found in patients carrying multiple PCSK9 LOF alleles suggest that the host response to sepsis in this group of patients has a favorable evolution associated with increased rates of infection resolution, and possibly decreased risk of future infection-related readmission. In addition, it has been hypothesized that PCSK9 LOF modulates the inflammatory response due to the increases in LDLR density, particularly in the liver, with subsequent increments in the clearance of endotoxins carried within LDL particles [18]. Interestingly, among sepsis survivors, patients carrying multiple PCSK9 genotype alleles demonstrated lower PCSK9 levels and concordantly lower post-sepsis LDL-C levels than our Ref. patients. It is plausible that this group of patients (multiple LOF) had greater hepatic LDLR density and hence cleared more efficiently endotoxins carried within LDL particles, represented here by decreased LDL-C levels and most importantly, by a risk reduction for infection-related readmission after sepsis. Based on these findings, we can speculate that the presence of multiple PCSK9 LOF alleles plays an important role in the process of resolution of infection and/or bacterial clearance, and therefore decreases the risk of infection-related readmission after sepsis.
Our inability to detect a protective effect by the presence of a single PCSK9 LOF allele may reflect the underlying biology of an additive genetic model, in which multiple PCSK9 LOF variants have recently been shown to confer greater LDL-C lowering and protection against cardiovascular events than a single PCSK9 LOF allele [40]. Frequency of combinations of alleles and HR per allele, per each outcome are described in Table S3.
The suggested benefits of PCSK9 LOF genotype described here were in disagreement with Berger et al. Even though they found that PCSK9 levels were greatly reduced in subjects carrying PCSK9 LOF alleles, no association was demonstrated between PCSK9 LOF genotype and plasma inflammatory markers [41]. These differences may be related to the fact that these authors analyzed different SNPs than us and/or due to subjects' characteristics, such as the presence of sepsis by itself in our population.
The strengths of our study include the assessment of PCSK9 genotype vs. long-term outcome, a novel composite of death or infection-related readmission over 1 year in 28-day sepsis survivors, the use of three independent cohorts and the biological plausibility based on plasma PCSK9 and LDL-C levels. The novel composite of death or infection-related readmission over 1 year is compelling because of the expansion of the sepsis survivors' population over recent years, and the growing evidence demonstrating that these patients experience greater risks of death, re-infection and hospital readmission when compared to patients who have never had a hospitalization for sepsis. Our study reinforces the relevance of genetics, particularly of PCSK9 genotype, for long-term outcomes in sepsis.
Our study has several limitations. First, due to its observational nature we were not able to assess potential mechanisms that explain the link between PCSK9 genotype and long-term outcomes of sepsis. We can only speculate that differences in the host response to sepsis and lipid metabolism may be involved. Second, considering an absolute risk reduction of 15% in our composite outcome in patients carrying multiple PCSK9 LOF alleles compared to the reference group, our statistical power was estimated in 68.1%. Nevertheless, the risk difference observed in our study between our two groups was greater than expected (19.4%), which makes our findings appealing. Third, other PCSK9 SNPs that might be associated with sepsis were not analyzed; however, a thorough review of PCSK9 GWAS literature [21] allowed us to conclude that the probability of finding additional known PCSK9 missense mutations in our cohort was extremely low. Fourth, we were not able to use the new definitions of sepsis [42] as our inclusion criteria in the Derivation cohort, given that some clinical variables required to an accurate assessment of qSOFA or SOFA scores were not available, such as altered mental score and bilirubin levels. However, according to the data available, 74% (N = 253) of participants from this cohort fulfilled the criteria for sepsis diagnosis using the new definitions, implying that this proportion may be underestimated. All Validation cohort patients had sepsis according to its updated definitions [42]. Fifth, the ethnicity data in our study was determined according to patients' phenotype and provided by the study coordinator, precluding us from adjusting our findings for genetic ancestry scores. However, a sensitivity analysis including the Caucasian ethnic group (phenotype-based) in our Cox regression models did not change our major findings (Table S4). Sixth, the reasons for the post sepsis LDL-C measurements being ordered were not assessed because plasma LDL-C levels were obtained by laboratory chart review. Finally, we could only assess 90-day mortality from sepsis admission in our meta-analysis as this time point was the longest follow-up applicable for all our sepsis cohorts. Data concerning 1-year death or readmission was not available in our Validation cohort.
5. Conclusions
PCSK9 LOF variants were associated with better long-term outcomes in 28-day survivors of sepsis. The presence of multiple PCSK9 LOF alleles decreased by >50% the composite risk of death or infection-related readmission, and this finding is driven entirely by reductions in infection-related readmission within one year after the index sepsis admission. PCSK9 genotype might be useful in the early recognition of septic patients at high risk of death or future IRR(s). Our study suggests a novel prognostic biomarker: genotyping septic patients could stratify 28-day sepsis survivors for risk of infection-related readmission.
Acknowledgments
Acknowledgements
The authors thank the patients and families who participated in the three Cohort studies. They have collaborated for improvements in the scientific and clinical understanding of sepsis. The authors also thank the Centre for Heart Lung Innovation's clinical research team for facilitating patient recruitment and data collection in the Derivation Cohort study.
Genga KR is sponsored by CNPq-Brazil (Science Without Borders Scholarship Program).
Competing interests
Dr. R. Genga, Mr. Lo, Mr. Cirstea, Dr. Leitao Filho, Dr. Linder, and Dr. Francis have nothing to disclose. Dr. Walley reports grants from CIHR, during the conduct of the study; other from Cyon Therapeutics, outside the submitted work; In addition, Dr. Walley has a patent PCSK9 inhibition in sepsis pending. Dr. Russell reports personal fees from AKPA, personal fees from SIB Therapeutics, personal fees from La Jolla Pharamceuticals, personal fees from Ferring Pharmaceuticals, personal fees from Cubist Pharmaceuticals, grants from Grifols, personal fees from CytoVale, outside the submitted work; In addition, Dr. Russell has a patent Vasopressin in septic shock issued, and a patent PCSK9 inhibition in sepsis issued. Dr. Boyd reports other from Cyon Therapeutics, outside the submitted work; In addition, Dr. Boyd has a patent PCT/CA2013/000488 issued.
Funding Source
This study was supported by the Canadian Institutes of Health Research – CIHR.
Authors' contributions
JHB conceived and designed the study, and helped to draft and revise the manuscript for important intellectual content. KRG participated in study conception and design, collected data, performed statistical analysis and interpretation, and drafted and revised the manuscript. KRW and JAR participated in study conception and design, and helped to draft and revise the manuscript. MC prepared and collected data and helped to draft the manuscript. CL acquired and prepared collected data. FSLF helped with statistical analysis and interpretation and revised the manuscript. GAF and AL contributed to the study conception and design, and helped to revise the manuscript. All authors made substantial contribution. All authors read and approved the final version of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.11.032.
Appendix A. Supplementary data
Supplementary material
References
- 1.Xu J., Murphy S.L., Kochanek K.D., Bastian B., Arias E. Deaths: Final Data for 2016. Natl Vital Stat Rep. 2018 Jul;67(5):1–76. [PubMed] [Google Scholar]
- 2.T N., S A., P Ax P., O P. Deaths involving sepsis in Canada. Canada. 2015. http://www.statcan.gc.ca/ Available from.
- 3.Kaukonen K.M., Bailey M., Suzuki S., Pilcher D., Bellomo R. Mortality related to severe sepsis and septic shock among critically ill patients in Australia and New Zealand, 2000-2012. JAMA. 2014;311(13):1308–1316. doi: 10.1001/jama.2014.2637. [DOI] [PubMed] [Google Scholar]
- 4.Levy M.M., Rhodes A., Phillips G.S. Surviving Sepsis Campaign: Association between performance metrics and outcomes in a 7.5-year study. Crit Care Med. 2015;43(1):3–12. doi: 10.1097/CCM.0000000000000723. [DOI] [PubMed] [Google Scholar]
- 5.Goodwin A.J., Rice D.A., Simpson K.N., Ford D.W. Frequency, cost, and risk factors of readmissions among severe sepsis survivors. Crit Care Med. 2015;43(4):738–746. doi: 10.1097/CCM.0000000000000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prescott H.C., Osterholzer J.J., Langa K.M., Angus D.C., Iwashyna T.J. Late mortality after sepsis: Propensity matched cohort study. BMJ. 2016;353 doi: 10.1136/bmj.i2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ou S.M., Chu H., Chao P.W., Lee Y.J., Kuo S.C., Chen T.J., Tseng C.M., Shih C.J., Chen Y.T. Long-Term Mortality and Major Adverse Cardiovascular Events in Sepsis Survivors. A Nationwide Population-based Study. Am J Respir Crit Care Med. 2016 Jul 15;194(2):209–217. doi: 10.1164/rccm.201510-2023OC. [DOI] [PubMed] [Google Scholar]
- 8.Griffiths J., Hatch R.A., Bishop J. An exploration of social and economic outcome and associated health-related quality of life after critical illness in general intensive care unit survivors: a 12-month follow-up study. Crit Care. 2013;17(3):R100. doi: 10.1186/cc12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delano M.J., Ward P.A. The immune system's role in sepsis progression, resolution, and long-term outcome. Immunol Rev. 2016;274(1):330–353. doi: 10.1111/imr.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linder A., Fjell C., Levin A., Walley K.R., Russell J.A., Boyd J.H. Small acute increases in serum creatinine are associated with decreased long-term survival in the critically ill. Am J Respir Crit Care Med. 2014;189(9):1075–1081. doi: 10.1164/rccm.201311-2097OC. [DOI] [PubMed] [Google Scholar]
- 11.Yende S., D'Angelo G., Kellum J.A. Inflammatory markers at hospital discharge predict subsequent mortality after pneumonia and sepsis. Am J Respir Crit Care Med. 2008;177(11):1242–1247. doi: 10.1164/rccm.200712-1777OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gentile L.F., Cuenca A.G., Efron P.A. Persistent inflammation and immunosuppression: a common syndrome and new horizon for surgical intensive care. J Trauma Acute Care Surg. 2012;72(6):1491–1501. doi: 10.1097/TA.0b013e318256e000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dwivedi D.J., Grin P.M., Khan M. Differential expression of PCSK9 modulates infection, inflammation, and coagulation in a murine model of sepsis. Shock. 2016;46(6):672–680. doi: 10.1097/SHK.0000000000000682. [DOI] [PubMed] [Google Scholar]
- 14.Tveten K., Strom T.B., Berge K.E., Leren T.P. PCSK9-mediated degradation of the LDL receptor generates a 17 kDa C-terminal LDL receptor fragment. J Lipid Res. 2013;54(6):1560–1566. doi: 10.1194/jlr.M034371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen J.C., Boerwinkle E., Mosley T.H., Jr., Hobbs H.H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354(12):1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 16.Kotowski I.K., Pertsemlidis A., Luke A. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am J Hum Genet. 2006;78(3):410–422. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azzam K.M., Fessler M.B. Crosstalk between reverse cholesterol transport and innate immunity. Trends Endocrinol Metab. 2012;23(4):169–178. doi: 10.1016/j.tem.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walley K.R., Francis G.A., Opal S.M., Stein E.A., Russell J.A., Boyd J.H. The central role of proprotein convertase subtilisin/kexin type 9 in septic pathogen lipid transport and clearance. Am J Respir Crit Care Med. 2015;192(11):1275–1286. doi: 10.1164/rccm.201505-0876CI. [DOI] [PubMed] [Google Scholar]
- 19.Walley K.R., Thain K.R., Russell J.A. PCSK9 is a critical regulator of the innate immune response and septic shock outcome. Sci Transl Med. 2014;6(258):258ra143. doi: 10.1126/scitranslmed.3008782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun A., Netzer G., Small D.S. Association between index hospitalization and hospital readmission in sepsis survivors. Crit Care Med. 2016;44(3):478–487. doi: 10.1097/CCM.0000000000001464. [DOI] [PubMed] [Google Scholar]
- 21.Abifadel M., Rabes J.P., Devillers M. Mutations and polymorphisms in the proprotein convertase subtilisin kexin 9 (PCSK9) gene in cholesterol metabolism and disease. Hum Mutat. 2009;30(4):520–529. doi: 10.1002/humu.20882. [DOI] [PubMed] [Google Scholar]
- 22.Russell J.A., Walley K.R., Singer J. Vasopressin versus norepinephrine infusion in patients with septic shock. N Engl J Med. 2008;358(9):877–887. doi: 10.1056/NEJMoa067373. [DOI] [PubMed] [Google Scholar]
- 23.Levy M.M., Fink M.P., Marshall J.C. 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Crit Care Med. 2003;31(4):1250–1256. doi: 10.1097/01.CCM.0000050454.01978.3B. [DOI] [PubMed] [Google Scholar]
- 24.Dubuc G., Chamberland A., Wassef H. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler Thromb Vasc Biol. 2004;24(8):1454–1459. doi: 10.1161/01.ATV.0000134621.14315.43. [DOI] [PubMed] [Google Scholar]
- 25.Knaus W.A., Draper E.A., Wagner D.P., Zimmerman J.E. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13(10):818–829. [PubMed] [Google Scholar]
- 26.Tibolla G., Norata G.D., Artali R., Meneghetti F., Catapano A.L. Proprotein convertase subtilisin/kexin type 9 (PCSK9): from structure-function relation to therapeutic inhibition. Nutr Metab Cardiovasc Dis. 2011;21(11):835–843. doi: 10.1016/j.numecd.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Liu L., Berger V.W., Hershberger S.L. Wiley statsref: Statistics reference online. John Wiley & Sons, Ltd; 2014. Trend tests for counts and proportions. [Google Scholar]
- 28.National Research Council . The National Academies Press; Washington, DC: 2010. The prevention and treatment of missing data in clinical trials. Panel on handling missing data in clinical trials. Committee on national statistics division of behavioral and social sciences and education. [Google Scholar]
- 29.Mayne J., Ooi T.C., Raymond A. Differential effects of PCSK9 loss of function variants on serum lipid and PCSK9 levels in caucasian and African Canadian populations. Lipids Health Dis. 2013;12:70. doi: 10.1186/1476-511X-12-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shioji K., Mannami T., Kokubo Y. Genetic variants in PCSK9 affect the cholesterol level in Japanese. J Hum Genet. 2004;49(2):109–114. doi: 10.1007/s10038-003-0114-3. [DOI] [PubMed] [Google Scholar]
- 31.Cameron J., Holla O.L., Ranheim T., Kulseth M.A., Berge K.E., Leren T.P. Effect of mutations in the PCSK9 gene on the cell surface LDL receptors. Hum Mol Genet. 2006;15(9):1551–1558. doi: 10.1093/hmg/ddl077. [DOI] [PubMed] [Google Scholar]
- 32.Holla O.L., Cameron J., Berge K.E., Ranheim T., Leren T.P. Degradation of the LDL receptors by PCSK9 is not mediated by a secreted protein acted upon by PCSK9 extracellularly. BMC Cell Biol. 2007;8:9. doi: 10.1186/1471-2121-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis J.S., He V., Anstey N.M., Condon J.R. Long term outcomes following hospital admission for sepsis using relative survival analysis: a prospective cohort study of 1,092 patients with 5 year follow up. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0112224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prescott H.C., Langa K.M., Liu V., Escobar G.J., Iwashyna T.J. Increased 1-year healthcare use in survivors of severe sepsis. Am J Respir Crit Care Med. 2014;190(1):62–69. doi: 10.1164/rccm.201403-0471OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linder A., Guh D., Boyd J.H., Walley K.R., Anis A.H., Russell J.A. Long-term (10-year) mortality of younger previously healthy patients with severe sepsis/septic shock is worse than that of patients with nonseptic critical illness and of the general population. Crit Care Med. 2014;42(10):2211–2218. doi: 10.1097/CCM.0000000000000503. [DOI] [PubMed] [Google Scholar]
- 36.Meyer N., Harhay M.O., Small D.S. Temporal trends in incidence, sepsis-related mortality, and hospital-based acute care after sepsis. Crit Care Med. 2018;46(3):354–360. doi: 10.1097/CCM.0000000000002872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang D.W., Tseng C.H., Shapiro M.F. Rehospitalizations following sepsis: Common and costly. Crit Care Med. 2015;43(10):2085–2093. doi: 10.1097/CCM.0000000000001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demerle K.M., Royer S.C., Mikkelsen M.E., Prescott H.C. Readmissions for recurrent sepsis: New or relapsed infection? Crit Care Med. 2017;45(10):1702–1708. doi: 10.1097/CCM.0000000000002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ortego A., Gaieski D.F., Fuchs B.D. Hospital-based acute care use in survivors of septic shock. Crit Care Med. 2015;43(4):729–737. doi: 10.1097/CCM.0000000000000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ference B.A., Robinson J.G., Brook R.D. Variation in PCSK9 and HMGCR and risk of cardiovascular disease and diabetes. N Engl J Med. 2016;375(22):2144–2153. doi: 10.1056/NEJMoa1604304. [DOI] [PubMed] [Google Scholar]
- 41.Berger J.M., Loza Valdes A., Gromada J., Anderson N., Horton J.D. Inhibition of PCSK9 does not improve lipopolysaccharide-induced mortality in mice. J Lipid Res. 2017;58(8):1661–1669. doi: 10.1194/jlr.M076844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singer M., Deutschman C.S., Seymour C.W. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material