Abstract
The complex and heterogeneous nature of traumatic brain injury (TBI) has rendered the identification of diagnostic and prognostic biomarkers elusive. A single acute biomarker may not be sufficient to categorize injury severity and/or predict outcome. Using multivariate dimension reduction analyses, we tested the sensitivity and specificity of a multi-analyte panel of proteins as an ensemble biomarker for TBI. Serum was collected within 24 h of injury in a cohort of 130 patients enrolled in the multi-center prospective Transforming Research and Clinical Knowledge in Traumatic Brain Injury Pilot (TRACK-TBI Pilot) study and run on an array that measured 72 proteins. Using unsupervised principal components analysis, we first identified the subset of protein changes accounting for the most variance across patients. This yielded a group of 21 proteins that reflected an inverse relationship between inflammatory cytokines and regulators of anti-inflammation, and generated an individual inflammatory profile score for each patient. We then tested the association between these scores and computed tomography (CT) findings at hospital admission, as well as their prognostic association with functional recovery at 3 and 6 months (Glasgow Outcome Scale-Extended), and cognitive recovery at 6 months (California Verbal Learning Test, Second Edition) after injury. Inflammatory signatures were significantly increased in patients with positive CT findings, as well as in those who showed poor or incomplete recovery. Inflammation biomarker scores also showed significant sensitivity and specificity as a discriminator of these outcome measures (all areas under the curve [AUCs] >0.62). This proof of concept for the feasibility of multivariate biomarker identification demonstrates the prognostic validity of using a proteomic panel as a potential biomarker for TBI.
Keywords: biomarkers, proteomics, TBI
Introduction
Traumatic brain injury (TBI) is a major public health issue, affecting 2,800,000 people per year in the United States alone, and costing an estimated $60 billion.1 Despite continuing advances in emergency medicine, surgical intervention, and rehabilitative care, there remains a dearth of accurate and robust tools to aid in diagnosis and management. Computed tomography (CT) imaging is crucial for identifying gross pathology, but is largely insensitive to mild TBI, which makes up roughly 90% of TBI cases.2,3 Likewise, although the Glasgow Coma Scale (GCS) provides a broad categorization of injury severity, its inter-rater reliability and consistency in predicting outcome has been questioned.4,5
The struggle to find reliable indicators of injury severity and outcome arises from the multifaceted nature of TBI. The acute biological response to TBI is a systemic process that involves, among other pathologies, complex inflammatory and immunomodulatory cascades. Therefore, detection of multiple protein concentration changes from plasma and/or serum in response to TBI can provide useful insight for diagnosis, and may help inform clinical management decisions. Work in the past decade has identified several promising candidate proteins as TBI blood biomarkers, most notably glial fibrillary acidic protein (GFAP), ubiquitin C-terminal hydrolase L1 (UCHL1), neurofilament light chain (NF-L), and microtubule associated protein tau.6–14 Although each of these proteins has been shown to adequately distinguish between TBI and healthy control patients, we have recently shown that assessing them in combination improved the sensitivity and specificity for identifying TBI.15 Others have found similar improvements in predictive validity when combing two or more biomarkers.16,17 These findings are a reminder that a single biomarker may not account for the heterogeneity in TBI processes, and suggest that integration of multiple biomarker measurements may be a more accurate approach for addressing TBI complexity.
We assessed the predictive value of a multi-analyte biomarker panel for use as a diagnostic and prognostic tool in patients with TBI. Acute (< 24 h after injury) plasma protein levels from patients enrolled in the prospective multi-center Transforming Research and Clinical Knowledge in Traumatic Brain Injury Pilot (TRACK-TBI Pilot) clinical study were assessed using a multiplexed array that allows for the simultaneous quantification of 72 proteins from one sample. This commercially available array (Rules Based Medicine, HumanMAP v2.0) covers a broad range of proteins involved in numerous biological processes including inflammation, angiogenesis, cellular metabolism, repair, and degeneration, and was specifically chosen in order to optimally survey the complex and heterogeneous nature of TBI. Therefore, the goal of the current analysis was to take an exploratory approach to biomarker discovery; similar approaches have been taken in the context of aging and neurodegenerative diseases, using this same multi-analyte assay.18–20
We used an analytic dimensional reduction approach to identify which cluster of proteins accounted for the most variance in the patient population, then cross-validated these findings to test the stability of the multi-analyte clusters. This approach allows the data to inform us which constellation of biomarkers is most likely to be relevant. We then tested the predictive validity of our identified protein ensemble against CT findings and 3 and 6 month functional, behavioral, and cognitive outcome measures.
Methods
Study population
A sample of 130 participants enrolled at three level I trauma centers for the TRACK-TBI Pilot study was included in this analysis. Patients were enrolled between April 2010 and January 2011. Inclusion criteria for enrollment in TRACK-TBI Pilot included patients who presented to the emergency department (ED) within 24 h of brain injury and were triaged to a clinically indicated non-contrast head CT scan. Following the evidence-based guidelines, informed consent was obtained, and all data collection procedures followed protocols by the institutional review boards of the participating centers (University of California, San Francisco/Zuckerberg San Francisco General Hospital, San Francisco, CA; University of Pittsburgh Medical Center, Pittsburgh, PA; and University Medical Center Brackenridge, Austin, TX). Patients were excluded if they were pregnant, could not speak English, or presented with a psychological disorder that precluded them from giving consent. Others were excluded because of major polytrauma that would interfere with follow-up tests, infectious conditions, or late-stage cancer.21
Sample collection
Blood samples were collected within 24 h of injury (see Fig. S1 for distribution of time from injury to blood draw for each patient), and plasma was prepared according to the TBI Common Data Elements (CDE) Biospecimens and Biomarkers Working Group guidelines (see online supplementary material at http://www.liebertpub.com).22 Samples were centrifuged, aliquoted, and frozen at −80°C for future batch processing. Frozen plasma samples were sent to Myriad Rules-Based Medicine (Myriad RBM, Austin, TX) for protein analysis. Proteins were quantified using a multiplexed fluorescent immunoassay profile (HumanMAP v2.0). Protein concentrations for a total of 72 targets were quantified for each sample. Lower limit of quantification and least detectable dose for each analyte can be found in Table S1 (see online supplementary material at http://www.liebertpub.com).
Initial CT scan
A brain CT was performed on all patients within 24 h of ED presentation. CT data collection and interpretation were performed in accordance with TBI-CDE Working Group.23 All CT scans were uploaded to a central database, from which a blinded board-certified neuroradiologist reviewed the de-identified scans.
Outcome measures
Subjects in TRACK-TBI Pilot underwent outcomes testing using measures from the TBI CDE core outcomes battery.24 The Glasgow Outcome Scale-Extended (GOS-E) was used to assess overall functional disability at 3 and 6 months post-injury.25 The GOS-E consists of eight categories of outcome: 1(Dead), 2(Vegetative State), 3(Lower Severe Disability), 4(Upper Severe Disability), 5(Lower Moderate Disability), 6(Upper Moderate Disability), 7(Lower Good Recovery), and 8(Upper Good Recovery).26 This 8-point GOS-E was dichotomized in two ways to assess (1) full recovery (GOS-E = 8, indicative of a return to pre-injury baseline), or not (GOS-E < 8), and (2) good recovery (GOS-E < 4) or poor recovery (GOS-E > 4), in keeping with the practice of previous TRACK-TBI studies and other clinical trials.15,21,27 Cognitive domain was assessed after injury using the California Verbal Learning Test-Second Edition (CVLT-II), a memory and verbal learning memory task. This test was only given at 6 months. The task consists of five learning trials, an interference trial, an immediate recall trial, and a delayed (20 min) recall trial. The CVLT-II trials 1– 5 Standard Score is a sum of the first five learning trials normed for age and sex, and provides a global index of verbal learning ability.
Statistical analysis
Statistical analyses were performed using Statistical Package for Social Sciences version 24 (IBM, Inc. Chicago, IL). Non-linear principal components analysis (NL-PCA) was used to determine covariance among proteins. To confirm the stability of the PC loadings, a bootstrapping procedure was employed using 1000 balanced iterations. To provide further cross-validation of PC loading patterns, the root mean squared difference between initial and bootstrapped PC loadings was assessed, as well the Pearson product correlation coefficient, coefficient of congruence, and Cattell's salient variable similarity index.27 Individual scores for each patient on the PCs were then calculated based on the weight of the loadings for all proteins. For CT and GOS-E measures, PC scores were compared with outcome measures using analysis of variance (ANOVA) and receiver operating characteristic (ROC) curves to determine the predictive validity of protein biomarker clusters. Linear regression was to test the first principal component (PC1) score as a predictor of CVLT, as well as to determine the variance in each outcome measure explained by PC1 score. Statistical significance level for all analyses was set to α = 0.05.
Results
Patient cohort, distribution of injury severity, CT, and GOS-E measures
Multi-analyte protein analysis was performed on 130 patients from the TRACK-TBI Pilot study (Fig. 1). Age in this cohort ranged from 16 to 79 years, with 94 male and 36 female participants. The majority (104 patients, 88.9%) were classified as having mild TBI according to an ED admission GCS score of 13–15. Seven patients (6.0%) were classified as having moderate TBI (GCS 9–12), and 19 patients (16.2%) were classified as having severe TBI (GCS 3–8). Positive initial CT findings were defined by the presence of any intracranial abnormality in accordance with TBI-CDE guidelines. Acute intracranial pathology was noted in 44.2% of mild TBI patients, and 100% of moderate and severe TBI patients. Functional recovery on the GOS-E was evaluated for 124 patients at 3 months, and for 130 patients at 6 months. At 3 months, 27.7% had made a full recovery to baseline (GOS-E = 8), and 16.9% showed poor recovery as evidenced by vegetative state or severe disability (GOS-E ≤ 4); by 6 months 30.8% patients had made a full recovery, with only 12.3% of patients showing poor recovery. A further breakdown of injury severity, injury type, and outcome distribution can be found in Table 1.
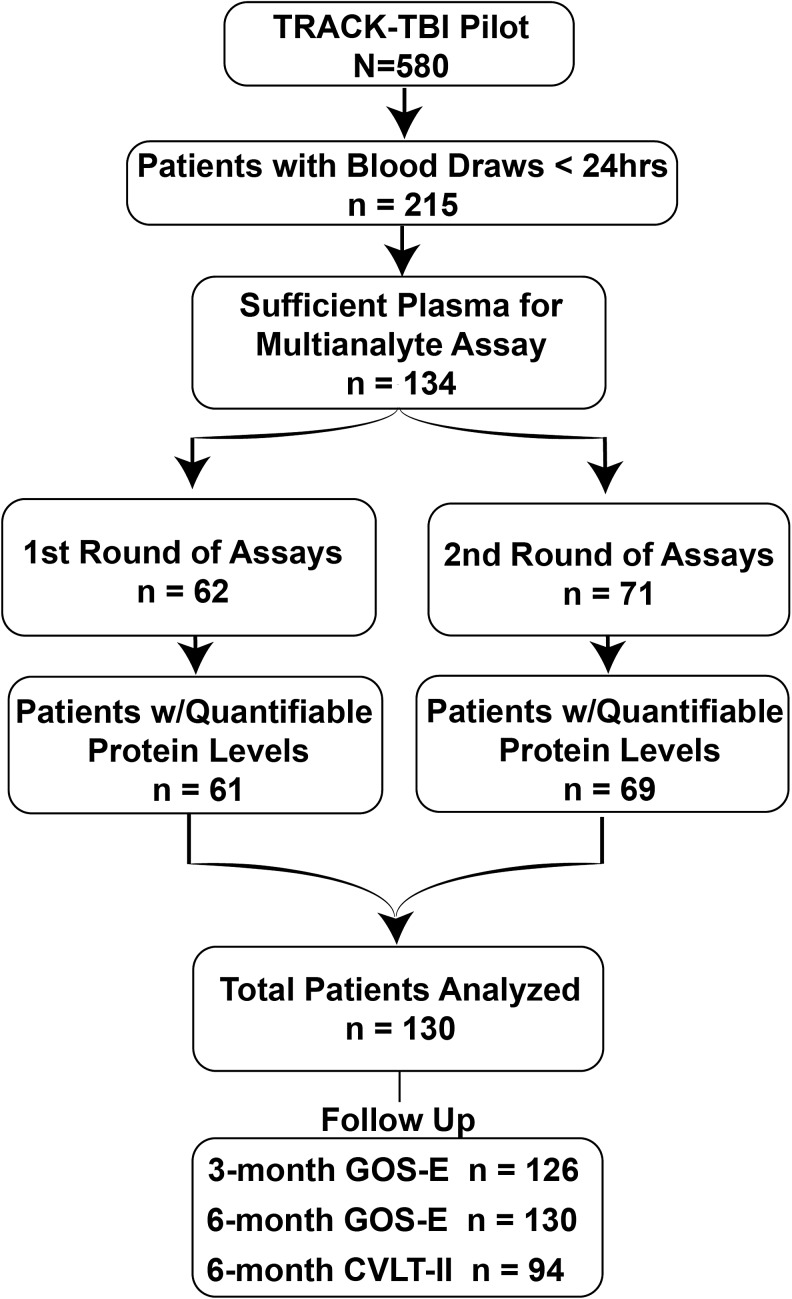
FIG. 1.
Flow chart of patients from the Transforming Research and Clinical Knowledge in Traumatic Brain Injury (TRACK-TBI) Pilot who were included in the current study. Patients were chosen based on sufficient volume of plasma available for multi-analyte assay. Assays were run in two rounds, with one patient from the first round, and two patients from the second round with insufficient plasma for detection. Total number of patients in the current study was 130.
Table 1.
Comparision of Patient Demographics
| n (%) | Mean ± SD | ||
|---|---|---|---|
| Sex | Age | 42.7 (18.1) | |
| Male | 94 (72.3) | ||
| Female | 36 (27.7) | CVLT-II | 49.6 (13.8) |
| Trial 1–5 Standard Score | |||
| Cause of Injury | |||
| Vehicle Accident | 22 (17.2) | ||
| Fall | 26 (20.3) | ||
| Assault | 13 (10.2) | ||
| Sport | 20 (15.6) | ||
| Explosion | 8 (6.3) | ||
| ED admission GCS | |||
| Severe (3–8) | 19 (14.6) | ||
| Moderate (9–12) | 7 (5.4) | ||
| Mild (13–15) | 104 (80.0) | ||
| ED admission head CT | |||
| Positive | 69 (53.1) | ||
| Negative | 61 (46.9) | ||
| Abbreviated Injury Scale (Extracranial Regions) | |||
| Abdomen/Pelvis AIS >2 | 2 (1.6) | ||
| Chest/Thorax AIS >2 | 17 (13.1) | ||
| Limbs/Pelvis/Girdle AIS >2 | 11 (8.5) | ||
| GOS-E | 3 months | ||
| 1 | 4 (3.1) | ||
| 2 | 1 (0.8) | ||
| 3 | 7 (5.4) | ||
| 4 | 9 (6.9) | ||
| 5 | 12 (9.2) | ||
| 6 | 20 (15.4) | ||
| 7 | 35 (26.9) | ||
| 8 | 36 (27.7) | ||
| GOS-E | 6 months | ||
| 1 | 4 (3.1) | ||
| 2 | 1 (0.8) | ||
| 3 | 7 (5.4) | ||
| 4 | 9 (6.9) | ||
| 5 | 12 (9.2) | ||
| 6 | 20 (15.4) | ||
| 7 | 35 (26.9) | ||
| 8 | 36 (27.7) |
ED, emergency department; GCS, Glasgow Coma Score; CT, computed tomography; GOS-E, Glasgow Outcome Score – Extended; CVLT-II, California Verbal Learning Test-Second Edition.
Dimension reduction and cross-validation of proteomic data
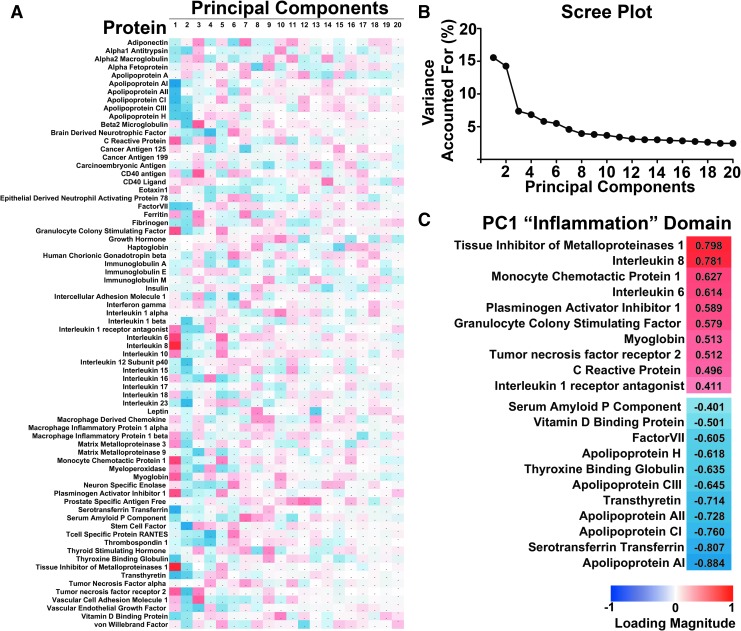
NL-PCA was used to partition the variance across patients into orthogonal components (Fig. 2A). PC1 accounted for 16.2% of the variance (eigenvalue = 10.4; Fig. 2B). The NL-PCA determined the unique correlation value between each protein and the variance explained within this PC. This value (protein “loading”) could then be used for face validation of the PC. The proteins were ranked by their loading, and loadings were then thresholded at an absolute value of |0.4| (determined a priori according to the threshold standard introduced by Stevens28), in order to “name” the PC based on only those markers that contributed most to the variance. This step yielded a set of 21 of the 72 analytes, and the known biological properties and processes that were common across this cluster were analyzed by domain experts from the TRACK-TBI team. Based on the known function of most proteins in PC1 and the inverse loading values between pro-inflammatory markers (e.g., C-reactive protein, Interleukin-6 [IL-6], TIMP-1, with high positive loadings) and anti-inflammatory apolipoproteins and regulatory markers with strong negative loadings (Fig. 2C), a consensus was formed that this PC could be identified as driven in large part by inflammatory markers.
FIG. 2.
Non-linear principal components analysis (PCA) of multi-analyte proteomic data from traumatic brain injury (TBI) patient plasma. (A) PCA matrix of 20 components by 72 proteins from the commercially available multi-analyte array (RBM, HumanMAP v2.0). Red indicates positive magnitude of loadings, blue indicates negative magnitude. (B) Scree plot showing variance accounted for by each orthogonal component. The first principal component (PC1) accounted for 16.2% of the variance. (C) After thresholding loadings at an absolute value of 0.4, the subset of proteins with strong loadings are identified as being predominantly associated with inflammation, with pro- and anti-inflammatory markers loading in opposite directions. Color image is available online at www.liebertpub.com/neu
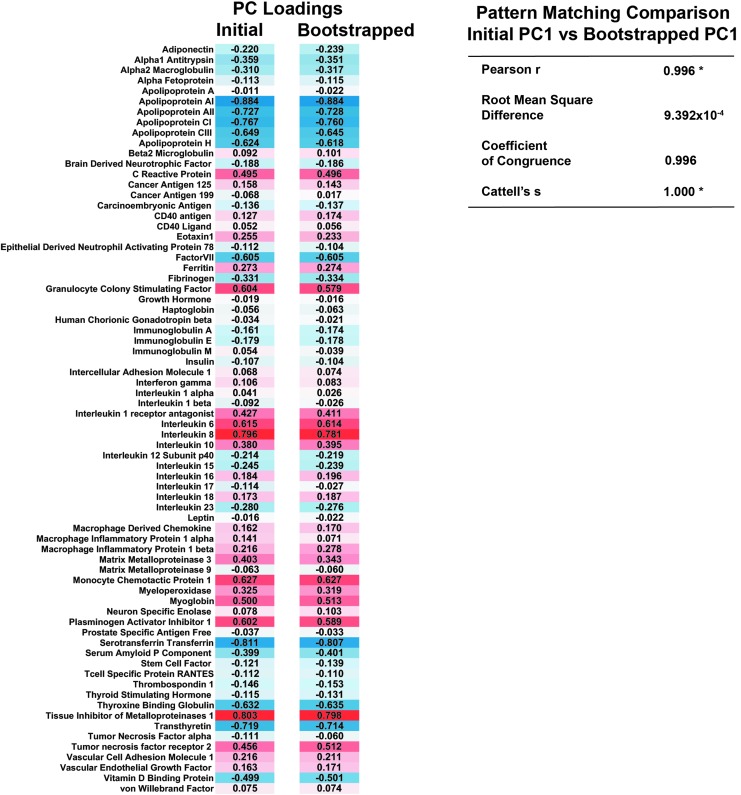
To cross-validate the loading pattern found across all proteins in PC1, we used an extensive bootstrapping method to simulate a much larger population.29 In this process, the data set was resampled 1000 times, with each iteration recalculating the protein loadings based on a slightly altered subset of the sample population. Pattern matching analysis was then used to determine the stability between the original and bootstrapped loadings. This analysis revealed a significant agreement between these loadings (Pearson product moment correlation coefficient = 0.996, root mean square difference = 0, Cattell salient variable similarity index), indicating that the PC loadings were robust and stable (Fig. 3). All subsequent analyses of predictive value were made using the individual PC1 scores, a normalized composite of all 72 markers weighted by their respective loadings.
FIG. 3.
Internal cross-validation of protein loading pattern. Comparison of initial first principal component (PC1) loadings for each protein against the mean loading value of 1000 bootstrapping iterations shows a very high degree of similarity, indicative of a robust and stable loading pattern (*p < 0.05). Color image is available online at www.liebertpub.com/neu
Association between inflammatory biomarker component and CT findings
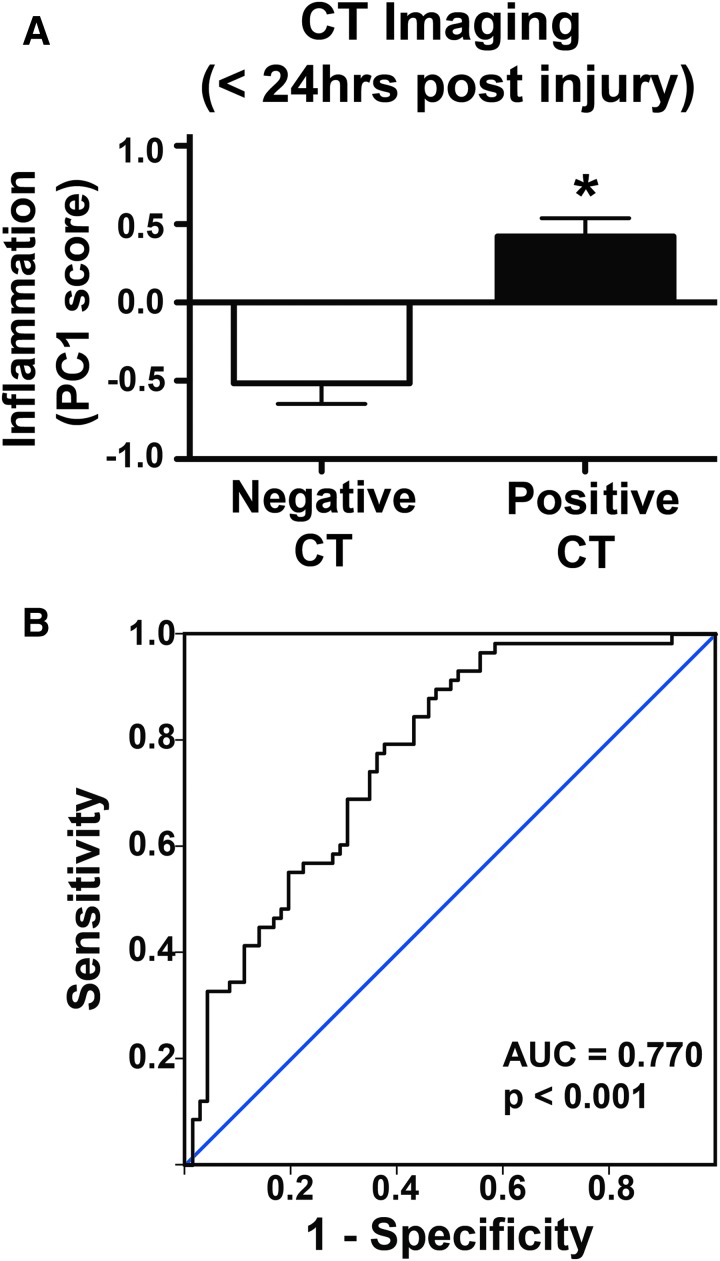
To assess the predictive validity of the biomarker ensemble, individual patient scores on PC1 (weighted by each of the 72 protein loading values) were first compared with the presence or absence of intracranial pathology findings on CT (Fig. 4). Patients with negative CT findings had significantly lower PC1 scores (indicating lower inflammation) than those with positive CT findings (p < 0.01; ηp2 = 0.18; 18.5% of CT variance accounted for by the PC1 score, Fig. 4A). ROC analysis showed Inflammation PC score to be significantly predictive of CT finding (area under the curve [AUC] = 0.77; 95% CI 0.69–0.85; p < 0.01; Fig. 4B).
FIG. 4.
Relationship between computed tomography (CT) findings and Inflammation biomarker component. (A) Patients categorized by either positive or negative CT pathology findings have significantly different mean Inflammation principal component (PC) scores, with patients who have positive CT findings exhibiting higher inflammation scores (p < 0.01). (B) Receiver operating characteristic (ROC) analysis shows that Inflammation PC score is significantly predictive of whether CT finding is positive or negative (area under the curve [AUC] = 0.770, p < 0.001). Color image is available online at www.liebertpub.com/neu
Association between inflammatory biomarker panel and outcome measures
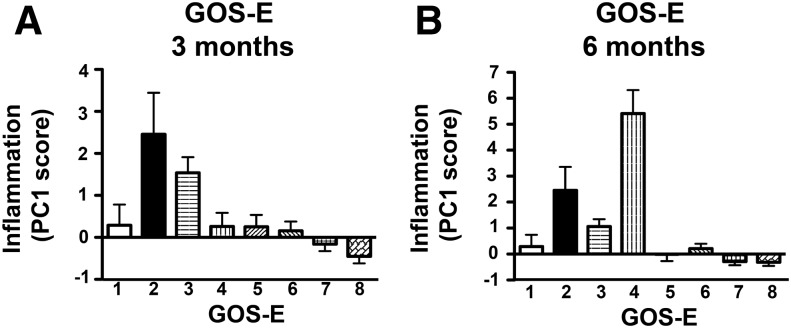
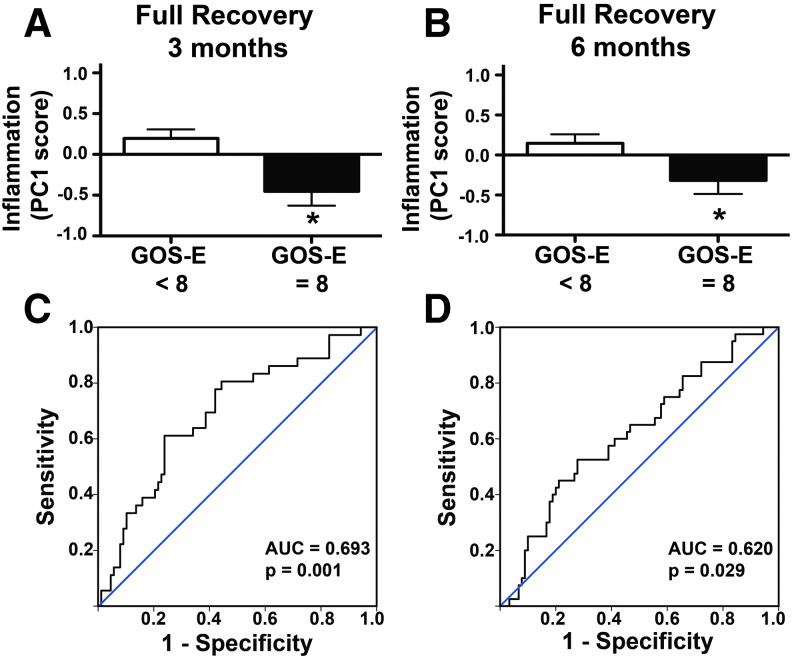
Inflammation PC1 score was then compared against the eight point GOS-E outcome scale at 3 and 6 months post-injury. ANOVA revealed a significant effect of GOS-E on PC1 score at both 3 months (Fig. 5A, p < 0.001, ηp2 = 0.23, 19.2% variance in 3 month GOS-E accounted for by PC1 score), and 6 months (Fig. 5B, p < 0.001, ηp2 = 0.36, 21.3% of variance in 6 month GOS-E accounted for by PC1 score), with the lowest mean PC1 scores found in patients with the greatest recovery (GOS-E >7). GOS-E was further subcategorized into two dichotomous sets: Full Recovery (GOS-E = 8) or not (Fig. 6), and Poor Recovery (GOS-E ≤ 4) or not (Fig. 7). Patients who achieved full recovery had significantly lower Inflammation PC scores, at either 3 months (p < 0.01, ηp2 = 0.07; Fig. 6A), or 6 months (p < 0.05, ηp2 = 0.04; Fig. 6B). Inflammation PC score was also modestly, yet significantly, predictive of full recovery by 3 months (AUC = 0.69; 95% CI 0.59–0.80; p < 0.05; Fig. 6C) and 6 months (AUC = 0.62; 95% CI 0.52–0.72; p < 0.05; Fig. 6D).
FIG. 5.
Relationship between inflammation component and Glasgow Outcome Scale-Extended (GOS-E). A significant main effect of principal component (PC) score was seen across GOS-E categories with patients with lower GOS-E (indicative of poor recovery) showing the highest PC scores (p < 0.01) at both 3 months (A) and 6 months (B) post-injury (p < 0.01). These findings indicate that lower acute ensemble biomarker scores may be predictive of improved future recovery.
FIG. 6.
Relationship between inflammation component and full recovery at 3 and 6 months after traumatic brain injury (TBI). Patient scores on the Glasgow Outcome Scale-Extended (GOS-E) were dichotomized into either full (GOS-E = 8) or incomplete (GOS-E < 8) recovery. (A) Patients who made a full recovery by 3 months had significantly lower Inflammation component scores (*p < 0.05). (B) Patients who made full recovery by 6 months also had significantly lower Inflammation component scores (*p < 0.05). (C) Receiver operating characteristic (ROC) analysis shows Inflammation principal component (PC) score to be significantly predictive of whether or not recovery at 3 months is complete (area under the curve [AUC] = 0.693, p < 0.01). (D) ROC analysis shows Inflammation PC score to be significantly predictive of whether or not recovery at 6 months is complete (AUC = 0.620, p < 0.05). Color image is available online at www.liebertpub.com/neu
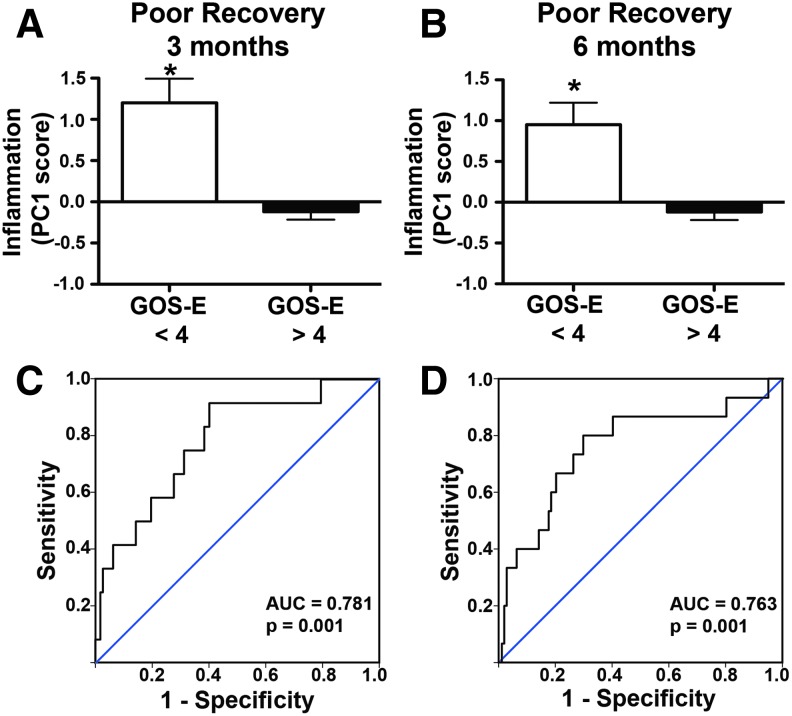
FIG. 7.
Relationship between inflammation component and poor recovery at 3 and 6 months after traumatic brain injury (TBI). To assess poor recovery, patient Glasgow Outcome Scale-Extended (GOS-E) scores were dichotomized into either poor recovery (GOS-E < 4) or not (GOS-E > 4). (A) Patients who had poor recovery at 3 months had significantly higher Inflammation component scores (*p < 0.05). (B) Patients who had poor recovery at 6 months also had significantly higher Inflammation component scores (*p < 0.05). (C) Receiver operating characteristic (ROC) analysis shows Inflammation principal component (PC) score to be significantly predictive of poor recovery at 3 months (area under the curve [AUC] = 0.781, p < 0.001). (D) ROC analysis shows Inflammation PC score to be significantly predictive of poor recovery at 6 months (AUC = 0.763, p < 0.05). Color image is available online at www.liebertpub.com/neu
Conversely, patients with poor recovery had significantly higher Inflammation PC scores at either 3 months (p < 0.01, ηp2 = 0.13; Fig. 7A) or 6 months (p < 0.01, ηp2 = 0.09; Fig. 7B), and Inflammation PC score was significantly predictive of poor recovery at 3 months (AUC = 0.78; 95% CI 0.65–0.92; p < 0.01; Fig. 7C) and 6 months (AUC = 0.76; 95% CI 0.62–0.91; p < 0.01; Fig. 7D).
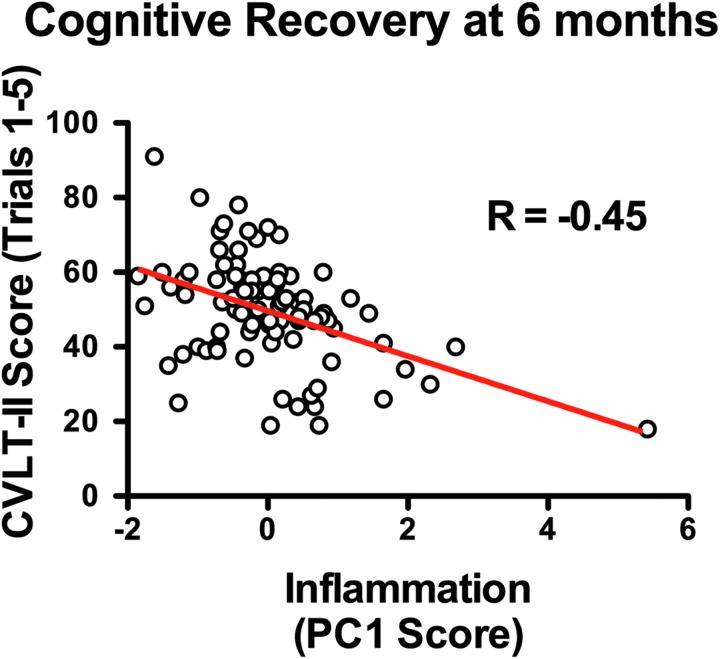
To assess the association between the inflammatory panel and cognitive recovery, Inflammation PC1 score was compared against scores on the CVLT-II at 6 months using linear regression. Inflammation PC1 score was shown to be significantly predictive of CVLT score at 6 months (R = -0.45, p < 0.01, Fig. 8), accounting for 20% of the variance in CVLT.
FIG. 8.
Relationship between inflammation component and cognitive recovery at 6 months after traumatic brain injury (TBI). Linear regression shows inflammation principal component (PC) score to be significantly predictive of California Verbal Learning Test-Second Edition (CVLT-II) score (R = -0.45, p < 0.01). Results indicate that higher acute inflammation PC score predicts lower cognitive recovery at 6 months post-TBI. Color image is available online at www.liebertpub.com/neu
Discussion
The current analysis from the prospective multi-center TRACK-TBI Pilot study used multivariate NL-PCA to identify a subset of proteins from a multi-analyte array, then tested the prognostic validity of this biomarker profile using individual PC scores. The first module identified by NL-PCA consisted primarily of inflammation-related targets. Hypothesis testing of individual patient PC scores from this inflammation component showed that patients with admission brain CT positive for acute intracranial pathology, and incomplete/poor recovery at 3 and 6 months, had a significantly increased Inflammation PC score. The Inflammation PC score was significantly sensitive and specific as a predictor for these outcome measures.
Although these findings highlight the feasibility of identifying and testing an ensemble of protein biomarkers, these markers were shown to be moderately sensitive and specific in predicting poor recovery (GOS-E < 4) at 3 and 6 months (AUC for ROC <0.70), compared with the performance of brain-specific biomarkers such as glial fibrillary acidic protein (GFAP) and ubiquitin C-terminal hydrolase L1 (UCHL1), which have recently been approved by the United States Food and Drug Administration (FDA) for TBI.15 Similarly, although this inflammatory protein ensemble significantly predicted positive CT findings, other individual markers have been shown to be highly robust predictors of CT positive versus CT negative findings in TBI patients.10,30 A recent TRACK-TBI Pilot demonstrated that the simultaneous detection of four candidate biomarkers (GFAP, UCHL1, neurofilament light chain [NF-L], and total tau) on a multiplex array was able to successfully distinguish patients with and without CT abnormalities.31 The current study provides the proof of concept that the predictive capacity of a broad range of biomarkers can be tested as a composite profile, a sophisticated multivariate approach that may help to model the complexity and heterogeneity of TBI. Further, this ensemble of biomarkers was sensitive to both acute diagnostic measures and long-term prognosis, suggesting that this data-driven approach may help to identify groups of markers that represent both acute and chronic latent features of TBI.
Data-driven biomarker discovery
The emergence of the “big-data” era in neurotrauma research has reinforced the need for analytical approaches that will adequately reduce data to manageable and interpretable results.32–34 Likewise, in order for findings to be readily shared and replicated across research groups, data standardization is necessary. To this end, the TRACK-TBI Pilot was the first federally funded prospective study to implement the TBI CDE mandated by the National Institutes of Health/National Institute of Neurological Disorders and Stroke (NIH-NINDS).35,36 Given that the data in the current study were collected over 3 years across multiple level I trauma centers, the standardization of fluid collection/preparation, imaging, and outcome measures allowed for data curation to be streamlined and, potentially, more easily replicable by other trials in the future.
Although the current study assessed interactions between a relatively small set of pre-curated proteins, a subset of proteins loaded highly together in PC1 and reflected a clear biological domain (inflammation). Although one might make an informed prediction that inflammation would be a prognostic indicator for TBI, the data-driven identification of this cluster without any prior stratification of patients highlights the utility of this type of approach; namely, as an unbiased method for uncovering promising avenues for further study. We have recently used another data-driven approach, topological data analysis, to uncover associations among imaging, outcomes, and high-dimensional genomic data from the TRACK-TBI Pilot.37 We used machine learning algorithms to identify specific multivariate phenotypes, and candidate single nucleotide polymorphism biomarkers that were uncovered by this data-driven approach were then shown to be predictive of outcome. Similarly, in the current study, NL-PCA identified an ensemble of proteins that could then be tested against other diagnostics and outcome measures. As the PC identifies and weighs these proteins according to their contribution to the variance in the data set, they likely reflect biological “universals”: multi-analyte signatures that are important. The fact that they predict both acute and long-term outcomes is consistent with this concept. By using these data-driven approaches as pre-screening tools that identify important domains for future hypothesis testing, we have the capacity to convert data complexity from an obstacle into an opportunity for discovery.
Acute inflammatory biomarkers of TBI
The ensemble of proteins identified by NL-PCA as accounting for the most variance was largely associated with the inflammatory response to TBI. TBI sets in motion a number of complex inflammatory cascades, as resident glial cells release cytokines and chemokines first in response to the disruptive mechanical force acting on central nervous system (CNS) tissue, as a part of the ongoing immunomodulatory response, and as key mediators of secondary injury processes. Here we found that a number of pro-inflammatory cytokines loaded highly together, notably TIMP1, Interleukin-6 (IL-6) and -8, monocyte chemotactic protein 1 (MCP1), and the classic marker of inflammation, C-reactive protein. Many of these markers have been previously shown to be associated with TBI. TIMP1 is a tissue inhibitor of matrix metalloproteinase, and has been shown to exert independent inflammatory cytokine-like activity.38 In a recent study of severe TBI patients, higher TIMP1 levels were found to be an indicator of increased mortality.39 Similarly, the cytokine IL-6 has been shown to be associated with acute blood–brain barrier dysfunction in severe TBI patients,40 and more recently, increased IL-6 was shown to be a robust predictor of poor short-term prognosis and development of infectious complications after TBI.41 MCP1 (also known as CCL2) is a chemokine produced by astrocytes,42 which attracts monocytes and macrophages to the injury site. Cerebral spinal fluid levels of MCP1 peak within the first 48 h after TBI admission,43 and in a protein array study of candidate plasma biomarkers, MCP1 was one of three markers (of 120 targets assayed) that had the most sensitivity and specificity for TBI.44
Conversely, several proteins that we identified by PCA as loading in the opposite direction have been shown to have regulatory or anti-inflammatory properties, including apolipoproteins and transferrin. The strongest negative loader, Apo A1, has been shown to be anti-inflammatory, with relatively high constitutive levels in healthy controls that are decreased in response to TBI.16 Further, it has been used in conjunction with S100B to improve the specificity of biomarkers for mild TBI, presumably because it is not susceptible to change in response to peripheral injuries, as has been shown with S100B.16,45 Similarly, the regulatory protein transferrin, which plays a role in iron metabolism, is downregulated in response to CNS injury, and has been shown to be a robust indicator of poor prognosis in patients with intracranial hemorrhage.46 This is further supported by the pre-clinical literature, which has shown that neuroinflammatory changes can predict both acute cell death and long-term neurocognitive recovery.
Considerations
The goal of the current study was to test the analytical feasibility of managing multi-analyte data coupled with NL-PCA dimension reduction as an approach for testing the diagnostic and prognostic validity of multiplexed proteins in ensemble. Although the biomarkers assessed in this study were not individually selected based on their specificity for TBI, one of the goals of this approach was to use the ensemble of proteins as a “composite biomarker” rather than assessing the specificity of each biomarker in isolation. The specific ensemble weighting of these biomarkers does indeed demonstrate reasonable sensitivity and specificity according to the data-driven analyses presented here; however, further work is needed to test their generality and reproducibility across different patient populations and analytical platforms.
The results are promising from an analytical workflow perspective, but we must keep in mind the clinical feasibility of running multi-analyte biomarker assays, in terms of cost and time. It is also important to consider that blood was collected for analysis within 24 h, and that the size of that window may affect the expression of proteins, as each marker is likely to have a distinct temporal profile of expression after injury. Future work will test the bounds of this approach; for example, the question of whether, if the arrays were reduced to only those candidate proteins that loaded highly in PC1, how few would be needed to maintain predictive validity. Also, there is the question of whether the strongest opposing biomarkers, as identified here by PCA, TIMP1, and Apo A1, could be assessed in conjunction as a biomarker pair.
It is also important to note the challenges in accurately and precisely categorizing TBI severity, especially in defining what is to be considered a mild TBI. In the current cohort, 44 of the 104 patients with a “mild” rating (GCS = 14–15) also had a positive head CT, a measure that others may use as selection criterion for categorizing an injury as non-mild. From that perspective, the patients in this study may not be seen as representing a typical mild TBI population, and may be skewed toward a more severe injury despite their mild categorization based on GCS score. The fact that 42% of those with “mild” GCS scores also had positive CT findings is consistent with the “complicated mild” category that has been proposed since the rise of brain imaging.47 This discrepancy between CT findings (and other measures) and severity categorization by GCS score is an issue that must be addressed in the TBI field. For example, although the majority of the patients in this study had an admission GCS that placed them in the mild TBI category, it is difficult to generalize findings to the broader population without better consensus on how best to define severity. We continue to push for a more reliable and valid multi-dimensional classification method that would better model the heterogeneity of TBI25 and, for example, would be based on pathoanatomic features.
Future work with the TRACK-TBI database will enfold biomarkers into a broader analytical context that may include genomic data, clinical information (blood pressure, surgical procedures, critical care data, medical history), pharmacological interventions, and neurorehabilitative treatment strategies. Prior work to this end, such as the International Mission for Prognostic and Analysis of Clinical Trials (IMPACT) has successfully used a multivariable regression approach to identify robust predictors of TBI, and have shown that integration of multiple fluid biomarkers with other core predictors such as age, GCS motor score, and pupil response, improves the power of the prognostic models.48,49 IMPACT has developed robust prognostic models that are focused on moderate and severe TBI; however, they are less well established in the mild and complicated mild populations featured in the present work.50,51
It is not likely that a single biomarker will be able to capture the breadth of TBI complexity. Likewise, any single domain may not be adequate to fully capture the complexity of the full-body response to TBI. Therefore, we hope that in the future an advanced multivariate approach such as the one presented in this study could be used to incorporate multimodal biomarker data as a robust tool for reducing the high dimensionality of multiple biomarker measures in order to develop and identify the most sensitive and specific biomarker profile possible. The TBI CDE effort has spearheaded improved clinical data collection standards, which will enable integration of biomarker analyses from multiple sites and trials. Combined with advances in rapid, multidimensional analyses, multivariate biomarker panels hold promise for precision medicine and critical clinical decision making in traumatic brain injury.
Supplementary Material
Acknowledgments
This work was supported by Department of Defense (DoD) grant W81XWH-13-1-0441 (G.T.M.), and NIH/NINDS grants NS069409 (G.T.M.), NS069409-02S1 (G.T.M.), NS086090 (G.T.M.), NS106899 (A.R.F.), and NS088475 (A.R.F.). TRACK-TBI investigators include (in alphabetical order): Opeolu M. Adeoye, MD, University of Cincinnati; Neeraj Badjatia, MD, University of Maryland, Baltimore; Kimberly D. Boase, BA, University of Washington; Yelena Bodien-Guller, PhD, Harvard/Spaulding Rehabilitation Hospital; Malcolm R. Bullock, MD, PhD, University of Miami; Randall M. Chesnut, MD, FCCM, FACS, University of Washington; John D. Corrigan, PhD, Ohio State University; Karen L. Crawford, MILS, University of Southern California; Ramon Diaz-Arrastia, MD, University of Pennsylvania; Sureyya S. Dikmen, PhD, University of Washington; Ann-Christine Duhaime, MD, Harvard/Massachusetts General Hospital; Richard G. Ellenbogen, MD, FACS, University of Washington; Frank Ezekiel, University of California, San Francisco; Venkata R. Feeser, MD, Virginia Commonwealth University; Joseph T. Giacino, PhD, Harvard/Spaulding Rehabilitation Hospital; Dana P. Goldman, PhD, University of Southern California; Luis Gonzales, BA, TIRR Memorial Hermann; Shankar P. Gopinath, MD, Baylor College of Medicine; Rao P. Gullapalli, PhD, MBA, University of Maryland, Baltimore; Jesse C. Hemphill, MD, University of California, San Francisco; Gillian A. Hotz, PhD, University of Miami; Joel H. Kramer, PsyD, University of California, San Francisco; Harvey Levin, PhD, Baylor College of Medicine; Christopher J. Lindsell, PhD, Vanderbilt University; Joan Machamer, MA, University of Washington; Christopher Madden, MD, UT Southwestern; Amy J. Markowitz, JD, University of California, San Francisco; Alastair Martin, PhD, University of California, San Francisco; Bruce E. Mathern, MD, Virginia Commonwealth University; Thomas W. McAllister, MD, Indiana University; Michael A. McCrea, PhD, Medical College of Wisconsin; Randall E. Merchant, PhD, Virginia Commonwealth University; Florence Noel, PhD, Baylor College of Medicine; Daniel P. Perl, MD, Uniformed Services University of the Health Sciences; Ava M. Puccio, RN, PhD, University of Pittsburgh; Miri Rabinowitz, PhD, University of Pittsburgh; Claudia S. Robertson, MD, Baylor College of Medicine; Jonathan Rosand, MD, MSC, Harvard/Massachusetts General Hospital; Angelle M. Sander, PhD, Baylor College of Medicine; Gabriela Satris, MSc, University of California, San Francisco; David M. Schnyer, PhD, University of Texas at Austin; Seth A. Seabury, PhD, University of Southern California; Paulina Sergot, MD, FACEP, Baylor College of Medicine; Mark Sherer, PhD, TIRR Memorial Hermann; Deborah M. Stein, MD, MPH, University of Maryland, Baltimore; Murray B. Stein, MD, MPH, FRCPC, University of California, San Diego; Sabrina R. Taylor, PhD, University of California, San Francisco; Nancy R. Temkin, PhD, University of Washington; Arthur W. Toga, PhD, University of Southern California; L. Christine Turtzo, MD, PhD, Uniformed Services University of the Health Sciences; Paul M. Vespa, MD, University of California, Los Angeles; Kevin K. Wang, PhD, University of Florida; Ross Zafonte, DO, Harvard/Spaulding Rehabilitation Hospital; Zhiqun Zhang, MD, University of Florida. The authors also give special thanks to Amy J. Markowitz, JD for editorial assistance.
Contributor Information
Collaborators: the TRACK-TBI Investigators, Opeolu M. Adeoye, Neeraj Badjatia, Kimberly D. Boase, Yelena Bodien-Guller, Malcolm R. Bullock, Randall M. Chesnut, John D. Corrigan, Karen L. Crawford, Ramon Diaz-Arrastia, Sureyya S. Dikmen, Ann-Christine Duhaime, Richard G. Ellenbogen, Frank Ezekiel, Venkata R. Feeser, Joseph T. Giacino, Dana P. Goldman, Luis Gonzales, Shankar P. Gopinath, Rao P. Gullapalli, Jesse C. Hemphill, Gillian A. Hotz, Joel H. Kramer, Harvey Levin, Christopher J. Lindsell, Joan Machamer, Christopher Madden, Amy J. Markowitz, Alastair Martin, Bruce E. Mathern, Thomas W. McAllister, Michael A. McCrea, Randall E. Merchant, Florence Noel, Daniel P. Perl, Ava M. Puccio, Miri Rabinowitz, Claudia S. Robertson, Jonathan Rosand, Angelle M. Sander, Gabriela Satris, David M. Schnyer, Seth A. Seabury, Paulina Sergot, Mark Sherer, Deborah M. Stein, Murray B. Stein, Sabrina R. Taylor, Nancy R. Temkin, Arthur W. Toga, L. Christine Turtzo, Paul M. Vespa, Kevin K. Wang, Ross Zafonte, and Zhiqun Zhang
Author Disclosure Statement
The TRACK-TBI authors declare no competing financial interests in Myriad/RBM including equity, consulting fees, or stock ownership.
References
- 1. Taylor C.A., Bell J.M., Breiding M.J., and Xu L. (2017). Traumatic brain injury–related emergency department visits, hospitalizations, and deaths — United States, 2007 and 2013. MMWR Surveill. Summ. 66, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mittl R.L., Grossman R.I., Hiehle J.F., Hurst R.W., Kauder D.R., Gennarelli T.A., and Alburger G.W. (1994). Prevalence of MR evidence of diffuse axonal injury in patients with mild head injury and normal head CT findings. AJNR Am. J. Neuroradiol. 15, 1583–1589 [PMC free article] [PubMed] [Google Scholar]
- 3. Thurman D.J., Alverson C., Browne D., Dunn K.A., Guerrero J., Johnson R., Johnson V., Longlois J.A., Pilkey D., and Sniezek J.E., (1999). Traumatic brain injury in the United States: A report to Congress. Division of Acute Care, Rehabilitation Research, and Disability Prevention, National Center for Injury Prevention and Control, Centers for Disease Control and Prevention, US Department of Health and Human Services [Google Scholar]
- 4. McNett M. (2007). A review of the predictive ability of Glasgow Coma Scale scores in head-injured patients. J. Neurosci. Nurs. 39, 68–75 [DOI] [PubMed] [Google Scholar]
- 5. Reith F.C., Synnot A., van den Brande R., Gruen R.L., and Maas A.I. (2017). Factors influencing the reliability of the Glasgow Coma Scale: a systematic review. Neurosurgery 80, 829–839 [DOI] [PubMed] [Google Scholar]
- 6. Berger R.P., Hayes R.L., Richichi R., Beers S.R., and Wang K.K.W. (2012). Serum concentrations of ubiquitin C-terminal hydrolase-L1 and αII-spectrin breakdown product 145 kDa correlate with outcome after pediatric TBI. J. Neurotrauma 29, 162–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brophy G.M., Mondello S., Papa L., Robicsek S.A., Gabrielli A., Joseph Tepas I., Buki A., Robertson C., Tortella F.C., Hayes R.L., and Wang K.K.W. (2011). Biokinetic analysis of ubiquitin C-terminal hydrolase-L1 (UCH-L1) in severe traumatic brain injury patient biofluids. J. Neurotrauma 28, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Metting Z., Wilczak N., Rodiger L.A., Schaaf J.M., and van der Naalt J. (2012). GFAP and S100B in the acute phase of mild traumatic brain injury. Neurology 78, 1428–1433 [DOI] [PubMed] [Google Scholar]
- 9. Okonkwo D.O., Yue J.K., Puccio A.M., Panczykowski D.M., Inoue T., McMahon P.J., Sorani M.D., Yuh E.L., Lingsma H.F., Maas A.I.R., Valadka A.B., Manley G.T., and Transforming Research and Clinical Knowledge in Traumatic Brain Injury (TRACK-TBI) Investigators. (2013). GFAP-BDP as an acute diagnostic marker in traumatic brain injury: results from the prospective transforming research and clinical knowledge in traumatic brain injury study. J. Neurotrauma 30, 1490–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Papa L., Lewis L.M., Falk J.L., Zhang Z., Silvestri S., Giordano P., Brophy G.M., Demery J.A., Dixit N.K., Ferguson I., Liu M.C., Mo J., Akinyi L., Schmid K., Mondello S., Robertson C.S., Tortella F.C., Hayes R.L., and Wang K.K.W. (2012). Elevated levels of serum glial fibrillary acidic protein breakdown products in mild and moderate traumatic brain injury are associated with intracranial lesions and neurosurgical intervention. Ann. Emerg. Med. 59, 471–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pelinka L.E., Kroepfl A., Leixnering M., Buchinger W., Raabe A., and Redl H. (2004). GFAP Versus S100B in serum after traumatic brain injury: relationship to brain damage and outcome. J. Neurotrauma 21, 1553–1561 [DOI] [PubMed] [Google Scholar]
- 12. Shahim P., Zetterberg H., Tegner Y., and Blennow K. (2017). Serum neurofilament light as a biomarker for mild traumatic brain injury in contact sports. Neurology 88, 1788–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shahim P., Gren M., Liman V., Andreasson U., Norgren N., Tegner Y., Mattsson N., Andreasen N., Öst M., Zetterberg H., Nellgård B., and Blennow K. (2016). Serum neurofilament light protein predicts clinical outcome in traumatic brain injury. Sci. Rep. 6, 36791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vos P.E., Jacobs B., Andriessen T.M.J.C., Lamers K.J.B., Borm G.F., Beems T., Edwards M., Rosmalen C.F., and Vissers J.L.M. (2010). GFAP and S100B are biomarkers of traumatic brain injury: an observational cohort study. Neurology 75, 1786–1793 [DOI] [PubMed] [Google Scholar]
- 15. Diaz-Arrastia R., Wang K.K., Papa L., Sorani M.D., Yue J.K., Puccio A.M., McMahon P.J., Inoue T., Yuh E.L., Lingsma H.F., Maas A.I., Valadka A.B., Okonkwo D.O., Manley G.T., and TRACK-TBI Investigators. (2014). Acute biomarkers of traumatic brain injury: relationship between plasma levels of ubiquitin C-terminal hydrolase-L1 and glial fibrillary acidic protein. J. Neurotrauma 31, 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bazarian J.J., Blyth B.J., He H., Mookerjee S., Jones C., Kiechle K., Moynihan R., Wojcik S.M., Grant W.D., Secreti L.M., Triner W., Moscati R., Leinhart A., Ellis G.L., and Khan J. (2013). Classification accuracy of serum Apo A-I and S100B for the diagnosis of mild traumatic brain injury and prediction of abnormal initial head computed tomography scan. J. Neurotrauma 30, 1747–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shan R., Szmydynger-Chodobska J., Mohammad Farah J.Z., and Chodobski A. (2015). A new panel of blood biomarkers for the diagnosis of mild traumatic brain injury/concussion in adults. J. Neurotrauma 33, 49–57 [DOI] [PubMed] [Google Scholar]
- 18. O'Bryant S.E., Xiao G., Barber R., Reisch J., Doody R., Fairchild T., Adams P., Waring S., and Diaz-Arrastia R. (2010). A serum protein–based algorithm for the detection of Alzheimer disease. Arch. Neurol. 67, 1077–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O'Bryant S.E., Xiao G., Barber R., Huebinger R., Wilhelmsen K., Edwards M., Graff-Radford N., Doody R., Diaz-Arrastia R. Texas Alzheimer's Research & Care Consortium, Alzheimer's Disease Neuroimaging Initiative. (2011). A blood-based screening tool for Alzheimer's disease that spans serum and plasma: findings from TARC and ADNI. PLoS One 6, e28092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barber R.C., Phillips N.R., Tilson J.L., Huebinger R.M., Shewale S.J., Koenig J.L., Mitchel J.S., O'Bryant S.E., Waring S.C., Diaz-Arrastia R., Chasse S., Wilhelmsen K.C., Initiative F.T.A.D.N., Research T.T.A., and Consortium C. (2015). Can genetic analysis of putative blood Alzheimer's disease biomarkers lead to identification of susceptibility loci? PLoS One 10, e0142360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bagiella E., Novack T.A., Ansel B., Diaz-Arrastia R., Dikmen S., Hart T., and Temkin N. (2010). Measuring outcome in traumatic brain injury treatment trials: recommendations from the traumatic brain injury clinical trials network. J. Head Trauma Rehabil. 25, 375–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Manley G.T., Diaz-Arrastia R., Brophy M., Engel D., Goodman C., Gwinn K., Veenstra T.D., Ling G., Ottens A.K., Tortella F., and Hayes R.L. (2010). Common Data Elements for Traumatic Brain Injury: recommendations from the Biospecimens and Biomarkers Working Group. Arch. Phys. Med. Rehabil. 91, 1667–1672 [DOI] [PubMed] [Google Scholar]
- 23. Duhaime A.-C., Gean A.D., Haacke E.M., Hicks R., Wintermark M., Mukherjee P., Brody D., Latour L., Riedy G., Common Data Elements Neuroimaging Working Group Members, and Pediatric Working Group Members. (2010). Common data elements in radiologic imaging of traumatic brain injury. Arch. Phys. Med. Rehabil. 91, 1661–1666 [DOI] [PubMed] [Google Scholar]
- 24. Wilde E.A., Whiteneck G.G., Bogner J., Bushnik T., Cifu D.X., Dikmen S., French L., Giacino J.T., Hart T., Malec J.F., Millis S.R., Novack T.A., Sherer M., Tulsky D.S., Vanderploeg R.D., and Steinbuechel von N. (2010). Recommendations for the use of common outcome measures in traumatic brain injury research. Arch. Phys. Med. Rehabil. 91, 1650–1660.e17 [DOI] [PubMed] [Google Scholar]
- 25. Teasdale G., and Jennett B. (1974). Assessment of coma and impaired consciousness. A practical scale. Lancet 2, 81–84 [DOI] [PubMed] [Google Scholar]
- 26. Wilson J.T.L., Pettigrew L.E.L., and Teesdale G.M. (1998). Structured interviews for the Glasgow Outcome Scale and the extended Glasgow Outcome Scale: guidelines for their use. J. Neurotrauma 15, 573–585 [DOI] [PubMed] [Google Scholar]
- 27. Alali A.S., Vavrek D., Barber J., Dikmen S., Nathens A.B., and Temkin N.R. (2015). Comparative study of outcome measures and analysis methods for traumatic brain injury trials. J. Neurotrauma 32, 581–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cattell R.B. (1969). Comparing factor trait and state scores across ages and cultures. J. Gerontol. 24, 348–& [DOI] [PubMed] [Google Scholar]
- 29. Stevens J.P. (2012). Applied Multivariate Statistics for the Social Sciences, Fifth Edition. New York: Routeledge [Google Scholar]
- 30. Efron B. (1979). (1979a). Bootstrap methods: another look at the jackknife. Ann. Stat. 7, 1–26 [Google Scholar]
- 31. Cervellin G., Benatti M., Carbucicchio A., Mattei L., Cerasti D., Aloe R., and Lippi G. (2012). Serum levels of protein S100B predict intracranial lesions in mild head injury. Clin. Biochem. 45, 408–411 [DOI] [PubMed] [Google Scholar]
- 32. Korley F.K., Yue J.K., Wilson D., Hrusovsky K., Diaz-Arrasta R., Ferguson A.R., Yuh E.L., Mukherjee P., Wang K.K.W., Valadka A., Puccio A., Okonkwo D.O., and Manley G. (2018). Performance evaluation of a multiplex assay for simultaneous detection of four clinically relevant TBI biomarkers. J. Neurotrauma [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ferguson A.R., Irvine K.-A., Gensel J.C., Nielson J.L., Lin A., Ly J., Segal M.R., Ratan R.R., Bresnahan J.C., and Beattie M.S. (2013). Derivation of multivariate syndromic outcome metrics for consistent testing across multiple models of cervical spinal cord injury in rats. PLoS One 8, e59712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ferguson A.R., Stück E.D., and Nielson J.L. (2011). Syndromics: a bioinformatics approach for neurotrauma research. Transl. Stroke Res. 2, 438–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nielson J.L., Paquette J., Liu A.W., Guandique C.F., Tovar C.A., Inoue T., Irvine K.-A., Gensel J.C., Kloke J., Petrossian T.C., Lum P.Y., Carlsson G.E., Manley G.T., Young W., Beattie M.S., Bresnahan J.C., and Ferguson A.R. (2015). Topological data analysis for discovery in preclinical spinal cord injury and traumatic brain injury. Nat. Commun. 6, 8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saatman K.E., Duhaime A.-C., Bullock R., Maas A.I.R., Valadka A., Manley G.T., and Workshop Scientific Team and Advisory Panel Members. (2008). Classification of Traumatic Brain Injury for Targeted Therapies. Mary Ann Liebert, Inc.: New Rochelle: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yue J.K., Vassar M.J., Lingsma H.F., Cooper S.R., Okonkwo D.O., Valadka A.B., Gordon W.A., Maas A.I.R., Mukherjee P., Yuh E.L., Puccio A.M., Schnyer D.M., Manley G.T., including T.-T.I., Casey S.S., Cheong M., Dams-O'Connor K., Hricik A.J., Knight E.E., Kulubya E.S., Menon D.K., Morabito D.J., Pacheco J.L., and Sinha T.K. (2013). Transforming research and clinical knowledge in traumatic brain injury pilot: multicenter implementation of the Common Data Elements for Traumatic Brain Injury. J. Neurotrauma 30, 1831–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nielson J.L., Cooper S.R., Yue J.K., Sorani M.D., Inoue T., Yuh E.L., Mukherjee P., Petrossian T.C., Paquette J., Lum P.Y., Carlsson G.E., Vassar M.J., Lingsma H.F., Gordon W.A., Valadka A.B., Okonkwo D.O., Manley G.T., Ferguson A.R., and TRACK-TBI Investigators. (2017). Uncovering precision phenotype-biomarker associations in traumatic brain injury using topological data analysis. PLoS One 12, e0169490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stetler-Stevenson W.G. (2008). Tissue inhibitors of metalloproteinases in cell signaling: metalloproteinase-independent biological activities. Sci. Signal. 1, re6–re6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lorente L., Martín M.M., López P., Ramos L., Blanquer J., Cáceres J.J., Solé-Violán J., Solera J., Cabrera J., Argueso M., Ortiz R., Mora M.L., Lubillo S., Jiménez A., Borreguero-León J.M., González A., Orbe J., Rodríguez J.A., and Páramo J.A. (2014). Association between serum tissue inhibitor of matrix metalloproteinase-1 levels and mortality in patients with severe brain trauma injury. PLoS One 9, e94370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kossmann T., Hans V.H.J., Imhof H.-G., Stocker R., Grob P., Trentz O., and Morganti-Kossmann M.C. (1995). Intrathecal and serum interleukin-6 and the acute-phase response in patients with severe traumatic brain injuries. Shock 4, 311. [DOI] [PubMed] [Google Scholar]
- 42. Woiciechowsky C., Schöning B., Cobanov J., Lanksch W.R., Volk H.-D., and Döcke W.-D. (2002). Early IL-6 plasma concentrations correlate with severity of brain injury and pneumonia in brain-injured patients. J. Trauma Acute Care Surg. 52, 339. [DOI] [PubMed] [Google Scholar]
- 43. Glabinski A.R., Balasingam V., Tani M., Kunkel S.L., Strieter R.M., Yong V.W., and Ransohoff R.M. (1996). Chemokine monocyte chemoattractant protein-1 is expressed by astrocytes after mechanical injury to the brain. J. Immunol. 156, 4363–4368 [PubMed] [Google Scholar]
- 44. Semple B.D., Bye N., Rancan M., Ziebell J.M., and Morganti-Kossmann M.C. (2009). Role of CCL2 (MCP-1) in traumatic brain injury (TBI): Evidence from severe TBI patients and CCL2−/− mice. J. Cereb. Blood Flow Metabol. 30, 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ho L., Zhao W., Dams-O'Connor K., Tang C.Y., Gordon W., Peskind E.R., Yemul S., Haroutunian V., and Pasinetti G.M. (2012). Elevated plasma MCP-1 concentration following traumatic brain injury as a potential “predisposition” factor associated with an increased risk for subsequent development of Alzheimer's disease. J. Alzheimers Dis. 31, 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Anderson R.E., Hansson L.-O., Nilsson O., Dijlai-Merzoug R., and Settergren G. (2001). High serum S100B levels for trauma patients without head injuries. Neurosurgery 48, 1255–1260 [DOI] [PubMed] [Google Scholar]
- 47. Yang G., Hu R., Zhang C., Qian C., Luo Q.-Q., Yung W.-H., Ke Y., Feng H., and Qian Z.-M. (2016). A combination of serum iron, ferritin and transferrin predicts outcome in patients with intracerebral hemorrhage. Sci. Rep. 6, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Williams D.H., Levin H.S., and Eisenberg H.M. (1990). Mild head injury classification. Neurosurgery 27, 422–428 [DOI] [PubMed] [Google Scholar]
- 49. Murray G.D., Butcher I., McHugh G.S., Lu J., Mushkudiani N.A., Maas A.I.R., Marmarou A., and Steyerberg E.W. (2007). Multivariable prognostic analysis in traumatic brain injury: results from the IMPACT study. J. Neurotrauma 24, 329–337 [DOI] [PubMed] [Google Scholar]
- 50. Maas A.I.R., Lingsma H.F., and IMPACT Study Group. (2008). New approaches to increase statistical power in TBI trials: insights from the IMPACT study. Acta Neurochir. Suppl. 101, 119–124 [DOI] [PubMed] [Google Scholar]
- 51. Lingsma H., Andriessen T.M.J.C., Haitsema I., Horn J., van der Naalt J., Franschman G., Maas A.I.R., Vos P.E., and Steyerberg E.W. (2013). Prognosis in moderate and severe traumatic brain injury: external validation of the IMPACT models and the role of extracranial injuries. J. Trauma Acute Care Surg. 74, 639–646 [DOI] [PubMed] [Google Scholar]
- 52. Roozenbeek B., Chiu Y.-L., Lingsma H.F., Gerber L.M., Steyerberg E.W., Ghajar J., and Maas A.I.R. (2012). Predicting 14-day mortality after severe traumatic brain injury: application of the IMPACT models in the Brain Trauma Foundation TBI-trac® New York State Database. J. Neurotrauma 29, 1306–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.