Abstract
The transmission of some schistosome parasites is dependent on the planorbid snail hosts. Bulinus truncatus is important in urinary schistosomiasis epidemiology in Africa. Hence, there is a need to define the snails’ phylogeography. This study assessed the population genetic structure of B. truncatus from Giza and Sharkia (Egypt), Barakat (Sudan) and Madziwa, Shamva District (Zimbabwe) using mitochondrial cytochrome oxidase subunit 1 gene (COI) and internal transcribed spacer 1 (ITS 1) markers. COI was sequenced from 94 B. truncatus samples including 38 (Egypt), 36 (Sudan) and 20 (Zimbabwe). However, only 51 ITS 1 sequences were identified from Egypt (28) and Sudan (23) (because of failure in either amplification or sequencing). The unique COI haplotypes of B. truncatus sequences observed were 6, 11, and 6 for Egypt, Sudan, and Zimbabwe, respectively. Also, 3 and 2 unique ITS 1 haplotypes were observed in sequences from Egypt and Sudan respectively. Mitochondrial DNA sequences from Sudan and Zimbabwe indicated high haplotype diversity with 0.768 and 0.784, respectively, while relatively low haplotype diversity was also observed for sequences from Egypt (0.334). The location of populations from Egypt and Sudan on the B. truncatus clade agrees with the location of both countries geographically. The clustering of the Zimbabwe sequences on different locations on the clade can be attributed to individuals with different genotypes within the population. No significant variation was observed within B. truncatus populations from Egypt and Sudan as indicated by the ITS 1 tree. This study investigated the genetic diversity of B. truncatus from Giza and Sharkia (Egypt), Barakat area (Sudan), and Madziwa (Zimbabwe), which is necessary for snail host surveillance in the study areas and also provided genomic data of this important snail species from the sampled countries.
Keywords: phylogeography, Bulinus truncatus, planorbidae, Africa
1. Background
The snail intermediate hosts of the genus Bulinus play active roles in the epidemiology of urinary schistosomiasis. The schistosome parasites depend on these snails for the development of the asexual phase of their life cycle before the cercariae are released into the water bodies to look for unsuspecting human hosts for penetration, where they continue the sexual phase of their development [1,2,3,4].
Members of the genus Bulinus are hermaphroditic planorbid snails, and this genus includes 37 recognized species distributed in the tropic and sub-tropic regions of the world including Africa, Mediterranean countries, and parts of the Middle East [5]. They differ in their interaction with schistosome parasites and some are involved in the transmission of human and animal schistosomiasis [6].
These important snail species inhabit various types of freshwater bodies such as streams, ponds, rivers, and irrigation canals [7]. The genetic structure of snail hosts is mostly determined by their habitat distribution, which is largely influenced by the spatial and temporal fluctuations in water availability [8,9] leading to population bottlenecks [5].
Snails belonging to the Bulinus group have a great capacity to rapidly increase their population size through cross- or self-fertilization, but B. truncatus has a preference for self-fertilization [10]. Selfing and population bottlenecks increase genetic differentiation among snail population but reduce the amount of genetic diversity within a population [11].
Whilst morphological identification of snails helps with identifying snails at group or genus level, it cannot give further insights about their interaction with the parasites [12].
Assessment of snail hosts population structure using molecular markers and other genetic tools creates a robust system for species identification and differentiation [8,12,13,14,15,16,17]. This provides useful information about their genetic diversity and detailed elucidation of the host–parasite relationship [4], which can be applied to target effective integrated schistosomiasis control strategies in most endemic areas [18].
The use of different markers such as COI, microsatellites and ITS 1, has helped to achieve identification of B. truncatus sampled from few African countries including Senegal, Niger, Tanzania, Burkina Faso, and Cameroon [11,12]. Studies have also observed strong population subdivision and low diversity for hermaphroditic freshwater snails including B. truncatus [11,19,20,21,22,23].
It is, therefore, imperative to provide information on the diversity of important snail hosts including B. truncatus through assessing their phylogenic status in most countries endemic for schistosomiasis across Africa, to further improve our understanding about their phylogenetic relationships as well as the disease epidemiology.
This study provided information on the phylogeography of B. truncatus populations from Giza and Sharkia (Egypt), Barakat area (Sudan), and Madziwa, Shamva District (Zimbabwe), using partial mitochondrial DNA cytochrome oxidase subunit I (COI) and internal transcribed spacer 1 (ITS 1) to determine their phylogenetic relationship, which is important for epidemiological investigation and snail hosts surveillance.
2. Materials and Methods
2.1. Sample Collection
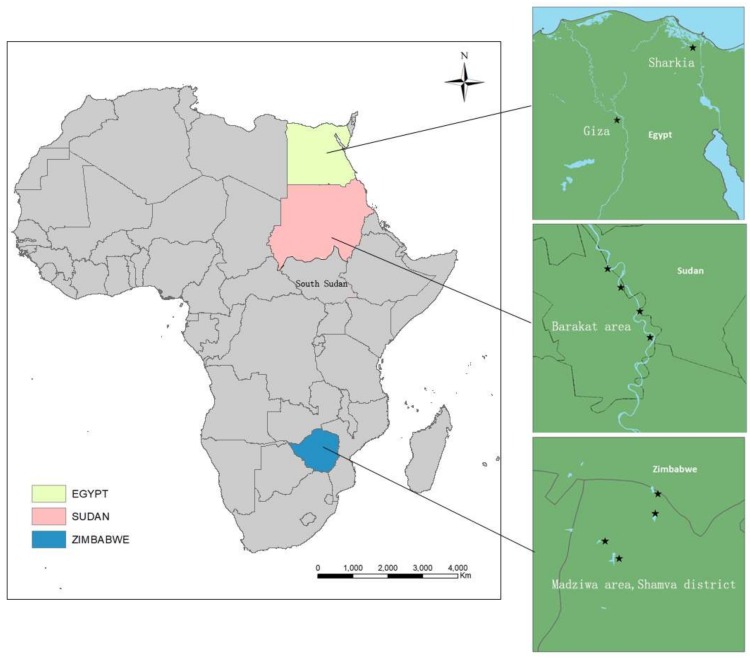
Bulinus snails were collected from different locations in freshwater bodies at Giza and Sharkia governorates (Egypt), Barakat area (Sudan) and Madziwa area, Shamva District (Zimbabwe). Snail sampling was done at selected sites along water bodies; these included water contact sites where people swim, carry out fishing activities, collect water for domestic purposes, bathing, and washing clothes and utensils. Sites with no apparent human water contact activities were also visited for snail collection. A total of 134 Bulinus snails was assessed from different locations across the three countries.
The snails were identified phenotypically using shell morphology [24]. Snails were then preserved in absolute ethanol. Information that includes snail collection and geographic coordinates of the study areas are shown in Table 1. A map of study areas is shown in Figure 1.
Table 1.
Geographic coordinates of the study areas.
| Country | Location | No. of Samples Collected | Time of Collection | Type of Water Body | Latitude | Longitude |
|---|---|---|---|---|---|---|
| Egypt | Giza (El-Nile river, Gezerite El-Warrak) | 25 | October, 2016 | River | 30.102 | 31.243 |
| Sharkia (El-Salam canal, El-Hesenia district) | 30 | November, 2016 | Canal | 31.258 | 32.270 | |
| Sudan | Barakat area, Wad Madani | 14 | July, 2016 | Canal | 14.33673 | 33.52736 |
| Barakat area, Wad Madani | 22 | August, 2016 | Canal | 14.31780 | 33.53167 | |
| Barakat area, Wad Madani | 5 | August, 2016 | Canal | 14.29210 | 33.55261 | |
| Barakat area, Wad Madani | 8 | August, 2016 | Canal | 14.25122 | 33.59070 | |
| Zimbabwe | Madziwa, Shamva District | 11 | March, 2016 | River | 16.93642 | 31.44603 |
| Madziwa | 6 | March, 2016 | River | 16.91498 | 31.42868 | |
| Madziwa | 10 | June, 2016 | River | 16.85695 | 31.49413 | |
| Madziwa | 3 | June, 2016 | River | 16.88070 | 31.49083 |
Figure 1.
Map of Africa showing countries where snail samples were collected.
2.2. Sample Preparation and DNA Extraction
The specimens were recounted, identified by morphological characters, and re-spirited (absolute ethanol) upon arrival at the National Institute of Parasitic Diseases, Shanghai, schistosomiasis laboratory [25]. Specimens were placed in TE buffer (10 mM Tris, 0.1 mM EDTA) pH 7.4 for 1 h to remove the remaining alcohol from within the tissue, which might interfere with subsequent extraction techniques. Total genomic DNA was isolated from head-foot snail tissue using the DNeasy Blood and Tissue kit (Qiagen, Crawley, UK) according to the manufacturer’s instructions. DNA was eluted into 200 μL AE buffer. The snails’ genomic DNA concentration was quantified using the Nanodrop ND-1000 Spectrophotometer (Nanodrop Technologies Inc., Thermo Fisher Scientific, Wilmington, DE, USA).
2.3. Polymerase Chain Reaction (PCR) Amplification of COI and ITS 1 Fragments
IllustraPuRe Taq Ready-To-Go PCR beads (GE Healthcare) were used for the amplification of the COI and ITS 1 fragments using the methods outlined in Kane et al. [12] with 0.4 µM each of Bulcox 5 (5′CCT TTA AGA GGN CCT ATT GC 3′) (forward primer) and Bulcox 14 (5′GGA AAT CAG TAM AYA AAA CCA GC 3′) (reverse primer) while ETTS10 (5′ GCA TAC TGC TTT GAA CAT CG 3′) (forward primer) and ETTS1(5′GC TTA AGT TCA GCG GGT 3′) (reverse primer) were used for B. truncatus amplification. A DNA template of 1 µL was added to each tube that contained 22 µL double distilled water, 1 µL each of forward and reverse primers. The total reaction volume was 25 µL. Double distilled water was used as the negative control. PCR amplification of snail genomic DNA was done using Applied Biosystems GeneAmp Thermal Cycler 2700 version 2.08. Cycling conditions for COI and ITS 1 reactions are as follows: one cycle of 95 °C for 5 min, 45 cycles of 95 °C for 30 s, 54.3 °C for 30 s, 72 °C for 45 s and 72 °C for 10 min and one cycle of 95 °C for 5 min, 45 cycles of 95 °C for 30 s, 42 °C for 30 s, 72 °C for 45 s and 72 °C for 10 min respectively. PCR fragments were separated on 1% agarose gel and visualization was performed using a gel documentation and analysis system (UVP, EpiChem II darkroom). Sequencing was performed on an Applied Biosystems 3730XL analyser (Life Technologies, Northumberland, UK).
2.4. Phylogenetic Analysis of Sequence Data
Nucleotide sequences were visually edited using Bioedit software v 7.0. [26]. BLAST searches via the National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) were performed for the obtained sequences against Genbank database to ensure that contaminant sequences had not been obtained by error [27] and aligned with the reference materials [12] using the Clustal W algorithm [28]. We performed the maximum-likelihood analyses for the COI and ITS 1 sequences using the program RAxML [29]. The maximum-likelihood estimates were bootstrapped for 1000 replicates based on the GTRGAMMA substitution model. Downloaded B. truncatus sequences deposited in Genbank from Niger (AM286316.2), Senegal (AM921807.1 and AM921806.1) Portugal (AM286314), Italy (AM286312.3), Burkina Faso (AM286315.2), and Tanzania (AM286313.2) [12] were used as reference isolates for COI sequences, while B. truncatus isolates from Tanzania (AM921983), Niger (AM921965) [16], and Cameroon (KJ157504.1, KJ157503.1, KJ157500.1, KJ157501.1, KJ157502.1) [13] were used as reference isolates for ITS 1 sequences. Sequence data from other Bulinus species on Genbank (detailed information on accession number and origin provided as Supplementary Data) were also included in constructing the maximum likelihood phylogenetic trees. Additionally, Bulinus forskalii (AM286306.2) was used as an outgroup for Bulinus truncatus group assessed with COI marker. Bulinus forskalii (AM921961.1) was used as the outgroup for the Bulinus truncatus group assessed with ITS 1 marker.
We also estimated the phylogenetic relationships of the COI and ITS 1 B. truncatus dataset using Bayesian inference in MrBayes version 3.2.0 programs [30] (Figures S1 and S2). Prior to Bayesian inference, the best fit nucleotide substitution models (HKY for COI and TrN for ITS 1) were determined using a hierarchical likelihood ratio test in jMODELTEST version 0.1.1 [31]. The posterior probabilities were calculated via 1,000,000 generations using Markov chain Monte Carlo (MCMC) simulations, and the chains were sampled every 1000 generations. At the end of this run, the average standard deviation of split frequencies was below 0.01, and the potential scale reduction factor was reasonably close to 1.0 for all parameters. A consensus tree was summarized and visualized in FigTree version 1.4.3 [32].
B. truncatus sequences from the three populations, reference isolates, and other Bulinus species sequences used for constructing ML trees were repeated for the construction of the Bayes ML trees Clade comprising B. forskalii (AM286308, AM286293.2, and AM286306.2) was used as the outgroup for COI B. truncatus sequences while B. forskalii (AM921961.1) was used the outgroup for ITS 1 B. truncatus sequences.
The minimum spanning tree was built using NETWORK 5.0.0.0 [33] (Figure S3). We built the network to support the COI ML tree and it showed the torso of the genetic structure.
DNA sequences have been submitted to the National Centre for Biotechnology Information Archive with accession numbers MG759386–MG759479 (B. truncatus group assessed with COI marker) and MG757840-757890 (B. truncatus group assessed with ITS 1 marker).
2.5. Determination of Haplotype and Nucleotide Diversity
The level of sequence diversity, which includes number of haplotype (h), haplotype diversity (hd), nucleotide diversity (π), Tajima’s D (D), and theta per site statistics, were calculated for B. truncatus populations assessed with both COI and ITS 1 markers in Arlequin software version 3.5 [34]. In addition, we compared Fst of B. truncatus studied populations using Arlequin software version 3.5 [34].
3. Results
3.1. Phylogeny
Altogether, 94 individual snail samples including 38 (Giza and Sharkia; Egypt), 36 (Barakat area; Sudan), and 20 (Madziwa, Shamva District; Zimbabwe) were successfully sequenced at the COI region (Table 2), and 51 including 28 (Egypt) and 23 (Sudan) were sequenced at the ITS 1 locus (Table 2), because of failure in either amplification or sequencing. Following the sequencing alignment and trimming of all the sequences, the final fragments of 737 bp (COI) and 580 bp (ITS 1) were obtained. Among these, 6, 11, and 6 unique COI haplotypes of B. truncatus sequences were observed from Giza and Sharkia (Egypt), Barakat area (Sudan) and Madziwa, Shamva district (Zimbabwe), respectively (Table 2). In Egypt and Sudan, respectively, 3 and 2 unique ITS 1 haplotypes of B. truncatus sequences were observed (Table 2). No information on B. truncatus ITS 1 sequences from Zimbabwe was recorded in this study due to failure in either amplification or sequencing.
Table 2.
Estimation of nucleotide diversity and summary statistics of Bulinus truncatus identified using COI I and ITS 1 markers.
| N | H | Hd | π | S.D.π | ΘS | s.d.S | Tajima’s D | p-Value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| COI | Egypt | 38 | 6 | 0.334 | 0.00205 | 0.001445 | 0.002916 | 0.001299 | −0.85621 | 0.24 |
| Sudan | 36 | 11 | 0.768 | 0.009359 | 0.005067 | 0.009602 | 0.003341 | −0.08761 | 0.538 | |
| Zimbabwe | 20 | 6 | 0.784 | 0.014701 | 0.007859 | 0.011655 | 0.004442 | 1.01745 | 0.898 | |
| ITS 1 | Egypt | 28 | 3 | 0.14 | 0.005589 | 0.003362 | 0.020107 | 0.006909 | −2.69592 | 0 |
| Sudan | 23 | 2 | 0.443 | 0.00169 | 0.001367 | 0.001034 | 0.000769 | 1.41416 | 0.923 |
Number of sequences (N), number of haplotypes (h), haplotype diversity (Hd), nucleotide diversity (pi), theta per site (ΘS), standard deviation (s.d.).
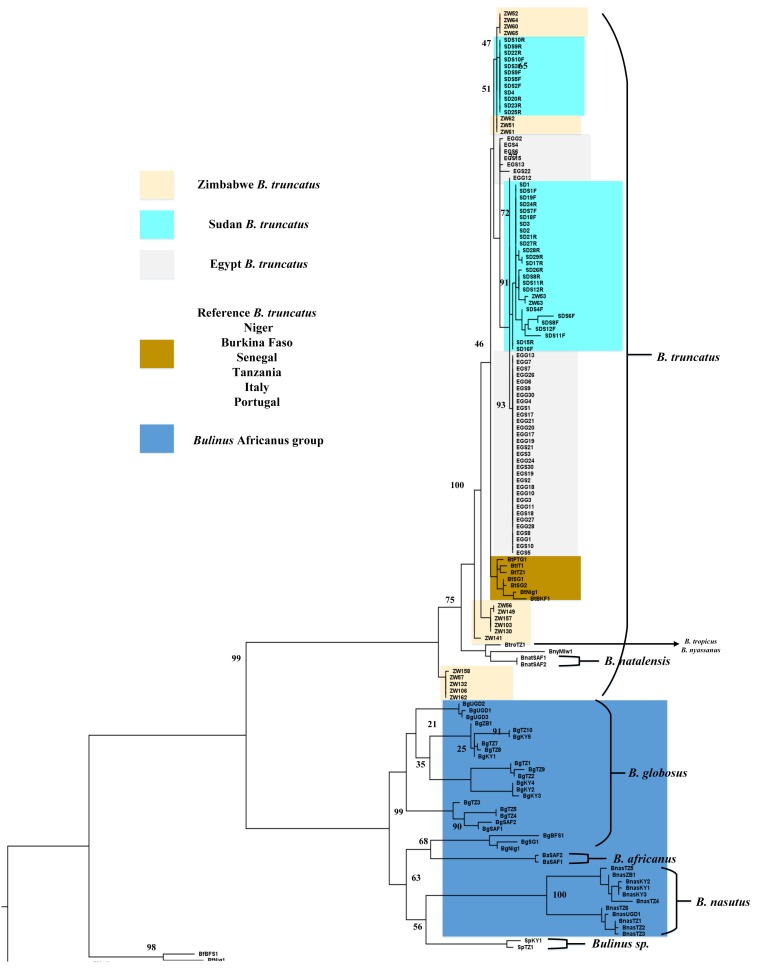
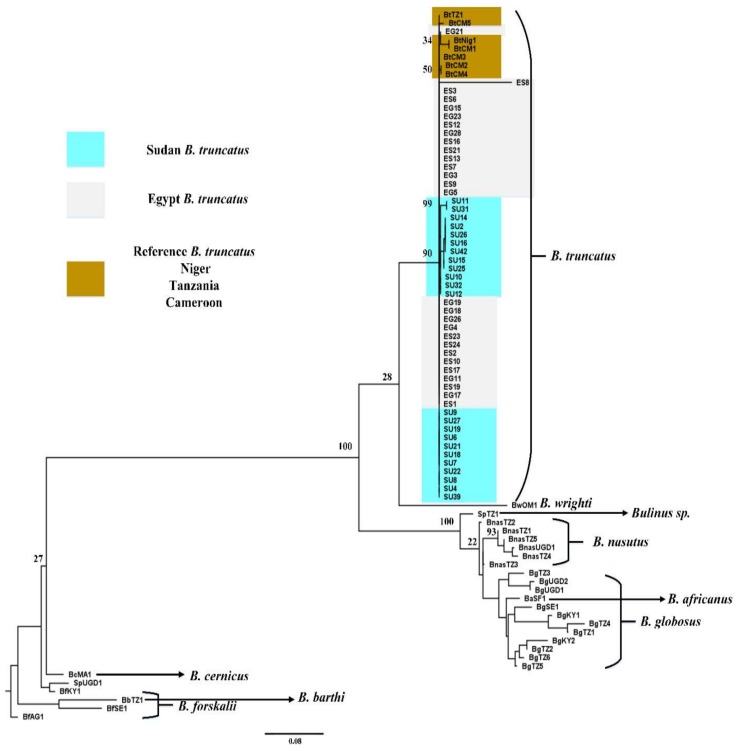
Phylogenetic analyses indicated some measures of variation in the genetic population structure of B. truncatus population from Giza and Sharkia (Egypt), Barakat area (Sudan), and Madziwa (Zimbabwe). A large quantity of COI sequence data from Giza and Sharkia (Egypt) and Barakat area (Sudan) cluster together on the B. truncatus clade but the COI sequence data of Madziwa (Zimbabwe) B. truncatus population cluster at different locations on the tree (Figure 2 and Figure S1). No significant variation was observed between B. truncatus populations from Giza and Sharkia (Egypt) and Barakat area (Sudan) (Figure 3 and Figure S2). The minimum spanning tree indicated the torso of B. truncatus populations genetic structure (Figure S3). The tree showed five haplotypes for B. truncatus obtained from Zimbabwe, while five and two haplotypes were indicated for Sudan and Egypt B. truncatus populations respectively. Cryptic lineages or other known species of B. truncatus were not detected.
Figure 2.
A rooted maximum likelihood tree of Bulinus truncatus for CO1 sequences. Maximum likelihood tree of a 737 bp fragment of the cytochrome oxidase subunit 1 (CO1) gene for B. truncatus in this study with an additional 51 published Genbank sequences including B. truncatus reference isolates. Values on the branches are bootstrap support based on 1000 replications. B. forskalii (AM286306.2) was defined as outgroup. * UGD-Uganda, TZ-Tanzania, SAF-South Africa, KY-Kenya, ZB-Zanzibar, SG-Senegal, BFS, Burkina Faso, Nig-Niger, Agl-Angola, Mlw-Malawi.
Figure 3.
A rooted maximum likelihood tree of Bulinus truncatus for ITS 1 sequences. Maximum likelihood tree of a 580 bp fragment of the internal transcribed spacer 1 (ITS 1) for B. truncatus in this study with an additional 33 published Genbank sequences including B. truncatus reference isolates. Values on the branches are bootstrap support based on 1000 replications. B. forslkalii (AM921961.1) was defined as outgroup. * UGD-Uganda, TZ-Tanzania, SF-South Africa, KY-Kenya, SE-Senegal, BFS, Burkina Faso, Nig-Niger, AG-Angola, MA-Mauritius, OM-Oman.
3.2. Haplotype and Nucleotide Diversity
Nucleotide polymorphism obtained for all B. truncatus sequences assessed in this study is shown in Table 2. Haplotype diversity observed of sequences from the three populations assessed with COI includes 0.334 (Giza and Sharkia, Egypt), 0.768 (Barakat area, Sudan), and 0.784 (Madziwa, Zimbabwe) while their nucleotide diversity includes 0.00205 (s.d. = 0.001445), 0.009359 (s.d. = 0.005067), and 0.014701 (s.d. = 0.007859) for Egypt, Sudan, and Zimbabwe, respectively (Table 2). Haplotype diversity for the two populations assessed with ITS 1 is 0.14 and 0.443 while nucleotide diversity includes 0.005589 (s.d. π 0.003362) and 0.00169 (s.d. π 0.001367) for (Giza and Sharkia, Egypt) and (Barakat area, Sudan), respectively. FST values in the pairwise population comparisons are shown in Table 3.
Table 3.
Population pairwise Φst.
| Egypt | Sudan | |
|---|---|---|
| Sudan | 0.34860 * | |
| Zimbabwe | 0.59653 * | 0.36160 * |
* Significant p value < 0.05.
4. Discussion
Urinary schistosomiasis burden is widely reported in Africa and this is a consequence of the unabated distribution of the important snail intermediate hosts of the genus Bulinus that serves as host to the schistosome parasite [4]. The Bulinus group is made up of about 37 recognized species and has been divided into four different groups for convenience [5].
This study assessed the genetic diversity of B. truncatus populations, the snail host implicated in the transmission of S. haematobium in Africa using mitochondrial cytochrome oxidase 1 (COI) gene and internal transcribed spacer (ITS 1).
Snail host identification using morphological characters is unreliable and sometimes ambiguous but the development and application of molecular techniques have been helpful, providing good species discrimination [4,5].
We observed that mitochondrial DNA sequences from Barakat area (Sudan) and Madziwa, Shamva (Zimbabwe) indicated high haplotype diversity including 0.768 and 0.784, respectively, a similar observation was earlier reported by Zein-Eddine et al. [14]. The level of haplotype diversity observed from the two populations in our study is less than the values reported by Zein-Eddine et al. [14]. However, relatively low haplotype diversity was also observed for sequences from Giza and Sharkia (Egypt), with 0.334. Nevertheless, the low levels of haplotype diversity observed for B. truncatus in this study were similar to findings by Zein-Eddine et al. [11] and those observed by Goodall-Copestake et al. [35,19], with low diversity species.
Our findings indicated some degree of variation in the Bulinus species population structure across Africa. Bulinus species populations on both COI and ITS 1 trees (Figure 2 and Figure 3, Figures S1 and S2) separated into populations that correspond to Bulinus species groups [12,36]. However, we observed that B. globosus from West Africa clusters separately from the East African species as indicated by the COI sequence data.
Kane et al. [12] reported the division between B. globosus from the two regions in Africa and that Bulinus africanus has a close affinity with West African B. globosus species. This is also evident from the information provided by our COI sequence data (Figure 2).
Some levels of segregation were observed within the COI B. truncatus populations. The location of populations from Giza and Sharkia (Egypt) and Barakat area (Sudan) on the B. truncatus clade agrees with the location of both countries geographically (Figure 2). The clustering of the Madziwa, Shamva (Zimbabwe) sequences in different locations on the B. truncatus clade can be attributed to individuals with different genotypes within the population (Figure 2).
Findings from this study using COI identified two reciprocally monophyletic B. truncatus sister subclades and this corresponds to B. truncatus and B. tropicus respectively [12,36]. Nalugwa et al. [36] also obtained similar results from findings on the B. truncatus/tropicus complex collected from Albertine Rift freshwater bodies in Uganda. Brown and Shaw [37] have shown that B. truncatus is a tetraploid and B. tropicus is diploid; however, it is difficult to distinguish B. truncatus and B. tropicus morphologically.
The minimum spanning network was constructed to support the ML tree and this informed our decision to include some outgroup sequences. The network did not indicate a substantial difference from the information on the tree (Figure S3).
Although Bulinus species populations separated distinctly into groups as earlier indicated [12], no significant variation was observed within B. truncatus populations from Giza and Sharkia (Egypt) and Barakat area (Sudan) as indicated by the ITS 1 tree (Figure 3). Kane et al. [12] stated that Bulinus wrighti has a characteristic COI sequence that positions the species and other members of the Bulinus reticulatus group close to the Bulinus truncatus complex. This was also observed from our ITS 1 result (Figure 3).
This study is not unique; however, it has investigated the genetic diversity of B. truncatus from Giza and Sharkia (Egypt), Barakat area (Sudan), and Madziwa (Zimbabwe), which is necessary for snail host surveillance in the study areas and it has also provided genomic data of this important snail species from the sampled countries.
Although no infection was detected from the snails when screened for patent and prepatent infection, the presence and distribution of B. truncatus in the studied areas poses a threat to the inhabitants of these areas should an infected person visit the water bodies and urinate inside or near enough for the schistosome eggs released with the urine to have contact with the water bodies, especially water contact sites such as the Nile River and El-Salam Canal (Egypt), where inhabitants engage in a lot of fishing activities, and the river at Madziwa (Zimbabwe) that people visit frequently to carry out their domestic chores as well as engage in activities such as swimming.
Previous studies implicated B. truncatus as the only Bulinus species that transmits S. haematobium in Egypt and other parts of northern Africa, while Bulinus globosus is implicated for schistosomiasis transmission in Zimbabwe [4]. The presence of B. truncatus in the southern African country can be attributed to the favorable environmental factors and migration of snail population; however, human activities have also increased the number of snail hosts of S. haematobium. This is a cause for concern and there is a need to improve measures for effective snail control strategies.
Differentiating snail host populations to assess their diversity should be prioritized in Africa, where host snails’ genome data is scarce for most schistosomiasis endemic countries [18]. Efforts should be made to initiate a continent-wide snail host genome project to help develop a more comprehensive and robust snail host genome database for the African continent.
5. Conclusions
This study identified B. truncatus, the snail host of S. haematobium obtained from Giza and Sharkia (Egypt), Barakat area (Sudan), and Madziwa, Shamva District (Zimbabwe) using COI and ITS 1 markers, as well as provided information on their genetic diversity.
With the increasing global call that effective schistosomiasis control programmes should target snail control, there is a need to prioritize snail studies for effective mapping of schistosomiasis transmission [38], as well as to strengthen surveillance strategies.
Acknowledgments
E.M.A. acknowledges the funding and support received from the National Institute of Parasitic Diseases, WHO Collaborating Centre for Tropical Diseases, and the China CDC for the postdoctoral fellowship.
Supplementary Materials
The following are available online at http://www.mdpi.com/2414-6366/3/4/127/s1. S1A–S1J, data were used to prepare Table 2, S1A: Egypt B. truncatus COI.out, S1B: Egypt B. truncatus COI Tajima D.out, S1C: Egypt B. truncatus Fu_Li Test.out, S1D: Egypt_SUD_ZWE B. truncatus COI final.out, S1E: Fu_Li Egypt_SUD_ZWE B. truncatus COI.out, S1F: Sudan B. truncatus COI.out, S1G: Sudan B. truncatus COI Tajima D.out, S1H: Sudan B. truncatus Fu_Li test.out, S1I: Zimbabwe B. truncatus.out, S1J: Zimbabwe B. truncatus COI Tajima D.out, S2A–S2F data were used to prepare Table 3, S2A: Egypt B. truncatus ITS Fu_Li test.out, S2B: Egypt B. truncatus ITS Tajima D.out, S2C: Egypt B. truncatus ITS.out, S2D: Sudan B. truncatus ITS.out, S2E: Sudan B. truncatus ITS Fu_Li test.out, S2F: Sudan B. truncatus ITS Tajima D.out, S3A–S3M were used to prepare Figure 2A,B, S3A: Egypt Bulinus truncatus GENBANK PROTEIN SEQ NEW.fas, S3B: Egypt Bulinus truncatus GENBANK SEQ NEW.d.txt, S3C: Egypt B. truncatus GENBANK SEQ NEW.fas, S3D: SUDAN B. truncatus COI Protein SEQ1.fas, S3E: SUDAN B. truncatus COI genbankSEQ.fas, S3F: SUDAN B. truncatus Genbank SEQ COI.txt, S3G: Zimbabwe B.truncatus COI protein SEQ for genbank submission.fas, S3H: Zimbabwe B. truncatus COI SEQ for genbank submission.fas, S3I: Zimbabwe B. truncatus COI SEQ for genbank submission.txt, S3J: ZWE_SUDAN_EGYPT_BT_REF_COI_OUTGROUP.txt, S3K.ZWE_SUDAN_EGYPT_BT_REF_COI_OUTGROUP2-v2NEW.fas, S3L: Summary of the STRUCTURE analysis for B. truncatus COI.docx, S3M: Table 1. Information on B. truncatus sequences submitted to genbank, S4A–S4H were used to prepare Figure 3A,B, S4A: Information B. truncatus ITS 1 sequences submitted to genbank.xls, S4B: Egypt B. truncatus ITS.fas, S4C: Egypt B. truncatus ITS.txt, S4D: Egypt_Sudan_REF B. truncatus ITS.fas, S4E: Egypt_Sudan_REF_Outgroup B. truncatus ITS.txt, S4F: SUDAN B. truncatus ITS.fas, S4G: SUDAN B. truncatus ITS.txt, S4H: Summary of STRUCTURE analysis for B truncatus.docx.
Author Contributions
E.M.A. and X.N.Z. conceived the study; E.M.A., M.J.M. and Y.H.G. carried out laboratory analysis; H.S., X.J.B. and E.M.A. carried out data analysis; M.M., M.H. and N.M. conducted fieldwork; J.X. and S.Z.L. contributed reagents, materials, and analysis tools; E.M.A., S.Z.L. and X.N.Z. wrote the paper.
Funding
This research received no external funding.
Conflicts of Interest
Authors declare there is no conflict of interest.
References
- 1.WHO . Country Profile: Preventive Chemotherapy and Transmission Control. World Health Organization; Geneva, Switzerland: 2010. [(accessed on 10 February 2018)]. Available online: http://www.who.int/neglected_diseases/preventive_chemotherapy/databank/CP. [Google Scholar]
- 2.Hanington P.C., Forys M.A., Loker E.S. A somatically diversified defense factor, FREP3, is a determinant of snail resistance to schistosome infection. PLoS Negl. Trop. Dis. 2012 doi: 10.1371/journal.pntd.0001591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordy M.A., Kish L., Tarrabain M., Hanington P.C. A comprehensive survey of larval digenean trematodes and their snail hosts in central Alberta, Canada. Parasitol. Res. 2016 doi: 10.1007/s00436-016-5152-9. [DOI] [PubMed] [Google Scholar]
- 4.Rollinson D., Stothard J.R., Southgate V.R. Interactions between intermediate snail hosts of the genus Bulinus and schistosomes of the Schistosoma haematobium group. Parasitology. 2001;123:S245–S260. doi: 10.1017/S0031182001008046. [DOI] [PubMed] [Google Scholar]
- 5.Brown D.S. Fresh Water Snails of Africa and Their Medical Importance. 2nd ed. Taylor and Francis; London, UK: 1994. p. 609. [Google Scholar]
- 6.Akinwale O.P., Kane R.A., Rollinson D., Stothard J.R., Ajayi M.B., Akande D.O., Ogungbemi M.O., Duker C., Gyang P.V., Adeleke M.A. Molecular approaches to the identification of Bulinus species in south-west Nigeria and observations on natural snail infections with schistosomes. J. Helminthol. 2010;85:283–293. doi: 10.1017/S0022149X10000568. [DOI] [PubMed] [Google Scholar]
- 7.Jarne P. Mating system, bottlenecks and genetic-polymorphism in hermaphroditic animals. Genet. Res. 1995;65:193–207. doi: 10.1017/S0016672300033279. [DOI] [Google Scholar]
- 8.Djuikwo-Teukenga F.F., Da Silva A., Njiokou F., Kamgang B., Same Ekobo A., Dreyfuss G. Significant population genetic structure of the Cameroonian freshwater snail, Bulinus globosus, (Gastropoda: Planorbidae) revealed by nuclear microsatellite loci analysis. Acta Trop. 2014;137:111–117. doi: 10.1016/j.actatropica.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 9.Brown D.S. Freshwater Snails of Africa and Their Medical Importance. Taylor and Francis; London, UK: 1980. p. 487. [Google Scholar]
- 10.Jarne P., Charlesworth D. The evolution of the selfing rate in functionally hermaphrodite plants and animals. Annu. Rev. Ecol. Syst. 1993;24:441–466. doi: 10.1146/annurev.es.24.110193.002301. [DOI] [Google Scholar]
- 11.Zein-Eddine R., Djuikwo-Teukeng F.F., Dar Y., Dreyfuss G., Van den Broeck F. Population genetics of the Schistosoma snail host Bulinus truncatus in Egypt. Acta Trop. 2017;172:36–43. doi: 10.1016/j.actatropica.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Kane R.A., Stothard J.R., Emery A.M., Rollinson D. Molecular characterization of freshwater snails in the genus Bulinus: A role for bar codes? Parasites Vectors. 2008;1:15. doi: 10.1186/1756-3305-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Standley C.J., Goodacre S.L., Wade C.M., Stothard J.R. The population genetic structure of Biomphalaria choanomphala in Lake Victoria, East Africa: Implications for schistosomiasis transmission. Parasites Vectors. 2014;7:524. doi: 10.1186/s13071-014-0524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zein-Eddine R., Djuikwo-Teukeng F.F., Al-Jawhari M., Senghor B., Huyse T., Dreyfuss G. Phylogeny of seven Bulinus species originating from endemic areas in three African countries, in relation to the human blood fluke Schistosoma haematobium. BMC Evol. Biol. 2014;14:271. doi: 10.1186/s12862-014-0271-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohamed A.H., Sharaf El-Din A.T., Mohamed A.M., Habib M.R. The relationship between genetic variability and the susceptibility of Biomphalaria alexandrina snails to Schistosoma mansoni infection. Mem. Inst. Oswaldo Cruz. 2012;107:326–337. doi: 10.1590/S0074-02762012000300006. [DOI] [PubMed] [Google Scholar]
- 16.Abdel-Hamid Z.A., Rawi S.M., Arafa A.F. Identification of a genetic marker associated with the resistance to Schistosoma mansoni infection using random amplified polymorphic DNA analysis. Mem. Inst. Oswaldo Cruz. 2006;101:863–868. doi: 10.1590/S0074-02762006000800007. [DOI] [PubMed] [Google Scholar]
- 17.Emery A.M., Loxton N.J., Stothard J.R., Jones C.S., Spinks J., Llewellyn-Hughes J., Noble L.R., Rollinson D. Microsatellites in the freshwater snail Bulinus globosus (Gastropoda: Planorbidae) from Zanzibar. Mol. Ecol. Notes. 2003;3:108–110. doi: 10.1046/j.1471-8286.2003.00368.x. [DOI] [Google Scholar]
- 18.Abe E.M., Guan W., Guo Y.H., Kassegne K., Qin Z.Q., Xu J., Chen J.H., Ekpo U.F., Li S.Z., Zhou X.N. Differentiating snail intermediate hosts of Schistosoma spp. using molecular approaches: Fundamental to successful integrated control mechanism in Africa. Infect. Dis. Poverty. 2018 doi: 10.1186/s40249-018-0401-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Djuikwo-Teukeng F.F., Njiokou F., Nkengazong L., De Meeus T., SameEkobo A., Dreyfuss G. Strong genetic structure in Cameroonian populations of Bulinus truncatus (Gastropoda: Planorbidae), intermediate host of Schistosoma haematobium. Infect. Genet. Evol. 2001;11:17–22. doi: 10.1016/j.meegid.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 20.Viard F., Bremond P., Labbo R., Justy F., Delay B., Jarne P. Microsatellites and the genetics of highly selfing populations in the freshwater snail Bulinus truncatus. Genetics. 1996;142:1237–1247. doi: 10.1093/genetics/142.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viard F., Justy F., Jarne P. Population dynamics inferred from temporal variation at microsatellite loci in the selfing snail Bulinus truncatus. Genetics. 1997;146:973–982. doi: 10.1093/genetics/146.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chlyeh G., Henry P.Y., Jarne P. Spatial and temporal variation of life-history traits documented using capture-mark-recapture methods in the vector snail Bulinus truncatus. Parasitology. 2003;127:243–251. doi: 10.1017/S0031182003003603. [DOI] [PubMed] [Google Scholar]
- 23.Jarne P., Viard F., Delay B., Cuny G. Variable microsatellites in the highly selfing snail Bulinus truncatus (Basommatophora: Planorbidae) Mol. Ecol. 1994;3:527–528. doi: 10.1111/j.1365-294X.1994.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 24.Brown D.S., Kristensen T.K. A Fish Guide to Fresh Water Snails. Danish Bilharziasis Laboratory; DK-2920 Charlottenlund, Denmark: 1993. [Google Scholar]
- 25.Emery A.M., Allan F.E., Rabone M.E., Rollinson D. Schistosomiasis collection at NHM (SCAN) Parasit Vectors. 2012;5:185. doi: 10.1186/1756-3305-5-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall T.A. Nucleic Acids Symposium Series. Information Retrieval Ltd.; London, UK: 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; pp. 95–98. [Google Scholar]
- 27.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ronquist F., Teslenko M., van der Mark P., Avres D.L., Darling A., Hohna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Posada D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 32.Rambaut A. Evolution and phylogenetics software. FigTree version 1.4.3. [(accessed on 8 December 2018)]; Available online: http://tree.bio.ed.ac.uk/software/figtree/
- 33.Polzin T., Daneshmand S.V. On Steiner trees and minimum spanning trees in hypergraphs. Oper. Res. Lett. 2003;31:12–20. doi: 10.1016/S0167-6377(02)00185-2. [DOI] [Google Scholar]
- 34.Excoffier L., Laval G., Schneider S. Arlequin ver.3.1: An integrated software package for population genetics data analysis. Evol. Bioinform. 2005;1:47–50. doi: 10.1177/117693430500100003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodall-Copestake W.P., Tarling G.A., Murphy E.J. On the comparison of population-level estimates of haplotype and nucleotide diversity: A case study using the gene cox1 in animals. Heredity. 2012;109:50–56. doi: 10.1038/hdy.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nalugwa A., Jørgensen A., Nyakaana S., Kristensen TK. Molecular phylogeny of Bulinus (Gastropoda: Planorbidae) reveals the presence of three species complexes in the Albertine Rift freshwater bodies. Int. J. Genet. Mol. Biol. 2010;2:130–139. [Google Scholar]
- 37.Brown D.S., Shaw K.M. Freshwater snails of thee Bulinus truncatus/tropicus complex in Kenya: Tetraploid species. J. Mollus. Stud. 1989;55:509–532. doi: 10.1093/mollus/55.4.509. [DOI] [Google Scholar]
- 38.Allan F., Sousa-Figueiredo J.C., Emery A.M., Paulo R., Mirante C., Sebastião A., Brito M., Rollinson D. Mapping freshwater snails in north-western Angola: Distribution, identity and molecular diversity of medically important taxa. Parasites Vectors. 2017;10:460. doi: 10.1186/s13071-017-2395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.